Drennan et al. use a new mouse to show that A20-deficient NKT cells are hyperresponsive to TCR-dependent stimuli and have severely impaired NKT cell development.
Abstract
Natural killer T (NKT) cells are innate lymphocytes that differentiate into NKT1, NKT2, and NKT17 sublineages during development. However, the signaling events that control NKT sublineage specification and differentiation remain poorly understood. Here, we demonstrate that the ubiquitin-modifying enzyme TNFAIP3/A20, an upstream regulator of T cell receptor (TCR) signaling in T cells, is an essential cell-intrinsic regulator of NKT differentiation. A20 is differentially expressed during NKT cell development, regulates NKT cell maturation, and specifically controls the differentiation and survival of NKT1 and NKT2, but not NKT17, sublineages. Remaining A20-deficient NKT1 and NKT2 thymocytes are hyperactivated in vivo and secrete elevated levels of Th1 and Th2 cytokines after TCR ligation in vitro. Defective NKT development was restored by compound deficiency of MALT1, a key downstream component of TCR signaling in T cells. These findings therefore show that negative regulation of TCR signaling during NKT development controls the differentiation and survival of NKT1 and NKT2 cells.
INTRODUCTION
NF-κB proteins comprise a family of transcription factors that can interact to form homo- and heterodimers with distinct regulatory functions (Sun et al., 2013). In unstimulated cells, inactive NF-κB dimers are sequestered in the cytosol by a family of inhibitory proteins called IκBs (Sun et al., 2013). After stimulation, phosphorylation of IκB inhibitors by the IκB kinase (IKK) complex results in their polyubiquitination and degradation, with concomitant translocation of active NF-κB to the nucleus. In T cells, NF-κB activation is induced after stimulation of the TCR and involves recruitment and assembly of the Carma1–Bcl10–MALT1 (CBM) complex (Rawlings et al., 2006), with the subsequent phosphorylation and degradation of IκB inhibitors (Schulze-Luehrmann and Ghosh, 2006). Ubiquitination of CBM complex proteins such as MALT1 and Bcl10 promotes association with regulatory components of the IKK complex (Oeckinghaus et al., 2007; Wu and Ashwell, 2008), thereby linking ubiquitination of the CBM complex to recruitment of IKKs and activation of NF-κB in T cells.
In this context, enzymes that disassemble ubiquitin chains from protein substrates have been shown to function as negative regulators of the NF-κB signaling pathway (Harhaj and Dixit, 2012). The zinc finger protein A20, also known as TNF-induced protein 3 (TNFAIP3), is characterized as a deubiquitinating enzyme (DUB) capable of negatively regulating activation of NF-κB in T cells (Coornaert et al., 2008; Stilo et al., 2008). Here, constitutive A20 DUB activity removes ubiquitin chains from protein substrates such as MALT1, thereby inhibiting inducible IKK activity in resting T cells. The rapid removal of A20 after TCR stimulation is therefore associated with elevated IKK activity and enhanced NF-κB signaling in activated T cells (Düwel et al., 2009). Such findings are reflected in a requirement for A20 during the development, activation, and survival of the CD4 and CD8 T cell compartments in mice (Giordano et al., 2014; Matsuzawa et al., 2015; Onizawa et al., 2015).
Invariant NKT (iNKT) cells are an innate T cell population derived from CD4+ T lineage–committed cells that recognize glycolipid antigens presented by the MHC class I–related molecule CD1d (Bendelac et al., 2007). Positive selection of iNKT precursors is associated with the initiation of a developmental program marked by the differential expression of CD24, CD44, and NK1.1 (Bendelac et al., 2007), which define sequential stages of maturation in the thymus and peripheral organs of mice. Inhibition of NF-κB in thymocytes arrests the development of iNKT cell precursors before the NK1.1+ stage (Stanic et al., 2004), presumably reflecting a threshold requirement for NF-κB activity during the earliest stages of iNKT cell differentiation. In contrast, NF-κB proteins, such as p50 (NF-κB1) and p65 (RelA), regulate maturation of the iNKT cell lineage (Sivakumar et al., 2003; Stankovic et al., 2011), and are proposed to have redundant functions during the initial stages of iNKT cell development. Interestingly, regulation of constitutive NF-κB activation by the deubiquitinase cylindromatosis (CYLD) controls iNKT cell maturation and hyperactivation in a manner dependent on ICOS expression and responsiveness toward the cytokine IL-7 (Lee et al., 2010). Whether A20 has a functional role during iNKT cell development has yet to be explored. In this study, we investigated the role of A20 during iNKT cell development and identify A20 as an essential regulator of iNKT cell differentiation and function in mice.
RESULTS AND DISCUSSION
A20GFP transgenic mice report increased expression of A20 mRNA within the peripheral iNKT cell compartment
Initial in situ hybridization studies revealed that A20 is constitutively expressed in subpopulations of thymocytes and peripheral T cells (Tewari et al., 1995; Giordano et al., 2014; Matsuzawa et al., 2015; Onizawa et al., 2015); however, it is currently unknown whether the iNKT cell lineage differentially expresses A20 mRNA in thymus and peripheral organs during development. To create a fluorescent reporter that would reflect endogenous expression of A20 mRNA in iNKT cells, we inserted GFP at the start codon of exon 2 of the A20 (tnfaip3) gene in a bacterial artificial chromosome (BAC) construct. Two of three C57BL/6J founders generated expressed identical levels of GFP in CD4+ and CD3ε+ thymocytes, respectively (Fig. 1, A and B; and not depicted). Flow cytometric analysis of lymphocytes isolated from the thymus, spleen, and liver of A20GFP reporters (founder 672) revealed differential expression of GFP within distinct T cell lineages during development. In thymus, A20GFP median fluorescence intensity (MFI) increased as thymocyte precursors matured from stages double-negative (DN) 1 to DN3 (Fig. 1 C), but was virtually extinguished within DN4 thymocytes. A20GFP was then reexpressed during positive selection of double-positive (DP) thymocytes (CD3ε− DP to CD3εhi DP; Fig. 1 C), and modestly up-regulated within the CD4 single-positive (SP), CD8 SP, and γδ T cell lineages during maturation. Similarly, developing iNKT thymocytes up-regulated A20GFP mRNA during maturation from immature stage 1 (CD44−NK1.1−) to mature stage 3 (CD44+NK1.1+) cells (Fig. 1 C). In spleen and liver, CD4, CD8, and γδ T cell lineages expressed roughly uniform levels of A20GFP (Fig. 1 D), with the highest comparative A20GFP MFI detected within splenic iNKT cells. In contrast, A20GFP expression within NKT1, 2, and 17 sublineages (Lee et al., 2013; Drennan et al., 2014) was comparable in all organs examined (Fig. 1 E), with the highest overall expression detected within splenic and popliteal LN (PLN) NKT sublineages, respectively. Thus, comparative levels of A20GFP are expressed in developing T and iNKT thymocytes, and peripheral iNKT cell maturation is associated with up-regulation of A20GFP, particularly within NKT sublineages present in the spleen and PLNs.
Figure 1.
Up-regulated A20 mRNA expression within peripheral iNKT cells in A20GFP reporter mice. A20GFP expression within (A) CD4+ and (B) CD3ε+ thymocytes isolated from Founder 672 (open histogram; n = 6) and C57BL/6J (filled histogram, n = 6) mice, respectively. (C and D) A20GFP median fluorescence intensity (MFI) within T cell subsets isolated from thymus, spleen, and liver of A20GFP mice (n = 6). (E) A20GFP MFI within NKT sublineages isolated from thymus, blood, spleen, liver, lung, and PLNs of A20GFP mice, respectively (n = 4). All data are from two independent experiments. Numbers in B–D represent A20GFP MFI within the indicated subsets. ST, stage; LP, lymphocyte precursor.
A20 is an essential regulator of iNKT cell development
A20 has recently been shown to regulate NF-κB–dependent development and activation of primary CD8 T cells in vivo (Giordano et al., 2014), thereby substantiating previous studies describing a regulatory role for A20 during activation of NF-κB in T cells in vitro (Coornaert et al., 2008; Stilo et al., 2008; Düwel et al., 2009). To examine the requirement for A20 during iNKT cell development, we generated CD4 T cell–specific, A20-deficient mice by intercrossing CD4-cre transgenic mice with mice carrying a LoxP-flanked A20 allele (Vereecke et al., 2010). Here, deletion of A20 DNA and protein in splenic CD4 T cells isolated from CD4-cre;A20F/F mice was confirmed by Southern and Western blot analysis, respectively (unpublished data). Strikingly, the frequency and absolute cell counts of iNKT cells present in the thymus, blood, spleen, liver, lung, and PLNs of CD4-cre;A20F/F mice were severely reduced when compared with littermate controls (Fig. 2, A and B). A subsequent analysis of iNKT cell maturation in thymus of CD4-cre;A20F/F mice using cell surface markers CD24, CD44, and NK1.1 (Bendelac et al., 2007) revealed similar percentages of stage 0 (CD24hiCD44loNK1.1−), stage 1 (CD24−CD44loNK1.1−), stage 2 (CD24−CD44hiNK1.1−), and stage 3 (CD24−CD44hiNK1.1hi) iNKT cells compared with littermate controls (Fig. 2 C); however, the absolute number of stage 3 iNKT thymocytes detected in CD4-cre;A20F/F thymi was significantly lower than that observed in control thymi (Fig. 2 D). These results suggest that A20 is not involved in the selection and expansion of iNKT cell precursors, but rather maturation of stage 3 NK1.1+ iNKT thymocytes. In this context, a previous study has shown that maturation of stage 3 iNKT thymocytes is unaffected in CYLD-deficient mice (Lee et al., 2010), suggesting that iNKT cell development is differentially regulated by CYLD. More recently, an alternate iNKT cell developmental model was proposed in which promyelocytic leukemia zinc finger–positive iNKT cell precursors give rise to three distinct iNKT cell sublineages, termed NKT1, NKT2, and NKT17 (Lee et al., 2013). NKT1, NKT2, and NKT17 sublineage cells are characterized by differential expression of the transcription factors T-bet, GATA-3, and RORγt and produce the cytokines IFN-γ, IL-4, and IL-17, respectively. Furthermore, NKT1 sublineage cells can be subdivided into two subsets, defined as NKT1a and NKT1b, which exhibit distinct developmental and tissue-specific distribution profiles (Drennan et al., 2014). An analysis of iNKT cell development in CD4-cre;A20F/F mice using the aforementioned alternate NKT cell developmental scheme revealed a selective reduction in NKT1a thymocytes in CD4-cre;A20F/F thymi relative to controls (Fig. 2, E and F), with no significant differences observed within the NKT1b, NKT2, or NKT17 populations in thymus. In contrast, reduced numbers of both NKT1 and NKT2 sublineages were detected in the blood, spleen, liver, lung, and PLNs of CD4-cre;A20F/F mice relative to controls (Fig. 2, E and F), whereas the frequency and distribution of NKT17 sublineage cells in all organs examined were no different between control and CD4-cre;A20F/F mice (Fig. 2, E and F). Thus, A20 regulates development of NKT1 cells in thymus and peripheral NKT1 and NKT2 sublineages but is not required for development of the NKT17 sublineage.
Figure 2.
iNKT cell development is regulated by A20. (A and B) Frequency and absolute iNKT cell counts in thymus (Thy), blood (Bld), spleen (Spl), liver (Lvr), lung (Lng), and PLN of control (n = 8) and CD4-cre;A20F/F (n = 9) mice, respectively. Data are from four independent experiments pooled. (C and D) Frequency and absolute cell counts of ST0, ST1, ST2, and ST3 iNKT cells isolated from thymi of control (n = 8) and CD4-cre;A20F/F (n = 9) mice, respectively. Data are pooled from two independent experiments. (E) Flow cytometric analysis and absolute cell counts (F) of NKT1a, NKT1b, NKT2, and NKT17 sublineages present in thymus, blood, spleen, liver, lung, and PLN of control (n = 8) and CD4-cre;A20F/F (n = 9) mice, respectively. Data are from four independent experiments pooled. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
A20 regulates activation and survival of NKT1 and NKT2 sublineages
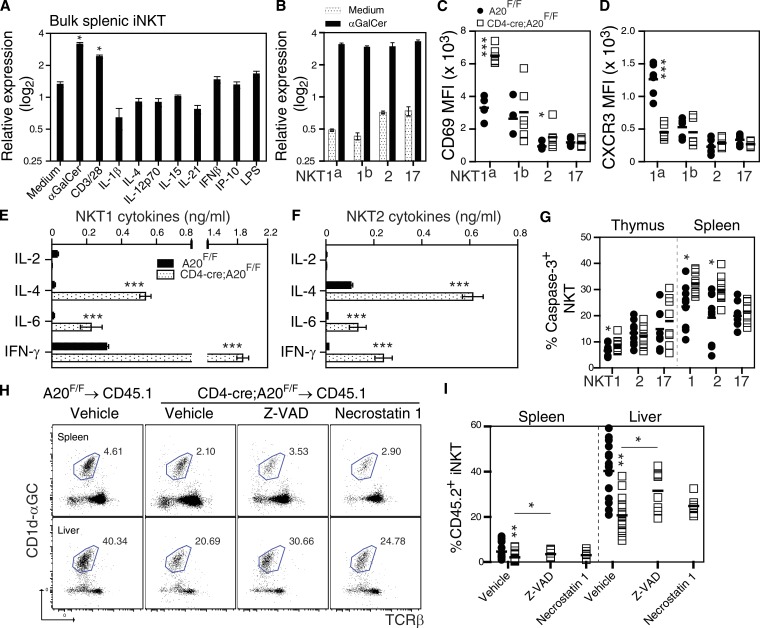
Because analysis of A20GFP reporter mice demonstrated that A20 is uniformly expressed within NKT sublineages in an organ-specific manner (Fig. 1 D), we examined whether A20 mRNA expression is differentially expressed in in vitro–expanded iNKT cells after stimulation. Here, A20 mRNA was significantly up-regulated in bulk splenic iNKT cells stimulated via their TCR, as opposed to iNKTs incubated in the presence of various cytokine or TLR-dependent stimuli (Fig. 3 A). Furthermore, TCR-dependent stimulation of in vitro–expanded NKT cell sublineages resulted in comparable induction of A20 mRNA relative to medium controls (Fig. 3 B), mirroring expression of A20 within NKT sublineages in A20GFP reporter mice (Fig. 1 D). A subsequent phenotypic analysis of NKT sublineage cells isolated from the thymi of CD4-cre;A20F/F mice revealed a significant up-regulation of CD69 within NKT1a and NKT2 thymocytes when compared with controls (Fig. 3 C), suggesting that A20 functions to restrict expression of this activation marker during development. In addition, A20-deficient NKT1a thymocytes failed to up-regulate the chemokine receptor CXCR3 (Fig. 3 D), whereas cell-surface expression of ICOS was not affected by deletion of A20 (unpublished data), suggesting that A20 and CYLD differentially regulate activation of NKT1 and 2 sublineages during development (Lee et al., 2010). We next examined whether A20 regulates TCR-induced cytokine production by in vitro–expanded NKT1 and NKT2 sublineage cells. For these experiments, NKT1 and NKT2 thymocytes were FACS sorted from control and CD4-cre;A20F/F thymi, expanded in vitro, and stimulated with plate-bound anti-CD3ε/CD28 as previously described (Govindarajan et al., 2015). As shown in Fig. 3 (E and F), A20-deficient NKT1 and NKT2 thymocytes secreted significantly higher levels of IL-4, IL-6, and IFN-γ after TCR stimulation than did control NKT cells, suggesting that A20 functions to limit TCR-dependent activation of NKT1 and 2 sublineages in thymus. Furthermore, up-regulated expression of CD69 and enhanced cytokine production by NKT1 and 2 thymocytes in the absence of A20 was associated with increased activity of intracellular caspase-3 in both thymus and spleen (Fig. 3 G), suggesting that A20 regulates activation-induced cell death (AICD) within the NKT1 and 2 compartments. To evaluate whether A20 limits AICD of iNKT cells via apoptosis or necroptosis (Onizawa et al., 2015) in vivo, we transferred control or A20-deficient thymocytes into irradiated congenic hosts pretreated with either the pan-caspase inhibitor Z-VAD-FMK or the necroptosis RIP1 kinase inhibitor necrostatin 1 (Onizawa et al., 2015). Analysis of the frequency of iNKT cells recovered from chimeras treated with vehicle revealed that fewer A20-deficient iNKT cells were detected in the spleen and liver when compared with those injected with wild-type iNKT cells (Fig. 3, H and I), and that chimeras treated with Z-VAD contained a significantly higher frequency of A20-deficient iNKT cells than did chimeras treated with necrostatin 1 (Fig. 3, H and I). We therefore conclude that A20 functions to regulate cytokine production by NKT1 and NKT2 sublineages in a TCR-dependent manner and limits AICD by apoptosis during development and maturation in the peripheral compartment. In contrast, we show a limited role for A20 during development and survival of the NKT17 sublineage.
Figure 3.
A20 regulates NKT1 and NKT2 sublineage activation and survival. qPCR analysis of A20 mRNA expression in bulk splenic iNKT (A) or NKT sublineage (B) cells after 24 h in vitro treatment with the indicated agents. Data are from two independent experiments. (C and D) CD69 and CXCR3 expression on NKT1a, NKT1b, NKT2, and NKT17 cells isolated from thymi of control (n = 6) and CD4-cre;A20F/F mice (n = 6), respectively. Data are from two independent experiments. (E and F) α-CD3/CD28-induced cytokine production by in vitro–expanded control and A20-deficient NKT1 and NKT2 thymocytes (n = 3). Data are from two independent experiments. (G) FACS analysis of caspase 3 activity in NKT1, 2, and 17 sublineages isolated from thymus and spleen of control (n = 9) and CD4-cre;A20F/F (n = 9) mice, respectively. Data are pooled from three independent experiments. (H and I) Frequency of control and A20-deficient iNKT cells recovered from CD45.1 congenic mice after in vivo treatment with vehicle (control, n = 16; CD4-cre;A20F/F, n = 17), Z-VAD (10 mg/kg; CD4-cre;A20F/F, n = 8), or Necrostatin-1 (10 mg/kg; CD4-cre;A20F/F, n = 7), respectively. FACS dot blots show CD45.2+ iNKT cells isolated from chimeras and represents pooled data from two independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.0005. n.s., not significant.
MALT1 threshold function regulates iNKT cell development in the absence of A20
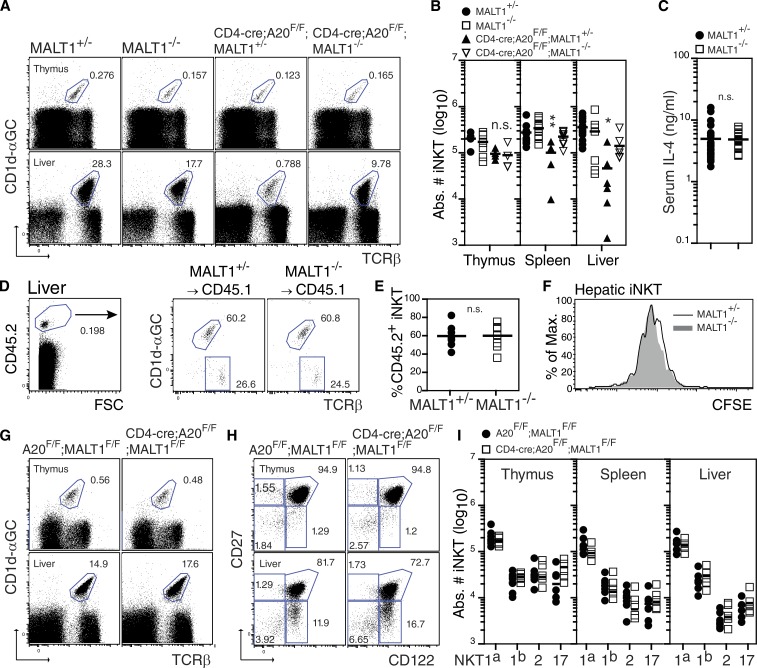
Previous studies have demonstrated that recruitment of MALT1 to the CBM complex is essential for TCR-induced activation of NF-κB within the conventional T cell compartment (Ruefli-Brasse et al., 2003; Kingeter and Schaefer, 2008). As a consequence, MALT1-deficient T cells exhibit impaired IL-2 production and diminished proliferation in response to TCR ligation. More recently, TCR ligation has been shown to induce the recruitment of A20 into a complex with MALT1 and Bcl10 (Coornaert et al., 2008; Rebeaud et al., 2008), resulting in MALT1-mediated cleavage of A20 and the induction of NF-κB. Thus, negative regulation of NF-κB in T cells also requires the proteolytic activity of MALT1 (Coornaert et al., 2008; Staal et al., 2011). Due to the central role of MALT1 during TCR signaling, we sought to determine whether a MALT1-dependent threshold was required during iNKT cell development and activation in CD4-cre;A20F/F mice (Figs. 2 and 3). Primary characterization showed no stringent requirement for MALT1 during iNKT cell development, as iNKT cell frequencies and absolute cell counts were comparable in thymus, spleen, and liver of germline MALT1-deficient (MALT1−/−) mice when compared with littermate controls (Fig. 4, A and B). Furthermore, MALT1-deficient and control iNKT cells produced equivalent levels of IL-4 after i.p. administration of α-Galactosylceramide (α-GalCer; Fig. 4 C), and transfer of MALT1-deficient iNKT thymocytes into irradiated congenic hosts revealed no significant differences in survival (Fig. 4, D and E) or proliferation (Fig. 4 F) when compared with control iNKT cells. These results therefore show that in iNKT cells that express A20, TCR-induced cytokine production and homeostasis of the iNKT cell lineage is not significantly altered by the absence of MALT1. To investigate MALT1 function during iNKT cell development in the absence of A20, we subsequently backcrossed CD4-cre;A20F/F mice onto a MALT1-deficient background. Strikingly, germline deletion of MALT1 in CD4-cre;A20F/F mice (CD4-cre;A20F/F;MALT1−/−) significantly restored iNKT cell development in the thymus, spleen, and liver when compared with CD4-cre;A20F/F;MALT1+/− control mice (Fig. 4, A and B). Furthermore, complete rescue of NKT1 and NKT2 sublineage development in the absence of A20 was achieved when MALT1 was conditionally deleted within iNKT cells using CD4-cre;A20F/F;MALT1F/F mice (Fig. 4, G–I). Thus, NKT sublineage differentiation and survival are regulated by a threshold function of MALT1 in the absence of A20.
Figure 4.
MALT1 deficiency rescues iNKT cell development in CD4-cre;A20F/F mice. (A) Frequency and absolute iNKT cell counts (B) in thymus and liver of MALT1+/− (n = 13), MALT1−/− (n = 12), CD4-cre;A20F/F;MALT1+/− (n = 6), and CD4-cre;A20F/F;MALT1−/− (n = 7) mice, respectively. Data are pooled from four independent experiments. (C) Serum IL-4 concentration in MALT1+/− (n = 26) and MALT−/− (n = 20) mice after i.p. injection of α-GalCer. Results are pooled from two independent experiments. (D and E) Frequency of MALT1+/− (n = 4) and MALT1−/− (n = 4) iNKT cells recovered from CD45.1 congenic mice after transfer of CFSE-labeled CD8-depleted thymocytes. (F) Homeostatic proliferation of MALT1+/− and MALT1−/− iNKT cells in the livers of congenic mice 8 d after transfer. Data are from two independent experiments pooled. (G–I) Frequency and absolute iNKT cell counts in thymus, spleen, and liver of control A20F/F;MALT1F/F (n = 6) and CD4-cre;A20F/F;MALT1F/F (n = 6) mice, respectively. FACS dot blots in H are gated on βTCR+CD1d-αGC+ iNKT cells from each organ. Data are pooled from three independent experiments.*, P < 0.05; **, P < 0.01. n.s., not significant.
To summarize, we define a regulatory role for A20 during differentiation and survival of NKT sublineages in mouse. Here, A20 functions to limit TCR-dependent activation of the NKT1 and NKT2 sublineages in thymus, thereby preventing AICD after stimulation of the TCR. Furthermore, we show that both A20 and MALT1 dually regulate NKT sublineage differentiation and survival in a cell-intrinsic manner. Our results therefore suggest that MALT1 may exert distinct threshold and/or modulatory functions in conjunction with A20 during iNKT cell development. Our findings also highlight that therapeutic inhibition of A20 within the iNKT cell compartment may have substantial clinical potential in modulating or limiting iNKT cell activation or survival in defined inflammatory and autoimmune diseases (Kumar and Delovitch, 2014).
MATERIALS AND METHODS
Transgenic mice
To generate A20 (tnfaip3)EGFP reporter mice on a C57BL/6J background, a BAC containing the entire genomic locus of mouse tnfaip3, RP23-453B18, was ordered from the BACPAC Resource Center (Oakland, CA). A cassette containing EGFP-stop-FRT-pgk/gb2-neo/kan-FRT was inserted in the ATG of exon 2 of tnfaip3 by Red/ET recombination. Subsequently, the selection cassette FRT-pgk/gb2-neo/kan-FRT was removed by Flpe recombination in E. coli, resulting in a reporter construct where EGFP expression is driven by the tnfaip3 promoter. A 193-kb NruI-digested BAC fragment was isolated by PFGE and injected into the pronuclei of C57BL/6J zygotes. Offspring was genotyped by PCR using primers specific for the EGFP transgene. To generate mice with a conditional MALT1 allele, Malt1tm1a(EUCOMM)Hmgu ES cells (129P2/OlaHsd) were ordered from IMPC, injected into 3.5-d blastocysts, and transferred to the uteri of pseudopregnant foster mothers. Male chimeras were mated with C57BL/6 females to obtain germline transmission of the MALT1-floxed allele containing the neomycine selection cassette (MALT1NFL). The Frt-flanked neomycin cassette was removed by crossing MALT1NFL mice with an Flp-deleter strain (Rodríguez et al., 2000), generating a MALT1-floxed allele (MALT1F/F). T cell–specific MALT1-deficient (CD4-cre;MALT1F/F) and A20-deficient (CD4-cre;A20F/F) mice were generated by intercrossing with C57BL/6 CD4-cre transgenic mice for 5 and >10 generations, respectively. Germline MALT1−/− mice on a sv129 background were provided by T. Mak (Princess Margaret Cancer Centre, Ontario, Canada) and have been backcrossed to C57BL/6 for >10 generations. Congenic B6.SJL-PtprcaPep3b/BoyJ breeders were purchased from Jackson ImmunoResearch Laboratories. All animal procedures were approved by the Institutional Animal Care and Ethics Committee.
Flow cytometry and antibodies
Cells were analyzed on FACSCantoII (BD) and FlowJo software (Tree Star), and sorted on FACS Aria II (BD). Antibodies used were CD122 PerCP-eFluor 710 (TM-b1), CD19-PerCP-Cy5.5 (eBio1D3), CD24-FITC (M1/69), CD25-PE (PC61.5), CD27-APC (LG.7F9), CD3ε-V500 (500A2), CD4-APC-eFluor 780 (RM4-5), CD44-FITC/APC/APC-eFluor 780 (IM7), CD45.1-PerCP-Cy5.5 (A20), CD45.2-APC-Cy7 (104), NK1.1-PE-Cy7/PerCP-Cy5.5 (PK136), CD62L-FITC/APC/PE-Cy7 (MEL-14), CD69-PE-Cy7 (H1.2F3), CD8α-APC (53–6.7), βTCR-APC-eFluor 780 (H57-597), Qa-2 FITC (69H1-9-9), and γδTCR PerCP-eFluor 710 (eBioGL3; all from eBioscience). iNKT cells were stained at room temperature using α-GalCer–loaded CD1d tetramers. Stainings for intracellular antigens were performed using the FoxP3/Transcription Factor staining buffer set (eBioscience). Active caspase-3+ iNKT cells were stained using PhiPhiLux G1D2 (Oncoimmunin) according to the manufacturer’s recommendations. Subsets are defined as DN1 (CD25−CD44+); DN2 (CD25+CD44+); DN3 (CD25+CD44−); DN4 (CD25−CD44−); ISP (CD25−CD44−CD3ε−CD4−CD8+); DP (CD4+CD8+); ST1 (CD44−NK1.1−); ST2 (CD44hiNK1.1−); ST3 (CD44hiNK1.1+); CD4naive (CD4+CD62LhiCD44−); CD4memory (CD4+CD62L−CD44hi); iNKT (TCRβ+CD1d-αGC+); NKT1a (CD27+IL-2Rβ+); NKT1b (CD27−IL-2Rβ+); NKT2 (CD27+IL-2Rβ−); NKT17 (CD27−IL-2Rβ−); and γδ (γδTCR+).
Cell suspension preparations
Single-cell suspensions from the thymus were resuspended in PBS containing 1 mM EDTA and 0.5% BSA, whereas spleen and liver suspensions were layered over Ficoll or 33% Percoll gradients, respectively. Peripheral blood mononuclear cells were isolated by cardiac puncture in 20 U/ml PBS/heparin and layered over a Ficoll gradient. iNKT thymocytes were enriched for using mouse CD8-Dynabeads (Invitrogen). Bone marrow mononuclear cells were flushed from the femur and tibia of mice and resuspended in PBS containing 1 mM EDTA and 0.5% BSA.
Quantitative real-time RT-PCR
Total RNA was extracted from lymphocytes isolated from the thymus, spleen, and liver using the miRNeasy Mini kit (QIAGEN) and analyzed using the Agilent 2100 Bioanalyzer and RNA 6000 LabChip kit (Agilent Technologies). cDNA was produced and amplified using the QuantiTect Whole Transcriptome kit (QIAGEN). A20 mRNA was detected using the following primer pair: forward, 5′-TTCAGGGCCAGCTAGTATCCTGGG-3′; reverse, 5′-ACCTGCTGGCAGGAAACCCC-3′. PCR products were amplified with the Fast Start SYBR Green Master mix (Roche) and detected with a LightCycler 480 System (Roche). Samples were normalized using qBasePlus (Biogazelle NV) against at least three of the following genes: Rpl13a, Eif4b, B2m, Actb, or Gapdh as described previously (Drennan et al., 2014).
In vitro expansion and stimulation of iNKT cells
iNKT cells were isolated and FACS sorted from the pooled livers of 5 C57BL/6J mice, as previously described (Govindarajan et al., 2015). In brief, iNKT cells were resuspended in complete RPMI-1640 medium containing 10 ng/ml recombinant mouse IL-2, 1 ng/ml recombinant mouse IL-12, and 1 µg/ml soluble anti-CD28 (all from eBioscience) and plated in α-CD3ε–coated (3 µg/ml) 96-well flat-bottom plates at 0.106 cells/well (BD). 2 d later, cells were rested for 2 d in complete RPMI-1640 medium, resuspended, and plated in complete RPMI-1640 medium containing 2 U/ml recombinant mouse IL-7 (eBioscience) until day 8. iNKT cells were then resuspended in complete RPMI-1640 and cultured in α-CD3ε–coated 96-well plates until day 11. The latter two culture steps were repeated and, after 20–21 d, fully expanded iNKT cells were used for experiments. For restimulation experiments, 105 iNKT cells were resuspended in RPMI-1640 medium containing 10% heat inactivated FCS, 100 U/ml penicillin/streptomycin, 2 mM glutamine, 0.1 mM nonessential amino acids, 5.5 × 102 µM µ-ME (all from GIBCO), and stimulated with either α-CD3ε/CD28 (3 µg.ml−1/5 µg.ml−1), α-GalCer/CD1d (10 µg/ml−1/100 ng.ml−1), IL-1β (20 ng/ml), IL-4 (20 ng/ml), IL-12p70 (20 ng/ml), IL-15 (20 ng/ml), IL-21 (20 ng/ml), IFN-β (100 U/ml), IP-10 (100 ng/ml), or LPS (20 ng/ml) for 24 h, after which cells were lysed and mRNA was isolated. Alternatively, 105 expanded NKT cell sublineage cells were restimulated with α-GalCer/CD1d (10 µg/ml−1/100 ng.ml−1) for 72 h, after which supernatants were collected and used for cytokine measurements by ELISA.
In vivo stimulation of iNKT cells
For serum IL-4 production, 5 µg of α-GalCer in 200ml PBS was injected i.p. Blood was subsequently collected after 4 h by retroorbital puncture and assayed for cytokine content by ELISA.
Generation of chimeras and transfer of CFSE-labeled thymocytes
Thymocytes isolated from MALT1+/− and MALT1−/− mice were depleted of CD8+cells using mouse CD8 Dynabeads (Invitrogen) and labeled with 3 µM CFSE (Molecular Probes). 1.5 × 107 CFSE-labeled thymocytes were subsequently transferred i.v. into irradiated (300 rad) 8-wk-old CD45.1+ host animals. Alternatively, animals treated with the pancaspase inhibitor Z-VAD-FMK (InvivoGen) or the necrosis inhibitor Necrostatin-1 (Sigma-Aldrich), 1.3 × 107 CD8-depleted thymocytes from A20F/F or CD4-cre;A20F/F mice were transferred i.v. into irradiated (300 rad) 12-wk-old congenic (CD45.1) host animals pretreated with either vehicle (DMSO in PBS), Z-VAD-FMK (10 mg/kg; InvivoGen) or Necrostatin-1 (10 mg/kg; Sigma-Aldrich) 30 min before thymocyte transfer. Intraperitoneal injections of vehicle, Z-VAD-FMK, or Necrostatin-1 were subsequently continued every 16 h for the following 6 d. Single-cell suspensions of spleen and liver were subsequently stained with FACS antibodies and analyzed by flow cytometry.
Statistical analysis
Unless stated otherwise, mean data are expressed as means ± 1 SD. P-values were calculated using the unpaired Student’s t test, and P < 0.05 was considered significant.
ACKNOWLEDGMENTS
Research was supported by grants from the Fund for Scientific Research-Flanders (FWO; to M.B. Drennan, D. Elewaut, R. Beyaert, and G. van Loo), the Group-ID Multidisciplinary Research Platform of Ghent University (to B.N. Lambrecht, D. Elewaut, R. Beyaert, and G. van Loo), the Interuniversity Attraction Poles Program (Project P7/07 to D. Elewaut and P7/32 to R. Beyaert), and the Strategic Basic Research program of the Instituut voor Innovatie door Wetenschap en Technologie (IWT; to D. Elewaut and R. Beyaert). J. Staal was supported as a postdoctoral fellow with the FWO.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AICD
- activation-induced cell death
- CYLD
- cylindromatosis
- DN
- double negative
- DP
- double positive
- IKK
- IκB kinase
- iNKT
- invariant NKT
- PLN
- popliteal LN
- SP
- single positive
- TNFAIP3
- TNF-induced protein 3
References
- Bendelac A., Savage P.B., and Teyton L.. 2007. The biology of NKT cells. Annu. Rev. Immunol. 25:297–336. 10.1146/annurev.immunol.25.022106.141711 [DOI] [PubMed] [Google Scholar]
- Coornaert B., Baens M., Heyninck K., Bekaert T., Haegman M., Staal J., Sun L., Chen Z.J., Marynen P., and Beyaert R.. 2008. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat. Immunol. 9:263–271. 10.1038/ni1561 [DOI] [PubMed] [Google Scholar]
- Drennan M.B., Govindarajan S., De Wilde K., Schlenner S.M., Ware C., Nedospasov S., Rodewald H.R., and Elewaut D.. 2014. The thymic microenvironment differentially regulates development and trafficking of invariant NKT cell sublineages. J. Immunol. 193:5960–5972. 10.4049/jimmunol.1401601 [DOI] [PubMed] [Google Scholar]
- Düwel M., Welteke V., Oeckinghaus A., Baens M., Kloo B., Ferch U., Darnay B.G., Ruland J., Marynen P., and Krappmann D.. 2009. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J. Immunol. 182:7718–7728. 10.4049/jimmunol.0803313 [DOI] [PubMed] [Google Scholar]
- Giordano M., Roncagalli R., Bourdely P., Chasson L., Buferne M., Yamasaki S., Beyaert R., van Loo G., Auphan-Anezin N., Schmitt-Verhulst A.M., and Verdeil G.. 2014. The tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) imposes a brake on antitumor activity of CD8 T cells. Proc. Natl. Acad. Sci. USA. 111:11115–11120. 10.1073/pnas.1406259111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan S., Elewaut D., and Drennan M.. 2015. An optimized method for isolating and expanding invariant natural killer T cells from mouse spleen. J. Vis. Exp. 105:e53256 10.3791/53256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harhaj E.W., and Dixit V.M.. 2012. Regulation of NF-κB by deubiquitinases. Immunol. Rev. 246:107–124. 10.1111/j.1600-065X.2012.01100.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingeter L.M., and Schaefer B.C.. 2008. Loss of protein kinase C theta, Bcl10, or Malt1 selectively impairs proliferation and NF-kappa B activation in the CD4+ T cell subset. J. Immunol. 181:6244–6254. 10.4049/jimmunol.181.9.6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., and Delovitch T.L.. 2014. Different subsets of natural killer T cells may vary in their roles in health and disease. Immunology. 142:321–336. 10.1111/imm.12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.J., Zhou X., Chang M., Hunzeker J., Bonneau R.H., Zhou D., and Sun S.C.. 2010. Regulation of natural killer T-cell development by deubiquitinase CYLD. EMBO J. 29:1600–1612. 10.1038/emboj.2010.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Holzapfel K.L., Zhu J., Jameson S.C., and Hogquist K.A.. 2013. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14:1146–1154. 10.1038/ni.2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa Y., Oshima S., Takahara M., Maeyashiki C., Nemoto Y., Kobayashi M., Nibe Y., Nozaki K., Nagaishi T., Okamoto R., et al. . 2015. TNFAIP3 promotes survival of CD4 T cells by restricting MTOR and promoting autophagy. Autophagy. 11:1052–1062. 10.1080/15548627.2015.1055439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A., Wegener E., Welteke V., Ferch U., Arslan S.C., Ruland J., Scheidereit C., and Krappmann D.. 2007. Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. EMBO J. 26:4634–4645. 10.1038/sj.emboj.7601897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onizawa M., Oshima S., Schulze-Topphoff U., Oses-Prieto J.A., Lu T., Tavares R., Prodhomme T., Duong B., Whang M.I., Advincula R., et al. . 2015. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat. Immunol. 16:618–627. 10.1038/ni.3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings D.J., Sommer K., and Moreno-García M.E.. 2006. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat. Rev. Immunol. 6:799–812. 10.1038/nri1944 [DOI] [PubMed] [Google Scholar]
- Rebeaud F., Hailfinger S., Posevitz-Fejfar A., Tapernoux M., Moser R., Rueda D., Gaide O., Guzzardi M., Iancu E.M., Rufer N., et al. . 2008. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat. Immunol. 9:272–281. 10.1038/ni1568 [DOI] [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., and Dymecki S.M.. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140. 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Ruefli-Brasse A.A., French D.M., and Dixit V.M.. 2003. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 302:1581–1584. 10.1126/science.1090769 [DOI] [PubMed] [Google Scholar]
- Schulze-Luehrmann J., and Ghosh S.. 2006. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 25:701–715. 10.1016/j.immuni.2006.10.010 [DOI] [PubMed] [Google Scholar]
- Sivakumar V., Hammond K.J., Howells N., Pfeffer K., and Weih F.. 2003. Differential requirement for Rel/nuclear factor κ B family members in natural killer T cell development. J. Exp. Med. 197:1613–1621. 10.1084/jem.20022234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staal J., Driege Y., Bekaert T., Demeyer A., Muyllaert D., Van Damme P., Gevaert K., and Beyaert R.. 2011. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 30:1742–1752. 10.1038/emboj.2011.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic A.K., Bezbradica J.S., Park J.J., Matsuki N., Mora A.L., Van Kaer L., Boothby M.R., and Joyce S.. 2004. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J. Immunol. 172:2265–2273. 10.4049/jimmunol.172.4.2265 [DOI] [PubMed] [Google Scholar]
- Stankovic S., Gugasyan R., Kyparissoudis K., Grumont R., Banerjee A., Tsichlis P., Gerondakis S., and Godfrey D.I.. 2011. Distinct roles in NKT cell maturation and function for the different transcription factors in the classical NF-κB pathway. Immunol. Cell Biol. 89:294–303. 10.1038/icb.2010.93 [DOI] [PubMed] [Google Scholar]
- Stilo R., Varricchio E., Liguoro D., Leonardi A., and Vito P.. 2008. A20 is a negative regulator of BCL10- and CARMA3-mediated activation of NF-kappaB. J. Cell Sci. 121:1165–1171. 10.1242/jcs.021105 [DOI] [PubMed] [Google Scholar]
- Sun S.C., Chang J.H., and Jin J.. 2013. Regulation of nuclear factor-κB in autoimmunity. Trends Immunol. 34:282–289. 10.1016/j.it.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari M., Wolf F.W., Seldin M.F., O’Shea K.S., Dixit V.M., and Turka L.A.. 1995. Lymphoid expression and regulation of A20, an inhibitor of programmed cell death. J. Immunol. 154:1699–1706. [PubMed] [Google Scholar]
- Vereecke L., Sze M., Mc Guire C., Rogiers B., Chu Y., Schmidt-Supprian M., Pasparakis M., Beyaert R., and van Loo G.. 2010. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J. Exp. Med. 207:1513–1523. 10.1084/jem.20092474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J., and Ashwell J.D.. 2008. NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-kappaB activation. Proc. Natl. Acad. Sci. USA. 105:3023–3028. 10.1073/pnas.0712313105 [DOI] [PMC free article] [PubMed] [Google Scholar]