Abstract
Background
Epithelial ovarian cancer (EOC) is the deadliest gynecological malignancy, and evidence is accumulating on how molecular markers may be associated with the origin and process of EOC. Sam68 (Src-associated in mitosis, of 68 kD), is a K homology domain RNA-binding protein that has been investigated as a risk factor in multiple types of tumors. The aim of the present study was to investigate the contribution of the Sam68 gene in the pathogenesis of EOC.
Material/Methods
Western blot assay and real-time quantitative PCR methods were performed to examine Sam68 expression in EOC tissue specimens. The association of Sam68 expression with clinic-pathologic variables of EOC was evaluated. Then gain-of-function and loss-of-function strategies were adopted to examine the regulation of Sam68 on the proliferation of EOC OVCAR-3 cells using CCK-8 and colony forming assays.
Results
Sam68 was overexpressed in both mRNA and protein levels in EOC tumor tissue (n=152) in an association with malignant factors of EOC such as International Federation of Gynecology and Obstetrics (FIGO) stage, residual tumor size (cm), histological grade, and lymph node metastasis. In vitro results demonstrated that Sam68 overexpression was upregulated while Sam68 knockdown downregulated the proliferation of EOC OVCAR-3 cells via regulation of cell growth and colony formation.
Conclusions
Sam68 was overexpressed in EOC tissue in association with such cancer malignant factors of FIGO stage, histological grade, and lymph node metastasis, and also positively regulated the proliferation of EOC cells. Our research suggests that Sam68 might accelerate cell cycle progression, and present as a prognostic marker for EOC.
MeSH Keywords: Biological Markers, Cell Proliferation, Ovarian Neoplasms
Background
Epithelial ovarian cancer (EOC) is the deadliest gynecological malignancy [1], with a median overall survival of only 2–4 years [2]. Multiple genetic variants in a number of genes may be involved in the origin and process of EOC [3], such as RAD51C [4], RAD51D [5], CASP8 [6], LIN28B [7], or BRCA [8]. However, only a few genetic variants exhibited strong evidence of an association with ovarian cancer, and the identification of genes associated with a predisposition to ovarian cancer requires further investigation [9]. Recently, molecular markers have been recognized to be oncogenic in EOC: lysophosphatidic acid (LPA)/Yes-associated protein (YAP) oncogenic signaling [10], Sirtuin 7 (SIRT7) [11], Interleukin-33 [12], Fli-1, the tyrosine kinase adaptor protein FRS2 [13] and other markers, high lightening the complexity of the oncology of EOC.
Sam68 (Src-associated in mitosis 68 kD) is a member of the signal transduction and activation of RNA metabolism (STAR) family of proteins, with a K homology RNA-binding domain [14]. Sam68 is involved in a wide range of cellular processes including signal transduction, transcription, RNA metabolism, cell cycle progression, and apoptosis [14–16]. Moreover, Sam68 has been recently recognized to be oncogenic in glioblastoma [15], lung cancer [17], breast cancer [18,19], prostate cancer [20], and cervical cancer [21]. Sam68 deregulates the cell cycle in tumorigenesis, by promoting the conversion of G1/S or G2/M phase [16,18,22] by modulating cancer-relevant splicing events, such as cyclin D1 and Bcl-xl, both of which are involve in cell cycle progression and apoptosis [23,24]. More recently, it was reported that the expression of Sam68 correlated with cell proliferation and survival in epithelial ovarian cancers [25].
The aim of this present study was to investigate the expression of Sam68 in EOC tissues, and to analyze the correlation of it with clinic-pathological characteristics of EOC patients. Then we examined the prognostic role of Sam68 for EOC patients, and determined the regulation of it on the proliferation of EOC OVCAR-3 cells. Our study confirmed the oncogenic role of Sam68 in EOC, and confirmed the positive regulation of it on the proliferation of EOC cells.
Material and Methods
Patients and samples
This study included 152 patients diagnosed with ovarian cancer (mean age of 48±9.82 years, from 31–62 years old, with a median age of 48 years) at Zaozhuang Municipal Hospital (Zaozhuang City, Shandong, China) between March 2011 and April 2014. Clinical characteristics, including age at diagnosis, International Federation of Gynecology and Obstetrics (FIGO) stage, residual tumor size, histological grade, lymph node metastasis, and histological type, were obtained from the patients’ medical records (Table 1). Most of the patients were of Han Chinese background and resided in Shandong Province, China. All of the participants provided written informed consent to participate in this study. The Ethical Committee of Zaozhuang Municipal Hospital approved this research (zzh2013-2-11). All 152 human EOC tissue specimens and the peritumor specimens were obtained by surgical resection, and were stored at −80°C before use.
Table 1.
Correlation of Sam68 expression with clinicopathological characteristics of EOC patients.
| Characteristics | No. (n=152) | High Sam68 | Low Sam68 | P value | ||
|---|---|---|---|---|---|---|
| N | % | N | % | |||
| Age at diagnosis (years) | 0.834629 | |||||
| <50 | 80 | 48 | 53.33 | 32 | 51.61 | |
| ≥50 | 72 | 42 | 46.67 | 30 | 48.39 | |
| FIGO stage | 0.009707 | |||||
| I | 25 | 11 | 12.22 | 14 | 22.58 | |
| II | 49 | 23 | 25.56 | 26 | 41.94 | |
| III | 56 | 42 | 46.67 | 14 | 22.58 | |
| IV | 22 | 14 | 15.56 | 8 | 12.90 | |
| Residual tumor size (cm)* | 0.035284 | |||||
| <2 | 80 | 41 | 45.56 | 39 | 62.90 | |
| ≥2 | 72 | 49 | 54.44 | 23 | 37.10 | |
| Histological grade | 0.012075 | |||||
| 1 | 40 | 19 | 21.11 | 21 | 33.87 | |
| 2 | 54 | 28 | 31.11 | 26 | 41.94 | |
| 3 | 58 | 43 | 47.78 | 15 | 24.19 | |
| LN metastasis | 90 | 0.020853 | ||||
| Positive | 76 | 52 | 57.78 | 24 | 38.71 | |
| Negative | 76 | 38 | 42.22 | 38 | 61.29 | |
| Histological type | 0.676589 | |||||
| Serous | 93 | 57 | 63.33 | 36 | 58.06 | |
| Endometrioid | 18 | 10 | 11.11 | 8 | 12.90 | P |
| Mucinous | 26 | 13 | 14.44 | 13 | 20.97 | |
| Clear cell | 15 | 10 | 11.11 | 5 | 8.06 | |
Cell lines and cell culture
The human EOC cell line OVCAR-3 was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). OVCAR-3 cells were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA, USA), which was supplemented with 10% fetal bovine serum (FBS) (Gibco, Rockville, MD, USA) and 1% streptomycin/penicillin (Sigma-Aldrich, St. Louis, MO, USA) in an incubator with 5% CO2 at 37°C. The medium was changed on alternate days and cells were split before they reached more than 90% confluence. Expression vectors pRC/CMV-Sam68-FLAG (Sam68 Up) and pcRC/CMV (Con Up, as control) were purchased from Promega (Promega, Madison, WI, USA). Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was utilized to transfect plasmids into OVCAR-3 cells. Sam68 knockdown was conducted by using siRNAi-Sam68 (5′-tga gtt aaa ata gat tta gga a-3′) (as control siRNA, 5′-uuc uca gaa cgu gug acg u-3′), which were transfected into OVCAR-3 cells with Lipofectamine RNAiMax (Invitrogen, Carlsbad, CA, USA).
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cancer tissues or OVCAR-3 cells using TRIzol reagent (Life Technologies, Grand Island, NY, USA) based on the product’s suggested protocol. Reverse transcription PCR was performed using a PrimeScript RT-PCR kit (TaKaRa, Dalian, China), and qRT-PCR was performed using the Applied Biosystems 7900HT machine (Applied Biosystems, Carlsbad, CA, USA). The following primers were used to amplify the Sam68 or β-actin. Forward primer for Sam68: 5′-tgc ccg aac tca tgg ccg ag-3′, reverse primer for Sam68: 5′-caa gta att ctc ctc atc at -3′. Forward primer for β-actin: 5′-tgc gtc tgg acc tgg ctg gc-3′, reverse primer for β-actin: 5′-acg tag cac agc ttc tcc tt -3′. The following primers were used for the qRT-PCR analysis of Sam68 and β-actin. Forward primer for Sam68: 5′-cct gcc cga act cat ggc cg-3′, and reverse primer for Sam68: 5′-tgc atg gcg tga gtg aag gac-3′. Forward primer for β-actin: 5′-cgg gac ctg act gac tac ct-3′, and reverse primer for β-actin: 5′-gct cgg ccg tgg tgg tga ag-3′. β-actin was used as the internal control. The one-step qRT-PCR was performed as following: 48°C for 15 minutes followed by 94°C for 5 minutes; and then 94°C for 20 seconds, 60°C for 30 seconds, with 40 cycles.
Western blot assay
Cellular protein samples were extracted from cancer tissues or OVCAR-3 cells with Nuclear/Cytosol Fractionation Kit (BioVision, San Diego, CA, USA), separated via 10% SDS-PAGE electrophoresis, and then transferred onto polyvinylidene fluoridehydrophobic membrane (Millipore, Bedford, MA, USA). Western blotting was performed using rabbit polyclone antibody against human Sam68 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), or rabbit polyclone antibody against β-actin antibody (Sigma-Aldrich, St. Louis, MO, USA). Horseradish peroxidase-linked goat anti-rabbit IgG (Pierce, Rockford, IL, USA) and the enhanced chemiluminescence (Thermo Scientific, Rockford, IL, USA) were utilized to analyze Sam68 level in cancer tissues or OVCAR-3 cells.
Cell proliferation and migration assay
Cell Counting Kit-8 (CCK-8) was utilized to examine the growth curve of the OVCAR-3 cells, in which Sam68 was overexpressed or knocked down. In brief, OVCAR-3 cells with Sam68 overexpressed or knocked down were incubated at 37 °C for 24, 48, or 72 hours, and then were incubated with CCK-8 solution (DOJINDO, Kumamoto, Japan). The 450 nm absorbance was detected after visual color occurrence. For cell colony formation assay, 4×102 OVCAR-3 cells, post Sam68 overexpressed or knocked down, were incubated in 6-well plates at 37°C containing 5% CO2. After an incubation of 48 hours (for Sam68 overexpression) or 96 hours (for Sam68 knockdown), cells were stained with crystal violet (0.005%) for 25 minutes, and the colony in each group was counted by imaging J software.
Statistical analysis
Quantitative data was presented as mean ± standard deviation. Student’s t-test was performed to compare differences between two groups. Comparisons among multiple samples were made by ANOVA analysis. Statistical analyses were conducted by SPSS 17.0 software (IBM SPSS, Armonk, NY, USA). Statistically significant difference was considered when p<0.05 or less.
Results
Sam68 was overexpressed in epithelial ovarian cancer (EOC) in association with cancer malignancy factors
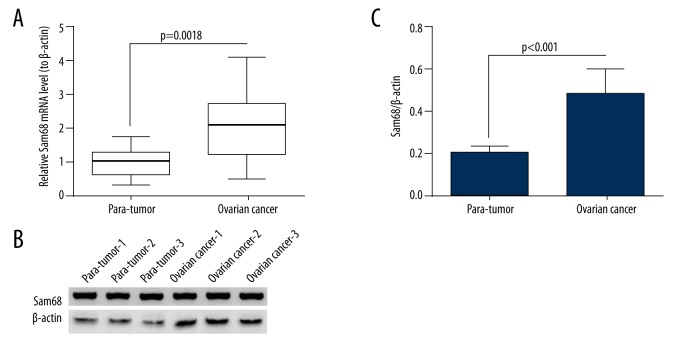
The Sam68 expression in mRNA levels in cancer tissue and in para-tumor tissue (more than 1 cm from tumor tissue) of each patient was examined via RT-qPCR method. As shown in Figure 1, the relative Sam68 mRNA level (compared to para-tumor sample) was significantly higher in the tumor specimens using the two-tailed paired t-test (p=0.0018). We then re-grouped those samples according to their Sam68 levels as high Sam68 group (n=90) and low Sam68 group (n=62). The patients with relative Sam68 mRNA levels higher than “1” in the tumor sample than in the para-tumor sample (“1” was the average Sam68 mRNA level in the para-tumor tissues) were classified into the high Sam68 group. The unpaired t-test indicated a significant difference between the two group (p<0.0001, data not shown). We examined the Sam68 expression in protein levels in the EOC tissues with Western blot assay. As shown in Figure 1B and 1C, the protein level of Sam68 was markedly higher in the EOC specimens than in the para-tumor specimens (p<0.001). Therefore, we found the overexpression of Sam68 in EOC tissues.
Figure 1.
Overexpression of Sam68 in ovarian cancers. (A) Relative mRNA level of Sam68 in the ovarian cancer specimens (n=152) with para-tumor tissues as control. (B) Representative Western blot analysis of Sam68 protein level in ovarian cancer and para-tumor specimens. (C) Relative protein level of Sam68 to β-actin in the ovarian cancer and para-tumor tissues (n=30). Paired t-test was performed to examine the difference in either mRNA or protein level of Sam68, and statistical significance was considered when p<0.05.
To recognize the association between the Sam68 high expression and each clinic-pathological characteristic, we analyzed the statistical difference in age, FIGO stage, residual tumor size, histological grade, lymph node metastasis, and histological type between the high Sam68 and low Sam68 groups. Univariate analysis (Table 2) found no significant difference in the age at diagnosis (<50 vs. ≥50) and histological type (serous vs. non serous); however, there was significant difference found in the FIGO stage (I–II vs. III–IV, p=0.0145), residual tumor size (cm) (<2 vs. ≥2, p=0.0431), histological grade (1+2 vs. 3, p=0.02874) and lymph node metastasis (positive vs. negative, p=0.0342). The hazard ratio (HR) and 95% confidence interval (CI) for each variable are shown in Table 2. Moreover, the Sam68 expression was also a significant prognostic factor for EOC patients (high vs. low, p=0.0013, HR 2.347, 95% CI 1.635–3.254). In addition, the multivariate analysis for each significant variable in the univariate analysis demonstrated that FIGO stage (p=0.0210, 95% CI 1.26–2.27), histological grade (p=0.0352, 95% CI 1.24–2.33) and Sam68 expression (p=0.0073, 95% CI 1.52–3.06) were independently associated with poor prognostic variables for EOC patients with EOC (Table 3). However, the residual tumor size (cm) was not an independent prognostic variable for EOC patients (p=0.0514).
Table 2.
Univariate analysis of prognostic factors in EOC patients.
| Variables | Univariable analysis | ||
|---|---|---|---|
| Hazard ratio | 95% CI | P value | |
| Age at diagnosis (years) | |||
| <50 vs. ≥50 | 0.641 | (0.326–1.264) | 0.9362 |
| FIGO stage | |||
| I–II vs. III–IV | 1.965 | (1.306–2.542) | 0.0145 |
| Residual tumor size (cm) | |||
| <2 vs. ≥2 | 1.730 | (1.135–2.438) | 0.0431 |
| Histological grade | |||
| 1+2 vs. 3 | 1.792 | (1.251–2.542) | 0.02874 |
| LN metastasis | |||
| Positive vs. Negative | 1.642 | (1.214–2.265) | 0.0342 |
| Histological type | |||
| Serous vs. others | 0.695 | (0.380–1.464) | 0.7522 |
| Sam68 expression | |||
| High vs. Low | 2.347 | (1.635–3.254) | 0.0013 |
Table 3.
Multivariate analysis of prognostic factors in OC patients.
| Variables | Multivariable analysis | ||
|---|---|---|---|
| Hazard ratio | 95% CI | P value | |
| FIGO stage | |||
| I–II vs. III–IV | 1.74 | (1.26–2.27) | 0.0210 |
| Residual tumor size (cm) | |||
| Positive vs. Negative | 1.56 | (1.13–1.88) | 0.0514 |
| Histological grade | |||
| 1+2 vs. 3 | 1.63 | (1.24–2.33) | 0.0352 |
| Sam68 expression | |||
| High vs. Low | 1.97 | (1.52–3.06) | 0.0073 |
Manipulation of Sam68 in ovarian cancer OVCAR-3 cells
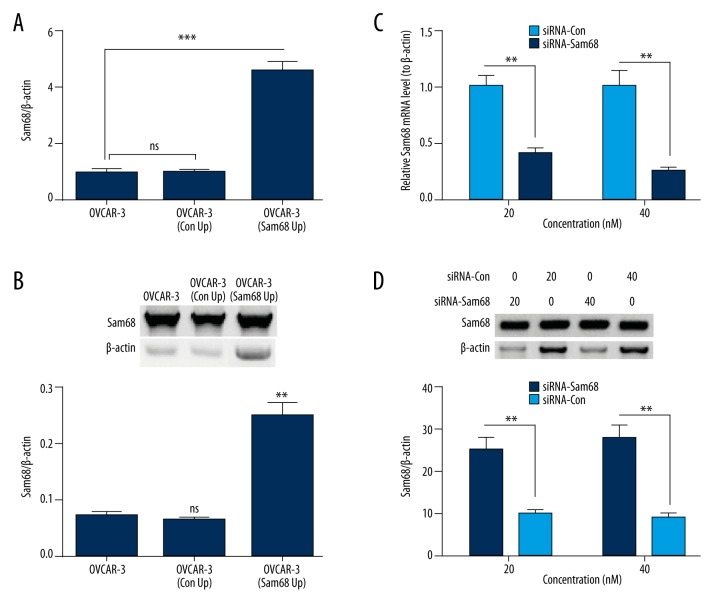
To investigate the regulatory role of Sam68 in ovarian cancer OVCAR-3 cells, we first manipulated the Sam68 expression with gain-of-function and loss-of-function strategies. We found that both mRNA (p<0.001, Figure 2A) and protein (p<0.01, Figure 2B) levels of Sam68 were significantly upregulated in the OVCAR-3 cells post transfection with pRC/CMV-Sam68-FLAG (Sam68 Up). However, the Sam68-specific siRNA was used to knock down the Sam68 expression in both mRNA and protein levels. Figure 2C and 2D demonstrate that the siRNA transfection (20 nM and 40 nM) reduced both mRNA (p<0.01) and protein (p<0.01) levels of Sam68 in the OVCAR-3 cells.
Figure 2.
pRC/CMV-mediated upregulation and siRNA-mediated knockdown of Sam68 in ovarian cancer OVCAR-3 cells (A) mRNA level of Sam68 in OVCAR-3 cells, which were transfected with pRC/CMV-Sam68-FLAG (Sam68 +) or pcRC/CMV (Con +, as control); (B) Western blot analysis of Sam68 in the blank, Sam68 +, or Con + OVCAR-3 cells; (C) mRNA level of Sam68 in the OVCAR-3 cells, which were transfected with siRNA-Con or siRNA-Sam68 (20 or 40 nM); (D) Western blot analysis of Sam68 in the siRNA-Con- or siRNA-Sam68-transfected (20 or 40 nM) OVCAR-3 cells. Data was averaged for triple independent results. ns represented no significance, ** represented p<0.01, and *** represented p<0.001.
Sam68 promotes the proliferation of OVCAR-3 cells
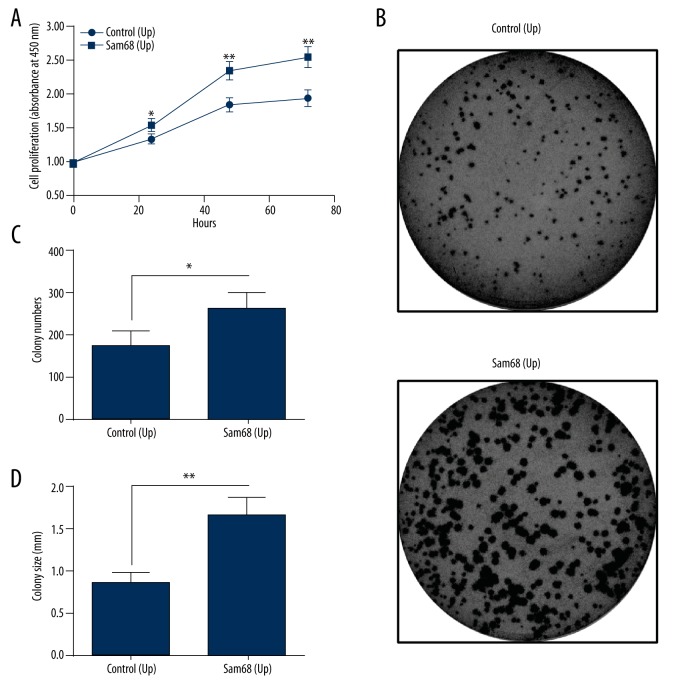
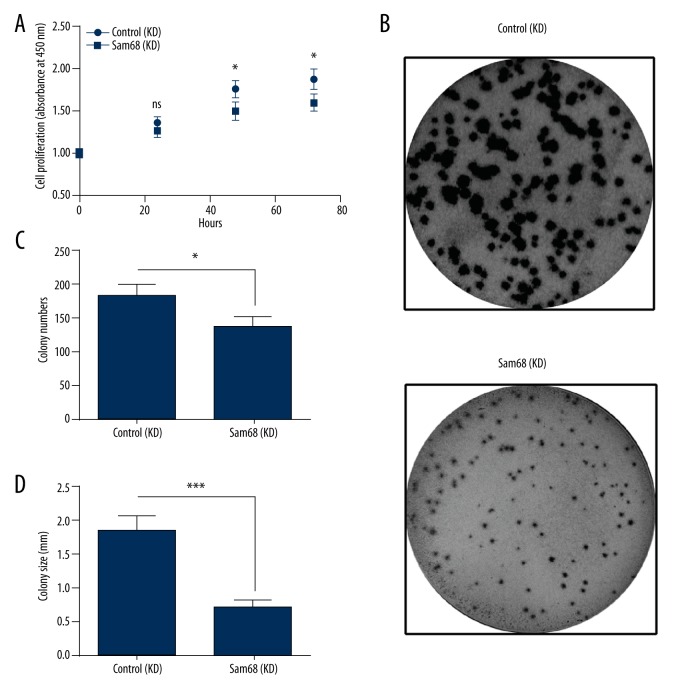
To investigate the regulation by Sam68 on tumor growth of EOC, we determined the proliferation of OVCAR-3 cells, in which Sam68 was upregulated or was knocked down. As shown in Figure 3A, there was significant difference in the growth curve between OVCAR-3 cells (Sam68 Up) and OVCAR-3 cells (Con Up), OVCAR-3 cells (Sam68 Up) grew more efficiently (p<0.05 or p<0.01). We also performed the colony formation of OVCAR-3 cells (Sam68 Up) and OVCAR-3 cells (Con Up) and found more colony numbers in the Sam68 Up group than in the Control group (p<0.05, Figure 3B, 3C). We also found that the OVCAR-3 cells (Sam68 Up) formed colonies with larger size (p<0.01, Figure 3B, 3C). In addition, we knocked down the Sam68 expression and re-examined the proliferation of OVCAR-3 cells. As shown in Figure 4A, there was a significant reduction in the growth efficiency of OVCAR-3 cells, post Sam68 knockdown (p<0.05). Colony forming results also indicated that the Sam68 (KD) OVCAR-3 cells formed less and smaller colonies than the Con (KD) OVCAR-3 cells (p<0.05 or p<0.001, Figure 4B, 4C). These results, taken together, confirmed that Sam68 promotes the proliferation of OVCAR-3 cells.
Figure 3.
Sam68 upregulation promotes the proliferation of ovarian cancer OVCAR-3 cells. (A) Growth curve of the OVCAR-3 cells, in which Sam68 was upregulated (Sam68 [Up]) or not (Control [Up]), as was determined by CCK-8 assay; (B) Colony forming assay of the Sam68 (Up) or Control (Up) OVCAR-3 cells, which were incubated for 48 hours; (C, D) Colony counting (C) and colony size (D) of the colony which was formed by the Sam68 (Up) or Control (Up) OVCAR-3 cells. Experiments were repeated in quadruplicate independently. ANOVA or unpaired t-test was performed to examine the difference in the proliferation of colony formation between the Sam68 (Up) and Control (Up) OVCAR-3 cells. Data were averaged for triple independent results. Statistical significance was considered when p<0.05. * represented p<0.05 and ** represented p<0.01.
Figure 4.
Sam68 knockdown inhibits the proliferation of ovarian cancer OVCAR-3 cells. (A) Growth curve of the OVCAR-3 cells, in which Sam68 was knocked down (Sam68 [KD]) or not (Control [KD]), as was determined by CCK-8 assay. (B) Colony forming assay of the Sam68 (KD) or Control (KD) OVCAR-3 cells, which were incubated for 96 hours. (C, D) Colony counting (C) and colony size (D) of the colony which was formed by the Sam68 (KD) or Control (KD) OVCAR-3 cells. Experiments were repeated in quadruplicate independently. ANOVA or unpaired t-test was performed to examine the difference in the proliferation of colony formation between the Sam68 (KD) and Control (KD) OVCAR-3 cells. Data was averaged for triple independent results. Statistical significance was considered when p<0.05. ns represented no significance, * represented p<0.05, and ** represented p<0.01.
Discussion
The poor prognostic role of Sam68 has recently been identified in non-small cell lung cancer [17], bladder cancer [26], esophageal squamous cell carcinoma [27], colorectal cancer [28], and aggressive neuroblastoma [29], and is associated with the tumor progression. Sam68-deficient mice have delayed development of sexual organs [30], and display a reduction in the number of developing ovarian follicles, alteration of estrous cycles, and impaired fertility [31]. More recently, the role of Sam68 in the tumorigenesis of ovarian cancers has been reported [25].
In the present study, we found overexpression of Sam68 in EOC tissues in association with the stage of malignancy, suggesting it is a significant prognostic factor for EOC patients, along with the FIGO stage, residual tumor size, histological grade, and lymph node metastasis of EOC. Our results also demonstrated that the upregulated Sam68 promoted tumor cell proliferation. Our results indicated that the level of Sam68 expression may serve as a clinical marker for the degree of tumor growth in vivo. Therefore, our findings add to the understanding of the mechanism of ovarian cancer tumorigenesis in association with Sam68.
The positive regulatory role of Sam68 has also been recognized in various other types of cancer. The Sam68 upregulation correlates with, and its downregulation inhibits, proliferation and tumorigenicity of breast cancer cells [18]. Sam68 also has been found to upregulate the proliferation and the aggression of other types of cancers; Sam68 promotes cellular proliferation and predicts poor prognosis in esophageal squamous cell carcinoma [27] and prostate tumor cells [32]. In the current study, the overexpressed Sam68 promoted growth and colony formation of ovarian OVCAR-3 cells, whereas the siRNA-medicated Sam68 knockdown markedly inhibited growth and colony formation of OVCAR-3 cells. Thus, we confirmed the positive regulation by Sam68 on the proliferation of ovarian cancer cells.
Conclusions
Sam68 is overexpressed in EOC tissues in association with cancer malignancy variables such as FIGO stage, histological grade, and lymph node metastasis, suggesting positive regulation of the proliferation of EOC cells. This research indicates that Sam68 might accelerate the cell cycle progression.
Footnotes
Conflict of interest
No conflict of interests.
Source of support: Departmental sources
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Moss C, Kaye SB. Ovarian cancer: Progress and continuing controversies in management. Eur J Cancer. 2002;38(13):1701–7. doi: 10.1016/s0959-8049(02)00161-2. [DOI] [PubMed] [Google Scholar]
- 3.Risch HA. Hormonal etiology of epithelial ovarian cancer, with a hypothesis concerning the role of androgens and progesterone. J Natl Cancer Inst. 1998;90(23):1774–86. doi: 10.1093/jnci/90.23.1774. [DOI] [PubMed] [Google Scholar]
- 4.Meindl A, Hellebrand H, Wiek C, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42(5):410–14. doi: 10.1038/ng.569. [DOI] [PubMed] [Google Scholar]
- 5.Loveday C, Turnbull C, Ramsay E, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43(9):879–82. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma X, Zhang J, Liu S, et al. Polymorphisms in the CASP8 gene and the risk of epithelial ovarian cancer. Gynecol Oncol. 2011;122(3):554–59. doi: 10.1016/j.ygyno.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 7.Permuth-Wey J, Kim D, Tsai YY, et al. LIN28B polymorphisms influence susceptibility to epithelial ovarian cancer. Cancer Res. 2011;71(11):3896–903. doi: 10.1158/0008-5472.CAN-10-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramus SJ, Antoniou AC, Kuchenbaecker KB, et al. Ovarian cancer susceptibility alleles and risk of ovarian cancer in BRCA1 and BRCA2 mutation carriers. Hum mutat. 2012;33(4):690–702. doi: 10.1002/humu.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braem MG, Schouten LJ, Peeters PH, et al. Genetic susceptibility to sporadic ovarian cancer: A systematic review. Biochim Biophys Acta. 2011;1816(2):132–46. doi: 10.1016/j.bbcan.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Cai H, Xu Y. The role of LPA and YAP signaling in long-term migration of human ovarian cancer cells. Cell Commun Signal. 2013;11(1):31. doi: 10.1186/1478-811X-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HL, Lu RQ, Xie SH, et al. SIRT7 exhibits oncogenic potential in human ovarian cancer cells. Asian Pac J Cancer Prev. 2015;16(8):3573–77. doi: 10.7314/apjcp.2015.16.8.3573. [DOI] [PubMed] [Google Scholar]
- 12.Tong X, Barbour M, Hou K, et al. Interleukin-33 predicts poor prognosis and promotes ovarian cancer cell growth and metastasis through regulating ERK and JNK signaling pathways. Mol Oncol. 2016;10(1):113–25. doi: 10.1016/j.molonc.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo LY, Kim E, Cheung HW, et al. The tyrosine kinase adaptor protein FRS2 is oncogenic and amplified in high-grade serous ovarian cancer. Mol Cancer Res. 2015;13(3):502–9. doi: 10.1158/1541-7786.MCR-14-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bielli P, Busa R, Paronetto MP, et al. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr Relat Cancer. 2011;18(4):R91–102. doi: 10.1530/ERC-11-0041. [DOI] [PubMed] [Google Scholar]
- 15.Modem S, Chinnakannu K, Bai U, et al. Hsp22 (HspB8/H11) knockdown induces Sam68 expression and stimulates proliferation of glioblastoma cells. J Cell Physiol. 2011;226(11):2747–51. doi: 10.1002/jcp.22868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor SJ, Resnick RJ, Shalloway D. Sam68 exerts separable effects on cell cycle progression and apoptosis. BMC Cell Biol. 2004;5:5. doi: 10.1186/1471-2121-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Xu Y, Sun N, et al. High Sam68 expression predicts poor prognosis in non-small cell lung cancer. Clin Transl Oncol. 2014;16(10):886–91. doi: 10.1007/s12094-014-1160-3. [DOI] [PubMed] [Google Scholar]
- 18.Song L, Wang L, Li Y, et al. Sam68 up-regulation correlates with, and its down-regulation inhibits, proliferation and tumourigenicity of breast cancer cells. J Pathol. 2010;222(3):227–37. doi: 10.1002/path.2751. [DOI] [PubMed] [Google Scholar]
- 19.Locatelli A, Lofgren KA, Daniel AR, et al. Mechanisms of HGF/Met signaling to Brk and Sam68 in breast cancer progression. Horm Cancer. 2012;3(1–2):14–25. doi: 10.1007/s12672-011-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busa R, Paronetto MP, Farini D, et al. The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene. 2007;26(30):4372–82. doi: 10.1038/sj.onc.1210224. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Yu CP, Zhong Y, et al. Sam68 expression and cytoplasmic localization is correlated with lymph node metastasis as well as prognosis in patients with early-stage cervical cancer. Ann Oncol. 2012;23(3):638–46. doi: 10.1093/annonc/mdr290. [DOI] [PubMed] [Google Scholar]
- 22.Li QH, Haga I, Shimizu T, et al. Retardation of the G2-M phase progression on gene disruption of RNA binding protein Sam68 in the DT40 cell line. FEBS Lett. 2002;525(1–3):145–50. doi: 10.1016/s0014-5793(02)03103-4. [DOI] [PubMed] [Google Scholar]
- 23.Chawla G, Lin CH, Han A, et al. Sam68 regulates a set of alternatively spliced exons during neurogenesis. mol cell biol. 2009;29(1):201–13. doi: 10.1128/MCB.01349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paronetto MP, Cappellari M, Busa R, et al. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 2010;70(1):229–39. doi: 10.1158/0008-5472.CAN-09-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Zhang W, Wang X, et al. Expression of Sam68 correlates with cell proliferation and survival in epithelial ovarian cancer. Reprod Sci. 2016 doi: 10.1177/1933719116650757. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Yu C, Li Y, et al. Utility of SAM68 in the progression and prognosis for bladder cancer. BMC Cancer. 2015;15:364. doi: 10.1186/s12885-015-1367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Liang L, Zhang J, et al. Sam68 promotes cellular proliferation and predicts poor prognosis in esophageal squamous cell carcinoma. Tumour Biol. 2015;36(11):8735–45. doi: 10.1007/s13277-015-3631-8. [DOI] [PubMed] [Google Scholar]
- 28.Liao WT, Liu JL, Wang ZG, et al. High expression level and nuclear localization of Sam68 are associated with progression and poor prognosis in colorectal cancer. BMC Gastroenterol. 2013;13:126. doi: 10.1186/1471-230X-13-126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Zhao X, Li Z, He B, et al. Sam68 is a novel marker for aggressive neuroblastoma. Onco Targets Ther. 2013;6:1751–60. doi: 10.2147/OTT.S52643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard S, Vogel G, Huot ME, et al. Sam68 haploinsufficiency delays onset of mammary tumorigenesis and metastasis. Oncogene. 2008;27(4):548–56. doi: 10.1038/sj.onc.1210652. [DOI] [PubMed] [Google Scholar]
- 31.Bianchi E, Barbagallo F, Valeri C, et al. Ablation of the Sam68 gene impairs female fertility and gonadotropin-dependent follicle development. Hum Mol Genet. 2010;19(24):4886–94. doi: 10.1093/hmg/ddq422. [DOI] [PubMed] [Google Scholar]
- 32.Brauer PM, Zheng Y, Wang L, et al. Cytoplasmic retention of protein tyrosine kinase 6 promotes growth of prostate tumor cells. Cell Cycle. 2010;9(20):4190–99. doi: 10.4161/cc.9.20.13518. [DOI] [PMC free article] [PubMed] [Google Scholar]