Abstract
3D tissue cultures provide a more physiologically relevant context for the screening of compounds, compared with 2D cell cultures. Cells cultured in 3D hydrogels also show complex phenotypes, increasing the scope for phenotypic profiling. Here we describe a high-content screening platform that uses invasive human prostate cancer cells cultured in 3D in standard 384-well assay plates to study the activity of potential therapeutic small molecules and antibody biologics. Image analysis tools were developed to process 3D image data to measure over 800 phenotypic parameters. Multiparametric analysis was used to evaluate the effect of compounds on tissue morphology. We applied this screening platform to measure the activity and selectivity of inhibitors of the c-Met and epidermal growth factor (EGF) receptor (EGFR) tyrosine kinases in 3D cultured prostate carcinoma cells. c-Met and EGFR activity was quantified based on the phenotypic profiles induced by their respective ligands, hepatocyte growth factor and EGF. The screening method was applied to a novel collection of 80 putative inhibitors of c-Met and EGFR. Compounds were identified that induced phenotypic profiles indicative of selective inhibition of c-Met, EGFR, or bispecific inhibition of both targets. In conclusion, we describe a fully scalable high-content screening platform that uses phenotypic profiling to discriminate selective and nonselective (off-target) inhibitors in a physiologically relevant 3D cell culture setting.
Keywords: 3D tissue culture, high-content screening, phenotypic profiling, c-Met, EGFR
抄録
化合物のスクリーニングでは、三次元組織培養の方が二次元細胞培養に比べてより生理学的な関連性を明らかにすることができる。また、三次元ヒドロゲルで培養した細胞に見られる複雑な表現型は、表現型プロファイリングの範囲を広げる。
本稿では、治療用小分子及び抗体生物製剤候補の活性を調べるために標準的な384ウェルアッセイプレートで三次元培養したヒト浸潤性前立腺癌細胞を使ったハイコンテントスクリーニングプラットフォームについて述べる。800を超える表現型パラメータを測定するため、三次元画像データを処理する画像解析ツールを開発した。化合物が組織形態に及ぼす影響の評価にはマルチパラメータ解析を使用した。
三次元培養した前立腺癌細胞におけるc-Met 及び上皮成長因子(以下「EGF」)受容体型(以下「EGFR」)チロシンキナーゼ阻害剤の活性と選択性を評価するために、本スクリーニングプラットフォームを採用した。c-Met及びEGFRの活性は、それぞれのリガンド、肝細胞成長因子(以下「HGF」)、EGFによって現れた表現型プロフィールに基づいて定量した。本スクリーニング法は、c-Met及びEGFRの推定阻害剤80種の新規収集に適用した。c-Met、EGFRの選択的阻害又は両標的の二重特異性阻害を示唆する表現型プロフィールを誘導する化合物が確認された。
結論として、著者らは、生理学的に関連性のある三次元細胞培養という設定において、選択的阻害剤と非選択的(非標的)阻害剤を識別するために表現型を使用する、十分に測定可能なハイコンテントスクリーニングプラットフォームについて述べた。
초록
3D 조직 배양은 2D 세포 배양에 비해 화합물의 스크리닝을 위한 더 많은 생리학적 관련성을 지닌 컨텍스트를 제공한다. 또한 3D 히드로겔에서 배양된 세포는 표현형 프로파일의 범위를 증가시키는 복잡한 표현형을 나타낸다.
여기서 우리는 표준 384정 분석 플레이트의 3D에서 배양된 침습성 전립선 암세포를 사용하여 잠재적 치료 소분자 및 항체 바이오로직스의 활동을 연구하는 하이콘텐츠 스크리닝 플랫폼을 설명한다. 이미지 분석 도구는 3D 화상 데이터를 처리하여 800개 이상의 표현형 파라미터를 측정하기 위해 개발되었다. 조직 형태학에서 화합물의 효과를 평가하기 위해 다중 파라미터 분석이 사용되었다.
우리는 3D 배양 전립선 암세포에서 c-Met와 EGF 수용체(EGFR) 티로신 인산화효소의 억제제 활동 및 선택성을 측정하기 위해 이 스크리닝 플랫폼을 적용했다. c-Met와 EGFR 활동은 각각의 리간드 HGF와 EGF에 의해 유도된 표현형 프로필을 기반으로 정량화되었다. 스크리닝 방법은 80개의 c-Met와 EGFR 추정 억제제의 신규 컬렉션에 적용되었다. 화합물은 c-Met, EGFR의 선택적 억제 또는 모든 대상의 이중특이성 억제를 나타내는 표현형 프로필을 유도하는 것으로 확인되었다.
결론적으로, 우리는 표현형 프로파일링을 사용하여 생리학적으로 관련된 3D 세포 배양 세팅에서 선택적 및 비선택적(오프타깃) 억제제를 구별하는 완벽하게 확장 가능한 하이콘텐츠 스크리닝 플랫폼을 설명한다.
摘要
與2D細胞培養相比,3D組織培養可為化合物的篩選提供更具生理學關聯性的菌肉。在3D水凝膠中培養的細胞同時顯現出了複雜的表型,從而擴大了表型分析的範圍。
在本文中,我們描述的是一種高含量篩選平台;此平台利用在標準384孔酶聯板中以3D模式培養的浸潤型人類前列腺癌細胞,對潛在治療性小分子和抗體生物製劑的活性進行研究。我們開發了用於處理3D影像資料的影像分析工具,繼而測量出了800多個表型參數。同時,採用了多參數分析法評估化合物對組織形態產生的作用。
我們運用這一篩選平台,對以3D形式培養的前列腺癌細胞c-Met以及EGF受體(EGFR)酪氨酸激酶抑制劑的活性和選擇性進行了測量;並根據表型分析(包括其各自的配體HGF和EGF)對c-Met以及EGF的活性進行了量化。針對80個新型c-Met以及EGFR假定抑制劑,我們運用了這種篩選方法。結果發現,鑒別出了一種化合物,此類化合物可誘發產生以c-Met或EGFR選擇性抑制或者兩個靶點雙特異性抑制為特徵的表型分佈。
綜上所述,我們描述了一種完全可衡量的高含量篩選平台;該平台可利用表型分析來鑒別具有生理學關聯性的3D細胞培養環境中的選擇性和非選擇性(非靶點)抑制劑。
摘要
与2D细胞培养相比,3D组织培养可为化合物的筛选提供更具生理学关联性的菌肉。在3D水凝胶中培养的细胞同时显现出了复杂的表型,从而扩大了表型分析的范围。
在本文中,我们描述的是一种高含量筛选平台;此平台利用在标准384孔酶联板中以3D模式培养的浸润型人类前列腺癌细胞,对潜在治疗性小分子和抗体生物制剂的活性进行研究。我们开发了用于处理3D影像数据的影像分析工具,继而测量出了800多个表型参数。同时,采用了多参数分析法评估化合物对组织形态产生的作用。
我们运用这一筛选平台,对以3D形式培养的前列腺癌细胞c-Met以及EGF受体(EGFR)酪氨酸激酶抑制剂的活性和选择性进行了测量;并根据表型分析(包括其各自的配体HGF和EGF)对c-Met以及EGF的活性进行了量化。针对80个新型c-Met以及EGFR假定抑制剂,我们运用了这种筛选方法。结果发现,鉴别出了一种化合物,此类化合物可诱发产生以c-Met或EGFR选择性抑制或者两个靶点双特异性抑制为特征的表型分布。
综上所述,我们描述了一种完全可衡量的高含量筛选平台;该平台可利用表型分析来鉴别具有生理学关联性的3D细胞培养环境中的选择性和非选择性(非靶点)抑制剂。
Introduction
While offering many practical advantages for cell-based screening, cells grown on tissue culture plastic lack normal cell-cell and cell-matrix interactions, resulting in deregulated growth, dedifferentiation, disruption of other processes, and a poor simulation of the in vivo (patho)physiology.1 3D cell culture methods have been developed that enable a more in vivo like tissue architecture and are therefore expected to represent a more physiologically relevant context for the evaluation of bioactive molecules.2–4 Although gaining in popularity for small-scale analyses, 3D cultures have yet to be widely adopted for screening. A number of factors account for this, although high-throughput 3D imaging and the analysis of large and complex image data sets can represent the most significant barrier to the implementation of high-content 3D culture-based screening.5 As a result, where 3D assays are used for screening, the most common end-point measurements tend to be biochemical determinations of cell viability, with the loss of potentially valuable phenotypic information.
Cells cultured in extracellular matrix (ECM)–containing hydrogels can generate complex multicellular tissues, supporting a higher level of tissue organization, such as formation of ductal structures by epithelial cells and complex networks of invasive tumor cells.6–10 These features may be relevant to the pathology of the disease being studied and offer a context for screening for therapeutic molecules. Complex phenotypes can also be exploited for compound profiling, allowing compounds that affect different targets and therefore induce different phenotypes to be discriminated, potentially providing additional information on the biological effects of compounds that cannot be obtained from single end-point measurements.
In many cultured cells, activation of receptor tyrosine kinases results in increased proliferation and motility.11 Cell motility has previously been used as a readout for high-throughput small-molecule and small interfering RNA (siRNA) screens for inhibitors of c-Met signaling.12–14 In 3D culture, activation of c-Met can result in invasion of tumor cells into the surrounding microenvironment,15 a process that closely resembles the in vivo pathophysiology. Similarly, activation of the epidermal growth factor (EGF) receptor is known to induce epithelial- to mesenchymal transition (EMT) and invasion in various cancer types.16,17 As expected, selective inhibition of the receptor tyrosine kinases inhibits the ligand-induced effects. However, compounds that inhibit multiple targets or are toxic may also inhibit ligand-induced effects. Here, we describe a 384-well screening assay that uses automated high-content analysis and profiling of 3D cultures of invasive prostate cancer tumor cells to identify selective inhibitors of the c-Met and EGF receptor (EGFR) tyrosine kinases.
Materials and Methods
Cell Lines
The human prostate adenocarcinoma cell line PC-3 was cultured in Dulbecco’s modified Eagle’s medium (DMEM)–F12 (Ham’s) growth medium (Gibco Fisher Scientific, Landsmeer, Netherlands), supplemented with 10% fetal bovine serum (FBS) (Gibco Fisher Scientific), 1 mM sodium pyruvate (Gibco Fisher Scientific), 1.5 g/L NaHCO3 (Merck, Schiphol-Rijk, Netherlands), 0.1 mM nonessential amino acids (Gibco Fisher Scientific), and 50 µg/mL gentamycin (Sigma Aldrich, Zwijndrecht, Netherlands). PC-3 cells were grown in 175-cm2 tissue culture flasks (Corning, Amsterdam, Netherlands) in culture medium as described above. Before reaching maximal density, cells were washed with 1× phosphate-buffered saline (PBS) (Sigma Aldrich) and trypsinized with 1× Trypsin (Gibco Fisher Scientific). Subsequently, medium was added and cells were pelleted by centrifugation and resuspended in FBS with 10% DMSO (Biosolve B.V., Valkenswaard, Netherlands) and stored in aliquots at −150 °C.
3D Invasion Assay
PC-3 cells were cultured in 384-well plates (Greiner Bio-One B.V., Alphen aan den Rijn, Netherlands) in 60% v/v growth factor–reduced Matrigel (>9 mg/mL) (Corning), which supported spheroid formation of the highly transformed PC-3 cells and invasion upon addition of motogenic cytokines. To generate gels, thawed cells were mixed with culture medium and growth factor–reduced Matrigel. Then, 14.5 µL of cell-gel mix was added to each well of a 384-well plate using a CyBi Selma 96/60 robotic liquid handler (Analytik Jena AG, Jena, Germany), at a final cell density of 2000 cells per well. After polymerization at 37 °C for 30 min, DMEM-F12 (Ham’s) growth medium containing growth factors (hepatocyte growth factor [HGF] or EGF) and compounds were added to the gel. The plate(s) were subsequently covered with a gas-permeable adhesive membrane (Thermo Scientific, Zwijndrecht, Netherlands). Compound exposures were performed for 96 h, after which gels were fixed with 3% formaldehyde (Sigma Aldrich), permeabilized with 0.2% Triton-X100 (Sigma Aldrich), and stained with 0.25 µM rhodamine-phalloidin (Sigma Aldrich) and 0.1% Hoechst 33258 (Sigma Aldrich) in PBS at 4 °C for 16 h. After staining, plates were washed in PBS and covered with a Greiner SilverSeal (Greiner Bio-One B.V., Alphen aan den Rijn, Netherlands).
Compounds
Anti c-Met and nontargeting control Fab antibodies were generously provided by Merus Biopharmaceuticals B.V. (Utrecht, Netherlands). c-Met inhibitor ArQ-197 (Tivantinib) was synthesized by Janssen Pharmaceutica NV (Beerse, Belgium), following the synthesis method published from ArQule, and subsequently sent to the authors as a gift from Souichi Ogata (Janssen Oncology R&D). Results were confirmed using a second source of ArQ-197 (SelleckChem/Bio-Connect B.V. Huissen, Huissen, Netherlands). EGFR inhibitor AG1478 was obtained from Santa Cruz (SC-200613; Bio-Connect B.V. Huissen). PHA-665752 was a gift from Pfizer, Inc. (La Jolla, CA, USA). A collection of 80 compounds selected to inhibit both EGFR and/or c-Met was generated by Vichem Chemie (Budapest, Hungary) (Suppl. Table S1).
Imaging
A BD Pathway 855 automated inverted fluorescence microscope (BD Biosciences, Breda, Netherlands) was used for automated imaging of 384-well plates (wide-field epifluorescence). This microscope was used to image both Hoechst 33258 and rhodamine-phalloidin staining, using a 4× Olympus objective, at focal planes spaced at intervals of 50 µm throughout the gel using Attovision software (BD Biosciences, Breda, Netherlands) accompanying the microscope. The gel was imaged through its entire depth (z-axis), requiring 25 images per well. Each image captured approximately 75% of the area of the well.
Data Analysis
Ominer image analysis software (OcellO, Leiden, Netherlands) and KNIME (https://www.knime.org) were used to extract in-focus information from the Z-stacks generated by the BD Pathway 855 (BD Biosciences) for both Hoechst 33258 (nuclei) and rhodamine-phalloidin (F-actin) using maximum-intensity projections.18 For image processing, a monochrome mask was created for both channels to define the regions of interest (ROIs). The in-focus images were used to quantify staining intensities and a set of Hu moments and Gabor wavelet-based features describing image intensity and texture. The Hoechst 33258–derived monochrome mask was used to determine number and area of nuclei and tumoroids. In addition, a detailed set of parameters was calculated to describe the shape of the rhodamine-phalloidin (F-actin) stained objects.18 First, for principal components analysis, data were Z score normalized to the negative control treatment median to account for plate-to-plate differences. The features derived from the image analysis and quantification steps were ranked based on their ability to distinguish positive and negative controls (Z′), and the features that showed greatest separation between control groups were retained (Z′ > –1.0). The median of treatment quadruplicates was used for a principal component analysis (PCA) that combined correlated features into (uncorrelated) principal components. This model was applied to all wells and summarized ~95% of all variation into principal components. The distance between treatment and control groups was quantified as a Z score in principal component 0 (PC0), which retained 60% of the variation of the data set. 2D density estimations and linear discriminant analysis (LDA) were performed using the MASS package (http://www.stats.ox.ac.uk/pub/MASS4/)19 for R-Studio 0.99.878 (https://www.rstudio.com/products/rstudio2/) with R 3.2.3 (https://www.r-project.org/). PCA was calculated in KNIME, and principal component plots were generated using ggplot2 (http://ggplot2.org/)20 with the scatterplot3D package for R-Studio 0.99878 (https://cran.r-project.org/web/packages/scatterplot3d/index.html).21 Other charts were generated using GraphPad Prism 6 software (GraphPad Software, La Jolla, CA). Results are displayed as mean ± standard deviation unless otherwise stated.
Western Blot Detection of Receptor Activation
Phosphorylation status of c-Met and EGFR in response to 20 ng/mL HGF or 20 ng/mL EGF was evaluated by Western blot. Briefly, 105 PC-3 cells were grown per well in 12-well plates. After 24 h, growth medium was replaced with starvation medium (without serum and antibiotics) containing test compounds. After 24 h of compound exposure, growth factors (20 ng/mL HGF/EGF) were added for 10 min. Protein was isolated using RIPA lysis buffer (1% w/w deoxycholate, 50 mM Tris [pH 7.5], 0.15 M NaCl, 0.1% sodium dodecyl sulfate [SDS], 1% NP-40, 2 mM EDTA, 1% protease inhibitor cocktail [Sigma Aldrich]) and quantified using the standard bicinchoninic acid (BCA) method according to the manufacturer’s instructions (Thermo Scientific). Western blot analysis was performed according to a standard protocol using phospho-c-Met (Tyr1234/1235) antibody (3077P; Cell Signaling) diluted 1:1000 in 5% BSA in TBS-T or phospho-EGFR (Tyr1173) (4407; Cell Signaling Technology B.V., Leiden, Netherlands) 1:1000 in 5% BSA in Tris-Buffered Saline with 0.05% Tween-20 (TBS-T). Phospho-c-Met and phospho-EGFR antibodies were detected using horseradish peroxidase (HRP)–conjugated anti–rabbit IgG secondary antibody (111-035-003; JacksonImmunoResearch, Suffolk, UK) and ECL Plus reagent (RPN2132; GE Healthcare Life Sciences, Eindhoven, Netherlands). Detection of antibody was performed using a LAS4000 scanner (GE Healthcare Life Sciences). Tubulin loading control was detected using an anti-Tubulin antibody (T-9026; Sigma-Aldrich) diluted 1:1000 in 5% BSA in TBS-T and an anti-mouse Alexa 647–linked IgG (115-605-006; JacksonImmunoResearch). The Alexa 647 signal was detected directly using the LAS4000.
In Vitro Kinase Activity Measurement
EGFR enzyme activity was assayed in 384 microtiter plates (Corning 3676) at three compound concentrations, in a total volume of 10 µL by the Transcreener ADP2 FP method (BellBrook Labs, Madison, WI). Assay buffer contained 20 mM HEPES (pH 7.5), 1 mM dithiothreitol (DTT), 10 mM MgCl2, 2 mM MnCl2, and 0.01% v/v NP40. The final EGFR concentration was 7 nM. Poly Glu-Tyr (4:1)/poly Glu-Tyr (4:1) was used as substrate at a final concentration of 0.01 mg/mL. The final adenosine triphosphate (ATP) concentration was at 7.52 µM. The enzyme reaction was incubated for 60 min and stopped by addition of 10 µL adenosine diphosphate (ADP) detection mixture (1×). Measurements were performed on a Tecan (Männedorf, Switzerland). Infinite M1000Pro reader. c-Met enzyme activity was assayed in 384 microtiter plates (Corning 3676) at three compound concentrations, in a total volume of 10 µL by the Transcreener ADP2 FP method (BellBrook Labs). Assay buffer contained 20 mM HEPES (pH 7.5), 1 mM DTT, 3 mM MgCl2, 3 mM MnCl2, and 0.01% v/v Tween-20. The final c-Met concentration was 8 nM. Poly Ala-Glu-Lys-Tyr (6:2:5:1) was used as substrate at a final concentration of 0.25 mg/mL. The final ATP concentration was at 9.6 µM. The enzyme reaction was incubated for 60 min and stopped by addition of 10 µL ADP detection mixture (1×). Measurements were performed using a Tecan Infinite M1000Pro reader.
Results
Quantification of Complex Phenotypic Changes of Prostate Cancer Cells Cultured in 3D in 384-Well Microplates
We developed a 3D cell culture screening method for tumor cell invasion driven by c-Met and EGFR. PC-3 cells were cultured in ECM protein-rich hydrogels in 384-well plates in the presence or absence of c-Met/EGFR agonists and antagonists. After fixation and staining with rhodamine-phalloidin to reveal the structure of prostate cancer tumoroids and Hoechst 33258 to label nuclei, gels were imaged in two fluorescence channels using a BD Pathway 855 (BD Biosciences), collecting paired stacks of xy images throughout the z-plane for each well. In-focus information from each image was extracted and a single xy image projection was generated (Suppl. Fig. S1).
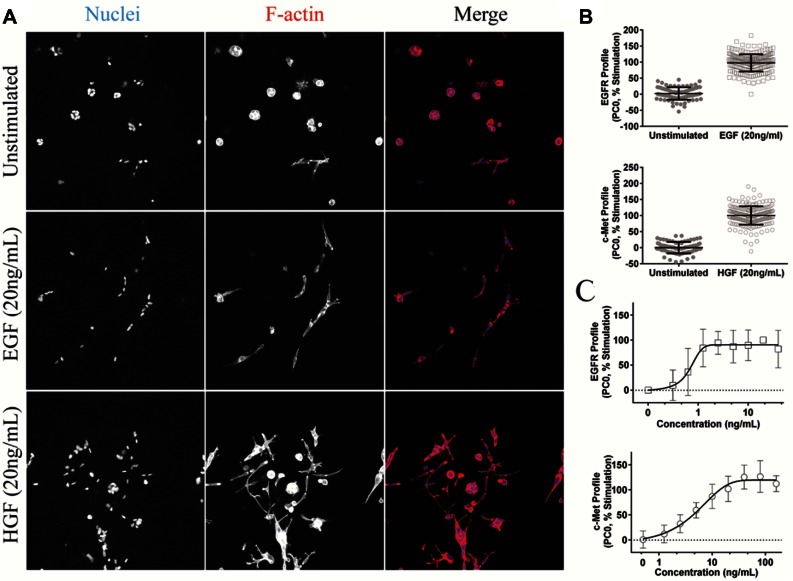
In the absence of added c-Met or EGFR agonists, PC-3 cells developed into spheroids over a period of 4 days ( Fig. 1A , upper panel). Treatment with the c-Met ligand (HGF) or the EGFR ligand (EGF) induced a highly invasive phenotype ( Fig. 1A , middle and lower panels). To quantify these phenotypic changes in detail, Ominer and KNIME software were used to identify and quantify different morphological features from the 2D projections of the cytoskeleton and nuclei-derived image stacks. To do this, a collection of different image analysis algorithms was applied to derive measurements of the shape and fluorescence intensity of individual tumoroids, wavelets, and Hu and Zernike moments as previously described.22 Supervised PCA was used to reduce measurements from approximately 800 phenotypic parameters to principal components (PCs). EGF-treated and untreated PC-3 cells were used to train a profile to represent active and inactive EGFR inhibitors, respectively, which was subsequently scaled to percent stimulation. Similarly, a separate supervised analysis was performed that was trained with HGF-treated and untreated cells to separate phenotypes representing active c-Met from unstimulated control phenotypes. The primary principal component (PC0) was plotted for both EGF- and HGF-induced phenotypic changes to visualize the responses ( Fig. 1B ). Both HGF- and EGF-induced phenotypes of PC-3–derived tumoroids could be clearly discriminated from untreated tissues. Many of the phenotypic features separating HGF- and EGF- induced changes in phenotype from unstimulated controls describe shape, circularity, and intensity. The distance in phenotypic space along PC0 to the unstimulated controls was presented graphically as a function of EGF or HGF concentration ( Fig. 1C ).
Figure 1.
Receptor activation induces a quantifiable change in phenotype. (A) Typical images of unstimulated, hepatocyte growth factor (HGF)–stimulated, and epidermal growth factor (EGF)–stimulated PC-3 cells taken using a Nikon TI Eclipse confocal microscope (Nikon, Tokyo, Japan). Receptor activation induced a change in phenotype characterized by invasion into the surrounding matrix. F-actin stained red; nuclei stained blue. (B) Phenotypic shift induced by HGF and EGF visualized using supervised (unstimulated-stimulated) principal components analysis (PCA). PC0 was scaled between 0% (unstimulated) and 100% (stimulated). Individual data points shown, as well as mean and standard deviations. (C) EGF and HGF cause a dose-dependent change in phenotype. Phenotypic change is derived from PC0 and scaled to percent response, as shown in B. Results are shown as means with standard deviation.
Phenotypic Profiling Can Be Used to Quantify c-Met Activation and Inactivation
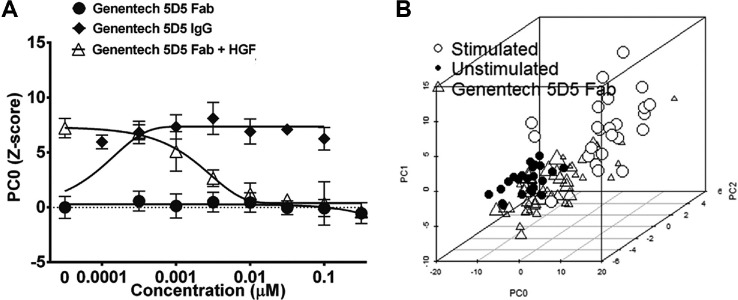
To test whether HGF-induced phenotypes could report c-Met activation, HGF was added in the presence of the c-Met function blocking Fab antibody, Genentech 5D5 Fab (Genentech, South San Francisco, CA). 5D5 antibody effectively inhibited the HGF-induced response (PC0, Fig. 2A ), resulting in phenotypes that projected in a similar location to unstimulated controls in phenotypic space (represented by three principal components), with an inhibition that was dose dependent ( Fig. 2A , B ). The c-Met inhibitory antibody had no quantifiable effect on the PC-3 phenotype in the absence of HGF ( Fig. 2A ). In contrast, treatment of PC-3 cells with a c-Met activating antibody, the bivalent IgG form of 5D5, which crosslinks c-Met, induced a phenotype that was indistinguishable from that induced by HGF. Together these results confirm the c-Met dependency of the HGF-induced phenotype and demonstrate that principal components can be used to quantify HGF-induced phenotypic changes correlating with c-Met activation and inhibition.
Figure 2.
c-Met–dependent phenotypic changes. (A) Inhibition of hepatocyte growth factor (HGF)–induced invasion in 3D-cultured PC-3 cells by function blocking c-Met Fab antibody 5D5 (Genentech, South San Francisco, CA) and stimulation of invasion by the crosslinking and activating bivalent IgG isoform. Principal components analysis (PCA) trained on unstimulated and stimulated control, PC0 shown and scaled as Z score to unstimulated control. Results are shown as means of quadruplicate wells and standard deviations. (B) 5D5 Fab (triangles) causes a shift in phenotype from stimulated (empty circles) back to unstimulated control (filled circles). Point size of markers for Fab treatment increases with concentration (seven concentrations; range, 0.316–316 nM). Individual data points are shown as a 3D scatterplot with PC0, PC1, and PC2.
Using Phenotype to Discriminate Compounds Based on Target Selectivity
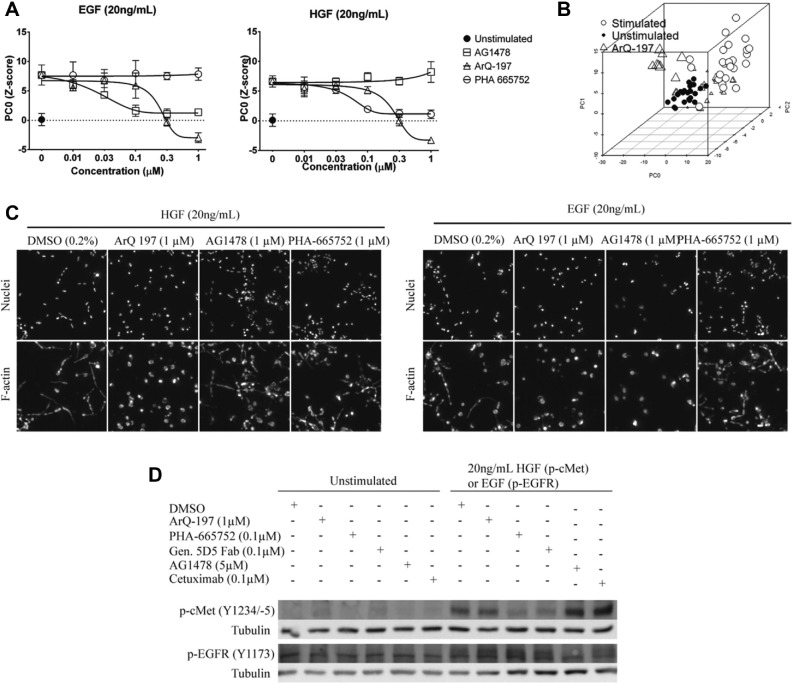
To determine whether the phenotypic assay can discriminate between selective c-Met and EGF inhibitors, PC-3 cells were cultured in the presence of either HGF or EGF, together with established small-molecule and antibody inhibitors of their cognate receptors. As expected, the small-molecule c-Met inhibitor PHA-66575223,24 selectively inhibited the phenotype induced by HGF. In contrast, the EGFR inhibitor AG147825 had no significant effect on the HGF-induced phenotype up to a concentration of 1 µM ( Fig. 3A , C , right panel). Conversely, AG1478 but not PHA-665752 inhibited the EGF-induced phenotype ( Fig. 3A , C , left panel). These findings suggest that residual c-Met and EGFR activity does not contribute to the 3D phenotype under the conditions used and that c-Met does not require EGFR for the ligand-induced response and vice versa. Interestingly, ArQ-197, which is reported to be a selective inhibitor of c-Met,22,26 inhibited both the HGF- and EGF-induced responses with a similar IC50, suggesting a more complex mechanism of action. At higher concentrations (>1 µΜ), this compound introduced a novel phenotype, shown by a shift away from the controls in a representation of phenotypic space ( Fig. 3B ).
Figure 3.
Discriminating on-target effects from off-target effects. (A) Hepatocyte growth factor (HGF)–induced (left panel) and epidermal growth factor (EGF)–induced (right panel) phenotypes can be inhibited by small molecules targeting c-Met and EGF receptor (EGFR), respectively. Responses are shown as PC0, Z score normalized to unstimulated. Values shown as means of quadruplicate wells with standard deviation. (B) Principal components analysis (PCA) plot identifies novel phenotype introduced by ArQ-197 (triangles) at high doses. Individual data points are shown as a 3D scatterplot with PC0, PC1, and PC2. Empty circles, stimulated controls; filled circles, unstimulated controls. (C) Representative images showing HGF- and EGF-induced invasion and inhibition by small molecules. Maximum intensity projections of both image channels shown. (D) Western blot of c-Met and EGFR phosphorylation in 2D-cultured PC-3 cells exposed to EGF or HGF and inhibitors.
To confirm that the interpretations based on phenotypic changes correlated with inhibition of receptor kinase activity, we performed Western blot analysis for phosphorylated c-Met (Y1234/-5) and phosphorylated EGFR (Y1173) ( Fig. 3D ). Results showed that PHA-665752 and the Genentech 5D5 Fab potently inhibited phosphorylation of c-Met but not EGFR phosphorylation. Conversely, AG1478 and cetuximab treatment inhibited phosphorylation of EGFR but not of c-Met. ArQ-197 did not show detectable inhibition of EGFR (Y1173) or c-Met (Y1234/-5) phosphorylation under these conditions ( Fig. 3D ). Therefore, inhibition of phenotypes in 3D culture by c-Met and EGFR inhibitors correlates with inhibition of the respective kinases.
Taken together, these results show that measurement of ligand-induced phenotypic changes can be used to discriminate and measure inhibition of c-Met and EGFR.
Using Phenotypic Profiles to Identify Novel Selective c-Met and EGFR Inhibitors
Using data obtained from c-Met and EGFR in vitro kinase assays, a collection of 80 putative single- and dual-specificity EGFR and c-Met inhibitors was compiled (Suppl. Table S1), which included several well-characterized reference compounds. These were screened in 3D-cultured PC-3 cells together with several published reference inhibitors of c-Met and EGFR. All compounds were screened in 384-well plates at three concentrations (10 µM, 3.16 µM, and 1 µM) in triplicate plates in both EGF- and HGF-stimulated conditions. Multiparametric analysis was performed as before, followed by Z score normalization of each parameter to the plate median. Supervised PCA was used to condense phenotypic measurements to one phenotypic descriptor (PC0), which was scaled to percent inhibition. PCA was found to perform superior to individual features in separating out stimulated and unstimulated controls (Suppl. Fig. S2).
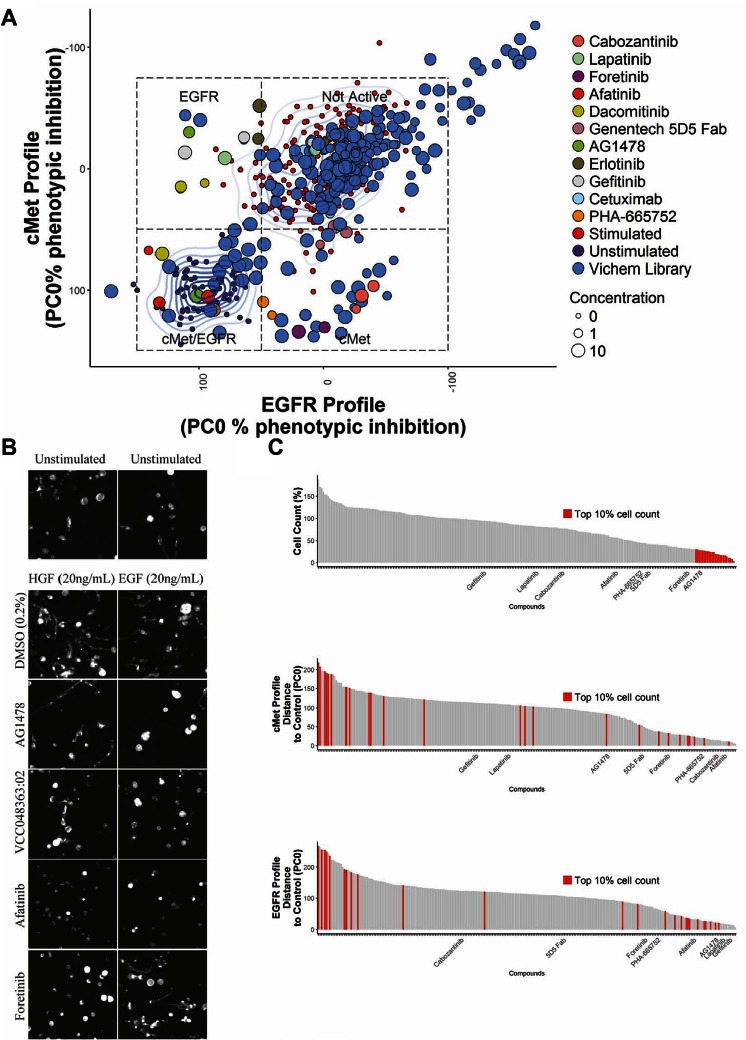
Figure 4A depicts the screening results combining the first principal component, PC0, for both EGF- and HGF-induced phenotypic changes in a single plot. Inhibition of c-Met is therefore represented by a decrease in the c-Met PC0 value, from the growth factor–treated to growth factor–untreated controls. Compounds that induced such a shift in the c-Met PC0 included various established c-Met reference inhibitors that were included in the screen, including the Genentech 5D5 Fab antibody, foretinib, and lower doses of PHA-665752. This confirmed that the phenotypic training using HGF-treated and untreated samples discriminated c-Met active and inactive conditions and was able to select compounds that induced a c-Met inhibitory phenotypic profile. Selective inhibition of the EGFR could be characterized by a horizontal shift to the left, from a profile associated with an EGF-stimulated phenotype to one associated with an unstimulated phenotype. This region contained several EGFR reference inhibitors, including erlotinib, gefitinib, and AG1478 (at 1 and 3.16 µM), which were included in the screen alongside test compounds. Compounds that clustered together with the unstimulated controls ( Fig. 4A , bottom left quadrant) were predicted to have a dual-inhibitory activity for both c-Met and EGFR. The compounds clustering in this area included afatinib and also a number of previously untested compounds of the Vichem library. The class to which each compound was attributed using this approach is shown in Table 1 and Supplemental Table S2. By using another form of supervised clustering, linear discriminant analysis (LDA), we could separate the highly similar EGF- and HGF-induced phenotypes into a multidimensional plot (Suppl. Fig. S3). However, this approach did not improve classification of reference c-Met and EGFR inhibitor compounds.
Figure 4.
Identification of selective c-Met/EGFR inhibitors in a Vichem epidermal growth factor (EGF) receptor (EGFR)/c-Met inhibiting compound library. (A) Compound screen of 80 Vichem compounds tested at 10, 3.16, and 1µM. Compounds were divided over triplicate plates and coexposed with either hepatocyte growth factor (HGF) (20 ng/mL) or EGF (20 ng/mL). Principal components analysis (PCA) was trained on unstimulated and stimulated controls separately for plates exposed to HGF or EGF, respectively, to obtain a principal component that could separate c-Met and EGFR responses. Data points represent mean determinations from three wells. Controls are color-coded as indicated in the legend. For stimulated (top middle) and unstimulated controls (bottom left), a 2D-density estimation (contour lines) is shown. (B) Representative 2D projected images derived from the F-actin staining after 96 h of compound treatment. Except for AG1478, which was 3.16 µM, shown treatment concentrations were 10 µM. Pictures were obtained using a BD Pathway 855 microscope (images trimmed to 300 × 300 pixels for presentation purposes). (C) Cell count (viability) is a poor criterion for selecting c-Met and EGFR inhibitors. Number of nuclei per well was percent normalized to stimulated control (100%) and lowest detected cell count and 0%, top panel. The top ranking 10% of compounds affecting cell count are color-coded in red. Middle and lower panels show the same compounds ranked on efficacy on c-Met and EGFR profiles (distance to unstimulated control). Mean values shown; each chart contains compounds at three different concentrations.
Table 1.
Hit Selection Phenotypic Screen.
| Vichem Compound ID | 1 µM | 3.16 µM | 10 µM |
|---|---|---|---|
| VCC030450:22 | EGFR | EGFR | EGFR |
| VCC048363:02 | EGFR | EGFR | EGFR/c-Met |
| VCC055393:01 | EGFR/c-Met | EGFR/c-Met | EGFR/c-Met |
| VCC109756:01 | c-Met | c-Met | c-Met |
| VCC155409:01 | None | None | EGFR/c-Met |
| VCC228833:01 | None | None | Top 10% cell count |
| VCC285946:01 | None | c-Met | c-Met |
| VCC376189:01 | c-Met | c-Met | EGFR/c-Met |
| VCC378728:01 | None | EGFR | None |
| VCC407451:10 | None | EGFR | EGFR |
| VCC415997:02 | c-Met | c-Met | c-Met |
| VCC429285:02 | None | None | EGFR/c-Met |
| VCC444414:01 | c-Met | c-Met | c-Met |
| VCC450892:17 | c-Met | c-Met | c-Met |
| VCC497510:01 | c-Met | c-Met | EGFR/c-Met |
| VCC502987:01 | None | None | EGFR/c-Met |
| VCC528301:01 | None | None | EGFR/c-Met |
| VCC656576:02 | None | None | None |
| VCC716837:01 | None | None | Top 10% cell count |
| VCC740005:11 | None | EGFR | EGFR |
| VCC744093:03 | None | None | None |
| VCC833029:24 | None | None | EGFR |
| VCC868449:01 | None | None | EGFR/c-Met |
| VCC960450:01 | c-Met | c-Met | c-Met |
EGFR, epidermal growth factor receptor.
Representative images of 3D-cultured PC-3 cells treated with EGFR and c-Met inhibitors are presented in Figure 4B and show a consistency between the automated classification of compounds to a specific inhibitory class and the phenotype that is induced in the presence of a specific growth factor. We compared the phenotypic classification of compounds with measurements of EGFR or c-Met enzyme inhibitory activity ( Table 2 and Suppl. Table S3). Phenotypic classification was found to frequently overlap with biochemical measurements. Inhibition of c-Met activity in enzymatic and phenotypic assays was closely correlated, with all compounds showing greater than 40% inhibition in an enzymatic assay being inhibitory in the phenotypic assay at the equivalent concentration. However, approximately half of the compounds that were active in EGFR enzymatic assays were inactive in the cell-based phenotypic assays. Furthermore, several of the compounds that were inhibitory at the lowest doses in the EGFR enzymatic assay only showed activity in the corresponding phenotypic assay at higher concentrations (e.g., VCC833029:24, VCC740005:11, and VCC407451:10), indicating lower sensitivity of the phenotypic assay compared to the enzymatic assay. Not all compounds that induced enzymatic inhibition of EGFR and/or c-Met could be identified as inhibitors from our screening results, suggesting that enzymatic inhibition does not directly translate to a phenotypic change under all conditions.
Table 2.
Enzyme Activity Measurement (Percent Inhibition).
| Inhibition (%) |
||||||
|---|---|---|---|---|---|---|
| Vichem Compound ID | EGFR 10 µM | EGFR 1 µM | EGFR 0.1 µM | c-Met 10 µM | c-Met 1 µM | c-Met 0.1 µM |
| VCC030450:22 | 101 | 99 | 97 | 4 | 2 | 0 |
| VCC048363:02 | 101 | 100 | 97 | 1 | −2 | −1 |
| VCC055393:01 | 99 | 100 | 100 | 3 | 3 | −4 |
| VCC109756:01 | 43 | 7 | 2 | 93 | 61 | 7 |
| VCC155409:01 | 76 | 34 | 10 | 25 | 2 | −2 |
| VCC228833:01 | 16 | 1 | 0 | 4 | 0 | −2 |
| VCC285946:01 | 32 | 0 | 9 | 84 | 49 | 8 |
| VCC376189:01 | 12 | 2 | −2 | 83 | 56 | 9 |
| VCC378728:01 | 85 | 41 | 8 | 72 | 16 | 0 |
| VCC407451:10 | 93 | 92 | 73 | 7 | 0 | −4 |
| VCC415997:02 | −11 | −11 | −7 | 95 | 77 | 14 |
| VCC429285:02 | 3 | 2 | 0 | 6 | −2 | 2 |
| VCC444414:01 | 7 | −4 | −6 | 90 | 47 | 3 |
| VCC450892:17 | 32 | 10 | 5 | 85 | 39 | 4 |
| VCC497510:01 | 20 | 10 | 2 | 90 | 54 | 9 |
| VCC502987:01 | 19 | 9 | 1 | 4 | 1 | 0 |
| VCC528301:01 | 94 | 82 | 29 | 37 | 7 | 1 |
| VCC656576:02 | −2 | −7 | −13 | 0 | −4 | −4 |
| VCC716837:01 | 48 | 10 | 1 | 41 | 6 | 0 |
| VCC740005:11 | 105 | 102 | 93 | 11 | 1 | 3 |
| VCC744093:03 | 77 | 25 | 3 | 27 | −1 | −1 |
| VCC833029:24 | 97 | 90 | 37 | −3 | 0 | 0 |
| VCC868449:01 | 95 | 89 | 37 | 56 | 14 | 3 |
| VCC960450:01 | −11 | −7 | −10 | 87 | 50 | 11 |
Scale of red to green shading used to visualise % inhibition. Red, no inhibition of target; Yellow, mild target inhibition; Green, potent inhibition of target. EGFR, epidermal growth factor receptor.
We then compared ranking of compounds based on phenotypic profiling with ranking based on cell count. Cell count was determined by the number of nuclei counted per well (one of the many features scored by the multiparametric analysis) and is a feature that correlates closely with biochemical measurements of cell proliferation (not shown). Using a ranking based on cell proliferation, inhibitors that were shown to potently inhibit the c-Met phenotype, including Genentech 5D5 Fab, did not score in the top 10% inhibitors of proliferation ( Fig. 4C and Suppl. Fig. S4). Consistent with the profiling results in Figure 4A , when compounds were ranked based on the difference in their phenotypic profiles to unstimulated controls (measured along PC0), a strong enrichment of c-Met and EGFR inhibitors was observed in the top 10% of ranked compounds in the presence of HGF and EGF, respectively (Suppl. Fig. S4). The most potent inhibitors of proliferation were found to perform poorly when ranked based on their phenotypic profiles ( Fig. 4C ). An analysis of the images from 3D cultures treated with these compounds showed that spheroid formation was disrupted, consistent with cytotoxicity of these compounds.
Discussion
Because tumor cells exist in a 3D ECM-rich environment, 3D matrix-embedded cell cultures provide a more physiologically relevant context in which to perform compound screening.2–4 The increased complexity of 3D cultures also offers increased potential for phenotypic profiling.27–29 We developed a fully scalable 3D tissue culture–based high-content screening platform that uses phenotypic profiling of cultured tumoroids derived from a prostate cancer cell line. This screening platform was used to identify several selective inhibitors for c-Met and EGFR, which represent important targets in many cancers and are known to be able to stimulate survival and invasive growth of tumours.15–17 We show that activation of c-Met with its ligand, HGF, induced a dose-dependent reorganization of PC-3 spheroids characterized by invasion of cells into the surrounding matrix. A similar change in phenotype was induced by the addition of EGF to the PC-3 cells. These phenotypic changes were not observed in 2D monolayers. Ultra-high-content multiparametric analysis allowed a clear discrimination of the phenotypes associated with active and inactive c-Met and EGFR, which could be quantified using a single PCA measurement. This approach allowed us to discriminate inhibitors of c-Met and EGFR and also putative biselective inhibitors of these receptor tyrosine kinases. The method also enabled nonselective compounds to be discriminated since they induced phenotypes that failed to match those induced by selective inhibition of c-Met or EGFR. The screening results were cross-validated with an in vitro measurement of enzyme activity and were found to largely correlate. Some compounds that inhibited EGFR or c-Met in the biochemical measurement were not identified by our phenotypic screen as inhibitors for these pathways, indicating that inhibition of purified enzyme does not always correlate with inhibition of the target enzyme in cells. These differences may be explained by poor compound stability over multiple days in aqueous solution (medium) or poor membrane permeability, resulting in lower cytoplasmic concentrations. It is also possible that these compounds induce mild phenotypic changes that are below the threshold of the phenotypic assay. A number of compounds were found to induce phenotypic changes in the EGFR and/or c-Met profiles, even though these failed to induce inhibition of either EGFR or c-Met activity. A possible explanation for this finding is that these molecules induced off-target phenotypic effects that could not be discriminated from inhibition of c-Met and EGFR.
Improvements in 3D cell culture reagents, methods, and automated microscopes that can capture 3D image data are making screening in 3D more accessible, including more complex models such as co-cultures.5,30 Here we describe how high-content analysis can maximize the information that can be extracted from the more complex phenotypes obtained in 3D cell culture. These include disease-relevant features, such as tumor cell invasion, but also a multitude of other features that can be exploited for profiling purposes. Thus, in addition to screening for inhibition of a disease phenotype, target-based screening can be performed. Traditionally, receptor activation has been measured using biochemical methods. While sensitive, these methods do not discriminate nonselective inhibition of the target. An advantage of the multiparametric profiling approach described here is that in addition to detecting inhibition of the c-Met and EGFR, off-target effects can also be detected if the off-targets have an impact on tissue phenotype.
Our method was used to determine inhibitory activity of both small molecules and antibodies. We applied the method to both c-Met and EGFR, but in principle, it can be applied to any target if a cell line can be identified in which the activity of the target influences cell phenotype. Combining the advantages of physiological relevance and phenotypic complexity, phenotypic screening and profiling with 3D cell cultures has the potential to improve the quality of hits from screens and make previously challenging targets more accessible, potentially leading to a higher success rate of molecules in clinical trials.
Supplementary Material
Acknowledgments
We thank Souichi Ogata (Janssen Oncology R&D) for kindly providing ArQ-197 and Rob Roovers (Merus) for providing the Genentech 5D5 Fab and IgG. Furthermore, we thank Dr. Marc Bickle, Max Planck, Dresden, for introducing us to the KNIME software.
Footnotes
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental.
Declaration of Conflicting Interests: L. S. Price is a founder and major share holder of OcellO B.V. The other authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: T. H. Booij is supported by the Dutch Technology Foundation STW (Project 11823), which is part of The Netherlands Organization for Scientific Research (NWO). The other authors received no financial support for the research, authorship, and/or publication of this article. The authors received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Mishra D. K., Sakamoto J. H., Thrall M. J., et al. Human Lung Cancer Cells Grown in an Ex Vivo 3D Lung Model Produce Matrix Metalloproteinases Not Produced in 2D Culture. PLoS One. 2012, 7, e45308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bradbury P., Fabry B., O’Neill G. M. Occupy Tissue: The Movement in Cancer Metastasis. Cell Adh. Migr. 2012, 6, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smalley K. S., Lioni M., Herlyn M. Life Isn’t Flat: Taking Cancer Biology to the Next Dimension. In Vitro Cell Dev. Biol. Anim. 2006, 42, 242–247. [DOI] [PubMed] [Google Scholar]
- 4. Breslin S., O’Driscoll L. Three-Dimensional Cell Culture: The Missing Link in Drug Discovery. Drug Discov. Today. 2013, 18, 240–249. [DOI] [PubMed] [Google Scholar]
- 5. Li L., Zhou Q., Voss T. C., et al. High-Throughput Imaging: Focusing in on Drug Discovery in 3D. Methods 2016, 96, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Taubenberger A. V., Bray L. J., Haller B., et al. 3D Extracellular Matrix Interactions Modulate Tumour Cell Growth, Invasion and Angiogenesis in Engineered Tumour Microenvironments. J. Act. Bio. 2016, 36, 73–85. [DOI] [PubMed] [Google Scholar]
- 7. Debnath J., Brugge J. S. Modelling Glandular Epithelial Cancers in Three-Dimensional Cultures. Nat. Rev. 2005, 5, 675–688. [DOI] [PubMed] [Google Scholar]
- 8. Schmeichel K. L., Bissell M. J. Modeling Tissue-Specific Signaling and Organ Function in Three Dimensions. J. Cell Sci. 2003, 116(Pt 12), 2377–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Brien L. E., Zegers M. M., Mostov K. E. Opinion: Building Epithelial Architecture: Insights from Three-Dimensional Culture Models. Nat. Rev. Mol. Cell Biol. 2002, 3, 531–537. [DOI] [PubMed] [Google Scholar]
- 10. Härmä V., Knuuttila M., Virtanen J., et al. Lysophosphatidic Acid and Sphingosine-1-Phosphate Promote Morphogenesis and Block Invasion of Prostate Cancer Cells in Three-Dimensional Organotypic Models. Oncogene 2014, 31, 2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ren Y., Cao B., Law S., et al. Hepatocyte Growth Factor Promotes Cancer Cell Migration and Angiogenic Factors Expression: A Prognostic Marker of Human Esophageal Squamous Cell Carcinomas. Clin. Cancer Res. 2005, 11, 6190–6197. [DOI] [PubMed] [Google Scholar]
- 12. Chan G. K. Y., Lutterbach B. A., Pan B.-S., et al. High-Throughput Analysis of HGF-Stimulated Cell Scattering. J. Biomol. Screen. 2008, 13, 847–854. [DOI] [PubMed] [Google Scholar]
- 13. Radtke S., Milanovic M., Rossé C., et al. ERK2 but Not ERK1 Mediates HGF-Induced Motility in Non–Small Cell Lung Carcinoma Cell Lines. J. Cell Sci. 2013, 126, 2381–2391. [DOI] [PubMed] [Google Scholar]
- 14. Zaritsky A., Natan S., Ben-Jacob E., et al. Emergence of HGF/SF Induced Coordinated Cellular Motility. PLoS One 2012, 7, e44671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Comoglio P. M., Trusolino L. Invasive Growth: From Development to Metastasis. J. Clin. Invest. 2002, 109, 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hardy K. M., Booth B. W., Hendrix M. J. C., et al. ErbB/EGF Signaling and EMT in Mammary Development and Breast Cancer. J. Mammary Gland. Biol. Neoplasia. 2010, 25, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Appert-Collin A., Hubert P., Crémel G., et al. Role of ErbB Receptors in Cancer Cell Migration and Invasion. Front. Pharmacol. 2015, 6, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zi D., Klop M. J., Rogkoti M., et al. Ultra High Content Analysis and Phenotype Profiling of 3D Cultured Micro-Tissues. PLoS One 2014, 9, e109688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Venables W. N., Ripley B. D. Modern Applied Statistics with S; Springer: New York, 2002; 4th ed. [Google Scholar]
- 20. Wickham H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, 2009. [Google Scholar]
- 21. Ligges U., Maechler M. Scatterplot3d—An R Package for Visualizing Multivariate Data. J. Stat. Softw. 2003, 8(11), 1–20. [Google Scholar]
- 22. Adjei A. A., Schwartz B., Garmey E. Early Clinical Development of ARQ 197, a Selective, Non-ATP-Competitive Inhibitor Targeting MET Tyrosine Kinase for the Treatment of Advanced Cancers. Oncologist 2011, 16, 788–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tu W. H., Zhu C., Clark C., et al. Efficacy of c-Met Inhibitor for Advanced Prostate Cancer. BMC Cancer 2010, 10, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Y., Wislez M., Fujimoto N., et al. A Selective Small Molecule Inhibitor of c-Met, PHA-665752, Reverses Lung Premalignancy Induced by Mutant K-ras. Mol. Cancer Ther. 2008, 7, 952–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caja L., Sancho P., Bertran E., et al. The Tyrphostin AG1478 Inhibits Proliferation and Induces Death of Liver Tumor Cells through EGF Receptor-Dependent and Independent Mechanisms. Biochem. Pharmacol. 2011, 82, 1583–1592. [DOI] [PubMed] [Google Scholar]
- 26. Feldman D. R., Einhorn L. H., Qiunn D. I., et al. A Phase 2 Multicenter Study of Tivantinib (ARQ 197) Monotherapy in Patients with Relapsed or Refractory Germ Cell Tumors. Invest. New Drugs, 2013, 31, 1016–1022. [DOI] [PubMed] [Google Scholar]
- 27. Sirenko O., Mitlo T., Hesley J., et al. High-Content Assays for Characterizing the Viability and Morphology of 3D Cancer Spheroid Cultures. Assay Drug Dev. Technol. 2015, 13, 402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Härmä V., Schukov H.-P., Happonen A., et al. Quantification of Dynamic Morphological Drug Responses in 3D Organotypic Cell Cultures by Automated Image Analysis. PLoS One. 2014, 9, e96426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Robinson S., Guyon L., Nevalainen J., et al. Segmentation of Image Data from Complex Organotypic 3D Models of Cancer Tissues with Markov Random Fields. PLoS One 2015, 10, e0143798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krausz E., de Hoogt R., Gustin E., et al. Translation of a Tumor Microenvironment Mimicking 3D Tumor Growth Co-Culture Assay Platform to High-Content Screening. J. Biomol. Screen. 2013, 18, 54–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.