Veillette and collaborators generate a mouse model with a deletion spanning the entire 400-kb Slam locus on chromosome 1 to show the overall role of SLAM proteins in NK cell development and function.
Abstract
Signaling lymphocytic activation molecule (SLAM) family receptors (SFRs) can mediate either activating or inhibitory effects during natural killer cell (NK cell) activation. In this study, we addressed the global role, regulation, and mechanism of action of the SLAM family in NK cells by analyzing a mouse lacking the entire ∼400-kilobase Slam locus, which encodes all six SFRs and CD48, the ligand of SFR 2B4. This mouse displayed enhanced NK cell activation responses toward hematopoietic target cells. Analyses of mice lacking individual SFRs showed that the inhibitory function of the Slam locus was due solely to 2B4 and was not influenced positively or negatively by other SFRs. Differences in NK cell responses between recognition of targets expressing or lacking ligands for SFRs were enhanced by IL-12 but suppressed by type I interferon. Cytokines also changed the levels of SLAM-associated protein adaptors, which prevent the inhibitory function of SFRs. The enhanced activation responses of SFR-deficient NK cells were dependent on integrin LFA-1 but not on DNAM-1 or NKG2D. SFR-mediated inhibition prevented the generation of activated forms of LFA-1. Hence, the Slam locus has an overall inhibitory role during NK cell activation that is solely dependent on 2B4. This effect is influenced by cytokines and leads to suppression of LFA-1 activity.
INTRODUCTION
NK cells play key roles in antitumor and antiviral immunity, as well as in normal immune regulation, through their capacity to kill abnormal or activated cells, in particular hematopoietic cells (Lanier, 2005; Gasser and Raulet, 2006; Waggoner et al., 2016). Activation of NK cells is determined by the balance between stimulation of various activating and inhibitory receptors and by ligands that may or may not be present on potential target cells. This activation is also influenced by cues received from surrounding cells before encounters with targets, in particular other hematopoietic cells. This effect can take place during or after NK cell maturation and is commonly termed NK cell education (Gasser and Raulet, 2006; Orr and Lanier, 2010; Elliott and Yokoyama, 2011; Narni-Mancinelli et al., 2013).
Signaling lymphocytic activation molecule (SLAM) family receptors (SFRs) include six transmembrane receptors named SLAM (SLAMF1; CD150), 2B4 (SLAMF4; CD244), Ly-9 (SLAMF3; CD229), CD84 (SLAMF5), SLAMF6 (Ly108; NTB-A), and SLAMF7 (CRACC; CS1) (Veillette, 2006, 2010; Calpe et al., 2008; Cannons et al., 2011). They are expressed only on hematopoietic cells. All SFRs except 2B4 are homotypic receptors, i.e., they recognize as a ligand another molecule of the same receptor expressed either on another cell (trans-interaction) or, in some cases, on the same cell (cis-interaction). 2B4 interacts with CD48 (SLAMF2), the expression of which is also restricted to hematopoietic cells. Although CD48 is related to SFRs, strictly speaking it is not an SFR, as, unlike SFRs, it is attached to the plasma membrane via a glycosylphosphatidylinositol moiety. Other receptors known as SLAMF8 and SLAMF9 are also not true members of the SLAM family, as they do not as yet have determined ligands, and they significantly differ from SFRs in their cytoplasmic domain. All bona fide SFRs except SLAM are expressed on NK cells.
By way of immunoreceptor tyrosine-based switch motifs located in their cytoplasmic domain, all SFRs associate with SLAM-associated protein (SAP) adaptors (Veillette, 2006, 2010; Calpe et al., 2008; Cannons et al., 2011). SAP adaptors include SAP, EAT-2 (Ewing’s sarcoma–associated transcript 2), and, in mice but not humans, EAT-2–related transducer (ERT). They are composed almost exclusively of a Src homology 2 (SH2) domain. All SAP adaptors are expressed in NK cells. Through their immunoreceptor tyrosine-based switch motifs, SFRs can also associate with SH2 domain–bearing inhibitory molecules such as protein tyrosine phosphatases SHP-1 and SHP-2 and inositol phosphatase SHIP-1 (SH2 domain–containing inositol phosphatase 1). When associated with SFRs, SAP adaptors prevent the interactions of SFRs with phosphatases.
SFRs and SAP adaptors have been clearly implicated in normal immune regulation and in immunological diseases (Veillette, 2006, 2010; Calpe et al., 2008; Cannons et al., 2011; Wu and Veillette, 2016). The SLAM locus (Slam in mice), which encompasses the genes coding for all SFRs and CD48 on chromosome 1, is highly polymorphic in humans and mice. Some of these polymorphisms have been linked to autoimmune diseases (Veillette, 2006, 2010; Calpe et al., 2008; Cannons et al., 2011; Wu and Veillette, 2016). In addition, the SAP-encoding gene SH2D1A is mutated and inactivated in a human primary immunodeficiency, X-linked lymphoproliferative disease (Veillette et al., 2013; Tangye, 2014). We and others showed that loss of SAP or other SAP adaptors converts SFRs into superinhibitory receptors because of enhanced coupling of SFRs to inhibitory effectors (Parolini et al., 2000; Dong et al., 2009, 2012; Kageyama et al., 2012; Zhao et al., 2012; Pérez-Quintero et al., 2014). This alteration compromises activation of NK cells and T cells, leading to multiple immune cell defects, including reduced NK cell cytotoxicity in response to hematopoietic target cells. These defects likely underlie the pathophysiology of X-linked lymphoproliferative disease.
Although SFRs are superinhibitory in NK cells lacking SAP adaptors, there is much debate about the functions of SFRs in normal NK cells, which contain SAP adaptors (Wu and Veillette, 2016). This is in part because of the fact that mice lacking individual SFRs usually exhibit minor phenotypes, possibly caused by redundancy between SFRs (Veillette, 2006, 2010; Calpe et al., 2008; Cannons et al., 2011). However, it has been difficult to address the issue of redundancy by breeding together mice lacking individual SFRs, given that all genes encoding SFRs are located in the same gene locus. Nonetheless, there is currently evidence that SFRs can be either activating or inhibitory in NK cells containing SAP adaptors depending on the context (Wu and Veillette, 2016).
For instance, it was reported that triggering of several SFRs on mouse or human NK cells, including 2B4, SLAMF7, SLAMF6, and Ly-9, using their ligands ectopically expressed on nonhematopoietic target cells or, in some cases, anti-SFR mAbs, results in enhanced NK cell activation (Veillette, 2006, 2010; Calpe et al., 2008; Cannons et al., 2011). Although the physiological relevance of these modes of stimulation is unclear, these data suggested that SFRs are positive regulators of NK cell activation. Paradoxically, however, NK cells from mice lacking the SFR 2B4 displayed enhanced activation in response to hematopoietic target cells in vitro and in vivo (Lee et al., 2004; Waggoner et al., 2010; Waggoner and Kumar, 2012). Hence, at least one SFR, 2B4, is primarily an inhibitory receptor in mouse NK cells expressing SAP adaptors. The roles of the other SFRs in normal NK cells are largely unknown.
Several important issues regarding the role, regulation, and mechanism of action of SFRs in normal NK cells remain elusive. As SFRs can be activating or inhibitory, whether the global function of the SLAM family in normal NK cells is activating, inhibitory, or neutral needs to be ascertained. In addition, the impact of the individual SFRs other than 2B4 in this setting has to be elucidated. Likewise, the potential effects of host factors, such as the cytokine milieu, on the impact of engagement of SLAM receptors during target cell recognition should be assessed. Lastly, the activating NK cell receptors whose functions are either enhanced or suppressed by SLAM receptors in normal NK cells must be identified.
Herein, we addressed these various issues by creating and analyzing a new mouse lacking the entire 400-kb Slam locus on chromosome 1. We found that Slam locus–deleted mice displayed enhanced NK cell activation in response to hematopoietic target cells. This effect was ascribed solely to the SLAM receptor 2B4 and was not influenced by other SLAM receptors. We also found that the impact of engagement of SFRs on NK cells by SFRs on targets was influenced by cytokines. Cytokines also controlled the levels of SAP adaptors, suggesting that they directly influenced SFR function. We also obtained evidence that SLAM receptors inhibited NK cell activation by interfering with the function of LFA-1 but not of other activating receptors such as DNAX accessory molecule 1 (DNAM-1) and NKG2D.
RESULTS
Generation of mouse lacking the entire Slam locus
To address the overall function of SFRs, a mouse lacking the entire 400-kb Slam locus on chromosome 1 was generated. A gene deletion approach was favored over multigene-targeted mutations, as the genes encoding SFRs frequently undergo alternative splicing that can create alternative protein products when point mutations or small deletions are created. Aside from the genes coding for all six SFRs and CD48, the ligand of 2B4, the Slam locus contains no other known genes with the exception of a short nonconserved open reading frame of unknown function named Gm10521. To address the possible impact of Gm10521, a Gm10521-disrupted mouse was also engineered (see The inhibitory effect of the Slam locus is mediated by 2B4 with no appreciable impact from other SFRs or Gm10521 section).
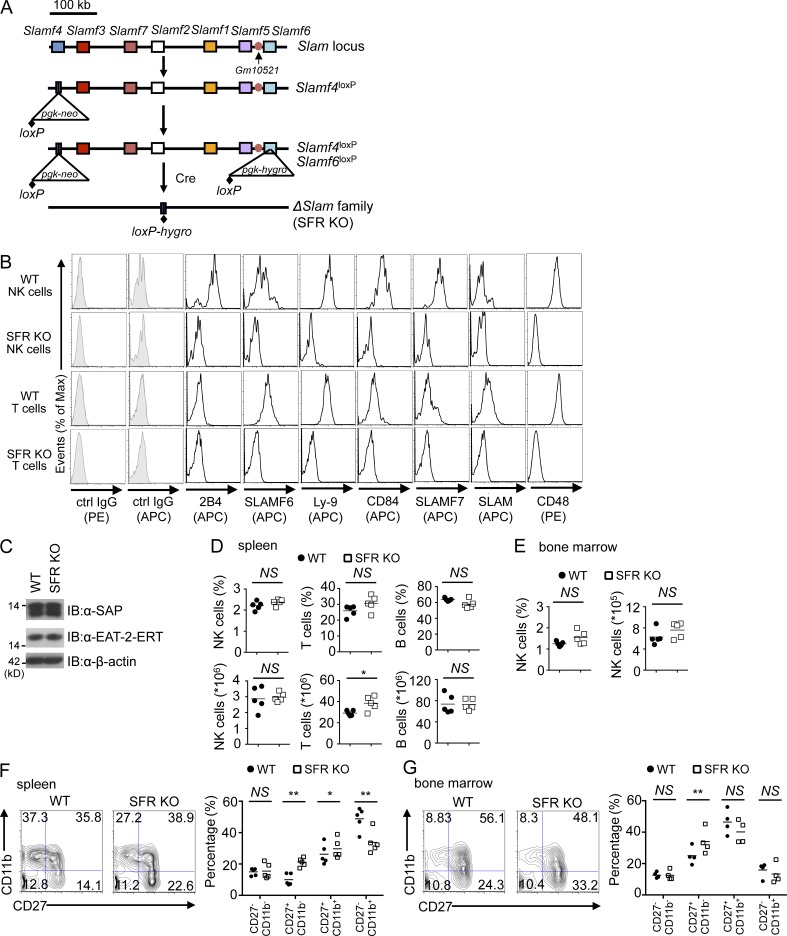
Deletion of the Slam locus was achieved by sequential gene targeting in C57BL/6 embryonic stem cells, in which loxP sites were inserted at the two extremities of the locus, followed by Cre-mediated deletion of the locus (Fig. 1 A). Resulting mice lacked all SFRs, as well as CD48, on immune cells, including NK cells and T cells, compared with wild-type littermates (Fig. 1 B). No effect was seen on the expression levels of SAP adaptors (Fig. 1 C). Mice were viable and fertile (not depicted). They were extensively backcrossed (6–10 generations) to the C57BL/6 background before experimentation.
Figure 1.
Deletion of the Slam locus in mice and its impact on NK cell development. (A) Schematic diagram of the strategy used to delete the Slam locus. In brief, the most 5′ gene of the locus, Slamf4, was disrupted using a targeting construct containing the neomycin (neo) resistance gene, flanked by one loxP site. Then, the most 3′ gene of the locus, Slamf6, was disrupted using a targeting construct that contains the hygromycin (hygro) resistance gene, also flanked by a single loxP site. After Cre-mediated recombination, the entire Slam locus was deleted. The position of the potential gene of unknown function, Gm10521, is also depicted. (B) Expression of SLAM receptors as well as CD48 was analyzed on splenic NK cells (CD3−NK1.1+) and splenic T cells (CD3+NK1.1−) from wild-type and Slam locus–deleted (SFR KO) mice. Isotype control (ctrl) is shown as filled histograms. Results are representative of at least eight experiments. (C) Expression of SAP and EAT-2–ERT was determined by immunoblotting (IB) of total cell lysates of IL-2–expanded NK cells from wild-type or SFR KO mice. β-actin was used as the loading control. Note that, as the sequences of EAT-2 and ERT proteins are very similar, the available antibodies do not distinguish them. The positions of prestained molecular mass markers are shown on the left. Results are representative of three experiments. (D and E) Percentages and absolute numbers of NK cells (CD3−NK1.1+) were analyzed in the spleen (D) and bone marrow (E) from wild-type and SFR KO mice. For bone marrow, cells were pooled from the two femurs and two tibias of each mouse. Percentages and absolute numbers of T cells (CD3+NK1.1−) and B cells (B220+CD3−) are also depicted (D). Symbols represent individual mice. Results are pooled from five mice. (F and G) Expression of CD27 and CD11b was analyzed on wild-type and SFR KO NK cells (CD3−NK1.1+) from the spleen (F, left) and bone marrow (G, left). The maturation stages of NK cells were divided into CD27−CD11b−, CD27+CD11b−, CD27+CD11b+, and CD27−CD11b+, and the percentages of each subset are shown in each quadrant. Data from multiple independent experiments are represented on the right. Symbols represent individual mice. Results are pooled from five (F) or four (G) mice. (D–G) *, P < 0.05; **, P < 0.01; unpaired Student’s t test.
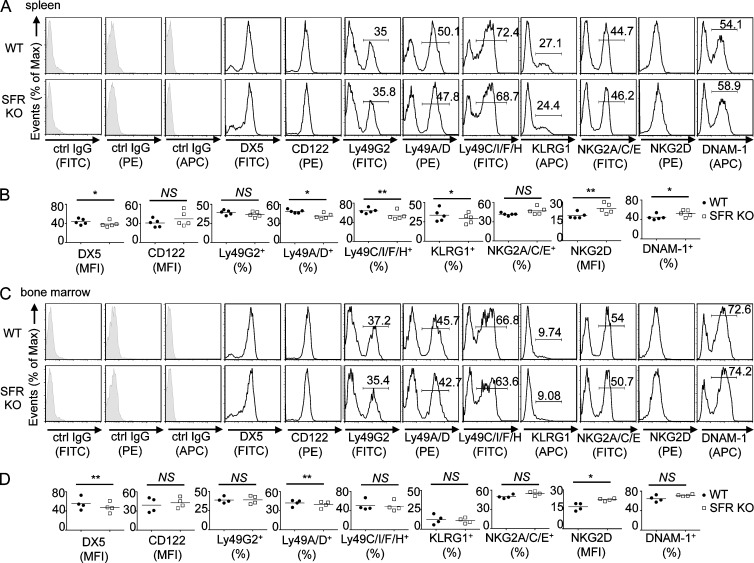
Compared with wild-type mice, mice lacking the Slam locus (SFR KO mice) had no alterations in NK cell numbers in spleen and bone marrow (Fig. 1, D and E). They also displayed little or no change in the total numbers of T cells and B cells in spleen (Fig. 1 D). However, SFR KO mice had a small alteration of NK cell maturation, particularly in spleen, e.g., an increased proportion of immature CD27+CD11b− and CD27+CD11b+ NK cells, with decreased proportions of mature CD27−CD11b+ NK cells (Fig. 1, F and G). SFR KO mice also had modest changes in NK cell repertoire (Fig. 2, A–D). Compared with wild-type mice, they displayed a small decrease (<5%) in the proportions of Ly49A/D+, Ly49C/I/F/H+, or KLRG1+ NK cells (Fig. 2, A–D). Levels of NKG2D and DNAM-1 were also slightly increased (Fig. 2, A–D). These effects were seen in spleen and, to a lesser extent, in bone marrow.
Figure 2.
Impact of Slam locus deletion on NK cell repertoire. (A–D) NK cell repertoire was analyzed by flow cytometry of NK cells (CD3−NK1.1+) from the spleen (A and B) or bone marrow (C and D) of wild-type and SFR KO mice. Isotype control (ctrl) is shown as filled histograms. Statistical analyses of mean fluorescence intensity (MFI) or percentage of positive populations from A and C are shown for multiple mice in B and D. Each symbol represents an independent mouse. *, P < 0.05; **, P < 0.01; paired Student’s t test. Data are representative of n = 5 (A and B) or n = 4 (C and D).
Hence, deletion of the Slam locus in mice resulted in loss of all SFRs and CD48. It had no appreciable impact on NK cell development, although it had a small impact on NK cell maturation and repertoire.
NK cells from Slam locus–deleted mice display enhanced responses toward hematopoietic target cells in vitro
Activation of SFR KO NK cells in response to various target cells, either hematopoietic or nonhematopoietic, was examined. In parallel, we studied NK cells from mice lacking the SAP adaptor (Sh2d1a−/; SAP KO) or lacking the Slam locus in addition to SAP (SFR-SAP double KO [dKO]) to test the previously proposed idea that SAP KO NK cells have reduced cytotoxicity toward hematopoietic target cells because SFRs become superinhibitory (Parolini et al., 2000; Dong et al., 2009, 2012; Pérez-Quintero et al., 2014). Either IL-2–activated or IL-15–activated NK cells were used for these experiments.
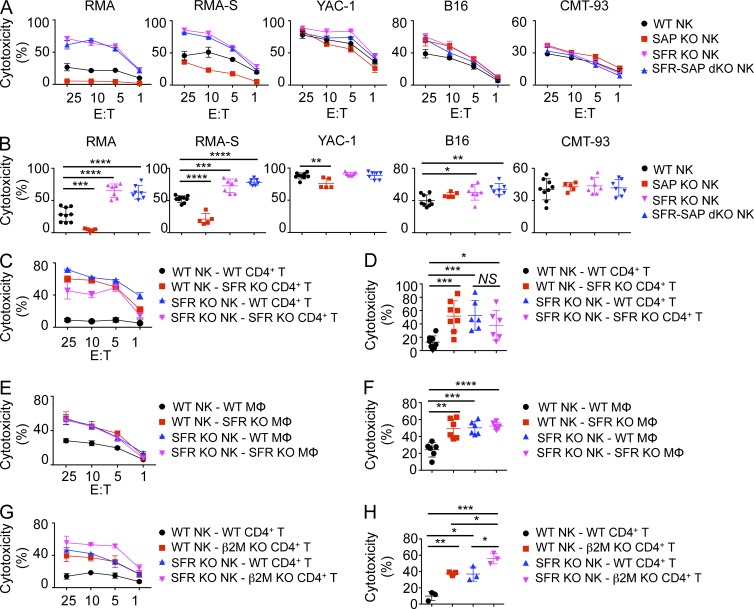
As reported previously (Dong et al., 2009, 2012; Pérez-Quintero et al., 2014), SAP KO NK cells exhibited compromised natural cytotoxicity toward hematopoietic target cells RMA (class I MHC–positive lymphoma), RMA-S (class I MHC–negative variant of RMA), and YAC-1 (class I MHC–negative thymoma), when compared with wild-type NK cells (Fig. 3, A and B). No defect was seen toward nonhematopoietic target cells B16 and CMT-93. In striking contrast, SFR KO NK cells displayed augmented killing of hematopoietic target cells, in comparison with wild-type NK cells. This effect was especially marked toward RMA and RMA-S but was also seen to a smaller extent toward YAC-1. A small increase in cytotoxicity was also seen toward B16 but not CMT-93. SFR-SAP dKO NK cells also displayed enhanced responses toward these various targets in a manner analogous to SFR KO NK cells.
Figure 3.
The SLAM family suppresses NK cell cytotoxicity toward hematopoietic target cells in vitro. (A) Natural cytotoxicity of IL-2–expanded NK cells purified from the spleen of wild-type, SFR KO, SAP KO, and SFR-SAP dKO mice was tested by 51Cr release assay using hematopoietic (RMA, RMA-S, and YAC-1) and nonhematopoietic (B16 and CMT-93) target cells at the indicated E/T ratio. Standard deviations of duplicates are shown by error bars. (B) Statistical analyses of results at the 25:1 E/T ratio for multiple independent experiments are shown. Each symbol represents an independent mouse. (A and B) Results are representative of at least five experiments (A) and are pooled from five experiments (B). (C and D) Natural cytotoxicity of IL-2–expanded NK cells from wild-type and SFR KO mice toward activated CD4+ T cells from wild-type or SFR KO mice was analyzed, as detailed for A and B. Each symbol represents an independent mouse. Results are representative of at least six experiments (C) and are pooled from six experiments (D). (E and F) Natural cytotoxicity of IL-2–expanded NK cells from wild-type and SFR KO mice toward bone marrow–derived macrophages (MΦ) from wild-type or SFR KO mice was tested, as detailed for A and B. Each symbol represents an independent mouse. Results are representative of six experiments (E) and are pooled from six experiments (F). (G and H) Natural cytotoxicity of IL-2–expanded NK cells from wild-type and SFR KO mice toward activated CD4+ T cells from wild-type or β2M KO mice was tested, as detailed for A and B. Each symbol represents an independent mouse. Results are representative of three experiments (G) and are pooled from three experiments (H). (C, E, and G) Mean values with standard deviations are depicted. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired Student’s t test.
All SFRs are self-ligands, with the exception of 2B4 which binds CD48 (Veillette, 2006, 2010; Calpe et al., 2008; Cannons et al., 2011). Therefore, to ensure that the increased responses of SFR KO NK cells were not caused by altered maturation or repertoire, similar experiments were conducted using NK cells, either wild type or SFR KO, incubated with activated CD4+ T cells or macrophages, which were either wild type or SFR KO. In keeping with the results obtained with hematopoietic cell lines (Fig. 3, A and B), SFR KO NK cells had enhanced cytotoxicity toward wild-type CD4+ T cells and macrophages when compared with wild-type NK cells (Fig. 3, C–F). Importantly, an increase in cytotoxicity was also seen when wild-type NK cells were activated by SFR KO CD4+ T cells and macrophages, in comparison with wild-type CD4+ T cells and macrophages. Similar results were obtained when SFR KO NK cells were used as effectors against SFR KO CD4+ T cells and macrophages. Of note, however, the ability of SFR KO NK cells to mediate cytotoxicity toward SFR KO CD4+ T cells was slightly lower than that of wild-type NK cells. However, this difference was not statistically significant.
Expression of class I MHC on targets suppresses NK cell activation by triggering inhibitory Ly49 or killer cell immunoglobulin-like receptors on NK cells. Thus, we tested the ability of SFR KO NK cells to kill CD4+ T cells deficient in class I MHC (β2 microglobulin [β2M] KO). Like wild-type NK cells, SFR KO NK cells showed increased killing of β2M KO CD4+ T cells, compared with wild-type CD4+ T cells (Fig. 3, G and H). The stimulatory effects of SFR deficiency in NK cells and class I MHC deficiency in targets on cytotoxicity were additive.
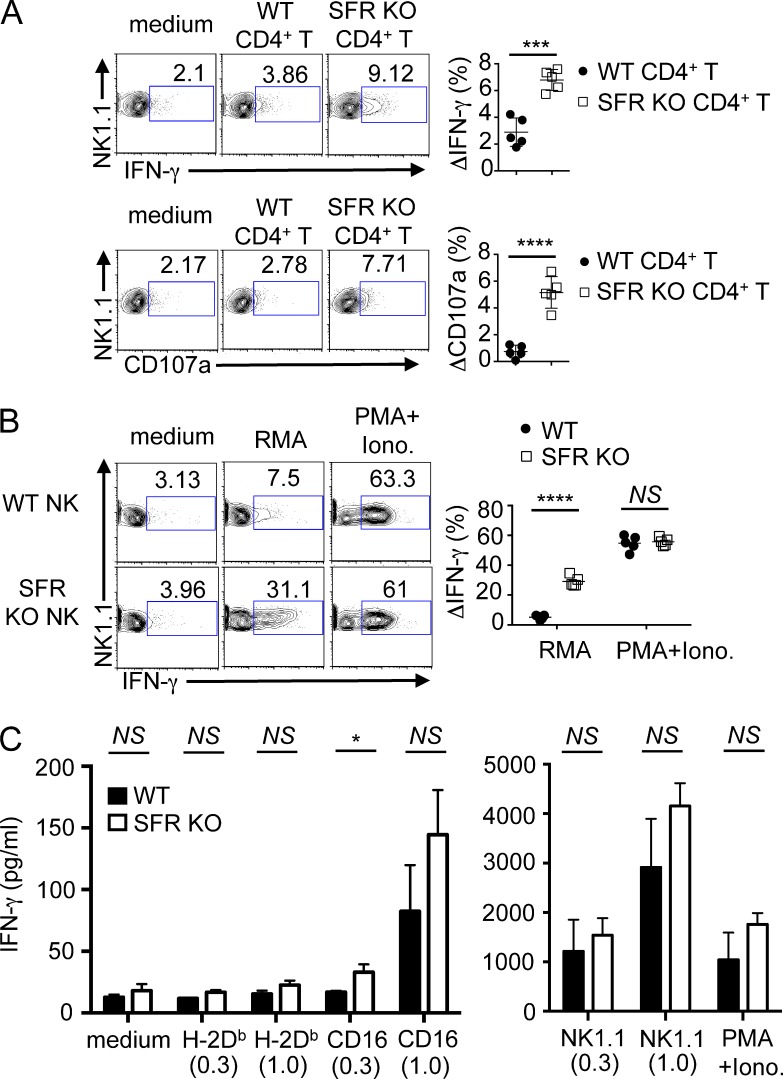
We also examined the impact of SFR deficiency on production of cytokines, in particular IFN-γ. Wild-type NK cells exhibited enhanced production of IFN-γ when stimulated with SFR KO CD4+ T cells, compared with wild-type CD4+ T cells (Fig. 4 A). A similar effect was seen on exposure of CD107a, a marker of degranulation. Moreover, compared with wild-type NK cells, SFR KO NK cells displayed increased IFN-γ production in response to RMA (Fig. 4 B). No difference was seen when cells were activated with PMA and ionomycin, which trigger cytokine production by bypassing activating receptors.
Figure 4.
The SLAM family suppresses NK cell–mediated cytokine production in response to hematopoietic target cells in vitro. (A) Splenocytes from wild-type mice were cultured with IL-2 for 1.5 d and then incubated for 4 h with medium alone, activated wild-type CD4+ T cells, or activated SFR KO CD4+ T cells. IFN-γ production was detected by intracellular staining (top), whereas degranulation was determined by surface staining of CD107a (bottom). NK cells were gated on CD3−NK1.1+. IFN-γ–producing or CD107a+ cells are boxed, and percentages are represented above each box. (Left) Representative experiments are shown. (Right) The increase in the percentage of IFN-γ–producing (ΔIFN-γ) or CD107a+ (ΔCD107a) cells between NK cells stimulated with medium alone or the indicated target for multiple independent mice is represented. Each symbol represents an independent mouse. Results are representative of five experiments. (B) Splenocytes from either wild-type or SFR KO mice were cultured in IL-2–containing medium for 1.5 d and then incubated with medium alone, RMA cells, or PMA + ionomycin (Iono.) for 4 h. IFN-γ production was detected by intracellular staining, as detailed for A. (Left) A representative experiment is shown. (Right) The increase in the percentage of IFN-γ–producing cells between NK cells stimulated with medium alone or the indicated stimulus for multiple independent mice is represented. Each symbol represents an independent mouse. Results are representative of five experiments. (C) Wild-type or SFR KO NK cells were expanded in IL-2 and stimulated for 24 h with the indicated concentrations (in µg/ml) of plate-bound antibodies against CD16, NKRp1c (NK1.1), or class I MHC (H-2Db). Cells were also stimulated with PMA and ionomycin. Secretion of IFN-γ was monitored by ELISA. Assays were done in duplicate and mean values of duplicates were used for statistical analyses. Data from three independent mice are included. Mean values for the three mice with standard deviations are depicted. Results are representative of two experiments with a total of five mice. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; unpaired Student’s t test.
We also tested the impact of SFR deficiency on NK cell responses triggered by activating receptors other than SFRs. Stimulation of NK cells with plate-bound antibodies against CD16 or NKRp1c (NK1.1) showed that SFR KO NK cells had a small (1.3- to 1.5-fold) increase in secretion of IFN-γ, compared with wild-type NK cells (Fig. 4 C). A similar effect was seen with PMA and ionomycin.
Thus, deletion of the Slam locus resulted in enhanced NK cell responses toward hematopoietic target cells, with little or no effect on nonhematopoietic target cells. This effect was seen whether the Slam locus was deleted in NK cells or in target cells. It was also seen whether target cells expressed or did not express class I MHC. Deletion of the Slam locus also restored the compromised responses of SAP KO NK cells toward hematopoietic target cells. The latter finding was in keeping with the idea that NK cell responses are compromised in SAP KO NK cells because SFRs become superinhibitory in the absence of SAP.
The inhibitory effect of the Slam locus is mediated by 2B4 with no appreciable impact from other SFRs or Gm10521
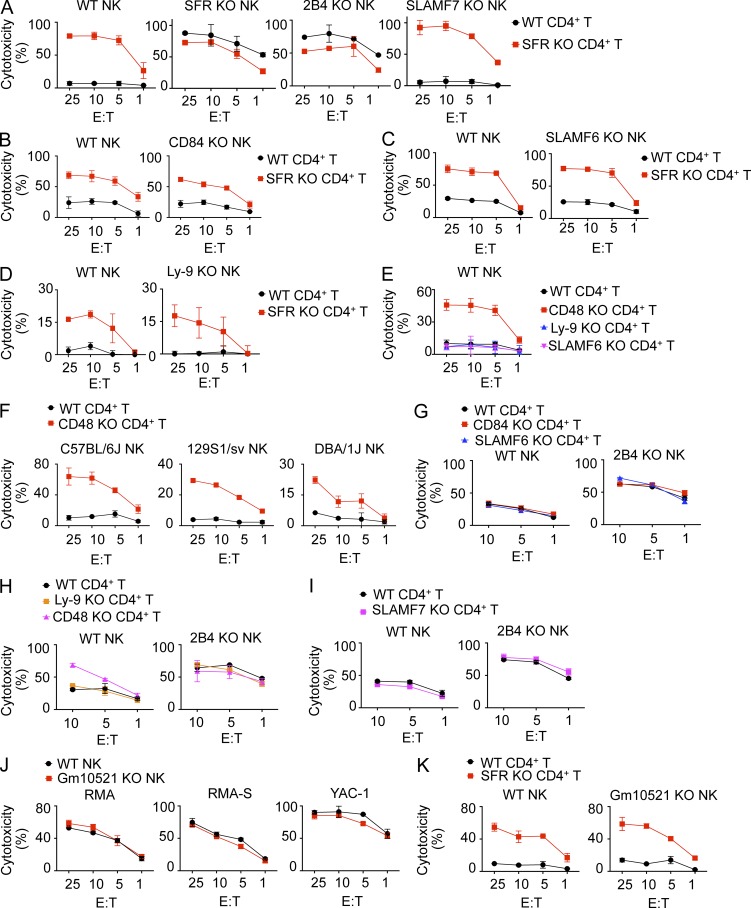
To ascertain the contributions of individual SFRs to the enhanced responses of SFR KO NK cells, the impact of removing single SFRs in NK cells was assessed. Like SFR KO NK cells, 2B4 KO NK cells displayed augmented killing of CD4+ T cells, in comparison with wild-type NK cells (Fig. 5 A). This effect was seen whether wild-type or SFR KO CD4+ T cells were used as targets. However, as shown in Fig. 3 (C and D) for SFR KO NK cells, this effect was slightly less toward SFR KO CD4+ T cells compared with wild-type CD4+ T cells. Little or no difference was seen with SLAMF7 KO, SLAMF6 KO, Ly-9 KO, or CD84 KO NK cells in comparison with wild-type NK cells (Fig. 5, A–D).
Figure 5.
The inhibitory effect of the Slam locus is mediated by 2B4 with no appreciable impact from other SLAM receptors or Gm10521. (A–D) Natural cytotoxicity of IL-2–expanded NK cells from wild-type, SFR KO, 2B4 KO, SLAMF7 KO (C57BL/6 background; A), CD84 KO (C57BL/6 background; B), SLAMF6 KO (C57BL/6 background; C), or Ly-9 KO (129S1/Sv background; D) mice was analyzed toward activated CD4+ T cells from either wild-type or SFR KO mice as targets, as detailed for Fig. 3 A. Results are representative of three (A), two (B and D), or four (C) experiments. (E) Natural cytotoxicity of IL-2–expanded NK cells from wild-type mice (C57BL/6) was tested against activated CD4+ T cells from wild-type, CD48 KO, Ly-9 KO, or SLAMF6 KO mice (129S1/Sv background), as detailed for Fig. 3 A. Results are representative of four experiments. (F) Natural cytotoxicity of IL-2–expanded NK cells from wild-type mice in C57BL/6, 129S1/Sv, or DBA/1J background was determined using activated CD4+ T cells from wild-type or CD48 KO mice as targets, as detailed for Fig. 3 A. Results are representative of three experiments. (G–I) Natural cytotoxicity of IL-15–expanded NK cells from either wild-type or 2B4 KO mice was analyzed against activated CD4+ T cells from wild-type, CD84 KO, SLAMF6 KO, Ly-9 KO, CD48 KO, or SLAMF7 KO mice, as detailed for Fig. 3 A. Results are representative of four (G), three (H), or two (I) experiments. (J and K) Natural cytotoxicity of IL-2–expanded NK cells from either wild-type or Gm10521 KO mice was tested at the indicated E/T ratio toward RMA, RMA-S, YAC-1, wild-type CD4+ T cells, and SFR KO CD4+ T cells. Results are representative of two experiments. Mean values of duplicates with standard deviations are depicted.
Similar experiments were conducted using wild-type NK cells and, as targets, CD4+ T cells from mice lacking individual SFRs or CD48. In keeping with the results described in the previous paragraph, wild-type NK cells displayed enhanced responses toward CD48 KO CD4+ T cells but not CD4+ T cells lacking Ly-9 or SLAMF6, compared with wild-type CD4+ T cells (Fig. 5 E). The increased responses toward CD48 KO CD4+ T cells were seen with wild-type NK cells from C57BL/6, 129S1/Sv, or DBA/1J mice (Fig. 5 F).
We also assessed whether SFRs other than 2B4 influenced the inhibitory function of 2B4. To this end, the ability of 2B4 KO NK cells to mediate cytotoxicity of CD4+ T cells lacking other SFRs was examined. The ability of 2B4 KO NK cells to kill Ly-9, SLAMF6, SLAMF7, or CD84 KO CD4+ T cells was neither increased nor decreased, compared with wild-type CD4+ T cells (Fig. 5, G–I).
Lastly, we examined the possible contribution of Gm10521 to the enhanced responses of SFR KO NK cells. For this purpose, we engineered Gm10521-disrupted KO mice using CRISPR-Cas–mediated gene editing, as detailed in the Mice section of Materials and methods. Gm10521-disrupted NK cells had no alteration of NK cell–mediated cytotoxicity toward RMA, RMA-S, YAC-1, or CD4+ T cells, compared with wild-type NK cells (Fig. 5, J and K).
Therefore, the inhibitory impact of the Slam locus on NK cell activation by hematopoietic target cells was solely mediated by 2B4. There was no appreciable contribution, either activating or inhibitory, from the other SFRs expressed in NK cells or Gm10521.
Mice lacking the Slam locus have augmented NK cell–mediated responses during lymphocytic choriomeningitis virus (LCMV) infection
A previous study showed that 2B4 KO mice displayed decreased numbers of virus-specific activated CD8+ T cells during infection by LCMV (Waggoner et al., 2010). These mice also had less extensive weight loss, which is believed to reflect less severe immunopathology, which can occur during LCMV infection. Importantly, these various effects were all dependent on NK cells and likely reflected an increased ability of cytokine-primed 2B4 KO NK cells to kill activated CD8+ T cells as well as activated CD4+ T cells that provide help to CD8+ T cells (Waggoner et al., 2011).
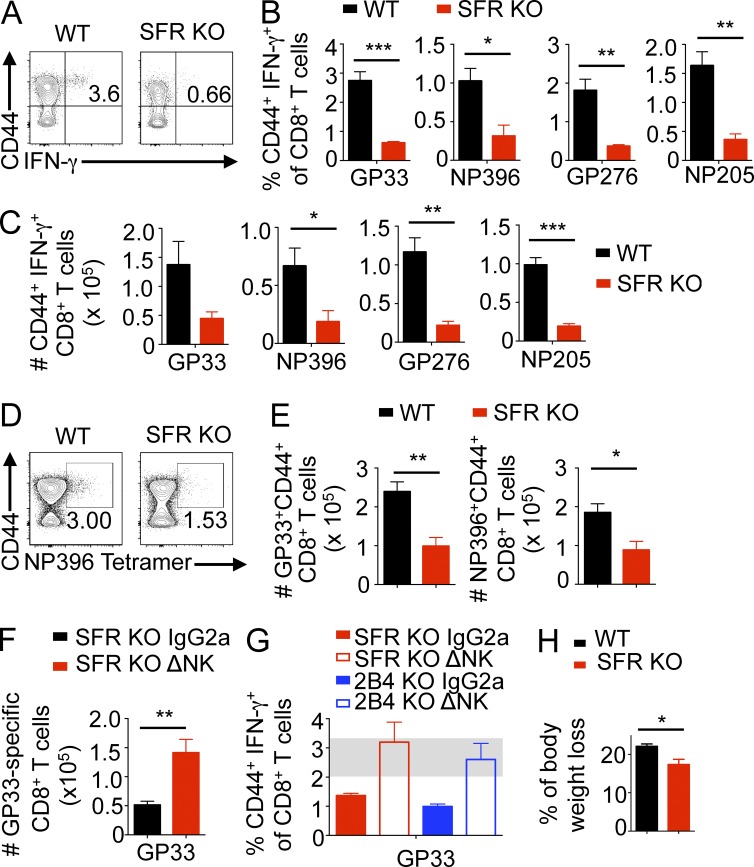
Thus, to address the role of the SLAM family in NK cell–mediated immune regulation in vivo, we examined the effect of SFR deficiency during infection by LCMV (clone 13). Mice were analyzed at day 8.5 after infection when control of virus-specific T cell responses by NK cells is near or at its peak. At day 8.5, SFR KO mice exhibited markedly reduced proportions and numbers of virus-specific IFN-γ–producing activated (CD44hi) CD8+ T cells in spleen, compared with wild-type mice (Fig. 6, A–C). This was shown with a variety of virus-derived antigenic peptides used in peptide restimulation assays (Fig. 6, B and C). Similar results were obtained when virus-specific activated CD8+ T cells were detected by staining splenocytes with viral peptide-loaded class I MHC tetramers (Fig. 6, D and E).
Figure 6.
Slam locus–deleted mice exhibit NK cell–dependent reduction of virus-specific T cell responses during LCMV infection. (A–C) Wild-type or SFR KO mice were infected with 2 × 106 PFU LCMV clone 13. After 8.5 d, splenic T cells were restimulated in vitro with the indicated LCMV peptides, and IFN-γ and CD44 expression by CD8+ T cells was monitored by flow cytometry. (A) A representative experiment using LCMV peptide GP33–41 is shown. Percentage of IFN-γ–producing CD44hi CD8+ T cells compared with total CD8+ T cells is shown in the top right quadrant. (B and C) The proportions (B) and total numbers (C) of LCMV GP33–41–, NP396–404–, GP276-286–, or NP205–212–responding CD44+ IFN-γ+ CD8+ T cells from four to five mice in each group are depicted. Numbers represent mean ± SEM. Results are representative of three independent experiments. (D and E) Data are as in A–C, except that virus-specific activated CD8+ T cells were detected by staining splenocytes with virus peptide-loaded class I MHC tetramers. (D) A representative experiment using NP396–404/Db tetramers is shown. Percentage of tetramer-binding CD44+ CD8+ T cells is indicated in the top right quadrant. (E) The total numbers of GP33–41/Db (left) or NP396–404/Db (right) tetramer-binding CD44+ CD8+ T cells are shown. Four to five mice were used per group. Results are representative of three independent experiments. (F) Data are as in A–C, except that SFR KO mice were injected with 25 µg of NK cell–depleting α-NKRp1c (NK1.1; ΔNK) antibody or IgG2a isotype control antibody 1 d before infection. Total numbers of GP33–41–specific CD44+ IFN-γ+ CD8+ T cells are shown. Seven to eight mice were used per group. Results are representative of two independent experiments. (G) Data are as in F, except that SFR KO and 2B4 KO mice were analyzed. Proportions of GP33–41–specific CD44+ IFN-γ+ CD8+ T cells are shown. Seven to eight mice were used per group. The gray bar represents the range for wild-type mice. Results are representative of two independent experiments. (H) Data are the same as A–C, except that the proportion of body weight loss between day −1 and day 8.5 of infection is depicted. Four to five mice per group were used. Results are representative of three independent experiments. Mean values from four to five mice (D, E, and H) or seven to eight mice (F and G) with standard deviations are depicted. *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test.
The reduction of virus-specific T cell responses in SFR KO mice, compared with wild-type mice, was markedly attenuated when mice were injected with anti-NKRp1c (NK1.1), which depletes NK cells (Fig. 6 F). This feature was analogous to that observed in 2B4 KO mice (Fig. 6 G), as previously reported (Waggoner et al., 2010). SFR KO mice also displayed statistically significant less weight loss, a feature of immunopathology, in comparison with wild-type mice (Fig. 6 H). In keeping with the results previously obtained with 2B4 KO mice (Waggoner et al., 2010), there was no difference in viral titers between wild-type and SFR KO mice at this time point (not depicted). Effects on viral control might become apparent at later time points, as suggested by the prior study of 2B4 KO mice (Waggoner et al., 2010).
Thus, SFR KO mice had reduced LCMV-specific CD8+ T cell responses in vivo. This effect was dependent on NK cells and resulted in less severe immunopathology. It was similar to the effect seen in 2B4 KO mice. These findings implied that, like the 2B4-encoding gene, the Slam locus had an inhibitory effect on NK cell–mediated control of activated T cell responses during LCMV infection in vivo.
The impact of the SLAM family requires NK cell priming by cytokines
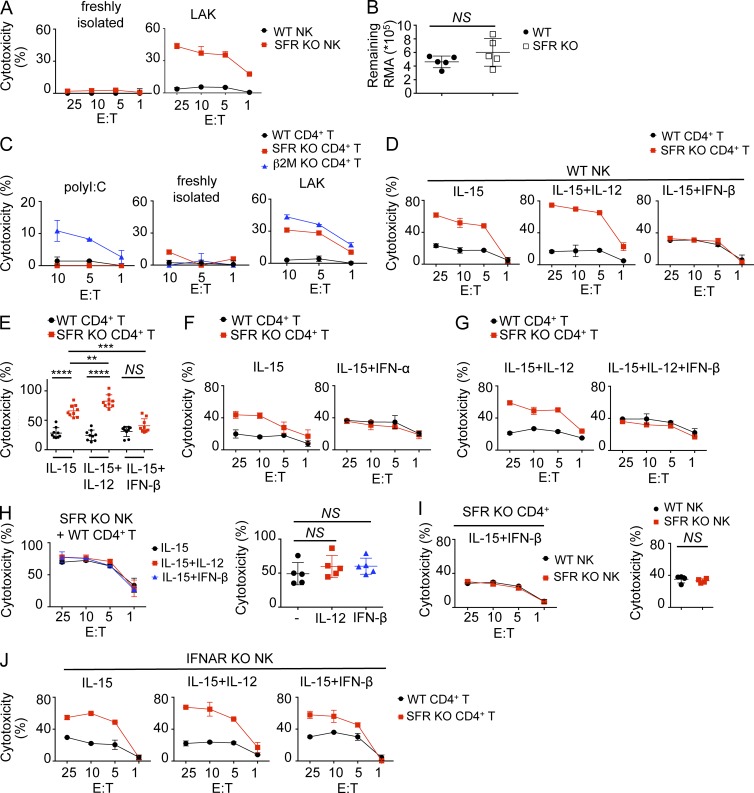
Multiple cytokines can influence NK cell functions, including IL-15, IL-12, IL-2, and type I IFN (Walzer et al., 2005; Cooper et al., 2009; Marçais et al., 2013). The experiments depicted in Fig. 3 were done using NK cells from nonprimed mice that were expanded in vitro either with IL-2 or with IL-15. To examine whether SFRs had an appreciable impact on NK cell functions in the absence of deliberate cytokine stimulation, similar experiments were conducted using freshly isolated NK cells that were not stimulated with any cytokine during the cytotoxicity assay. Unlike IL-2–activated SFR KO NK cells, freshly isolated SFR KO NK cells displayed no increase in cytotoxicity toward WT CD4+ T cells, compared with wild-type NK cells (Fig. 7 A). We also tested the ability of nonprimed mice to eliminate in vivo RMA, the tumor target cell line showing the largest increase in killing by cytokine-stimulated SFR KO NK cells in vitro (Fig. 3 A), using a peritoneal clearance assay. There was no difference in the ability of wild-type or SFR KO mice to eliminate RMA cells in this assay (Fig. 7 B). These observations suggested that at the steady state and in the absence of added cytokines, there was no detectable increase in NK cell responses toward hematopoietic target cells in the absence of SFRs. Thus, it appeared that the impact of the SLAM family on hematopoietic target cell killing required cytokine priming.
Figure 7.
Cytokines influence the functions of SLAM receptors. (A) Natural cytotoxicity of either freshly isolated or IL-2–expanded NK cells (lymphokine-activated killer [LAK]) from wild-type or SFR KO mice was tested toward activated CD4+ T cells from wild-type mice, as detailed for Fig. 3 A. Results are representative of at least three experiments. (B) The ability of nonprimed wild-type or SFR KO mice to eliminate RMA cells (106 cells injected) expressing GFP was evaluated in a peritoneal clearance assay. Residual RMA cells were counted 18 h after injection. Each symbol represents a different mouse. Unpaired Student’s t test was used. Results are representative of three experiments. (C) Wild-type mice were primed or not primed for 36 h with polyI:C. Cytotoxicity of freshly isolated NK cells toward activated CD4+ T cells from wild-type, SFR KO, or β2M KO mice was evaluated, as detailed for Fig. 3 A. IL-2–activated NK cells were analyzed in parallel as control. Results are representative of at least three experiments. (D and E) IL-15–expanded NK cells from wild-type mice were treated or not treated overnight with either IL-12 or IFN-β. (D) Natural cytotoxicity toward activated CD4+ T cells from wild-type or SFR KO mice was then examined, as detailed for Fig. 3 A. (E) Statistical analyses of results at the 10:1 E/T ratio from multiple independent experiments are shown. Results are representative of nine experiments (D) and are pooled from nine experiments (E). (B and E) **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired Student’s t test. (F) Data are as in D, except that IL-15–expanded NK cells from wild-type mice were treated or not treated with IFN-α. Results are representative of three experiments. (G) Data are as in D, except that IL-15–expanded NK cells from wild-type mice were treated with IL-12 alone or together with IFN-β. Results are representative of three experiments. (H, left) NK cells from SFR KO mice were expanded in IL-15 and stimulated overnight with or without IL-12 or IFN-β. Natural cytotoxicity toward wild-type CD4+ T cells was determined at the indicated E/T ratio. (Right) Statistical analyses of results at the 10:1 E/T ratio are shown. Unpaired Student’s t test was used. Results are representative of five experiments. (I) Data are as in D, except that NK cells from wild-type or SFR KO mice were expanded in IL-15 and stimulated overnight with IFN-β. (Left) Natural cytotoxicity toward CD4+ T cells from SFR KO mice was determined at the indicated E/T ratio. (Right) Statistical analyses of results at the 10:1 E/T ratio from several independent experiments are shown. Unpaired Student’s t test was used. Results are representative of four experiments. (J) Data are as in D, except that IL-15–expanded NK cells from IFNAR KO mice were treated or not treated with IL-12 or IFN-β. Results are representative of three experiments. (A, C, F, G, and J) Mean values of duplicates with standard deviations are depicted. (B, D, H, and I) Mean values with standard deviations are depicted.
Inhibition of NK cell responses by SLAM family is differentially influenced by IL-12 and type I IFN
We also performed similar experiments using NK cells isolated from mice treated with polyinosinic:polycytidylic acid (polyI:C), a TLR3 receptor agonist classically used to enhance NK cell functions in vivo (Beuneu et al., 2011). Surprisingly, freshly isolated NK cells from polyI:C-primed wild-type mice did not display an increase in cytotoxicity toward SFR KO CD4+ T cells, in comparison with wild-type CD4+ T cells (Fig. 7 C). However, they had increased cytotoxicity toward β2M KO CD4+ T cells. These findings implied that polyI:C enhanced NK cell responses toward targets lacking class I MHC but not toward targets lacking SFRs.
As polyI:C is a potent inducer of type I IFN (Beuneu et al., 2011), these findings raised the possibility that the impact of SFRs on NK cell activation was regulated by type I IFN. To address directly this possibility, wild-type NK cells were purified from nonprimed mice, expanded for 3 d in IL-15, and treated overnight with IL-15 alone, IL-15 plus IFN-β (a type I IFN), or IL-15 plus IL-12, another cytokine known to regulate NK cell functions. IL-15 is critical to enable survival of NK cells ex vivo. After extensive washing to remove the cytokines, NK cells were incubated with wild-type or SFR KO CD4+ T cells, and cytotoxicity was measured.
Wild-type NK cells treated with IL-15 demonstrated augmented killing toward SFR KO CD4+ T cells compared with wild-type CD4+ T cells (Fig. 7, D and E), in keeping with the results described in Fig. 3. This differential response was further enhanced when IL-12 was added (Fig. 7, D and E). Strikingly, however, it was markedly diminished when IFN-β was added. A similar effect was seen with IFN-α, another type I IFN (Fig. 7 F). Addition of both IL-12 and IFN-β had the same impact as addition of IFN-β alone (Fig. 7 G). IFN-β had no effect on the responses of SFR KO NK cells toward wild-type CD4+ T cells, suggesting that the impact of the cytokine was mediated by an effect on SFRs in NK cells (Fig. 7 H).
We examined the possibility that type I IFN suppressed the differential responses of NK cells toward SFR KO CD4+ T cells, compared with wild-type CD4+ T cells, by rendering SFRs more inhibitory in NK cells when targets lacked SFRs. This could result from increased cis-interactions between SFRs on NK cells. To this end, we compared the ability of SFR KO NK cells and wild-type NK cells to kill SFR KO CD4+ T cells. SFR KO NK cells and wild-type NK cells treated with IFN-β displayed equivalent cytotoxicity toward SFR KO CD4+ T cells (Fig. 7 I). This finding suggested that type I IFN was not rendering SFRs more inhibitory in NK cells activated by targets lacking SFRs.
To ensure that the effect of type I IFN was dependent on IFN receptor signaling, similar experiments were conducted using NK cells from IFN-α/β receptor 1 (IFNAR1) KO mice. IFNAR1 is a critical component of the type I IFN receptor, IFNAR. Compared with wild-type NK cells, IFNAR1 KO NK cells did not display the reduction of differential responses toward SFR KO CD4+ T cells, compared with wild-type CD4+ T cells, in the presence of IFN-β (Fig. 7 J). Loss of IFNAR1 had no impact on expression of SFRs on NK cells (not depicted).
Thus, stimulation with IL-12 further augmented the differential responses of WT NK cells toward targets lacking SFRs compared with targets expressing SFRs. The opposite effect was seen with type I IFN in a manner dependent on expression of IFNAR1.
Cytokines regulate expression of SAP adaptors
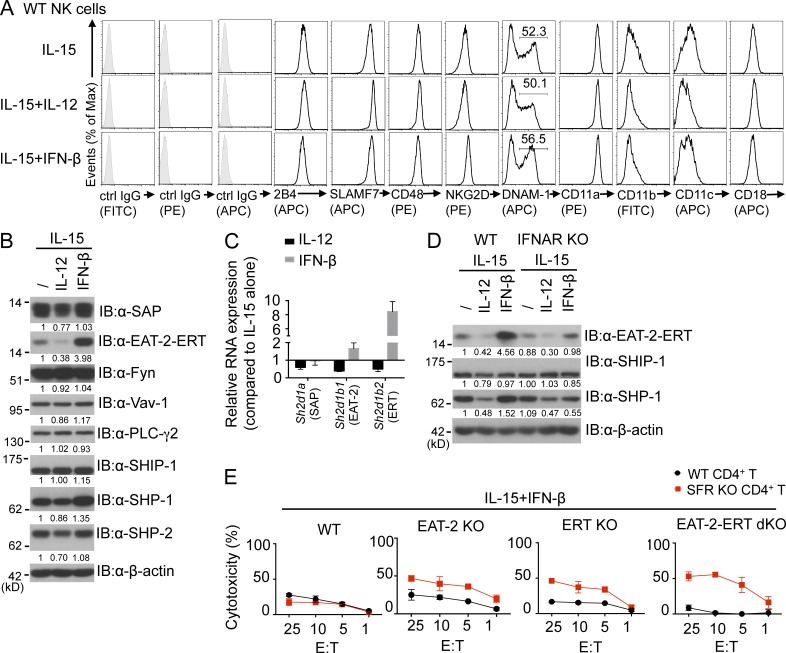
Several possible mechanisms, either direct or indirect, could explain the impact of the cytokines on the differential responses of NK cells toward targets lacking SFRs compared with targets expressing SFRs (see Discussion). To test the possibility that cytokines were directly influencing the function of SFRs, the effect of cytokines on expression of various molecules regulating SFR functions was first examined. Addition of IL-12 or IFN-β to IL-15–stimulated NK cells had little or no impact on the expression levels of SFRs when compared with cells treated with IL-15 alone (Fig. 8 A). However, IL-12 caused a reproducible decrease in the expression levels of SAP (by ∼25%) and EAT-2–ERT (by ∼60–70%; Fig. 8 B). As EAT-2 and ERT are very similar, the available antibodies do not distinguish between the two proteins (Pérez-Quintero et al., 2014). In contrast, IFN-β caused an increase (about fourfold) in expression of EAT-2–ERT, although it did not affect SAP.
Figure 8.
Evidence that cytokines influence SFR-mediated inhibition by regulating expression of SAP adaptors. (A) Expression of SLAM receptors, CD48, NKG2D, DNAM-1, and integrin subunits on IL-15–expanded wild-type NK cells treated with or without IL-12 or IFN-β was examined. NK cells were gated on TCR-β−NK1.1+ cells. Isotype control (ctrl) is shown in filled histograms. Results are representative of four experiments. (B) Expression of various positive or negative effectors of SLAM receptor signaling was analyzed by immunoblotting (IB) of total cell lysates with the indicated antibodies. IL-15–expanded NK cells treated or not treated with IL-12 or IFN-β were used. Numbers below each lane indicate levels of protein expression relative to those of cells cultured with IL-15 alone. Quantification was normalized to β-actin. Results are representative of seven experiments. (C) Data are the same as B, except that levels of RNA encoding SAP (Sh2d1a), EAT-2 (Sh2d1b1), and ERT (Sh2d1b2) were analyzed by quantitative real-time PCR. Levels were compared with those of cells expanded in IL-15 alone. Levels of Gapdh RNA were used for normalization. Results are representative of three experiments. (D) Data are the same as B, except that wild-type or IFNAR KO NK cells were analyzed. Results are representative of three experiments. (E) Data are the same as Fig. 7 D, except that NK cells from wild-type, EAT-2 KO, ERT KO, or EAT-2–ERT dKO mice expanded in IL-15 and IFN-β were used. Results are representative of three experiments. (C and E) Mean values of duplicates with standard deviations are depicted.
The changes in expression of SAP, EAT-2, and ERT were also shown at the RNA level using real-time PCR (Fig. 8 C). As PCR can distinguish the transcripts encoding EAT-2 and ERT, it also demonstrated that expression of both EAT-2– and ERT-encoding RNAs was affected by IL-12 and IFN-β. Of note, though, IFN-β had a more pronounced effect on expression of RNAs encoding ERT (about eightfold) compared with EAT-2 (about twofold). This may relate to the fact that ERT, but not EAT-2, is nearly absent in freshly isolated mouse NK cells (Pérez-Quintero et al., 2014).
IL-12 and IFN-β had little or no impact on other effectors of SFR signaling, including the kinase Fyn, the exchange factor Vav-1, phospholipase C–γ2, and the inhibitory effectors SHIP-1 and SHP-2 (Fig. 8 B). The only reproducible exception was SHP-1, which was decreased (by ∼15–50% depending on the experiment) by IL-12 and enhanced (∼1.5-fold) by IFN-β. There was little or no effect on expression of NKG2D, DNAM-1, LFA-1 (CD11a), CD18, and CD11b (Fig. 8 A).
To confirm that the increase in EAT-2–ERT expression in response to type I IFN was caused by IFNAR signaling, similar experiments were conducted using IFNAR1 KO NK cells. Unlike wild-type NK cells, IFNAR1 KO NK cells displayed little or no increase in EAT-2–ERT expression in response to IFN-β, compared with cells treated with IL-15 alone (Fig. 8 D). However, the decrease in EAT-2–ERT expression in response to IL-12 was still observed. The increase in SHP-1 expression in response to IFN-β was also abrogated.
As SAP adaptors prevent the inhibitory function of SFRs (Parolini et al., 2000; Dong et al., 2009, 2012; Pérez-Quintero et al., 2014), these results suggested that cytokines were acting at least in part by directly influencing the function of SFRs through changes in the expression levels of SAP adaptors. To address further this idea, we focused on IFN-β, which caused the most pronounced effect on SFR-mediated inhibition. Thus, the effect of IFN-β on differential NK cell responses toward SFR KO CD4+ T cells compared with wild-type CD4+ T cells was tested using NK cells from EAT-2 KO, ERT KO, or EAT-2–ERT dKO mice. Unlike wild-type NK cells, the EAT-2 KO, ERT KO, and EAT-2–ERT dKO NK cells displayed appreciable differences in NK cell responses toward SFR KO CD4+ T cells, compared with wild-type CD4+ T cells, in the presence of IFN-β (Fig. 8 E).
Therefore, treatment of NK cells with IFN-β or IL-12 resulted in changes in the expression levels of SAP adaptors. These effects might explain at least in part the impact of IFN-β on the differential outcome of NK cell activation by targets expressing or lacking SFRs.
Integrin LFA-1 is required for enhancement of NK cell responses by loss of SLAM family
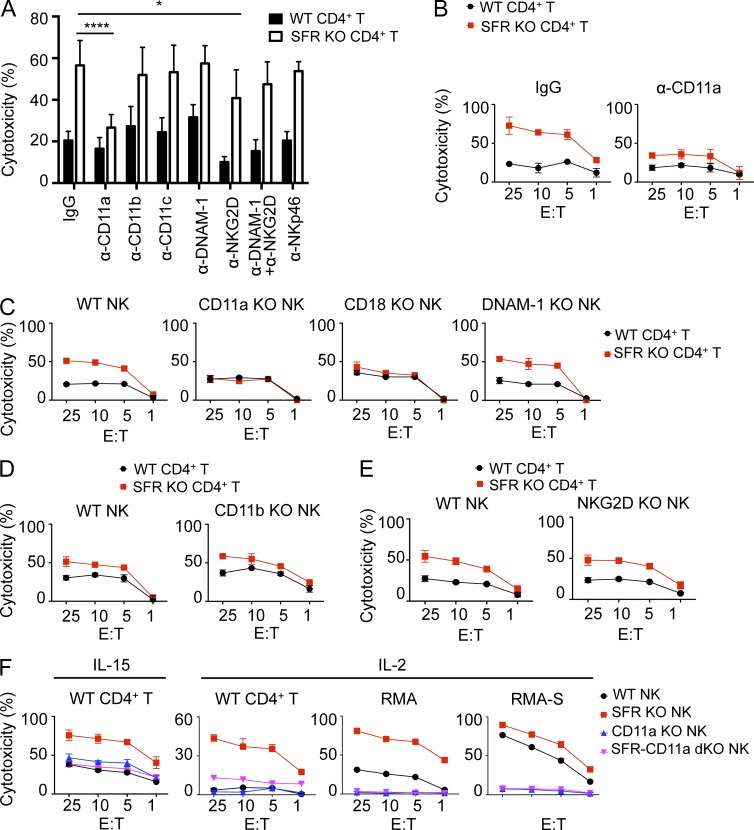
Based on analyses of SAP adaptor–deficient NK cells, we previously proposed that SFRs played a key role in the ability of NK cells to recognize and kill hematopoietic target cells (Dong et al., 2009). However, the results described in Figs. 3, 4, 5, 6, 7, and 8 clearly showed that this was not the case. Rather, SFRs inhibited this function. To identify the activating receptors mediating NK cell activation by hematopoietic target cells in the absence of SFRs, the impact of blocking antibodies toward various activating receptors was tested. Addition of blocking antibodies against NKG2D, DNAM-1, or NKp46 had little or no impact on the enhanced killing of wild-type NK cells toward SFR KO CD4+ T cells, compared with wild-type CD4+ T cells (Fig. 9 A). Similar effects were seen when the antibodies against NKG2D and DNAM-1 were combined. However, antibodies against the integrin LFA-1 (anti-CD11a) nearly abrogated the enhanced responses toward SFR KO CD4+ T cells (Fig. 9, A and B). No effect was seen when antibodies against other integrin components, CD11b and CD11c, were used.
Figure 9.
Integrin LFA-1, but not other activating receptors, is required for enhancement of NK cell responses upon loss of SLAM receptors. (A) Data are as in Fig. 7 D, except that natural cytotoxicity of IL-15–expanded wild-type NK cells toward activated CD4+ T cells from wild-type or SFR KO mice was tested at the 10:1 E/T ratio in the presence or the absence of various blocking antibodies during incubation (final concentration of 10 µg/ml). *, P < 0.05; ****, P < 0.0001; unpaired Student’s t test. Results are pooled from six experiments. (B) Data are as in A, except that natural cytotoxicity of IL-15–expanded wild-type NK cells toward activated CD4+ T cells from wild-type or SFR KO mice was tested at the indicated E/T ratio in the presence of blocking anti-CD11a antibody or control IgG antibody. Results are representative of six experiments. (C–E) Data are as in Fig. 7 D, except that natural cytotoxicity of IL-15–expanded NK cells from wild-type, CD11a KO, CD18 KO, DNAM-1 KO, CD11b KO, and NKG2D KO mice was determined. Results are representative of three experiments. (F) Data are as in Fig. 7 D, except that natural cytotoxicity of IL-15–expanded or IL-2–expanded NK cells from wild-type, SFR KO, CD11a KO, or SFR-CD11a dKO mice toward activated CD4+ T cells from wild-type mice, RMA, or RMA-S was determined. Results are representative of three experiments. Mean values of duplicates with standard deviations are depicted.
To validate these observations, the function of NK cells from mice lacking activating receptors was examined. Unlike wild-type NK cells, the CD11a KO NK cells, but not DNAM-1 KO NK cells, CD11b KO NK cells, or NKG2D KO NK cells, failed to demonstrate augmented cytotoxicity toward SFR KO CD4+ T cells, compared with wild-type CD4+ T cells (Fig. 9, C–E). A similar defect was seen with CD18 KO NK cells, which lack expression of β2 integrin (CD18), another component of LFA-1 (Fig. 9 C). CD11a KO NK cells displayed no alteration of expression of SFRs or other activating receptors (not depicted).
We also examined the contribution of LFA-1 to the enhanced ability of SFR KO NK cells to kill hematopoietic cells. To this end, SFR KO mice were bred with CD11a KO mice to generate SFR-CD11a dKO mice. Compared with SFR KO NK cells, SFR-CD11a dKO NK cells had a lower capacity to kill wild-type CD4+ T cells (Fig. 9 F). This was seen whether IL-15–stimulated or IL-2–stimulated NK cells were used. Similar results were obtained when RMA or RMA-S cells were used as targets (Fig. 9 F).
Together, these data implied that LFA-1 but not DNAM-1, NKG2D, NKp46, or other integrins played a key role in the enhanced ability of NK cells lacking SLAM receptors to kill hematopoietic target cells.
Evidence that 2B4 inhibits induction of the activated form of LFA-1
These results raised the possibility that SFRs suppressed NK cell activation at least partly by inhibiting the function of LFA-1, either directly or indirectly. LFA-1 has a dual role in NK cell activation (Barber et al., 2004; Gross et al., 2010; Zhang et al., 2014). On the one hand, it functions as an adhesion molecule that enhances conjugate formation between NK cells and target cells. On the other hand, it triggers activating signals that promote NK cell effector responses. The activating function of LFA-1 is dependent on a modification of its conformation, which becomes activated and can be assessed by conformation-sensitive anti–LFA-1 antibodies.
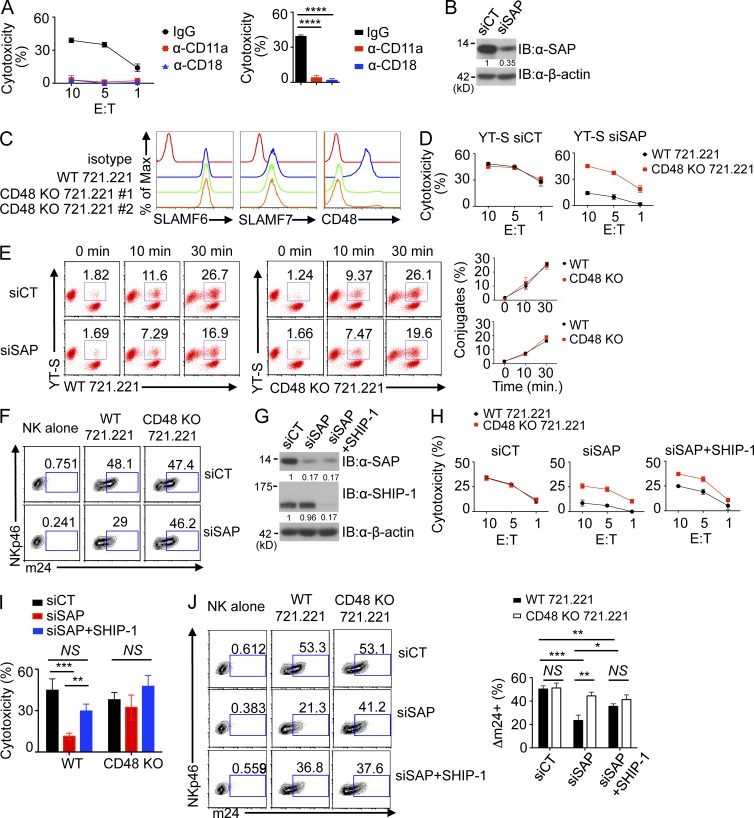
It is nearly impossible to monitor activation of LFA-1 in mice because of lack of suitable reagents, but there are available antibodies that recognize the activated conformation of human LFA-1. Thus, to test the impact of SFRs on LFA-1 function, we developed a LFA-1–dependent human NK cell activation system using the human NK cell line YT-S, which expresses 2B4 and SAP, and the human B cell target 721.221, which expresses CD48. The ability of YT-S cells to kill 721.221 was blocked by anti–LFA-1 (CD11a) or anti-CD18 antibodies, implying that it is LFA-1 dependent (Fig. 10 A).
Figure 10.
2B4-mediated inhibition results in decreased activation of LFA-1. (A, left) The ability of the human NK cell line YT-S to mediate cytotoxicity of the B cell line 721.221 was assessed at the indicated E/T ratio in the presence of blocking antibodies against CD11a (LFA-1) or CD18 or control IgG. Standard deviations of duplicates are shown by error bars. (Right) Statistical analyses of results at the 10:1 E/T ratio for three independent experiments are shown. ****, P < 0.0001. (B) Expression of SAP in YT-S cells transfected with scrambled siRNA (control [siCT]) or siRNAs against SAP (siSAP) was analyzed by immunoblot (IB) analysis. Results are representative of four experiments. (C) Expression of SLAMF6, SLAMF7, and CD48 was analyzed on parental (WT) or CD48 KO 721.221 cells. Two different targeting sequences were used to generate two different CD48 KO derivatives of 721.221 through CRISPR-Cas–mediated genome editing. Isotype control is shown in red. Results are representative of three experiments. (D) Natural cytotoxicity of YT-S cells transfected with control siRNAs (left) or siRNAs against SAP (right) was analyzed toward wild-type or CD48 KO 721.221 cells at the indicated E/T ratio. For CD48 KO 721.221 cells, a pool of CD48 KO 721.221 no. 1 and no. 2 cells was used. Mean values of duplicates with standard deviations are depicted. Results are representative of four experiments. (E) YT-S cells transfected with control or SAP-specific siRNAs were labeled with Brilliant violet 421–conjugated anti–human NKp46 and then incubated for the indicated times at 37°C with parental (WT) or CD48-deficient 721.221 cells labeled with PE-conjugated anti–human CD19. Conjugate formation was assessed by flow cytometry. A representative experiment is depicted on the left, whereas a statistical analysis of duplicates with mean values and standard deviations is shown on the right. Data are representative of three experiments. (F) YT-S cells transfected with control siRNAs or SAP-specific siRNAs were incubated with medium alone, wild-type 721.221, or CD48 KO 721.221 for 5 min at 37°C. After stimulation, cells were stained with anti-CD11a conformational antibody (m24) for 30 min at room temperature, followed by anti-NKp46 staining for 15 min at room temperature. YT-S cells were gated on NKp46+ cells and analyzed for m24 staining. Results are representative of three experiments. (G) Expression of SAP and SHIP-1 in YT-S cells transfected with control siRNA, siRNAs against SAP, or SHIP-1 was analyzed by immunoblot analysis. β-actin was used as the control. Results are representative of three experiments. (H) Natural cytotoxicity of YT-S cells transfected with the siRNAs detailed in G was determined at the indicated E/T ratio toward wild-type or CD48 KO 721.221 cells. (I) Statistical analyses of the results at the 10:1 E/T ratio from multiple independent experiments are shown. (J, left) YT-S cells transfected with the siRNAs detailed in G were incubated with medium alone, WT 721.221, or CD48 KO 721.221 and stained with the anti-CD11a conformational antibody (m24), as outlined for F. (Right) Statistical analyses of the results from multiple independent experiments are shown. (H–J) *, P < 0.05; **, P < 0.01; ***, P < 0.001; unpaired Student’s t test. Mean values with standard deviations are depicted. Results are representative of four experiments.
To favor 2B4-mediated inhibition in YT-S cells, SAP-deficient (SAP knockdown [KD]) YT-S cells were generated using SAP-specific siRNAs (Fig. 10 B). Moreover, to address the impact of 2B4-mediated inhibition in these cells, 721.221 derivatives lacking CD48 were created using CRISPR-Cas genome editing (Fig. 10 C). Two independent CD48 KO 721.221 derivatives were generated using two unrelated guide RNA targeting sequences. Control siRNA-transduced YT-S showed little or no difference in cytotoxicity toward parental and CD48 KO 721.221 (Fig. 10 D). This finding suggested that, in control YT-S cells exposed to 721.221, 2B4 was neither inhibitory nor activating. However, SAP KD YT-S displayed reduced cytotoxicity toward parental 721.221, compared with CD48 KO 721.221, in keeping with the inhibitory effect of 2B4 (Fig. 10 D). Hence, this system was suitable to test the impact of 2B4-mediated inhibition on LFA-1 function.
We first wanted to ascertain whether 2B4-mediated inhibition resulted in diminished conjugate formation, a function dependent on LFA-1–mediated adhesion. As reported for SAP KO mouse NK cells (Dong et al., 2012), SAP KD YT-S cells displayed reduced conjugate formation with 721.221 compared with control YT-S cells (Fig. 10 E). However, this effect was of similar magnitude toward parental and CD48 KO 721.221 cells. The latter finding implied that 2B4 was not responsible for the reduced conjugate formation of SAP KD YT-S cells. Presumably, one or more other SFRs expressed in YT-S cells were responsible. To address whether 2B4-mediated inhibition interfered with activation of LFA-1, LFA-1 activation was monitored using an antibody (m24) recognizing the activated conformation of LFA-1. SAP KD YT-S had reduced LFA-1 activation in response to parental 721.221, compared with CD48 KO 721.221 (Fig. 10 F).
To understand the mechanism by which 2B4-mediated inhibition suppressed LFA-1 activation, similar experiments were conducted using SAP KD YT-S cells, in which expression of the lipid phosphatase SHIP-1, which was reported to mediate the inhibitory function of 2B4 (Dong et al., 2012), was suppressed by siRNAs (Fig. 10 G). Down-regulation of expression of SHIP-1 corrected the lower cytotoxicity of SAP KD YT-S cells toward parental 721.221, compared with CD48 KO 721.221 (Fig. 10, H and I). Importantly, it also ameliorated the defect in LFA-1 activation (Fig. 10 J). Similar effects were seen with two other SHIP-1–specific siRNAs (not depicted).
These data implied that 2B4-mediated inhibition did not result in reduced conjugate formation with targets. However, it caused a diminution of the activation of LFA-1. The latter inhibitory effect was mediated by SHIP-1.
DISCUSSION
Herein, we created a mouse in the C57BL/6 background bearing a deletion of the entire Slam locus, which includes the genes coding for all six SFRs and CD48, the ligand of 2B4 (Calpe et al., 2008; Veillette, 2010; Cannons et al., 2011). NK cells isolated from SFR KO mice displayed markedly enhanced activation responses toward hematopoietic target cells, including tumor cells such as RMA and RMA-S, and normal hematopoietic cells, such as activated CD4+ T cells and macrophages, in vitro. The increase in activation responses was seen toward either class I MHC–negative or class I MHC–positive targets. Both cytotoxicity and IFN-γ production were augmented. Likewise, SFR KO mice infected with LCMV had decreased numbers of LCMV-specific activated CD8+ T cells and less severe immunopathology compared with wild-type mice. These effects were attenuated by depletion of NK cells, suggesting that they were caused by enhanced killing of activated T cells by NK cells.
Although deletion of the Slam locus had a detectable, albeit small, impact on NK cell maturation and repertoire, it is unlikely that these changes were responsible for the enhanced responses of SFR KO NK cells. Indeed, an analogous increase in NK cell activation was seen when wild-type NK cells were incubated with SFR KO CD4+ T cells or macrophages. However, it should also be noted that a small increase in cytotoxicity toward the nonhematopoietic target cell B16 melanoma was seen. B16 lacks ligands for SFRs (Dong et al., 2009). Similarly, a small increase in IFN-γ secretion was seen when SFR KO NK cells were stimulated through CD16 or NKRp1c or with PMA and ionomycin. These various effects may be caused by altered NK cell maturation, repertoire, or education. Additional studies will be needed to address this possibility.
Analyses of mice lacking individual SFRs showed that only 2B4 KO NK cells displayed augmented responses toward hematopoietic target cells in vitro. Conversely, only CD48 KO CD4+ T cells exhibited enhanced killing by wild-type NK cells. To ascertain the possibility that other SFRs attenuated or enhanced the impact of loss of 2B4, we examined activation of 2B4 KO NK cells by CD4+ T cells lacking the other SFRs. Loss of other SFRs on targets had no influence on the enhanced responses of 2B4 KO NK cells. Thus, no SFR other than 2B4 had any appreciable effect, either inhibitory or activating, on NK cell activation. This notion was also in agreement with the observation that the reduction of LCMV-induced CD8+ T cell responses in SFR KO mice was analogous to that noted in 2B4 KO mice (Waggoner et al., 2010).
Although SFRs other than 2B4 had no appreciable impact on NK cell activation, it is conceivable that some have alternative functions in NK cells. Perhaps, they regulate NK cell proliferation, survival, memory, or migration. These possibilities deserve future consideration. It should be mentioned that we previously showed that another SFR, SLAMF6, plays a critical role in NK cell education, thereby promoting NK cell responses toward nonhematopoietic cells (Wu et al., 2016). Whereas this effect was seen in 129S1/Sv and DBA/1J mice (and in human NK cells), it was not observed in C57BL/6 mice. Hence, it is not surprising that it was not seen in the current study.
Previous studies showed that loss of SAP adaptors strongly diminished the ability of NK cells to kill hematopoietic target cells, both in mice and in humans (Parolini et al., 2000; Dong et al., 2009, 2012; Pérez-Quintero et al., 2014). Although the basis for this effect was not formally established, it was proposed to result from an increase in the inhibitory function of SFRs in the absence of SAP adaptors, which prevent inhibition by SFRs both by interfering with coupling of SFRs to inhibitory effectors and by mediating activating signals (Parolini et al., 2000; Dong et al., 2009, 2012; Pérez-Quintero et al., 2014). The availability of an SFR KO mouse enabled us to test this idea. We found that breeding of SFR KO mice with SAP KO mice strongly improved the compromised NK cell responses of SAP KO NK cells to the same level as that observed in SFR KO NK cells. This observation implied that the reduced responses of SAP adaptor–deficient NK cells toward hematopoietic target cells are most probably caused by an increase in the inhibitory function of SFRs in the absence of SAP adaptors.
Whereas studies of NK cells lacking SAP adaptors had also implied that SFRs plays key roles in the ability of NK cells to eliminate hematopoietic target cells (Parolini et al., 2000; Calpe et al., 2008; Orr and Lanier, 2010; Pérez-Quintero et al., 2014), the findings with SFR KO NK cells reported herein showed that this is not the case. Interestingly, experiments with blocking antibodies revealed that LFA-1 was critical for the enhanced responses of NK cells toward SFR KO CD4+ T cells. The pivotal role of LFA-1 was confirmed by analyses of NK cells from CD11a KO and CD18 KO mice. As hematopoietic cells express high levels of ICAM-1 and ICAM-2, the ligands of LFA-1, it is plausible that LFA-1 plays a central function in NK cell–mediated responses against hematopoietic target cells.
Using the human NK cell line YT-S and the B cell target 721.221 expressing or not expressing CD48, we obtained evidence that 2B4-mediated inhibition was accompanied by decreased induction of the activated form of LFA-1, as suggested by reactivity with the anti–LFA-1 mAb m24. This was mediated at least in part by the lipid phosphatase SHIP-1. However, 2B4-mediated inhibition had no impact on conjugate formation between NK cells and target cells, implying that the decreased activation of LFA-1 did not compromise conjugate formation. Nonetheless, it should be pointed out that activation of LFA-1 is also implicated in LFA-1–dependent signaling leading to effector functions. Hence, it is plausible that 2B4 was compromising effector functions by inhibiting the signaling potential of LFA-1. Such an effect could be caused by a direct impact of 2B4 on LFA-1. Alternatively, it could reflect the influence of 2B4 on one or more other activating receptors that regulate LFA-1, although DNAM-1, NKG2D, and NKp46 did not appear to be responsible. Future studies will be needed to resolve these various possibilities.
Although NK cells stimulated in vitro with cytokines such as IL-15, IL-12, and IL-2 displayed enhanced activation responses in the absence of engagement of SFRs by hematopoietic cell targets, no obvious increase was seen when freshly isolated NK cells were analyzed. Similarly, SFR KO mice did not display any increase in elimination of RMA cells in a peritoneal clearance assay. In contrast, SFR KO mice had a better NK cell–dependent suppression of virus-specific T cell responses during LCMV infection. Coupled with the finding that SFR KO mice did not have any reduction of the steady-state numbers of T cells and B cells, these observations implied that immune stimulation by cytokines or viral infection was needed to observe increased activation responses in the absence of SFRs.
Interestingly, however, NK cells isolated from wild-type mice injected with polyI:C, another immune stimulant, failed to display enhanced responses toward SFR KO CD4+ T cells, compared with wild-type CD4+ T cells. This was not caused by lack of adequate priming by polyI:C, as these NK cells had increased responses toward β2M KO CD4+ T cells. Treatment of NK cells in vitro with type I IFN, either IFN-α or IFN-β, dramatically attenuated the increased responses of wild-type NK cells toward targets lacking SFRs compared with targets expressing SFRs. This effect was dependent on IFNAR1.
The impact of type I IFN could be caused by several effects that directly or indirectly influenced the function of SFRs. Our results suggested that the influence of type I IFN was mediated at least in part by a decrease in the inhibitory function of SFRs when engaged by SFRs on targets. This would be caused by increased expression of EAT-2 and ERT. However, it is also possible that type I IFN altered the impact of SFRs when they were engaged in cis on NK cells, in particular if SFRs were lacking on targets. This could lead to changes in NK cell education or to some other form of preconditioning of NK cell functions. This possibility might seem less likely though, given that SFR KO NK cells and wild-type NK cells treated with type I IFN displayed similar cytotoxicity toward SFR KO CD4+ T cells. Lastly, it is conceivable that cytokines influenced the function of the activating receptors repressed by SFRs or that of inhibitory receptors cooperating with SFRs to suppress NK cell activation. Additional studies will be needed to address these various possibilities.
In summary, we found that the Slam locus in mice has an overall inhibitory role in NK cell activation by hematopoietic target cells. This inhibitory effect is mediated by 2B4, with no appreciable impact from any other SFR expressed in NK cells. It is influenced by cytokines such as IL-12, IL-15, and type I IFN, perhaps because of a direct effect of cytokines on SFR functions, although additional mechanisms may be involved. In addition to enhancing our understanding of the role and mechanism of action of the SLAM family in NK cell activation, these findings imply that the impact of SFRs in NK cells is likely to be dynamically regulated in vivo, depending on the cytokine milieu. This may be especially important in the context of immune stimulation by viruses and other pathogens.
So far, an activating role of SFRs in NK cell activation has been demonstrated primarily in experiments in which SFR ligands were ectopically expressed on nonhematopoietic target cells or in which SFRs on NK cells were triggered with anti-SFR antibodies (Wu and Veillette, 2016). This is true both for mouse NK cells and for human NK cells. Whereas the data herein firmly establish the overall inhibitory role of SFRs in NK cell activation against hematopoietic target cells in mice, it is possible that SFRs are activating in mouse NK cells under physiological conditions. For instance, this may occur when levels of SFR ligands on hematopoietic target cells or the amounts of SAP adaptors in NK cells are increased. As a corollary to our findings, it is also plausible that, under physiological circumstances of stimulation, 2B4 has an inhibitory role in human NK cells.
MATERIALS AND METHODS
Mice
To generate a mouse lacking the entire Slam locus (ΔSlam), the following constructs and strategy were used. The first construct contained a 5′ arm bearing part of intron 1 and a 3′ arm bearing exons 4–6 of the 2B4-encoding gene Slamf4. Slamf4 is the most 5′ gene in the Slam locus. These arms flanked a single loxP site and the neo gene. This construct was transfected in C57BL/6-derived Bruce 4 embryonic stem cells, and transfected cells were selected with G418. Cells with successful homologous recombination of Slamf4 were then used for blastocyst injection to generate a 2B4-deficient (Slamf4−/−) mouse. They were also transfected with a second construct that contained a 5′ arm bearing exons 4–6 and a 3′ arm bearing exon 8 of Slamf6. Slamf6 is the most 3′ gene in the Slam locus. These arms flanked a single loxP site and the hygro gene. Transfected cells were selected with hygromycin. Cells with homologous recombination in both Slamf4 and Slamf6 were then transiently transfected with the pIC-Cre plasmid (a gift from H. Gu, Institut de recherches cliniques de Montréal, Montréal, Québec, Canada). This resulted in deletion of the entire Slam locus, except for exon 1 of Slamf4 and exons 8 and 9 of Slamf6. One loxP site and the hygro gene remained. Clones were screened by PCR to detect deletion of the Slam locus. Deletion of the Slam locus was confirmed by genomic sequencing. Cells were then injected into blastocysts to generate chimeric mice. After germline transmission, mice were backcrossed for 6–10 generations with C57BL/6J mice (The Jackson Laboratory). Deletion of the Slam locus was confirmed by genomic sequencing of mouse DNA. Gm10521-disrupted mice were generated by CRISPR-Cas–mediated genome editing, using the plasmid pSpCas9(BB) (formerly pX330; Addgene) and the guide RNA sequence 5′-TATACCAGGGCACCTGTACCTGG-3′. DNA was injected in fertilized oocytes of C57BL/6J mice. Mice were then screened by PCR and sequencing of the Gm10521 locus. Mice bearing a frameshift deletion of five nucleotides at the 5′ end of the potential coding sequence of Gm10521 locus were bred to homozygosity. Mice lacking Ly-9 (Slamf3−/−), SLAMF6 (Slamf6−/−), CD48 (Slamf2−/−), or EAT-2–ERT (Sh2b1b1−/−Sh2d1b2−/−) in the 129S1/Sv background were described previously (Roncagalli et al., 2005; Wu et al., 2016). Mice lacking β2M (B2m−/−), CD84 (Slamf5−/−), or SLAMF6 (Slamf6−/−) in the C57BL/6 background were obtained from The Jackson Laboratory. The 2B4-deficient mouse (Slamf4−/−) used for LCMV infection experiments was previously reported (Vaidya et al., 2005). Mice lacking SLAMF7 (Slamf7−/−) in the C57BL/6 background were generated by L.-A. Pérez-Quintero and A. Veillette (Institut de recherches cliniques de Montréal) by Cre-mediated deletion of Slamf7 exon 2 (which was flanked by loxP sites and contains the initiating ATG) in the mouse germ line. Mice lacking LFA-1 (CD11a; Itgal−/−), CD18 (Itgb2−/−), CD11b (Itgam−/−), or IFNAR1 (Ifnar1−/−) were obtained from The Jackson Laboratory. Mice lacking SAP (Sh2d1a−/), DNAM-1 (Cd226−/−), or NKG2D (Klrk1−/−) were provided by L. Yin (International Agency for Research on Cancer, Lyon, France), M. Colonna (Washington University, St. Louis, MO), and D. Raulet (University of California, Berkeley, Berkeley, CA), respectively (Gilfillan et al., 2008; Guerra et al., 2008; Dong et al., 2012). All mice were maintained in the C57BL/6J background, unless specified. They were also kept in a specific pathogen-free environment. Littermates were used as controls in all experiments. Animal experimentation was approved by the Animal Care Committee of Institut de recherches cliniques de Montréal and performed as defined by the Canadian Council of Animal Care or by the Institutional Animal Use and Care Committees of Cincinnati Children's Hospital Medical Center.
Cells
Spleen NK cells were purified by positive or negative selection (STEMCELL Technologies). In some cases, mice were injected i.p. with 250 µg polyI:C (Sigma-Aldrich) 24–36 h before purification. After purification, NK cells were either used immediately for experimentation or cultured for 3 d with IL-15 (catalog no. 210-15; PeproTech), followed or not followed by addition of IL-12 (catalog no. 210-12; PeproTech), IFN-α (catalog no. 12100-1; PBL Assay Science), or IFN-β (catalog no. 12405-1; PBL Assay Science) overnight. Alternatively, they were expanded in 1,000 U/ml IL-2 for 5 d. Cells treated with cytokines were washed in culture medium at least three times before the functional assays to eliminate carry over cytokines. RMA and RMA-S (mouse lymphoma), YAC-1 (mouse thymoma), B16F10 (mouse melanoma), CMT-93 (mouse rectal carcinoma), and YT-S (human NK cell line) have been previously described (Dong et al., 2009, 2012; Pérez-Quintero et al., 2014; Wu et al., 2016). Typically, NK cell purity was >90%. 721.221 (EBV-transformed human B cell line) was obtained from the International Histocompatibility Working Group. Variants lacking CD48 were generated by CRISPR-Cas–mediated gene editing using the plasmid pSpCas9(BB)-2A-GFP (Addgene) and the sequence 5′-TCACTTGGTACATATGACCGTGG-3′ or 5′-GTACATATGACCGTGGTCTCCGG-3′ as guide RNA. Activated CD4+ T cells and bone marrow–derived macrophages were generated as described previously (Dong et al., 2009; Rhee et al., 2013). In brief, activated CD4+ T cells were produced by stimulation of purified splenic CD4+ T cells in the presence of plate-bound hamster anti-CD3 mAb 145-2C11 and soluble hamster anti-CD28 mAb 37.51 for 2 d, followed by expansion in 50 U/ml of IL-2–containing medium for 2 d.
siRNAs
Previously validated siRNAs against human SAP and SHIP-1 as well as scrambled siRNAs were purchased from QIAGEN (Wu et al., 2016). They were transfected in YT-S cells using the AMAXA human NK cell Nucleofector kit (Lonza), as previously described (Wu et al., 2016). The following siRNAs were used: control siRNA, AllStars Negative Control siRNA (1027280); SAP-specific siRNAs, Hs_SH2D1A_4 (SI00036568) and Hs_SH2D1A_5 (SI03039897); and SHIP-1–specific siRNAs, Hs_INPP5D_1 (SI00078575), Hs_INPP5D_2 (SI00078582), and Hs_INPP5D_5 (SI03029005).
Antibodies
mAbs recognizing SFRs (except 2B4), SAP, and EAT-2–ERT were produced in our laboratory, as described previously (Roncagalli et al., 2005; Dong et al., 2009). mAbs against CD3 (145-2C11), NK1.1 (PK136), CD11b (M1/70), CD27 (LG 7F9), IFN-γ (XMG1.2), CD107a (1D4B), Ly49A/D (11A8), Ly49G2 (LGL-1), Ly49C/I/F/H (14B11), CD122 (TMb1), DNAM-1 (10E5; 480.2; for blocking antibody), NKG2A/C/E (20d5), NKG2D (CX5), CD11a (M17/4), CD11c (N418), CD18 (M18/2), 2B4 (244F4), CD48 (HM48-1), CD49b (DX5), KLRG1 (2F1), NKp46 (29A1.4), Rae-1 (CX1), and matched isotype controls were from eBioscience. Antibodies against human NKp46 (9E2), human SLAMF6 (NT-7), human SLAMF7 (162.1), human CD48 (BJ40), human CD19 (HIB19), and human CD18 (TS1/18) were from BioLegend. Antibodies against activated human LFA-1 (m24) were obtained from BioLegend. Antibodies against human CD11a (TS1/22) were from Thermo Fisher Scientific. Rabbit polyclonal antibodies against Fyn, Vav-1, phospholipase C–γ2, SHIP-1, SHP-1, and SHP-2 were described previously (Dong et al., 2009, 2012; Pérez-Quintero et al., 2014; Wu et al., 2016). Antibodies against β-actin (C4) were from Santa Cruz Biotechnology, Inc.
In vitro NK cell assays
51Cr release assays were performed as outlined previously using either mouse NK cells or YT-S cells (Dong et al., 2009, 2012; Pérez-Quintero et al., 2014; Wu et al., 2016). Antibody-blocking experiments were also previously described (Wu et al., 2016). For measurement of IFN-γ production and CD107a exposure, 106 splenocytes were stimulated with IL-2 for 1.5 d and incubated for 4 h with 106 activated CD4+ T cells or 106 RMA cells. Assays were then performed as previously described (Dong et al., 2012; Wu et al., 2016). Splenocytes stimulated with 50 ng/ml PMA plus 1 µM ionomycin served as controls. Antibody-induced secretion of IFN-γ was monitored as detailed previously (Roncagalli et al., 2005). Conjugate formation assays were performed as described previously (Dong et al., 2012). To measure LFA-1 activation, YT-S cells were stimulated with 721.221 cells for 5 min at 37°C. Cells were then stained with anti–LFA-1 mAb m24 followed by anti-NKp46 to identify YT-S cells. Immunoblots and real-time PCR assays to detect expression of RNA encoding SAP adaptors were performed as previously described (Dong et al., 2009, 2012; Pérez-Quintero et al., 2014; Wu et al., 2016).
LCMV infection
The clone 13 strain of LCMV was propagated in BHK21 cells (Welsh et al., 1976) and titrated by plaque assay on Vero cells from ATCC. SFR KO and their wild-type littermate controls or 2B4 KO mice (Vaidya et al., 2005) were inoculated with 2 × 106 PFU LCMV clone 13 via intravenous tail vein injection. They were then analyzed on day 8.5 after infection, as previously described (Waggoner et al., 2010, 2011). Mice were weighed the day before infection and the day of harvest. For experiments involving in vivo NK cell depletion, depletion was achieved via one i.p. injection of 25 µg α-NKRp1c (NK1.1) mAb PK136 per mouse, whereas control mice received one i.p. injection of 25 µg of mouse IgG2a isotype control antibody (C1.18A) produced by Bio X Cell (Waggoner et al., 2011). Synthetic peptides representing LCMV-encoded epitopes were purchased from 21st Century Biochemicals. The peptides used for in vitro stimulation and analysis of cytokine release were as follows: Db-presented LCMV GP33–41 (KAVYNFATC), LCMV GP276–286 (SGVENPGGYCL), and LCMV NP396–404 (FQPQNGQFI) and Kb-presented LCMV NP205–212 (YTVKYPNI). LCMV GP33–41 (KAVYNFATM)– and NP396–404 (FQPQNGQFI)–loaded H-2Db class I MHC tetramers were produced at the National Institutes of Health Tetramer Facility. Class I tetramer staining was performed at 4°C for 60 min.
Statistical analyses
Paired or unpaired two-tailed Student’s t tests were performed using Prism 6 (GraphPad Software).
ACKNOWLEDGMENTS
We thank Zhongjun Dong (Institut de recherches cliniques de Montréal; present address: Tsinghua University, Beijing, China) for making the constructs used for targeting of the Slam locus. We also acknowledge the National Institutes of Health Tetramer Core Facility (contract HHSN272201300006C) for provision of MHC tetramers.
This work was supported by grants from the Canadian Institutes of Health Research (MOP-82906 and FDN-143338) and the Canadian Cancer Society Research Institute (018114) to A. Veillette. Additional funding came from the Ellison Medical Foundation (grant AG-NS-0959-12), Cincinnati Children’s Research Foundation, and the National Institutes of Health (grant DA038017 to S.N. Waggoner). S.A. Cranert is an Arnold Strauss Fellow and is supported by funds from CancerFree KIDS. N. Wu was a recipient of a fellowship from Fonds de la recherché du Québec – Santé (award no. 29163). A. Veillette holds the Canada Research Chair in Signaling in the Immune System.
The authors declare no competing financial interests.
Author contributions: H. Guo planned experiments, performed experiments, interpreted data, and wrote the manuscript. Y. Lu, S. Zhang, and M.-C. Zhong generated and characterized reagents, performed experiments, and interpreted data. J. Chen, R. Li, N. Wu, and D. Davidson characterized reagents and interpreted data. S.A. Cranert and S.E. Mahl planned experiments, performed experiments, and interpreted data. S.N. Waggoner planned experiments, interpreted data, and provided funding. A. Veillette planned experiments, generated reagents, interpreted data, wrote the manuscript, and obtained funding.
Footnotes
Abbreviations used:
- β2M
- β2 microglobulin
- dKO
- double KO
- ERT
- EAT-2–related transducer
- KD
- knockdown
- LCMV
- lymphocytic choriomeningitis virus
- polyI:C
- polyinosinic:polycytidylic acid
- SAP
- SLAM-associated protein
- SFR
- SLAM family receptor
- SH2
- Src homology 2
- SLAM
- signaling lymphocytic activation molecule
References
- Barber D.F., Faure M., and Long E.O.. 2004. LFA-1 contributes an early signal for NK cell cytotoxicity. J. Immunol. 173:3653–3659. 10.4049/jimmunol.173.6.3653 [DOI] [PubMed] [Google Scholar]
- Beuneu H., Deguine J., Bouvier I., Di Santo J.P., Albert M.L., and Bousso P.. 2011. Cutting edge: A dual role for type I IFNs during polyinosinic-polycytidylic acid-induced NK cell activation. J. Immunol. 187:2084–2088. 10.4049/jimmunol.1004210 [DOI] [PubMed] [Google Scholar]
- Calpe S., Wang N., Romero X., Berger S.B., Lanyi A., Engel P., and Terhorst C.. 2008. The SLAM and SAP gene families control innate and adaptive immune responses. Adv. Immunol. 97:177–250. 10.1016/S0065-2776(08)00004-7 [DOI] [PubMed] [Google Scholar]
- Cannons J.L., Tangye S.G., and Schwartzberg P.L.. 2011. SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol. 29:665–705. 10.1146/annurev-immunol-030409-101302 [DOI] [PubMed] [Google Scholar]
- Cooper M.A., Elliott J.M., Keyel P.A., Yang L., Carrero J.A., and Yokoyama W.M.. 2009. Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. USA. 106:1915–1919. 10.1073/pnas.0813192106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Cruz-Munoz M.E., Zhong M.C., Chen R., Latour S., and Veillette A.. 2009. Essential function for SAP family adaptors in the surveillance of hematopoietic cells by natural killer cells. Nat. Immunol. 10:973–980. 10.1038/ni.1763 [DOI] [PubMed] [Google Scholar]
- Dong Z., Davidson D., Pérez-Quintero L.A., Kurosaki T., Swat W., and Veillette A.. 2012. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity. 36:974–985. 10.1016/j.immuni.2012.03.023 [DOI] [PubMed] [Google Scholar]
- Elliott J.M., and Yokoyama W.M.. 2011. Unifying concepts of MHC-dependent natural killer cell education. Trends Immunol. 32:364–372. 10.1016/j.it.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S., and Raulet D.H.. 2006. Activation and self-tolerance of natural killer cells. Immunol. Rev. 214:130–142. 10.1111/j.1600-065X.2006.00460.x [DOI] [PubMed] [Google Scholar]
- Gilfillan S., Chan C.J., Cella M., Haynes N.M., Rapaport A.S., Boles K.S., Andrews D.M., Smyth M.J., and Colonna M.. 2008. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J. Exp. Med. 205:2965–2973. 10.1084/jem.20081752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross C.C., Brzostowski J.A., Liu D., and Long E.O.. 2010. Tethering of intercellular adhesion molecule on target cells is required for LFA-1–dependent NK cell adhesion and granule polarization. J. Immunol. 185:2918–2926. 10.4049/jimmunol.1000761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra N., Tan Y.X., Joncker N.T., Choy A., Gallardo F., Xiong N., Knoblaugh S., Cado D., Greenberg N.M., and Raulet D.H.. 2008. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 28:571–580. 10.1016/j.immuni.2008.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R., Cannons J.L., Zhao F., Yusuf I., Lao C., Locci M., Schwartzberg P.L., and Crotty S.. 2012. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 36:986–1002. 10.1016/j.immuni.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier L.L. 2005. NK cell recognition. Annu. Rev. Immunol. 23:225–274. 10.1146/annurev.immunol.23.021704.115526 [DOI] [PubMed] [Google Scholar]
- Lee K.M., McNerney M.E., Stepp S.E., Mathew P.A., Schatzle J.D., Bennett M., and Kumar V.. 2004. 2B4 acts as a non–major histocompatibility complex binding inhibitory receptor on mouse natural killer cells. J. Exp. Med. 199:1245–1254. 10.1084/jem.20031989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais A., Viel S., Grau M., Henry T., Marvel J., and Walzer T.. 2013. Regulation of mouse NK cell development and function by cytokines. Front. Immunol. 4:450 10.3389/fimmu.2013.00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli E., Ugolini S., and Vivier E.. 2013. Tuning the threshold of natural killer cell responses. Curr. Opin. Immunol. 25:53–58. 10.1016/j.coi.2012.11.005 [DOI] [PubMed] [Google Scholar]
- Orr M.T., and Lanier L.L.. 2010. Natural killer cell education and tolerance. Cell. 142:847–856. 10.1016/j.cell.2010.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolini S., Bottino C., Falco M., Augugliaro R., Giliani S., Franceschini R., Ochs H.D., Wolf H., Bonnefoy J.Y., Biassoni R., et al. 2000. X-linked lymphoproliferative disease. 2B4 molecules displaying inhibitory rather than activating function are responsible for the inability of natural killer cells to kill Epstein-Barr virus–infected cells. J. Exp. Med. 192:337–346. 10.1084/jem.192.3.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Quintero L.A., Roncagalli R., Guo H., Latour S., Davidson D., and Veillette A.. 2014. EAT-2, a SAP-like adaptor, controls NK cell activation through phospholipase Cγ, Ca++, and Erk, leading to granule polarization. J. Exp. Med. 211:727–742. 10.1084/jem.20132038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I., Davidson D., Souza C.M., Vacher J., and Veillette A.. 2013. Macrophage fusion is controlled by the cytoplasmic protein tyrosine phosphatase PTP-PEST/PTPN12. Mol. Cell. Biol. 33:2458–2469. 10.1128/MCB.00197-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roncagalli R., Taylor J.E., Zhang S., Shi X., Chen R., Cruz-Munoz M.E., Yin L., Latour S., and Veillette A.. 2005. Negative regulation of natural killer cell function by EAT-2, a SAP-related adaptor. Nat. Immunol. 6:1002–1010. 10.1038/ni1242 [DOI] [PubMed] [Google Scholar]
- Tangye S.G. 2014. XLP: clinical features and molecular etiology due to mutations in SH2D1A encoding SAP. J. Clin. Immunol. 34:772–779. 10.1007/s10875-014-0083-7 [DOI] [PubMed] [Google Scholar]
- Vaidya S.V., Stepp S.E., McNerney M.E., Lee J.K., Bennett M., Lee K.M., Stewart C.L., Kumar V., and Mathew P.A.. 2005. Targeted disruption of the 2B4 gene in mice reveals an in vivo role of 2B4 (CD244) in the rejection of B16 melanoma cells. J. Immunol. 174:800–807. 10.4049/jimmunol.174.2.800 [DOI] [PubMed] [Google Scholar]
- Veillette A. 2006. NK cell regulation by SLAM family receptors and SAP-related adapters. Immunol. Rev. 214:22–34. 10.1111/j.1600-065X.2006.00453.x [DOI] [PubMed] [Google Scholar]
- Veillette A. 2010. SLAM-family receptors: immune regulators with or without SAP-family adaptors. Cold Spring Harb. Perspect. Biol. 2:a002469 10.1101/cshperspect.a002469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veillette A., Pérez-Quintero L.A., and Latour S.. 2013. X-linked lymphoproliferative syndromes and related autosomal recessive disorders. Curr. Opin. Allergy Clin. Immunol. 13:614–622. 10.1097/ACI.0000000000000008 [DOI] [PubMed] [Google Scholar]
- Waggoner S.N., and Kumar V.. 2012. Evolving role of 2B4/CD244 in T and NK cell responses during virus infection. Front. Immunol. 3:377 10.3389/fimmu.2012.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner S.N., Taniguchi R.T., Mathew P.A., Kumar V., and Welsh R.M.. 2010. Absence of mouse 2B4 promotes NK cell-mediated killing of activated CD8+ T cells, leading to prolonged viral persistence and altered pathogenesis. J. Clin. Invest. 120:1925–1938. 10.1172/JCI41264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner S.N., Cornberg M., Selin L.K., and Welsh R.M.. 2011. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 481:394–398. 10.1038/nature10624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner S.N., Reighard S.D., Gyurova I.E., Cranert S.A., Mahl S.E., Karmele E.P., McNally J.P., Moran M.T., Brooks T.R., Yaqoob F., and Rydyznski C.E.. 2016. Roles of natural killer cells in antiviral immunity. Curr. Opin. Virol. 16:15–23. 10.1016/j.coviro.2015.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer T., Dalod M., Robbins S.H., Zitvogel L., and Vivier E.. 2005. Natural-killer cells and dendritic cells: “l’union fait la force”. Blood. 106:2252–2258. 10.1182/blood-2005-03-1154 [DOI] [PubMed] [Google Scholar]
- Welsh R.M. Jr., Lampert P.W., Burner P.A., and Oldstone M.B.. 1976. Antibody-complement interactions with purified lymphocytic choriomeningitis virus. Virology. 73:59–71. 10.1016/0042-6822(76)90060-X [DOI] [PubMed] [Google Scholar]
- Wu N., and Veillette A.. 2016. SLAM family receptors in normal immunity and immune pathologies. Curr. Opin. Immunol. 38:45–51. 10.1016/j.coi.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Wu N., Zhong M.C., Roncagalli R., Pérez-Quintero L.A., Guo H., Zhang Z., Lenoir C., Dong Z., Latour S., and Veillette A.. 2016. A hematopoietic cell-driven mechanism involving SLAMF6 receptor, SAP adaptors and SHP-1 phosphatase regulates NK cell education. Nat. Immunol. 17:387–396. 10.1038/ni.3369 [DOI] [PubMed] [Google Scholar]
- Zhang M., March M.E., Lane W.S., and Long E.O.. 2014. A signaling network stimulated by β2 integrin promotes the polarization of lytic granules in cytotoxic cells. Sci. Signal. 7:ra96 10.1126/scisignal.2005629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Cannons J.L., Dutta M., Griffiths G.M., and Schwartzberg P.L.. 2012. Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis. Immunity. 36:1003–1016. 10.1016/j.immuni.2012.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]