Abstract
Rat tail collagen solutions have been used as polymerizable in vitro three dimensional (3D) extracellular matrix (ECM) gels for single and collective cell migration assays as well as spheroid formation. Factors such as ECM concentration, pH, ionic concentration, and temperature can alter collagen polymerization and ECM architecture. In this unit, we demonstrate how to generate 3D collagen gels that have distinct architectures ranging from a highly reticular meshwork of short thin fibrils with small pores to a loose matrix consisting of stiff, parallel-bundled long fibrils by changing collagen polymerization temperature. This permits the analysis of 3D cell migration in different ECM architectures found in vivo while maintaining a similar ECM concentration. This chapter also includes collagen labeling techniques helpful for ECM visualization during live fluorescence imaging.
Keywords: Rat-tail collagen, 3D matrix, ECM architecture, fluorescent labelling
Introduction
Since the late 1990's, the study of cells and their ability to migrate through in vivo-like 3D microenvironments has become of great importance to the general cell biology community. It is well documented that ECM dependent factors such as rigidity, protein composition, ECM ligand type, and dimensionality all contribute to the regulation of intracellular signaling pathways and a cell's migratory phenotype(Doyle et al., 2013). Recent evidence suggests that the topography 3D ECMs can also affect cell migration rate as well as the dynamics of cell adhesions(Doyle et al., 2015).
Several in vitro 3D ECMs have become invaluable in our understanding of how cells migrate in 3D, including cell-derived matrices (3D CDM, unit 10.8) and collagen type I gels (units 10.3 and 10.18). 3D CDMs closely represent developmental ECMs containing high amounts of fibronectin, perlecan, and collagen and often demonstrate highly linearized uniaxial fibers(Doyle et al., 2009; Kutys et al., 2013). The caveat with 3D CDMs is they are thin (15-30 μm in thickness) and being secreted and formed directly by cells themselves there are no current methods to easily change the ECM architecture. On the other hand collagen type I gels are polymerized from neutralized collagen solutions that can be readily altered: Changing the collagen concentration directly impacts matrix pore size (the distance between fibers in three dimensional space) which affects cell motility(Wolf et al., 2009).
Previous investigations determined that besides collagen concentration, ionic strength, pH, and temperature all alter collagen's polymerization properties. More recently several studies have utilized these properties, namely pH and temperature, to generate 3D collagen gels with distinctly different ECM architectures(Doyle et al., 2015; Raub et al., 2007; Raub et al., 2008; Williams et al., 1978; Wolf et al., 2013). Under normal polymerization conditions (37°C and neutral pH ∼7.4) a 2 mg/ml concentration collagen gel demonstrates a small pore size (∼ 1 μm diameter), with a homogeneous, highly reticular mesh of small diameter (>300 nm) individual fibrils. By reducing either polymerization temperature or pH, ECM pore size increases, the matrix becomes heterogeneous, and collagen fibrils become thicker(Doyle et al., 2015; Raub et al., 2007; Raub et al., 2008; Wolf et al., 2013). For temperature, super resolution imaging indicates the increased thickness is due to the parallel bundling of multiple fibrils together(Doyle et al., 2015). Collagen gels with these latter characteristics are also physically stiffer and directly affect 3D cellular mechanotransduction. The usefulness of these distinctly different matrix types (homogeneous network of thin fibrils versus loose heterogeneous networks of stiff bundled fibrils) is that both exist in human and mouse dermis(Wolf et al., 2009). The protocols in this unit together define in detail how to generate collagen gels with distinctly different ECM through changing polymerization temperature. In addition, support protocols describe how to label collagen gels with fluorescent dyes, useful in determining and defining the ECM architecture, as well as how to properly incorporate fluorescently labeled collagen into collagen solutions that are suitable to sustain long term cell viability and migration.
Basic Protocol 1
Rat Tail Collagen Polymerization at Different Temperatures
This protocol describes how to generate collagen gels that have distinctly different architectures. Here we utilize temperature to easily change the gel architecture. Because changes to any of these factors will alter the overall gel architecture, care must be taken to insure reproducibility. With changing pH, for example, similar architectures can be created; however this requires a separate batch for each condition that could vary in composition due to pipetting errors made between conditions. We chose to vary polymerization temperature because a single batch can then be used to generate different architectures. The general protocol for collagen gel generation closely resembles that of Artym and Matsumoto's found in unit 10.18, support protocol 1, with a few variations and especially temperature.
Materials
Rat tail collagen solution dissolved in 20 mM acetic acid (commercial brands are fine, but in-house preparations are usually cleaner and polymerize faster) at a concentration greater than 5 mg/ml (6 mg/ml used here)
10X DMEM (see recipe)
10X reconstitution buffer (10X RB; see recipe)
1N NaOH (500 μl in a microfuge tube; see recipe)
1N HCl (500 μl in a microfuge tube)
Dulbecco's PBS with Calcium and Magnesium (PBS++) chilled to 4°C and at room temperature
MatTek Dishes (35mm, #1.5 coverslip, 20 mm opening: Part # P35G-1.5-20-C)
10 and 1000 μl pipettes and tips
ColorpHast pH-indicator-strips with pH range 6.5-10 (EMD Biosciences)
10-100 and 1000 μl Gilson positive displacement pipettes and tips (these are optional but really worth the money to get consistent results) www.gilson.com/en/Pipette/Products/44.224/Default.aspx#.VIm6raNOmRs
Rectangular ice bucket packed with ice
Parafilm
Chilling/heating dry bath (Echotherm from Torrey Pines Scientific). This is optional; an alternative is to use a heated dry block in a cold room (4°C) and adjust the temperature to your temperature specifications.
Rectangular glazed ceramic tile: 4×8 inches (purchase from a local hardware store. This is optional; however, it provides you with a smooth working surface that transmits cold efficiently when working with glass-bottomed dishes on ice).
1.5 ml microfuge tubes
Mini centrifuge
Cell culture incubator set to 37°C
Lab timer
Calculating collagen concentrations and volumes, and preparing reagents
It is advised to calculate the collagen concentration and volumes needed prior to beginning work in the tissue-culture hood.
-
For a 20 mm diameter glass bottomed dish 150 μls of collagen solution will generate a 300 μm-thick gel when polymerized at 37°C (at lower temperatures gels will be thinner, ∼200-250 μm). Due to the high viscosity of collagen solutions add 20 μl extra for each dish. Always polymerize a minimum of 1 extra dish for each temperature condition. Below is an example calculation for 4 different conditions (37 °C, 21 °C, 16 °C, and 4°C) with 2 dishes per condition and at a final concentration of 3 mg/ml:
-
a
8 dishes * 170 (μl collagen per dish) = 1360 μl of collagen solution
-
bCalculate amount of collagen:
Divide by 10 to calculate the amount of reconstitution buffer and 10X DMEM: 68.0 μl
-
Calculate the amount of 1N NaOH to neutralize the collagen and bring the collagen to a pH of approximately 7.4: A rough estimate is 0.55 μl is needed for each dish or 4.44 μl in total
Note: This will vary greatly depending on the original pH of your collagen and can even vary between commercial lots and preparations. Initial testing maybe required for determining the actual amount needed per dish
Calculate PBS++ volume: PBS++= Total volume-(Collagen+10X DMEM+10X RB+1N NaOH), or PBS++= 1360-(680+68+68+4.44), PBS++=539.56 μl
-
a
- Pre-chill all gel components on ice (collagen stock solution, 10X DMEM, 10X RB, 1N NaOH, 1N HCl, PBS++) and pre cool a microfuge tube. Leave MatTek dishes at room temperature until the gel solution has been mixed.Chilling dishes too far in advance will cause water in the air to condense and form droplets on the glass surface that can alter the collagen concentration. Place the ceramic tile on top of the ice to provide a level surface for your dishes.
Turn on and set the cooling dry block to the appropriate temperatures (the Echotherm has two such blocks), or use a heating dry block in a cold room set to your specifications.
Label the bottom edge of the MatTek dishes based on the polymerization temperature, for example 37, 21, 16, and 9.
Preparing collagen
It is highly recommended to invest in 10-100 and a 100-1000 μl positive displacement pipettes which greatly help accurate pipetting of viscous solutions such as collagen. The greater your accuracy in pipetting the better the overall experimental reproducibility.
-
5
Pipette the proper calculated amount of stock collagen solution into the pre-cooled microfuge tube. Be sure to keep the tube in the ice and avoid touching to reduce warming the bottom of the chilled tube.
-
6
Add the 10X DMEM and then the 10X RB. The RB will temporarily turn the solution red. After adding each slowly triturate with the pipette making sure not to introduce any bubbles. Because of the phenol red in the DMEM you will observe a color change from red to yellowish-orange. pH is likely between 5 and 6.
-
7Add the calculated 1N NaOH to the solution and triturate with the 100 ul positive displacement pipette set to ∼50 μl. Once the solution is mixed well and shows a consistent, single color take 1 μl and test the pH with a pH strip, waiting 1-2 minutes to determine the pH. The pH should be between 7.0 and 7.4 and the solution color should be peach (pinkish orange). If the solution is over a pH of 7.4, adjust with 1N HCl, 0.5 μl at a time. Be sure to note the changes and adjust the PBS++ accordingly (i.e., if 2 μl of HCl were added, reduce the PBS++ by the same amount).If bubbles form at any point a quick 5-10 second spin on a mini centrifuge will clear the solution, however be sure to immediately transport it back into the ice so as to not allow the collagen to increase temperature.If you are using these for 3D cell culture, see Support Protocol 2. It is recommended that the desired ECM architecture be determined prior to using cells in experiments.
-
8Add PBS++ (adjusted if necessary) to the microfuge tube and triturate again until fully mixed and place again in the ice.Note: If you have added cells to the collagen at this point you can no longer spin down the sample to remove air bubbles.
-
9
Place the labeled MatTek dishes onto the ceramic tile and let cool for 1-2 minutes.
-
10
Set your 10-100 μl positive displacement pipette to 75 μl and add to the middle of each dish. Be sure to only submerge the very tip of the pipette into the solution so as to not lose collagen that will cling to the sides of the pipette. Repeat this so the final volume per dish is 150 μl of the neutralized collagen solution. For the last dish or two you may be required to briefly centrifuge the remaining collagen to the bottom of the tube.
-
11Using the tip of the pipette, gently smear the thick neutralized collagen solution over the glass bottom to the edges of the dish. It is helpful to allow gravity to do the work by tilting the dish to 45 degrees and rotating the dish while using the pipette tip to cover the surface of the glass with the collagen solution.It is important to have the collagen touch every portion of the plastic-glass interface; a thicker meniscus will form and this aids in keeping the gel attached to the underlying surface. We have found that gels lift off more frequently without complete coverage to this edge.
-
12Once all dishes have been covered, place the dishes in/on their respective temperature controlled areas (heated incubator for 37 °C, cooling dry block for at or below room temperature, in our case 21°C, 16 °C, and 9 °C). Start a lab timer counting up.If cooling to 4 °C it is recommended to use a refrigerator, but pre-check the temperature at the location since shelf-to-shelf temperature differences are common. For polymerization at 9 °C or lower it is recommended to wrap each dish with parafilm to reduce any fluid loss via evaporation.
-
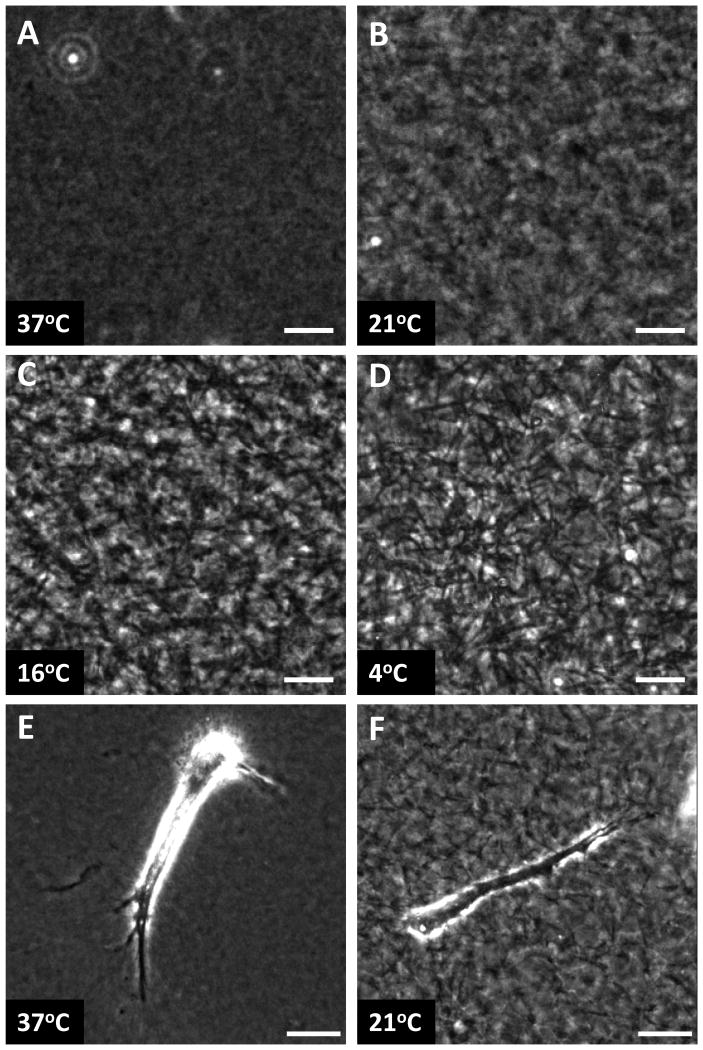
13In general, collagen solutions should be checked every 15 minutes to determine the degree of polymerization. This is done empirically with a standard phase contrast microscope using a 20X phase objective (Figure 1A-F). Be sure to note the time for polymerization at the different temperature conditions. If you are unsure of the polymerization time it is feasible to utilize the light scattering properties of collagen gels and use a temperature-controlled fluorimeter measuring the same emitted wavelength that was used for excitation (Artym and Matsumoto, 2010).After the first hour solutions polymerized at or above 16 °C should be polymerized and show varying degrees of phase-dense fibers. For 37 °C gels fiber will not be observable. Polymerization of collagen solutions below 16 °C can take considerable time: at 4 °C overnight polymerization is recommended.
-
14Once you have determined that the collagen is polymerized fully remove the MatTek dish and bring to room temperature for 5 to 10 minutes. To each dish add 2 ml of PBS++ to the dish. Add the PBS++ to the corner of the plastic area of the dish away from the glass interface very slowly; ejecting 1 ml should take approximately 10 seconds. The PBS++ should build up towards the collagen and once in contact will gently flow over the surface. Add the second 1 ml to the same corner of the dish and gently rotate the dish to cover the entire surface. Adding the PBS++ directly to the gel surface even gently can lead to the gel lifting off the glass.It should be noted that making collagen gels does require practice so the gels do not release from the dish and float. Always make extra dishes. If your gels consistently release and detach, activating the underlying glass surface with an aldehyde-terminated silane such as triethoxysilylbutraldehyde (TESBA) prior to gel polymerization can help keep gels firmly attached (See Basic Protocol 1 of unit 10.15 for glass activation).
-
15
Finally, you can visualize your sample in several ways to observe the architectural differences at the different polymerization temperatures: using backscatter or reflection microscopy using a confocal microscope (detailed in unit 10.18 and 4.5), or directly labeling the collagen fibrils using an NHS-ester based dye, detailed in Support protocol 2 in this unit.
Figure 1. Collagen gels polymerized at different temperatures.
(A-D) Phase contrast images taken with a 10X objective to demonstrate the observable differences in gel architecture for 3 mg/ml collagen polymerized at 37°C (A), 21 °C (B), 16 °C (C), and 4 °C (D). (E and F) 20X phase contrast images of NIH/3T3 fibroblasts imbedded in 3 mg/ml collagen gel polymerized at 37°C (E) and 21°C. Note that fibrils are not observable by light microscopy in 37°C polymerized collagen. Scale bars are 25 μm.
Support Protocol 1
Direct Labeling of Rat Tail Collagen Gels with NHS-Ester Dyes to Visualize ECM Architecture
This is an easy method to directly label collagen for observing the ECM architecture. It is not recommended for use with live cells. N-hydroxysuccinimide (NHS) ester dyes are organic compounds that react specifically with primary amines at the N-terminus of proteins and the side chains of lysine residues. NHS-ester dyes are water insoluble and require inorganic solvents such as DMSO for dilution. Once in aqueous medium they become active and react within 30 minutes, therefore small aliquots are normally stored and kept frozen at -20°C until use. NHS-esters are temperature and pH sensitive; raising either increases the rate of the reaction with amines, hence we perform these reactions at room temperature and use a buffer at pH of 9.0. It should be noted that this protocol is only for visualization of the gel and not for cell-based experimentation (blocking of all amines will disrupt cellular attachment). An alternative protocol follows for properly labeling collagen gels for use with cells.
Materials
1 mg/ml Atto-488 NHS-ester dye in DMSO (see recipe)
50 mM Borate buffer pH 9.0 (see recipe)
PBS++
15 ml conical tubes
-
Defrost an aliquot of Atto-488 NHS-ester in DMSO at room temperature. Be sure to not open the tube until the DMSO has fully liquefied.
NOTE: upon defrosting dye aliquots be sure to let them come to room temperature prior to opening. Being very hygroscopic, NHS-esters start reacting with the moisture in the air and will become inactive within 30 minutes.
-
Aspirate off the PBS++ and add 2 mls of room temperature borate buffer to each 35 mm dish of polymerized collagen for fluorescent labeling. Let dishes acclimate for 5 minutes.
When aspirating with a vacuum source be sure to place the pipette tip in corner of the dish and tilt at approximately 30 degrees, keeping the pipette tip at the bottom. This will insure you will not accidentally suction off the fragile collagen gel.
Meanwhile, make a solution containing 2 μls of NHS-ester dye for every 1 ml of borate buffer (1:500 ratio). Prepare 1 ml for each collagen gel.
Remove the borate buffer from step two and replace with 1 ml of the dye buffer.
Incubate for 15-30 minutes at room temperature in the dark (a drawer works well).
Next rinse with PBS++ 5 times over 15 minutes.
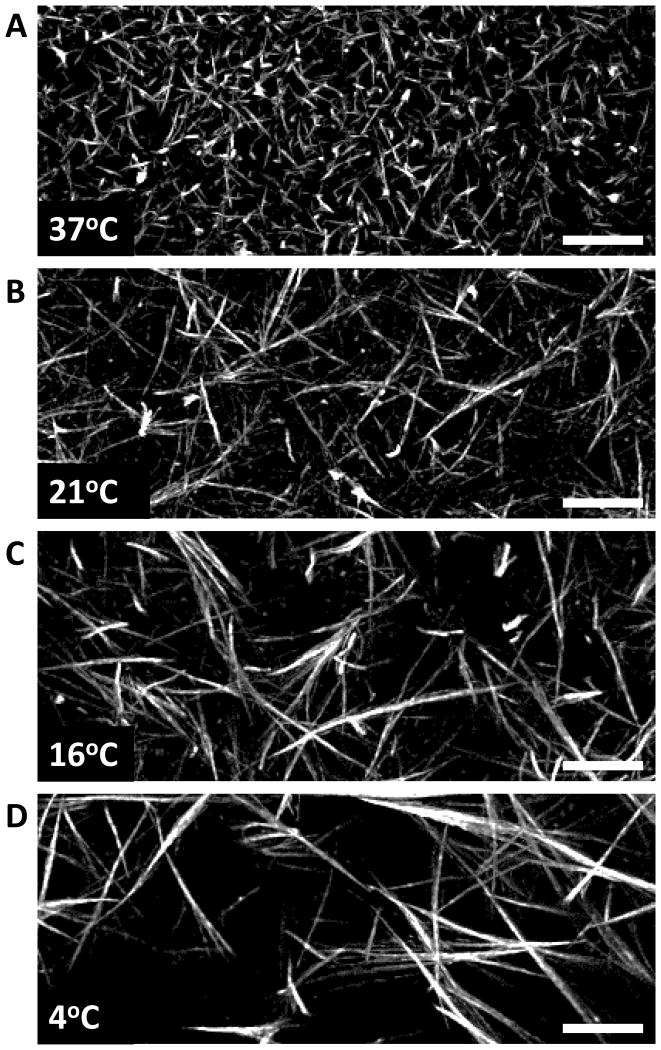
Image immediately using a laser scanning confocal or spinning disk microscope. Examples of the expected results are shown in Figure 2. If waiting for more than one day, add the appropriate amount of penicillin/streptomycin to the PBS++ to inhibit bacterial contamination.
Figure 2. Fluorescent labeling of collagen gels demonstrates diverse architecture at different polymerization temperatures.
(A-D) 10 μm thick maximum intensity projection images of 3 mg/ml collagen gels polymerized at 37°C (A), 21 °C (B), 16 °C (C), and 4 °C (D) following labeling with Atto 488 NHS-esters. Note the differences in collage fibril thickness as well as the change in ECM pore size between conditions. Images were taken at 60X on a spinning disk confocal microscope and then deconvolved. Scale bars are 10 μm.
Support Protocol 2
Addition of Cells to Neutralized Collagen Solutions for 3D Culture
3D collagen gels are highly suited for cell migration and 3D cell culture work, being a more in vivo-like environment compared to rigid 2D tissue culture plastic. Once the ECM architecture has been determined, cells that are viable in collagen, such as fibroblasts and many cancer cell lines, can easily be mixed into the neutralized collagen and plated. Below describes how to correctly mix cells into the PBS++ and then allow for polymerization at the different temperatures. It should be noted that polymerizing at extremely low temperatures (less than 14°C) could have an adverse effect on cell viability. Luckily, ECM pore size increases as polymerizing temperature is decreased. For these latter polymerization conditions it is recommended that cells be plated on top and allowed to invade into a previously polymerized collagen gel.
Materials
Neutralized rat tail collagen solution from Basic Protocol 1 (through step 7)
Minimum of 1.0×106 cells (e.g., human dermal fibroblast, HT-1080, etc.) in a tissue culture dish
PBS++ chilled on ice
Appropriate cell culture medium
Detach fibroblasts or cancer cells of choice using preferred methodology (trypsin-EDTA solution, etc.) and centrifuge cells briefly (2-4 minutes) using a swinging bucket rotor to pellet the cells.
- Re-suspend cells in 2 mls of ice cold PBS++. Count cells and adjust your previous calculations for PBS++ so your final cell count is 5.0 × 104 per ml of collagen.For example, using the final volume of 1360 μl from Basic Protocol 1, add 6.8 X104 cells into the PBS++. For a cell count of 5.0 × 105 per ml, add 136 μls of the PBS++/cell suspension to 439.47 μl of PBS.
Continue with steps 8 through 14 from Basic Protocol 1, substituting cell culture medium for PBS++ in step 14.
Place cells in a 37°C incubator with 10% CO2. Culture overnight before imaging.
Basic Protocol 2
Labeling Rat Tail Collagen Gels with NHS-Ester Dyes for Use with Live Cells
As mentioned in Support Protocol 2, complete labeling of a collagen gel with NHS-ester dyes can only be used to quickly assess the overall ECM architecture and not be used with cells. While laser scanning confocal microscopes can utilize reflection microscopy (also known as backscatter, unit 4.18) or second harmonic generation (unit 4.15) to visualize collagen fibers, we have found both techniques do not always properly depict the ECM architecture. A fluorescently-tagged ECM allows imaging of the smallest individual fibers. Unlike most proteins, collagen cannot be labeled when in solution because the numerous lysine residues are required for alpha helix formation with other monomers during polymerization(Chandrakasan et al., 1976). For this reason the labeling must be accomplished on preformed gels. This protocol describes how to label a polymerized gel, bring the collagen back into solution with acetic acid, and properly mix a minimal amount (2-4% of total protein) of labeled collagen with an unlabeled fraction to generate a bright, fluorescent collagen gel capable sustaining cell viability while allowing observation of ECM architecture over multiple hours of fluorescence imaging.
Materials
Rat tail collagen solution dissolved in 20 mM acetic acid (commercial brands are fine, but in house preparations are usually cleaner and polymerize faster) at a concentration greater than 5 mg/ml (6 mg/ml used here)
10X DMEM (see recipe)
10X reconstitution buffer (10X RB; see recipe)
1N NaOH (500 μl in a microfuge tube)
1N HCl (500 μl in a microfuge tube)
PBS ++ chilled to 4°C
MatTek Dishes (35mm, #1.5 coverslip, 20 mm opening: Part # P35G-1.5-20-C)
ColorpHast pH-indicator-strips with pH range 6.5-10 (EMD Biosciences)
10-100 and 1000 μl Gilson positive displacement pipettes and tips
Rectangular ice bucket packed with ice
10 cm tissue culture dish
Cell scrapers (3-4: Costar 3008)
Aluminum foil
50 mM Tris buffer (see recipe)
Scintillation vial or small 50 ml beaker
500 mM acetic acid
20 mM acetic acid chilled to 4°C
2-4 liter beaker
Magnetic stir bar to large beaker
Magnetic stir bar for scintillation vial
5 mg/ml Atto-488 NHS-ester dye in DMSO (see recipe)
50 mM Borate buffer pH 9.0 (see recipe)
3 ml Slide-A-Lyzer Dialysis cassette (G2) with 20,000 MW cutoff (Pierce: no syringe required)
Plastic wrap
Sircol Collagen Assay kit (available from accuratechemical.com)
Calculating collagen concentrations and volumes
-
Calculate reagent volumes for 5 mls of a 3 mg/ml collagen gel (see step 1 of Basic Protocol 1 for sample calculation):
6 mg/ml Rat tail collagen: 2.5 ml
10X DMEM: 250 μl
10X RB: 250 μl
1N NaOH: ∼16 μl
PBS++: ∼1984 μl
- Pre-chill all gel components on ice (collagen stock solution, 10X DMEM, 10X RB, 1N NaOH, 1N HCl, PBS++) and the 10 cm tissue culture dish.In order to reduce collagen solution loss when transferring from a conical tube to the 10 cm tissue culture dish we find it helpful to mix directly in the dish.
Preparing collagen
This portion of the protocol is identical to Basic Protocol 1 with the minor difference of mixing all components directly in the tissue culture dish.
-
3
Pipette the proper calculated amount of stock collagen solution into the pre-cooled tissue culture dish. Be sure to keep the dish on ice.
-
4
Add the 10X DMEM and then the 10X RB. After adding each slowly triturate with the pipette making sure not to introduce any bubbles. Alternatively, you can use a cell scraper to homogenize.
-
5
Add the calculated 1N NaOH to the solution and triturate/mix with a positive displacement pipette or cell scraper. Once the solution is mixed well and shows a consistent, single color take 1 μl and test the pH with a pH strip, waiting 1-2 minutes to determine the pH. The pH should be between 7.0 and 7.4 and the solution color should be peach (pinkish orange). If the solution is over a pH of 7.4 adjust with 1N HCl, 0.5 μl at a time. Be sure to note the changes and adjust the PBS++ accordingly (i.e. if 2 μl of HCl were added, reduce the PBS++ by the same amount.
-
6
Add PBS++ (adjusted if necessary) to the tissue culture dish until fully mixed and place again in the ice.
-
7
Use a cell scraper to coat the entire bottom of the dish with the collagen solution. It is helpful to allow gravity to do the work by tilting the dish to 45 degrees and rotating the dish while using the scraper to cover the dish surface.
-
8
Cover the dish and place the dish on the bench top and allow the collagen to polymerize at room temperature (approximately 21°C). Check the collagen gel after 30 minutes.
Collagen gel labeling with NHS-ester dye
-
9
Once the collagen has gelled add 10 ml of 50 mM borate buffer (pH 9.0) and incubate for 15 minutes.
-
10
Meanwhile, calculate the amount of dye needed to properly label the amount of protein within the gel using the following equation:
The above equation is for 1mg of Atto-488 NHS-ester diluted in 200 μl of DMSO using a 2-molar excess which is recommended by the company. NOTE: each dye has a different molar-excess that works the best for NHS-conjugation. Do not assume the above will work for all dyes. Over-labeling can lead to issues with gel formation later on. -
11
Add 45.28 ul of Atto-488 NHS-ester dye and bring the volume up to 5 mls with 50 mM borate buffer and vortex quickly.
-
12
Carefully aspirate the borate buffer from the tissue culture dish (bring the dish to a 45 degree angle and siphon off at the bottom edge with an aspiration pipette).
-
13Pipette the dye solution to the collagen and wrap the culture dish with aluminum foil to protect from light. Allow the dye to conjugate to the collagen gel for 1 hour at room temperature or 4 hours at 4°C (can do overnight) while rocking.At room temperature the majority of the dye will conjugate within the first 20 minutes.
-
14
Aspirate dye and add 10 ml of 50mM Tris buffer (pH 7.5) to quench the dye reaction. Incubate with rocking for 10 minutes. Keep the gel covered with foil to exclude light.
-
15
Add 10 mls of PBS++. Rinse gel with PBS++ 6X times over the next 4 hours to wash out the excess dye.
-
16Aspirate PBS++ and invert dish with one side raised on its lid in a tissue culture hood for 10-15 minutes to reduce the amount of fluid within the gel.All of the following steps (17-24) should be performed at 4°C.
-
17Add 500 to 1000 μl of 500 mM acetic acid to the gel. Bring the gel into a cold room and rock slowly for 1 hour.Note: the larger the volume of acetic acid you add, the easier it is to get the gel to go into solution. However, this will decrease your final concentration. We suggest starting with 500 μl and adding extra incrementally over time if needed.
-
18
After 1 hour use a cell scraper to mix the gel. Scrape gel to one side of the dish (on an angle) and pipette up gel solution with a 1000 μl positive displacement pipette set to 750 μl. If using a regular pipette, you may need to cut off the pipette tip at about the 100 μl mark for a larger tip opening because of the gel viscosity.
-
19
Transfer the collagen solution to a scintillation vial wrapped in foil and add a small magnetic stir bar. Stir gel for 2-4 hours at 4°C or overnight. Check periodically to see if the gel has gone into solution.
-
20
Check the volume of the gel. Based on the starting volume (5 ml) and concentration (3 mg/ml) you can guess at the concentration. If you need a more concentrated solution proceed to the salt precipitation protocol (Support Protocol 3). If the guesstimate is fine, then continue to the next step below.
Dialyzing the dye-labeled collagen solution
-
21
Add 2-4 liters of 20 mM acetic acid to a large beaker. Wet the Slide-A-Lyzer cassette in the acid. Pipette the collagen into the cassette being sure to remove any air bubbles before closing.
-
22
Add a large stir bar and the cassette containing the labeled collagen to the beaker and cover with plastic wrap. Stir for 4 hours.
-
23
Change acetic acid once and let dialyze further overnight.
-
24
Remove collagen solution from the cassette to several 1.5 ml centrifuge tubes. Place tubes in a cooling centrifuge. Spin at 15,000 RPM for 1 hour at 4°C. Remove and save the supernatant, being careful not to pull up any of the pellet.
-
25
Perform a Sircol collagen assay using the manufacturer's instructions to determine the collagen concentration.
-
26
Store at 4°C protected from light.
-
27
Proceed to Support Protocol 4 to determine the proper amount of fluorescently-labeled collagen to mix with an unlabeled fraction.
Support Protocol 3
Concentration of Collagen Solutions Using Salt Precipitation
Salt precipitation was originally used by Chandrakasan et al(Chandrakasan et al., 1976) in 1976 for preparing monomeric solutions of rat collagen. This allows easy concentration of the solution while reducing the amount of covalently crosslinked collagen in the form of dimers, trimers, or larger.
Materials
8% NaCl solution (see recipe)
500 mM acetic acid
15 ml conical tube capable of high-speed centrifugation (12,000 RPM)
Small magnetic stir bar
-
Estimate the current volume of the collagen to be concentrated (should still be in the scintillation vial) and add an equal volume of 8% NaCl.
A precipitate should begin forming with several minutes.
Stir the solution at 4°C 4 hours or overnight.
Once the liquid portion is clear with no apparent collagen remaining, transfer the collagen to a 15 ml centrifuge tube.
Spin down the solution at 12,000 RPM for 20 minutes at 4°C.
Aspirate the supernatant and keep the pellet. Add 2 ml 500 mM acetic acid (approximately concentrating by two times).
Stir the solution at 4°C until collagen goes into solution, 3-4 hours.
Go to step 21 in Basic Protocol 2 for dialysis.
Support Protocol 4
Calculating and Mixing Labeled and Unlabeled Rat Tail Collagen for Live Cell Fluorescence Imaging
Many researchers have mixed fluorescently-labeled and unlabeled collagen solutions together at specific ratios (1:5, 1:4, etc.) in order to generate a fluorescently visible collagen gel capable of sustaining cell life. Collagen that is completely labeled will not polymerize and often is too bright for actual imaging. Too often the final collagen concentration is incorrectly determined because ratios do not take into consideration differences in collagen concentration between labeled and unlabeled fractions. Furthermore, batch-to-batch differences in the labeled collagen concentration make gel consistency an issue. Here is described how to calculate and mix a 2-4% fluorescently-labeled gel based on protein weight. For simplicity sake we show calculations for 4% only to make up a 6 ml volume. Formulas are shown on the left, calculations are shown on the right.
6 mg/ml rat tail collagen solution dissolved in 20 mM acetic acid
5 mg/ml fluorescently labeled rat tail collagen solution dissolved in 20 mM acetic acid
10-100 and 1000 μl Gilson positive displacement pipettes and tips
15 ml conical tubes
Calculating and mixing labeled rat tail collagen with unlabeled and getting the proper amounts
- Calculate what 4% of the unlabeled collagen (ULC) is for the amount you want to mix:
- Multiple X by 0.04 to get 4% of this amount (Y).
- Use this number to then calculate the volume (V) you need to take out from the unlabeled.
- Perform the same for calculation for the fluorescently labeled collagen (FLC) as you did in #3 above.
Mix these together in a 15 ml conical tube slowly over 1 hour. Transfer to 1.5 ml centrifuge tubes and spin at 15,000 RPM for 30 minutes to remove any debris.
-
Calculate the “new” amount (C; in mg) since the volumes removed and added in 3 and 4 will likely not be the same.
((FLC added) * (concentration of FLC)) + ((ULC added) * (concentration of ULC)) = cNote that protein (in mg) should be identical to step 1
- Then divide the new volume C by the total volume (TV= (total UL RTC- removed) + (added F RTC).
Label the tube with the concentration and store at 4°C.
This 4% fluorescently-labeled collagen can now be used for generating collagen gels using Basic Protocol 1.
Reagents and Solutions
10X DMEM
1 powdered DMEM with phenol red (Sigma Aldrich: catalog #D2429) packet
50 ml distilled water
Stir mixture with heat (approximately 50°C) until DMEM powder goes into solution.
Sterile filter (0.2 μm Steriflip from Millipore or similar) while warm. Make four 10 ml and twenty 0.5 ml aliquots. 0.5 ml sizes are for daily experiments.
Store both sizes at -20°C until use. Upon defrosting heat aliquots in a 37°C waterbath, then vortex and cool on ice.
Can be kept indefinitely at -20°C and 1 month at 4°C.
10X reconstitution buffer (10X RB)
2.2g Sodium bicarbonate
4.8g HEPES -or- 20mL 1M HEPES stock solution for [0.2M] final
Distilled water up to 100 ml
Filter sterilize and store at -20°C in aliquots similar to 10X DMEM
Can be kept indefinitely at -20°C and 1 month at 4°C.
Fluorescent NHS-ester dye in DMSO (for Support Protocol1)
Atto-488 NHS-ester 1 mg (Sigma) Note that any NHS-ester dye is suitable and can be substituted
1 ml DMSO (Sigma)
Add DMSO to dye tube and mix (wrapped in foil) on a rotational mixer for 1 hour.
Split into 5-10 ul aliquots.
Store at -20°C or -80°C.
Can be kept indefinitely at -80°C.
Fluorescent NHS-ester dye in DMSO (for Basic Protocol 2)
Atto-488 NHS-ester 1 mg (Sigma)
200 μl DMSO (Sigma)
Add DMSO to dye tube and mix (wrapped in foil) on a rotational mixer for 1 hour.
Use immediately and store remaining dye in aliquots at -20°C or -80°C.
Can be kept indefinitely at -80°C.
50 mM Borate buffer (pH 9.0)
1.55 g boric acid (powder 99.5%, Sigma)
Add distilled water to 400 ml
Add several solid NaOH pellets at a time while mixing until pH is ∼9.0
Add distilled water to 500 ml
Filter sterilize using a 0.2 μm filter
Store up to 1 year at room temperature
Sodium Hydroxide solution, (NaOH), 1N
0.5 g NaOH pellets
Distilled water to 12.5 ml
Mix well
Filter sterilize, divide into 500 μl aliquots and store indefinitely at -20°C
Sodium Chloride (NaCl), 8% solution
4 g NaCl
Distilled water to 50 ml
Mix well
Filter sterilize using a 0.2 μm filter
Store up to 6 months at 4°C
Commentary
Background Information
In the middle of the 20th century it was demonstrated that collagen from intact skin and rat tail tendon sources can be brought into solution using weak acids or neutral salts without protein denaturation (Fitch et al., 1955; Gross et al., 1955). Weak acids such as acetic acid can effectively solubilize collagen monomers that have not undergone extensive chemical crosslinking. This crosslinking increases in all mammals with age and is attributed to an increase in lysl oxidase-based crosslinking of lysine residues between individual collagen molecules(Wolf et al., 2009). Research by Chandrakasan et al.(Chandrakasan et al., 1976) showed salt precipitation (4% solution) can efficiently reduce non-monomeric or crosslinked forms of type I collagen. The monomeric form of type I collagen is what is commercially available. Other extraction methods include treatment with pepsin (notably used to extract collagen from bovine skin) removes nearly all crosslinks and greatly reduces the polymerization rate of these solutions(Wolf et al., 2009).
The original Work by Williams et al.(Williams et al., 1978) demonstrated that several critical parameters other than collagen concentration can effect collagen polymerization: 1) phosphate concentration, 2) ionic strength, 3) pH, and 4) temperature. This and other studies performing biochemical/biophysical analysis on acidified collagen solution has led to collagen being readily commercially available and preferentially used for 3D cell culture assays. A series of papers by the Friedl and Grinnell laboratories have documented the cellular phenotype of primary fibroblasts as well as numerous cancer cell lines(Friedl and Wolf, 2010; Grinnell, 2003; Grinnell et al., 2003; Grinnell and Petroll, 2010; Wolf et al., 2009; Wolf et al., 2013; Wolf et al., 2007). More recently, with the interest in mechanobiology and how the cellular microenvironment can regulate cellular function, polymerizing collagen gels into different ECM architectures has demonstrated how the surrounding ECM is involved in control of intracellular events such as focal adhesion dynamics(Doyle et al., 2015). These different ECM architectures are currently considered a controllable alternative to in vivo microenvironments.
Critical Parameters and Troubleshooting
As mentioned several times throughout this unit, the formation of collagen type I 3D gels is highly dependent on 1) collagen concentration, 2) the ionic concentration, 3) pH, and of course, 4) temperature. Because each of these can affect the time to polymerization, which inevitably leads to the differences in ECM architecture, it is vital that each of the above parameters be consistent between experiments. pH is by far the most variable since it needs to be tested and adjusted to neutralize each batch of collagen made. It is suggested that pH is checked for each batch.Do not assume that the amount of 1N NaOH will always be the same. Small aliquots of stock solutions are used throughout the protocol in order to cut down on changes in concentration due to evaporation that can occur over time. Yet another critical issue to bear in mind is maintaining the consistent ionic concentration of 10X DMEM solution. Due to high salinity of this solution, a precipitate will always be present at 4°C or when on ice. Hence trituration of the 10X DMEM solution immediately before addition to the mixture is important to keep a consistent ionic concentration.
Another critical parameter and likely a reason for unexpected results is the variability between collagen lots being used. Lot-to-lot variability of commercial or in house rat tail collagen preps is common. Animal age can play a role since intramolecular crosslinking increases with age and will effect gel polymerization. While Basic Protocol 1 gives specific pH, temperatures, and collagen concentrations to use, it is likely that these parameters may need to be altered to get the desired ECM architecture. An example of this is if collagen polymerization at 16°C is taking an excessive amount of time (greater than 1.5 hours) at a pH of 7.2, increasing the pH to 8.0 will further promote polymerization. It is suggested that if issues with polymerization below 37°C do not give the anticipated results (distinctly different ECM architectures), test different collagen concentrations, the final pH of the neutralized collagen, and possibly the collagen itself. As mentioned previously, pepsin-digested collagen, which has been tested, will not polymerize at low temperatures, hence we do not recommend its use here.
Anticipated Results
It is expected that by varying the polymerization temperature of neutralized collagen described in Basic Protocol 1 the different ECM architectures will be observed by comparison of phase contrast microscopy or confocal reflection microscopy (see Figure 1). The direct labeling of collagen gels with NHS-ester dyes (Support protocol 1) will further help in defining the differences in the collagen gels beyond what can be determined by aforementioned microscopy techniques. After overnight incubation, it is expected that fibroblasts should demonstrate different cellular phenotypes during migration that are directly related to the ECM architecture (See Figure 1E and F). The combination of Basic Protocol 2 with Support Protocols 3 and 4 will allow the direct fluorescent visualization of the collagen fibrils while supporting cellular function including motility and growth.
Time Considerations
Basic Protocol 1
Performing the calculations and preparing the neutralized collagen solution should take 30-60 minutes, depending on your familiarity with the protocol. Initially it takes a little getting used to working with the viscous collagen and not generate bubbles that greatly disrupt the ECM structure. The time for polymerization will be highly variable and is dependent on the temperature(s) you choose: 37°C, 21 °C, 16 °C, 9 °C, and 4 °C should polymerize in 20 minutes, 40 minutes, 60 minutes, 3 hours, and overnight, respectively.
Support Protocol 1
The direct labeling of collagen gels generated in Basic Protocol 1 should take approximately 1 hour until the samples can be imaged.
Support Protocol 2
The addition of cells to the collagen gel should take an added 15 minutes, and overall 45-75 minutes, not including polymerization time. Imaging should not be performed until the following day (approximately 18 hours).
Basic Protocol 2
The labeling of collagen for use in live cell imaging experiments will likely take 3 to 4 days and can be highly variable, depending on the use of support protocol 2 to concentrate the collagen solution. Gel formation should take 1.5-2 hours and the initial conjugation of the NHS-ester dye to the polymerized gels 1 hour. Gel quenching and rinsing 4 hours. Gel acidification with acetic acid should take between 6 and 18 hours (overnight; beginning of day 2). It is best to dialyze for a minimum of 8 hours to overnight (end of day 2 or beginning of day 3). Sircol collagen assay takes 2.5 hours.
Support Protocol 3
Salt precipitation of the collagen out of solution can again vary between 4 to 18 hours (overnight, beginning of day 2). Getting the collagen back into a 500 mM acetic acid solution will take 3 to 4 hours.
Support Protocol 4
Calculations should take 10 minutes and the mixing and centrifugation 1.5 hours.
Acknowledgments
This work was supported by the NIDCR Division of Intramural Research.
Literature Cited
- Artym VV, Matsumoto K. Imaging cells in three-dimensional collagen matrix. Curr Protoc Cell Biol. 2010;18:11–20. doi: 10.1002/0471143030.cb1018s48. Chapter 10: Unit 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrakasan G, Torchia DA, Piez KA. Preparation of intact monomeric collagen from rat tail tendon and skin and the structure of the nonhelical ends in solution. The Journal of biological chemistry. 1976;251:6062–6087. [PubMed] [Google Scholar]
- Doyle AD, Carvajal N, Jin A, Matsumoto K, Yamada KM. Local 3D matrix microenvironment regulates cell migration through spatiotemporal dynamics of contractility-dependent adhesions. Nature communications. 2015;6:8720. doi: 10.1038/ncomms9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Petrie RJ, Kutys ML, Yamada KM. Dimensions in cell migration. Current opinion in cell biology. 2013;25:642–649. doi: 10.1016/j.ceb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AD, Wang FW, Matsumoto K, Yamada KM. One-dimensional topography underlies three-dimensional fibrillar cell migration. The Journal of cell biology. 2009;184:481–490. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch SM, Harkness ML, Harkness RD. Extraction of collagen from tissues. Nature. 1955;176:163. doi: 10.1038/176163a0. [DOI] [PubMed] [Google Scholar]
- Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. The Journal of cell biology. 2010;188:11–19. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnell F. Fibroblast biology in three-dimensional collagen matrices. Trends in Cell Biology. 2003;13:264–269. doi: 10.1016/s0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- Grinnell F, Ho CH, Tamariz E, Lee DJ, Skuta G. Dendritic fibroblasts in three-dimensional collagen matrices. Molecular biology of the cell. 2003;14:384–395. doi: 10.1091/mbc.E02-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinnell F, Petroll WM. Cell motility and mechanics in three-dimensional collagen matrices. Annual review of cell and developmental biology. 2010;26:335–361. doi: 10.1146/annurev.cellbio.042308.113318. [DOI] [PubMed] [Google Scholar]
- Gross J, Highberger JH, Schmitt FO. Extraction of Collagen from Connective Tissue by Neutral Salt Solutions. Proceedings of the National Academy of Sciences of the United States of America. 1955;41:1–7. doi: 10.1073/pnas.41.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutys ML, Doyle AD, Yamada KM. Regulation of cell adhesion and migration by cell-derived matrices. Experimental cell research. 2013;319:2434–2439. doi: 10.1016/j.yexcr.2013.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raub CB, Suresh V, Krasieva T, Lyubovitsky J, Mih JD, Putnam AJ, Tromberg BJ, George SC. Noninvasive assessment of collagen gel microstructure and mechanics using multiphoton microscopy. Biophysical journal. 2007;92:2212–2222. doi: 10.1529/biophysj.106.097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raub CB, Unruh J, Suresh V, Krasieva T, Lindmo T, Gratton E, Tromberg BJ, George SC. Image correlation spectroscopy of multiphoton images correlates with collagen mechanical properties. Biophysical journal. 2008;94:2361–2373. doi: 10.1529/biophysj.107.120006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Gelman RA, Poppke DC, Piez KA. Collagen fibril formation. Optimal in vitro conditions and preliminary kinetic results. The Journal of biological chemistry. 1978;253:6578–6585. [PubMed] [Google Scholar]
- Wolf K, Alexander S, Schacht V, Coussens LM, von Andrian UH, van Rheenen J, Deryugina E, Friedl P. Collagen-based cell migration models in vitro and in vivo. Seminars in cell & developmental biology. 2009;20:931–941. doi: 10.1016/j.semcdb.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Te Lindert M, Krause M, Alexander S, Te Riet J, Willis AL, Hoffman RM, Figdor CG, Weiss SJ, Friedl P. Physical limits of cell migration: control by ECM space and nuclear deformation and tuning by proteolysis and traction force. The Journal of cell biology. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nature cell biology. 2007;9:893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]