Kanneganti and collaborators propose that the lysosomal protease cathepsin B provides a checkpoint for activation of the transcription factor TFEB and lysosomal biogenesis and explore the impact of this pathway on host defense against bacterial infection.
Abstract
Lysosomal cathepsins regulate an exquisite range of biological functions, and their deregulation is associated with inflammatory, metabolic, and degenerative diseases in humans. In this study, we identified a key cell-intrinsic role for cathepsin B as a negative feedback regulator of lysosomal biogenesis and autophagy. Mice and macrophages lacking cathepsin B activity had increased resistance to the cytosolic bacterial pathogen Francisella novicida. Genetic deletion or pharmacological inhibition of cathepsin B down-regulated mechanistic target of rapamycin activity and prevented cleavage of the lysosomal calcium channel TRPML1. These events drove transcription of lysosomal and autophagy genes via transcription factor EB, which increased lysosomal biogenesis and activation of autophagy initiation kinase ULK1 for clearance of the bacteria. Our results identified a fundamental biological function of cathepsin B in providing a checkpoint for homeostatic maintenance of lysosome populations and basic recycling functions in the cell.
INTRODUCTION
Lysosomes are cytoplasmic membrane–enclosed organelles filled with numerous acidic hydrolases. These organelles are dynamic and can fuse with a variety of vesicles to mediate degradation of extracellular materials that have been endocytosed or intracellular components that have been sequestered by autophagy. Furthermore, lysosomes are recycled and reformed after fusion with cargo-containing vacuoles (Luzio et al., 2007; Yu et al., 2010). Fundamental cellular processes orchestrated by lysosomes require cathepsin proteases; dysregulation of these activities contributes to the development of malignancy and Alzheimer’s disease (Gocheva et al., 2006; Mueller-Steiner et al., 2006). Secretion of cathepsins from lysosomes is involved in lysosomal membrane permeabilization–mediated cell death via caspase-dependent and caspase-independent pathways (Kreuzaler et al., 2011; Aits and Jäättelä, 2013). In addition, lysosomal rupture and cytoplasmic release of cathepsin B triggered by phagocytosis of crystals lead to activation of the NLRP3 inflammasome and maturation of the proinflammatory cytokines IL-1β and IL-18, which are key events linked to the development of atherosclerosis and Alzheimer’s disease (Halle et al., 2008; Duewell et al., 2010).
Certain lysosomal proteases also provide antimicrobial host defense against infection. Lysosomal acidic hydrolases, especially cathepsins, are executioner proteases used for degradation of intracellular bacteria within lysosomes. Macrophages lacking cathepsin D have a reduced capacity to kill bacteria during pneumococcal infection (Bewley et al., 2011). Similarly, mice with a genomic deletion of cathepsin E have increased susceptibility to infection by Staphylococcus aureus and Porphyromonas gingivalis (Tsukuba et al., 2006). Furthermore, a protective role for cathepsin L has been reported for Mycoplasma pulmonis infection (Xu et al., 2013). However, the relative contribution of different members within this large protease family in antibacterial host defense is largely underappreciated.
Here, we identified an important role for cathepsin B in driving increased susceptibility to the cytosolic pathogen Francisella novicida but not to the vacuolar-restricted pathogen Salmonella enterica serovar Typhimurium (S. Typhimurium). Genetic deletion or pharmacological inhibition of cathepsin B triggered signaling via down-regulation of mechanistic target of rapamycin (mTOR) and the lysosomal calcium channel TRPML1 to activate transcription factor EB (TFEB), which in turn results in increased lysosomal biogenesis and autophagy to mediate clearance of the bacteria. This phenomenon was not observed with genetic deletion of cathepsin G, elastase, or elastase and neutrophil proteinase 3. Our results, therefore, identified a fundamental negative regulatory function of cathepsin B in the host cell against F. novicida infection.
RESULTS
Cathepsin B negatively regulates host defense against F. novicida infection
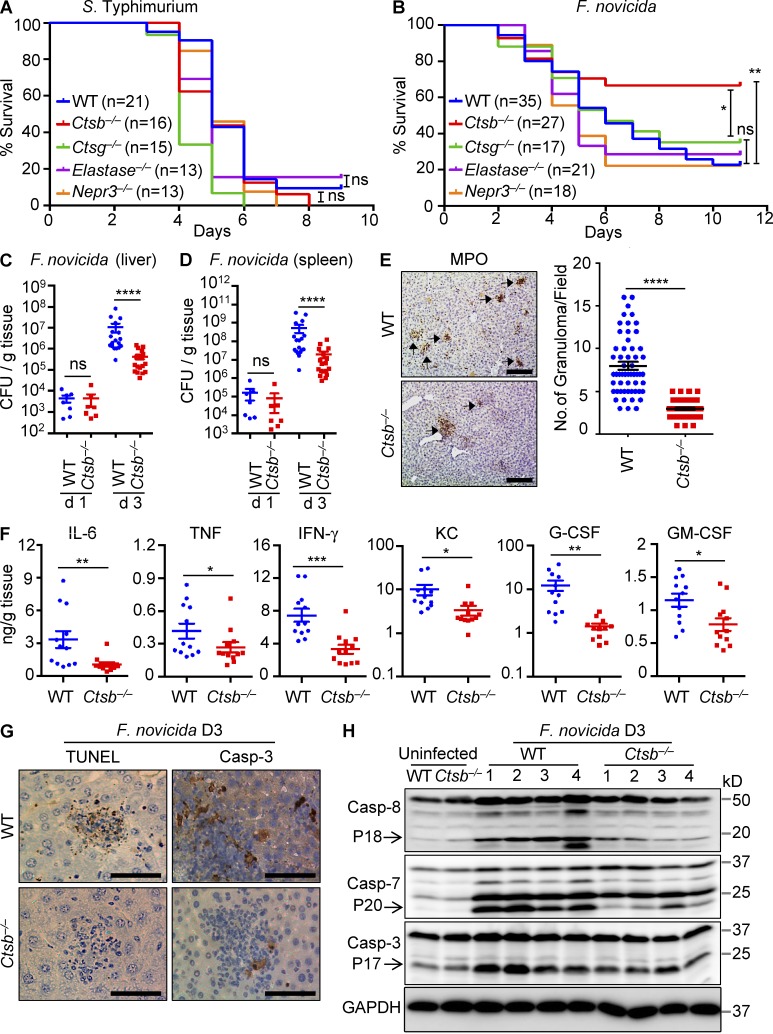
Intracellularly replicating bacterial pathogens have evolved strategies to evade degradation by lysosomes to survive and replicate in the hostile environment of the host cell (Ray et al., 2009; Huang and Brumell, 2014). F. novicida escapes the vacuole to undergo replication in the cytoplasm of infected cells, whereas S. Typhimurium replicates inside S. enterica–containing vacuoles. Despite decades of research on cathepsin biology, the role of lysosomal cathepsins in host defense against intracellularly replicating bacterial pathogens with distinct intracellular lifestyles has remained enigmatic. To address this, WT mice and mice deficient in the lysosomal cathepsin B, cathepsin G, or the extralysosomal proteases elastase and neutrophil proteinase 3 were infected with F. novicida or S. Typhimurium, and survival was monitored over time. Mice of all genotypes succumbed with similar kinetics to S. Typhimurium infection with most animals dying within 8 d after infection (Fig. 1 A). In contrast, when infected with F. novicida, cathepsin B–deficient mice (Ctsb−/− mice) were significantly protected from lethality, with 65% of animals from this group surviving beyond day 10, compared with only 25% of WT mice (Fig. 1 B). Enhanced resistance to F. novicida infection was specific to Ctsb−/− mice because mice lacking cathepsin G, elastase, or elastase and neutrophil proteinase 3 died with similar kinetics as WT mice (Fig. 1 B).
Figure 1.
Ctsb−/−mice are resistant to F. novicida infection. (A) WT and mutant mice were infected i.p. with 5 × 103 CFU of S. Typhimurium, and survival was monitored. (B) WT and mutant mice were infected subcutaneously with 1.5 × 105 CFU of F. novicida, and survival was monitored. (C and D) WT and Ctsb−/− mice were infected with 1.5 × 105 CFU of F. novicida. Bacterial burden in liver (C) and spleen (D) on days 1 and 3 was measured. (E) Liver sections from WT and Ctsb−/− mice infected with F. novicida (day 3) were stained with MPO. Quantification of MPO-stained granuloma and the number of granuloma per field are shown. (F) Proinflammatory cytokine and chemokine levels were measured in the liver from WT and Ctsb−/− mice on day 3 after infection with F. novicida. G-CSF, granulocyte CSF; KC, keratinocyte-derived chemokine. (G) TUNEL staining and cleaved caspase 3 (Casp-3) staining of liver sections from WT and Ctsb−/− mice after infection with F. novicida for 3 d (D3). (H) Liver tissue samples from WT and Ctsb−/− mice after infection with F. novicida for 3 d were homogenized, and lysates were analyzed for caspases 8, 3, and 7 activation by immunoblotting analysis. Each symbol indicates an individual mouse for C, D, and F. Data represent means ± SEM. Results are pooled from three independent experiments for A–D and are representative of two independent experiments for E–H. Bars, 100 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. ns, not significant.
Reduced mortality in Ctsb−/− mice could be caused by reduced bacterial burden. Therefore, we determined the number of bacteria in the spleen and liver of WT and Ctsb−/− mice after 1 and 3 d of infection with F. novicida. Bacterial loads in the liver and spleen of Ctsb−/− mice were significantly lower than those in WT mice on day 3 but not on day 1 after infection (Fig. 1, C and D). In agreement, less granulomas were formed in the liver of infected Ctsb−/− mice relative to WT controls (Fig. 1 E). Reduced bacterial burden of Ctsb−/− mice on day 3 after infection was also associated with reduced production of proinflammatory cytokines and chemokines in the liver (Fig. 1 F). Moreover, cell death and associated activation of the apoptotic caspases 3, 7, and 8 was diminished in the liver tissue of Ctsb−/− mice 3 d after infection (Fig. 1, G and H). These results suggested cathepsin B plays negative roles in host defense against F. novicida infection.
Intracellular growth of F. novicida is suppressed by the lack of cathepsin B
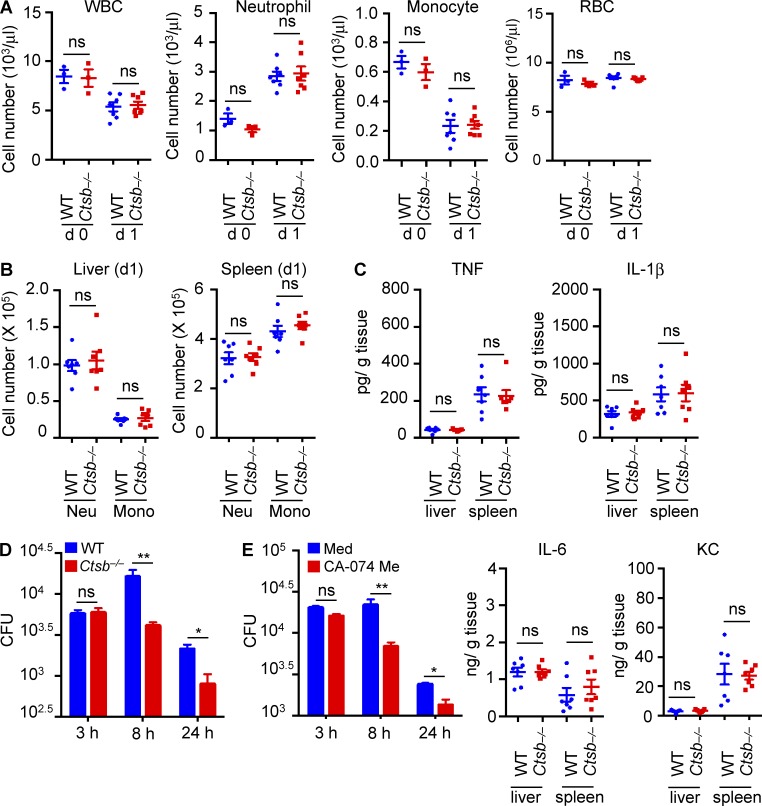
Recruitment of immune cells and production of proinflammatory cytokines are hallmarks of effective innate immune responses to infection (Cowley and Elkins, 2011). One possibility is that the improved early innate immune response in Ctsb−/− mice might restrict bacterial replication or increase bacterial killing and clearance. Therefore, we investigated the number of circulating total white blood cells, red blood cells, neutrophils, and monocytes in the blood of uninfected WT and Ctsb−/− mice and WT and Ctsb−/− mice infected with F. novicida. Relative to their uninfected controls, both WT and Ctsb−/− mice exhibited a global reduction in the numbers of circulating white blood cells (lymphocytes and myeloid cells) and monocytes and an increase in the number of neutrophils 24 h after F. novicida infection, whereas total red blood cell counts remained unchanged (Fig. 2 A). These results suggested that certain populations of circulating immune cells might have migrated from the BM to the sites of infection. However, no significant differences were noted in the prevalence of specific immune cell populations in circulation between infected WT and Ctsb−/− mice (Fig. 2 A). We also did not observe any difference in the number of CD11b+ Ly6g+ neutrophils and CD11b+ Ly6g− monocyte populations in the liver and spleen of WT and Ctsb−/− mice 24 h after F. novicida infection when WT and Ctsb−/− mice have comparable bacterial burden in the liver and spleen (Fig. 2 B). Finally, the levels of the proinflammatory cytokines TNF, IL-1β, and IL-6 and the keratinocyte-derived chemokine (CXCL1) in liver and spleen of infected WT and Ctsb−/− mice were similar after 24 h of infection (Fig. 2 C), suggesting that cathepsin B controlled host defense against F. novicida independently of immune cell recruitment and cytokine or chemokine production.
Figure 2.
Macrophages lacking cathepsin B have enhanced bactericidal activity. (A) Blood cells from uninfected mice (d 0) or mice infected with F. novicida (d 1) were analyzed for different cell populations with a Forcyte hematology analyzer. WBC, white blood cell. (B) On day 1 after F. novicida infection, neutrophil (CD11b+ Ly6g+; Neu) and monocyte (CD11b+ Ly6g−; Mono) infiltration into the liver and spleen were analyzed. (C) Cytokine levels were measured in the liver and spleen from WT and Ctsb−/− mice on day 1 after infection with F. novicida. (D) BMDMs from WT and Ctsb−/− mice were infected with F. novicida (MOI 10) for 3 h, and numbers of intracellular bacteria were enumerated at the indicated times. (E) BMDMs from WT mice were infected with F. novicida (MOI 10) for 3 h in media (Med) with or without the cathepsin B inhibitor CA-074 Me (5 µM). The number of intracellular bacteria was enumerated at the indicated times. KC, keratinocyte-derived chemokine. Each symbol indicates an individual mouse and means ± SEM are shown. Data are representative of two independent experiments for A–C or three independent experiments for D and E. *, P < 0.05; **, P < 0.01. ns, not significant.
Macrophages play a key role in controlling intracellular replication and dissemination of F. novicida (Hall et al., 2008). To investigate the role of cathepsin B in the control of F. novicida replication in macrophages, we infected primary BMDMs from WT and Ctsb−/− mice with F. novicida and determined the number of intracellular bacteria over time. Notably, Ctsb−/− BMDMs cleared bacteria more efficiently relative to WT BMDMs at 8 and 24 h after infection (Fig. 2 D). In agreement, inhibition of the cathepsin B protease activity with CA-074 methyl ester (Me) also enhanced the bactericidal activity of WT BMDMs (Fig. 2 E).
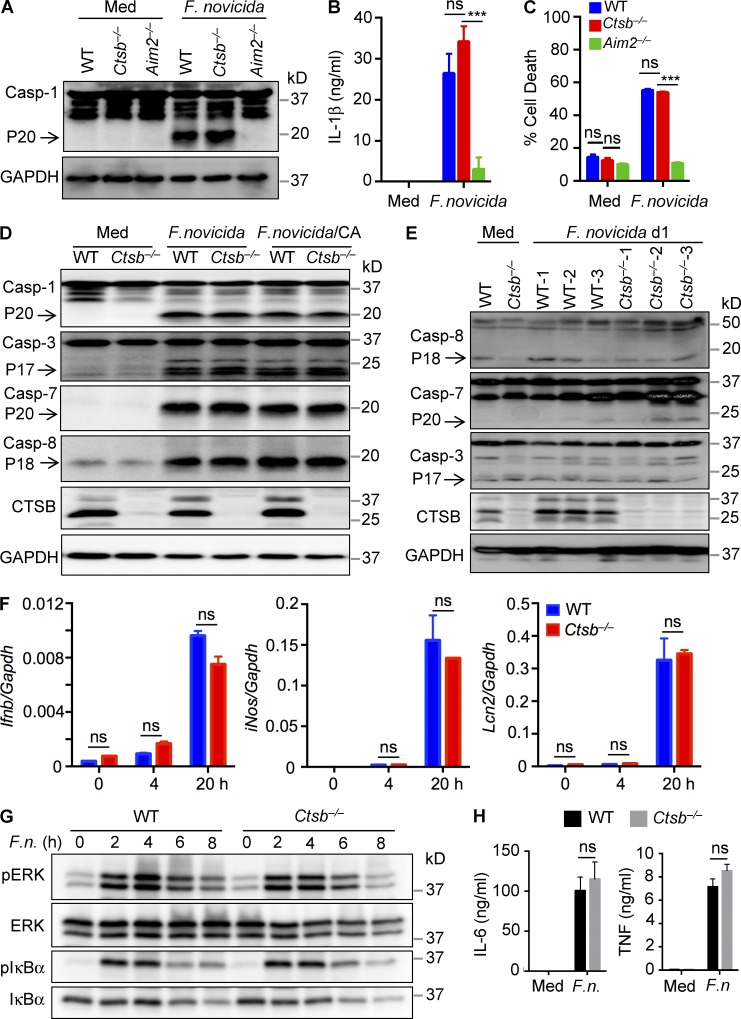
F. novicida infection in macrophages is detected by the DNA-sensing AIM2 inflammasome (Fernandes-Alnemri et al., 2010; Jones et al., 2010; Rathinam et al., 2010; Man et al., 2016), but cathepsin B was not required for AIM2-dependent activation of caspase 1, the subsequent release of the caspase 1 substrate IL-1β, and pyroptotic cell death in BMDMs infected with F. novicida (Fig. 3, A–C). Other members of the caspase family also contribute to innate immunity during infection (Man and Kanneganti, 2016). However, infected Ctsb−/− BMDMs displayed similar levels of caspases 3, 7, and 8 activation compared with infected WT BMDMs, in agreement with results from CA-074 Me–pretreated WT BMDMs (Fig. 3 D). Differential activation of apoptotic caspases was also not observed in liver tissues from WT and Ctsb−/− mice after 24 h of infection (Fig. 3 E).
Figure 3.
Cathepsin B is dispensable for caspase activation and NF-κB and ERK signaling in response to F. novicida infection. (A–C) Immunoblot analysis of caspase 1 (Casp-1) and its subunit P20 (A) and analysis of IL-1β release (B) and cell death (C) in WT, Ctsb−/−, and Aim2−/− BMDMs infected with F. novicida (MOI 100) for 20 h. (D) Immunoblot analysis of caspases 1, 3, 7, and 8 in WT and Ctsb−/− BMDMs infected with F. novicida (MOI 100) for 20 h with or without 5 µM CA-074 Me (CA). Uninfected cells were used as negative controls. Arrows indicate the cleaved forms. (E) Liver homogenates from WT and Ctsb−/− mice after infection with F. novicida for 24 h were analyzed for caspases 8, 7, and 3 activation by immunoblotting analysis. Arrows indicate the cleaved forms. (F) Expression of the genes encoding IFN-β, iNOS, and Lcn2 was analyzed in WT and Ctsb−/− BMDMs infected with F. novicida for the indicated times by quantitative real-time PCR. (G) BMDMs from WT and Ctsb−/− mice were infected with F. novicida (MOI 100; F.n.) for the indicated times, and cell lysates were analyzed for phosphorylated and total ERK and IκBα proteins by immunoblotting analysis. (H) Production of IL-6 and TNF was detected in uninfected or F. novicida–infected WT and Ctsb−/− BMDMs. Data are representative of three independent experiments and are means ± SEM. ***, P < 0.001. Med, uninfected; ns, not significant.
Apart from inflammasome signaling, type I interferons, inducible nitric oxide synthase (iNOS), and antimicrobial peptides regulate host protection against F. novicida infection (Lindgren et al., 2005; Henry et al., 2007). However, WT and Ctsb−/− BMDMs that have been infected with F. novicida expressed similar transcript levels of Ifnb, iNos, and the antimicrobial glycoprotein lipocalin 2 (Lcn2; Fig. 3 F). Also, the levels of activated NF-κB and extracellular signal–regulated kinase (ERK) as well as production of IL-6 and TNF were not differentially regulated in F. novicida–infected WT and Ctsb−/− BMDMs (Fig. 3, G and H). These data collectively suggested that the role of cathepsin B in the regulation of F. novicida replication is uncoupled from macrophage cell death, inflammasome activation, and inflammatory cytokine production.
Lysosomes target F. novicida in the absence of cathepsin B
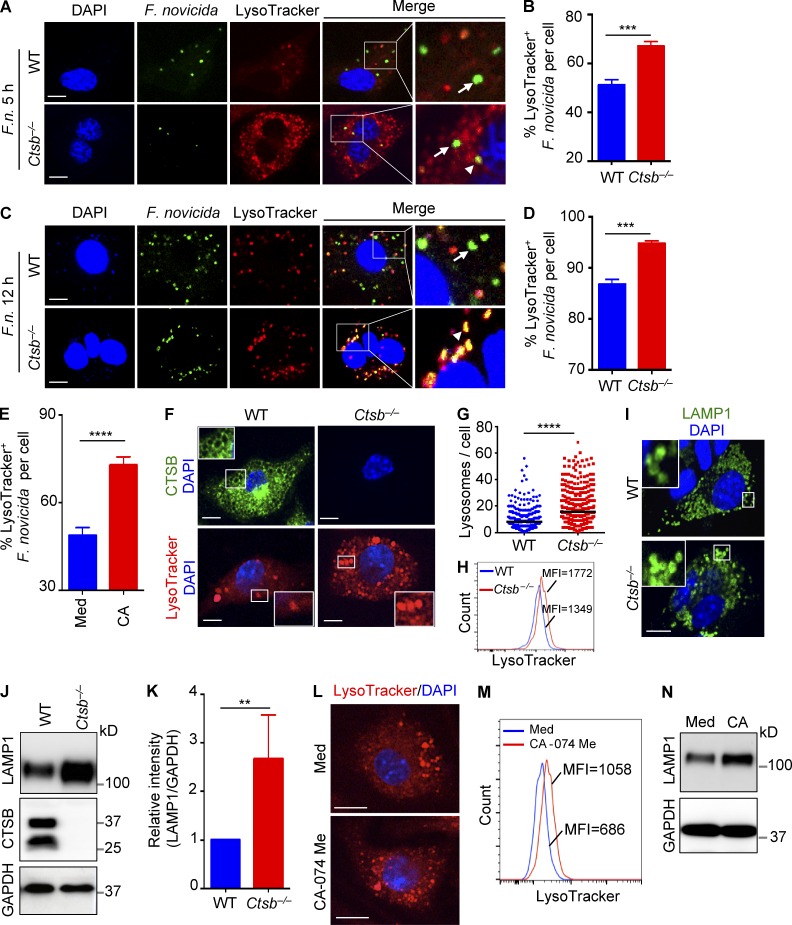
Fusion of lysosomes with pathogen-occupied vacuoles is a major bactericidal mechanism in macrophages (Roy et al., 2006). In response, bacterial pathogens have evolved mechanisms to evade the lysosomal degradation pathway. During F. novicida infection, bacteria escape the vacuoles of human macrophages after 3–4 h of infection (Santic et al., 2005; Clemens et al., 2009). To investigate whether improved control of F. novicida replication in the absence of cathepsin B was caused by increased fusion of F. novicida–containing vacuoles with lysosomes, we examined colocalization of a GFP-expressing strain of F. novicida with LysoTracker in infected WT and Ctsb−/− BMDMs. We found a significantly elevated frequency of F. novicida colocalizing with lysosomes in Ctsb−/− BMDMs compared with the numbers observed in WT BMDMs at 5 h (67.2 ± 1.8% vs. 51.1 ± 2.2%) and 12 h (94.8 ± 0.4% vs. 86.8 ± 0.9%) after infection (Fig. 4, A–D). These results suggested that the prevalence of lysosome-associated F. novicida increased in Ctsb−/− BMDMs compared with WT BMDMs. In agreement, treatment of WT BMDMs with a cathepsin B protease inhibitor CA-074 Me significantly increased the prevalence of LysoTracker-positive F. novicida (Fig. 4 E). Notably, we observed that Ctsb−/− BMDMs contained significantly more LysoTracker-positive vacuoles before infection, and the number of LysoTracker puncta in uninfected Ctsb−/− BMDMs was nearly twofold that of WT BMDMs (15.9 ± 0.5 vs. 8.3 ± 0.3 per cell; Fig. 4, F and G). The increased prevalence of LysoTracker-positive vacuoles and increased expression of the lysosomal membrane protein LAMP1 in Ctsb−/− BMDMs was further confirmed by flow cytometry and immunoblotting analysis, respectively (Fig. 4, H–K). Similar to Ctsb−/− BMDMs, the cathepsin B inhibitor CA-074 Me increased the number of LysoTracker puncta and expression of LAMP1 in WT BMDMs (Fig. 4, L–N).
Figure 4.
Recruitment of the lysosome to F. novicida is accelerated in the absence of cathepsin B. (A and C) BMDMs from WT and Ctsb−/− mice were infected with GFP-expressing F. novicida (MOI 20; F.n.) for 5 h (A) or 12 h (C) followed by labeling with 100 nM LysoTracker for 30 min. Fixed samples were processed for confocal microscopy. Arrowheads indicate lysosome-targeted F. novicida; arrows indicate F. novicida free from lysosome colocalization. (B and D) Quantification of A (B) and C (D) are shown. At least 220 cells were analyzed for each group. (E) Quantification of the percentage of F. novicida colocalized with lysosomes in WT BMDMs infected with GFP-expressing F. novicida (MOI 20) for 5 h in the presence or absence (Med) of 5 µM CA-074 Me (CA). At least 200 cells were analyzed for each group. (F) Confocal microscopy analysis of WT and Ctsb−/− BMDMs stained with cathepsin B antibody (top) or labeled with LysoTracker (bottom). Insets indicate enlarged images. (G) Quantification of lysosomes in WT and Ctsb−/− BMDMs. Each symbol indicates an individual cell. At least 500 cells were analyzed for each group. (H) FACS analysis of WT and Ctsb−/− BMDMs stained with LysoTracker. (I) Confocal microscopy showing LAMP1 staining in WT and Ctsb−/− BMDMs. Insets indicate enlarged images. (J) Cell lysates from WT and Ctsb−/− BMDMs were analyzed for LAMP1 protein expression by immunoblotting. (K) Quantification of the relative protein level of LAMP1 in J. (L) Representative images of WT BMDMs treated with or without cathepsin B inhibitor (5 µM CA-074 Me) for 12 h and labeled with LysoTracker. (M) FACS analysis of LysoTracker staining in WT BMDMs in the presence or absence of 5 µM CA-074 Me for 12 h. (N) Immunoblot analysis of LAMP1 expression in WT BMDMs in media or treated with 5 µM CA-074 Me for 12 h. Data are representative of three independent experiments and are means ± SEM. Bars, 10 µm. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. MFI, mean fluorescence intensity.
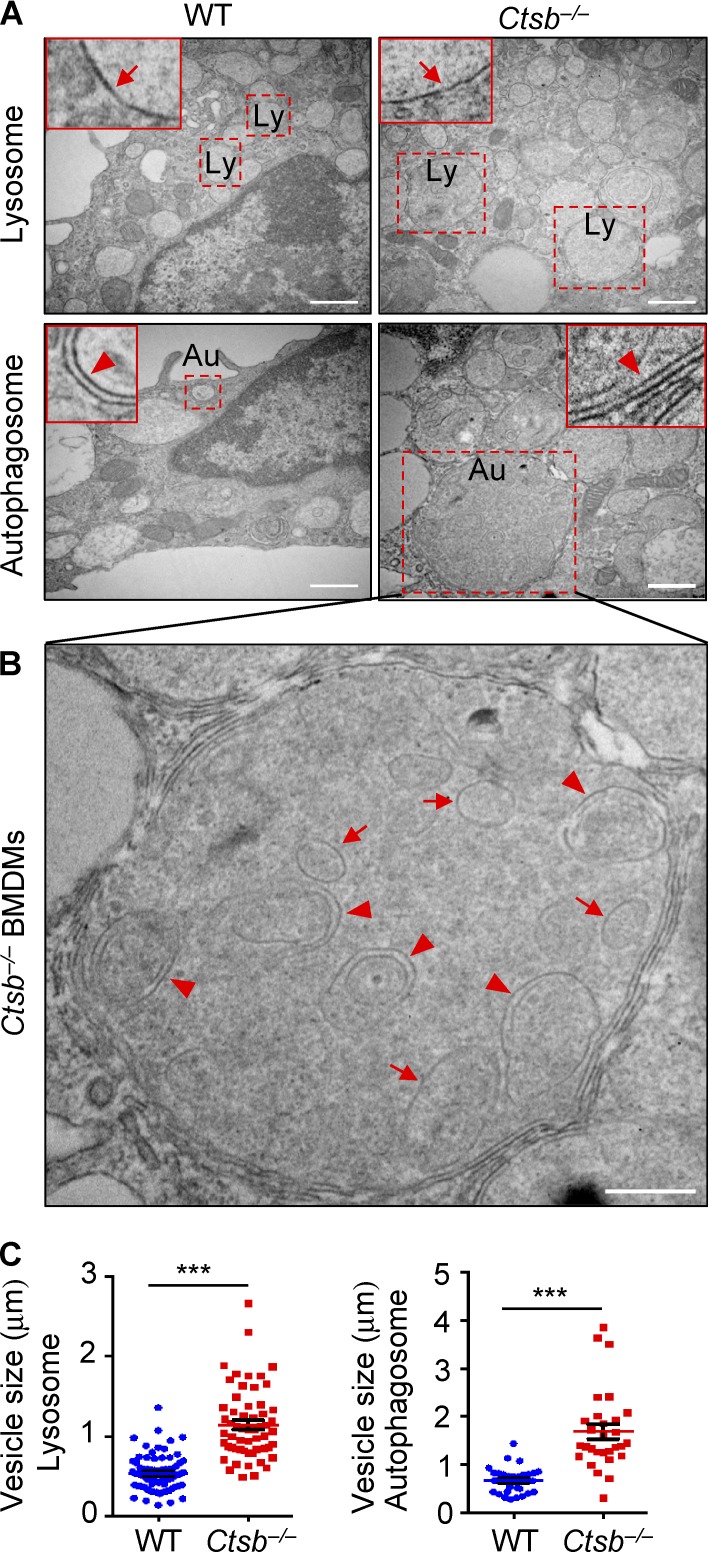
Transmission electron microscopy reveled that uninfected Ctsb−/− BMDMs contained larger single-membrane lysosomes and double-membrane autophagosomes compared with that in uninfected WT BMDMs (Fig. 5, A and C). Remarkably, higher resolution analysis of these micrographs revealed increased accumulation of partially digested vesicles in autolysosomes of Ctsb−/− BMDMs (Fig. 5 B), suggesting impaired nutrient recycling by Ctsb−/− lysosomes. Collectively, these results indicated that the enhanced antimicrobial activity of Ctsb−/− BMDMs to F. novicida might be attributed to their increased lysosomal biogenesis, which may result in increased efficiency in delivering the bacteria to lysosomes for killing.
Figure 5.
Cathepsin B deficiency results in impaired lysosomal recycling. (A) Transmission electron microscopy analysis of WT and Ctsb−/− BMDMs. Arrows in each inset indicate the single membrane lysosome– or double membrane autophagosome–associated vesicles. Dashed squares indicate a lysosome (Ly) or autophagosome (Au). (B) Undigested vesicles are accumulated in the autolysosomes of Ctsb−/− BMDMs. Arrows indicate double membrane–enclosed vesicles, and arrowheads indicate single membrane–enclosed vesicles. (C) Quantification of the size of lysosomes and autophagosomes from WT and Ctsb−/− BMDMs. The diameters of more than 60 lysosomes and 30 autophagosomes were analyzed. Data are representative of two independent experiments and are means ± SEM. Bars: (A) 1 µm; (B) 0.5 µm. ***, P < 0.001.
Lysosomal biogenesis and autophagy are elevated in the absence of cathepsin B
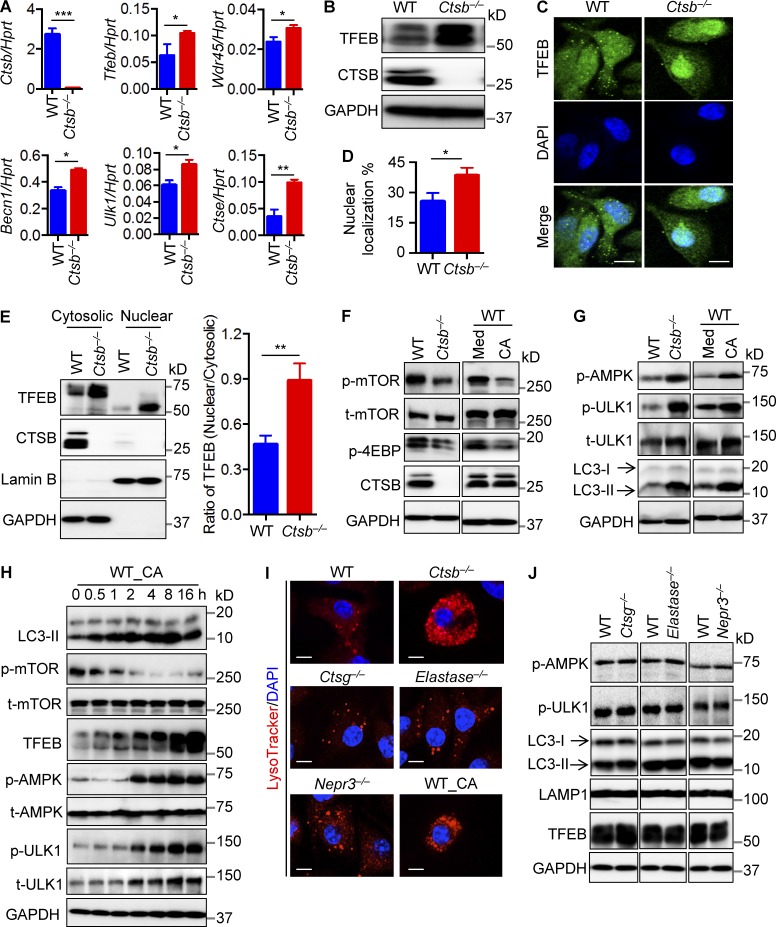
We performed genome-wide expression analysis to profile differentially expressed genes in uninfected WT and Ctsb−/− BMDMs to examine whether the increased number of lysosomes in Ctsb−/− BMDMs is caused by enhanced lysosomal biogenesis. We identified increased expression of the gene encoding the TFEB and differentially regulated genes involved in the Coordinated Lysosomal Expression and Regulation (CLEAR) network as well as genes encoding lysosomal hydrolases, lysosomal membrane proteins, lysosomal acidification proteins, and autophagy proteins (Fig. 6 A and Fig. S1; Palmieri et al., 2011). Importantly, autophagy and lysosomal biogenesis in response to starvation signals are up-regulated by the CLEAR gene network and its regulatory transcription factor TFEB (Sardiello et al., 2009; Settembre et al., 2011). Consistent with the increased expression of TFEB mRNA, we found a substantial increase in the protein expression level of TFEB in uninfected Ctsb−/− BMDMs relative to those of WT controls (Fig. 6 B). Activated TFEB translocates into the nucleus to drive transcription of genes encoding lysosomal proteins containing the CLEAR element (Sardiello et al., 2009). We indeed observed increased nuclear localization of TFEB in Ctsb−/− BMDMs by immunofluorescence microscopy and immunoblotting analysis of TFEB (Fig. 6, C–E). These findings indicated that Ctsb−/− BMDMs have a superior capacity to drive lysosomal biogenesis by unleashing the activity of TFEB.
Figure 6.
Cathepsin B restricts lysosomal biogenesis and autophagy. (A) Expression analysis of genes encoding cathepsin B, TFEB, WDR45, BECN1, ULK1, and cathepsin E in WT and Ctsb−/− BMDMs by quantitative real-time PCR. Ctse, cathepsin E; Hprt, hypoxanthine-guanine phosphoribosyltransferase. (B) Immunoblot analysis of TFEB, cathepsin B, and GAPDH (loading control) in uninfected WT and Ctsb−/− BMDMs. (C) Confocal microscopy analysis of TFEB staining in uninfected WT and Ctsb−/− BMDMs. (D) Percentage of nuclear localization of TFEB in uninfected WT and Ctsb−/− BMDMs. At least 200 cells were analyzed for each group. (E) Immunoblot analysis of TFEB in the nuclear and cytoplasmic fractions of uninfected WT and Ctsb−/− BMDMs (left) and the relative intensity of TFEB in nuclear versus cytoplasmic (right). Lamin B and GAPDH were used as quality controls for nuclear and cytoplasmic fractions separation, respectively. Quantification of the relative protein expression was processed using ImageJ. (F and G) Immunoblot analysis of phosphorylation of mTOR, 4EBP, AMPK, ULK1, and LC3-II in uninfected WT and Ctsb−/− BMDMs (left) or WT BMDMs in media (Med) with or without 5 µM CA-074 Me (CA) for 2 h. (H) Immunoblot analysis of proteins in F and G in WT BMDMs treated with 5 µM CA-074 Me for the indicated times. (I) Representative images of LysoTracker-labeled WT, Ctsb−/−, Ctsg−/−, Elastase−/−, Nepr3−/−, and cathepsin B inhibitor treated WT (WT_CA) BMDMs. (J) Immunoblot analysis of AMPK, ULK1, LC3, LAMP1, TFEB, and GAPDH (loading control) in uninfected WT, Ctsg−/−, Elastase−/−, and Nepr3−/− BMDMs. (C and I) Bars, 10 µm. Data are representative of three independent experiments and are means ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. t-AMPK, total AMPK; t-mTOR, total mTOR.
Activation and nuclear localization of TFEB are tightly regulated by signaling via mTOR (Settembre et al., 2012). Activated mTOR retains TFEB in the cytoplasm by inducing phosphorylation of TFEB, whereas inhibition of mTOR results in activation and nuclear translocation of TFEB. We investigated whether the mechanism of increased TFEB activation in the absence of cathepsin B was caused by down-regulated mTOR activation. Remarkably, phosphorylation of both mTOR and its direct target 4E-BP were significantly diminished in uninfected Ctsb−/− BMDMs compared with uninfected WT BMDMs (Fig. 6 F). We further confirmed this observation and found that WT BMDMs treated with CA-074 Me recapitulated the results obtained in Ctsb−/− BMDMs (Fig. 6 F).
Inhibition of mTOR is mediated by the conserved sensor of cellular energy and nutrient status, known as adenosine monophosphate–activated protein kinase (AMPK; Alers et al., 2012). AMPK also directly activates ULK1 to induce the autophagy pathway under stressed conditions (Kim et al., 2011). Notably, phosphorylation of both AMPK and ULK1 were substantially elevated in Ctsb−/− BMDMs (Fig. 6 G). Moreover, WT BMDMs treated with CA-074 Me phenocopied Ctsb−/− BMDMs (Fig. 6 G). To further confirm alterations in autophagy, we examined conversion of the autophagy marker LC3-I to the lipidated form of LC3-II. We indeed observed increased level of LC3-II in untreated Ctsb−/− BMDMs or in WT BMDMs treated with the cathepsin B inhibitor compared with untreated WT BMDMs (Fig. 6 G).
Next, we examined which pathway governed by mTOR, AMPK, and TFEB was the apical signal initiating the increase in lysosomal biogenesis and autophagy. A time course experiment assessing WT BMDMs treated with CA-074 Me showed that LC3-II accumulation and reduction of phosphorylated mTOR occurred within 30 min of cathepsin B inhibition (Fig. 6 H). In line with reduced phosphorylation of mTOR, the expression of TFEB was induced after treatment with the inhibitor for 30 min (Fig. 6 H). Phosphorylation of AMPK and ULK1 increased after 2 h of treatment with the cathepsin B inhibitor (Fig. 6 H), suggesting that AMPK and ULK1 activation succeeded LC3-II accumulation, reduction of phosphorylated mTOR, and activation of TFEB. These results suggested that impaired lysosomal degradation and recycling contributed to lysosomal biogenesis, whereas signaling via AMPK preferentially regulated the autophagy pathway and is not the apical signal driving increased lysosomal biogenesis in Ctsb−/− BMDMs. Importantly, we did not observe substantial differences in the number of lysosomes, phosphorylation of AMPK and ULK1, and protein expression of LC3-II, TFEB, and LAMP1 in Cathepsin G−/− (Ctsg−/−), Elastase−/−, or Nepr3−/− BMDMs compared with WT BMDMs (Fig. 6, I and J), suggesting that enhanced lysosomal biogenesis and autophagy are specific to Ctsb−/− BMDMs. Collectively, these results provided evidence to support that cathepsin B negatively regulated lysosomal biogenesis and autophagy at both the transcriptional and posttranslational levels through a mechanism that involved TFEB activation and ULK1 phosphorylation.
Cathepsin B down-regulates the TRPML1 channel activity in the lysosome
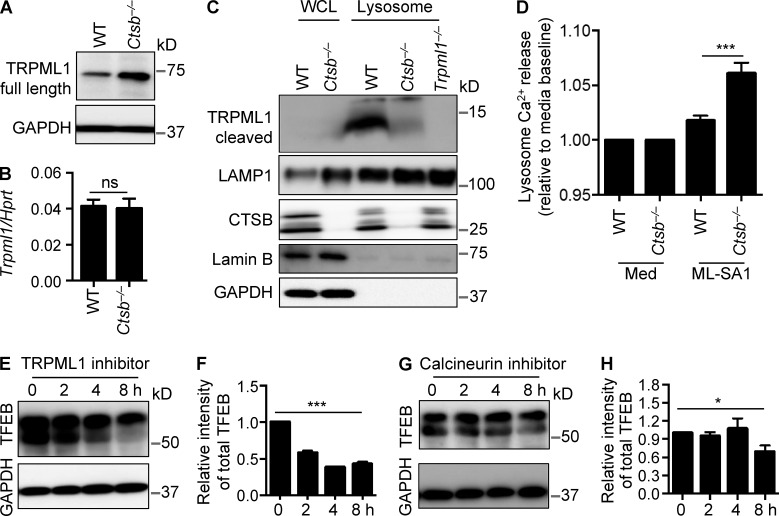
Lysosomal ion channels and transporters play essential roles in regulating signals from the lysosome (Xu and Ren, 2015). For instance, the vacuolar ATPase, a key player in lysosome nutrient sensing machinery, can sense lysosomal nutrient levels and regulate mTORC1 activity (Settembre et al., 2013). Calcium signaling mediated by the lysosomal calcium channel TRPML1 (MCOLN1) regulates TFEB activity from the lysosome through the phosphatase calcineurin. Local calcineurin activation via TRPML1 is essential for nuclear translocation and activation of TFEB. The pathway regulating lysosomal biogenesis and autophagy seems to operate independently of mTOR (Medina et al., 2015). Previous studies have shown that cathepsin B mediates proteolytic cleavage of the lysosomal calcium flux channel TRPML1 (Kiselyov et al., 2005) and that a loss of TRPML1 induces leakage of cathepsin B into the cytoplasm (Colletti et al., 2012). It is possible that cathepsin B might mediate TRPML1 channel activity to regulate TFEB activity. Indeed, the protein level of the full-length TRPML1 was increased in Ctsb−/− BMDMs, whereas differential levels of the transcription of the gene encoding TRPML1 was not observed between untreated WT and Ctsb−/− BMDMs (Fig. 7, A and B), suggesting decreased cleavage of TRPML1 in Ctsb−/− BMDMs. We enriched lysosome fractions from BMDMs to investigate the level of TRPML1 cleavage. Cleavage of TRPML1 in the lysosome was substantially impaired in untreated Ctsb−/− BMDMs compared with untreated WT BMDMs (Fig. 7 C). We further examined whether an increased level of the full-length TRPML1 protein in Ctsb−/− BMDMs resulted in altered Ca2+ efflux from the lysosome. WT and Ctsb−/− BMDMs were treated with the TRPML1 agonist ML-SA1 (Shen et al., 2012). We observed increased Ca2+ release from the lysosome in Ctsb−/− BMDMs stimulated with ML-SA1 compared with treated WT BMDMs (Fig. 7 D). To further confirm whether lysosomal calcium signaling supported the increased TFEB activity in Ctsb−/− BMDMs, we treated Ctsb−/− BMDMs with the lysosomal TRPML1 inhibitor PI(4,5)P2 (Zhang et al., 2012) and found that, over 8 h of inhibitor treatment, the expression of TFEB was substantially reduced in Ctsb−/− BMDMs (Fig. 7, E and F). Similar effects were observed in Ctsb−/− BMDMs treated with the calcineurin inhibitor cyclosporine A (Fig. 7, G and H). Collectively, these results suggested that cathepsin B negatively regulated TFEB activity, in part through lysosomal calcium signaling and degradation of TRPML1.
Figure 7.
Cathepsin B regulates TRPML1 channel activity. (A) Immunoblot analysis of full-length TRPML1 in WT and Ctsb−/− BMDMs. (B) Quantitative real-time PCR analysis of Trpml1 expression in WT and Ctsb−/− BMDMs. Hprt, hypoxanthine-guanine phosphoribosyltransferase; ns, not significant. (C) Immunoblot analysis of cleaved TRPML1 in the whole cell lysate (WCL) or lysosomes purified from WT, Ctsb−/−, and immortalized Trpml1−/− BMDMs. LAMP1, cathepsin B, lamin B, and GAPDH were used as quality controls. (D) Lysosome calcium release was analyzed in WT and Ctsb−/− BMDMs stimulated with 20 µM ML-SA1. Med, media. (E) Immunoblot analysis of TFEB in Ctsb−/− BMDMs treated with 0.1 µM TRPML1 inhibitor PI(4,5)P2 for the indicated times. (F) Quantification of data in E. (G) Immunoblot analysis of TFEB in Ctsb−/− BMDMs treated with 1 µM calcineurin inhibitor cyclosporin A for the indicated times. (H) Quantification of data in G. Data are representative of three independent experiments and are means ± SEM. *, P < 0.05; ***, P < 0.001.
Autophagy is up-regulated in the absence of cathepsin B during F. novicida infection
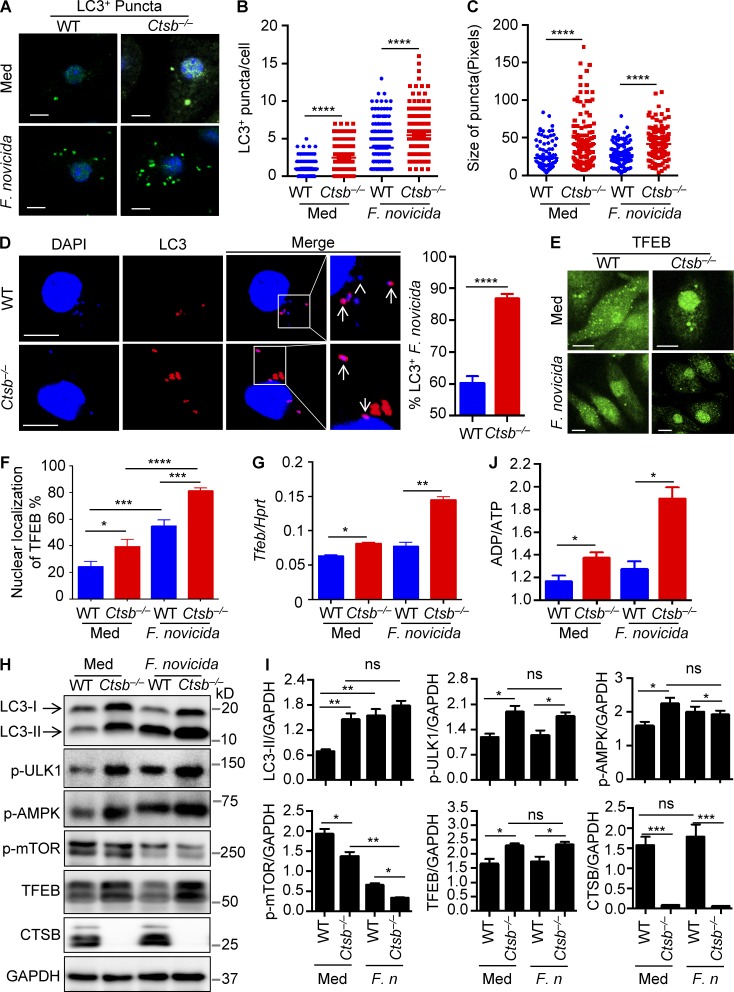
Autophagy has been previously shown to mediate reentry of cytosolic-dwelling F. novicida to the endocytic pathway and fusion with lysosomes (Checroun et al., 2006). To investigate how cathepsin B modulates lysosomal biogenesis and autophagy during F. novicida infection, WT and Ctsb−/− BMDMs were infected with F. novicida and analyzed for LC3+ puncta formation and TFEB expression. We found that F. novicida infection induced autophagy formation and significantly elevated the number of LC3+ puncta per cell in Ctsb−/− BMDMs compared with infected WT BMDMs (Fig. 8, A and B). Moreover, the size of LC3+ puncta in infected Ctsb−/− BMDMs was markedly increased relative to those observed in infected WT BMDMs (Fig. 8 C). Notably, the frequency of autophagosome-associated F. novicida was significantly higher in Ctsb−/− BMDMs than in WT BMDMs (Fig. 8 D). Furthermore, we found an increased frequency of nuclear-localized TFEB in Ctsb−/− BMDMs compared with WT BMDMs after F. novicida infection (Fig. 8, E and F). Accordingly, both mRNA and protein levels of TFEB in infected Ctsb−/− BMDMs were higher than that of infected WT BMDMs (Fig. 8, G and H). We observed elevated phosphorylation of AMPK and ULK1 in Ctsb−/− BMDMs compared with their WT counterparts after F. novicida infection (Fig. 8, H and I). In line with the observation of increased phosphorylation of AMPK, the ratio of ADP to ATP in Ctsb−/− BMDMs was significantly higher than that of WT BMDMs after F. novicida infection (Fig. 8 J). Phosphorylated mTOR was further reduced in Ctsb−/− BMDMs infected with F. novicida (Fig. 8, H and I).
Figure 8.
F. novicida infection induces autophagy formation and TFEB activation in macrophages. (A) Representative images of LC3-II+ puncta in uninfected (Med) WT and Ctsb−/− BMDMs or BMDMs infected with F. novicida for 4 h. (B) Quantification of the numbers of LC3-II+ puncta in WT and Ctsb−/− BMDMs with or without F. novicida infection. At least 200 cells were analyzed for each group. (C) Size of LC3-II+ puncta in WT and Ctsb−/− BMDMs in B. At least 100 cells were analyzed for each group. (D) Representative images of LC3-II+ puncta in WT and Ctsb−/− BMDMs infected with F. novicida for 4 h. Arrows indicate LC3-associated F. novicida; the arrowhead indicates LC3-II+ puncta free from F. novicida colocalization. At least 100 cells were analyzed in each group. (E) Confocal microscopy analysis of TFEB nuclear localization in WT and Ctsb−/− BMDMs with or without F. novicida infection. (F) Quantification of the percentage of TFEB nuclear localization. At least 200 cells were analyzed for each group. (G) Expression of the gene encoding TFEB was analyzed by quantitative real-time PCR. Hprt, hypoxanthine-guanine phosphoribosyltransferase. (H) Immunoblot analysis of LC3-II and TFEB and phosphorylation of ULK1, AMPK, and mTOR in uninfected WT and Ctsb−/− BMDMs or BMDMs infected with F. novicida for 3 h. (I) Quantification of the data in H. F. n, F. novicida; ns, not significant. (J) ADP to ATP ratio analysis in uninfected WT and Ctsb−/− BMDMs or BMDMs infected with F. novicida for 1 h. Data are representative of two independent experiments (A–D) or three independent experiments (E–J). Data represent means ± SEM. Bars, 10 µm. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. ns, not significant.
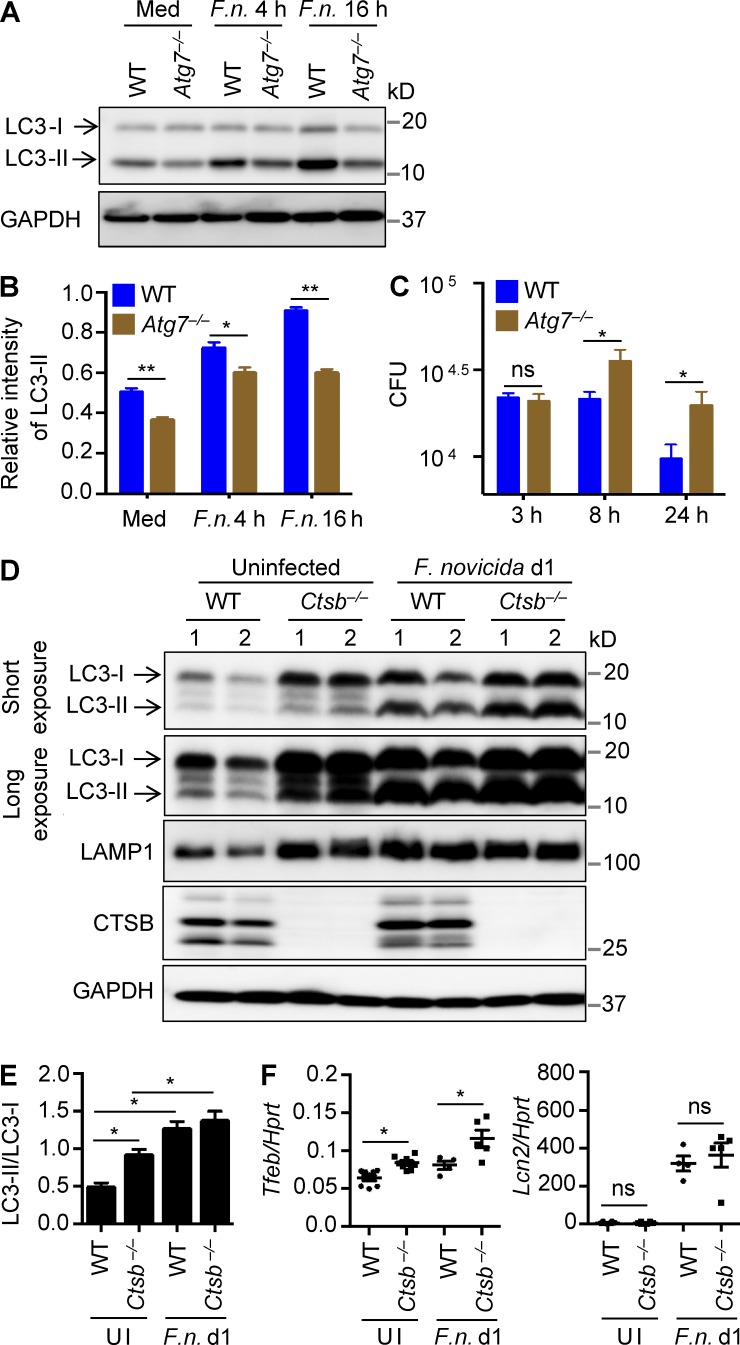
To further confirm that autophagy could play a protective role in the host defense against F. novicida infection, BMDMs from WT and autophagy-defective Atg7−/− mice (Martinez et al., 2011) were infected with F. novicida and analyzed for LC3 conversion and intracellular bacterial burden. Reduced LC3-II was observed in infected Atg7−/− BMDMs relative to infected WT BMDMs (Fig. 9, A and B), which was associated with increased numbers of F. novicida bacteria recovered from infected Atg7−/− BMDMs compared with the number recovered from infected WT BMDMs (Fig. 9 C).
Figure 9.
Ctsb−/− mice have a greater capacity to induce autophagy and lysosomal biogenesis. (A) Immunoblot analysis of LC3 in uninfected and F. novicida (F.n.)–infected WT and Atg7−/− BMDMs. (B) Quantification of data in A. (C) BMDMs from WT and LysM-Cre-Atg7f/f mice were infected with F. novicida (MOI 10) for 3 h, and numbers of intracellular bacteria were enumerated at the indicated times. (D) Immunoblot analysis of LC3, LAMP1, cathepsin B, and GAPDH (loading control) in liver homogenates of uninfected WT and Ctsb−/− mice and mice infected with F. novicida for 1 d. (E) Quantification of data for LC3-II conversion in D. (F) Expression of the genes encoding TFEB and LCN2 in the liver of uninfected mice and mice infected with F. novicida for 1 d. Data are representative of three independent experiments (A–C) or two independent experiments (D–F). Data represent means ± SEM. *, P < 0.05; **, P < 0.01. Hprt, hypoxanthine-guanine phosphoribosyltransferase; Med, media; ns, not significant; UI, uninfected.
To investigate whether the molecular mechanism orchestrated by cathepsin B to regulate autophagy and lysosomal biogenesis is recapitulated in a physiological setting, we analyzed signaling pathways in liver tissue homogenates from WT and Ctsb−/− mice. In agreement with our cellular analyses, uninfected Ctsb−/− mice or those infected with F. novicida exhibited a higher level of LC3-II compared with the corresponding WT controls (Fig. 9, D and E). Remarkably, LAMP1 expression was dramatically up-regulated in the liver of uninfected Ctsb−/− mice compared with uninfected WT mice, and both infected WT and Ctsb−/− mice maintained elevated levels of LAMP1 protein expression (Fig. 9 D). Moreover, expression of TFEB was significantly increased in Ctsb−/− mice (Fig. 9 F). In contrast, expression of Lcn2, a bacteriostatic factor produced during the innate immune response to bacterial infection, was similar in WT and Ctsb−/− mice after F. novicida infection (Fig. 9 F). Collectively, we identified a central role for cathepsin B in governing activities of TFEB and the kinase ULK1, resulting in suppression of lysosomal biogenesis and autophagy, respectively. Thus, the coordination of autophagy and lysosomal biogenesis induced by inhibition of cathepsin B facilitates host defense against F. novicida infection (Fig. 10).
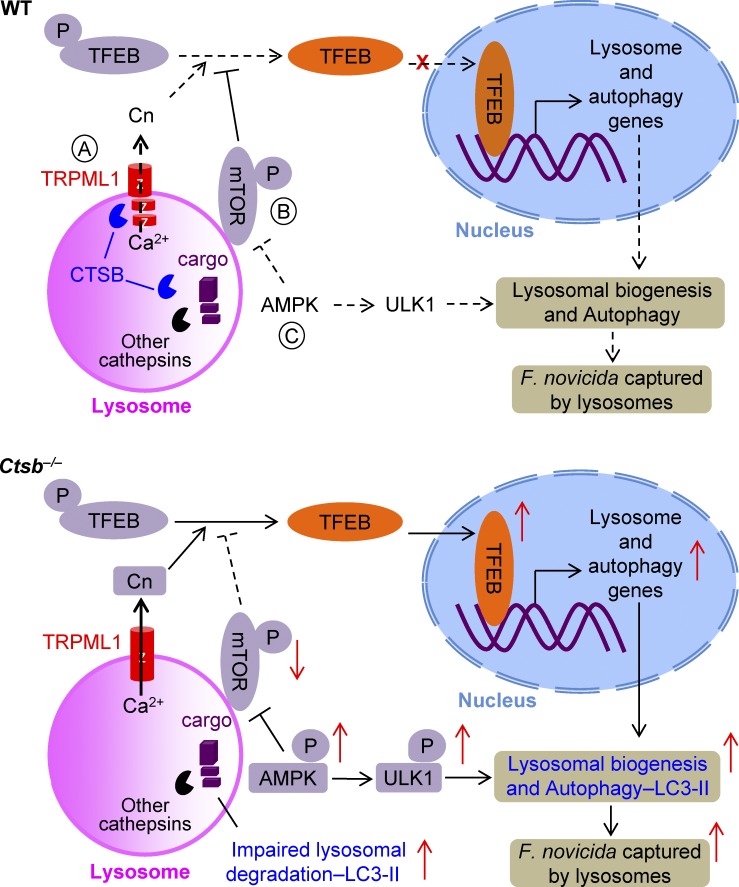
Figure 10.
A model of the regulation of lysosomal biogenesis and autophagy by cathepsin B in response to infection with F. novicida. (A) Cathepsin B mediates cleavage of the TRPML1 channel and negatively regulates calcineurin (Cn) activity. (B) Deficiency in cathepsin B results in inhibition of mTOR activation owing to a stressed signal from the impaired lysosomal degradation. Both increased calcineurin and decreased mTOR activity in Ctsb−/− BMDMs drive nuclear translocation of TFEB, which in turn activates the transcription of genes associated with lysosome biogenesis and autophagy. (C) Stressed signals from the lysosome or a feedback signal from increased lysosomal biogenesis induces AMPK and ULK1 phosphorylation, which further activates the autophagy pathway. Thus, cathepsin B negatively regulates lysosomal biogenesis and autophagy at both the transcriptional and posttranslational levels. Increased lysosomal biogenesis and autophagy induced by cathepsin B deficiency results in effective clearance of the intracellular bacterial pathogen F. novicida. Red arrow, induced or reduced activity; black arrow, activation; black line with bar head, inhibition; solid line, activated; dash line, not activated.
DISCUSSION
Lysosomal cathepsins have been conventionally recognized as executive proteases for cargo degradation and cell death. Our study identified a central role for cathepsin B in governing activities of TFEB and the kinase ULK1, resulting in suppression of lysosomal biogenesis and autophagy, respectively. The involvement of cathepsin B in the lysosomal cell death pathway and activation of the NLRP3 inflammasome under certain physiological settings have attracted scientific interests (Tschopp and Schroder, 2010; Man and Kanneganti, 2015). Our results, however, indicated that genetic deletion of cathepsin B is dispensable for activation of the AIM2 inflammasome, cytokine production, and cell death typically associated with F. novicida infection (Henry et al., 2007; Fernandes-Alnemri et al., 2010; Jones et al., 2010; Rathinam et al., 2010; Man et al., 2015; Meunier et al., 2015), suggesting that there were additional roles of cathepsin B in host defense beyond inflammasome activation and cell death.
In this study, we found a role for cathepsin B in negatively regulating lysosomal biogenesis and autophagy, such that cells lacking cathepsin B possessed a superior ability to induce fusion of their lysosomes with F. novicida. It is also interesting to speculate that F. novicida might be engulfed by autophagosomes and require cathepsin B to mediate entry to the cytoplasm for further replication. In addition, it is possible that the localization of cathepsin B proteins to a particular location within the host cell might contribute to their ability to influence the number of bacteria in the host cell.
We found that genomic deletion of cathepsin B in mice did not confer protection against the vacuolar-adapted pathogen S. Typhimurium, which could be because of differences in the mechanisms used by this bacterium to evade the lysosomal degradation pathway or to the functional redundancy among cathepsin proteases that are capable of inducing degradation of S. Typhimurium (Diacovich and Gorvel, 2010; Huang and Brumell, 2014). Further studies investigating the role of cytosolic pathogens, including both bacterial and viral pathogens, would yield insight into the biology of cathepsin B in infectious diseases. In particular, examining the effect of a range of doses of F. novicida infection in vivo would provide information on the strength of the impact of cathepsin B deletion (Nano et al., 2004; Kanistanon et al., 2012; Chu et al., 2014).
The role of autophagy in the host defense against members of the Francisella genus is controversial. The ability of different species or strains of the Francisella genus to evade the autophagy pathway might determine their intracellular survival (Chiu et al., 2009; Cremer et al., 2009; Chong et al., 2012; Steele et al., 2013). Results from the ATG7-deficient BMDMs from our study suggested that canonical autophagy is protective in the host defense against F. novicida infection. Lysosomes play critical roles in nutrient sensing and energy homeostasis (Settembre et al., 2013). During infection, neutralization of lysosomal pH by uropathogenic Escherichia coli in infected bladder epithelial cells can be sensed by the lysosomal cation channel TRPML3, subsequently triggering expulsion of exosome-encased bacteria (Miao et al., 2015). Our study now reveals a critical role of cathepsin B in controlling activity of the key lysosomal Ca2+ channel TRPML1. A lack of cathepsin B would leave the activity of TRPML1 unchecked, and a previous study has demonstrated that Ca2+ release by lysosomes via TRPML1 activates the phosphatase calcineurin, which, in turn, drives nuclear translocation of TFEB (Medina et al., 2015). Stressed signals from the lysosome itself might also inhibit mTOR activity and reduce phosphorylation of TFEB, leading to TFEB-dependent transcription of genes encoding molecules involved in lysosomal biogenesis and autophagy. The down-regulation of mTOR activity in Ctsb−/− BMDMs is consistent with a recent study showing that the mTORC1 pathway is activated by lysosomal degradation (Palm et al., 2015). It is not clear which specific signal elicited in the absence of cathepsin B down-regulated the phosphorylation of mTOR. However, enhanced AMPK phosphorylation in the absence of cathepsin B might provide a positive feedback reflecting increased lysosomal biogenesis, as this event occurred subsequent to TFEB and mTOR activation.
Our results unveiled a fundamental biological function of cathepsin B in providing a checkpoint for homeostatic maintenance of the lysosome population and basic recycling functions in the cell. Importantly, deficiency in cathepsin proteases causes lysosome storage diseases in humans owing to accumulation of partially digested macromolecules and enlarged lysosomes (Alroy and Lyons, 2014). TFEB-mediated lysosomal biogenesis and autophagy are crucial in exogenous antigen presentation and cancer metabolism (Perera et al., 2015; Samie and Cresswell, 2015). Cathepsin B can also mediate trypsinogen activation to promote the development of acute pancreatitis (van Acker et al., 2002). Our findings suggested that cathepsin B is a negative regulator of lysosomal biogenesis and autophagy in the intracellular milieu during a microbial infection, which broadens the biological repertoire of this protease in the context of health and disease. Our study, therefore, raises the intriguing possibility that modulating cathepsin B activity may hold therapeutic potential as a mean to increase host immunity against lethal F. novicida infection and to offer treatment options to those with debilitating lysosome storage diseases.
MATERIALS AND METHODS
Mice
WT C57BL/6J mice were purchased from The Jackson Laboratory and bred in house. Ctsb−/− mice were provided by T. Reinheckel (Albert-Ludwigs-University Freiburg, Freiburg, Germany), generated as described previously (Halangk et al., 2000), and backcrossed to the C57BL/6J background. Ctsg−/− (MacIvor et al., 1999) and Nepr3−/− (Kessenbrock et al., 2008) mice were provided by C. Pham (Washington University School of Medicine, St. Louis, MO). Aim2−/− (Jones et al., 2010) mice were provided by V.M. Dixit (Genentech, San Francisco, CA). LysM-Cre-Atg7f/f mice were described previously (Martinez et al., 2011). Elastase−/− mice were purchased from The Jackson Laboratory (stock no. 006112; Belaaouaj et al., 1998). All mutant mice were crossed with the C57BL/6J mice for >10 generations. All mice were kept in specific pathogen–free conditions within the Animal Resource Center at St. Jude Children’s Research Hospital. Animal studies were conducted under protocols approved by the Institutional Animal Care and Use Committee of St. Jude Children’s Research Hospital.
Bacterial culture and infection of mice
The bacterial strains used in this study were F. novicida strain U112, GFP-expressing F. novicida (from D.M. Monack, Stanford University, Stanford, CA), and S. Typhimurium strain SL1344. F. novicida and S. Typhimurium were grown overnight with shaking in tryptic soy broth (TSB) and Luria-Bertani medium, respectively, subcultured for an additional 3–4 h, and resuspended in PBS. 8–10-wk-old mice were injected subcutaneously with F. novicida U112 (1.5 × 105 CFU per mouse) as described previously (Man et al., 2015). Mice were weighted and monitored daily over a period of 3 wk. Mice were euthanized at days 1 and 3 after infection, and liver and spleen were harvested to determine the bacterial burden. Liver and spleen were homogenized, serially diluted, plated onto TSB agar plates, and incubated overnight at 37°C as described previously (Man et al., 2015). For S. Typhimurium infection, mice were infected i.p. with 5 × 103 CFU per mouse. For immune cell infiltration analysis, whole blood from WT and Ctsb−/− mice was analyzed on a hematology analyzer (Forcyte; Oxford Sciences) by a hematologist blinded to the experimental groups.
Bacterial infection of macrophages
To generate BMDMs, BM cells were cultured in L929 cell–conditioned IMDM supplemented with 10% FBS, 1% nonessential amino acids, and 1% penicillin-streptomycin for 5 d. Immortalized BM-derived cells from Trpml1−/− mice used for controls were from H. Fares (University of Arizona, Tucson, AZ; Miller et al., 2015). BMDMs were seeded in 12-well plates (1 million cells per well) and cultured overnight. The next day, cells were washed and supplied with fresh media without antibiotics. BMDMs were infected with F. novicida for the indicated times. For inhibitor treatment, BMDMs were treated with the cathepsin B inhibitor CA-074 Me (sc-214647; Santa Cruz Biotechnology, Inc.), AMPK inhibitor compound C (171260; EMD Millipore), calcineurin inhibitor cyclosporine A (PHR1092; Sigma-Aldrich), or lysosomal TRPML1 inhibitor PI(4,5)P2 (R4504; Echelon Biosciences; Zhang et al., 2012). BMDMs were lysed in radioimmunoprecipitation assay buffer with protease and phosphatase inhibitors (Roche) for immunoblot analysis.
Bacterial killing assay
BMDMs were infected with F. novicida with a multiplicity of infection (MOI) of 10 for 2 h and washed, and 50 µg/ml gentamicin was added to kill extracellular bacteria. After 1 h, cells were washed twice and cultured in fresh media. BMDMs were lysed in sterile water at the indicated times after infection, serially diluted, plated onto TSB agar plates, and incubated overnight for CFU enumeration.
Preparation of liver sample for immunohistochemical staining and immunoblot analysis
Liver tissues were fixed in 10% formalin and embedded in paraffin for immunochemical staining. 4-µm–thick liver sections were stained with anti–cleaved caspase 3 or anti-myeloperoxidase (MPO) followed by incubation for 30 min with Rabbit-on-Rodent HRP polymer (RMR622; BioCare Medical). TUNEL (terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling) staining was performed with the Dead End kit (G7130) according to the manufacturer’s instructions (Promega). Liver tissues were homogenized in radioimmunoprecipitation assay buffer with protease and phosphatase inhibitors for immunoblot analysis. Protein concentrations were determined using a bicinchoninic acid assay kit (Thermo Fisher Scientific) according to manufacturer’s instructions.
Immunoblot analysis and antibodies
Samples were separated by 12% SDS-PAGE and followed by electrophoretic transfer onto polyvinylidene fluoride membranes as described previously (Karki et al., 2015). Membranes were blocked with 5% nonfat milk and further incubated overnight in primary antibody at 4°C. The following primary antibodies were used: mouse IκBα, phosphorylated IκBα (p-IκBα; S32), ERK, p-ERK (T202Y204 [Cell signaling Technology], 9242L, 2859L, 9101L, and 9102L), anti–caspase 1 (AG-20B-0042; Adipogen), anti-LC3B (NB600-1384; Novus Biologicals), anti-LAMP1 (14-1071-85; eBioscience), antibodies for caspase 3, 7, and 8 (9661S, 9491S, and 8592S; Cell Signaling Technology), anti–cathepsin B (AF965; R&D Systems), antibodies for p-ULK1 (S555), total mTOR, p-mTOR (S2448), p–4E-BP1 (T37/46), total AMPK, p-AMPK (T172 [Cell Signaling Technology], 5869S, 2972S, 5536S, 2855P, 2793S, and 2535P), anti-ULK1 (A7481; Sigma-Aldrich), anti-TFEB (A303-673A; Bethyl Laboratories, Inc.), anti-TRPML1 (M1571; Sigma-Aldrich), anti-ATP6V1B2 (ab73404; Abcam), anti-MARCKS (myristoylated alanine-rich C kinase substrate; ab51100; Abcam), anti–lamin B (SC-6216; Santa Cruz Biotechnology, Inc.), and anti-GAPDH (5174S; Cell Signaling Technology). The secondary antibodies used were HRP-labeled anti–rabbit, anti–mouse, or anti–goat antibodies (Jackson ImmunoResearch Laboratories, Inc.). Quantification of the relative protein expression was processed using ImageJ (National Institutes of Health).
Immunofluorescence staining and microscopy
For LysoTracker staining and colocalization with F. novicida, WT and Ctsb−/− BMDMs were infected with GFP-expressing F. novicida and followed by labeling with 100 nM LysoTracker (L-7528; Molecular Probes) for the last 30 min. Cells were washed and fixed with 4% paraformaldehyde for 15 min at room temperature. Cells were washed three times and mounted using mounting medium (H-1200; Vector Laboratories). For LC3B, LAMP1, and TFEB immunostaining, uninfected and infected BMDMs were fixed in 4% paraformaldehyde for 15 min at room temperature. Cells were then permeabilized with cold methanol for 5 min, washed with PBS, and blocked in 1× ELISA buffer with 0.1% saponin for 1 h. Cells were stained with anti-LC3B (NB600-1384; Novus Biologicals), anti-LAMP1 (14-1071-85; eBioscience), or anti-TFEB (A303-673A; Bethyl Laboratories, Inc.), all at 1:500 dilution, overnight at 4°C. Cells were washed, stained with fluorescence-conjugated secondary antibody for 1 h, and mounted using mounting medium (H-1200; Vector Laboratories). Cells were observed on a confocal microscope (Axio Observer; Z1; SlideBook 6 software; ZEISS) for image acquisition and data analysis.
Transmission electron microscopy
BMDMs were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 1 h at 37°C. Cells were embedded and sectioned for transmission electron microscopy by the Cell and Tissue Imaging Core Facility of St. Jude Children’s Research Hospital. Quantification of lysosome and autophagosome size was performed with ImageJ.
Lysosome isolation
Lysosomes from BMDMs were isolated using a lysosome enrichment kit with cultured cell sonication and density gradient centrifugation, followed by lysosome precipitation according to the manufacturer’s manual (89839; Thermo Fisher Scientific). The purity of isolated lysosomes was determined by detecting the lysosomal membrane protein LAMP1, luminal lysosomal protease cathepsin B, nuclear protein lamin B, and GAPDH using immunoblotting analysis.
Lysosome calcium release measurement
WT and Ctsb−/− BMDMs were labeled with a calcium indicator (O-6807; Thermo Fisher Scientific) in calcium-free media for 15 min. Calcium release from the lysosome induced by 20 µM ML-SA1 and 200 µM glycylphenylalanine 2-naphthylamide was analyzed according to fluorescence intensity at 488 nm recorded with a confocal imaging system as described previously (Shen et al., 2012).
ADP/ATP ratio assay
Both infected and uninfected WT and Ctsb−/− BMDMs were lysed in assay buffer, and ATP and ADP content were measured using an ADP/ATP ratio assay kit according to the manufacturer’s protocols (MAK135; Sigma-Aldrich).
Microarray analysis
Total RNA was extracted from WT and Ctsb−/− BMDMs, reverse transcribed into biotin-labeled cRNA with an Ambion WT Expression kit (Thermo Fisher Scientific), and hybridized to a Mouse Gene 2.0 ST GeneChip (Affymetrix). Gene expression data were normalized and transformed into log2 transcript expression values with the robust multiarray average algorithm (version 6.6; Partek Genomics Suite; Irizarry et al., 2003). Differential expression was defined by application of a difference in expression of 0.5-fold (log2 signal) between conditions. The dataset has been deposited in the GEO database under accession no. GSE79508.
Lactate dehydrogenase release assay
Cell culture supernatants were collected at the indicated times, and lactate dehydrogenase activity was measured by using the Promega cytotoxicity kit according to the manufacturer’s protocols.
ELISA
Cell culture supernatant or sera from mice were analyzed for cytokine and chemokine release using a 22-multiplex assay (EMD Millipore) following the manufacturer’s instructions.
Real-time quantitative PCR
Total RNA was isolated from BMDMs using TRIzol (Invitrogen). cDNA was reverse transcribed by using Superscript III reverse transcriptase (Invitrogen). Real-time quantitative PCR was performed on the ABI Prism 7500 sequence detection system (Applied Biosystems). Primer sequences are listed in Table S1.
Statistical analysis
Data are given as means ± SEM. Statistical analyses were performed using one-way ANOVA with multiple comparisons, two-tailed Student t tests, and log-rank tests. P-values ≤0.05 were considered significant.
Online supplemental material
Fig. S1 shows the expression of genes encoding TFEB, and its targets are differentially regulated in the absence of cathepsin B. Table S1 shows real-time quantitative PCR primer sequences. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151938/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Reinheckel for the Ctsb−/− mice, C. Pham for Ctsg−/− and Nepr3−/− mice, V.M. Dixit for the Aim2−/− mice, D.M. Monack for GFP-expressing F. novicida, H. Fares for the immortalized Trpml1−/− cells, and M. Barr, R. Johnson, A. Burton, and S. Frase (St. Jude Children’s Research Hospital) for technical assistance.
This work was supported by the US National Institutes of Health (AI101935, AI124346, AR056296, and CA163507 to T.-D. Kanneganti), the American Lebanese Syrian Associated Charities (grant to T.-D. Kanneganti), the European Research Council (281600 to M. Lamkanfi), the Fund for Scientific Research-Flanders (G030212N to M. Lamkanfi), and the National Health and Medical Research Council of Australia (R.G. Menzies Early Career Fellowship APP1091544 to S.M. Man).
The authors declare no competing financial interests.
Author contributions: X. Qi, S.M. Man, and T.-D. Kanneganti designed the study; X. Qi, S.M. Man, R.K.S. Malireddi, R. Karki, C. Lupfer, P. Gurung, C.S. Guy, and G. Neale performed experiments and analyzed the data; and X. Qi, S.M. Man, M. Lamkanfi, and T.-D. Kanneganti wrote the manuscript.
Footnotes
Abbreviations used:
- AMPK
- adenosine monophosphate–activated protein kinase
- CA-074 Me
- CA-074 methyl ester
- ERK
- extracellular signal–regulated kinase
- iNOS
- inducible nitric oxide synthase
- MOI
- multiplicity of infection
- MPO
- myeloperoxidase
- mTOR
- mechanistic target of rapamycin
- TFEB
- transcription factor EB
- TSB
- tryptic soy broth
References
- Aits S., and Jäättelä M.. 2013. Lysosomal cell death at a glance. J. Cell Sci. 126:1905–1912. 10.1242/jcs.091181 [DOI] [PubMed] [Google Scholar]
- Alers S., Löffler A.S., Wesselborg S., and Stork B.. 2012. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 32:2–11. 10.1128/MCB.06159-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alroy J., and Lyons J.A.. 2014. Lysosomal storage diseases. Journal of Inborn Errors of Metabolism & Screening. 79:619–636. 10.1134/S0006297914070049 [DOI] [Google Scholar]
- Belaaouaj A., McCarthy R., Baumann M., Gao Z., Ley T.J., Abraham S.N., and Shapiro S.D.. 1998. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat. Med. 4:615–618. 10.1038/nm0598-615 [DOI] [PubMed] [Google Scholar]
- Bewley M.A., Marriott H.M., Tulone C., Francis S.E., Mitchell T.J., Read R.C., Chain B., Kroemer G., Whyte M.K.B., and Dockrell D.H.. 2011. A cardinal role for cathepsin D in co-ordinating the host-mediated apoptosis of macrophages and killing of pneumococci. PLoS Pathog. 7:e1001262 10.1371/journal.ppat.1001262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checroun C., Wehrly T.D., Fischer E.R., Hayes S.F., and Celli J.. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. USA. 103:14578–14583. 10.1073/pnas.0601838103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu H.-C., Soni S., Kulp S.K., Curry H., Wang D., Gunn J.S., Schlesinger L.S., and Chen C.-S.. 2009. Eradication of intracellular Francisella tularensis in THP-1 human macrophages with a novel autophagy inducing agent. J. Biomed. Sci. 16:110 10.1186/1423-0127-16-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A., Wehrly T.D., Child R., Hansen B., Hwang S., Virgin H.W., and Celli J.. 2012. Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy. 8:1342–1356. 10.4161/auto.20808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu P., Cunningham A.L., Yu J.J., Nguyen J.Q., Barker J.R., Lyons C.R., Wilder J., Valderas M., Sherwood R.L., Arulanandam B.P., and Klose K.E.. 2014. Live attenuated Francisella novicida vaccine protects against Francisella tularensis pulmonary challenge in rats and non-human primates. PLoS Pathog. 10:e1004439 10.1371/journal.ppat.1004439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens D.L., Lee B.-Y., and Horwitz M.A.. 2009. Francisella tularensis phagosomal escape does not require acidification of the phagosome. Infect. Immun. 77:1757–1773. 10.1128/IAI.01485-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colletti G.A., Miedel M.T., Quinn J., Andharia N., Weisz O.A., and Kiselyov K.. 2012. Loss of lysosomal ion channel transient receptor potential channel mucolipin-1 (TRPML1) leads to cathepsin B-dependent apoptosis. J. Biol. Chem. 287:8082–8091. 10.1074/jbc.M111.285536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley S.C., and Elkins K.L.. 2011. Immunity to Francisella. Front. Microbiol. 2:26 10.3389/fmicb.2011.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T.J., Amer A., Tridandapani S., and Butchar J.P.. 2009. Francisella tularensis regulates autophagy-related host cell signaling pathways. Autophagy. 5:125–128. 10.4161/auto.5.1.7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diacovich L., and Gorvel J.-P.. 2010. Bacterial manipulation of innate immunity to promote infection. Nat. Rev. Microbiol. 8:117–128. 10.1038/nrmicro2295 [DOI] [PubMed] [Google Scholar]
- Duewell P., Kono H., Rayner K.J., Sirois C.M., Vladimer G., Bauernfeind F.G., Abela G.S., Franchi L., Nuñez G., Schnurr M., et al. . 2010. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 464:1357–1361. 10.1038/nature08938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J.W., Juliana C., Solorzano L., Kang S., Wu J., Datta P., McCormick M., Huang L., McDermott E., et al. . 2010. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11:385–393. 10.1038/ni.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocheva V., Zeng W., Ke D., Klimstra D., Reinheckel T., Peters C., Hanahan D., and Joyce J.A.. 2006. Distinct roles for cysteine cathepsin genes in multistage tumorigenesis. Genes Dev. 20:543–556. 10.1101/gad.1407406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halangk W., Lerch M.M., Brandt-Nedelev B., Roth W., Ruthenbuerger M., Reinheckel T., Domschke W., Lippert H., Peters C., and Deussing J.. 2000. Role of cathepsin B in intracellular trypsinogen activation and the onset of acute pancreatitis. J. Clin. Invest. 106:773–781. 10.1172/JCI9411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J.D., Woolard M.D., Gunn B.M., Craven R.R., Taft-Benz S., Frelinger J.A., and Kawula T.H.. 2008. Infected-host-cell repertoire and cellular response in the lung following inhalation of Francisella tularensis Schu S4, LVS, or U112. Infect. Immun. 76:5843–5852. 10.1128/IAI.01176-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle A., Hornung V., Petzold G.C., Stewart C.R., Monks B.G., Reinheckel T., Fitzgerald K.A., Latz E., Moore K.J., and Golenbock D.T.. 2008. The NALP3 inflammasome is involved in the innate immune response to amyloid-β. Nat. Immunol. 9:857–865. 10.1038/ni.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry T., Brotcke A., Weiss D.S., Thompson L.J., and Monack D.M.. 2007. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 204:987–994. 10.1084/jem.20062665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., and Brumell J.H.. 2014. Bacteria-autophagy interplay: a battle for survival. Nat. Rev. Microbiol. 12:101–114. 10.1038/nrmicro3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., and Speed T.P.. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 4:249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Jones J.W., Kayagaki N., Broz P., Henry T., Newton K., O’Rourke K., Chan S., Dong J., Qu Y., Roose-Girma M., et al. . 2010. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. USA. 107:9771–9776. 10.1073/pnas.1003738107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanistanon D., Powell D.A., Hajjar A.M., Pelletier M.R., Cohen I.E., Way S.S., Skerrett S.J., Wang X., Raetz C.R., and Ernst R.K.. 2012. Role of Francisella lipid A phosphate modification in virulence and long-term protective immune responses. Infect. Immun. 80:943–951. 10.1128/IAI.06109-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki R., Man S.M., Malireddi R.K., Gurung P., Vogel P., Lamkanfi M., and Kanneganti T.D.. 2015. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe. 17:357–368. 10.1016/j.chom.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessenbrock K., Fröhlich L., Sixt M., Lämmermann T., Pfister H., Bateman A., Belaaouaj A., Ring J., Ollert M., Fässler R., and Jenne D.E.. 2008. Proteinase 3 and neutrophil elastase enhance inflammation in mice by inactivating antiinflammatory progranulin. J. Clin. Invest. 118:2438–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., and Guan K.-L.. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13:132–141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselyov K., Chen J., Rbaibi Y., Oberdick D., Tjon-Kon-Sang S., Shcheynikov N., Muallem S., and Soyombo A.. 2005. TRP-ML1 is a lysosomal monovalent cation channel that undergoes proteolytic cleavage. J. Biol. Chem. 280:43218–43223. 10.1074/jbc.M508210200 [DOI] [PubMed] [Google Scholar]
- Kreuzaler P.A., Staniszewska A.D., Li W., Omidvar N., Kedjouar B., Turkson J., Poli V., Flavell R.A., Clarkson R.W.E., and Watson C.J.. 2011. Stat3 controls lysosomal-mediated cell death in vivo. Nat. Cell Biol. 13:303–309. 10.1038/ncb2171 [DOI] [PubMed] [Google Scholar]
- Lindgren H., Stenman L., Tärnvik A., and Sjöstedt A.. 2005. The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect. 7:467–475. 10.1016/j.micinf.2004.11.020 [DOI] [PubMed] [Google Scholar]
- Luzio J.P., Pryor P.R., and Bright N.A.. 2007. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8:622–632. 10.1038/nrm2217 [DOI] [PubMed] [Google Scholar]
- MacIvor D.M., Shapiro S.D., Pham C.T., Belaaouaj A., Abraham S.N., and Ley T.J.. 1999. Normal neutrophil function in cathepsin G-deficient mice. Blood. 94:4282–4293. [PubMed] [Google Scholar]
- Man S.M., and Kanneganti T.D.. 2015. Regulation of inflammasome activation. Immunol. Rev. 265:6–21. 10.1111/imr.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., and Kanneganti T.D.. 2016. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16:7–21. 10.1038/nri.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Karki R., Malireddi R.K., Neale G., Vogel P., Yamamoto M., Lamkanfi M., and Kanneganti T.D.. 2015. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16:467–475. 10.1038/ni.3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Karki R., and Kanneganti T.D.. 2016. AIM2 inflammasome in infection, cancer, and autoimmunity: Role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 46:269–280. 10.1002/eji.201545839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J., Almendinger J., Oberst A., Ness R., Dillon C.P., Fitzgerald P., Hengartner M.O., and Green D.R.. 2011. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. USA. 108:17396–17401. 10.1073/pnas.1113421108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D.L., Di Paola S., Peluso I., Armani A., De Stefani D., Venditti R., Montefusco S., Scotto-Rosato A., Prezioso C., Forrester A., et al. . 2015. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 17:288–299. 10.1038/ncb3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E., Wallet P., Dreier R.F., Costanzo S., Anton L., Rühl S., Dussurgey S., Dick M.S., Kistner A., Rigard M., et al. . 2015. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16:476–484. 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Li G., Zhang X., Xu H., and Abraham S.N.. 2015. A TRP channel senses lysosome neutralization by pathogens to trigger their expulsion. Cell. 161:1306–1319. 10.1016/j.cell.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A., Schafer J., Upchurch C., Spooner E., Huynh J., Hernandez S., McLaughlin B., Oden L., and Fares H.. 2015. Mucolipidosis type IV protein TRPML1-dependent lysosome formation. Traffic. 16:284–297. 10.1111/tra.12249 [DOI] [PubMed] [Google Scholar]
- Mueller-Steiner S., Zhou Y., Arai H., Roberson E.D., Sun B., Chen J., Wang X., Yu G., Esposito L., Mucke L., and Gan L.. 2006. Antiamyloidogenic and neuroprotective functions of cathepsin B: implications for Alzheimer’s disease. Neuron. 51:703–714. 10.1016/j.neuron.2006.07.027 [DOI] [PubMed] [Google Scholar]
- Nano F.E., Zhang N., Cowley S.C., Klose K.E., Cheung K.K., Roberts M.J., Ludu J.S., Letendre G.W., Meierovics A.I., Stephens G., and Elkins K.L.. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186:6430–6436. 10.1128/JB.186.19.6430-6436.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W., Park Y., Wright K., Pavlova N.N., Tuveson D.A., and Thompson C.B.. 2015. The utilization of extracellular proteins as nutrients is suppressed by mTORC1. Cell. 162:259–270. 10.1016/j.cell.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., and Ballabio A.. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20:3852–3866. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Perera R.M., Stoykova S., Nicolay B.N., Ross K.N., Fitamant J., Boukhali M., Lengrand J., Deshpande V., Selig M.K., Ferrone C.R., et al. . 2015. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 524:361–365. 10.1038/nature14587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A., Jiang Z., Waggoner S.N., Sharma S., Cole L.E., Waggoner L., Vanaja S.K., Monks B.G., Ganesan S., Latz E., et al. . 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11:395–402. 10.1038/ni.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K., Marteyn B., Sansonetti P.J., and Tang C.M.. 2009. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat. Rev. Microbiol. 7:333–340. 10.1038/nrmicro2112 [DOI] [PubMed] [Google Scholar]
- Roy C.R., Salcedo S.P., and Gorvel J.P.. 2006. Pathogen-endoplasmic-reticulum interactions: in through the out door. Nat. Rev. Immunol. 6:136–147. 10.1038/nri1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samie M., and Cresswell P.. 2015. The transcription factor TFEB acts as a molecular switch that regulates exogenous antigen-presentation pathways. Nat. Immunol. 16:729–736. 10.1038/ni.3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M., Molmeret M., and Abu Kwaik Y.. 2005. Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-γ. Cell. Microbiol. 7:957–967. 10.1111/j.1462-5822.2005.00529.x [DOI] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. . 2009. A gene network regulating lysosomal biogenesis and function. Science. 325:473–477. [DOI] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. . 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. . 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Fraldi A., Medina D.L., and Ballabio A.. 2013. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14:283–296. 10.1038/nrm3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Wang X., Li X., Zhang X., Yao Z., Dibble S., Dong X.P., Yu T., Lieberman A.P., Showalter H.D., and Xu H.. 2012. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 3:731 10.1038/ncomms1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele S., Brunton J., Ziehr B., Taft-Benz S., Moorman N., and Kawula T.. 2013. Francisella tularensis harvests nutrients derived via ATG5-independent autophagy to support intracellular growth. PLoS Pathog. 9:e1003562 10.1371/journal.ppat.1003562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J., and Schroder K.. 2010. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 10:210–215. 10.1038/nri2725 [DOI] [PubMed] [Google Scholar]
- Tsukuba T., Yamamoto S., Yanagawa M., Okamoto K., Okamoto Y., Nakayama K.I., Kadowaki T., and Yamamoto K.. 2006. Cathepsin E–deficient mice show increased susceptibility to bacterial infection associated with the decreased expression of multiple cell surface Toll-like receptors. J. Biochem. 140:57–66. 10.1093/jb/mvj132 [DOI] [PubMed] [Google Scholar]
- van Acker G.J.D., Saluja A.K., Bhagat L., Singh V.P., Song A.M., and Steer M.L.. 2002. Cathepsin B inhibition prevents trypsinogen activation and reduces pancreatitis severity. Am. J. Physiol. Gastrointest. Liver Physiol. 283:G794–G800. 10.1152/ajpgi.00363.2001 [DOI] [PubMed] [Google Scholar]
- Xu H., and Ren D.. 2015. Lysosomal physiology. Annu. Rev. Physiol. 77:57–80. 10.1146/annurev-physiol-021014-071649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Greenland J., Baluk P., Adams A., Bose O., McDonald D.M., and Caughey G.H.. 2013. Cathepsin L protects mice from mycoplasmal infection and is essential for airway lymphangiogenesis. Am. J. Respir. Cell Mol. Biol. 49:437–444. 10.1165/rcmb.2013-0016OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., McPhee C.K., Zheng L., Mardones G.A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., et al. . 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 465:942–946. 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Li X., and Xu H.. 2012. Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc. Natl. Acad. Sci. USA. 109:11384–11389. 10.1073/pnas.1202194109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.