Wen et al. show that there are gut microbial antigens sharing significant homology with the host pancreatic protein peptide IGRP and that can drive self-reactive T cell activation and accelerate diabetes in NOD mice.
Abstract
Both animal model and human studies indicate that commensal bacteria may modify type 1 diabetes (T1D) development. However, the underlying mechanisms by which gut microbes could trigger or protect from diabetes are not fully understood, especially the interaction of commensal bacteria with pathogenic CD8 T cells. In this study, using islet-specific glucose-6-phosphatase catalytic subunit–related protein (IGRP)–reactive CD8 T cell receptor NY8.3 transgenic nonobese diabetic mice, we demonstrated that MyD88 strongly modulates CD8+ T cell–mediated T1D development via the gut microbiota. Some microbial protein peptides share significant homology with IGRP. Both the microbial peptide mimic of Fusobacteria and the bacteria directly activate IGRP-specific NY8.3 T cells and promote diabetes development. Thus, we provide evidence of molecular mimicry between microbial antigens and an islet autoantigen and a novel mechanism by which the diabetogenicity of CD8+ T cells can be regulated by innate immunity and the gut microbiota.
INTRODUCTION
Type 1 diabetes (T1D) is an autoimmune disease characterized by T cell–mediated destruction of insulin-producing pancreatic β cells (Bluestone et al., 2010). Interacting genetic and environmental factors eventually lead to the loss of functional β cell mass and hyperglycemia (Bluestone et al., 2010; Polychronakos and Li, 2011; Nielsen et al., 2014). The discordant diabetes incidence in monozygotic twins (<50% developed T1D) strongly suggests that nongenetically determined factors regulate T1D development (Kaprio et al., 1992). Recent studies in mouse models and humans have shown that gut microbiota play an important role in disease development.
We previously showed that commensal bacteria modified diabetes development in nonobese diabetic (NOD) mice via myeloid differentiation primary response 88 (MyD88). Specific pathogen-free MyD88-deficient (MyD88−/−) NOD mice were protected from T1D development, whereas germ-free MyD88−/−NOD mice developed normal diabetes (Wen et al., 2008). Gender bias in T1D in NOD mice is influenced by microbiota (Markle et al., 2013; Yurkovetskiy et al., 2013). Studies in humans also indicate that the gut microbiome plays an important role in T1D development. Gut microbial communities in high-risk children are characterized as less diverse, distinct from those of healthy controls (Brown et al., 2011; Kostic et al., 2015). Furthermore, a low abundance of lactate- and butyrate-producing bacteria, reduced Bifidobacterium species, and increased bacteria of the Bacteroides genus were found in islet autoantibody–positive children (de Goffau et al., 2013). Individuals with islet autoantibodies, sero-negative first-degree relatives, and new-onset patients had different abundances of Lactobacillus and Staphylococcus (both Firmicutes) compared with healthy control subjects (Alkanani et al., 2015). Thus, altered gut microbiota are associated with β cell autoimmunity in humans at risk of developing T1D, underscoring a role for gut microbiota in the pathogenesis of antiislet cell autoimmunity and T1D development.
CD8+ T cells play an important role in the development of T1D in both NOD mice and humans (Wong et al., 1999, 2009; Liblau et al., 2002; Lieberman et al., 2003; Tsai et al., 2008). CD8+ T cells predominate in islet infiltrates in patients with T1D (Willcox et al., 2009), and islet autoantigen–specific CD8+ T cell clones were generated from the peripheral blood of T1D patients (Peakman et al., 1994; Unger et al., 2007; Skowera et al., 2008). Other studies have demonstrated heightened expression of MHC class I and CD8+ T cells in the islets of patients with T1D (Foulis et al., 1987; Coppieters et al., 2012; Pugliese et al., 2014; Rodriguez-Calvo et al., 2015). However, there is a substantial gap in our knowledge of how islet autoimmunity mediated by CD8+ T cells is shaped by innate immunity and the gut microbiota. To investigate the link of innate immunity and gut microbiota to CD8+ T cells in T1D development, we generated MyD88−/−NOD mice carrying diabetogenic CD8+ TCR NY8.3, specific for islet-specific glucose-6-phosphatase catalytic subunit–related protein (IGRP; Verdaguer et al., 1997; Lieberman et al., 2003). To our surprise, the mice developed highly accelerated diabetes with altered gut microbiota enriched in Fusobacteria that express a magnesium transporter (Mgt) encompassing a microbial peptide mimic of IGRP. The mimic peptide directly activated IGRP-specific CD8+ T cells and, importantly, induced robust diabetes in vivo. Supercolonization of the mice with this strain of bacteria accelerated diabetes in NY8.3NOD mice. Lastly, increased fecal Fusobacteria were also associated with diabetes progression in NOD mice. Therefore, our study provides direct evidence that molecular mimicry by microbial peptides of islet autoantigen contributes to T1D.
RESULTS
Accelerated diabetes in MyD88−/−NY8.3NOD mice is MyD88 dependent, IGRP reactive, and TCR specific
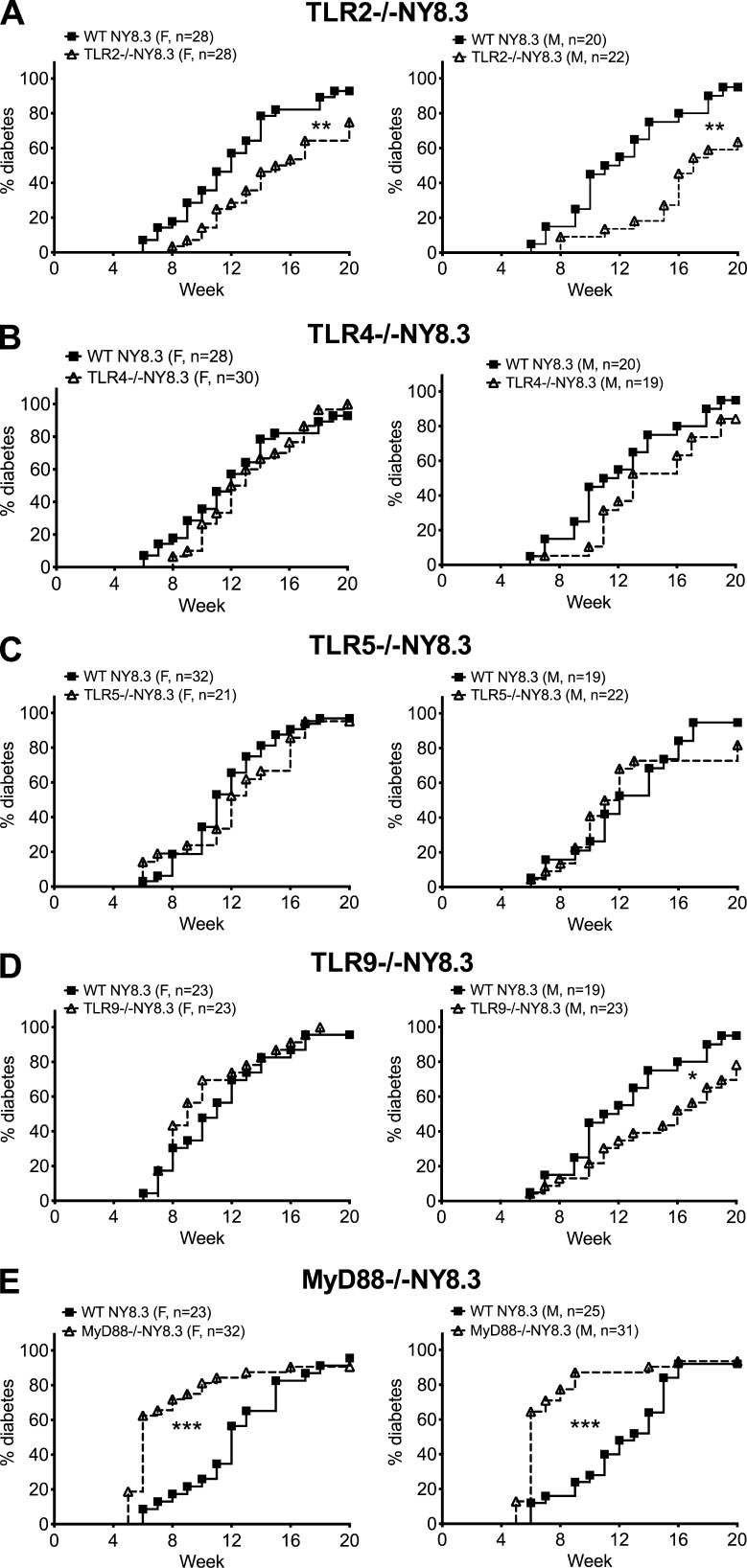
To better understand the interplay among innate immunity, gut microbes, and diabetogenic CD8+ T cells, we generated several lines of TLR-deficient (TLR−/−) and MyD88−/− NY8.3NOD mice. TLR2−/− and male TLR9−/−NY8.3 mice had significantly delayed diabetes onset (Fig. 1, A and D), whereas TLR4−/−, TLR5−/−, and female TLR9−/−NY8.3 mice were not affected by the loss of these TLRs (Fig. 1, B–D). In contrast, MyD88−/−NY8.3 mice (both sexes) developed markedly accelerated diabetes (Fig. 1 E). This also contrasts with the protected phenotype in polyclonal MyD88−/−NOD mice in specific pathogen-free conditions (Wen et al., 2008). Interestingly, there was no gender difference in diabetes incidence in either WT NY8.3NOD or MyD88−/−NY8.3NOD mice (not depicted). Collectively, our data suggest that the accelerated diabetes in MyD88−/−NY8.3NOD mice is MyD88 dependent.
Figure 1.
MyD88 deficiency has different effect on diabetes development. (A–E) Individual TLR- or MyD88−/−NY8.3NOD mice were generated by breeding different TLR−/− or MyD88−/−NOD mice with NY8.3 NOD mice. Diabetes development was observed, and data were pooled from at least five independent experiments. (A) TLR2−/−NY8.3. (B) TLR4−/−NY8.3. (C) TLR5−/−NY8.3. (D) TLR9−/−NY8.3. (E) MyD88−/−NY8.3. Wilcoxon test for survival was used for analysis of diabetes incidence. *, P < 0.05; **, P < 0.01; ***, P < 0.001. F, female. M, male.
CD8+ T cells are more activated in MyD88−/−NY8.3 mice
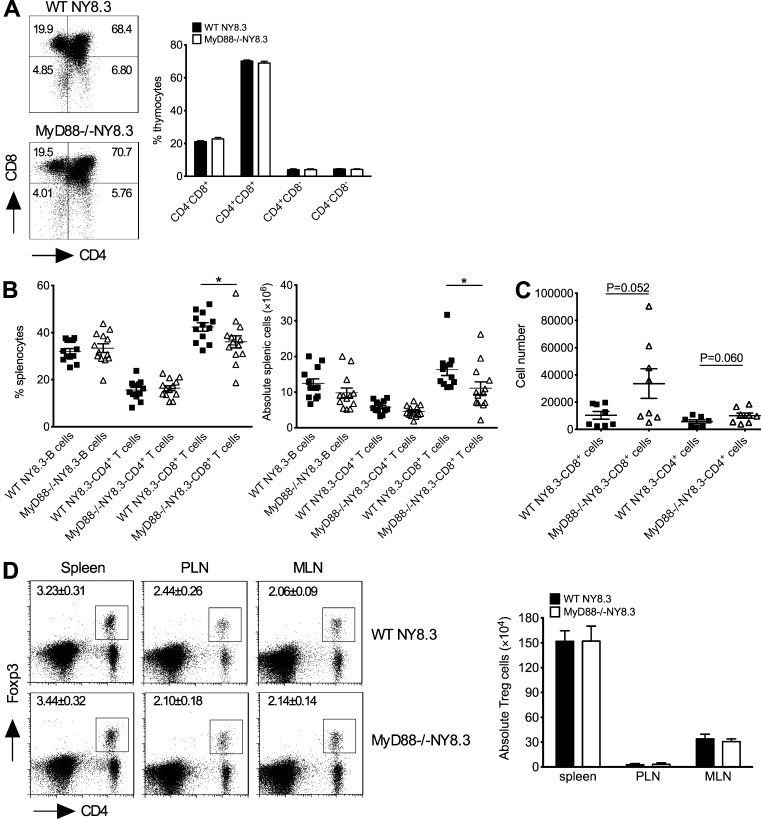
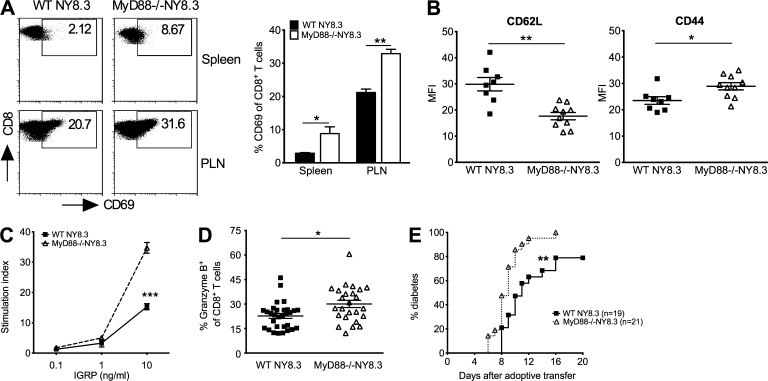
We next examined the phenotype and function of NY8.3 CD8+ T cells. MyD88 deficiency does not affect thymic selection of NY8.3 T cells (Fig. 2 A); however, the number of splenic CD8+ T cells was lower in MyD88−/−NY8.3NOD mice compared with the NY8.3NOD mice (Fig. 2 B). There were no differences in the number of CD8+ T cells in pancreatic LN or mesenteric LN (MLN; not depicted). In contrast, we found more CD8+ T cells in islet infiltrates of MyD88−/−NY8.3NOD mice compared with WT NY8.3NOD mice (Fig. 2 C). Furthermore, there was no difference in Foxp3+CD4+ T regulatory cells in the lymphoid tissues examined (Fig. 2 D), but there were more activated (CD69+) and memory/effector (CD62Llow/CD44high) NY8.3 CD8+ T cells in MyD88−/− hosts (Fig. 3, A and B). CD8+ T cells from MyD88−/−NY8.3NOD mice also showed stronger responses to their native autoantigen, IGRP206–214 peptide, and expressed more granzyme B upon anti-CD3 stimulation in vitro, compared with WT mice (Fig. 3, C and D). Importantly, they induced more aggressive diabetes in vivo (Fig. 3 E). These data demonstrated that MyD88 deficiency resulted in stronger activation of CD8+ T cells in NY8.3NOD mice.
Figure 2.
MyD88 deficiency has no effect on T cell development in thymus and regulatory T cell differentiation. (A) Thymocytes from 6–7-wk-old MyD88−/−NY8.3NOD and WT NY8.3NOD mice (sex matched) were stained with fluorochrome-conjugated anti-TCRβ, anti-CD4, and anti-CD8 antibodies. n = 8–10 mice/group from two independent experiments. (B) CD8+ T cells in the spleen of MyD88−/−NY8.3NOD mice. The experiment was performed in three independent experiments, and two-tailed Mann-Whitney test was used for statistical analysis. *, P < 0.05. (C) The absolute number of infiltrated CD4+ and CD8+ T cells in the islets of MyD88−/−NY8.3NOD mice after gating with TCRβ-positive cells is shown. The data were from two independent experiments. (D) Natural regulatory T cells in MyD88−/−NY8.3NOD mice. n ≥ 6 mice/group/experiment from two independent experiments. PLN, pancreatic LN. Data are mean ± SEM.
Figure 3.
MyD88 deficiency promotes highly pathogenic NY8.3 CD8+ T cells. (A) CD69 expressing CD8+ T cells in MyD88−/−NY8.3 mice. n = 4 mice/group/experiment from more than three experiments. PLN, pancreatic LN. (B) Mean fluorescence intensity (MFI) of CD62L and CD44 of gated splenic CD8+ T cells from MyD88−/−NY8.3 and WT NY8.3 mice was determined by flow cytometric analysis from three independent experiments. (C) Splenocytes from MyD88−/−NY8.3 or WT NY8.3 mice were tested upon antigen stimulation by [3H]thymidine incorporation assay. n = 3 mice/group/experiment from more than three experiments. Two-way ANOVA was used for the comparison with Bonferroni correction. (D) Granzyme B–expressing CD8+ T cells in the spleen of MyD88−/−NY8.3NOD mice stimulated by anti-CD3 and anti-CD28 followed by ICC staining. Data are from five independent experiments. (E) Diabetes incidence after adoptive transfer. 106 purified IGRP-stimulated splenic CD8+ T cells were injected i.v. into recipient NOD mice in two independent experiments. Wilcoxon test for survival was used for analysis. (A, B, and D) Two-tailed Student’s t test was used for analysis. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
MyD88 deficiency changes gut microbiota composition in NY8.3 mice and modifies diabetes development
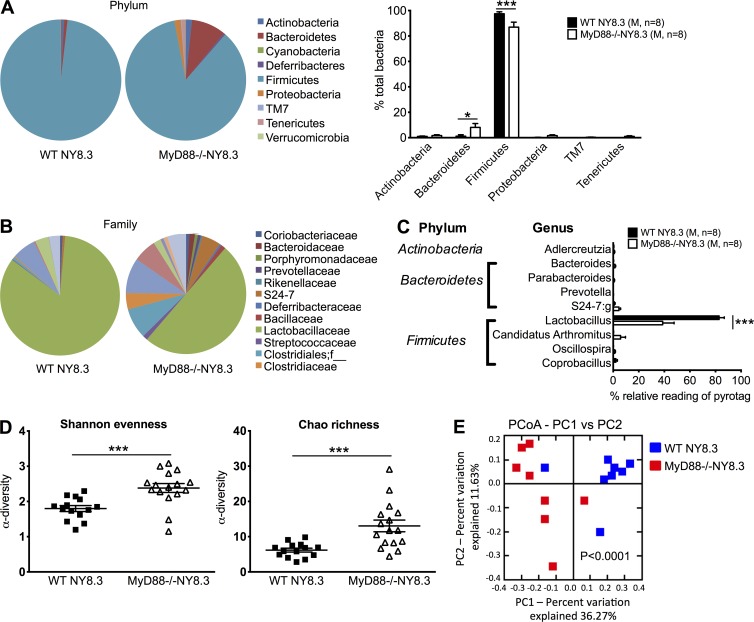
We tested whether MyD88 deficiency also affected the composition of gut microbiota in MyD88−/−NY8.3NOD mice as in polyclonal NOD mice (Wen et al., 2008) by 16S rRNA pyrosequencing of the fecal samples from MyD88−/− and WT NY8.3NOD mice. MyD88 deficiency indeed altered gut microbiota composition (Fig. 4, A–C) with a much greater α and β diversity in the bacterial community of MyD88−/−NY8.3NOD mice compared with WT NY8.3NOD mice (Fig. 4, D and E). Actinobacteria, Bacteroidetes, and Proteobacteria were increased, and Firmicutes was reduced at the phylum level, whereas Lactobacillus was significantly less abundant at the genus level in MyD88−/−NY8.3NOD mice compared with WT NY8.3NOD mice.
Figure 4.
MyD88 deficiency results in distinct gut microbiota in NY8.3NOD mice. (A–C) Fecal samples from WT NY8.3NOD and MyD88−/−NY8.3NOD mice were used for taxonomic analysis by 16S rRNA sequencing. The taxonomic compositions of gut bacteria were shown in phylum (left, pies; right, bars; A) family (B), and genus (C). (A and C) Two-way ANOVA with Bonferroni correction was used. (D) α-Diversity of gut microbiota of WT NY8.3NOD and MyD88−/−NY8.3NOD mice was shown by Shannon evenness and Chao richness. Two-tailed Student’s t test was used. (E) β-Diversity, shown by principal component analysis (PCoA; unweighted) of taxonomic families of gut microbiota. ANOSIM was used for statistical analysis. Two independent sequencing experiments with different cohorts were performed, and one representative (A, B, C, and E) and the pooled data (D) are shown. Data are mean ± SEM. *, P < 0.05; ***, P < 0.001. M, male.
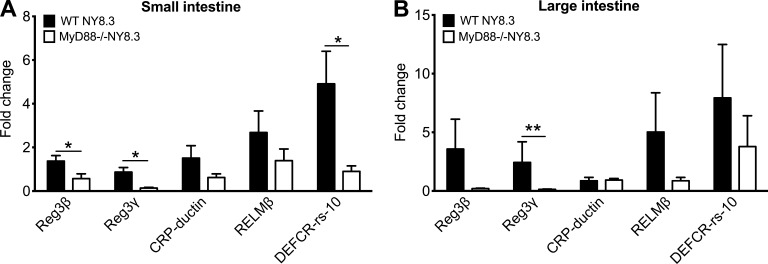
Next, we quantified the levels of antimicrobial peptides, which affect gut bacterial growth, in the intestines. Expression of Reg3β, Reg3γ, RELMβ, and C-reactive protein–ductin were reduced in MyD88−/−NY8.3NOD mice (Fig. 5, A and B), which may have contributed to the altered gut microbiome in MyD88−/−NY8.3NOD mice, affecting diabetes development.
Figure 5.
Reduced intestinal antimicrobial peptide expression in NY8.3NOD mice in the absence of MyD88. (A and B) Gene expression for the intestinal antimicrobial peptides. Fragments (∼3 cm) of the small intestines from ileum and large intestine adjacent to the cecum were harvested from 6–7-wk-old sex-matched WT NY8.3NOD and MyD88−/−NY8.3NOD mice. After intensive washing with sterile PBS, RNA was extracted from the fragments using TRIzol, followed by reverse transcription. qPCR was performed to determine the mRNA levels of antimicrobial peptides, which were normalized to the housekeeping gene GAPDH. (A) Small intestine. (B) Large intestine. Five to eight mice/group from two independent experiments are shown. One-tailed Mann-Whitney test was used for statistical analysis. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01. CRP, C-reactive protein.
Manipulation of gut microbiota alters diabetes development in MyD88−/−NY8.3NOD mice
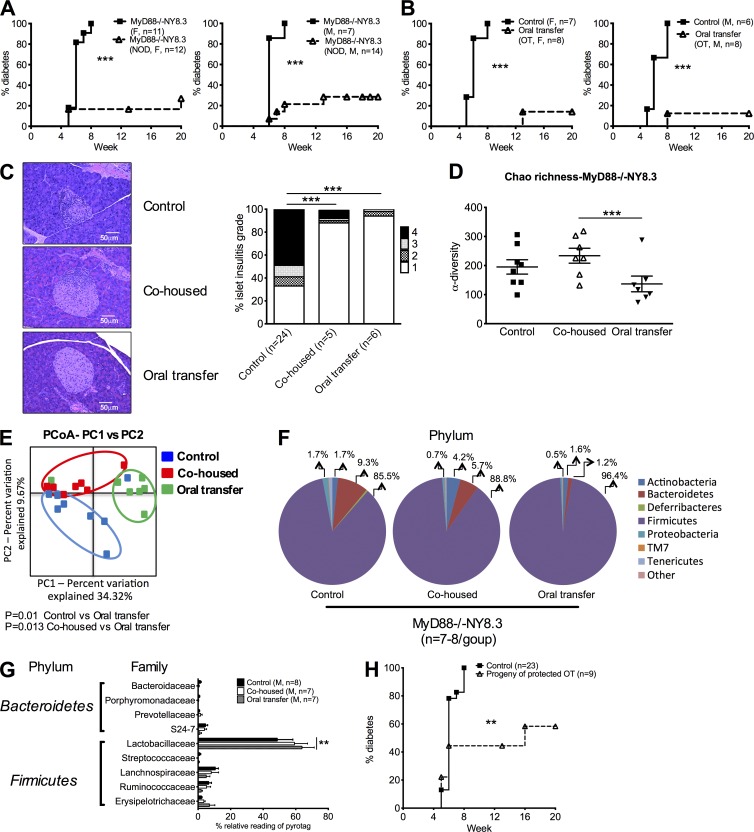
To test whether the altered gut microbiota in MyD88−/−NY8.3NOD mice was responsible for the accelerated diabetes, we cohoused the MyD88−/−NY8.3NOD mice with WT NOD mice. Remarkably, ∼80% of MyD88−/−NY8.3NOD mice were completely protected from diabetes development when cohoused with age- and sex-matched WT NOD mice (Fig. 6 A). This cohousing neither changed the activation of CD4+ or CD8+ T cells nor affected the insulitis in WT NOD mice (not depicted), and there was no modulation of the diabetes development in WT NOD mice (not depicted). This implies that the WT microbiota had a dominant effect on the MyD88−/−NY8.3NOD mice rather than vice versa. As a control, we also cohoused WT NY8.3NOD mice with WT NOD mice, which did not affect diabetes incidence in WT NY8.3NOD (not depicted). To test whether the protected phenotype was horizontally transferable to a second host, we performed fecal transfer by orally gavaging the fecal suspension from diabetes-free cohoused MyD88−/−NY8.3NOD mice to another set of naïve MyD88−/−NY8.3NOD mice. Interestingly, oral transfer (OT) of gut microbes from diabetes-protected MyD88−/−NY8.3NOD donors conferred even greater protection against diabetes, accompanied by a marked reduction in insulitis (Fig. 6, B and C) and an alteration of gut microbiota α-diversity, β-diversity, or composition ascertained by 16S rRNA pyrosequencing (Fig. 6, D–G). Interestingly, in the mice that were protected from diabetes by cohousing or fecal transfer, there was a clear reduction in Bacteroidetes and an increase of Firmicutes at the phylum level of the microbiota community in MyD88−/−NY8.3 mice compared with the control counterparts (Fig. 6 F). To investigate whether the protected phenotype was vertically transferable to the next generation, we used horizontally protected MyD88−/−NY8.3NOD mice as breeders and observed diabetes development in their progeny. Strikingly, the disease protection was indeed vertically transferable (Fig. 6 H). Collectively, our data suggest that MyD88 deficiency in NY8.3NOD mice results in dysbiosis of the gut microbiota, leading to accelerated diabetes. However, cohousing completely reversed the diabetic phenotype, and importantly, diabetes protection was transferable to new hosts and to the next generation.
Figure 6.
Manipulation of gut microbiota protects from diabetes development in MyD88−/−NY8.3 mice. (A) Diabetes incidence in MyD88−/−NY8.3NOD mice after 4-wk-old MyD88−/−NY8.3NOD mice were cohoused with an equal number and age- and sex-matched NOD mice. F, female; M, male. (B) Diabetes development of MyD88−/−NY8.3NOD mice after fecal OT. The fresh gut bacteria (∼108–109 culturable bacteria/mouse) from diabetes-free cohoused MyD88−/−NY8.3 mice (from A) were fed orally to sex-matched 4-wk-old naive MyD88−/−NY8.3NOD mice. (C) Insulitis of MyD88−/−NY8.3NOD mice after cohousing with WT NOD mice or OT (from A and B). More than120 islets from five to six treated mice/group were examined. Representative pancreatic sections are shown on the left, and the summary of the insulitis scores is shown on the right. χ2 test was used for analysis. (D) α-Diversity of gut microbiota is shown by Chao richness. Fecal samples from the control, cohoused, and OT MyD88−/−NY8.3NOD mice were used for taxonomic analysis by 16S rRNA sequencing. One- or two-tailed Mann-Whitney test was used for analysis. Data are shown as individual samples. (E) β-Diversity is shown. Assessment of the differences in the bacterial composition of the gut microbiota in the mice that were treated either by cohousing or OT (red or green symbols) compared with the control mice (blue symbols) is shown by principal component analysis (PCoA). ANOSIM was used for statistical analysis. (F and G) Comparison of the abundance of gut bacteria in the control, cohoused, and OT MyD88−/−NY8.3NOD mice is shown by phylum (F) and family (G). Two-way ANOVA was performed with Bonferroni correction. (H) Diabetes incidence in the progeny of MyD88−/−NY8.3NOD mice that were protected from diabetes after fecal OT (shown in B). (A, B, and G) The data were pooled from at least two independent experiments. (D–G) The results are from one of two independent experiments. Data are shown as mean ± SEM. **, P < 0.01; ***, P < 0.001.
Microbial mimic peptides can stimulate diabetogenic CD8+ T cells in vitro and induce diabetes in vivo
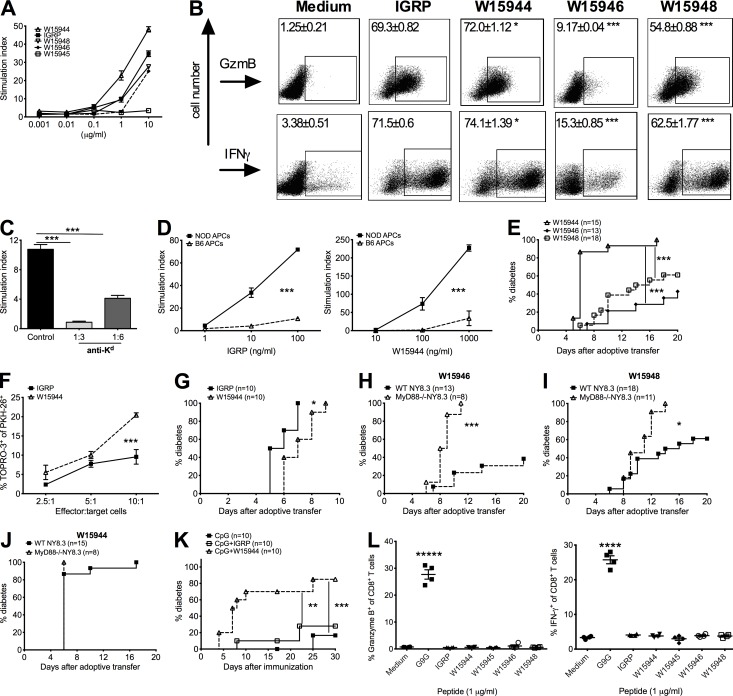
To probe the mechanism by which dysbiosis of gut commensals, in the absence of MyD88, leads to highly accelerated diabetes development in NY8.3NOD mice, we hypothesized that gut bacterial products may activate NY8.3 CD8+ T cells. We compared the IGRP206–214 peptide sequence against bacterial protein sequences in the nonredundant protein sequence database and found several peptides from oral or gut bacteria that share strong homology with IGRP206–214, the native autoantigen of NY8.3 CD8+ T cells (Table 1). Three of these IGRP mimic peptides, designated as W15944, W15946, and W15948, activated NY8.3 CD8+ T cells with a comparable potency with IGRP native peptide using: (a) proliferation of NY8.3 CD8+ T cells (Fig. 7 A), (b) IFN-γ production, and (c) induction of granzyme B (Fig. 7 B). Furthermore, all the responses were restricted by MHC class I–Kd (Fig. 7, C and D). Importantly, the microbial mimic peptides, which activated NY8.3 CD8+ T cells, induced these NY8.3 CD8+ T cells to rapidly transfer diabetes in vivo (Fig. 7 E). The peptide mimic W15945 was not stimulated NY8.3 CD8+ T cells in vitro (Fig. 7 A) and did not induce diabetes in vivo (not depicted). Comparatively, W15944 stimulated the NY8.3 T cells most strongly both in vitro and in vivo (Fig. 7, A, B, and E). When compared with IGRP206–214, W15944 induced stronger CTL function of NY8.3 CD8+ T cells in vitro and similar diabetogenicity in vivo (Fig. 7, F and G). W15944-, W15946-, and W15948-activated CD8+ T cells from MyD88−/−NY8.3NOD mice all induced significantly higher diabetes incidence in irradiated young NOD mice compared with their WT counterparts that were activated in parallel (Fig. 7, H–J). 100% of NOD recipients developed diabetes by day 6 after adoptive transfer of W15944-activated MyD88−/−NY8.3 CD8+ T cells (Fig. 7 J). Furthermore, immunization with W15944 induced significantly accelerated diabetes in WT NY8.3NOD mice compared with IGRP206–214 or CpG alone (Fig. 7 K). To further test that the microbial peptides specifically stimulated NY8.3 CD8+ T cells, we stimulated insulin-reactive G9 CD8+ T cells with the mimic peptides. As expected, all the microbial peptides tested failed to activate G9 CD8+ T cells that are specific for insulin B15-23 (Wong et al., 1996, 2009), suggesting the specificity of the microbial peptides to IGRP (Fig. 7 L). Collectively, our data strongly suggest that microbial antigen mimics can act as agonists that not only activate islet antigen–specific pathogenic CD8+ T cells in vitro, but also induce robust diabetes in vivo.
Table 1. The origin and sequence of the peptides used in the study.
| Name of the peptides | Protein name | Origin | Position | Peptide sequence |
|---|---|---|---|---|
| IGRP | IGRP | Mouse | 206–214 | VYLKTNVFL |
| W15944 | Mgt | L. goodfellowii (gram negative, mainly oral cavity) | 267–275 | TYLKTNVFT |
| W15945 | glycosyl transferase, group 2 | uncultured Flavobacteriia bacterium (gram negative, commensal) | 303–311 | FYLKTSVFL |
| W15946 | hypothetical protein IEM_00289 | Bacillus cereus (gram positive, gut flora) | 252–260 | VYLKVNVFK |
| W15948 | NAD synthetase | Enterobacter mori LMG 25706 (gram negative) | 32–40 | SYLKTNAFL |
Bold letters represent the amino acids different from those in the same positions in the control IGRP peptide.
Figure 7.
Microbial peptide mimics can activate NY8.3 CD8+ T cells in vitro and induce diabetes in vivo. (A) NY8.3 CD8+ T cell response to microbial peptide mimics shown in proliferation assay by [3H]thymidine incorporation. The experiment was repeated at least four times. (B) Representative FACS plots for expression of granzyme B (GzmB) and IFN-γ in NY8.3 CD8+ T cells stimulated with mimic peptides or IGRP206–214 peptide (1 µg/ml). Student’s t test was used for statistical analysis to compare individual peptide mimics with IGRP206–214 peptide. Data are representative of four independent experiments. n ≥ 10. (C) MHC class I blocking assay. Two-tailed Student’s t test was used for statistical analysis. n = 3/group/experiment, and the experiment was performed three times. (D) MHC restriction assay. n = 3/group/experiment, and the experiment was performed twice. Two-way ANOVA with Bonferroni correction was used for analysis. (E) Diabetes development induced by NY8.3 CD8+ T cells activated with microbial peptide mimics from two independent experiments. Log-rank test for survival was used. (F) CTL activity of CD8+ T cells induced by W15944 peptide. Two-way ANOVA with Bonferroni correction was used for analysis. n = 4/group/experiment, and the experiment was performed three times. (G) Pathogenic effect of W15944-stimulated NY8.3 CD8+ T cells. Log-rank test for survival was used for statistical analysis. The results are from two independent experiments (n = 5/group/experiment). (H–J) MyD88−/−NY8.3 and WT NY8.3NOD CD8 T cells activated by microbial peptide mimics induced diabetes by adoptive transfer into irradiated NOD mice. The experiments were repeated twice. (H) W15946. (I) W15948. (J) W15944. Log-rank test for survival was used for analysis. (K) Incidence of diabetes in WT NY83.NOD mice after immunization with mimic peptide from L. goodfellowii. The data were pooled from two independent experiments. Log-rank test for survival was used for analysis. (L) Response of insulin-specific G9 T cells to the microbial peptides. (Left) Granzyme B. (Right) IFN-γ. n = 2/group/experiment, and the experiment was performed twice. One-way ANOVA was used for statistical analysis. Data are mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; *****, P < 0.00001.
Increased abundance of Fusobacteria and Leptotrichia goodfellowii is associated with accelerated diabetes
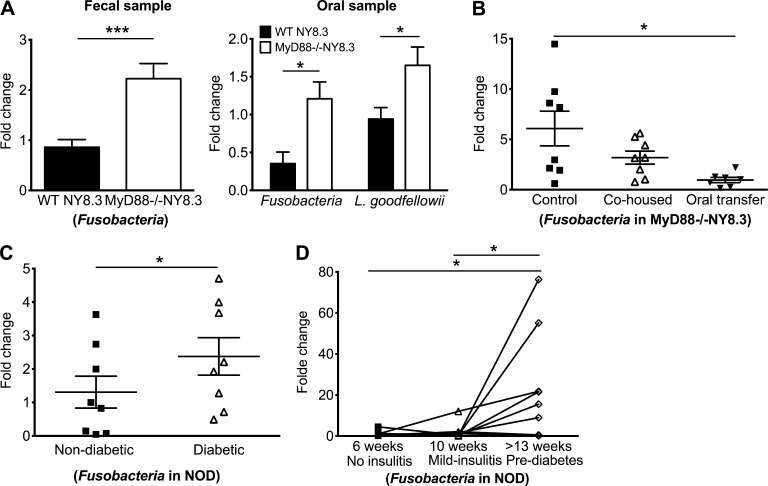
The microbial peptide mimic W15944 derives from an Mgt (GenePept accession no. WP_006806773) of L. goodfellowii, a member of the phylum Fusobacteria (gram-negative anaerobe), a human and NOD mouse oral commensal (not depicted). Fusobacteria are associated with inflammatory disorders including skin ulcers (Legaria et al., 2005) and inflammatory bowel diseases (Ohkusa et al., 2003; Strauss et al., 2011). We found that the Fusobacteria represent <0.1% of sequencing reads of fecal samples of MyD88−/− and WT NY8.3NOD mice. To quantify the presence of Fusobacteria or L. goodfellowii, we performed quantitative PCR (qPCR) with specific primers using bacterial DNA from fecal and oral samples of prediabetic MyD88−/−NY8.3NOD and sex- and age-matched WT NY8.3NOD mice. MyD88−/−NY8.3NOD mice harbored significantly more Fusobacteria in both fecal and oral samples and more L. goodfellowii in oral samples (Fig. 8 A). Interestingly, a significant reduction of the abundance of Fusobacteria was observed in the fecal samples of the protected cohoused or orally transferred MyD88−/−NY8.3NOD mice compared with control mice, indicating that the reduced abundance of Fusobacteria could be associated with the reduced diabetes incidence in these mice (Fig. 8 B).
Figure 8.
Increased abundance of Fusobacteria is associated with diabetes development. (A) Abundance of Fusobacteria and L. goodfellowii in WT NY8.3NOD and MyD88−/−NY8.3NOD mice. Two-tailed Mann-Whitney test (fecal samples) or two-tailed unpaired Student’s t test with Welch’s correction (oral samples) was used. n = 7–16 mice/group from two independent experiments. Data are mean ± SEM. (B) Abundance of Fusobacteria in fecal samples from the control, cohoused, and OT MyD88−/−NY8.3NOD mice. Repeated measures ANOVA with Tukey’s posthoc test was used for analysis. (C) Abundance of Fusobacteria in diabetic and nondiabetic female NOD mice. Student’s t test was used. (D) Bacterial DNA was extracted from fecal samples of female NOD mice every two weeks from 6 wk old until diabetes onset. Results from three time points are shown. Repeated measures ANOVA with Tukey’s posthoc test was used. (B–D) Data were pooled from at least two independent experiments. *, P < 0.05; ***, P < 0.001.
Next, we investigated whether the abundance of Fusobacteria is associated with diabetes development in WT NOD mice. We found a significantly higher abundance of Fusobacteria in the fecal samples of new-onset diabetic compared with age-matched nondiabetic female NOD mice (Fig. 8 C). Furthermore, we collected serial fecal samples from 6-wk-old female NOD mice until diabetes onset. Of note, we observed a significant increase in the abundance of Fusobacteria in the fecal samples from prediabetic (13–16 wk old) compared with young mice (no insulitis 6 wk or mid-insulitis 10 wk), indicating a possible role of Fusobacteria in diabetes development, as these mice all became diabetic later (Fig. 8 D). Collectively, these data suggest a possible association between the abundance of Fusobacteria and diabetes development.
L. goodfellowii activates NY8.3 CD8+ T cells in vitro and accelerates diabetes in WT NY8.3NOD mice in vivo
As the peptide mimic W15944 exerted pathogenic effects by strongly stimulating NY8.3 CD8+ T cells in vitro and inducing rapid diabetes in vivo, we tested whether APCs can process and present W15944 from L. goodfellowii. First, we stimulated splenocytes from MyD88−/− or WT NY8.3NOD mice with the whole cell lysates of L. goodfellowii. Bacteroides thetaiotaomicron (B. theta), also a gram-negative anaerobe, was used as a control. Bacterial lysates of L. goodfellowii induced significantly higher expression of IFN-γ in MyD88−/−NY8.3NOD CD8+ T cells compared with WT NY8.3NOD CD8+ T cells (Fig. 9 A). This was confirmed using purified BMDCs or enriched APCs from spleens and gut-associated lymphoid tissue (GALT), including MLN and Peyer’s patches (PP), of NOD mice (not depicted). Second, using a cell-free Escherichia coli S30 extraction system to express recombinant Mgt (rMgt) protein, we stimulated NY8.3 CD8+ T cells and induced a significantly larger amount of MIP-1β (early CD8+ T cell activation indicator) production compared with the negative control, E. coli S30 with an empty vector (Fig. 9 B). We also used truncated rMgt protein as an antigen in the stimulation assay. CD8+ T cells from both WT NY8.3 NOD and MyD88−/−NY8.3NOD mice responded strongly to the antigen presented by purified NOD splenic APCs (Fig. 9 C). Third, we tested the capacity of GALT APCs, in vitro pulsed with L. goodfellowii, to process and present the IGRP mimic in the absence of exogenous antigen. L. goodfellowii–pulsed GALT APC induced significantly higher expression of IFN-γ and TNF by NY8.3 CD8+ T cells than those pulsed with B. theta (Fig. 9 D). Fourth, we supercolonized 2–3-mo-old female NOD mice by oral gavage with either L. goodfellowii or B. theta daily for five consecutive days. Splenic or GALT APCs from the L. goodfellowii–gavaged NOD mice readily induced a significantly larger amount of MIP-1β and a higher expression of IFN-γ in NY8.3 CD8+ T cells compared with the splenic or GALT APCs from B. theta–treated mice (Fig. 9, E and F). To further confirm the specific recognition of L. goodfellowii by NY8.3 CD8+ T cells, we tested two more bacterial strains, Bacteroides dorei and Lactobacillus reuteri, in addition to B. theta. All the strains tested, except L. goodfellowii, failed to stimulate NY8.3 CD8+ T cells (Fig. 9 G). Finally, we orally gavaged the young WT NY8.3NOD mice with L. goodfellowii or B. theta to test the diabetogenicity directly. The supercolonization of L. goodfellowii significantly accelerated diabetes development in WT NY8.3NOD mice compared with B. theta–colonized mice or untreated WT NY8.3NOD mice (Fig. 9 H). Collectively, these data demonstrated that L. goodfellowii, as well as the purified Mgt protein, can be processed by splenic or GALT APCs from NOD mice to stimulate the NY8.3 cells. Most importantly, the bacteria can trigger early onset of diabetes.
Figure 9.
L. goodfellowiican activate NY8.3 CD8+ T cells in vitro, and supercolonization accelerates diabetes in naive young WT NY8.3NOD mice in vivo. (A) Expression of IFN-γ (ICC staining) in NY8.3 CD8+ T cells stimulated with the L. goodfellowii lysate. (B) MIP-1β secretion (ELISA) from NY8.3 CD8+ T cells stimulated with the translated rMgt protein. (C) MIP-1β secretion (ELISA) from NY8.3 CD8+ T cells stimulated with the purified truncated rMgt protein. (D) Inflammatory cytokine induction (ICC staining) in NY8.3 CD8+ T cells stimulated by APCs pulsed with heat-killed bacteria lysates. (E) MIP-1β (ELISA) and IFN-γ production (ICC staining) of NY8.3 CD8+ T cells stimulated with the APCs from NOD mice supercolonized with L. goodfellowii or control B. theta. (F) Secreted IFN-γ (ELISA) from NY8.3 CD8+ T cells stimulated with APCs from NOD mice supercolonized with L. goodfellowii or control B. theta. (G) 106 splenocytes from NY8.3 mice were stimulated with different heat-killed bacterial strains. (Left) IFN-γ expression (ICC staining). (Right) IFN-γ secretion (ELISA). One-way ANOVA was used for statistical analysis. n = 2/group/experiment, and the experiment was performed twice. (H) Diabetes incidence in NY8.3NOD mice after supercolonization with L. goodfellowii or control B. theta. Log-rank test for survival was used for analysis. The results are combined from two independent experiments (n = 5–8/group/experiment). (A–F) Two-tailed Student’s t test was used. n = 3–6/group/experiment from three independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
DISCUSSION
In this study, we present evidence that a transporter protein peptide from a strain of Fusobacteria can be processed and presented by gut-associated antigen-presenting cells and stimulate IGRP-reactive CD8 T cells to accelerate diabetes. These bacteria are increased in the gut microbiome in the absence of MyD88. Using IGRP-reactive NY8.3 TCR transgenic mice as a model, MyD88−/−NY8.3NOD mice developed highly accelerated diabetes. The gut microbiota play a critical role in the disease acceleration, confirmed by disease reduction when the gut microbiome are altered via cohousing or fecal transfer from nondiabetes-accelerated mice. Moreover, supercolonization with the live bacteria or immunization with the microbial peptide in NY8.3 mice accelerated diabetes, and importantly, these bacteria also increase in polyclonal NOD mice during diabetes progression and at diabetes onset.
Although there are several studies showing gut commensals influence autoimmunity, none have demonstrated direct influence through stimulation of the autoreactive T cells in diabetes. Molecular mimicry has long been suggested as a pathogenic mechanism for the development of T1D, but to date, although demonstrated in some autoimmune diseases, there has been no convincing direct evidence for T1D. To probe the mechanism, a defined setting is required. We used TCR transgenic mice that develop spontaneous diabetes, allowing us to study the known TCR clonotypes and the possible mimic agonists. The NY8.3 cells are highly promiscuous as demonstrated by the initial difficulty of identifying the autoantigen by the screening of a peptide library (Anderson et al., 1999), and indeed this is the case for many low-affinity autoreactive T cells (Wooldridge et al., 2012). This promiscuity of T cell antigen recognition makes it likely that more than one naturally occurring peptide product can stimulate the T cells, implying that the gut bacteria could be a particularly rich source of potential antigens. Several microbial peptides are highly homologous to islet autoantigen IGRP206–214, the ligand of NY8.3 TCR, and can stimulate the diabetogenic NY8.3 cells. One of these is from the Mgt protein of L. goodfellowii, a strain of Fusobacteria, enriched in the MyD88−/−NY8.3NOD mice. IGRP is a key target for diabetogenic CD8 T cells in both the NOD mouse as well as in humans (Lieberman et al., 2003; Jarchum et al., 2008; Ko et al., 2014). NY8.3-like CD8 T cells increase in NOD mice with increasing avidity for their target antigen along with the progression to diabetes development in unmanipulated NOD mice (Amrani et al., 2000), and this coincides with the increase of fecal Fusobacteria in NOD mice shown in this study. To our knowledge, this is the first direct demonstration of a specific gut bacterium to stimulate a TCR that increases in avidity with time.
Previous studies have shown TCR cross-reactivity between MHC II–binding self-antigen and foreign peptides including microbial products, which could cause autoimmunity by stimulating autoreactive T cells (Birnbaum et al., 2014; Horai et al., 2015; Nelson et al., 2015). Most interestingly, a newly published study used a human CD8+ T cell clone (1E6), which is MHC class I restricted and human preproinsulin reactive, to generate high-resolution structures of the 1E6 TCR bound to seven altered peptide ligands, including a microbe-derived peptide (Cole et al., 2016). The authors found that this clonal TCR could stably and functionally bind with multiple peptide–major histocompatibility complexes through a conserved lock-and-key–like minimal binding footprint. This enables 1E6 TCR to tolerate vast numbers of substitutions outside of these so-called hotspots, which were found in foreign antigens compared with the natural autoimmune ligand, indicating that T cell cross-reactivity with microbe-derived antigens could break self-tolerance to induce autoimmune disease. Viral proteins with homology to autoantigens have been identified in diabetes. However, there is no direct evidence of their diabetogenicity. It is important to show that not only is this a theoretical possibility, but also that the microbial peptides can actually be processed and presented appropriately and that there is a direct interaction of the bacteria with the autoreactive T cells. Our data clearly demonstrated that bacterial lysates as well as the purified protein from the L. goodfellowii could be processed by DCs and GALT APC and presented the IGRP mimic peptides to NY8.3 T cells inducing inflammatory chemokine or cytokine MIP-1β, IFN-γ, and TNF production. Most importantly, we have shown that after supercolonizing the mice with L. goodfellowii, the extracted GALT APC stimulated NY8.3 cells in the absence of exogenous antigen. This clearly demonstrated that not only could the peptide be processed by APC in vitro, but also that the L. goodfellowii in the gut could be presented to NY8.3 cells in vivo.
Ultimately, however, the question arises as to whether these bacteria could have a direct effect on diabetes in vivo, as this is a major test of whether these bacteria have physiological and/or immunological relevance. We have shown this in several ways. First, immunizing NY8.3 TCR transgenic mice with the bacterial mimic peptide accelerated spontaneous diabetes, demonstrating the ability of this particular peptide to be presented in vivo and activate IGRP reactive cells to induce disease. Second, we have shown that disease acceleration can occur by increasing L. goodfellowii after supercolonization of the bacteria in the gut, indicating that the bacterial proteins and peptides can be presented in vivo to the IGRP-reactive T cells in the NY8.3 mice.
The gut microbiota comprises a large number of genera of bacteria that are very sensitive to the environment, including influences from diet, water, and antibiotics. Host factors including MyD88, a key adaptor for innate immunity, are also important (Wen et al., 2008). Identifying commensal gut bacteria that could trigger or increase autoreactive responses will considerably advance our understanding of environmental influences in pathogenesis of disease and provide important targets for therapy. The increase in Fusobacteria in the MyD88−/−NY8.3NOD mice increases the abundance of the bacterial protein peptide that can stimulate the NY8.3 T cells. When the gut microbiota composition in MyD88−/−NY8.3NOD mice was altered through cohousing or fecal transfer, this completely reversed the diabetes acceleration in MyD88−/−NY8.3NOD mice and was associated with the reduction of the Fusobacteria. Moreover, L. goodfellowii, a strain of Fusobacteria, can directly stimulate IGRP-reactive T cells in this pathogenic process. Fusobacteria are normally present in low abundance in NOD mice; however, there was a strong correlation between the abundance of Fusobacteria and progression to diabetes in polyclonal NOD mice. Furthermore, there were also significantly more Fusobacteria in diabetic NOD mice compared with the age- and gender-matched nondiabetic NOD littermates. Thus, the expansion of NY8.3-like CD8 T cells correlates with an increase in gut Fusobacteria and could conceivably be caused by the increase of Fusobacteria. It should be noted that IGRP-reactive T cells are not the only autoreactive CD8 T cells involved in the development of diabetes, and there is heterogeneity of early abundance of different TCR clonotypes (Lieberman et al., 2004). Furthermore, IGRP reactivity is dependent on an autoimmune response to insulin (Krishnamurthy et al., 2006, 2008), but these cells also increase in avidity and abundance with time (Amrani et al., 2000). Thus, it is likely, in the polyclonal setting, that other factors will also interact to increase the abundance of these bacteria or other microbial antigens to contribute to the development of diabetes. Fusobacteria have been reported to be associated with inflammatory disorders including inflammatory bowel diseases (Ohkusa et al., 2003; Strauss et al., 2011). Whether Fusobacteria are also associated with human T1D development remains to be elucidated.
The increase of Firmicutes and decrease of Bacteroidetes are correlated with delay of or protection from diabetes development in our current study, which support the findings in other animal models and/or human studies (Giongo et al., 2011; de Goffau et al., 2013; Murri et al., 2013; Wolf et al., 2014). However, our previous study using non-TCR transgenic MyD88−/−NOD mice showed a lower ratio of Firmicutes/Bacteroidetes, suggesting that T cell repertoire may play a role in the composition of gut microbiota of the host (Wen et al., 2008). It is not clear, at this stage, how the T cell repertoire affects the bacterial community and the possible biological consequences. However, it is clear that changing gut microbiota will have a significant impact on T1D development.
Our study therefore, provides direct evidence of molecular mimicry of microbial peptides with islet autoantigen and a novel mechanism by which the diabetogenicity of CD8+ T cells can be regulated by innate immunity and the gut microbiota in T1D. We believe that these data will be not only of great interest to investigators involved in studies of the pathogenesis and therapy of T1D, but also will be of general importance in providing evidence that molecular mimicry, originating in the gut microbiome, can indeed be a causative factor in the development of autoimmune disease.
MATERIALS AND METHODS
Mice
NOD/Caj mice have been maintained at Yale University for >25 yr. TLR2−/−NOD, TLR4−/−NOD, TLR9−/−NOD, and MyD88−/−NOD mice were generated as previously described (Wen et al., 2008; Wong et al., 2008). TLR5−/−NOD was generated by backcrossing TLR5−/− C57BL/6 (provided by R. Flavell, Yale University, New Haven, CT) with NOD mice for >12 generations. NY8.3NOD mice (The Jackson Laboratory) were bred with TLR or MyD88-deficient NOD mice to obtain TLR2−/−NY8.3, TLR4−/−NY8.3, TLR5−/−NY8.3, TLR9−/−NY8.3, and MyD88−/−NY8.3NOD mice. G9Cα−/−NOD mice were generated as reported previously (Wong et al., 2009). All the mice were kept in specific pathogen-free conditions in a 12-h dark/light cycle and housed in individually ventilated filter cages with autoclaved food and bedding. The use of animals and all procedures were approved by the Institutional Animal Care and Use Committee of Yale University and the Home Office, Wales, UK.
Antibodies and reagents
All the fluorochrome-conjugated mAbs used in this study were purchased from eBioscience or BioLegend unless otherwise stated. Hybridoma supernatants containing mAbs, used for cell purification or stimulation, were generously provided by C. Janeway Jr. (Yale University). Magnetic beads conjugated with goat anti–mouse IgG, goat anti–mouse IgM, or goat anti–rat IgG were purchased from QIAGEN. RPMI 1640 medium and heat-inactivated FCS were purchased from Invitrogen and Gemini, respectively.
Isolation of infiltrated immune cells from islets
The pancreata were removed from 6–7-wk-old MyD88−/−NY8.3NOD or WT NY8.3NOD mice and incubated with 10 ml HBSS containing 10 mM Hepes and 1.5 mg/ml collagenase V (Sigma-Aldrich) at 37°C for 12 min with vigorous shaking. The digestion was terminated by adding 10 ml RPMI 1640 containing 5% FBS, and the supernatant was discarded, followed by two more washes with HBSS buffer. The islets were spun down and treated with 0.25% trypsin–0.02% EDTA at 37°C for 5–10 min. The islets were then passed through a syringe with a 27-gauge needle to disperse into a cell suspension. After washing with PBS, the isolated cells were stained with fluorochrome-conjugated mAbs for flow cytometric analysis.
Histology of pancreatic tissues
Pancreatic tissues were fixed with 10% buffered formalin, embedded in paraffin, and sectioned at a thickness of 5 µm, stained with hematoxylin and eosin, and examined under light microscopy.
Diagnosis of diabetes
Mice were screened for glycosuria and confirmed by a blood glucose of >250 mg/dl (13.9 mmol/L).
Cohousing
MyD88−/−NY8.3NOD mice or WT NY8.3NOD mice (4 wk old) were cohoused with an equal number of age- and sex-matched NOD mice. All the mice were monitored for glycosuria twice weekly, and the experiments were terminated at 20 and 30 wk after birth for MyD88−/−NY8.3NOD or WT NY8.3NOD and WT NOD mice, respectively, unless the mice developed diabetes.
Bacteria OT
Fecal samples were collected from diabetes-protected MyD88−/−NY8.3NOD mice, cohoused with NOD mice for >3 mo, and orally fed to the sex-matched 4-wk-old MyD88−/−NY8.3NOD mice, two to three times weekly for three consecutive weeks. All the mice were monitored for glycosuria twice weekly. We randomly selected diabetes-free MyD88−/−NY8.3NOD mice (Fig. 6 B) as breeders and observed for diabetes in their progeny, also using the progeny from the breeders without OT as controls. In some experiments, NOD mice (1.5–2-mo-old females) were gavaged with L. goodfellowii or B. theta (∼109 bacteria/mouse) daily for 5 d. APCs from spleens or MLNs, PPs and the GALT were used for antigen-presentation assays ex vivo.
Extraction of gut bacterial DNA
Fecal samples from WT NY8.3NOD and MyD88−/−NY8.3NOD mice were collected biweekly starting from 1-mo-old mice and stored at −20°C. Bacterial DNA was extracted as previously described (Peng et al., 2014). In brief, the fecal samples (∼15–25 mg/mouse) were resuspended in 300 µl TE (10 mM Tris and 1 mM EDTA, pH 8) and incubated for 1 h at 37°C in the presence of 7.5 µl SDS (0.5%) and 3 µl proteinase K (20 mg/ml). One volume of phenol/chloroform/isoamyl alcohol (25:24:1), 200 µl of 20% SDS, and 0.3 g of zirconium/silica beads (0.1 mm; Biospec Inc.) were added, and samples were mixed with a Mini-bead-beater (Biospec) for 2 min. The sample was then mixed with 820 µl phenol/chloroform/isoamyl alcohol (25:24:1) and centrifuged, and the aqueous layer was collected into a new tube. The bacterial DNA was precipitated with 0.6 volume of isopropanol, washed with 70% ethanol, air dried, and resuspended in 100 µl of sterile water.
16S rRNA sequencing and microbiota classification
The V4 region of the bacterial 16S ribosomal gene was amplified from each DNA sample with barcoded broadly conserved bacterial primers (Table 2). The PCR products were purified with a gel extraction kit (QIAGEN) and quantified with a spectrophotometer (Nanodrop), and equimolar amounts of each sample were pooled and pyrosequenced on an Ion Torrent personal genome machine sequencing system (Thermo Fisher Scientific). The results were analyzed using the QIIME software package (version 1.8) and UPARSE pipeline (version 7.0). After removing the primer sequences, the sequences were demultiplexed, quality filtered using QIIME, and further quality and chimera filtered in UPARSE pipeline. Operational taxonomic units were picked with 97% identity in UPARSE pipeline. In QIIME, the Greengenes reference database was used for taxonomy assignment, which was performed at various levels using representative sequences of each operational taxonomic unit. β-Diversity was calculated to compare differences between microbial community profiles, and the data are shown as a principal coordinate analysis. α-Diversity was measured by Shannon evenness or Chao richness in the bacterial communities.
Table 2. List of primers.
| Application | Gene name | Sequence | |
|---|---|---|---|
| Sequencing | 16S | Forward | 5′-GTGCCAGCMGCCGCGGTAA-3′ |
| Reverse | 5′-GGACTACHVGGGTWTCTAAT-3′ | ||
| qPCR | Fusobacteria | Forward | 5′-CCCTTCAGTGCCGCAGT-3′ |
| Reverse | 5′-GTCGCAGGATGTCAAGAC-3′ | ||
| Mgt | Forward | 5′-GGCTTGCCGTACTTATGATTTC-3′ | |
| Reverse | 5′-TTTAAAGGCATCTTTGGGAGAA-3′ | ||
| 16S | Forward | 5′-ACTCCTACGGGAGGCAGCAGT-3′ | |
| Reverse | 5′-ATTACCGCGGCTGCTGGC-3′ | ||
| GAPDH | Forward | 5′-TGACATCAAGAAGGTGGTGAAG-3′ | |
| Reverse | 5′-TGCTGTAGCCGTATTCATTGTC-3′ | ||
| Cloning | Mgt | Forward | 5′-ATCCGGAATTCCATGAAAGAAATAATAGAAACTCTTA-3′ |
| Reverse | 5′-ATCCGCTCGAGAATTTTCAAAAACAATGTAGCAATT-3′ | ||
Quantitation of Fusobacteria and L. goodfellowii by qPCR
Bacterial DNA was extracted as described in the Extraction of gut bacterial DNA section from fecal pellet or oral swab samples from 6–7-wk-old MyD88−/−NY8.3NOD and WT NY8.3NOD mice. qPCR was performed using a qPCR detection system (iQ5; Bio-Rad Laboratories) according to the manufacturer’s instructions with the specific primers for Fusobacteria or L. goodfellowii’s Mgt (Table 2). The relative abundance of these bacteria was determined using the 2−ΔΔCt method by normalization with total bacteria.
In another experiment, bacterial DNA was extracted from fecal samples of female NOD mice (biweekly from 6 wk old until diabetic). The abundance of Fusobacteria in the selected samples from three time points (no insulitis at −6 wk, mid-insulitis at −10 wk, and insulitis at >13 wk before diabetic) was quantified by qPCR.
Quantitation of intestinal antimicrobial peptides by qPCR
Approximately 3-cm fragments from the ileum of small intestine and the area adjacent to the cecum of large intestines were obtained from 6–7-wk-old MyD88−/−NY8.3NOD and WT NY8.3NOD mice. Total RNA was extracted from the gut fragments with TRIzol and reverse transcribed to cDNA using a SuperScript III First-strand synthesis kit with random hexamers (Invitrogen). qPCR was performed with the specific primers for Reg3β, Reg3γ, C-reactive protein–ductin, RELMβ, and DEFCR-rs-10 (Vaishnava et al., 2008). The relative mRNA levels of these peptides were determined using the 2−ΔΔCt method by normalization with the housekeeping gene GAPDH (Table 2).
Identification and synthesis of mimic peptides
The IGRP206–214 peptide sequence was pBLAST searched against the nonredundant protein sequences database within bacterial families, and peptides with high homology were identified. The microbial peptide mimics (Table 1) were synthesized and purified with HPLC by the Yale Keck Facility.
Culture of anaerobic bacteria
L. goodfellowii (HM-12; BEI Resources), B. theta (provided by A. Goodman and A. Wexler, Yale University, New Haven, CT), B. dorei (BEI Resources), and L. reuteri (BEI Resources) were grown in MTGE Anaerobic Enrichment Broth (Anaerobe Systems) overnight. For heat-killed bacteria, the bacterial pellet was resuspended in sterile 1× PBS and heated at 65°C for 60 min. The bacterial lysates were then used to stimulate the NY8.3 CD8+ T cells followed by intracellular cytokine (ICC) analysis.
Translation of rMgt and expression and purification of truncated rMgt protein
The coding region of Mgt (GenPept accession no. WP_006806773) from L. goodfellowii was amplified with PCR from genomic DNA from L. goodfellowii with specific primers (Table 2) and then cloned into EcoR I/Xho I digested pET23b+ (EMD Millipore). The recombinant vector was confirmed with DNA sequencing by the Yale Keck Facility. The in vitro translation of Mgt was performed in an E. coli High-yield Protein Expression System (S30 T7; Promega) according to the manufacturer’s instructions. Empty pET23b+ was used as a negative control. The truncated rMgt was expressed and purified by Detai Biologics. In brief, the coding region of Mgt was modified by removing two transmembrane fragments (355–374 and 382–404) and replacing the rare codons for E. coli and cloned into pET43.1a+. The recombinant protein was obtained by refolding insoluble inclusion bodies and purifying by affinity column chromatography. The single-band purity was confirmed by both SDS-PAGE and Western blotting.
Generation of BMDCs, APC isolation, and NY8.3 CD8+ T cell purification
BMDCs were generated from BM cells of 2-mo-old NOD mice. The BM cells were cultured in the presence of 25 ng/ml GM-CSF and 25 ng/ml IL-4 in RPMI 1640 complete medium supplemented with 5% heat-inactivated FBS. Culture medium was replenished every 2 d. On day 5, adherent and loosely adherent cells were harvested. These cells were ∼70% CD11c+ DCs by FACS analysis.
Peripheral APCs in this study refer to T cell–depleted immune cells (CD11c+, CD11b+, and CD19+ cells) and were isolated from spleen, MLN, and PP of 2-mo-old female NOD mice by depleting CD4+ and CD8+ T cells with hybridoma supernatant and magnetic beads.
CD8+ T cells were purified from spleen of 5–8-wk-old WT NY8.3NOD or MyD88−/−NY8.3NOD mice by negative selection with magnetic beads according to the manufacturer’s instructions (QIAGEN). The purity was routinely 90–95% as verified by flow cytometry.
Cell proliferation assay
Splenocytes or purified CD8+ T cells from WT NY8.3NOD or MyD88−/−NY8.3NOD mice were cultured in triplicate (105 cells/well) in the presence or absence of IGRP206–214 peptide or bacterial peptide mimics at different concentrations for 4 d. [3H]thymidine was added during the last 18 h of a 4-d culture. Proliferation was measured by [3H]thymidine incorporation, and the data are presented as a stimulation index, which is the mean cpm in the presence of antigen/the mean cpm in the absence of antigen.
MHC blocking assay
Splenocytes from WT NY8.3NOD mice were cultured in triplicate (105 cells/well) in the presence or absence of 100 ng/ml W15944 together with different concentrations of anti-Kd (hybridoma supernatant of clone HB159) or control IgG. [3H]thymidine was added in the last 16 h of 4-d culture. Proliferation was measured by [3H]thymidine incorporation, and the data are presented as a stimulation index, which is the mean cpm in the presence of antigen/the mean cpm in the absence of antigen.
MHC restriction assay
Purified splenic NY8.3 CD8+ T cells were cultured in triplicate (105 cells/well) with different concentrations of IGRP or W15944 peptide in the presence of irradiated splenic APCs from NOD (Kd) or C57BL/6 (Kb) mice. [3H]thymidine was added in the last 16 h of 4-d culture. Proliferation was measured by [3H]thymidine incorporation, and the data are presented as a stimulation index, which is the mean cpm in the presence of antigen/the mean cpm in the absence of antigen.
CTL killing assays
Target P815 cells (104 cells/tube) were labeled with PKH-26 followed by co-culturing with purified NY8.3 CD8+ T cells, as effector cells, from 6–7-wk-old WT NY8.3 mice at different ratios in the absence or presence of IGRP or W15944 (0.2 µg/ml) in 200 µl at 37°C for 16–18 h. Cells were stained with TOPRO-3 iodide before analysis by flow cytometry. The cytotoxicity was calculated as the percentage of apoptotic cells shown as double positive for TOPRO-3 iodide and PKH-26.
Supercolonization of WT NY8.3 mice with B. theta or L. goodfellowii
1-mo-old WT NY8.3NOD mice were orally gavaged with either B. theta or L. goodfellowii (∼109 bacteria/mouse) daily for 30 consecutive days, unless the mice developed diabetes.
ICC, granzyme B, MIP-1β, and IFN-γ secretion assay
ICC staining was performed according to the protocol provided by eBioscience. In brief, 1–2 × 106 cells/ml splenocytes from WT NY8.3 or MyD88−/−NY8.3 mice were stimulated with anti-CD3 (clone 2C-11) and anti-CD28 (clone 37N51) antibodies, IGRP206–214, peptide mimics, heat-killed bacteria, and APCs either pulsed with heat-killed bacteria or directly isolated from bacteria-supercolonized NOD mice or rMgt overnight, followed by further stimulation with 50 ng/ml PMA (Sigma-Aldrich) and 500 ng/ml ionomycin (Sigma-Aldrich) in the presence of Golgi-plug (eBioscience) for an additional 4 h. 106 cells/ml splenocytes from G9Cα−/−NOD mice were stimulated with insulin B15-23 peptide (G9G) and the bacterial peptide mimics in a similar assay. The cells were then stained with antibodies to surface markers before fixation and permeabilization. Fc receptors were blocked with 2.4G2 Fc-blocking antibody before staining with fluorochrome-labeled antibody to detect ICCs or cytolytic protein (IFN-γ and granzyme B). Some of the supernatants were used for ELISA to measure the concentration of MIP-1β or IFN-γ.
In some experiments, 1–2 × 105 purified NY8.3 CD8+ T cells were incubated with 105 cells BMDCs or 1–5 × 105 APCs, in 300 µl, in the presence of 107 CFU/500 µl heat-killed bacterial lysates (L. goodfellowii or B. theta), translated Mgt protein (1 µl of the protein lysate; empty vector as control), or 1 µg of purified truncated rMgt protein obtained from E. coli by the pET43.1a+ expression vector (carrier protein BSA as control) overnight, followed by further stimulation with PMA and ionomycin in the presence or absence of Golgi-plug for an additional 4 h. The cells were used for ICC staining in the cell culture in the presence of Golgi-plug, and the culture supernatants without Golgi-plug were tested for MIP-1β secretion by an ELISA kit (R&D Systems).
To test whether APCs could present antigens from the bacteria, 2 × 105 APCs from spleen, MLN, and PP were isolated from 6–8-wk-old naive NOD mice and pulsed with heat-killed L. goodfellowii or control B. theta lysates for 3 h at 37°C. After washing, the APCs were incubated with 105 purified NY8.3 CD8+ T cells from WT NY8.3 overnight in 300 µl, and the cells were stained for use for ICC staining in the cell culture in the presence of Golgi-plug.
After supercolonization with the gut bacteria, 105 purified NY8.3 CD8+ T cells were incubated with purified APCs from spleen (106 cells), MLN (3 × 105 cells), or PP (1–2 × 105 cells) overnight in 300 µl without the addition of exogenous antigens. The supernatants before the addition of Golgi-plug were tested for both MIP-1β and IFN-γ secretion by ELISA (R&D Systems), and after 4-h stimulation with PMA/ionomycin in the presence of Golgi-plug, the cells were used for ICC staining.
Adoptive transfer
Irradiated (600 rads) 1-mo-old NOD mice (both genders) were used as recipients in adoptive transfer experiments. Splenocytes from 6–7-wk-old WT NY8.3 or MyD88−/−NY8.3 mice were stimulated with IGRP206–214 (10 ng/ml), W15944 (10 ng/ml or 500 ng/ml), or other peptide mimics (500 ng/ml) for 3 d at 37°C. 106 activated CD8+ T cells, purified with magnetic-activated cell-sorting beads, were i.v. injected into age- and sex-matched recipients. Recipients were screened for diabetes development by testing for glycosuria daily, and the experiments were terminated 20 d after the cell transfer unless the mice developed diabetes.
Immunization of mimic peptide in WT NY8.3NOD mice
1-mo-old WT NY8.3NOD mice were i.p. immunized once with W15944 or IGRP peptide (25 µg/mouse) together with CpG (25 µg/mouse) as adjuvant. Control mice were injected with CpG (25 µg/mouse) only. Mice were screened for glycosuria daily, and the experiments were terminated 30 d after immunization unless the mice developed diabetes.
Statistical analysis
Statistical analysis was performed using Prism 6 (GraphPad Software). Diabetes incidence was compared using Wilcoxon or log-rank tests. Insulitis was analyzed by χ2 test. In vitro assays were analyzed with two-tailed Student’s t test or one- or two-tailed Mann Whitney test for statistical significance. Analysis of similarities (ANOSIM) was used to analyze β-diversity of taxonomic families of gut microbiota. For multiple comparisons, one-way or two-way ANOVA followed by Bonferroni test or Tukey’s posthoc test was performed. P-values <0.05 were considered significant.
ACKNOWLEDGEMENTS
We thank Drs. A. Wexler and A. Goodman for initial molecular modification of L. goodfellowii and for providing B. theta; Drs. F. Li, X. Yu, and H. Zhao for help with statistical analysis; G.M. Vijay for experimental assistance; and Dr. H. Ding for help with the truncated Mgt protein.
This work was supported by grants from the National Institutes of Health (DK088181, DK092882, DK100500, and P30 DK945735), the Iacocca Family Foundation, and the Juvenile Diabetes Research Foundation (2015-136).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ANOSIM
- analysis of similarities
- GALT
- gut-associated lymphoid tissue
- ICC
- intracellular cytokine
- IGRP
- islet-specific glucose-6-phosphatase catalytic subunit–related protein
- Mgt
- magnesium transporter
- MLN
- mesenteric LN
- MyD88
- myeloid differentiation primary response 88
- NOD
- nonobese diabetic
- OT
- oral transfer
- PP
- Peyer’s patches
- qPCR
- quantitative PCR
- rMgt
- recombinant Mgt
- T1D
- type 1 diabetes
References
- Alkanani A.K., Hara N., Gottlieb P.A., Ir D., Robertson C.E., Wagner B.D., Frank D.N., and Zipris D.. 2015. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. 64:3510–3520. 10.2337/db14-1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani A., Verdaguer J., Serra P., Tafuro S., Tan R., and Santamaria P.. 2000. Progression of autoimmune diabetes driven by avidity maturation of a T-cell population. Nature. 406:739–742. 10.1038/35021081 [DOI] [PubMed] [Google Scholar]
- Anderson B., Park B.J., Verdaguer J., Amrani A., and Santamaria P.. 1999. Prevalent CD8+ T cell response against one peptide/MHC complex in autoimmune diabetes. Proc. Natl. Acad. Sci. USA. 96:9311–9316. 10.1073/pnas.96.16.9311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum M.E., Mendoza J.L., Sethi D.K., Dong S., Glanville J., Dobbins J., Ozkan E., Davis M.M., Wucherpfennig K.W., and Garcia K.C.. 2014. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 157:1073–1087. 10.1016/j.cell.2014.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bluestone J.A., Herold K., and Eisenbarth G.. 2010. Genetics, pathogenesis and clinical interventions in type 1 diabetes. Nature. 464:1293–1300. 10.1038/nature08933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.T., Davis-Richardson A.G., Giongo A., Gano K.A., Crabb D.B., Mukherjee N., Casella G., Drew J.C., Ilonen J., Knip M., et al. . 2011. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 6:e25792 10.1371/journal.pone.0025792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D.K., Bulek A.M., Dolton G., Schauenberg A.J., Szomolay B., Rittase W., Trimby A., Jothikumar P., Fuller A., Skowera A., et al. . 2016. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J. Clin. Invest. 126:2191–2204. 10.1172/JCI85679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppieters K.T., Dotta F., Amirian N., Campbell P.D., Kay T.W., Atkinson M.A., Roep B.O., and von Herrath M.G.. 2012. Demonstration of islet-autoreactive CD8 T cells in insulitic lesions from recent onset and long-term type 1 diabetes patients. J. Exp. Med. 209:51–60. 10.1084/jem.20111187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Goffau M.C., Luopajärvi K., Knip M., Ilonen J., Ruohtula T., Härkönen T., Orivuori L., Hakala S., Welling G.W., Harmsen H.J., and Vaarala O.. 2013. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 62:1238–1244. 10.2337/db12-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulis A.K., Farquharson M.A., and Hardman R.. 1987. Aberrant expression of class II major histocompatibility complex molecules by B cells and hyperexpression of class I major histocompatibility complex molecules by insulin containing islets in type 1 (insulin-dependent) diabetes mellitus. Diabetologia. 30:333–343. 10.1007/BF00299027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giongo A., Gano K.A., Crabb D.B., Mukherjee N., Novelo L.L., Casella G., Drew J.C., Ilonen J., Knip M., Hyöty H., et al. . 2011. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 5:82–91. 10.1038/ismej.2010.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horai R., Zárate-Bladés C.R., Dillenburg-Pilla P., Chen J., Kielczewski J.L., Silver P.B., Jittayasothorn Y., Chan C.C., Yamane H., Honda K., and Caspi R.R.. 2015. Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity. 43:343–353. 10.1016/j.immuni.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarchum I., Nichol L., Trucco M., Santamaria P., and DiLorenzo T.P.. 2008. Identification of novel IGRP epitopes targeted in type 1 diabetes patients. Clin. Immunol. 127:359–365. 10.1016/j.clim.2008.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaprio J., Tuomilehto J., Koskenvuo M., Romanov K., Reunanen A., Eriksson J., Stengård J., and Kesäniemi Y.A.. 1992. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. 35:1060–1067. 10.1007/BF02221682 [DOI] [PubMed] [Google Scholar]
- Ko H.J., Chee J., Sutherland R.M., Thomas H.E., Zhan Y., Krishnamurthy B., Kay T.W., and Lew A.M.. 2014. Functional cytotoxic T lymphocytes against IGRP206-214 predict diabetes in the non-obese diabetic mouse. Immunol. Cell Biol. 92:640–644. 10.1038/icb.2014.29 [DOI] [PubMed] [Google Scholar]
- Kostic A.D., Gevers D., Siljander H., Vatanen T., Hyötyläinen T., Hämäläinen A.M., Peet A., Tillmann V., Pöhö P., Mattila I., et al. DIABIMMUNE Study Group . 2015. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 17:260–273. 10.1016/j.chom.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy B., Dudek N.L., McKenzie M.D., Purcell A.W., Brooks A.G., Gellert S., Colman P.G., Harrison L.C., Lew A.M., Thomas H.E., and Kay T.W.. 2006. Responses against islet antigens in NOD mice are prevented by tolerance to proinsulin but not IGRP. J. Clin. Invest. 116:3258–3265. 10.1172/JCI29602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy B., Mariana L., Gellert S.A., Colman P.G., Harrison L.C., Lew A.M., Santamaria P., Thomas H.E., and Kay T.W.. 2008. Autoimmunity to both proinsulin and IGRP is required for diabetes in nonobese diabetic 8.3 TCR transgenic mice. J. Immunol. 180:4458–4464. 10.4049/jimmunol.180.7.4458 [DOI] [PubMed] [Google Scholar]
- Legaria M.C., Lumelsky G., Rodriguez V., and Rosetti S.. 2005. Clindamycin-resistant Fusobacterium varium bacteremia and decubitus ulcer infection. J. Clin. Microbiol. 43:4293–4295. 10.1128/JCM.43.8.4293-4295.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liblau R.S., Wong F.S., Mars L.T., and Santamaria P.. 2002. Autoreactive CD8 T cells in organ-specific autoimmunity: emerging targets for therapeutic intervention. Immunity. 17:1–6. 10.1016/S1074-7613(02)00338-2 [DOI] [PubMed] [Google Scholar]
- Lieberman S.M., Evans A.M., Han B., Takaki T., Vinnitskaya Y., Caldwell J.A., Serreze D.V., Shabanowitz J., Hunt D.F., Nathenson S.G., et al. . 2003. Identification of the β cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc. Natl. Acad. Sci. USA. 100:8384–8388. 10.1073/pnas.0932778100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman S.M., Takaki T., Han B., Santamaria P., Serreze D.V., and DiLorenzo T.P.. 2004. Individual nonobese diabetic mice exhibit unique patterns of CD8+ T cell reactivity to three islet antigens, including the newly identified widely expressed dystrophia myotonica kinase. J. Immunol. 173:6727–6734. 10.4049/jimmunol.173.11.6727 [DOI] [PubMed] [Google Scholar]
- Markle J.G., Frank D.N., Mortin-Toth S., Robertson C.E., Feazel L.M., Rolle-Kampczyk U., von Bergen M., McCoy K.D., Macpherson A.J., and Danska J.S.. 2013. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 339:1084–1088. 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- Murri M., Leiva I., Gomez-Zumaquero J.M., Tinahones F.J., Cardona F., Soriguer F., and Queipo-Ortuño M.I.. 2013. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 11:46 10.1186/1741-7015-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R.W., Beisang D., Tubo N.J., Dileepan T., Wiesner D.L., Nielsen K., Wüthrich M., Klein B.S., Kotov D.I., Spanier J.A., et al. . 2015. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity. 42:95–107. 10.1016/j.immuni.2014.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen D.S., Krych Ł., Buschard K., Hansen C.H., and Hansen A.K.. 2014. Beyond genetics. Influence of dietary factors and gut microbiota on type 1 diabetes. FEBS Lett. 588:4234–4243. 10.1016/j.febslet.2014.04.010 [DOI] [PubMed] [Google Scholar]
- Ohkusa T., Okayasu I., Ogihara T., Morita K., Ogawa M., and Sato N.. 2003. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 52:79–83. 10.1136/gut.52.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakman M., Wen L., McNab G.L., Watkins P.J., Tan K.C., and Vergani D.. 1994. T cell clones generated from patients with type 1 diabetes using interleukin-2 proliferate to human islet antigens. Autoimmunity. 17:31–39. 10.3109/08916939409014656 [DOI] [PubMed] [Google Scholar]
- Peng J., Narasimhan S., Marchesi J.R., Benson A., Wong F.S., and Wen L.. 2014. Long term effect of gut microbiota transfer on diabetes development. J. Autoimmun. 53:85–94. 10.1016/j.jaut.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polychronakos C., and Li Q.. 2011. Understanding type 1 diabetes through genetics: advances and prospects. Nat. Rev. Genet. 12:781–792. 10.1038/nrg3069 [DOI] [PubMed] [Google Scholar]
- Pugliese A., Vendrame F., Reijonen H., Atkinson M.A., Campbell-Thompson M., and Burke G.W.. 2014. New insight on human type 1 diabetes biology: nPOD and nPOD-transplantation. Curr. Diab. Rep. 14:530 10.1007/s11892-014-0530-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Calvo T., Suwandi J.S., Amirian N., Zapardiel-Gonzalo J., Anquetil F., Sabouri S., and von Herrath M.G.. 2015. Heterogeneity and lobularity of pancreatic pathology in type 1 diabetes during the prediabetic phase. J. Histochem. Cytochem. 63:626–636. 10.1369/0022155415576543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowera A., Ellis R.J., Varela-Calviño R., Arif S., Huang G.C., Van-Krinks C., Zaremba A., Rackham C., Allen J.S., Tree T.I., et al. . 2008. CTLs are targeted to kill β cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J. Clin. Invest. 118:3390–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J., Kaplan G.G., Beck P.L., Rioux K., Panaccione R., Devinney R., Lynch T., and Allen-Vercoe E.. 2011. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 17:1971–1978. 10.1002/ibd.21606 [DOI] [PubMed] [Google Scholar]
- Tsai S., Shameli A., and Santamaria P.. 2008. CD8+ T cells in type 1 diabetes. Adv. Immunol. 100:79–124. 10.1016/S0065-2776(08)00804-3 [DOI] [PubMed] [Google Scholar]
- Unger W.W., Pinkse G.G., Mulder-van der Kracht S., van der Slik A.R., Kester M.G., Ossendorp F., Drijfhout J.W., Serreze D.V., and Roep B.O.. 2007. Human clonal CD8 autoreactivity to an IGRP islet epitope shared between mice and men. Ann. N. Y. Acad. Sci. 1103:192–195. 10.1196/annals.1394.024 [DOI] [PubMed] [Google Scholar]
- Vaishnava S., Behrendt C.L., Ismail A.S., Eckmann L., and Hooper L.V.. 2008. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA. 105:20858–20863. 10.1073/pnas.0808723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaguer J., Schmidt D., Amrani A., Anderson B., Averill N., and Santamaria P.. 1997. Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J. Exp. Med. 186:1663–1676. 10.1084/jem.186.10.1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Ley R.E., Volchkov P.Y., Stranges P.B., Avanesyan L., Stonebraker A.C., Hu C., Wong F.S., Szot G.L., Bluestone J.A., et al. . 2008. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 455:1109–1113. 10.1038/nature07336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox A., Richardson S.J., Bone A.J., Foulis A.K., and Morgan N.G.. 2009. Analysis of islet inflammation in human type 1 diabetes. Clin. Exp. Immunol. 155:173–181. 10.1111/j.1365-2249.2008.03860.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf K.J., Daft J.G., Tanner S.M., Hartmann R., Khafipour E., and Lorenz R.G.. 2014. Consumption of acidic water alters the gut microbiome and decreases the risk of diabetes in NOD mice. J. Histochem. Cytochem. 62:237–250. 10.1369/0022155413519650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F.S., Visintin I., Wen L., Flavell R.A., and Janeway C.A. Jr. 1996. CD8 T cell clones from young nonobese diabetic (NOD) islets can transfer rapid onset of diabetes in NOD mice in the absence of CD4 cells. J. Exp. Med. 183:67–76. 10.1084/jem.183.1.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F.S., Karttunen J., Dumont C., Wen L., Visintin I., Pilip I.M., Shastri N., Pamer E.G., and Janeway C.A. Jr. 1999. Identification of an MHC class I-restricted autoantigen in type 1 diabetes by screening an organ-specific cDNA library. Nat. Med. 5:1026–1031. 10.1038/12465 [DOI] [PubMed] [Google Scholar]
- Wong F.S., Hu C., Zhang L., Du W., Alexopoulou L., Flavell R.A., and Wen L.. 2008. The role of Toll-like receptors 3 and 9 in the development of autoimmune diabetes in NOD mice. Ann. N. Y. Acad. Sci. 1150:146–148. 10.1196/annals.1447.039 [DOI] [PubMed] [Google Scholar]
- Wong F.S., Siew L.K., Scott G., Thomas I.J., Chapman S., Viret C., and Wen L.. 2009. Activation of insulin-reactive CD8 T-cells for development of autoimmune diabetes. Diabetes. 58:1156–1164. 10.2337/db08-0800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooldridge L., Ekeruche-Makinde J., van den Berg H.A., Skowera A., Miles J.J., Tan M.P., Dolton G., Clement M., Llewellyn-Lacey S., Price D.A., et al. . 2012. A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 287:1168–1177. 10.1074/jbc.M111.289488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Burrows M., Khan A.A., Graham L., Volchkov P., Becker L., Antonopoulos D., Umesaki Y., and Chervonsky A.V.. 2013. Gender bias in autoimmunity is influenced by microbiota. Immunity. 39:400–412. 10.1016/j.immuni.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]