Abstract
Catalase is a tetrameric heme-containing enzyme with essential antioxidant functions in biology. Multiple factors including nitric oxide (NO) have been shown to attenuate its activity. However, the possible impact of NO in relation to the maturation of active catalase, including its heme acquisition and tetramer formation, has not been investigated. We found that NO attenuates heme insertion into catalase in both short-term and long-term incubations. The NO inhibition in catalase heme incorporation was associated with defective oligomerization of catalase, such that inactive catalase monomers and dimers accumulated in place of the mature tetrameric enzyme. We also found that GAPDH plays a key role in mediating these NO effects on the structure and activity of catalase. Moreover, the NO sensitivity of catalase maturation could be altered up or down by manipulating the cellular expression level or activity of thioredoxin-1, a known protein-SNO denitrosylase enzyme. In a mouse model of allergic inflammatory asthma, we found that lungs from allergen-challenged mice contained a greater percentage of dimeric catalase relative to tetrameric catalase in the unchallenged control, suggesting that the mechanisms described here are in play in the allergic asthma model. Together, our study shows how maturation of active catalase can be influenced by NO, S-nitrosylated GAPDH, and thioredoxin-1, and how maturation may become compromised in inflammatory conditions such as asthma.
Keywords: catalase, nitric oxide, heme, thioredoxin-1, S-nitrosylated GAPDH, asthma, oligomerization
Introduction
Catalase (EC 1.11.1.6) is a heme containing enzyme capable of destroying high levels of H2O2 produced inside the cells under oxidative stress (1,2). Mammalian catalases are typically homotetramers that bind 4 Fe-protoporphyrin IX (heme) and 2 to 4 NADPH molecules (1-4). The heme groups are deeply buried in the structure using tyrosine, histidine and asparagine (Y415, H75 and N201) as ligands, and thin channels provide H2O2 access to heme and allow release of product(4,5). In addition to H2O2 dismutase activity, catalase also acts as peroxidase. Both the H2O2 decomposition and peroxidase activities of catalase are heme dependent (1,6-8) and these two activities correlate with each other (6) with a few exceptions (9,10).
Cellular mechanisms that influence catalase maturation are not very clear. The presence of apo-catalase or intermediates of catalase maturation have been reported (11-13). In rat liver apo-catalase is present in 3 fold excess (1.6% compared to 0.5%) of holo-monomeric catalase (13). Lack of heme can increase the pool of apo-catalase and may promote its turnover (14,15). Exposure to oxidative stress can inactivate catalase (16-19), but whether this affects its structure is not completely understood.
Nitric oxide (NO) is an important signal and effector molecule in biology, and is generated in animals by the NO synthases (NOSs) (20). NO can competitively inhibit catalase by direct binding to its heme or limiting its heme content (21-25). NO can also inhibit catalase at both translational as well as post-translational level (25-28).
Our previous work showed that NO can inhibit heme insertion into a broad range of cytosolic heme proteins, including apo-catalase (23). Follow-up studies on the inducible NOS (iNOS) showed: (i) buildup of S-nitrosated glyceraldehyde-3-phosphate dehydrogenase (SNO-GAPDH) was required for NO to inhibit iNOS heme insertion(29), and (ii) the sensitivity of iNOS heme insertion toward NO inhibition could be modulated by altering the cellular thioredoxin-1 (Trx-1) activity, due to Trx-1 controlling the level of SNO-GAPDH in the cell (30). Although this information shed light on how NO can control iNOS maturation, it is still unclear if NO inhibition of heme insertion into other hemeproteins follows these principles, or if it may occur naturally in animals during inflammation. To address these questions, we investigated if NO inhibition of catalase heme maturation is regulated by SNO-GAPDH and Trx-1 in cells, and also looked for evidence of an impact on catalase maturation in lungs from mice that were treated to develop an inflammatory asthma, as a model of chronic inflammation. Our results suggest that the impact of NO on catalase heme insertion is under the control of SNO-GAPDH and Trx1 activity, and may impact catalase maturation both in cultured cells and in the inflamed lung.
Experimental Procedures
Reagents
All chemicals were purchased from Sigma (St Louis, MO) unless stated otherwise. Antibodies were purchased from following companies: mouse anti-GAPDH from Fitzgerald (Fitzgerald Industries Inc., MA, USA), rabbit polyclonal anti-Trx-1 from Cell signaling (Cell Signaling Technology Inc., MA, USA) and anti-Catalase from Sigma (Sigma Aldrich, USA). NOC18 was bought from Alexis Biochemicals (San Diego, USA). HA-tagged wild type and C152S-GAPDH genes in pRK5 expression vector were gifts from Solomon H. Snyder (John Hopkins University School of Medicine, MD).
Cell culture
HEK293T (HEK) cells (ATCC, Rockville, MD) were cultured as described elsewhere(31). Cells were transfected with 4μg of his-tagged catalase expression vector using lipofectamine (Life Technologies Inc.) and incubated for 24h at 37°C and 5% CO2 in a humidified incubator. For generation of apo-catalase, cells were pre-incubated with 250μM of Succinyl Acetone (SA) for 2 days. For heme insertion assay, heme was added directly to the media containing cycloheximide for 3h in presence or absence of NOC18 or deprenyl. Cells were returned to incubator (37°C and 5% CO2) for the desired amount of time. Cells were then quickly washed with PBS and immediately frozen on a sheet of dry ice for 5min. Later, cells were thawed on ice and supernatants were prepared as described before (29). Lentiviral particles over-expressing Trx-1 were generated and used as described previously(30).
Smooth Muscle Cell isolation and culture
BALB/c mice were purchased from Jackson Labs. All mice were maintained at the Cleveland Clinic Lerner Research Institute Biological Research unit in a temperature controlled facility with an automatic 12-hour light-dark cycle and were given free access to food and water. All animal protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the Cleveland Clinic. Murine airway SMCs were obtained enzymatically from excised tracheas of BALB/c mice. Briefly, the tracheas were carefully cleaned under a dissecting microscope, opened longitudinally and digested overnight at 4°C with 0.15% Pronase (Roche, Indianapolis, IN) in HAM's F-12 medium (Gibco) with 2X antibiotic/antimycotic (Gibco) to remove the epithelial cells. The remaining tissue containing the SMCs was washed carefully with Dulbecco's modified Eagle's/Ham's F12 (DMEM/F12) medium containing an antiobiotic/antimycotic, cut into small (<1 mm) pieces and digested in serum-free DMEM/F12 with 0.1% type 4 collagenase from Clostridium histolyticum (Worthington Biochemical Corporation, Lakewood, NJ), 0.05% porcine pancreatic elastase (Worthington Biochemical Corporation) and antibiotic/antimycotic for up to 6 hours at 37°C. Following digestion, the cells were thoroughly rinsed and were grown in DMEM/F12 medium with HEPES, 10% fetal bovine serum (FBS) (Bio-Whitaker, Walkersville, MD), and an antibiotic/antimycotic mixture (complete medium). All cultures were maintained at 37°C in 95% air and 5% CO2 and split at a maximal ratio of 1:3. Passages 2-3 were employed in these studies.
Biotin switch assay
The biotin switch assay was performed as mentioned(32) and pull-downs from NeutrAvidin™ resin (Pierce, IL) were probed by standard western blotting.
Catalytic activity assay
Catalase activity was measured as described elsewhere(33). Briefly, decrease in absorbance of 30μM solution of H2O2 was monitored at 240nm for 60 sec. The extinction coefficient of 43.6 M−1cm−1 was used for calculating activity. Total cellular protein, measured by using Bradford assay(34), was used for calculation of activity.
Peroxidase activity assay
Conversion of methanol to formaldehyde in the presence of added H2O2 (44mM) was measured spectrophotometrically (at 550nm) using 4-amino-3-hydrazine-5-mercapto-1,2,4 trizole (Purplad) as chromogen(35,36). Commercial formaldehyde solution (4.25M) was used to determine standard curve. Total protein concentration was used for the activity calculation.
Thioredoxin activity
Thioredoxin activity was measured as per the kit instructions (Caymen Chemicals Company, USA).
Oligomerization detection
Cell lysates were mixed with non-reducing sample buffer and loaded onto 8% SDS-PAGE. Gel was run at 4°C and 90V. Proteins from gel were then transferred on to PVDF membrane using semi-dry transfer apparatus and processed as standard western blotting for detection of catalase.
Heme staining
The detailed protocol of in-gel staining is mentioned elsewhere(23,29). Briefly, cell lysates were mixed with non-reducing sample buffer and run on 8% SDS-PAGE. Gel was then washed, fixed and processed for o-dianisidine staining for heme detection.
OVA-challenge of mice
Airway inflammation was induced as previously described(37). Briefly, 6 week old BALB/c mice received an intraperitoneal injection of 10 μg of chicken ovalbumin (OVA) (grade V, Sigma-Aldrich) adsorbed to 20 mg of Al(OH)3 (Sigma-Aldrich) in PBS in a volume of 100 μl. Two weeks later the mice were exposed for approximately 45 min to an aerosol of 1% OVA in PBS produced by an Ultrasonic 2000 nebulizer. The mice were treated with OVA daily for one or four days. Twenty-four hours after the last OVA aerosol treatment the mice were sacrificed by an intraperitoneal injection of sodium pentobarbital. Lungs and trachea were excised and frozen in liquid nitrogen.
Fractionation of cytosols
200μg of lung lysates or cell cytosols were fractionated onto gel-filtration column (Superdex 30/300) coupled with FPLC at 4°C. Column was washed with 5X water and equilibrated with 2X lysis buffer before loading the sample. After the passage of bed volume (7.4ml), fractions of 0.5ml were collected. Equal volume from each fraction was used to measure catalytic activity and western blots.
Immunogold-TEM staining
Lungs from untreated and 1d OVA-challenged mice were harvested and fixed with glutaraldehyde based fixative. Sections were made and immunostained with Trx-1 antibody followed by gold conjugated secondary antibody. Staining and images were processed at Image core facility at Lerner Research Institute.
Statistical analysis
Two tailed Student's t-test at equal variance was used to calculate p value between the groups. P value of 0.05 or less was considered significant.
Results
NO inhibits heme insertion into catalase
We treated catalase transfected HEK cells with SA to generate heme-deficient catalase, and then followed heme insertion for 3h into apo-catalase as previously described (23). Addition of SA to catalase expressing cells resulted in 50% reduction in its catalytic activity, which later showed 100% recovery upon incubation with heme for 3hrs (Fig. 1A). Concurrent NO exposure during 3 h incubation with heme, inhibited the heme insertion into catalase, as measured by catalytic activity (Fig.1A) confirming our original findings.
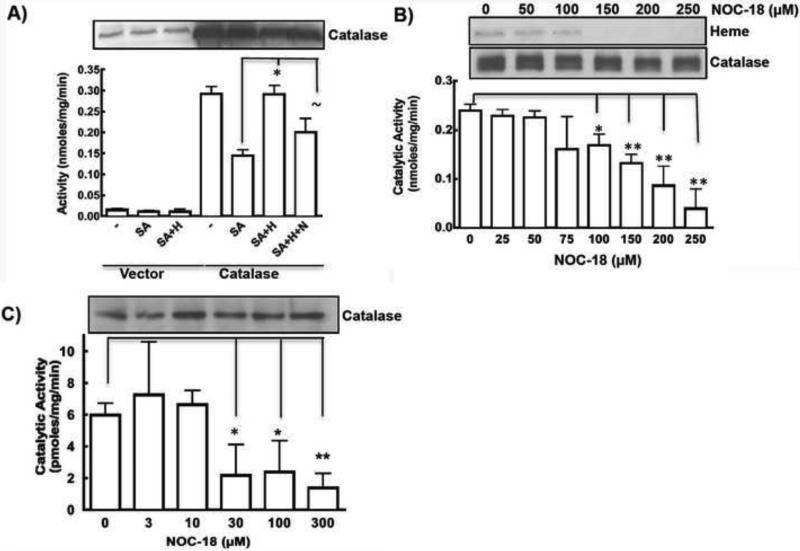
Figure 1. Exogeneous NO decreases catalase activity by decreasing the heme content of enzyme.
A) Heme insertion in SA pretreated catalase was studied by incubating heme (H) in absence or presence of 250μM of NOC18 (N), and measuring its catalytic activity by H2O2 decomposition assay. Representative catalase expression is shown. B) Effect of long-term (18h) NOC18 on heme content and activity of catalase as measured by in-gel heme staining and H2O2 decomposition activity. Loss of heme content increases at higher NO concentrations. C) Catalase activity in the primary airway smooth muscle cells (ASMCs) from mice was found to decrease when incubated with increased NO concentrations (0-300μM) for 20h. Each experiment repeated at-least 3 times. *indicates p <0.01, ** indicates p <0.001 and ~ indicates non-significant difference.
To understand how a more chronic exposure to NO may affect the maturation of catalase, we subjected catalase-transfected HEK cells (6h post-transfection) to different doses of the NO donor NOC18 (0-250μM) for another 18h. This resulted in a dose-dependent decrease in catalase activity with no effect on the catalase protein expression level (Fig. 1B). Due to the his-tag construct of catalase used in this study, over-expressed catalase ran slightly higher in MW compared to endogeneous catalase thereby giving doublet impression in western blots. The decrease in activity correlated with a decreased heme content of catalase as evident by in-gel heme staining (Fig. 1B). Similar to over-expressed catalase in HEK cells, endogenous catalase in primary human airway smooth muscle cells (ASMCs) also showed a dose-dependent decrease in catalase activity with increasing NO donor concentration after 18 h exposure, without any significant effect on the catalase protein expression level (Fig. 1C). Exposing ASMCs to a higher dose of NOC18 (250μM) for 18h did not decrease the total cell heme content relative to controls (data not shown). However, catalase activity in these cells was more sensitive to NOC18 and was significantly inhibited even at concentrations < 30μM.
Heme and NO impact oligomerization of catalase
Catalase that was expressed in SA treated HEK cells had lower heme content and lower percentage of tetrameric catalase (Fig. 2A), consistent with heme insertion being required for catalase tetramerization. Overnight incubation of catalase transfected HEK cells with increasing concentrations of NOC18 also caused a dose-dependent decrease in tetrameric catalase (Fig. 2B). In heme-deficient cells expressing apo-catalase, the addition of heme for 3 h led to formation of tetrameric catalase, and this was prevented if NOC18 was added (Fig. 2C). This coincided with changes in activity as shown in previous Fig 1A. However, treatment of cells with deprenyl protected against the NO-mediated inhibition of catalase tetramer formation. Because deprenyl is a pharmacological inhibitor of SNO-GAPDH formation in cells (38), this result implies that SNO-GAPDH is involved in mediating the NO inhibition of catalase maturation.
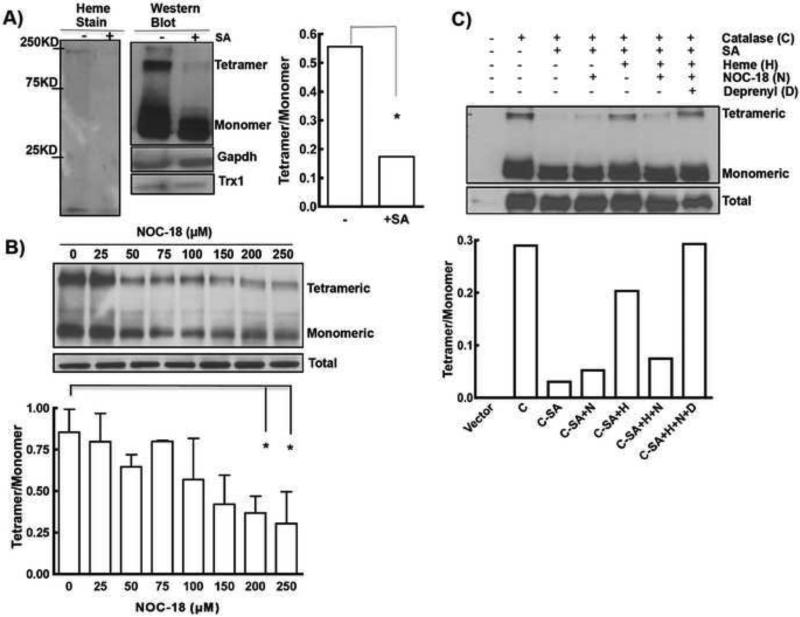
Figure 2. Oligomerization of catalase is heme and NO-sensitive.
A) HEK-Catalase cells incubated with SA, to create heme deficiency, resulted in loss of heme stain as well as tetrameric structure of catalase as evident by heme stain (left) and western blot (right). Difference in tetramer to monomer ratio upon SA treatment was quantified using Image J. No effect on GAPDH or Trx-1 expression was observed. B) Tetrameric catalase content is reduced with increasing concentration of NOC18 (0-250μM) (n=2-3). Representative western blot and its quantitation are shown. Each experiment repeated 3-4 times. Presence of higher-order of oligomers that remained stuck in the wells of SDS-PAGE and did not migrate through the gel are not shown. * indicates p <0.01 and ** indicates p <0.001. C) Heme (H, 7.5μM) insertion promotes tetramerization of catalase © which is sensitive to NO (N, 250μM) and reversed if deprenyl (D, 5μM) was present.
SNO-GAPDH formation limits catalase activity
To investigate if NO-mediated inhibition of catalase heme maturation routes via cellular SNO-GAPDH levels, we examined SNO-GAPDH buildup in primary ASMCs and in catalase over-expressing HEK cells exposed to NOC18 under conditions where we had observed significant impact on the activity and oligomeric structure of catalase. Cells were exposed to different doses of NOC18 (as in Fig. 1B) for 18h followed by detection of SNO-GAPDH by a biotin switch method(30). We observed a concentration-dependent increase in cell SNO-GAPDH level in both the catalase over-expressing HEK cells and endogenous catalase expressing ASMCs when exposed to NOC18 (Fig. 3A-B), which correlated with the loss of catalase activity and its tetrameric assembly (Fig. 1 B & C). In the heme insertion assay (3 h), deprenyl prevented the buildup of SNO-GAPDH that otherwise occurred in the cells exposed to NOC18 (Fig. 3C). Our results suggest that the SNO-GAPDH buildup is associated with poor catalase heme insertion and tetramer formation.
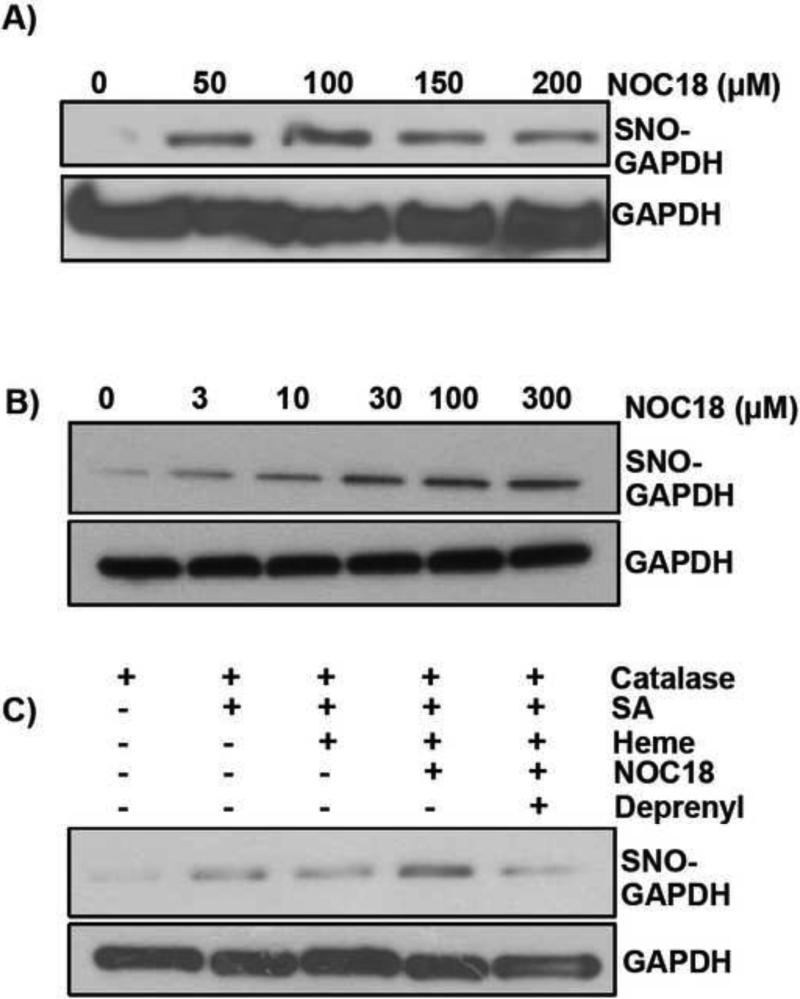
Figure 3. SNO-GAPDH levels are increased upon NO treatment.
A) HEK cell transiently overexpressing catalase, or B) Primary ASMCs, when treated with different NOC18 concentrations for 20h show dose-dependent increase in cellular SNO-GAPDH levels as measured by biotin-switch pull-downs. C) Increase in SNO-GAPDH levels in the 3h heme insertion assay was evident when NO (250μM) was present which was prevented in the presence of deprenyl (5μM).
C152S GAPDH protects catalase from the NO inhibition
To gauge the importance of SNO-GAPDH in limiting catalase maturation, we co-transfected either wild-type GAPDH or the C152S mutant (which does not undergo S-nitrosation (29,39)) along with catalase in HEK cells. We obtained similar levels of expression and activity when catalase was co-transfected with either of the two GAPDH constructs (Fig. 4A). When done in the SA treated cells, a 3h heme addition resulted in similar increases in catalase activity as well as peroxidase activity for both the WT- and C152S-GAPDH. When NO donor (NOC18) was added during the 3 h heme insertion, it prevented the increase in activity in the cells expressing wild-type GAPDH, but in the cells expressing C152S GAPDH, the heme-dependent gain in catalase activities was insensitive toward the NO donor (Fig. 4A). This suggests that S-nitrosation of GAPDH at cys152 was required for NO to block heme insertion into catalase over the 3 h period. We also next tested if C152S-GAPDH expression would protect catalase maturation during an overnight exposure to graded concentrations of NO donor. HEK cells co-transfected with catalase and C152S-GAPDH did show an increased resistance toward NO compared to cells co-transfected with wild-type GAPDH, as measured by catalase activity, tetramer formation and heme content (Fig. 4B-C, Supplemental Fig. S1). This indicates that inhibition of catalase maturation under chronic NO exposure occurred through SNO-GAPDH formation as well.
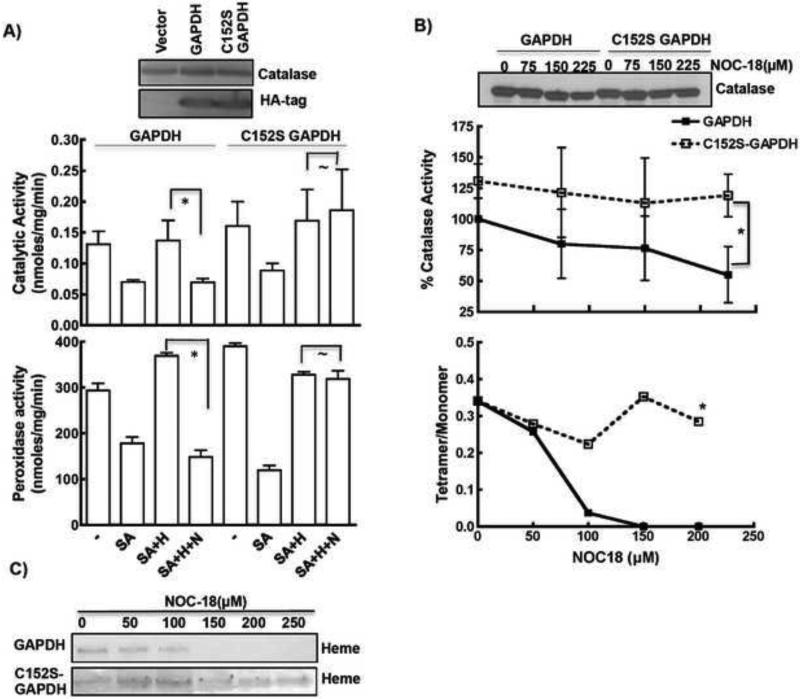
Figure 4. SNO-GAPDH regulates oligomerization and catalase activity.
A) Over-expression of C152S GAPDH protected NO mediated inhibition of heme insertion into catalase as measured by both catalytic and peroxidase activity. No effect on the expression of catalase was observed. Both C152S GAPDH and wild-type GAPDH were expressed at equal level as shown by western blot of anti-HA tag antibody. B) C152S GAPDH over-expression protected catalase activity and tetramerization from long-term NO effects, especially at lower doses of NOC18. C) Over-expression of C152S GAPDH mutant (lower panel) helped in retaining heme content of catalase in presence of NOC18 (top panel from Fig. 1B). * indicates p <0.01, and ~ indicates non-significant.
Thioredoxin-1 regulates catalase activity and tetrameric structure
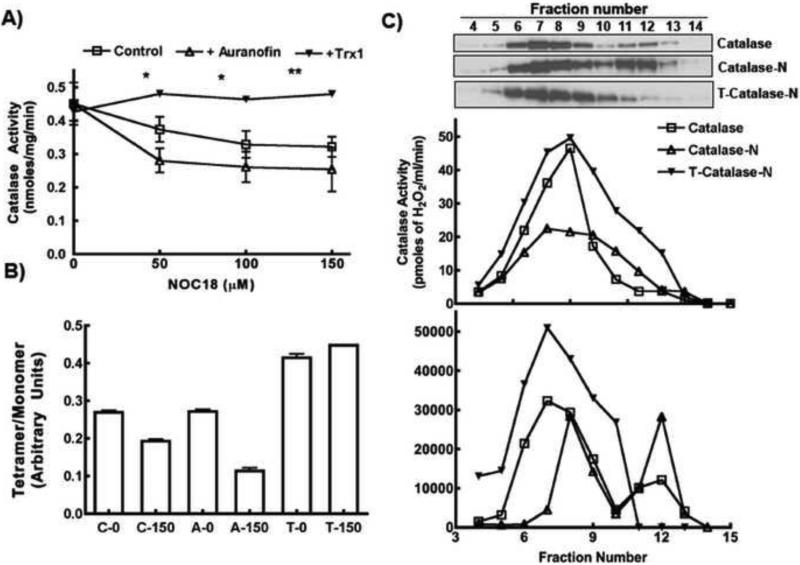
To manipulate cellular SNO-GAPDH levels, we targeted Trx-1, a known denitrosylase of SNO-GAPDH (30,40,41). We over-expressed Trx-1 using lentiviral transduction, or inhibited its activity using a pharmacological inhibitor of Trx-1 reductase (auranofin)(40). Trx-1 over-expression made cellular catalase activity less sensitive to graded concentrations of NOC18, whereas auranofin made the catalase activity more sensitive toward the NO donor (Fig. 5A). These responses toward NO coincided with a lower percentage of tetrameric catalase in the auranofin-treated cells and a higher percentage in the Trx-1 over-expressing cells (Fig. 5B).
Figure 5. Thioredoxin can regulate catalase activity in response to NO.
A) Treatment with 2μM of auranofin or over-expression of Trx-1 significantly affected catalase activity in response to NOC18. Presence of auronofin made catalase activity more susceptible to NO, in opposite Trx-1 overexpression provided protection against NO and retained catalase activity. B) Corresponding tetramer/monomer ratio of catalase at 0 or 150μM of NOC18 treatment in presence of Trx-1 (T) or auroanofin (A) compared to control (C). C) Cell lysates, from HEK cells over-expressing catalase with/without Trx-1, and treated with NOC18 were separated using gel filtration column on FPLC and catalase activity was measured (upper). Fractions were tested for the presence of catalase protein by western blots and the expression was quantified by densitometric analysis using ImageJ (lower). Representative blots are shown. Distribution of catalytic activity and protein expression vs. MW indicate most of the activity resides in the tetrameric structure corresponding to 240KD. * indicates p <0.01, ** indicates p <0.001 and ~ indicates non-significant.
To confirm our findings by an independent approach, we fractionated cytosols of HEK cells over-expressing catalase on a gel filtration column and analyzed each fraction for catalase activity and catalase protein content (Fig. 5C). Based upon the standard molecular weight fractionation, we observed that catalase expressed in HEK cells was predominantly (75%) tetrameric and its activity was associated with the tetramer fractions corresponding to 240 kDa. Cells incubated overnight with NO donor had a lower percentage of tetrameric catalase, a lower associated activity, and an increased percentage of catalase monomer whose elution time indicated a MW of 70 kDa. Trx-1 over-expression caused the NO-treated cells to have a greater percentage of active tetrameric catalase and it prevented buildup of monomeric catalase in response to the NO donor. This confirms results we obtained with non-denaturing gels, and establishes that cellular Trx-1 activity can modulate the NO sensitivity of catalase maturation.
Inflamed airway contains dimeric and less active catalase
Because increased NO production is associated with allergic inflammation in asthma(26,42), we explored if changes in catalase maturation occurred in an established murine model of allergic asthma. We studied catalase activity in the lungs of mice that had been sensitized to and then exposed to ovalbumin (OVA) for 4 days, which causes them to develop a pulmonary inflammation that mimics asthma(26).
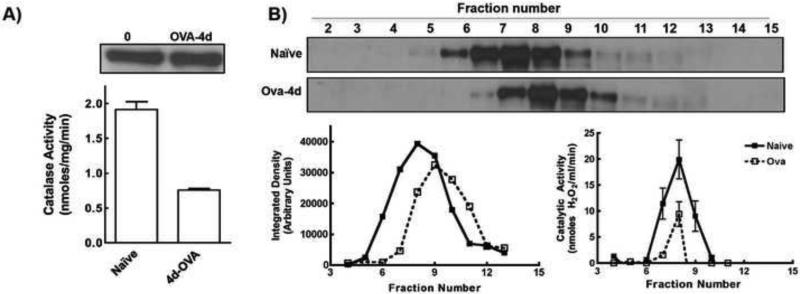
Lung lysates from the 4d OVA-challenged mice had less catalase activity per mg protein than the naïve mice, although they had equivalent catalase protein expression levels (Fig. 6A). Fractionating the lysates on a gel filtration column showed that their catalase activities were maximal in fractions corresponding to an approximate Mr of 240 kDa, indicating the active catalase tetramer was present in both naïve and OVA-challenged lung lysates (Fig. 6B). However, in the OVA-challenged lung lysates, a significant fraction of catalase protein eluted in later fractions corresponding to a lower Mr of approximately 188 kDa (Fig. 6B, supplementary table S1 and figure S2). This suggests that catalase tetramerization and activity were reduced in the inflamed lungs of OVA-challenged mice.
Figure 6. Catalase is altered in the lungs of OVA-challenged mice.
A) Catalase activities were significantly reduced after 4d of OVA challenge while no change in total protein expression was detected. B) lung cytosols of Naïve and 4d-OVA challenged mice were fractionated using FPLC gel filtration column. Fractions were analyzed for catalase activity and expression. Top Panel Most of catalase activity in both samples was observed in 8th fraction. Lower panel Fractions were tested for presence of catalase by western blot and expression was quantified by densitometry.
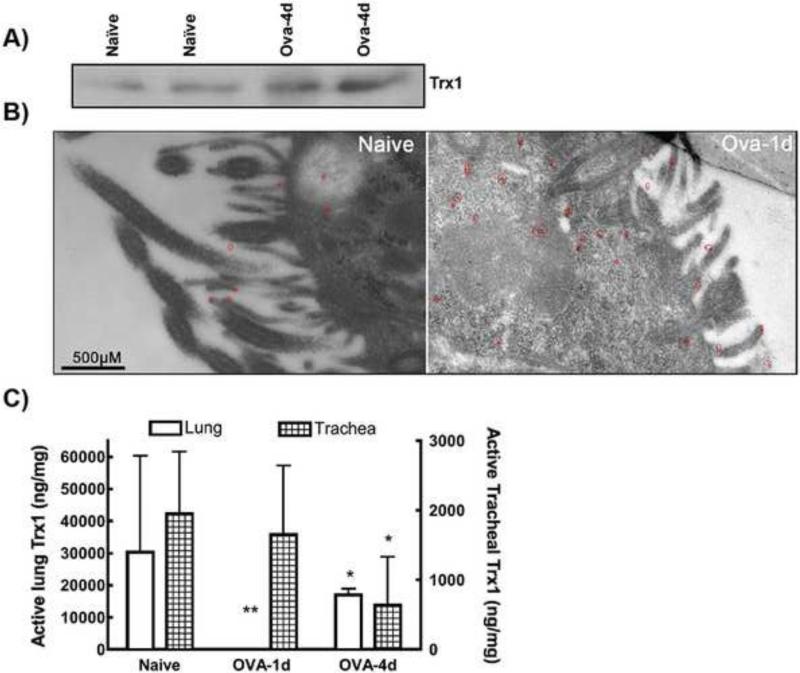
Interestingly, the lungs of OVA-challenged mice had a higher Trx-1 protein expression (Fig. 7A & B). Immuno-electron microscopy of lung sections after 1 d of OVA-challenge confirmed an increase in Trx-1 expression level in the lung epithelial cells, particularly near the plasma membrane and around cilia. Despite this, supernatants prepared from the lungs and trachea of OVA-challenged mice had lower Trx-1 activity compared to the sham-treated control (Fig. 7C), implying that a significant proportion of their airway Trx-1 became inactivated during the inflammation.
Figure 7. Trx-1 expression and activity in the lungs of OVA-challenged mice.
A) Thioredoxin-1 expression level in the lung lysates of two Naïve or 4d OVA-challenged mice is shown. B) Immuno-EM of Trx-1 in lungs from 1d OVA-challenged mice indicate increased expression upon inflammation. At 49000X magnification, gold staining (shown as clusters of black spots circled by red) representing Trx-1 was observed. Scale bar=500nM. C) Trx-1 activity was measured in the lung & Trachea lysates during the different time points of OVA-challenge in mice. Each experiment was repeated 2 times using 3 mice per condition. * indicates p <0.01, ** indicates p <0.001 and ~ indicates non-significant.
Discussion
Catalase was initially thought to primarily be a peroxisomal protein, but subsequent studies indicated a significant cytosolic pool as well (11,43-46). A number of studies have focused on understanding catalase maturation with respect to its sub-cellular compartmentalization. Catalase has a noncanonical peroxisome targeting sequence at the C-terminus(47), and a shuttle receptor (PEX5) is required for peroxisomal translocation of catalase, which is reported to interact only with monomeric catalase (48). This interaction is also shown to inhibit catalase tetramerization, suggesting tetramerization of catalase is a peroxisomal event(45,48). However, several other reports suggest that cytosolic assembly of catalase can occur independent of peroxisomes (15,49,50). Four amino acids on the N-terminal end of Candida Tropicalis catalase are shown to regulate tetramerization of the protein(51). In addition to tetramer, catalase is also known to be present as a high order oligomer, a dimer, or a monomer in different systems including liver cytosols and erythrocytes(52,53) (54). In some cases, dimeric catalase was also shown to be active (8,55). However it is well established that apo-catalase is neither active nor tetrameric in structure (1,3,51,55). Several reports indicate catalase monomers accumulate in rat liver cytosol (11-13). The physiological relevance of such heme-free or heme-containing monomers of catalase is not known.
Despite the important role heme plays in catalase activity, our understanding of heme insertion into apo-catalase is very limited. One recent in vitro study using Enterococcus faecalis catalase demonstrated the importance of heme insertion in formation of tetrameric active catalase(14). To understand heme maturation of catalase in mammalian cells, we utilized our previous finding of NO-mediated inhibition of heme maturation into catalase to identify factors regulating its heme incorporation and therefore tetramerization to make the active enzyme.
Our results suggest that NO inhibits catalase maturation by causing build-up of SNO-GAPDH in the cells. This is supported by our finding that (i) over-expression of a denitrosylase enzyme (Trx-1) that decreased cellular SNO-GAPDH levels, (ii) pharmacologic prevention of SNO-GAPDH formation with deprenyl, or (iii) expression of the S-nitrosation-resistant GAPDH variant (C152S), all made catalase activity, and/or its heme insertion and tetramer formation, resistant to the NO inhibition. Indeed, the degree of protection afforded to catalase by these manipulations was high enough to suggest that the SNO-GAPDH pathway may be the primary mechanism by which NO inhibits catalase in the cellular systems that we utilized. Thus, catalase maturation appears to be regulated through the same mechanisms that regulate cell iNOS maturation during NO exposure(29), with SNO-GAPDH buildup as a key required feature. We have also identified a cellular mechanism, based upon Trx-1, that can protect catalase maturation from the deleterious effect of NO.
In our current study and previously, we found that NO can block heme insertion into apo-catalase when this form of the protein is already present in cells at the point of addition of heme and an NO donor. But in our current experiments where cells underwent a prolonged, continuous low level NO exposure for 12-24 h, additional mechanisms may play a role. For example, upon initiation of the prolonged NO exposure the cells contained tetrameric, heme-replete catalase, and so the loss of bound heme from this population should be considered. However, NO binding to the catalase heme does not result in any structural disturbance that could cause heme loss (22). Catalase heme groups are also sequestered in the protein 25 Å from the surface of the enzyme (56), and tetrameric catalase is quite stable (4). On the other hand, the holo-catalase protein has a half-life in rat liver of about 40 h(57), and consistent with this, inhibiting cellular heme synthesis caused apo-catalase to build up within 18 h in our hands, presumably due to holo-protein turnover and new apo-protein synthesis and accumulation. Thus, during a longer exposure, NO may still act primarily to prevent heme insertion into apo-catalase, rather than cause heme loss from the holoenzyme. In addition to catalase, other cellular components that can destroy H2O2, but do not require heme, seems to be unaffected by SA or NO treatments as evident by our observing about 50% reduction in total H2O2 decomposition activity as shown in Fig. 1A. Therefore, incubation with SA and/or NO can provide a measure of the catalase-specific H2O2 decomposition in cells.
Catalase can undergo several post-translational modifications that impact its activity, including nitration, oxidation, chlorination, ubiquitinylation, S-nitrosation, phosphorylation (21,26,28,58,59), and modification of the enzyme catalytic site in vitro (21,22,24). An inhibition of cellular catalase activity after 9 h of NO exposure has also been attributed to a decrease in cell heme availability brought on by NO-induced changes in heme biosynthesis and degradation(21). In addition, direct NO-mediated S-nitrosation or oxidative damage within catalase are also possible, and may affect its activity. Whether such modification pathways or changes in heme availability play a role was not tested in our study. However, our findings do imply that their involvement would still have to rely on SNO-GAPDH buildup in some way in order to impact catalase maturation.
There is a consensus that heme needs to be inserted into apo-catalase before its tetramerization can occur(1,2). Initially, maturation of catalase was thought to occur in the peroxisomes, but catalase expression studies in a peroxisome-deficient dermal fibroblast, in S. cerevisiae, or in bacterial expression systems, all indicate that its maturation can be peroxisome-independent (7,44,45,50,60). Thus, NO effects on catalase heme insertion probably take place before the tetramer forms. Our results are also consistent with heme-free or otherwise not fully-mature forms of catalase existing and being fairly stable in cells (13). Apo-catalase was also found in rat liver extracts under various conditions, and different oligomeric states of catalase have been reported to exist in equilibrium under in vitro as well as in vivo circumstances (54,55). Thus, natural conditions may exist that would allow NO to limit maturation of apo-catalase by limiting its heme insertion, and thus preventing its tetramerization. An NO-mediated limitation on heme insertion that impacts tetramerization of catalase has not been previously considered.
The tetrameric status of catalase has not been investigated before in physiologically-relevant models of inflammation, although a decrease in catalase activity has been observed (25,26). In different models of diabetes and asthma, loss in catalase activity was not correlated with any decrease in its protein expression level (25,26). The presence of inactive dimeric erythrocytic catalase in patients with Swiss Type Acatalasemia has also been indicated (54). Our observing a dimeric catalase in the diseased mouse lung compared to only finding tetrameric catalase in the healthy lung implies that its change in oligomerization status might be an epiphenomenon of inflammation and the increased NO exposure or oxidant stress as described in our cell culture studies. Although we observed an increase in Trx-1 expression in the allergen challenged lungs, it was not sufficient to fully protect catalase activity and tetramer structure. This could reflect an insufficient increase in Trx-1 to counter the stress during the pulmonary inflammation, or alternatively, it could reflect a decreased activity of the Trx-1. Indeed, in both lung and tracheal lysates from the OVA-challenged mice, we found a significant decrease in Trx-1 catalytic activity. Thus, a loss of Trx-1 activity, despite increased Trx-1 protein expression, is consistent with the poor catalase maturation in this circumstance.
Summary
Defects in catalase activity under various diseases that involve oxidative stress and inflammation have been widely reported. Here we show that there is destabilization of catalase structure, possibly contributing to a decrease in its activity. Under circumstances of chronic NO exposure, the cellular Trx-1 and SNO-GAPDH levels are key regulatory factors that determine catalase heme content, oligomeric structure, and enzyme activity.
Supplementary Material
Highlights.
Heme is essential for catalase tetramerization and activity.
NO antagonizes heme insertion and formation of an active catalase tetramer.
Cellular SNO-GAPDH and TRX1 levels determine NO sensitivity of catalase.
Chronic inflammation of lungs in asthma affects catalase oligomerizaton and activity.
Acknowledgements
We express our gratitude to Dr. S.J. Haque for sharing catalase expression plasmid. We are thankful to Mei Yin at Image core (Lerner Research institute) for help in capturing lung-TEM images. This work was supported by National Institute of Health Grants GM097041 and HL076491.
Abbreviations
- NO
Nitric oxide
- OVA
Ovalbumin
- Trx1
Thioredoxin1
- SNO GAPDH
S-nitrosated GAPDH
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goyal MM, Basak A. Human catalase: looking for complete identity. Protein Cell. 2010;1:888–897. doi: 10.1007/s13238-010-0113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholls P. Classical catalase: ancient and modern. Archives of biochemistry and biophysics. 2012;525:95–101. doi: 10.1016/j.abb.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Kirkman HN, Gaetani GF. Catalase: a tetrameric enzyme with four tightly bound molecules of NADPH. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:4343–4347. doi: 10.1073/pnas.81.14.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz A, Loewen PC, Fita I, Carpena X. Thirty years of heme catalases structural biology. Archives of biochemistry and biophysics. 2012;525:102–110. doi: 10.1016/j.abb.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Reid TJ, Murthy MRN, Sicignano A, Tanaka N, Musick WDL, Rossmann MG. Structure and Heme Environment of Beef-Liver Catalase at 2.5 a Resolution. Proceedings of the National Academy of Sciences of the United States of America-Biological Sciences. 1981;78:4767–4771. doi: 10.1073/pnas.78.8.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sichak SP, Dounce AL. Analysis of the peroxidatic mode of action of catalase. Archives of biochemistry and biophysics. 1986;249:286–295. doi: 10.1016/0003-9861(86)90004-4. [DOI] [PubMed] [Google Scholar]
- 7.Seah TC, Kaplan JG. Purification and properties of the catalase of bakers' yeast. The Journal of biological chemistry. 1973;248:2889–2893. [PubMed] [Google Scholar]
- 8.Aebi H, Scherz B, Ben-Yoseph Y, Wyss SR. Dissociation of erythrocyte catalase into subunits and their re-association. Experientia. 1975;31:397–399. doi: 10.1007/BF02026338. [DOI] [PubMed] [Google Scholar]
- 9.Havir EA. The in Vivo and in Vitro Inhibition of Catalase from Leaves of Nicotiana sylvestris by 3-Amino-1,2,4-Triazole. Plant Physiol. 1992;99:533–537. doi: 10.1104/pp.99.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Inada Y, Kurozumi T, Shibata K. Peroxidase activity of hemoproteins. I. Generation of activity by acid or alkali denaturation of methemoglobin and catalase. Archives of biochemistry and biophysics. 1961;93:30–36. doi: 10.1016/0003-9861(61)90311-3. [DOI] [PubMed] [Google Scholar]
- 11.Sugita Y, Higashi T. Biosynthesis of liver catalase in rats treated with allylisopropylacetylcarbamide. III. Occurrence of a possible precursor to catalase. Journal of biochemistry. 1981;89:999–1004. [PubMed] [Google Scholar]
- 12.Sugita Y, Tobe T, Sakamoto T, Higashi T. Immature precursor catalase in subcellular fractions of rat liver. Journal of biochemistry. 1982;92:509–515. doi: 10.1093/oxfordjournals.jbchem.a133958. [DOI] [PubMed] [Google Scholar]
- 13.Lazarow PB, de Duve C. The synthesis and turnover of rat liver of rat liver peroxisomes. IV. Biochemical pathway of catalase synthesis. The Journal of cell biology. 1973;59:491–506. doi: 10.1083/jcb.59.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baureder M, Barane E, Hederstedt L. In Vitro Assembly of Catalase. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M114.596148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eising R, Suselbeck B. Turnover of catalase heme and apoprotein moieties in cotyledons of sunflower seedlings. Plant Physiol. 1991;97:1422–1429. doi: 10.1104/pp.97.4.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ando T, Mimura K, Johansson CC, Hanson MG, Mougiakakos D, Larsson C, Martins da Palma T, Sakurai D, Norell H, Li M, Nishimura MI, Kiessling R. Transduction with the antioxidant enzyme catalase protects human T cells against oxidative stress. J Immunol. 2008;181:8382–8390. doi: 10.4049/jimmunol.181.12.8382. [DOI] [PubMed] [Google Scholar]
- 17.Shi X, Shi Z, Huang H, Zhu H, Zhou P, Zhu H, Ju D. Ability of Recombinant Human Catalase to Suppress Inflammation of the Murine Lung Induced by Influenza A. Inflammation. 2014 doi: 10.1007/s10753-013-9800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pigeolet E, Corbisier P, Houbion A, Lambert D, Michiels C, Raes M, Zachary MD, Remacle J. Glutathione peroxidase, superoxide dismutase, and catalase inactivation by peroxides and oxygen derived free radicals. Mechanisms of ageing and development. 1990;51:283–297. doi: 10.1016/0047-6374(90)90078-t. [DOI] [PubMed] [Google Scholar]
- 19.Escobar JA, Rubio MA, Lissi EA. Sod and catalase inactivation by singlet oxygen and peroxyl radicals. Free radical biology & medicine. 1996;20:285–290. doi: 10.1016/0891-5849(95)02037-3. [DOI] [PubMed] [Google Scholar]
- 20.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. The chemical biology of nitric oxide: implications in cellular signaling. Free radical biology & medicine. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YM, Bergonia HA, Muller C, Pitt BR, Watkins WD, Lancaster JR., Jr. Loss and degradation of enzyme-bound heme induced by cellular nitric oxide synthesis. The Journal of biological chemistry. 1995;270:5710–5713. doi: 10.1074/jbc.270.11.5710. [DOI] [PubMed] [Google Scholar]
- 22.Purwar N, McGarry JM, Kostera J, Pacheco AA, Schmidt M. Interaction of nitric oxide with catalase: structural and kinetic analysis. Biochemistry. 2011;50:4491–4503. doi: 10.1021/bi200130r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waheed SM, Ghosh A, Chakravarti R, Biswas A, Haque MM, Panda K, Stuehr DJ. Nitric oxide blocks cellular heme insertion into a broad range of heme proteins. Free radical biology & medicine. 2010;48:1548–1558. doi: 10.1016/j.freeradbiomed.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown GC. Reversible binding and inhibition of catalase by nitric oxide. Eur J Biochem. 1995;232:188–191. doi: 10.1111/j.1432-1033.1995.tb20798.x. [DOI] [PubMed] [Google Scholar]
- 25.Sigfrid LA, Cunningham JM, Beeharry N, Lortz S, Tiedge M, Lenzen S, Carlsson C, Green IC. Cytokines and nitric oxide inhibit the enzyme activity of catalase but not its protein or mRNA expression in insulin-producing cells. Journal of molecular endocrinology. 2003;31:509–518. doi: 10.1677/jme.0.0310509. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh S, Janocha AJ, Aronica MA, Swaidani S, Comhair SA, Xu W, Zheng L, Kaveti S, Kinter M, Hazen SL, Erzurum SC. Nitrotyrosine proteome survey in asthma identifies oxidative mechanism of catalase inactivation. J Immunol. 2006;176:5587–5597. doi: 10.4049/jimmunol.176.9.5587. [DOI] [PubMed] [Google Scholar]
- 27.Begara-Morales JC, Lopez-Jaramillo FJ, Sanchez-Calvo B, Carreras A, Ortega-Munoz M, Santoyo-Gonzalez F, Corpas FJ, Barroso JB. Vinyl sulfone silica: application of an open preactivated support to the study of transnitrosylation of plant proteins by S-nitrosoglutathione. BMC Plant Biol. 2013;13:61. doi: 10.1186/1471-2229-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foster MW, Stamler JS. New insights into protein S-nitrosylation. Mitochondria as a model system. The Journal of biological chemistry. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 29.Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakravarti R, Stuehr DJ. Thioredoxin-1 regulates cellular heme insertion by controlling S-nitrosation of glyceraldehyde-3-phosphate dehydrogenase. The Journal of biological chemistry. 2012;287:16179–16186. doi: 10.1074/jbc.M112.342758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panda K, Chawla-Sarkar M, Santos C, Koeck T, Erzurum SC, Parkinson JF, Stuehr DJ. Visualizing inducible nitric-oxide synthase in living cells with a heme-binding fluorescent inhibitor. Proc Natl Acad Sci U S A. 2005;102:10117–10122. doi: 10.1073/pnas.0408972102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE. 2001;2001:pl1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]
- 33.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 34.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 35.Wheeler CR, Salzman JA, Elsayed NM, Omaye ST, Korte DW., Jr. Automated assays for superoxide dismutase, catalase, glutathione peroxidase, and glutathione reductase activity. Analytical biochemistry. 1990;184:193–199. doi: 10.1016/0003-2697(90)90668-y. [DOI] [PubMed] [Google Scholar]
- 36.Johansson LH, Borg LA. A spectrophotometric method for determination of catalase activity in small tissue samples. Analytical biochemistry. 1988;174:331–336. doi: 10.1016/0003-2697(88)90554-4. [DOI] [PubMed] [Google Scholar]
- 37.Cheng G, Swaidani S, Sharma M, Lauer ME, Hascall VC, Aronica MA. Hyaluronan deposition and correlation with inflammation in a murine ovalbumin model of asthma. Matrix Biol. 2011;30:126–134. doi: 10.1016/j.matbio.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hara MR, Thomas B, Cascio MB, Bae BI, Hester LD, Dawson VL, Dawson TM, Sawa A, Snyder SH. Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3887–3889. doi: 10.1073/pnas.0511321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen N, Hara MR, Kornberg MD, Cascio MB, Bae BI, Shahani N, Thomas B, Dawson TM, Dawson VL, Snyder SH, Sawa A. Nitric oxide-induced nuclear GAPDH activates p300/CBP and mediates apoptosis. Nat Cell Biol. 2008;10:866–873. doi: 10.1038/ncb1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ito T, Yamakuchi M, Lowenstein CJ. Thioredoxin increases exocytosis by denitrosylating N-ethylmaleimide-sensitive factor. The Journal of biological chemistry. 2011;286:11179–11184. doi: 10.1074/jbc.M110.201780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dweik RA, Boggs PB, Erzurum SC, Irvin CG, Leigh MW, Lundberg JO, Olin AC, Plummer AL, Taylor DR. An official ATS clinical practice guideline: interpretation of exhaled nitric oxide levels (FENO) for clinical applications. Am J Respir Crit Care Med. 2011;184:602–615. doi: 10.1164/rccm.9120-11ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Geerts A, De Prest B, Roels F. On the topology of the catalase biosynthesis and -degradation in the guinea pig liver. A cytochemical study. Histochemistry. 1984;80:339–345. doi: 10.1007/BF00495414. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto K, Volkl A, Hashimoto T, Fahimi HD. Catalase in guinea pig hepatocytes is localized in cytoplasm, nuclear matrix and peroxisomes. European journal of cell biology. 1988;46:129–135. [PubMed] [Google Scholar]
- 45.Lazarow PB, de Duve C. The synthesis and turnover of rat liver peroxisomes. V. Intracellular pathway of catalase synthesis. The Journal of cell biology. 1973;59:507–524. doi: 10.1083/jcb.59.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tobe T, Higashi T. Studies on rat liver catalase. XI. Site of synthesis and segregation by stripped ER membranes. Journal of biochemistry. 1980;88:1341–1347. doi: 10.1093/oxfordjournals.jbchem.a133102. [DOI] [PubMed] [Google Scholar]
- 47.Purdue PE, Lazarow PB. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. The Journal of cell biology. 1996;134:849–862. doi: 10.1083/jcb.134.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Freitas MO, Francisco T, Rodrigues TA, Alencastre IS, Pinto MP, Grou CP, Carvalho AF, Fransen M, Sa-Miranda C, Azevedo JE. PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. The Journal of biological chemistry. 2011;286:40509–40519. doi: 10.1074/jbc.M111.287201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robbi M, Lazarow PB. Synthesis of catalase in two cell-free protein-synthesizing systems and in rat liver. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:4344–4348. doi: 10.1073/pnas.75.9.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wanders RJ, Strijland A, van Roermund CW, van den Bosch H, Schutgens RB, Tager JM, Schram AW. Catalase in cultured skin fibroblasts from patients with the cerebro-hepato-renal (Zellweger) syndrome: normal maturation in peroxisome-deficient cells. Biochimica et biophysica acta. 1987;923:478–482. doi: 10.1016/0304-4165(87)90057-2. [DOI] [PubMed] [Google Scholar]
- 51.Ueda M, Kinoshita H, Maeda SI, Zou W, Tanaka A. Structure-function study of the amino-terminal stretch of the catalase subunit molecule in oligomerization, heme binding, and activity expression. Appl Microbiol Biotechnol. 2003;61:488–494. doi: 10.1007/s00253-003-1251-5. [DOI] [PubMed] [Google Scholar]
- 52.Middelkoop E, Wiemer EA, Schoenmaker DE, Strijland A, Tager JM. Topology of catalase assembly in human skin fibroblasts. Biochimica et biophysica acta. 1993;1220:15–20. doi: 10.1016/0167-4889(93)90091-3. [DOI] [PubMed] [Google Scholar]
- 53.Takeda A, Samejima T. On the characterization of porcine erythrocyte catalase and its disulfide-linked dimer. Journal of biochemistry. 1977;82:1025–1033. doi: 10.1093/oxfordjournals.jbchem.a131773. [DOI] [PubMed] [Google Scholar]
- 54.Aebi H, Wyss SR, Scherz B, Skvaril F. Heterogeneity of erythrocyte catalase II. Isolation and characterization of normal and variant erythrocyte catalase and their subunits. Eur J Biochem. 1974;48:137–145. doi: 10.1111/j.1432-1033.1974.tb03751.x. [DOI] [PubMed] [Google Scholar]
- 55.Prakash K, Prajapati S, Ahmad A, Jain SK, Bhakuni V. Unique oligomeric intermediates of bovine liver catalase. Protein Sci. 2002;11:46–57. doi: 10.1110/ps.20102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mate MJ, Ortiz-Lombardia M, Marina A, Fita I. Crystallization and preliminary structural results of catalase from human erythrocytes. Acta Crystallogr D Biol Crystallogr. 1999;55:1066–1068. doi: 10.1107/s0907444999002747. [DOI] [PubMed] [Google Scholar]
- 57.Hayashi H. Degradation of peroxisomal catalase and urate oxidase of rat liver. Biochimica et biophysica acta. 1979;585:220–228. doi: 10.1016/0304-4165(79)90022-9. [DOI] [PubMed] [Google Scholar]
- 58.He X, Kermode AR. Programmed cell death of the megagametophyte during post-germinative growth of white spruce (Picea glauca) seeds is regulated by reactive oxygen species and the ubiquitin-mediated proteolytic system. Plant Cell Physiol. 2010;51:1707–1720. doi: 10.1093/pcp/pcq130. [DOI] [PubMed] [Google Scholar]
- 59.Rafikov R, Kumar S, Aggarwal S, Hou Y, Kangath A, Pardo D, Fineman JR, Black SM. Endothelin-1 stimulates catalase activity through the PKCdelta-mediated phosphorylation of serine 167. Free radical biology & medicine. 2013;67C:255–264. doi: 10.1016/j.freeradbiomed.2013.10.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walton PA, Hill PE, Subramani S. Import of stably folded proteins into peroxisomes. Mol Biol Cell. 1995;6:675–683. doi: 10.1091/mbc.6.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.