In a cohort of Zambian heterosexual transmission pairs, the authors show that HIV-1–transmitted variants already exhibit a significant degree of preadaptation to the new host's HLA alleles, which, modulated by polymorphisms that decrease viral fitness, determines early set-point VL and the rate of disease progression in the newly infected individual.
Abstract
HIV-1 adapts to a new host through mutations that facilitate immune escape. Here, we evaluate the impact on viral control and disease progression of transmitted polymorphisms that were either preadapted to or nonassociated with the new host’s HLA. In a cohort of 169 Zambian heterosexual transmission pairs, we found that almost one-third of possible HLA-linked target sites in the transmitted virus Gag protein are already adapted, and that this transmitted preadaptation significantly reduced early immune recognition of epitopes. Transmitted preadapted and nonassociated polymorphisms showed opposing effects on set-point VL and the balance between the two was significantly associated with higher set-point VLs in a multivariable model including other risk factors. Transmitted preadaptation was also significantly associated with faster CD4 decline (<350 cells/µl) and this association was stronger after accounting for nonassociated polymorphisms, which were linked with slower CD4 decline. Overall, the relative ratio of the two classes of polymorphisms was found to be the major determinant of CD4 decline in a multivariable model including other risk factors. This study reveals that, even before an immune response is mounted in the new host, the balance of these opposing factors can significantly influence the outcome of HIV-1 infection.
INTRODUCTION
In HIV-1 infection, cytotoxic immune response plays a central role in controlling viral replication. When an HIV-1 variant is transmitted to a new recipient, the founder virus or viruses face an immune response mediated by mostly novel HLA alleles (i.e., not present in the previous host) and CTL escape mutations to the recipient’s HLA alleles are selected, whereas escape mutations to the donor’s HLA revert, leading to a “new virus” that is now adapted to the new host. This current view is supported by previous studies showing that the presence of multiple HIV-1 polymorphisms in a particular individual is statistically linked to their HLA alleles (Moore et al., 2002; Bhattacharya et al., 2007; Avila-Rios et al., 2009; Brumme et al., 2009; Carlson et al., 2012), that rapid selection of escape mutations occurs in the early weeks of infection (Brockman et al., 2007, 2010; Brumme et al., 2008; Goonetilleke et al., 2009; Fischer et al., 2010; Henn et al., 2012), and that certain CTL escape mutations are deleterious to the viral fitness (Troyer et al., 2009; Brockman et al., 2010; Song et al., 2012; Wright et al., 2012; Liu et al., 2014a,b; Adland et al., 2015; Shahid et al., 2015), imposing pressure to revert to WT after being transmitted to a new host lacking the selecting HLA allele (Leslie et al., 2004; Matthews et al., 2008; Crawford et al., 2009). Despite the fact that some CTL escape mutations are deleterious to in vivo fitness, selection of these mutations still occurs, likely because the benefit of escaping immune pressure is more important. Moreover, this model is supported by the observation that population-level consensus sequences of HIV-1 are relatively stable over time, suggesting that individuals are infected with viruses in which a majority of epitopes relevant to their immune response are consensus (Dilernia et al., 2008; Carlson et al., 2014; Cotton et al., 2014).
HIV-1 evolving to readapt to a new host’s immunogenetic background is the basis for the rationale implemented for vaccine design in which epitopes designed based on population-level consensus sequences are selected as immunogens (Fischer et al., 2007; Létourneau et al., 2007; McMichael et al., 2010; Borthwick et al., 2014; Kulkarni et al., 2014; Haynes, 2015; Mothe et al., 2015). However, the consequences of wrongly assuming that all epitopes in the transmitted virus will be consensus could have a significant impact on the success of current and future vaccine candidates, leading vaccine development in the wrong direction. In fact, transmission of CTL escape mutations has been previously reported by us (Goepfert et al., 2008; Crawford et al., 2009; Carlson et al., 2014) and others (Goulder et al., 2001; Leslie et al., 2004; Matthews et al., 2008; Schneidewind et al., 2009; Roberts et al., 2015) and, despite evidence for reversion, there is also evidence for accumulation of these mutations at the population level (Leslie et al., 2005; Dilernia et al., 2008; Kawashima et al., 2009; Cotton et al., 2014). Moreover, besides the fact that viral escape during an established infection has been widely studied (Goulder et al., 1997; Kelleher et al., 2001; Leslie et al., 2004, 2005; Brockman et al., 2007; Matthews et al., 2008; Crawford et al., 2009), the impact of transmission of CTL escape mutations relevant to a new host that has not yet mounted a cytotoxic immune response on disease progression remains unclear.
In the present study, we performed an in-depth analysis of transmitted Gag polymorphisms in a cohort of subtype C HIV-1 heterosexual transmission pairs from Zambia in which plasma samples from both the transmitting partner (donor) and the newly infected individual (recipient) were collected near the time of transmission. We show here that a majority of the mutations found in the chronic virus are likely the result of immune selection that occurred in prior rounds of infection, arguing in favor of the transmission and stability of certain escape mutations. We also show that almost a third of available HLA-linked sites present in the founder virus already harbor mutations that are associated with escape. Finally, we demonstrate that it is the delicate balance between the number of transmitted mutations relevant to the recipient’s HLA alleles (preadapted) and the remaining mutations transmitted from the donor partner (nonassociated) that ultimately determines the rate of disease progression within a newly infected individual.
RESULTS
Transmitted viruses exhibit a large degree of preadaptation to the new host
Estimation of the actual degree of adaptation of the transmitted virus to the recipient’s immunogenetic background (preadaptation) has been difficult to determine due to the lack of information on the quasispecies present in the donor at the time of transmission, leading to uncertainty in determining if the mutations observed in the newly infecting virus were transmitted from the donor or rapidly selected in the recipient. This is because, in general, newly infected individuals identified for previous studies have been insufficiently close to transmission to unambiguously define the transmitted/founder virus. In this study, we were able to determine the degree of preadaptation of the transmitted virus to the HLA Class I alleles present in the recipient by simultaneously collecting plasma samples from 169 donors and their virologically linked recipient within 3 mo after the estimated date of infection (EDI; median [interquartile range; IQR] = 45.5 [39–49] d) and performing population PCR amplification and sequencing of the gag gene.
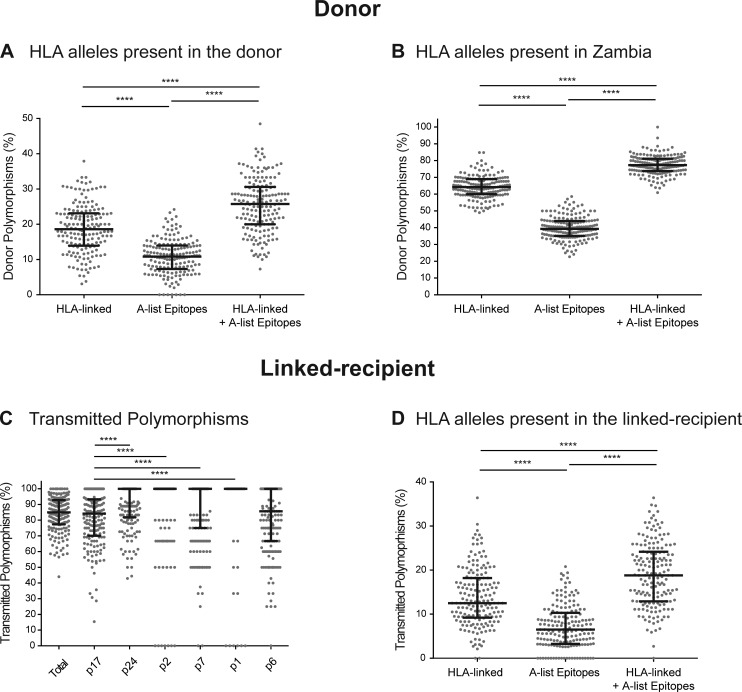
Initially, we determined the degree of adaptation of the viral population from the chronically infected partner (donor) relative to their own HLA Class I alleles. For this, we determined the number of polymorphisms (defined as amino acids not present in the Zambian consensus sequence) that could be attributed to immune escape from the donor’s HLA alleles. This number was obtained by counting polymorphisms that were located in positions statistically linked to the particular HLA Class I alleles present in each donor (HLA-linked, considering positions identified with a q-value <0.2 on the correction for multiple comparisons in a larger cohort of subtype C–infected individuals from Zambia, Botswana, and South Africa; Carlson et al., 2014), as well as polymorphisms located in well-defined CTL epitopes (A-list epitopes; Llano et al., 2013) restricted by the HLA alleles present in the donor. By obtaining the sum of these two previous numbers (after removing redundant cases), we determined that a median 25.8% (IQR, 20.0–30.6%) of the polymorphisms found in the donor sequence could be associated with immune escape to the HLA Class I–mediated immune response in that host (HLA-linked + A-list epitopes; Fig. 1 A). As shown in Fig. 1 A, the percentages of HLA-linked and epitope-located polymorphisms incompletely overlapped, with HLA-linked polymorphisms located outside known epitopes as well as polymorphisms located in epitopes that were not statistically linked to the selecting HLA Class I allele. This relatively low percentage of HLA-associated polymorphisms in the donor cohort indicates that the bulk of polymorphisms present in the circulating chronic viruses are not associated with viral adaptation to the current host’s immune response. Indeed, when we expanded this analysis to any HLA allele present in the study population, we found that a majority (median [IQR] = 77.3% [73.8–81.1%]) of polymorphisms could be attributed to selection by some HLA Class I allele present in Zambia (Fig. 1 B), suggesting they had been selected in previous rounds of infection and subsequently transmitted.
Figure 1.
A high proportion of transmitted polymorphisms are already adapted to the newly infected individual’s HLA alleles. The degree of adaptation of the donor virus to the HLA alleles of the donor (A) and the degree of preadaptation of the recipient virus (considering only polymorphisms also present in the donor virus) to the HLA alleles of the recipient (D) were measured based on the number of HLA-linked polymorphisms relevant to the host (HLA-linked), the number of polymorphisms located within well-defined CTL epitopes restricted by its HLA alleles (A-list epitopes) and the nonredundant sum of these two variables (HLA-linked + A-list epitopes). We also measured the degree of adaptation of the donor virus with respect to all the HLA alleles present in the Zambia population (B) using the same strategy. Transmission of polymorphisms (C) was calculated as the percentage of donor viral polymorphisms that were also found on the recipient virus. n = 168 (because HLA-C typing was missing in one patient) for donors and n = 169 for recipients, except when breaking down Gag into p2 (n = 161), p7 (n = 167), p1 (n = 71), and p6 (n = 168). In every case, each data point represents a single individual. ****, P < 0.0001, Dunn's test for multiple comparisons.
The fact that circulating chronic viruses exhibit large numbers of polymorphisms that are adapted to multiple HLA Class I alleles present in the Zambian population suggests that transmitted variants might harbor significant numbers of preadapted polymorphisms relevant to the HLA Class I allele repertoire of the recipient. This is supported by the fact that we observed that a majority (median [IQR] = 85% [77.6–92.7%]) of polymorphisms found in the donor sequence were also present in the transmitted virus, indicating a high rate of transmission of polymorphisms (Fig. 1 C). Interestingly, as also shown also in Fig. 1 C, transmission of polymorphisms was not homogeneous between all the proteins encoded by gag (P < 0.0001, Kruskal-Wallis test), with p17 and p6 transmitting a significantly smaller proportion of polymorphisms than p24, p2, p7, and p1. Therefore, to estimate the degree of preadaptation of the transmitted virus to the HLA alleles of the recipient, we determined the number of transmitted polymorphisms (defined as polymorphisms present in the corresponding donor quasispecies sequence and the recipient virus sequence at the time of transmission) that were associated with the HLA Class I alleles present in the recipient, considering both HLA-linked as well as those located in well-described epitopes, as described previously. We found that overall, 18.8% of transmitted polymorphisms (median [IQR], 12.9–24.1%) were already adapted to the HLA Class I–mediated immune response in the recipient (Fig. 1 D).
Because our study population is infected with subtype C variants and most of the well-defined (A-list) CTL epitopes have only been described for subtype B, we identified putative CTL epitopes across the Zambian subtype C consensus sequence. A total of 120 9-mer peptides were predicted to bind toHLA alleles present in the population (IC50 < 500 nM) and a median of 25 (IQR, 16–35) of those 9-mer peptides were predicted to constitute the putative repertoire of epitopes targeted per patient. Across these putative epitopes, we found that 40% (median [IQR], 34.1–47.6%) of these potential immune targets harbored at least one transmitted polymorphism that could impact their recognition by the CTL immune response.
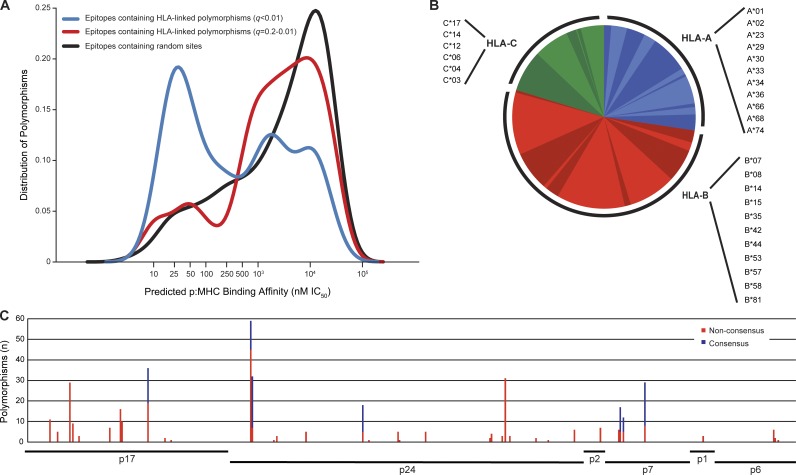
If transmitted preadapted polymorphisms affect the capacity of the recipient to mount an immune response against the transmitted/founder virus, the degree of preadaptation found in the transmitted virus will have a significant impact on the course of the infection in the newly infected individual. To test this hypothesis, we restricted further analysis to the HLA associations with a q-value <0.01, because they represent those with the highest potential for biological relevance. Also, compared with the set of HLA associations with a q-value between 0.2 and 0.01, transmitted polymorphisms linked to HLA with a q-value <0.01 were significantly more represented in predicted epitopes exhibiting strong binding affinities for the selecting HLA allele (<50 nM), whereas they were less represented in predicted epitopes exhibiting binding affinities for the selecting HLA above the biologically relevant range (>1,100 nM; Fig. 2 A).
Figure 2.
Transmitted preadapted residues are located mostly in predicted epitopes with strong binding affinity, are selected by a wide variety of HLA-A, -B, and -C alleles, and are found all across the Gag protein. Transmitted HLA-linked polymorphisms identified using HLA associations with a q-value <0.01 were selected to estimate the degree of preadaptation of the recipient sequence. A putative CD8+ T cell epitope overlapping each of the transmitted HLA-linked polymorphisms was identified as the 8-, 9-, 10-, or 11-mer peptide exhibiting the lowest IC50 to the associated HLA allele as predicted by NetMHCpan (A). Associated HLA alleles for the preadapted residues (nonconsensus and consensus) are shown in low resolution, though in some cases it represented more than one high-resolution allele (B). The sum of these polymorphisms for the 169 recipients (n = 394) was mapped relative to the HXB2 reference sequence, classifying them as nonconsensus (red) and consensus (blue) residues based on the Zambian cohort consensus sequence (C).
Moreover, because some escape mutations selected by the CTL immune response can accumulate to >50% (Kawashima et al., 2009), but nevertheless can add to the level of preadaptation of the transmitted virus, we also included adapted consensus residues in our estimation of the preadaptation of the transmitted virus. This was observed in 7 out of the 50 positions with HLA associations (n = 131 associations). The full set of preadapted residues (consensus and nonconsensus) were linked to a wide variety of HLA-A, B, and C alleles and were distributed across all of the proteins encoded by the gag gene including the less studied p7, p2, p6, and p1. Overall, most were linked to HLA-B alleles (52.4%), although 27.4% of them were identified to be associated with HLA-A and 20.3% with HLA-C (Fig. 2 B). Preadapted residues were particularly frequent in certain positions, such as A146NPSTV (59/169 individuals), Y79FH (36/169 individuals), I147LM (32/169 individuals), and D312E (31/169 individuals). Interestingly, these preadapted residues were linked to multiple different HLA alleles, indicating that these positions constitute hot spots of immune recognition. For each of the remaining positions with HLA associations (n = 46), preadapted residues were observed <20% of the time in the study population (Fig. 2 C).
In summary, the overall degree of transmitted viral preadaptation was estimated by calculating the number of HLA-linked positions that could be targeted in each individual according to their HLA alleles repertoire and then determining the proportion of these sites that was preadapted (harbored a preadapted nonconsensus or consensus residue) for each individual. We found that 28.6% (median [IQR], 18.8–40.0%) of a median of 8 (IQR, 5–10) HLA-linked positions/individual harbored a transmitted residue that could be associated with escape. This proportion of preadapted sites was used in the analyses described below to determine the impact of transmitted preadaptation on viral control and disease progression.
The balance between preadaptation and nonassociated polymorphisms in the transmitted virus defines early set-point VL in the newly infected individual
We analyzed the impact of preadaptation and nonassociated polymorphisms (defined as polymorphisms not linked to the HLA alleles present in the recipient) of the transmitted virus on the early control of viremia in a subset of recipients for whom we had data on early set-point viral load (VL), transmitted virus replicative capacity (RC; measured using Gag-MJ4 chimeras; Prince et al., 2012) and longitudinal CD4 counts (n = 74). An analysis of the correlation between the proportion of preadapted sites in the transmitted virus and early set-point VL (defined as the earliest nadir VL value after peak viremia) showed a positive association (P = 0.0009; Spearman test; Fig. 3 A). In contrast, nonassociated polymorphisms were inversely correlated with early set-point VL (P = 0.016; Spearman test; Fig. 3 B).
Figure 3.
A greater ratio of preadapted to nonassociated sites in the transmitted virus is correlated with a higher early set-point VL in the newly infected individual. The univariate correlation between proportion of preadapted sites (A), nonassociated polymorphisms (B) and the ratio between the two (C), and early set-point VL was analyzed using a Spearman test. The amount of transmitted nonassociated polymorphisms in male and female (D) and the proportion of preadapted sites in individuals that do and do not share HLA-B alleles (E) were compared using a Mann-Whitney U test (P = 0.017 and P = 0.019, respectively). n = 74.
Because several host-genetic and viral factors have been previously reported to have an independent significant contribution to early set-point VL (Yue et al., 2013), we further assessed the biological effect of transmitted preadaptation and nonassociated polymorphisms by performing a multivariable analysis in which we adjusted for other risk factors, including gender, B*57 allele, HLA-B allele sharing between donor and recipient, and RC. All factors included were significantly associated with early set-point VL in a univariate analysis (gender, P = 0.002; B*57, P = 0.001; HLA-B sharing, P = 0.032; RC, P = 0.021, Mann-Whitney test) and, with the exception of HLA-B sharing (P = 0.092), remained significant in a multivariable model (Table 1). After adding transmitted preadaptation and nonassociated polymorphisms to this model, we observed that both were significant with opposing effects (β = 0.009, P = 0.032 and β = −0.035, P = 0.001, respectively; Table 1), whereas the influence of gender and sharing of HLA-B alleles was lost. This may reflect the fact that, consistent with our previous observation (Carlson et al., 2014), females had a significantly higher number of nonassociated polymorphisms transmitted to them (P = 0.017, Mann-Whitney test; Fig. 3 D) and also with the observation that individuals who share HLA-B alleles with their donor have a higher proportion of preadapted sites (P = 0.019, Mann-Whitney test; Fig. 3 E).
Table 1. Multivariable models evaluating the role of transmitted proportion of preadapted sites, nonassociated polymorphisms, and the ratio between the two in determining early set-point VL in the context of other risk factors.
| Feature | 95% Confidence interval | ||||
|---|---|---|---|---|---|
| β | Lower | Upper | Wald Chi-Square | P-value | |
| Early set-point VL | |||||
| Gender (male) | 0.404 | 0.063 | 0.745 | 5.399 | 0.020 |
| B*57 | −0.871 | −1.392 | −0.350 | 10.750 | 0.001 |
| HLA-B sharing | 0.325 | −0.053 | 0.703 | 2.834 | 0.092 |
| RCa | −0.370 | −0.725 | −0.014 | 4.152 | 0.042 |
| Early set-point VL (including preadapted sites and nonassociated polymorphisms) | |||||
| Gender (male) | 0.170 | −0.168 | 0.509 | 0.970 | 0.325 |
| B*57 | −0.878 | −1.357 | −0.399 | 12.914 | 3.26 × 10−4 |
| HLA-B sharing | 0.137 | −0.228 | 0.503 | 0.544 | 0.461 |
| RCa | −0.435 | −0.765 | −0.106 | 6.700 | 0.010 |
| Proportion of preadapted sites | 0.009 | 0.001 | 0.017 | 4.611 | 0.032 |
| Nonassociated polymorphisms | −0.035 | −0.056 | −0.014 | 10.518 | 0.001 |
| Early set-point VL (including ratio preadapted/nonassociated) | |||||
| Gender (male) | 0.253 | −0.094 | 0.599 | 2.038 | 0.153 |
| B*57 | −0.866 | −1.365 | −0.367 | 11.579 | 0.001 |
| HLA-B sharing | 0.173 | −0.207 | 0.554 | 0.797 | 0.372 |
| RCa | −0.380 | −0.721 | −0.040 | 4.786 | 0.029 |
| Ratio preadapted/nonassociated | 0.306 | 0.072 | 0.540 | 6.578 | 0.010 |
A generalized linear model (IBM SPSS 21) was use to analyze the independent contribution of each of the variables included in defining early set-point VL. n = 74 in every model.
Dichotomized variable. RC was dichotomized on the lowest tercile as shown previously in Prince et al. (2012), representing individuals with low RC (n = 25).
Because the impact of transmitted preadaptation and nonassociated polymorphisms on early set-point VL is not strictly a 1:1 relationship, because these two values varied independently of each other, one might expect that individuals infected by a virus with a greater degree of preadaptation and a smaller number of nonassociated polymorphisms would fare worse than one infected with these factors reversed. Thus, we sought to investigate further their contribution by using a ratio of the two that summarized the opposing effects into one single variable for each individual. An analysis of the correlation between this ratio and early set-point VL showed a highly significant positive association (P < 0.0001, Spearman test; Fig. 3 C), that remained significant in a multivariable analysis (β = 0.306; P = 0.010; Table 1). Thus, early set-point VL reflects the balance between the positive influence of preadaptation and a negative impact of nonassociated polymorphisms.
A greater ratio of transmitted preadaptation/nonassociated polymorphisms is associated with a faster CD4 decline in the newly infected individual
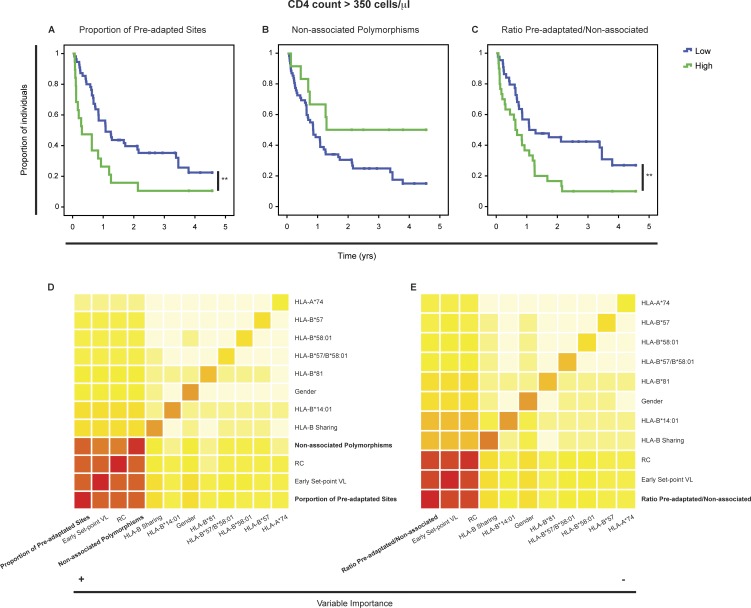
To assess the impact of preadaptation and nonassociated polymorphisms in the transmitted virus on the course of infection in a newly infected individual, we analyzed the association between these two variables and CD4 decline to <350 cells/µl (n = 74). In a univariate Cox analysis, we found that the proportion of preadapted sites in the transmitted virus was near significant for CD4 decline (P = 0.052). However, recipients infected with viruses in which one half or more of their HLA-linked sites were preadapted showed a significantly faster CD4 decline (P = 0.006, log-rank test; Fig. 4 A), consistent with the detrimental role observed for preadaptation on early set-point VL. In contrast, nonassociated polymorphisms were not significantly associated with CD4 decline in a univariate Cox analysis (P = 0.58); however, for those individuals in the upper quintile, we found a trend toward significance for slower CD4 decline (P = 0.062, log-rank test; Fig. 4 B), consistent with the negative impact of these polymorphisms on early set-point VL.
Figure 4.
A greater ratio of preadapted to nonassociated sites in the transmitted virus is associated with faster disease progression. The univariate association between proportion of preadapted sites (A), nonassociated polymorphisms (B), and the ratio between the two (C), and the rate of CD4 decline to <350 cells/µl was analyzed by the log-rank test (P = 0.006, P = 0.062, and P = 0.006, respectively). All three variables were dichotomized using a cut-off consistent with that obtained in an independent analysis (RSF). A RSF test was used to determine the contribution of the proportion of preadapted sites and nonassociated polymorphisms (D), as well as the ratio between the two (E) to the rate of CD4 decline in the context of other risk factors. In this analysis, the order of the variables from left to right, as well as the color in the diagonal, indicate their importance whereas the color of the squares outside of the diagonal indicates the strength of the interaction between the two variables (dark red to light yellow). n = 74.
To further investigate the relative contributions of transmitted preadaptation and nonassociated polymorphisms to the rate of disease progression in the context of other baseline risk factors, we used the random survival forest (RSF) method. Compared with the traditional Cox regression model, the RSF is less restrictive in terms of the assumptions it makes, is more resistant to nuisance variables and is able to detect interactions between variables. By implementing this method, we found that the proportion of preadapted sites was the variable most strongly associated with CD4 decline, followed by early set-point VL, RC, and nonassociated polymorphisms (indicated by the dark red color in the diagonal in Fig. 4 D). Also, we found that these two opposing aspects of the transmitted virus (preadaptation and polymorphisms that likely reduce viral replicative fitness) interact with early set-point VL and RC in defining the rate of CD4 decline (indicated by the darker shades of color outside the diagonal in Fig. 4 D). Consistent with this result, a Cox regression analysis that included both transmitted preadaptation and nonassociated polymorphisms (dichotomized accordingly with the RSF analysis) in the context of RC, showed that preadaptation was significantly associated with more rapid CD4 decline (HR, 2.07; P = 0.015) and nonassociated polymorphisms with slower CD4 decline (HR, 0.42; P = 0.046; Table 2).
Table 2. Multivariable models evaluating the role of transmitted proportion of preadapted sites, nonassociated polymorphisms, and the ratio between the two in determining CD4 decline in the context of other risk factors.
| Feature | 95% Confidence interval | ||||
|---|---|---|---|---|---|
| HR | Lower | Upper | Wald Chi-Square | P-value | |
| Risk of CD4 count <350 cells/μl (including preadapted sites and nonassociated polymorphisms) | |||||
| RCa | 0.59 | 0.33 | 1.07 | 3.013 | 0.083 |
| Proportion of preadapted sitesa | 2.07 | 1.15 | 3.73 | 5.932 | 0.015 |
| Nonassociated polymoprhismsa | 0.42 | 0.18 | 0.99 | 3.983 | 0.046 |
| Risk of CD4 count <350 cells/μl (including ratio preadapted/nonassociated) | |||||
| RCa | 0.53 | 0.30 | 0.95 | 4.479 | 0.034 |
| Ratio preadapted/nonassociated | 1.70 | 1.15 | 2.51 | 6.943 | 0.008 |
A Cox regression (IBM SPSS 21) was used to analyze the independent contribution of each of the variables included in defining CD4 decline. Proportion of preadapted sites was dichotomized on the highest quintile, which corresponded to individuals where the transmitted virus had half or more of its HLA-linked sites adapted (n = 19). Nonassociated polymorphisms were also dichotomized in the highest quintile, which corresponded to individuals where the transmitted virus harbored 38 or more nonassociated polymorphisms (n = 12). RC was dichotomized on the lowest tercile as shown previously in Prince et al. (2012), representing individuals with low RC (n = 25). All these three cut-offs were consistent with the cut-offs used in an independent RSF analysis. n = 74 in every model.
Dichotomized variable.
Given the opposing effects of transmitted preadaptation and nonassociated polymorphisms in CD4 decline, we again anticipated that the relative abundance of one versus the other within a single virus might better define their contribution. Analyzing this ratio (preadapted/nonassociated), we found a stronger statistical association compared with the analysis performed with each variable independently in a Cox univariate analysis (P = 0.007). Moreover, individuals infected with virus exhibiting a high ratio showed a significantly higher rate of CD4 decline (P = 0.006, log-rank test; Fig. 4 C). This result was confirmed in a RSF analysis where the ratio showed again the strongest contribution to CD4 decline followed by early set-point VL and RC, with each of these factors demonstrating interactions (Fig. 4 E). Consistent with these results, a Cox regression analysis of this ratio in the context of RC showed independent contributions of both variables (HR = 1.70, P = 0.008 and HR = 0.53, P = 0.034, respectively; Table 2), but we were not able to test the impact of these variables in the context of early set-point VL as it was highly correlated with the ratio (Fig. 3 C). Therefore, the balance between transmitted preadaptation and nonassociated polymorphisms significantly determines CD4 decline independently of RC, to a large extent through its impact on early set-point VL. However, the RSF analysis suggests that this ratio, having the highest contribution, may also influence CD4 decline through an additional independent mechanism.
A greater transmitted preadaptation is associated with a narrower and diminished CTL immune response in the newly infected individual
One mechanistic explanation for the association between preadaptation and high VL would be that individuals infected with a virus exhibiting a high proportion of preadapted sites induces a less robust immune response. We, therefore, evaluated early CTL immune responses in linked recipients infected with a virus exhibiting either a high (≥50% and an early set-point VL ≥ 40,000 copies/ml) or a low (≤20% and an early set-point VL<40,000 copies/ml) proportion of preadapted sites by stimulating cryopreserved PBMCs (median of 68 d after EDI) with autologous peptides using an IFN-γ ELISpot assay. Autologous peptides were synthesized based on the putative epitopes predicted on the Zambian consensus sequence and covered every HLA-linked position that could be targeted in each individual according to their HLA allele repertoire. We found that individuals infected with a virus exhibiting a high proportion of preadapted sites responded against a lower number (P = 0.004, Mann-Whitney U test; Fig. 5 A) and percentage (P = 0.004, Mann-Whitney U test; Fig. 5 B) of epitopes compared with individuals infected with a virus exhibiting a low proportion of preadapted sites, even though both groups had a similar number of target sites per individual (High vs. Low, median [IQR] 6 [4.5–6.8] vs. 7.5 [7–9.8]; P = 0.059; Mann-Whitney U test). We also evaluated the IFN-γ–producing response against a pool of Consensus C Gag peptides in both groups and observed a lower magnitude in individuals infected with a virus exhibiting a high proportion of preadapted sites (P = 0.007, Mann-Whitney U test; Fig. 5 C).
Figure 5.
Individuals with a high proportion of preadapted sites have a narrower and diminished CTL immune response. Autologous peptides for each linked-recipient (n = 20), based on the putative epitopes predicted on the Zambian consensus sequence that contained every HLA-linked site that could be targeted in each individual according to their HLA alleles repertoire, were used for stimulation of PBMCs in an ELISpot assay. Individuals with a high (≥50% and an early set-point VL≥40,000 copies/ml) and a low (≤20% and an early set-point VL<40,000 copies/ml) proportion of preadapted sites were compared in both the number (A) and the percentage (B) of responding peptides using a Mann-Whitney U test (P = 0.004 for both). Stimulation with a pool of Consensus C Gag peptides (C) was also performed (P = 0.007). Peptides were classified as adapted (containing an HLA-linked residue) or nonadapted and compared in their ability to elicit an IFN-γ secreting response (D, proportion of responding peptides, P = 0.013, Fisher's exact test; E, magnitude of the positive responses, P = 0.009, Mann-Whitney U test).
To confirm that preadapted epitopes elicit either no or low-magnitude CTL immune responses, we classified all the autologous peptides as adapted (when they contained an HLA-associated residue) or nonadapted (which included consensus peptides as well as all variants where polymorphisms were located outside of the target sites). We observed that only 21% of adapted peptides elicited a positive immune response in at least one individual, whereas 46% of nonadapted peptides did (P = 0.013, Fisher’s exact test; Fig. 5 D). When comparing the magnitude of the positive responses (Fig. 5 E), we found that nonadapted epitopes induced higher numbers of IFN-γ–producing cells than adapted epitopes (P = 0.009, Mann-Whitney U test).
Interestingly, out of the 61 different predicted epitopes tested, 19.7% (12/61) were previously described in the best-defined CTL epitopes, also known as A list epitopes (Llano et al., 2013), whereas 80.3% (49/61) constituted novel targets, that were either located in novel regions (23/49) or restricted by a different HLA allele (26/49). Of these 49 epitopes tested, 53.1% (26/49) elicited a CTL immune response in at least one subtype C infected individual, confirming them as novel epitopes (Table S2). In fact, this response rate was similar to that observed for the 12 A list epitopes tested (50%; 6/12). Also important, these autologous peptides (as well as the consensus C Gag peptide pool) did not elicit any positive responses when tested in HIV-negative donors (n = 11).
Together, these results support our hypothesis that transmission of preadapted epitopes limits the ability of the immune response to target the transmitted variant, contributing to higher early set-point VLs and faster CD4 decline in the newly infected individual.
DISCUSSION
During HIV-1 transmission, despite a level of selection bias that favors consensus amino acids (Carlson et al., 2014), we show that a majority (85%) of the nonconsensus polymorphisms encoded by the donor virus quasispecies are present in the transmitted virus. Surprisingly, while only a minority (25.8%) of donor polymorphisms can be attributed to selection within the donor, a large majority (77.3%) is associated with selection by Zambian HLA allele-mediated immune responses. Moreover, we show that HIV-1–transmitted variants already exhibit a significant degree of preadaptation to the new host’s HLA alleles that determines early set-point VL and the rate of disease progression in the newly infected individual.
Several studies have focused on analyzing the role of transmitted viral characteristics and host genetics in disease progression (Migueles et al., 2000; Gao et al., 2005, 2010; Thobakgale et al., 2009; Brennan et al., 2012; Prince et al., 2012; Casado et al., 2013; Dalmau et al., 2014; Adland et al., 2015; Claiborne et al., 2015). However, the role of transmitted preadaptation, even when implicated in disease progression, has been under appreciated, likely because without prior knowledge of the sequence of the virus quasispecies in the transmitting partner, it is difficult to distinguish transmitted adaptation from mutations that arise early in the newly infected individual (Dalmau et al., 2014). In the present study, we obtained the donor virus sequence near the time of transmission and focused our analysis only on polymorphisms that were present in both the donor and the recipient, which allowed us to determine that on average, almost one-fifth of transmitted nonconsensus polymorphisms were preadapted to the new host’s HLA alleles. Although it is possible that there were cases in which a minor variant in the donor was not detected and the polymorphism was still transmitted to the newly infected individual, only a small percentage (median [IQR] = 6.5% [2.9–11.4%]) of polymorphisms present in the recipient sequence were absent in the donor sequence. It is more likely that this small subset of polymorphisms is actually the result of early escape selected by the developing CTL immune response (Fischer et al., 2010; Henn et al., 2012), and this possibility is supported by the fact that the proportion of these polymorphisms present in the recipient was positively correlated with the time from EDI to sampling date (P = 0.018, Spearman test).
A surprising observation was that the fraction of polymorphisms associated with CTL escape in the virus quasispecies of chronically infected donors (25.8%) is only 7% higher than the fraction of preadapted polymorphisms in the variant transmitted to the newly infected individual (18.8%). This suggests that, even though the virus undergoes a dynamic process of selection and reversion after transmission, around three-quarters of the viral adaptation observed in the chronic donors is acquired at the moment of transmission. This result is also consistent with the small number of newly selected HLA-linked polymorphisms observed in Gag during the course of infection (Gounder et al., 2015; Roberts et al., 2015). Moreover, this study also shows that the high degree of preadaptation found in transmitted viruses is not simply a consequence of allele sharing between donor and recipient, but is instead a result of the high proportion of donor polymorphisms that can be associated with HLA alleles that are not necessarily the donor’s but that are nonetheless present in the Zambian population.
It is also interesting to note that the large proportion of polymorphisms adapted to HLA alleles not present in the donor belies the concept that a majority of polymorphisms are detrimental to the virus and will revert shortly after transmission. In fact, our finding is consistent with our recent report of a limited reversion rate in newly infected individuals during the first two years of infection (2% per yr; Carlson et al., 2014). This is also consistent with evidence of accumulation of CTL escape mutations at the population level (Leslie et al., 2005; Dilernia et al., 2008; Kawashima et al., 2009; Cotton et al., 2014) and likely means that these mutations either have a limited impact on virus fitness or have evolved in the presence of compensatory mutations (Liu et al., 2014a; Shahid et al., 2015). It is important to note, however, that none of these studies have found a large increase in the prevalence of CTL escape mutations over time, suggesting a limit to their accumulation at the population level. Although we did find recipients infected with variants harboring as many as 47 nonassociated transmitted polymorphisms, these larger numbers were associated with lower early set-point VLs and slower CD4 decline. Overall, this evidence suggests a limited impact of individual polymorphisms on virus fitness but a detrimental effect derived from their accumulation.
A key finding of this study is that the contribution of individual polymorphisms to HIV-1 pathogenesis depends on the genetic context presented by the new host. We found that there is a balance of effects from polymorphisms that reduce immune recognition of the transmitted-founder virus, as shown by the reduced number of responsive epitopes as well as the lower proportion of responsive target sites observed in individuals with a high proportion of preadapted sites, and those that might negatively impact in vivo replicative fitness. Specifically, a higher proportion of preadapted HLA-linked sites (preadaptation) is positively associated with higher early set-point VL, whereas increasing numbers of transmitted nonassociated polymorphisms are associated with a lower early set-point VL. The former result is consistent with an analysis using an alternative probabilistic model for defining adaptation of a viral sequence to an individual’s HLA alleles (Carlson et al., 2016), whereas the latter result is consistent with our previous report on a smaller number of individuals (n = 35) where HLA-B associated polymorphisms transmitted to recipients lacking the selecting alleles were independently associated with lower early VLs (Goepfert et al., 2008). However, here we take into account the impact of all polymorphisms that are not relevant to the recipient’s HLA-alleles irrespective of whether or not they are statistically linked to other HLA alleles, and analyze the relative impact of these antagonistic effects.
The balance between the opposing effects of preadaptation and viral replicative fitness is further highlighted by the much more significant impact on early set-point VL we observed when the ratio of preadaptation to nonassociated polymorphisms within each transmitted virus was analyzed. Importantly, we were able to observe this effect in a multivariable model that included other risk factors, such as gender, protective alleles, B-allele sharing, and RC. This ratio appeared to capture the relevance of both gender and HLA-B allele sharing, as these two parameters lost significance in the model, whereas B*57 and RC remained significant.
Similarly, both transmitted preadaptation and nonassociated polymorphisms are directly associated with the rate of CD4 decline but with opposing effects, and their impact on disease progression is even larger when both are considered together. The ratio of transmitted preadapted sites to nonassociated polymorphisms for each virus shows the strongest effect on CD4 decline (<350 cells/µl), even when early set-point VL, RC, gender, HLA-B allele sharing, and protective HLA alleles are taken into account. In other words, the balance between the degree of preadaptation and the overall divergence from consensus of the transmitted variant is a major determinant of the rate of disease progression. Moreover, in the RSF analyses, it is clear that there are strong interactions between preadaptation, nonassociated polymorphisms, early set-point VL and RC that also play a role in this process. It seems likely that both preadaptation and nonassociated polymorphisms may be exerting their impact on CD4 decline, at least in part through their influence on early set-point VL. Nevertheless, the results of the RSF analysis, which included all three independent variables, showed that preadaptation provided a stronger predictive ability than early set-point VL itself. This may reflect the reduced capacity of the immune system to target epitopes in viruses with a higher degree of transmitted adaptation later in infection, as the virus continues to escape from the CTL immune response. Indeed, because transmitted preadapted polymorphisms that impact disease progression are preferentially located in epitopes predicted to have a strong binding affinity for the linked HLA allele, this may require targeting of suboptimal epitopes that are less effective in immune control.
Relevant for the development of an HIV vaccine, it is important to note that almost a third of predicted epitopes in the transmitted virus were already adapted to the newly infected individual HLA allele repertoire. Preadaptation was associated with impaired recognition or weaker IFN-γ responses elicited by these epitopes. This suggests that population-level consensus sequences are not the optimal source of information for immunogen design, as many of these epitopes may be absent in the transmitted virus, arguing for immunogens that represent conserved regions of the HIV-1 proteome lacking significant adaptation.
Collectively, these results show that a large number of CTL escape mutations persist and circulate in the population, leading to the transmission of preadapted viral variants. We present evidence indicating that it is the balance between the effects of transmitted preadaptation, which determines the number and quality of epitopes effectively targeted during the course of infection, and nonassociated polymorphisms, which are likely linked to a reduced viral replicative fitness, that dictates HIV-1 disease progression. Therefore, transmitted polymorphisms represent a fundamental predictor of disease outcome and could inform HIV vaccine design strategies by focusing the development of immunogens on regions with limited evidence of selection and transmission of CTL escape mutations.
MATERIALS AND METHODS
Study subjects
All participants in the Zambia-Emory HIV Research Project (ZEHRP) discordant couples cohort in Lusaka and Ndola, Zambia were enrolled in human subject protocols approved by both the University of Zambia Research Ethics Committee and the Emory University Institutional Review Board. Before enrollment, individuals received counseling and signed a written informed consent form. The subjects selected from the cohort (n = 169 couples) were initially HIV-1 serodiscordant partners in cohabiting heterosexual couples with subsequent intracouple (epidemiologically linked) HIV-1 transmission (McKenna et al., 1997; Allen et al., 2007; Kempf et al., 2008). Epidemiological linkage was defined by phylogenetic analyses of HIV-1 gp41 sequences from both partners (Trask et al., 2002). The estimated time since infection (ETI) was calculated as the time between the EDI (Haaland et al., 2009) and the time of the first available sample. Only couples with an ETI for the linked-recipient <3 mo were included in the study (median [IQR] = 45.5 [38.5–49.0] days). Blood samples were obtained from both the transmitting source partner (donor) and the linked seroconverting partner (linked-recipient) at the earliest time point. All the patients included in this study were infected with subtype C HIV-1 strains and were antiretroviral therapy naive.
VL and CD4 count determinations
HIV-1 plasma VL was determined at the Emory Center for AIDS Research Virology Core Laboratory using the Amplicor HIV-1 Monitor Test (version 1.5; Roche). A mean of 15 determinations were performed during the course of infection per linked recipients. Early set-point VL for the linked recipients was defined as the earliest nadir VL value measured after peak viremia (between 1 and 12 mo after EDI) that did not show a significant increase in value within a 1-yr window.
CD4 count was based on T cell immunophenotyping, with assays done using the FACScount System (Beckman Coulter) in collaboration with the International AIDS Vaccine Initiative. This determination was performed for the linked recipients every 3 mo during the first 2 yr of infection, and every 6 mo afterward. Time to CD4 count <350 cells/µl was used to analyze the rate of disease progression. This variable was coded as the time in days between EDI and the first CD4 count below that specific value, or the date when antiretroviral treatment was initiated. CD4 count values were censored after ART initiation.
HLA class I genotyping
Genomic DNA was extracted from whole blood or buffy coats (QIAamp blood kit; QIAGEN). HLA class I genotyping relied on a combination of PCR-based techniques, involving sequence-specific primers (Invitrogen) and sequence-specific oligonucleotide probes (Innogenetics), as described previously (Tang et al., 2002). Ambiguities were resolved by direct sequencing of three exons in each gene, using kits (Abbott Molecular, Inc.) designed for capillary electrophoresis and the ABI 3130xl DNA Analyzer (Applied Biosystems).
Amplification and sequencing of the gag gene from donors and linked-recipients
Viral RNA was extracted from 140 µl of plasma using a viral RNA extraction kit (QIAGEN). Gag-Pol population sequences were generated using nested gene-specific primers. Combined RT-PCR and first round synthesis was performed using SuperScript III Platinum One Step RT-PCR (Invitrogen) and 5 µl viral RNA template. RT-PCR and first round primers included GOF (forward) 5′-ATTTGACTAGCGGAGGCTAGAA-3′ and VifOR (RT-PCR and reverse) 5′-TTCTACGGAGACTCCATGACCC-3′. Second round PCR was performed using Expand High Fidelity Enzyme (Roche) and 1 µl of the first round PCR product. Nested second round primers include GIF (forward) 5′-TTTGACTAGCGGAGGCTAGAAGGA-3′ and VifIR (reverse) 5′-TCCTCTAATGGGATGTGTACTTCTGAAC-3′. Three positive amplicons per individual were pooled and purified via the PCR purification kit (QIAGEN). Purified products were Sanger sequenced by the University of Alabama at Birmingham DNA Sequencing Core. Sequence chromatograms were analyzed using Sequencher 5.0 (Gene Codes Corp.) and degenerate bases were denoted using the International Union of Pure and Applied Chemistry (IUPAC) codes when minor peaks exceeded 25% of the total peak height in both forward and reverse reads. Codons containing degenerate bases that corresponded to more than one amino acid were defined as mixtures, whereas those with no evidence of degenerate bases or with minor peaks comprising <25% of the total were defined as dominant variants. Sequences were codon aligned to the HXB2 reference sequence using HIVAlign (http://www.hiv.lanl.gov/content/sequence/VIRALIGN/viralign.html), followed by hand-editing. All viral sequences have been submitted to GenBank, accession nos. FJ606137-FJ606433, JQ219842, KM048382-KM048989, KP715728-KP715844, and KU043117-KU043162.
For every given couple, the total amount of nonconsensus polymorphisms present in the donor sequence was determined by comparing each sequence against the Zambian consensus sequence. The Zambian cohort consensus was defined using sequences from 375 chronically infected individuals, as described in Carlson et al. (2014). However, the total amount of nonconsensus polymorphisms transmitted to the linked recipient was defined as the total amount of these polymorphisms present in the donor sequence that were also present in the recipient sequence. In this case, every polymorphism that was present in the recipient sequence but absent in the donor sequence was disregarded and considered as a newly selected polymorphism in the linked-recipient.
Identification of viral polymorphisms associated with CTL escape
Viral polymorphisms were considered to be associated with CTL escape if they belonged to any of the following three categories: (1) HLA-linked polymorphisms, which are defined as polymorphisms located at an amino acid position for which a significant statistical association between a specific residue and a specific HLA class I allele has been found. The methodology implemented to identify these associations was described before and a complete list of significant associations at a q-value <0.2 (corresponding to a false discovery rate of 20%) is available in the External Database S1 (Carlson et al., 2014). This analysis was done using the complete list of significant associations at a q-value <0.2, as well as limiting it to the associations identified at a q-value <0.01, which indicates that 99% of them are expected to be nonspurious. (2) Epitope-located polymorphisms, which are defined as polymorphisms located within the best-defined CTL epitopes, also known as A list epitopes (Llano et al., 2013). (3) HLA-linked + Epitope-located polymorphisms, which are defined as the sum of polymorphisms that belong to any of the previous two categories, HLA-linked or epitope-located, excluding repeated cases. In this case, two sums were performed using either the q-value <0.2 or the q-value <0.01 cut-off for the count of HLA-linked polymorphisms.
Estimation of viral adaptation to the host CTL immune response
The degree of adaptation of the viral sequence to the CTL immune response of the host was measured based on the number of HLA-linked polymorphisms relevant to the host in terms of its HLA alleles repertoire, as well as the number of polymorphisms located within well-defined CTL epitopes restricted by its HLA alleles. Specifically, for the HLA-linked analysis, a viral polymorphism at a given position of the Gag protein in a given individual, was defined to be consistent with viral adaptation to that individual if: (1) the individual harbored an HLA allele that is statistically linked to a particular residue in that position; and (2) either the residue was positively correlated (referred to as adapted in the literature) with the HLA, or the residue was any other residue than the one negatively correlated (referred to as nonadapted in the literature) with that HLA.
In the case of the donor, all nonconsensus polymorphisms in the donor viral sequence were subjected to this classification, and the number obtained was considered as an estimation of the degree of adaptation of the chronic viral population to the CTL immune response of the donor, achieved by the virus at the moment it got transmitted to the recipient. In the case of the linked recipient, only recipient viral nonconsensus polymorphisms that were confirmed to be transmitted by comparison with the donor viral sequence were subjected to this classification, and the number obtained was considered to be an estimation of the degree of preadaptation of the transmitted virus to the new host. Alternatively, the degree of preadaptation of the transmitted virus was estimated by calculating the proportion of HLA-linked sites that could be targeted by each individual according to their HLA alleles that harbored a transmitted adapted residue (nonconsensus or consensus). Finally, the number of transmitted nonassociated polymorphisms was calculated for each linked-recipient by subtracting the number of preadapted HLA-linked polymorphisms from the total number of transmitted polymorphisms in a way that [total transmitted polymorphisms] = [nonassociated polymorphisms] + [HLA-linked polymorphisms relevant to the host].
Putative epitope identification
Total putative CD8+ T cell epitopes were identified in the Zambian consensus sequence using computational HLA/peptide binding predictors to evaluate every possible 9-mer (n = 514). The binding affinity of each peptide to each HLA allele present in the Zambian population was predicted using NetMHCpan (version 2.8). An IC50 < 500 nM was used as the criteria to identify all putative epitopes.
To identify the putative epitopes that contained all HLA-linked sites, we considered every overlapping 8-, 9-, 10-, and 11-mer peptide from the Zambian consensus sequence. Again, we evaluated the binding affinity of each peptide to the associated HLA allele and we selected the peptide with the lowest IC50 as the putative epitope. The distribution of IC50 values associated with three groups of sites was compared: (1) HLA-linked polymorphisms identified with a q-value <0.01; (2) HLA-linked polymorphisms identified with a q-value between 0.2 and 0.01; and (3) random sites. The set of random sites was chosen from the sequence alignment with uniform probability. Next, the epitope identification algorithm was repeated using the same set of HLA alleles. This set of putative epitopes was representative of a null hypothesis in which transmitted HLA-linked polymorphisms are no different from random sites.
Peptide synthesis
Autologous peptides for 20 linked-recipients were synthesized in a 96-well array format (New England Peptide). These peptides were based on the putative epitopes previously predicted on the Zambian consensus sequence that contained every HLA-linked site that could be targeted in each individual according to their HLA alleles repertoire. A list of all autologous peptides evaluated per linked recipient is provided in Table S1.
IFN-γ ELISpot assays
IFN-γ ELISpot assays were performed using PBMCs obtained with a median of 67.8 (IQR, 47.6–87.5) d after EDI and a median of 24.5 (IQR, 6.5–57.5) d after the viral sequence was derived from plasma, as described previously (Bansal et al., 2007, 2010). In brief, cryopreserved PBMCs (n = 20) were thawed and allowed to rest overnight at 37°C. Cells were plated at 105/well in triplicate and incubated with the antigens for 18 h. Autologous peptides and a pool of Consensus C Gag peptides (HIV-1 Consensus C Gag Peptide Pool, catalog number 12756, National Institutes of Health AIDS Reagent Program, Division of AIDS, NIAID, National Institutes of Health) were used at a final concentration of 10 µM and 2 µg/ml, respectively. Unstimulated and PHA stimulated cells (10 µg/ml) plated in quadruplicate and duplicate, respectively, were used as negative and positive controls. Spot numbers were counted using an automated plate reader (CTL ImmunoSpot) and normalized to 106 PBMCs (spot-forming units [SFU]/106 PBMCs). All PBMCs samples showed a PHA response >500 SFU/106 PBMCs. Positive responses were defined as 50 SFU/106 PBMCs or >3 times the media-only negative control, whichever was highest.
Assays using the aforementioned antigens were also performed in HLA Class I–matched HIV-negative donors (n = 11) recruited from the Alabama Vaccine Research Clinic as a control for nonspecific reactivity.
Statistical analysis
Differences in percentages of viral polymorphisms between groups were assessed using the Kruskal-Wallis test and the Dunn's test for multiple comparisons, as implemented in Prism 6 (GraphPad).
The unadjusted correlation between transmitted proportion of preadapted sites and nonassociated polymorphisms as well as the ratio between these two variables (preadapted/nonassociated) and early set-point VL (n = 74) was assessed by the Spearman test, as implemented in Prism 6. Generalized linear models (IBM SPSS 21) were applied to assess the impact of each of these three variables on early set-point VL (n = 74) while adjusting for gender (n = 39 for males and n = 35 for females), B*57 (n = 9), HLA-B allele sharing between donor and linked-recipient (n = 20), and RC of the transmitted virus, which was dichotomized on the lowest tercile as previously reported (n = 25; Prince et al., 2012). All VL data were log10 transformed.
The unadjusted association between transmitted proportion of preadapted sites and nonassociated polymorphisms as well as the ratio between these two variables (preadapted/nonassociated) and the rate of CD4 decline to <350 cells/µl (n = 74) was assessed by a Cox regression, as implemented on IBM SPSS 21. A log-rank test (IBM SPSS 21) was implemented also to analyze the rate of CD4 decline in individuals with high and low values for each of these three variables that were dichotomized using cut-off values that were consistent with those obtained from the multivariable analysis (RSF).
Multivariable models were applied to assess the impact of each of these three variables on the rate of CD4 decline while adjusting for several host genetic and viral characteristics known to influence this rate. These variables included: gender (n = 39 for males and n = 35 for females), expression of protective alleles such as A*74 (n = 9), B*14:01 (n = 5), B*57 (n = 9), B*58:01 (n = 8), B*57/B*58:01 (n = 17) and B*81 (n = 4), HLA-B allele sharing between donor and linked-recipient (n = 20), RC of the transmitted virus (dichotomized on the lowest tercile; n = 25), and early set-point VL. Initially, we applied the RSF method using the R package “randomForestSRC” (Ishwaran et al., 2008; Ishwaran and Kogalur, 2010). This is a relatively new statistical technique for the analysis of right-censored survival data that does not make the proportional hazard assumption. It is an ensemble tree method based on growing a large number of survival trees using bootstrap samples. For each tree, at each split, the variable is chosen from a random subset of variables by maximizing survival difference between daughter nodes. The final prediction is obtained by combining the outcome of all the trees. Here, the interest was mainly in the capability of selecting important variables and identifying variable interactions. Afterward, we confirmed these results by performing a multivariable Cox regression (IBM SPSS 21) including the main predictive variables identified by the RSF analysis that consisted in the three variables of interest along with RC. We were not able to include early set-point VL in this multivariable model due to the highly significant correlation observed with the three variables of interest.
Differences in the number and percentage of responsive epitopes as well as in the magnitude of the IFN-γ responses between groups were assessed using the Mann-Whitney U test, as implemented in Prism 6. A Fisher’s exact test was used to compare the proportion of responsive/nonresponsive adapted and nonadapted epitopes, as implemented in Prism 6.
Online supplemental material
A list of all autologous peptides evaluated per linked recipient is provided in Table S1. A list of the novel epitopes identified is provided in Table S2. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20151984/DC1.
Supplementary Material
ACKNOWLEDGMENTS
The investigators thank all the volunteers in Zambia who participated in this study and all the staff at the Zambia-Emory HIV Research Project in Lusaka who made this study possible. The investigators would like to thank Jonathan Carlson for in depth statistical discussions and manuscript review, and Victor Du, Jon Allen, and Mackenzie Hurlston for technical assistance and sample management.
This study was funded by grants R01 AI64060, R01 AI112566, and R37 AI51231 (E. Hunter and P. Goepfert) from the National Institutes of Allergy and Infectious Diseases, National Institutes of Health, and by the International AIDS Vaccine Initiative (S. Allen), and made possible in part by the generous support of the American people through the United States Agency for International Development (USAID). The contents are the responsibility of the study authors and do not necessarily reflect the views of USAID or the United States Government. This work was also supported, in part, by the Biostatistics and Biomedical Informatics and Virology Cores at the Emory Center for AIDS Research (grant P30 AI050409), the Yerkes National Primate Research Center (base grant 2P51RR000165-51), the National Center for Research Resources (grant P51RR165), and the Office of Research Infrastructure Programs (grant P51OD11132). D.C. Mónaco was supported in part by an Action Cycling Fellowship.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- EDI
- estimated date of infection
- IQR
- interquartile range
- RC
- replicative capacity
- RSF
- random survival forest
- SFU
- spot-forming unit
- VL
- viral load
References
- Adland E., Paioni P., Thobakgale C., Laker L., Mori L., Muenchhoff M., Csala A., Clapson M., Flynn J., Novelli V., et al. 2015. Discordant impact of HLA on viral replicative capacity and disease progression in pediatric and adult HIV infection. PLoS Pathog. 11:e1004954 10.1371/journal.ppat.1004954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S., Karita E., Chomba E., Roth D.L., Telfair J., Zulu I., Clark L., Kancheya N., Conkling M., Stephenson R., et al. 2007. Promotion of couples’ voluntary counselling and testing for HIV through influential networks in two African capital cities. BMC Public Health. 7:349 10.1186/1471-2458-7-349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Rios S., Ormsby C.E., Carlson J.M., Valenzuela-Ponce H., Blanco-Heredia J., Garrido-Rodriguez D., Garcia-Morales C., Heckerman D., Brumme Z.L., Mallal S., et al. 2009. Unique features of HLA-mediated HIV evolution in a Mexican cohort: a comparative study. Retrovirology. 6:72 10.1186/1742-4690-6-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A., Yue L., Conway J., Yusim K., Tang J., Kappes J., Kaslow R.A., Wilson C.M., and Goepfert P.A.. 2007. Immunological control of chronic HIV-1 infection: HLA-mediated immune function and viral evolution in adolescents. AIDS. 21:2387–2397. 10.1097/QAD.0b013e3282f13823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal A., Carlson J., Yan J., Akinsiku O.T., Schaefer M., Sabbaj S., Bet A., Levy D.N., Heath S., Tang J., et al. 2010. CD8 T cell response and evolutionary pressure to HIV-1 cryptic epitopes derived from antisense transcription. J. Exp. Med. 207:51–59. 10.1084/jem.20092060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya T., Daniels M., Heckerman D., Foley B., Frahm N., Kadie C., Carlson J., Yusim K., McMahon B., Gaschen B., et al. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 315:1583–1586. 10.1126/science.1131528 [DOI] [PubMed] [Google Scholar]
- Borthwick N., Ahmed T., Ondondo B., Hayes P., Rose A., Ebrahimsa U., Hayton E.J., Black A., Bridgeman A., Rosario M., et al. 2014. Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol. Ther. 22:464–475. 10.1038/mt.2013.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.A., Ibarrondo F.J., Sugar C.A., Hausner M.A., Shih R., Ng H.L., Detels R., Margolick J.B., Rinaldo C.R., Phair J., et al. 2012. Early HLA-B*57-restricted CD8+ T lymphocyte responses predict HIV-1 disease progression. J. Virol. 86:10505–10516. 10.1128/JVI.00102-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman M.A., Schneidewind A., Lahaie M., Schmidt A., Miura T., Desouza I., Ryvkin F., Derdeyn C.A., Allen S., Hunter E., et al. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608–12618. 10.1128/JVI.01369-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockman M.A., Brumme Z.L., Brumme C.J., Miura T., Sela J., Rosato P.C., Kadie C.M., Carlson J.M., Markle T.J., Streeck H., et al. 2010. Early selection in Gag by protective HLA alleles contributes to reduced HIV-1 replication capacity that may be largely compensated for in chronic infection. J. Virol. 84:11937–11949. 10.1128/JVI.01086-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme Z.L., Brumme C.J., Carlson J., Streeck H., John M., Eichbaum Q., Block B.L., Baker B., Kadie C., Markowitz M., et al. 2008. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J. Virol. 82:9216–9227. 10.1128/JVI.01041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumme Z.L., John M., Carlson J.M., Brumme C.J., Chan D., Brockman M.A., Swenson L.C., Tao I., Szeto S., Rosato P., et al. 2009. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One. 4:e6687 10.1371/journal.pone.0006687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.M., Listgarten J., Pfeifer N., Tan V., Kadie C., Walker B.D., Ndung’u T., Shapiro R., Frater J., Brumme Z.L., et al. 2012. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J. Virol. 86:5230–5243. 10.1128/JVI.06728-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.M., Schaefer M., Monaco D.C., Batorsky R., Claiborne D.T., Prince J., Deymier M.J., Ende Z.S., Klatt N.R., DeZiel C.E., et al. 2014. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 345:1254031 10.1126/science.1254031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J.M., Du V.Y., Pfeifer N., Bansal A., Tan V.Y., Power K., Brumme C.J., Kreimer A., DeZiel C.E., Fusi N., et al. 2016. Impact of pre-adapted HIV transmission. Nat. Med. 22:606–613. 10.1038/nm.4100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casado C., Pernas M., Sandonis V., Alvaro-Cifuentes T., Olivares I., Fuentes R., Martínez-Prats L., Grau E., Ruiz L., Delgado R., et al. 2013. Identification of a cluster of HIV-1 controllers infected with low replicating viruses. PLoS One. 8:e77663 10.1371/journal.pone.0077663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claiborne D.T., Prince J.L., Scully E., Macharia G., Micci L., Lawson B., Kopycinski J., Deymier M.J., Vanderford T.H., Nganou-Makamdop K., et al. 2015. Replicative fitness of transmitted HIV-1 drives acute immune activation, proviral load in memory CD4+ T cells, and disease progression. Proc. Natl. Acad. Sci. U.S.A. 112:E1480–E1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton L.A., Kuang X.T., Le A.Q., Carlson J.M., Chan B., Chopera D.R., Brumme C.J., Markle T.J., Martin E., Shahid A., et al. 2014. Genotypic and functional impact of HIV-1 adaptation to its host population during the North American epidemic. PLoS Genet. 10:e1004295 10.1371/journal.pgen.1004295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford H., Lumm W., Leslie A., Schaefer M., Boeras D., Prado J.G., Tang J., Farmer P., Ndung’u T., Lakhi S., et al. 2009. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206:909–921. 10.1084/jem.20081984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmau J., Rotger M., Erkizia I., Rauch A., Reche P., Pino M., Esteve A., Palou E., Brander C., Paredes R., et al. CoRP Study Group . 2014. Highly pathogenic adapted HIV-1 strains limit host immunity and dictate rapid disease progression. AIDS. 28:1261–1272. 10.1097/QAD.0000000000000293 [DOI] [PubMed] [Google Scholar]
- Dilernia D.A., Jones L., Rodriguez S., Turk G., Rubio A.E., Pampuro S., Gomez-Carrillo M., Bautista C.T., Deluchi G., Benetucci J., et al. 2008. HLA-driven convergence of HIV-1 viral subtypes B and F toward the adaptation to immune responses in human populations. PLoS One. 3:e3429 10.1371/journal.pone.0003429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer W., Perkins S., Theiler J., Bhattacharya T., Yusim K., Funkhouser R., Kuiken C., Haynes B., Letvin N.L., Walker B.D., et al. 2007. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 13:100–106. 10.1038/nm1461 [DOI] [PubMed] [Google Scholar]
- Fischer W., Ganusov V.V., Giorgi E.E., Hraber P.T., Keele B.F., Leitner T., Han C.S., Gleasner C.D., Green L., Lo C.C., et al. 2010. Transmission of single HIV-1 genomes and dynamics of early immune escape revealed by ultra-deep sequencing. PLoS One. 5:e12303 10.1371/journal.pone.0012303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Bashirova A., Iversen A.K., Phair J., Goedert J.J., Buchbinder S., Hoots K., Vlahov D., Altfeld M., O’Brien S.J., and Carrington M.. 2005. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat. Med. 11:1290–1292. 10.1038/nm1333 [DOI] [PubMed] [Google Scholar]
- Gao X., O’Brien T.R., Welzel T.M., Marti D., Qi Y., Goedert J.J., Phair J., Pfeiffer R., and Carrington M.. 2010. HLA-B alleles associate consistently with HIV heterosexual transmission, viral load, and progression to AIDS, but not susceptibility to infection. AIDS. 24:1835–1840. 10.1097/QAD.0b013e32833c3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert P.A., Lumm W., Farmer P., Matthews P., Prendergast A., Carlson J.M., Derdeyn C.A., Tang J., Kaslow R.A., Bansal A., et al. 2008. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J. Exp. Med. 205:1009–1017. 10.1084/jem.20072457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goonetilleke N., Liu M.K., Salazar-Gonzalez J.F., Ferrari G., Giorgi E., Ganusov V.V., Keele B.F., Learn G.H., Turnbull E.L., Salazar M.G., et al. CHAVI Clinical Core B . 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206:1253–1272. 10.1084/jem.20090365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder P.J., Phillips R.E., Colbert R.A., McAdam S., Ogg G., Nowak M.A., Giangrande P., Luzzi G., Morgan B., Edwards A., et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217. 10.1038/nm0297-212 [DOI] [PubMed] [Google Scholar]
- Goulder P.J., Brander C., Tang Y., Tremblay C., Colbert R.A., Addo M.M., Rosenberg E.S., Nguyen T., Allen R., Trocha A., et al. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 412:334–338. 10.1038/35085576 [DOI] [PubMed] [Google Scholar]
- Gounder K., Padayachi N., Mann J.K., Radebe M., Mokgoro M., van der Stok M., Mkhize L., Mncube Z., Jaggernath M., Reddy T., et al. 2015. High frequency of transmitted HIV-1 Gag HLA class I-driven immune escape variants but minimal immune selection over the first year of clade C infection. PLoS One. 10:e0119886 10.1371/journal.pone.0119886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaland R.E., Hawkins P.A., Salazar-Gonzalez J., Johnson A., Tichacek A., Karita E., Manigart O., Mulenga J., Keele B.F., Shaw G.M., et al. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274 10.1371/journal.ppat.1000274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes B.F. 2015. New approaches to HIV vaccine development. Curr. Opin. Immunol. 35:39–47. 10.1016/j.coi.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn M.R., Boutwell C.L., Charlebois P., Lennon N.J., Power K.A., Macalalad A.R., Berlin A.M., Malboeuf C.M., Ryan E.M., Gnerre S., et al. 2012. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 8:e1002529 10.1371/journal.ppat.1002529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishwaran H., and Kogalur U.B.. 2010. Consistency of random survival forests. Stat. Probab. Lett. 80:1056–1064. 10.1016/j.spl.2010.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishwaran H., Kogalur U.B., Blackstone E.H., and Lauer M.S.. 2008. Random Survival Forests. Ann. Appl. Stat. 2:841–860. 10.1214/08-AOAS169 [DOI] [Google Scholar]
- Kawashima Y., Pfafferott K., Frater J., Matthews P., Payne R., Addo M., Gatanaga H., Fujiwara M., Hachiya A., Koizumi H., et al. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 458:641–645. 10.1038/nature07746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher A.D., Long C., Holmes E.C., Allen R.L., Wilson J., Conlon C., Workman C., Shaunak S., Olson K., Goulder P., et al. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375–386. 10.1084/jem.193.3.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf M.C., Allen S., Zulu I., Kancheya N., Stephenson R., Brill I., Tichacek A., Haworth A., and Chomba E.; Rwanda Zambia HIV Research Group . 2008. Enrollment and retention of HIV discordant couples in Lusaka, Zambia. J. Acquir. Immune Defic. Syndr. 47:116–125. 10.1097/QAI.0b013e31815d2f3f [DOI] [PubMed] [Google Scholar]
- Kulkarni V., Valentin A., Rosati M., Alicea C., Singh A.K., Jalah R., Broderick K.E., Sardesai N.Y., Le Gall S., Mothe B., et al. 2014. Altered response hierarchy and increased T-cell breadth upon HIV-1 conserved element DNA vaccination in macaques. PLoS One. 9:e86254 10.1371/journal.pone.0086254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A.J., Pfafferott K.J., Chetty P., Draenert R., Addo M.M., Feeney M., Tang Y., Holmes E.C., Allen T., Prado J.G., et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289. 10.1038/nm992 [DOI] [PubMed] [Google Scholar]
- Leslie A., Kavanagh D., Honeyborne I., Pfafferott K., Edwards C., Pillay T., Hilton L., Thobakgale C., Ramduth D., Draenert R., et al. 2005. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J. Exp. Med. 201:891–902. 10.1084/jem.20041455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Létourneau S., Im E.J., Mashishi T., Brereton C., Bridgeman A., Yang H., Dorrell L., Dong T., Korber B., McMichael A.J., and Hanke T.. 2007. Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One. 2:e984 10.1371/journal.pone.0000984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Zuo T., Hora B., Song H., Kong W., Yu X., Goonetilleke N., Bhattacharya T., Perelson A.S., Haynes B.F., et al. 2014a Preexisting compensatory amino acids compromise fitness costs of a HIV-1 T cell escape mutation. Retrovirology. 11:101 10.1186/s12977-014-0101-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Rao U., McClure J., Konopa P., Manocheewa S., Kim M., Chen L., Troyer R.M., Tebit D.M., Holte S., et al. 2014b Impact of mutations in highly conserved amino acids of the HIV-1 Gag-p24 and Env-gp120 proteins on viral replication in different genetic backgrounds. PLoS One. 9:e94240 10.1371/journal.pone.0094240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llano A., Olvera A.W.A., Silva-Arrieta S., and Brander C.. 2013. Best-Characterized HIV-1 CTL Epitopes: The 2013 Update. In HIV Molecular Immunology 2013. 2013. Yusim B.T.M.K.K., Brander C., Barouch D., de Boer R., Haynes B.F., Koup R., Moore J.P., Walker B.D., and Watkins D.I., editors. Los Alamos National Laboratory, Los Alamos, New Mexico: pp. 3–25. [Google Scholar]
- Matthews P.C., Prendergast A., Leslie A., Crawford H., Payne R., Rousseau C., Rolland M., Honeyborne I., Carlson J., Kadie C., et al. 2008. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J. Virol. 82:8548–8559. 10.1128/JVI.00580-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna S.L., Muyinda G.K., Roth D., Mwali M., Ng’andu N., Myrick A., Luo C., Priddy F.H., Hall V.M., von Lieven A.A., et al. 1997. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS. 11(Suppl 1):S103–S110. [PubMed] [Google Scholar]
- McMichael A.J., Borrow P., Tomaras G.D., Goonetilleke N., and Haynes B.F.. 2010. The immune response during acute HIV-1 infection: clues for vaccine development. Nat. Rev. Immunol. 10:11–23. 10.1038/nri2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueles S.A., Sabbaghian M.S., Shupert W.L., Bettinotti M.P., Marincola F.M., Martino L., Hallahan C.W., Selig S.M., Schwartz D., Sullivan J., and Connors M.. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA. 97:2709–2714. 10.1073/pnas.050567397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C.B., John M., James I.R., Christiansen F.T., Witt C.S., and Mallal S.A.. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 296:1439–1443. 10.1126/science.1069660 [DOI] [PubMed] [Google Scholar]
- Mothe B., Hu X., Llano A., Rosati M., Olvera A., Kulkarni V., Valentin A., Alicea C., Pilkington G.R., Sardesai N.Y., et al. 2015. A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques. J. Transl. Med. 13:60 10.1186/s12967-015-0392-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince J.L., Claiborne D.T., Carlson J.M., Schaefer M., Yu T., Lahki S., Prentice H.A., Yue L., Vishwanathan S.A., Kilembe W., et al. 2012. Role of transmitted Gag CTL polymorphisms in defining replicative capacity and early HIV-1 pathogenesis. PLoS Pathog. 8:e1003041 10.1371/journal.ppat.1003041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts H.E., Hurst J., Robinson N., Brown H., Flanagan P., Vass L., Fidler S., Weber J., Babiker A., Phillips R.E., et al. SPARTAC trial investigators . 2015. Structured observations reveal slow HIV-1 CTL escape. PLoS Genet. 11:e1004914 10.1371/journal.pgen.1004914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidewind A., Brumme Z.L., Brumme C.J., Power K.A., Reyor L.L., O’Sullivan K., Gladden A., Hempel U., Kuntzen T., Wang Y.E., et al. 2009. Transmission and long-term stability of compensated CD8 escape mutations. J. Virol. 83:3993–3997. 10.1128/JVI.01108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid A., Olvera A., Anmole G., Kuang X.T., Cotton L.A., Plana M., Brander C., Brockman M.A., and Brumme Z.L.. 2015. Consequences of HLA-B*13-associated escape mutations on HIV-1 replication and Nef function. J. Virol. 89:11557–11571. 10.1128/JVI.01955-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Pavlicek J.W., Cai F., Bhattacharya T., Li H., Iyer S.S., Bar K.J., Decker J.M., Goonetilleke N., Liu M.K., et al. 2012. Impact of immune escape mutations on HIV-1 fitness in the context of the cognate transmitted/founder genome. Retrovirology. 9:89 10.1186/1742-4690-9-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Tang S., Lobashevsky E., Myracle A.D., Fideli U., Aldrovandi G., Allen S., Musonda R., Kaslow R.A., and Zambia U.A.B.H.I.V.R.P.. Zambia-UAB HIV Research Project . 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276–8284. 10.1128/JVI.76.16.8276-8284.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thobakgale C.F., Prendergast A., Crawford H., Mkhwanazi N., Ramduth D., Reddy S., Molina C., Mncube Z., Leslie A., Prado J., et al. 2009. Impact of HLA in mother and child on disease progression of pediatric human immunodeficiency virus type 1 infection. J. Virol. 83:10234–10244. 10.1128/JVI.00921-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trask S.A., Derdeyn C.A., Fideli U., Chen Y., Meleth S., Kasolo F., Musonda R., Hunter E., Gao F., Allen S., and Hahn B.H.. 2002. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J. Virol. 76:397–405. 10.1128/JVI.76.1.397-405.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyer R.M., McNevin J., Liu Y., Zhang S.C., Krizan R.W., Abraha A., Tebit D.M., Zhao H., Avila S., Lobritz M.A., et al. 2009. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 5:e1000365 10.1371/journal.ppat.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright J.K., Naidoo V.L., Brumme Z.L., Prince J.L., Claiborne D.T., Goulder P.J., Brockman M.A., Hunter E., and Ndung’u T.. 2012. Impact of HLA-B*81-associated mutations in HIV-1 Gag on viral replication capacity. J. Virol. 86:3193–3199. 10.1128/JVI.06682-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue L., Prentice H.A., Farmer P., Song W., He D., Lakhi S., Goepfert P., Gilmour J., Allen S., Tang J., et al. 2013. Cumulative impact of host and viral factors on HIV-1 viral-load control during early infection. J. Virol. 87:708–715. 10.1128/JVI.02118-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.