Abstract
Leishmania has a plastic genome, and drug pressure can select for gene copy number variation (CNV). CNVs can apply either to whole chromosomes, leading to aneuploidy, or to specific genomic regions. For the latter, the amplification of chromosomal regions occurs at the level of homologous direct or inverted repeated sequences leading to extrachromosomal circular or linear amplified DNAs. This ability of Leishmania to respond to drug pressure by CNVs has led to the development of genomic screens such as Cos-Seq, which has the potential of expediting the discovery of drug targets for novel promising drug candidates.
Keywords: Leishmania, Ploidy, Drug Resistance, Mode of action, Cos-Seq
Introduction
Leishmania are dimorphic parasites living as extracellular promastigotes in the digestive tract of Phlebotomus or Lutzomyia sandflies and as intracellular amastigotes within phagocytic cells (mainly macrophages) of the vertebrate hosts. Leishmania species cause leishmaniasis, the second largest parasite killer; there are 1.3 million new cases annually, 12 million people are affected worldwide, and 350 million people are currently at risk 1. The Leishmania genus includes several species, among which more than 20 are pathogenic to humans 2. Leishmania can be divided into two subgenera: the Leishmania Leishmania subgenus responsible for visceral and cutaneous leishmaniasis and the Leishmania Viannia subgenus often associated with either cutaneous or muco-cutaneous forms of the disease 2. Visceral leishmaniasis is mainly caused by L. donovani and L. infantum and is characterized by fever, hepatosplenomegaly, and pancytopenia 3, making it the most severe and deadly form of the disease when compared with the self-healing but nonetheless debilitating skin lesions of cutaneous leishmaniasis 4, 5.
No effective human vaccine is currently available against Leishmania (a canine Leishmania vaccine was recently registered in Europe 6), and control measures mainly involve chemotherapy 7– 9. Pentavalent antimony (Sb) has been the standard drug for 70 years and remains the mainstay in many endemic regions, apart from Northern India, where antimonial formulations have been rendered obsolete because of widespread parasite resistance. Other first-line therapies include the polyene antibiotic amphotericin B (AMB) for which a single dose was shown to be 95% effective against visceral leishmaniasis in India 10. Liposomal AMB has become a standard treatment in many countries 11 but requires administration by the intravenous route. Geographical differences in the response to liposomal AMB were reported, and visceral leishmaniasis cases in India were more responsive than those from East Africa or South America 12. Clinical AMB resistance is scarce and parasites remained susceptible even after multiple rounds of treatment in the same patient 13. The alkyl-lysophospholipid analogue miltefosine (MTF) is the only oral drug against Leishmania 14, 15. It has been successfully used for the treatment of visceral leishmaniasis since its registration in 2002 in India 16. However, relapse rates are on the rise in India 17, 18 and Nepal 19, 20, making MTF resistance a likely problem. The aminoglycoside paromomycin (PMM) is also approved for the treatment of visceral leishmaniasis in India 21, 22. So far, the scarce use of PMM has limited the emergence of resistance, but geographical variations in PMM efficacy against visceral leishmaniasis were noted between East Africa (especially Sudan) and India 23, 24. Lastly, pentamidine (PTD) has been abandoned for the treatment of visceral leishmaniasis because of serious toxicities and is mainly restricted to patients with cutaneous leishmaniasis in South America 25– 27.
Despite six decades of use, the mode of action (MOA) of antimonials is not known. It has been shown to lead to the production of reactive oxygen species 28– 31, the depletion of trypanothione 32, and apoptosis-like death 33– 36, but an exact MOA is still awaited. The same applies for MTF, AMB, PTD, and PMM, with the possible exception of AMB, which kills Leishmania by forming pores in ergosterol-containing membranes. New molecules with well-defined drug targets are clearly needed.
Leishmania and its genome
The Leishmania genome is around 32 Mb and displays over 8,300 coding genes 37, 38. Within the Leishmania genus, gene synteny is conserved for more than 99% of genes between L. major, L. infantum, and L. braziliensis, and only few species-specific genes were found 38. Leishmania species have between 34 and 36 chromosomes ranging in size from 0.3 to 2.8 Mb 37– 41. One unique feature characterizing trypanosomatid parasites lies in their genome architecture, their protein-coding genes being organized as large polycistronic units 42, 43. In the absence of defined RNA polymerase II promoters, transcription of the long polycistronic units occurs in a bidirectional fashion from transcriptional start sites located at strand switch regions 43, 44. Processing into individual messenger RNAs (mRNAs) occurs by the addition through trans-splicing of a spliced leader RNA (39 nt) to the 5′ ends of each mRNA, coupled to 3′ end polyadenylation 45, 46. Because of its lack of transcriptional control, Leishmania uses several adaptive mechanisms to regulate gene expression when facing changing environmental conditions during its development. 3′ untranslated regions (3′ UTRs) were shown to be major players in monitoring mRNA stability and translation rates in this parasite 47– 53. To overcome stressful conditions like drug pressure, Leishmania also often relies on DNA copy number variations (CNVs) (aneuploidy, gene amplification, or gene deletion) for regulating the expression of drug targets, drug transporters, or other determinants of resistance. This is not restricted to Leishmania, however, and variations in gene dosage or chromosome copy numbers also influence drug susceptibility, adaptability, and proliferation in fungi and cancer cells 54– 57. In addition to CNVs, single-nucleotide polymorphisms (SNPs) in drug targets or in transporters can lead to drug resistance without the need for altering gene expression.
Copy number variations
During the last few decades, Leishmania parasites were considered to be essentially diploid but recent data have shown that aneuploidy seems to be the norm 58– 64. Within populations of Leishmania parasites, distinct aneuploidy patterns were shown to occur at the level of individual cells. This phenomenon was called mosaic aneuploidy and can translate into a seemingly average diploid population when the cumulative ploidy is derived from next-generation DNA sequencing data but in which few parasites actually share the same ploidy for individual chromosomes 62, 63, 65. Interestingly, variations in the size and content of chromosomes have also been observed between different strains of the related trypanosomatid parasite Trypanosoma cruzi 66, 67. In the case of Leishmania, circumstantial links between the presence of supernumerary chromosomes or chromosomal losses and drug resistance have been observed 58– 60, 64, 68– 71, suggesting that a particular group of genes on the variant chromosomes may possibly act together in establishing resistance, but this has yet to be demonstrated.
Aneuploidy is generally linked to developmental abnormalities as best exemplified by the trisomy 21 syndrome in humans. However, Leishmania uses aneuploidy as a lifestyle. This is raising a number of questions about aneuploidy generation, stability, transmission, and biological significance (reviewed elsewhere 60, 62, 72). In the absence of transcription initiation control, increases (or decreases) in chromosome copy number may serve as a strategy for regulating expression under environmental cues. This can happen at the level of whole chromosomes, and indeed there was a good correlation between chromosome ploidy and the level of DNA and RNA expression 59. Increasing the copy numbers of entire chromosomes may lead to the overexpression of toxic genes, but the Leishmania genome (32 Mb) is spread in 34 to 36 chromosomes, thus reducing the co-expression of many genes. However, as explained in detail below, Leishmania also has the ability to amplify (or delete) specific smaller regions of DNA as part of extrachromosomal elements by recombination/rearrangements at the level of homologous repeated sequences (RSs). RNA levels derived from these amplifications are correlated to DNA copy number.
The genome of Leishmania is populated with repeated DNA sequences. A recent study highlighted the entire set of RSs in different Leishmania species, and it was found that the whole Leishmania genome has the potential to be rearranged at the level of those RSs for generating extrachromosomal elements 73. Indeed, almost 2,000 RSs are distributed over the genome of L. infantum and these potentially support the formation of more than 3,000 extrachromosomal DNA elements 73. Short interspersed degenerate retroposons (SIDERs) account for up to 65% of all RSs. SIDERs are truncated versions (~0.55 kb) of formerly active retroposons that are predominantly located in 3′ UTRs and have been associated with post-transcriptional regulation at the levels of both mRNA stability and translation 47, 48, 52, 53, 74, 75. Because SIDERs are degenerated, they were found in different RS groups. Remarkably, SIDER elements would have dual roles: one functional by regulating gene expression and a second one structural, providing the backbone to facilitate gene rearrangements for changing copy number of chromosomal DNA regions.
Extrachromosomal DNA amplifications are frequently detected in Leishmania parasites challenged with drugs or other stressful conditions 58, 59, 64, 76– 87. The episomes are amplified as either circular or linear extrachromosomal DNA and formed through rearrangements at the level of direct or inverted homologous RSs, respectively ( Figure 1) 73, 80, 88. Interestingly, between 60% and 80% of the predicted amplicons appear to be already present in the population in the absence of selection and these pre-existing stochastic gene amplifications were shown to foster the selection of adaptive traits in response to drug pressure 73. Beneficial amplicons were shown to increase in abundance upon higher drug pressure and to decrease when the drug is removed, allowing parasites to respond to a changing environment 73.
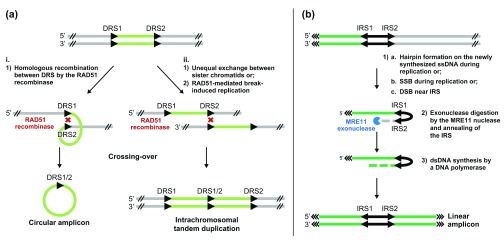
Figure 1. Potential mechanisms for gene amplification in Leishmania.
( a) The RAD51 recombinase mediates homologous recombination between direct repeated sequences (DRS) and leads to (i) extrachromosomal circular amplicon or (ii) intrachromosomal tandem duplication by unequal sister chromatid exchange or RAD51-mediated break-induced replication. Black arrows represent DRS. ( b) The MRE11 nuclease processes DNA ends after single-strand break (SSB), double-strand break (DSB), or hairpin formation during replication and leads to extrachromosomal linear amplification. Black arrows represent inverted repeated sequences (IRS). The green segments represent the amplified DNA regions. dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
Since gene rearrangements through RSs are primary responses to drug pressure, a reasonable hypothesis was that identifying recombinase proteins involved in these rearrangements could lead to strategies to prevent the emergence of resistance. A first candidate was the RAD51 DNA repair protein, a key protein involved in homologous recombination (HR), a mechanism evolutionarily conserved in trypanosomatids 89. Interestingly, the expression of RAD51 was induced in Leishmania by DNA double-strand breaks (DSBs) 90, 91. Inactivating RAD51 led to viable parasites unable to generate circular extrachromosomal elements but still capable of producing linear amplicons upon drug pressure 73. Leishmania has three RAD51 paralogs (RAD51-3, RAD51-4, and RAD51-6) that were shown to work as a complex in promoting HR through their capacity to stimulate RAD51 activity 92. Inactivation of RAD51-4 was also shown to prevent the formation of circular amplicons in L. infantum exposed to drugs, but not of linear amplicons 92.
Linear amplicons are formed by the annealing of RSs found in an inverted orientation 64, 73, 93. MRE11 is a DNA repair nuclease that interacts with RAD50 and NBS1 to form the MRN complex 94, 95 and is important for DSB repair by HR 96, 97 or for non-homologous end joining 89 ( Figure 1b). Inactivation of MRE11 impaired the ability of L. infantum to form linear amplicons upon drug selection at the level of inverted RSs, although the capacity to generate circular amplicons was similar to that of wild-type parasites 93. Moreover, a fully functional MRE11 is important for linear amplification, as parasites expressing DNA-binding-proficient but nuclease-deficient MRE11 exclusively generated circular amplicons during drug selection 93. Interestingly, inactivation of MRE11 alone or along with its partner RAD50 led to extensive chromosomal translocation in L. infantum 98, showing that the MRE11/RAD50 complex is important for the maintenance of genome integrity in addition to its role in gene rearrangements. The number of enzymes involved in the formation of extrachromosomal elements suggests that targeting this pathway may not be a viable strategy for preventing the emergence of resistance, although this remains to be experimentally tested.
Single-nucleotide polymorphisms and small nucleotide insertions or deletions
Although CNVs are important contributors of drug resistance, SNPs and small nucleotide insertions or deletions (indels) can also contribute to resistance. This was proven with experimental drugs 99, 100 and was highlighted with MTF where amino acid substitutions or non-sense mutations were observed in the MTF transporter (MT) 101 or in its Ros3 subunit 102. This was further confirmed in additional mutants using whole genome sequencing 103, 104 or by deep sequencing of MT 105. Mutations detected in the MT gene of L. infantum isolates serially collected from an MTF-treated patient who had multiple relapses were shown to correlate with resistance 106, suggesting that MTF resistance could become a clinical reality in the near future.
Genome-wide surveys of genetic variations in L. donovani isolates from the Indian subcontinent supported the notion that resistance to antimonials emerged on several distinct occasions 58, 107. Isolates could also be clustered on the basis of their genetic structure and haplotypes, with some groups being enriched for non-responsive strains 58, 107, 108. Interestingly, a particular group of highly resistant isolates that clustered together were found to share genomic features associated with resistance 107. Among these were a higher copy number for the H-locus, coding for the well-characterized ABC transporter MRPA 109, and a homozygous two-base-pair insertion in the aquaglyceroporin 1 (AQP1) gene involved in Sb uptake and whose inactivation or downregulation is strongly correlated with resistance 71, 107, 110– 117. The potential for these genomic variants in predicting treatment outcome is exciting, given the lack of molecular markers for Sb resistance, but will need to be thoroughly evaluated by using larger and geographically diversified sets of well-defined isolates.
Exploiting copy number variations for understanding drug mode of action and resistance mechanisms
Target-based assays and phenotypic whole-cell-based assays are the cornerstones of drug discovery. The current trend for anti-parasitic agents is for whole cell assays. A drawback of phenotype-based assays is the lack of knowledge about the targets of hit compounds. Although the molecules could be brought to the clinic without further knowledge about their MOA, a clear understanding of the molecular targets will facilitate the improvement of a candidate drug through lead optimization. Characterization of drug-resistant mutants, which often revealed mutations or CNVs in drug targets or in proteins responsible for drug transport, is one strategy to pinpoint drug targets. However, it is salient to point out that this strategy has not yet led to targets against the current anti-leishmanials, although amplification of gene targets was observed in mutants made resistant to experimental drugs 64, 99. Since CNVs are often associated with resistance, forward genetic tools can experimentally mimic this. One such gain-of-function screen was based on functional cloning where Leishmania cosmid libraries were electroporated into Leishmania and these transfectants were selected for a specific phenotype 118. Selection is possible because of the high copy number (and gene expression) of the cosmids. This screen was successfully applied while selecting for drug resistance or susceptibility 101, 114, 119– 123. This technique selects for cosmids conferring dominant phenotypes (leaving out less enriched cosmids) and is not easily amenable to high-throughput screening. The sensitivity of cosmid-based functional screening was enhanced by its recent coupling to next-generation sequencing in an approach termed Cos-Seq 124. The proportion of parasites with cosmids providing a selective advantage is expected to rise with increasing drug pressure, and these can be tracked and quantified at each drug increment by Illumina sequencing 124, 125. Thus, the dynamics of cosmid enrichment can be followed over the entire course of selection instead of being monitored only at endpoint. Published or ongoing Cos-Seq screens using experimental drugs with known targets (for example, methotrexate, terbinafine, and 5-fluorouracil) confirmed the recovery of the relevant target genes by Cos-Seq 124. Interestingly, Cos-Seq supported the hypothesis that the current anti-leishmanials (MF, AMB, Sb, PMM, and PTD) may not act via specific major protein targets 124. Indeed, although an unprecedented number of resistance genes (known and novel) were isolated using Cos-Seq, none emerged as a clear target candidate and it is conceivable that these antiquated drugs are broadly cytotoxic by disrupting multiple minor targets. Whether some of the genes are genuine drug targets remains to be established, and non-protein targets represent another possibility as these would not be detected by Cos-Seq.
The advent of high-content screening for intracellular L. donovani amastigotes 126 is also key in the search for novel molecules having favourable anti-leishmanial properties directly on the intracellular stage of the parasite. This allowed the discovery of 192 new leads against visceral leishmaniasis from an initial set of 1.8 million compounds from GlaxoSmithKline 127. An MOA could be hypothesized for 80 of the lead compounds using prior proprietary knowledge and bioinformatics analyses of TriTryp genomes, which revealed an over-representation of putative kinase inhibitors 127. Cos-Seq was initially carried out with the insect form of the parasite but this could easily be adapted to intracellular parasites and this technique could be used to find the targets of these promising novel molecules or of other drugs repurposed against Leishmania 128.
The Cos-Seq technique does not allow the isolation of loss-of-function mutations such as those found in the aquaglyceroporin AQP1 or in the MT transporter genes (see above). These require high-throughput dominant negative screening approaches, like inducible RNA interference target sequencing (RIT-Seq), which proved instrumental in elucidating mechanisms of drug uptake in trypanosomes 129. Although RNA interference is absent from the L. Leishmania subgenus, it is active in species of the L. Viannia subgenus 130. The lack of inducible expression in Leishmania was also a limitation of this technique, but two recent reports have shown the feasibility of inducible expression in Leishmania 131, 132. Thus, it is theoretically possible to develop a technology similar to RIT-Seq in L. Viannia parasites. An alternative approach to RIT-Seq would be to rely on RNA-guided nuclease systems using clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) enzymes, as these have proven very efficient for achieving targeted genomic modifications in a wide range of genomes 133– 135. In trypanosomatid parasites, the CRISPR/Cas9 system derived from Streptococcus pyogenes has been used for disrupting genes in L. major 136, L. donovani 137, and T. cruzi 138 and in principle could be used for generating whole-genome Cas9-mediated gene deletion libraries.
Concluding remarks
The toolkit for drug target discovery and resistance mechanism elucidation for Leishmania is expanding. With new promising drug candidates in the pipeline and further technological developments, it should now be possible to find new targets which should further help in the control of this important neglected tropical disease.
Abbreviations
3′ UTR, 3′ untranslated region; AMB, amphotericin B; AQP1, aquaglyceroporin 1; Cas, clustered regularly interspaced short palindromic repeat-associated; CNV, copy number variation; CRISPR, clustered regularly interspaced short palindromic repeat; DSB, double-strand break; HR, homologous recombination; MOA, mode of action; mRNA, messenger RNA; MT, miltefosine transporter; MTF, miltefosine; PMM, paromomycin; PTD, pentamidine; RIT-Seq, RNA interference target sequencing; RS, repeated sequence; Sb, antimony; SIDER, short interspersed degenerate retroposon; SNP, single-nucleotide polymorphism.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Gerald Spaeth, Unité de Parasitologie moléculaire et Signalisation, Department of Parasites and Insect Vectors, Institut Pasteur, INSERM U1201, Paris, France
Yvon Sterkers, UMR MIVEGEC (CNRS 5290 - IRD 224 - University Montpellier), University Regional Hospital of Montpellier, Montpellier, France
Richard McCulloch, The Wellcome Trust Centre for Molecular Parasitology, Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow, UK
Joachim Clos, Bernhard Nocht Institute for Tropical Medicine, Hamburg, Germany
Francisco Gamarro, Instituto de Parasitología Lopez-Neyra, IPBLN-CSIC, Granada, Spain
Funding Statement
BP is supported by grants from the Canadian Institutes of Health Research, the Natural Sciences and Engineering Research Council of Canada, and the Ministère du Developpement Économique de l’Innovation et de l’Exportation du Québec. MO is supported by grants from the Canadian Institutes of Health Research and holds a Canada Research Chair on Antimicrobial Resistance.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 5 approved]
References
- 1. Alvar J, Vélez ID, Bern C, et al. : Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7(5):e35671. 10.1371/journal.pone.0035671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bañuls AL, Hide M, Prugnolle F: Leishmania and the leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv Parasitol. 2007;64:1–109. 10.1016/S0065-308X(06)64001-3 [DOI] [PubMed] [Google Scholar]
- 3. Guerin PJ, Olliaro P, Sundar S, et al. : Visceral leishmaniasis: current status of control, diagnosis, and treatment, and a proposed research and development agenda. Lancet Infect Dis. 2002;2(8):494–501. 10.1016/S1473-3099(02)00347-X [DOI] [PubMed] [Google Scholar]
- 4. David CV, Craft N: Cutaneous and mucocutaneous leishmaniasis. Dermatol Ther. 2009;22(6):491–502. 10.1111/j.1529-8019.2009.01272.x [DOI] [PubMed] [Google Scholar]
- 5. Herwaldt BL: Leishmaniasis. Lancet. 1999;354(9185):1191–9. 10.1016/S0140-6736(98)10178-2 [DOI] [PubMed] [Google Scholar]
- 6. Gradoni L: Canine Leishmania vaccines: still a long way to go. Vet Parasitol. 2015;208(1–2):94–100. 10.1016/j.vetpar.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 7. Handman E: Leishmaniasis: current status of vaccine development. Clin Microbiol Rev. 2001;14(2):229–43. 10.1128/CMR.14.2.229-243.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murray HW, Berman JD, Davies CR, et al. : Advances in leishmaniasis. Lancet. 2005;366(9496):1561–77. 10.1016/S0140-6736(05)67629-5 [DOI] [PubMed] [Google Scholar]
- 9. Palatnik-de-Sousa CB: Vaccines for leishmaniasis in the fore coming 25 years. Vaccine. 2008;26(14):1709–24. 10.1016/j.vaccine.2008.01.023 [DOI] [PubMed] [Google Scholar]
- 10. Sundar S, Chakravarty J, Agarwal D, et al. : Single-dose liposomal amphotericin B for visceral leishmaniasis in India. N Engl J Med. 2010;362(6):504–12. 10.1056/NEJMoa0903627 [DOI] [PubMed] [Google Scholar]
- 11. Bern C, Adler-Moore J, Berenguer J, et al. : Liposomal amphotericin B for the treatment of visceral leishmaniasis. Clin Infect Dis. 2006;43(7):917–24. 10.1086/507530 [DOI] [PubMed] [Google Scholar]
- 12. Berman JD, Badaro R, Thakur CP, et al. : Efficacy and safety of liposomal amphotericin B (AmBisome) for visceral leishmaniasis in endemic developing countries. Bull World Health Organ. 1998;76(1):25–32. [PMC free article] [PubMed] [Google Scholar]
- 13. Lachaud L, Bourgeois N, Plourde M, et al. : Parasite susceptibility to amphotericin B in failures of treatment for visceral leishmaniasis in patients coinfected with HIV type 1 and Leishmania infantum. Clin Infect Dis. 2009;48(2):e16–22. 10.1086/595710 [DOI] [PubMed] [Google Scholar]
- 14. Croft SL, Neal RA, Pendergast W, et al. : The activity of alkyl phosphorylcholines and related derivatives against Leishmania donovani. Biochem Pharmacol. 1987;36(16):2633–6. 10.1016/0006-2952(87)90543-0 [DOI] [PubMed] [Google Scholar]
- 15. Jha TK, Sundar S, Thakur CP, et al. : Miltefosine, an oral agent, for the treatment of Indian visceral leishmaniasis. N Engl J Med. 1999;341(24):1795–800. 10.1056/NEJM199912093412403 [DOI] [PubMed] [Google Scholar]
- 16. Sundar S, Jha TK, Thakur CP, et al. : Oral miltefosine for Indian visceral leishmaniasis. N Engl J Med. 2002;347(22):1739–46. 10.1056/NEJMoa021556 [DOI] [PubMed] [Google Scholar]
- 17. Burza S, Nabi E, Mahajan R, et al. : One-year follow-up of immunocompetent male patients treated with miltefosine for primary visceral leishmaniasis in Bihar, India. Clin Infect Dis. 2013;57(9):1363–4. 10.1093/cid/cit508 [DOI] [PubMed] [Google Scholar]
- 18. Sundar S, Singh A, Rai M, et al. : Efficacy of miltefosine in the treatment of visceral leishmaniasis in India after a decade of use. Clin Infect Dis. 2012;55(4):543–50. 10.1093/cid/cis474 [DOI] [PubMed] [Google Scholar]
- 19. Pandey BD, Pandey K, Kaneko O, et al. : Relapse of visceral leishmaniasis after miltefosine treatment in a Nepalese patient. Am J Trop Med Hyg. 2009;80(4):580–2. [PubMed] [Google Scholar]
- 20. Rijal S, Ostyn B, Uranw S, et al. : Increasing failure of miltefosine in the treatment of Kala-azar in Nepal and the potential role of parasite drug resistance, reinfection, or noncompliance. Clin Infect Dis. 2013;56(11):1530–8. 10.1093/cid/cit102 [DOI] [PubMed] [Google Scholar]
- 21. Jha TK, Olliaro P, Thakur CP, et al. : Randomised controlled trial of aminosidine (paromomycin) v sodium stibogluconate for treating visceral leishmaniasis in North Bihar, India. BMJ. 1998;316(7139):1200–5. 10.1136/bmj.316.7139.1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sundar S, Jha TK, Thakur CP, et al. : Injectable paromomycin for Visceral leishmaniasis in India. N Engl J Med. 2007;356(25):2571–81. 10.1056/NEJMoa066536 [DOI] [PubMed] [Google Scholar]
- 23. Hailu A, Musa A, Wasunna M, et al. : Geographical variation in the response of visceral leishmaniasis to paromomycin in East Africa: a multicentre, open-label, randomized trial. PLoS Negl Trop Dis. 2010;4(10):e709. 10.1371/journal.pntd.0000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Musa AM, Younis B, Fadlalla A, et al. : Paromomycin for the treatment of visceral leishmaniasis in Sudan: a randomized, open-label, dose-finding study. PLoS Negl Trop Dis. 2010;4(10):e855. 10.1371/journal.pntd.0000855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lai A Fat EJ, Vrede MA, Soetosenojo RM, et al. : Pentamidine, the drug of choice for the treatment of cutaneous leishmaniasis in Surinam. Int J Dermatol. 2002;41(11):796–800. 10.1046/j.1365-4362.2002.01633.x [DOI] [PubMed] [Google Scholar]
- 26. Roussel M, Nacher M, Frémont G, et al. : Comparison between one and two injections of pentamidine isethionate, at 7 mg/kg in each injection, in the treatment of cutaneous leishmaniasis in French Guiana. Ann Trop Med Parasitol. 2006;100(4):307–14. 10.1179/136485906X105561 [DOI] [PubMed] [Google Scholar]
- 27. Soto J, Buffet P, Grogl M, et al. : Successful treatment of Colombian cutaneous leishmaniasis with four injections of pentamidine. Am J Trop Med Hyg. 1994;50(1):107–11. [DOI] [PubMed] [Google Scholar]
- 28. Mandal G, Wyllie S, Singh N, et al. : Increased levels of thiols protect antimony unresponsive Leishmania donovani field isolates against reactive oxygen species generated by trivalent antimony. Parasitology. 2007;134(Pt 12):1679–87. 10.1017/S0031182007003150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mehta A, Shaha C: Mechanism of metalloid-induced death in Leishmania spp.: role of iron, reactive oxygen species, Ca 2+, and glutathione. Free Radic Biol Med. 2006;40(10):1857–68. 10.1016/j.freeradbiomed.2006.01.024 [DOI] [PubMed] [Google Scholar]
- 30. Mookerjee Basu J, Mookerjee A, Sen P, et al. : Sodium antimony gluconate induces generation of reactive oxygen species and nitric oxide via phosphoinositide 3-kinase and mitogen-activated protein kinase activation in Leishmania donovani-infected macrophages. Antimicrob Agents Chemother. 2006;50(5):1788–97. 10.1128/AAC.50.5.1788-1797.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moreira W, Leprohon P, Ouellette M: Tolerance to drug-induced cell death favours the acquisition of multidrug resistance in Leishmania. Cell Death Dis. 2011;2:e201. 10.1038/cddis.2011.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wyllie S, Cunningham ML, Fairlamb AH: Dual action of antimonial drugs on thiol redox metabolism in the human pathogen Leishmania donovani. J Biol Chem. 2004;279(38):39925–32. 10.1074/jbc.M405635200 [DOI] [PubMed] [Google Scholar]
- 33. Lee N, Bertholet S, Debrabant A, et al. : Programmed cell death in the unicellular protozoan parasite Leishmania. Cell Death Differ. 2002;9(1):53–64. 10.1038/sj.cdd.4400952 [DOI] [PubMed] [Google Scholar]
- 34. Sereno D, Holzmuller P, Mangot I, et al. : Antimonial-mediated DNA fragmentation in Leishmania infantum amastigotes. Antimicrob Agents Chemother. 2001;45(7):2064–9. 10.1128/AAC.45.7.2064-2069.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sudhandiran G, Shaha C: Antimonial-induced increase in intracellular Ca 2+ through non-selective cation channels in the host and the parasite is responsible for apoptosis of intracellular Leishmania donovani amastigotes. J Biol Chem. 2003;278(27):25120–32. 10.1074/jbc.M301975200 [DOI] [PubMed] [Google Scholar]
- 36. Vergnes B, Gourbal B, Girard I, et al. : A proteomics screen implicates HSP83 and a small kinetoplastid calpain-related protein in drug resistance in Leishmania donovani clinical field isolates by modulating drug-induced programmed cell death. Mol Cell Proteomics. 2007;6(1):88–101. 10.1074/mcp.M600319-MCP200 [DOI] [PubMed] [Google Scholar]
- 37. Ivens AC, Peacock CS, Worthey EA, et al. : The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309(5733):436–42. 10.1126/science.1112680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peacock CS, Seeger K, Harris D, et al. : Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet. 2007;39(7):839–47. 10.1038/ng2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Britto C, Ravel C, Bastien P, et al. : Conserved linkage groups associated with large-scale chromosomal rearrangements between Old World and New World Leishmania genomes. Gene. 1998;222(1):107–17. 10.1016/S0378-1119(98)00472-7 [DOI] [PubMed] [Google Scholar]
- 40. Raymond F, Boisvert S, Roy G, et al. : Genome sequencing of the lizard parasite Leishmania tarentolae reveals loss of genes associated to the intracellular stage of human pathogenic species. Nucleic Acids Res. 2012;40(3):1131–47. 10.1093/nar/gkr834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wincker P, Ravel C, Blaineau C, et al. : The Leishmania genome comprises 36 chromosomes conserved across widely divergent human pathogenic species. Nucleic Acids Res. 1996;24(9):1688–94. 10.1093/nar/24.9.1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. El-Sayed NM, Myler PJ, Blandin G, et al. : Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–9. 10.1126/science.1112181 [DOI] [PubMed] [Google Scholar]
- 43. Martinez-Calvillo S, Yan S, Nguyen D, et al. : Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol Cell. 2003;11(5):1291–9. 10.1016/S1097-2765(03)00143-6 [DOI] [PubMed] [Google Scholar]
- 44. Martinez-Calvillo S, Nguyen D, Stuart K, et al. : Transcription initiation and termination on Leishmania major chromosome 3. Eukaryotic Cell. 2004;3(2):506–17. 10.1128/EC.3.2.506-517.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Haile S, Papadopoulou B: Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr Opin Microbiol. 2007;10(6):569–77. 10.1016/j.mib.2007.10.001 [DOI] [PubMed] [Google Scholar]
- 46. Matthews KR, Tschudi C, Ullu E: A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8(4):491–501. 10.1101/gad.8.4.491 [DOI] [PubMed] [Google Scholar]
- 47. Boucher N, Wu Y, Dumas C, et al. : A common mechanism of stage-regulated gene expression in Leishmania mediated by a conserved 3'-untranslated region element. J Biol Chem. 2002;277(22):19511–20. 10.1074/jbc.M200500200 [DOI] [PubMed] [Google Scholar]
- 48. Bringaud F, Muller M, Cerqueira GC, et al. : Members of a large retroposon family are determinants of post-transcriptional gene expression in Leishmania. PLoS Pathog. 2007;3(9):1291–307, e136. 10.1371/journal.ppat.0030136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dupe A, Dumas C, Papadopoulou B: An Alba-domain protein contributes to the stage-regulated stability of amastin transcripts in Leishmania. Mol Microbiol. 2014;91(3):548–61. 10.1111/mmi.12478 [DOI] [PubMed] [Google Scholar]
- 50. Haile S, Dupe A, Papadopoulou B: Deadenylation-independent stage-specific mRNA degradation in Leishmania. Nucleic Acids Res. 2008;36(5):1634–44. 10.1093/nar/gkn019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McNicoll F, Muller M, Cloutier S, et al. : Distinct 3'-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J Biol Chem. 2005;280(42):35238–46. 10.1074/jbc.M507511200 [DOI] [PubMed] [Google Scholar]
- 52. Muller M, Padmanabhan PK, Papadopoulou B: Selective inactivation of SIDER2 retroposon-mediated mRNA decay contributes to stage- and species-specific gene expression in Leishmania. Mol Microbiol. 2010;77(2):471–91. 10.1111/j.1365-2958.2010.07226.x [DOI] [PubMed] [Google Scholar]
- 53. Muller M, Padmanabhan PK, Rochette A, et al. : Rapid decay of unstable Leishmania mRNAs bearing a conserved retroposon signature 3'-UTR motif is initiated by a site-specific endonucleolytic cleavage without prior deadenylation. Nucleic Acids Res. 2010;38(17):5867–83. 10.1093/nar/gkq349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gordon DJ, Resio B, Pellman D: Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13(3):189–203. 10.1038/nrg3123 [DOI] [PubMed] [Google Scholar]
- 55. Pfau SJ, Amon A: Chromosomal instability and aneuploidy in cancer: from yeast to man. EMBO Rep. 2012;13(6):515–27. 10.1038/embor.2012.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rutledge SD, Cimini D: Consequences of aneuploidy in sickness and in health. Curr Opin Cell Biol. 2016;40:41–6. 10.1016/j.ceb.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 57. Shor E, Perlin DS: Coping with stress and the emergence of multidrug resistance in fungi. PLoS Pathog. 2015;11(3):e1004668. 10.1371/journal.ppat.1004668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Downing T, Imamura H, Decuypere S, et al. : Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21(12):2143–56. 10.1101/gr.123430.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Leprohon P, Legare D, Raymond F, et al. : Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 2009;37(5):1387–99. 10.1093/nar/gkn1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mannaert A, Downing T, Imamura H, et al. : Adaptive mechanisms in pathogens: universal aneuploidy in Leishmania. Trends Parasitol. 2012;28(9):370–6. 10.1016/j.pt.2012.06.003 [DOI] [PubMed] [Google Scholar]
- 61. Rogers MB, Hilley JD, Dickens NJ, et al. : Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21(12):2129–42. 10.1101/gr.122945.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sterkers Y, Lachaud L, Bourgeois N, et al. : Novel insights into genome plasticity in Eukaryotes: mosaic aneuploidy in Leishmania. Mol Microbiol. 2012;86(1):15–23. 10.1111/j.1365-2958.2012.08185.x [DOI] [PubMed] [Google Scholar]
- 63. Sterkers Y, Lachaud L, Crobu L, et al. : FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell Microbiol. 2011;13(2):274–83. 10.1111/j.1462-5822.2010.01534.x [DOI] [PubMed] [Google Scholar]
- 64. Ubeda JM, Légaré D, Raymond F, et al. : Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9(7):R115. 10.1186/gb-2008-9-7-r115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lachaud L, Bourgeois N, Kuk N, et al. : Constitutive mosaic aneuploidy is a unique genetic feature widespread in the Leishmania genus. Microbes Infect. 2014;16(1):61–6. 10.1016/j.micinf.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 66. Minning TA, Weatherly DB, Flibotte S, et al. : Widespread, focal copy number variations (CNV) and whole chromosome aneuploidies in Trypanosoma cruzi strains revealed by array comparative genomic hybridization. BMC Genomics. 2011;12:139. 10.1186/1471-2164-12-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Reis-Cunha JL, Rodrigues-Luiz GF, Valdivia HO, et al. : Chromosomal copy number variation reveals differential levels of genomic plasticity in distinct Trypanosoma cruzi strains. BMC Genomics. 2015;16:499. 10.1186/s12864-015-1680-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Brotherton MC, Bourassa S, Leprohon P, et al. : Proteomic and genomic analyses of antimony resistant Leishmania infantum mutant. PLoS One. 2013;8(11):e81899. 10.1371/journal.pone.0081899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. do Monte-Neto RL, Coelho AC, Raymond F, et al. : Gene expression profiling and molecular characterization of antimony resistance in Leishmania amazonensis. PLoS Negl Trop Dis. 2011;5(5):e1167. 10.1371/journal.pntd.0001167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kumar P, Lodge R, Raymond F, et al. : Gene expression modulation and the molecular mechanisms involved in Nelfinavir resistance in Leishmania donovani axenic amastigotes. Mol Microbiol. 2013;89(3):565–82. 10.1111/mmi.12298 [DOI] [PubMed] [Google Scholar]
- 71. Mukherjee A, Boisvert S, Monte-Neto RL, et al. : Telomeric gene deletion and intrachromosomal amplification in antimony-resistant Leishmania. Mol Microbiol. 2013;88(1):189–202. 10.1111/mmi.12178 [DOI] [PubMed] [Google Scholar]
- 72. Sterkers Y, Crobu L, Lachaud L, et al. : Parasexuality and mosaic aneuploidy in Leishmania: alternative genetics. Trends Parasitol. 2014;30(9):429–35. 10.1016/j.pt.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 73. Ubeda JM, Raymond F, Mukherjee A, et al. : Genome-wide stochastic adaptive DNA amplification at direct and inverted DNA repeats in the parasite Leishmania. PLoS Biol. 2014;12(5):e1001868. 10.1371/journal.pbio.1001868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ouellette M, Papadopoulou B: Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J Biol. 2009;8(11):100. 10.1186/jbiol203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Smith M, Bringaud F, Papadopoulou B: Organization and evolution of two SIDER retroposon subfamilies and their impact on the Leishmania genome. BMC Genomics. 2009;10:240. 10.1186/1471-2164-10-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Beverley SM: Gene amplification in Leishmania. Annu Rev Microbiol. 1991;45:417–44. 10.1146/annurev.mi.45.100191.002221 [DOI] [PubMed] [Google Scholar]
- 77. Beverley SM, Coderre JA, Santi DV, et al. : Unstable DNA amplifications in methotrexate-resistant Leishmania consist of extrachromosomal circles which relocalize during stabilization. Cell. 1984;38(2):431–9. 10.1016/0092-8674(84)90498-7 [DOI] [PubMed] [Google Scholar]
- 78. Garvey EP, Santi DV: Stable amplified DNA in drug-resistant Leishmania exists as extrachromosomal circles. Science. 1986;233(4763):535–40. 10.1126/science.3726545 [DOI] [PubMed] [Google Scholar]
- 79. Grondin K, Haimeur A, Mukhopadhyay R, et al. : Co-amplification of the gamma-glutamylcysteine synthetase gene gsh1 and of the ABC transporter gene pgpA in arsenit e-resistant Leishmania tarentolae. EMBO J. 1997;16(11):3057–65. 10.1093/emboj/16.11.3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Grondin K, Papadopoulou B, Ouellette M: Homologous recombination between direct repeat sequences yields P-glycoprotein containing amplicons in arsenite resistant Leishmania. Nucleic Acids Res. 1993;21(8):1895–901. 10.1093/nar/21.8.1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Haimeur A, Brochu C, Genest P, et al. : Amplification of the ABC transporter gene PGPA and increased trypanothione levels in potassium antimonyl tartrate (SbIII) resistant Leishmania tarentolae. Mol Biochem Parasitol. 2000;108(1):131–5. 10.1016/S0166-6851(00)00187-0 [DOI] [PubMed] [Google Scholar]
- 82. Kundig C, Leblanc E, Papadopoulou B, et al. : Role of the locus and of the resistance gene on gene amplification frequency in methotrexate resistant Leishmania tarentolae. Nucleic Acids Res. 1999;27(18):3653–9. 10.1093/nar/27.18.3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mittal MK, Rai S, Ashutosh, et al. : Characterization of natural antimony resistance in Leishmania donovani isolates. Am J Trop Med Hyg. 2007;76(4):681–8. [PubMed] [Google Scholar]
- 84. Mukherjee A, Padmanabhan PK, Singh S, et al. : Role of ABC transporter MRPA, gamma-glutamylcysteine synthetase and ornithine decarboxylase in natural antimony-resistant isolates of Leishmania donovani. J Antimicrob Chemother. 2007;59(2):204–11. 10.1093/jac/dkl494 [DOI] [PubMed] [Google Scholar]
- 85. Ouellette M, Hettema E, Wüst D, et al. : Direct and inverted DNA repeats associated with P-glycoprotein gene amplification in drug resistant Leishmania. EMBO J. 1991;10(4):1009–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Papadopoulou B, Roy G, Ouellette M: Frequent amplification of a short chain dehydrogenase gene as part of circular and linear amplicons in methotrexate resistant Leishmania. Nucleic Acids Res. 1993;21(18):4305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. White TC, Fase-Fowler F, van Luenen H, et al. : The H circles of Leishmania tarentolae are a unique amplifiable system of oligomeric DNAs associated with drug resistance. J Biol Chem. 1988;263(32):16977–83. [PubMed] [Google Scholar]
- 88. Navarro M, Liu J, Muthui D, et al. : Inverted repeat structure and homologous sequences in the LD 1 amplicons of Leishmania spp. Mol Biochem Parasitol. 1994;68(1):69–80. 10.1016/0166-6851(94)00147-2 [DOI] [PubMed] [Google Scholar]
- 89. Genois MM, Paquet ER, Laffitte MC, et al. : DNA repair pathways in trypanosomatids: from DNA repair to drug resistance. Microbiol Mol Biol Rev. 2014;78(1):40–73. 10.1128/MMBR.00045-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Genois MM, Mukherjee A, Ubeda JM, et al. : Interactions between BRCA2 and RAD51 for promoting homologous recombination in Leishmania infantum. Nucleic Acids Res. 2012;40(14):6570–84. 10.1093/nar/gks306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. McKean PG, Keen JK, Smith DF, et al. : Identification and characterisation of a RAD51 gene from Leishmania major. Mol Biochem Parasitol. 2001;115(2):209–16. 10.1016/S0166-6851(01)00288-2 [DOI] [PubMed] [Google Scholar]
- 92. Genois MM, Plourde M, Éthier C, et al. : Roles of Rad51 paralogs for promoting homologous recombination in Leishmania infantum. Nucleic Acids Res. 2015;43(5):2701–15. 10.1093/nar/gkv118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Laffitte MC, Genois MM, Mukherjee A, et al. : Formation of linear amplicons with inverted duplications in Leishmania requires the MRE11 nuclease. PLoS Genet. 2014;10(12):e1004805. 10.1371/journal.pgen.1004805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Assenmacher N, Hopfner KP: MRE11/RAD50/NBS1: complex activities. Chromosoma. 2004;113(4):157–66. 10.1007/s00412-004-0306-4 [DOI] [PubMed] [Google Scholar]
- 95. Stracker TH, Petrini JH: The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol. 2011;12(2):90–103. 10.1038/nrm3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Shibata A, Moiani D, Arvai AS, et al. : DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell. 2014;53(1):7–18. 10.1016/j.molcel.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Mimitou EP, Symington LS: DNA end resection--unraveling the tail. DNA Repair (Amst ). 2011;10(3):344–8. 10.1016/j.dnarep.2010.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Laffitte MC, Leprohon P, Hainse M, et al. : Chromosomal Translocations in the Parasite Leishmania by a MRE11/RAD50-Independent Microhomology-Mediated End Joining Mechanism. PLoS Genet. 2016;12(6):e1006117. 10.1371/journal.pgen.1006117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ritt JF, Raymond F, Leprohon P, et al. : Gene amplification and point mutations in pyrimidine metabolic genes in 5-fluorouracil resistant Leishmania infantum. PLoS Negl Trop Dis. 2013;7(11):e2564. 10.1371/journal.pntd.0002564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Vasudevan G, Ullman B, Landfear SM: Point mutations in a nucleoside transporter gene from Leishmania donovani confer drug resistance and alter substrate selectivity. Proc Natl Acad Sci USA. 2001;98(11):6092–7. 10.1073/pnas.101537298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Pérez-Victoria FJ, Gamarro F, Ouellette M, et al. : Functional cloning of the miltefosine transporter. A novel P-type phospholipid translocase from Leishmania involved in drug resistance. J Biol Chem. 2003;278(50):49965–71. 10.1074/jbc.M308352200 [DOI] [PubMed] [Google Scholar]
- 102. Pérez-Victoria FJ, Sánchez-Cañete MP, Castanys S, et al. : Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J Biol Chem. 2006;281(33):23766–75. 10.1074/jbc.M605214200 [DOI] [PubMed] [Google Scholar]
- 103. Coelho AC, Boisvert S, Mukherjee A, et al. : Multiple mutations in heterogeneous miltefosine-resistant Leishmania major population as determined by whole genome sequencing. PLoS Negl Trop Dis. 2012;6(2):e1512. 10.1371/journal.pntd.0001512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Mondelaers A, Sanchez-Cañete MP, Hendrickx S, et al. : Genomic and Molecular Characterization of Miltefosine Resistance in Leishmania infantum Strains with Either Natural or Acquired Resistance through Experimental Selection of Intracellular Amastigotes. PLoS One. 2016;11(4):e0154101. 10.1371/journal.pone.0154101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Laffitte MN, Leprohon P, Légaré D, et al. : Deep-sequencing revealing mutation dynamics in the miltefosine transporter gene in Leishmania infantum selected for miltefosine resistance. Parasitol Res. 2016;1–5. 10.1007/s00436-016-5195-y [DOI] [PubMed] [Google Scholar]
- 106. Cojean S, Houzé S, Haouchine D, et al. : Leishmania resistance to miltefosine associated with genetic marker. Emerging Infect Dis. 2012;18(4):704–6. 10.3201/eid1804.110841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Imamura H, Downing T, Van den Broeck F, et al. : Evolutionary genomics of epidemic visceral leishmaniasis in the Indian subcontinent. eLife. 2016;5: pii: e12613. 10.7554/eLife.12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Vanaerschot M, Decuypere S, Downing T, et al. : Genetic markers for SSG resistance in Leishmania donovani and SSG treatment failure in visceral leishmaniasis patients of the Indian subcontinent. J Infect Dis. 2012;206(5):752–5. 10.1093/infdis/jis424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Légaré D, Richard D, Mukhopadhyay R, et al. : The Leishmania ATP-binding cassette protein PGPA is an intracellular metal-thiol transporter ATPase. J Biol Chem. 2001;276(28):26301–7. 10.1074/jbc.M102351200 [DOI] [PubMed] [Google Scholar]
- 110. Decuypere S, Rijal S, Yardley V, et al. : Gene expression analysis of the mechanism of natural Sb(V) resistance in Leishmania donovani isolates from Nepal. Antimicrob Agents Chemother. 2005;49(11):4616–21. 10.1128/AAC.49.11.4616-4621.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Gourbal B, Sonuc N, Bhattacharjee H, et al. : Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J Biol Chem. 2004;279(30):31010–7. 10.1074/jbc.M403959200 [DOI] [PubMed] [Google Scholar]
- 112. Mandal G, Mandal S, Sharma M, et al. : Species-specific antimonial sensitivity in Leishmania is driven by post-transcriptional regulation of AQP1. PLoS Negl Trop Dis. 2015;9(2):e0003500. 10.1371/journal.pntd.0003500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mandal S, Maharjan M, Singh S, et al. : Assessing aquaglyceroporin gene status and expression profile in antimony-susceptible and -resistant clinical isolates of Leishmania donovani from India. J Antimicrob Chemother. 2010;65(3):496–507. 10.1093/jac/dkp468 [DOI] [PubMed] [Google Scholar]
- 114. Marquis N, Gourbal B, Rosen BP, et al. : Modulation in aquaglyceroporin AQP1 gene transcript levels in drug-resistant Leishmania. Mol Microbiol. 2005;57(6):1690–9. 10.1111/j.1365-2958.2005.04782.x [DOI] [PubMed] [Google Scholar]
- 115. Monte-Neto R, Laffitte MC, Leprohon P, et al. : Intrachromosomal amplification, locus deletion and point mutation in the aquaglyceroporin AQP1 gene in antimony resistant Leishmania (Viannia) guyanensis. PLoS Negl Trop Dis. 2015;9:e0003476. 10.1371/journal.pntd.0003476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Plourde M, Ubeda JM, Mandal G, et al. : Generation of an aquaglyceroporin AQP1 null mutant in Leishmania major. Mol Biochem Parasitol. 2015;201(2):108–11. 10.1016/j.molbiopara.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 117. Uzcategui NL, Zhou Y, Figarella K, et al. : Alteration in glycerol and metalloid permeability by a single mutation in the extracellular C-loop of Leishmania major aquaglyceroporin LmAQP1. Mol Microbiol. 2008;70(6):1477–86. 10.1111/j.1365-2958.2008.06494.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ryan KA, Garraway LA, Descoteaux A, et al. : Isolation of virulence genes directing surface glycosyl-phosphatidylinositol synthesis by functional complementation of Leishmania. Proc Natl Acad Sci USA. 1993;90(18):8609–13. 10.1073/pnas.90.18.8609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Carter NS, Drew ME, Sanchez M, et al. : Cloning of a novel inosine-guanosine transporter gene from Leishmania donovani by functional rescue of a transport-deficient mutant. J Biol Chem. 2000;275(27):20935–41. 10.1074/jbc.M002418200 [DOI] [PubMed] [Google Scholar]
- 120. Coelho AC, Beverley SM, Cotrim PC: Functional genetic identification of PRP1, an ABC transporter superfamily member conferring pentamidine resistance in Leishmania major. Mol Biochem Parasitol. 2003;130(2):83–90. 10.1016/S0166-6851(03)00162-2 [DOI] [PubMed] [Google Scholar]
- 121. Cotrim PC, Garrity LK, Beverley SM: Isolation of genes mediating resistance to inhibitors of nucleoside and ergosterol metabolism in Leishmania by overexpression/selection. J Biol Chem. 1999;274(53):37723–30. 10.1074/jbc.274.53.37723 [DOI] [PubMed] [Google Scholar]
- 122. Kundig C, Haimeur A, Legare D, et al. : Increased transport of pteridines compensates for mutations in the high affinity folate transporter and contributes to methotrexate resistance in the protozoan parasite Leishmania tarentolae. EMBO J. 1999;18(9):2342–51. 10.1093/emboj/18.9.2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Vasudevan G, Carter NS, Drew ME, et al. : Cloning of Leishmania nucleoside transporter genes by rescue of a transport-deficient mutant. Proc Natl Acad Sci U S A. 1998;95(17):9873–8. 10.1073/pnas.95.17.9873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gazanion E, Fernandez-Prada C, Papadopoulou B, et al. : Cos-Seq for high-throughput identification of drug target and resistance mechanisms in the protozoan parasite Leishmania. Proc Natl Acad Sci U S A. 2016;113(21):E3012–21. 10.1073/pnas.1520693113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Tejera Nevado P, Bifeld E, Höhn K, et al. : A Telomeric Cluster of Antimony Resistance Genes on Chromosome 34 of Leishmania infantum. Antimicrob Agents Chemother. 2016;60(9):5262–75. 10.1128/AAC.00544-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. De Rycker M, Hallyburton I, Thomas J, et al. : Comparison of a high-throughput high-content intracellular Leishmania donovani assay with an axenic amastigote assay. Antimicrob Agents Chemother. 2013;57(7):2913–22. 10.1128/AAC.02398-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Peña I, Pilar Manzano M, Cantizani J, et al. : New compound sets identified from high throughput phenotypic screening against three kinetoplastid parasites: an open resource. Sci Rep. 2015;5:8771. 10.1038/srep08771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Khraiwesh M, Leed S, Roncal N, et al. : Antileishmanial Activity of Compounds Derived from the Medicines for Malaria Venture Open Access Box Against Intracellular Leishmania major Amastigotes. Am J Trop Med Hyg. 2016;94(2):340–7. 10.4269/ajtmh.15-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Alsford S, Eckert S, Baker N, et al. : High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482(7384):232–6. 10.1038/nature10771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Lye LF, Owens K, Shi H, et al. : Retention and loss of RNA interference pathways in trypanosomatid protozoans. PLoS Pathog. 2010;6(10):e1001161. 10.1371/journal.ppat.1001161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Duncan SM, Myburgh E, Philipon C, et al. : Conditional gene deletion with DiCre demonstrates an essential role for CRK3 in Leishmania mexicana cell cycle regulation. Mol Microbiol. 2016;100(6):931–44. 10.1111/mmi.13375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Kraeva N, Ishemgulova A, Lukeš J, et al. : Tetracycline-inducible gene expression system in Leishmania mexicana. Mol Biochem Parasitol. 2014;198(1):11–3. 10.1016/j.molbiopara.2014.11.002 [DOI] [PubMed] [Google Scholar]
- 133. Sander JD, Joung JK: CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–55. 10.1038/nbt.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Selle K, Barrangou R: Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol. 2015;23(4):225–32. 10.1016/j.tim.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 135. Wright AV, Nuñez JK, Doudna JA: Biology and Applications of CRISPR Systems: Harnessing Nature's Toolbox for Genome Engineering. Cell. 2016;164(1–2):29–44. 10.1016/j.cell.2015.12.035 [DOI] [PubMed] [Google Scholar]
- 136. Sollelis L, Ghorbal M, MacPherson CR, et al. : First efficient CRISPR-Cas9-mediated genome editing in Leishmania parasites. Cell Microbiol. 2015;17(10):1405–12. 10.1111/cmi.12456 [DOI] [PubMed] [Google Scholar]
- 137. Zhang WW, Matlashewski G: CRISPR-Cas9-Mediated Genome Editing in Leishmania donovani. MBio. 2015;6(4):e00861–15. 10.1128/mBio.00861-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Peng D, Kurup SP, Yao PY, et al. : CRISPR-Cas9-mediated single-gene and gene family disruption in Trypanosoma cruzi. MBio. 2015;6(1):e02097–14. 10.1128/mBio.02097-14 [DOI] [PMC free article] [PubMed] [Google Scholar]