Abstract
Neuroimaging-based single subject prediction of brain disorders has gained increasing attention in recent years. Using a variety of neuroimaging modalities such as structural, functional and diffusion MRI, along with machine learning techniques, hundreds of studies have been carried out for accurate classification of patients with heterogeneous mental and neurodegenerative disorders such as schizophrenia and Alzheimer's disease. More than 500 studies have been published during the past quarter century on single subject prediction focused on a multiple brain disorders. In the first part of this study, we provide a survey of more than 200 reports in this field with a focus on schizophrenia, mild cognitive impairment (MCI), Alzheimer's disease (AD), depressive disorders, autism spectrum disease (ASD) and attention-deficit hyperactivity disorder (ADHD). Detailed information about those studies such as sample size, type and number of extracted features and reported accuracy are summarized and discussed. To our knowledge, this is by far the most comprehensive review of neuroimaging-based single subject prediction of brain disorders. In the second part, we present our opinion on major pitfalls of those studies from a machine learning point of view. Common biases are discussed and suggestions are provided. Moreover, emerging trends such as decentralized data sharing, multimodal brain imaging, differential diagnosis, disease subtype classification and deep learning are also discussed. Based on this survey, there are extensive evidences showing the great potential of neuroimaging data for single subject prediction of various disorders. However, the main bottleneck of this exciting field is still the limited sample size, which could be potentially addressed by modern data sharing models such as the ones discussed in this paper. Emerging big data technologies and advanced data-intensive machine learning methodologies such as deep learning have coincided with an increasing need for accurate, robust and generalizable single subject prediction of brain disorders during an exciting time. In this report, we survey the past and offer some opinions regarding the road ahead.
Keywords: Neuroimaging, Machine Learning, Classification, Brain Disorders, Prediction
1. Introduction
Neuroimaging has opened up an exciting non-invasive window into the human brain over the past few decades. This interdisciplinary field has attracted scientists from areas such as medicine, engineering, mathematics, physics, statistics, computer science, and psychology (Epstein et al., 2001). Imaging modalities such as magnetic resonance imaging (MRI) and magnetoencephalography (MEG) along with more traditional methods such as electroencephalography (EEG) have made it possible to noninvasively study various aspects of the human brain with unprecedented accuracy. MRI-related techniques such as structural MRI (sMRI), functional MRI (fMRI) and diffusion MRI (dMRI) have the benefit of providing localized spatial information about the brain structure and function as well as functional and structural connectivity. These techniques have provided new insight into the human brain and have brought hope to researchers trying to unravel the secrets of one of the most complex systems in the universe, the human brain.
Structural MRI has made it possible to visualize the brain at high spatial resolution (one cubic millimeter or less) (Liang and Lauterbur, 2000). SMRI high resolution images of the brain are ideal for studying various brain structures and also for detecting physical abnormalities, lesions and damages. DMRI is an imaging technique for visualization of anatomical connections between different brain regions (Le Bihan et al., 2001; Merboldt et al., 1985). Functional MRI measures brain activity by detecting changes in the blood oxygenation (DeYoe et al., 1994; Ogawa et al., 1990). FMRI makes it possible to study functional regions and networks of the brain as well as temporal associations among them.
Unfortunately, brain disorders are major health problems in US and the rest of the world that not only impair lives of millions of people but also impose huge financial burdens on societies (DiLuca and Olesen, 2014; Ernst and Hay, 1994; Rice, 1999). Moreover, there are no clinical tests to identify many brain disorders such as schizophrenia. One of the major hopes underlying the advanced neuroimaging tools mentioned above is to provide new understanding of brain disorders such as schizophrenia, bipolar disorder, autism spectrum disorder (ASD), Alzheimer's disease (AD), major depressive disorders, attention-deficit hyperactivity disorder (ADHD) and mild cognitive impairment (MCI). Brain disorder research aims at understanding the impact of each disease on the brain's function and structure from the cellular to system level, as well as the pathogenesis of these complex disorders. As a result, thousands of studies have been published on different aspects of brain disorders to show aberrations of some features (structural or functional) in a patient group usually in comparison with a healthy cohort (Jack et al., 1997; Jafri et al., 2008; Lorenzetti et al., 2009; McAlonan et al., 2005). While these studies are valuable in terms of finding relevant disease biomarkers, they are not sufficient for direct clinical diagnostic/prognostic adoption. The main reason is that many of these findings are statistically significant at the group level, but the individual discrimination ability of the proposed biomarkers is not typically evaluated. Since classification provides information for each individual subject, it is considered a much harder task than reporting group differences.
In recent years, there has been a growing trend in designing neuroimaging-based prognostic/diagnostic tools. As a result, there have been a lot of efforts using neuroimaging tools to automatically discriminate patients with brain disorders from healthy control or from each other (Klöppel et al., 2012). Many of these studies have reported promising prediction performances with the claim that complex diseases can be diagnosed robustly, accurately and rapidly in a automatic fashion. However, until now, these tools have not been integrated into the clinical realm. We believe the main reason for this is that many of the studies of this nature, despite the promising results on a specific research dataset, are not designed to generalize to other datasets, specifically the clinical ones.
The purpose of this study is two-fold. First, we reviewed a large number of MRI-based brain disorder diagnostic/prognostic studies in schizophrenia, ASD, ADHD, depressive disorder, MCI and Alzheimer's disease. These studies are compared in a number of key aspects such as type of features, classifier and reported accuracies. Next, we formed our opinion on the issues associated with how machine learning is applied in neuroimaging and have suggested solutions that might address these pitfalls. Considering the immense potential of neuroimaging tools for clinical adoption, careful implementation and interpretation of machine learning in neuroimaging is crucial. Machine learning is a relatively new domain for many neuroimaging researchers coming from other fields and therefore pitfalls are unfortunately not rare. We attempt to identify and emphasize some common mistakes that result in these shortcomings and biases. At the end, we discuss emerging trends in neuroimaging such as data sharing, multimodal brain imaging and differential diagnosis.
1.1 Group Difference vs. Classification
As pointed out in the introduction, many brain disorder studies have shown abnormality in the average sense in one or more brain features in a patient cohort in comparison with a healthy group using statistical tests. The success of such methodology is usually measured by the means of p-values. On the other hand, the goal of single subject prediction is to automatically classify each subject into one of the groups in the study (e.g., healthy vs. patient). The success of classification studies is usually measured by accuracy.
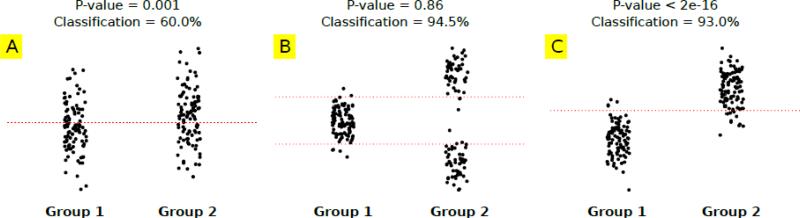
These two problems are very different in essence as they try to address varying research questions. In general, showing group differences is much easier compared to single subject prediction. To better illustrate the difference between these types of analysis, we show an example in Figure 1. Suppose there are two groups each with 100 samples (subjects) and we have measurements of one brain feature for each subject. Figure 1A shows a case where the mean of two groups is different as measured by a two-sample t-test. The difference is statistically significant (p-value=0.001). However, if one tries to classify subjects based on a threshold on this brain feature (the dotted red line placed between the mean of two groups), a weak classification rate of 60.0% will be achieved. The reason for this is the range of values for that specific feature is highly overlapping for the two groups. So, a highly significant group difference does not necessarily translate into a strong classification result. But the opposite is also true, as high classification based on a feature doesn't necessarily mean that group-level mean differences exist. Figure 1B shows a case where the two-sample t-test on the two groups is not significant (p-value= 0.86) but the classification based on two thresholds (red dotted lines placed between each mode of group 2 and mean of group 1) is very strong (94.5%). In this case, the abnormality is bidirectional, which does not cause significant mean differences but makes it possible to separate the groups with two thresholds (dotted lines). Interestingly, bidirectional abnormalities are observed in neuroimaging studies (Mohammad R. Arbabshirani and Calhoun, 2011; Calhoun et al., 2006b). Figure 1C shows a case where strong group differences and successful classification go hand in hand. The abnormality is one-directional and the mean difference is very significant (p-value<2e-16). The mean of two groups is so far apart that the values of most of the samples of the two groups do not overlap. Therefore, a strong classification rate of 93.5% is achieved (based on one threshold).
Figure 1.
Comparison of group difference analysis and classification in three different scenarios using toy data. Group difference is analyzed by two-sample t-tests and classification is performed by simple thresholding (red dotted lines). Each group/class has 100 samples. A: Significant group difference (p-value<0.001) but poor classification (60.0%). B: Insignificant group difference (p-value=0.865) but high classification accuracy (94.5%). C: Significant group difference (p-value<2e-16) and high classification accuracy (93.0%). Significant group difference doesn't necessarily cause high classification and vice versa.
The main purpose of example in Figure 1 is to show that group level analysis and classification are two different methods for different problems. We will return to this example later for criticism of selecting features based on p-value.
2. Survey of MRI-based Single-Subject Prediction of Brain Disorders
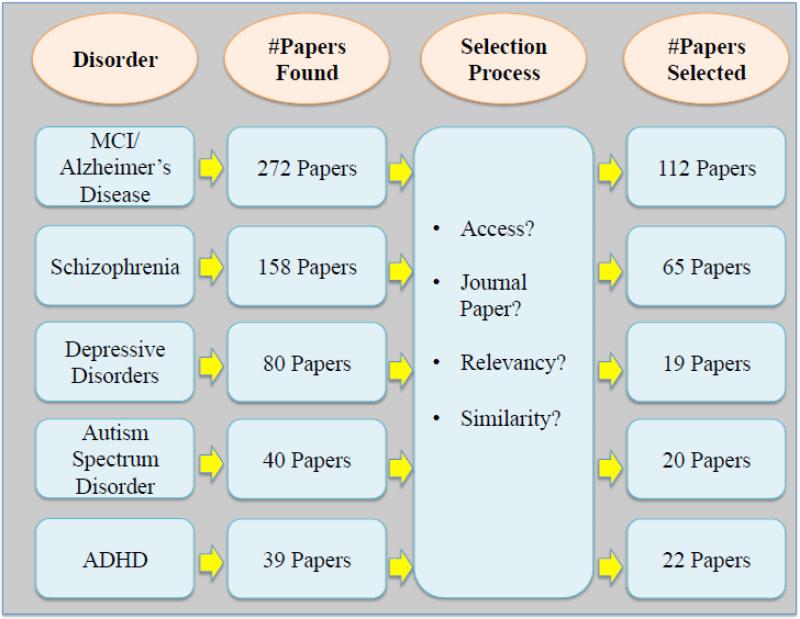
Based on a search on Pubmed from 1990 to 20151, more than 500 papers on MRI-based single subject prediction of brain disorders were found. Figure 2 summarizes the paper selection procedure for this study. More than 200 papers were eventually selected for this survey (112 AD/MCI, 63 schizophrenia, 19 depressive disorders, 20 ASD and 22 ADHD papers).
Figure 2.
The literature review procedure, the inclusion criteria and the number of surveyed studies for each modality.
We limited our search to journal papers in English published up to December 2015. In a few instances, the full paper was not found and therefore those studies were excluded from this survey. Also, in cases of very similar papers from the same authors, only one was selected. Key aspects of each study such as modality, machine learning method, sample size and type features were investigated. A list of all abbreviations used in the tables and the manuscript itself is provided in Table 1.
Table 1.
Glossary
| Abbreviation | Full Term |
|---|---|
| AAL | Automated anatomical Labeling |
| ABIDE | Autism brain imaging data exchange |
| AD | Alzheimer's disease |
| ADAS | Alzheimer's Disease Assessment Scale |
| ADHD | Attention Deficit Hyperactivity Disorder |
| ADHD-C | ADHD Combined |
| ADHD-HI | Hyperactive/impulsive ADHD |
| ADHD-IA | Inattentive ADHD |
| ADNI | Alzheimer's disease neuroimaging initiative |
| ADOS | Autism Diagnostic Observation Schedule |
| AG | Angular Gyrus |
| ALFF | Amplitude of low frequency fluctuations |
| aMCI | amnestic MCI |
| ANN | Artificial Neural Network |
| ANOVA | Analysis of variance |
| AOD | Auditory Oddball |
| ASD | Autism Spectrum Disease |
| AUC | Area under curve |
| AX-CPT | AX version of continuous performance task |
| BOLD | Blood-Oxygen Level Dependent |
| BP | Bipolar Disorder |
| CFT | Complex Figure Test |
| cMCI | MCI converter |
| CN | Cognitively normal |
| CSF | Cerebrospinal fluid |
| DA | Axial Diffusion |
| DAT | Dementia of the Alzheimer's Type |
| DLPFC | Dorsolateral prefrontal cortex |
| DMN | Default-Mode network |
| dMRI | Diffusion Magnetic Resonance Imaging |
| DR | Radial Diffusion |
| DRS | Dementia Rating Scale |
| EC | Elderly Controls |
| EEG | Electroencephalography |
| ELM | Extreme Learning Machines |
| EMCI | Early MCI |
| ERC | Entorhinal Cortex |
| FA | Fractional anisotropy |
| FALLF | Fractional Amplitude of low frequency fluctuations |
| FBIRN | Functional Biomedical Informatics Research Network |
| FC | Functional Connectivity |
| FDG | Fluorodeoxyglucose |
| FDG-PET | Fluorodeoxyglucose Positron Emission Tomography |
| FFT | Fast Fourier Transform |
| fMRI | Functional Magnetic Resonance Imaging |
| FNC | Functional Network Connectivity |
| FTD | Frontotemporal Dementia |
| GLM | General Linear Modeling |
| GM | Gray matter |
| GMD | Gray Matter Density |
| HC | Healthy controls |
| ICA | Independent Component Analyses |
| ITG | Inferior Temporal Gyrus |
| jICA | Joint Independent Component Analysis |
| LBD | Lewy body dementia |
| LDA | Linear Discriminant Analysis |
| LDDMM | Large Deformation Diffeomorphic Metric Mapping |
| LLD | Late-life Depression |
| LLE | Locally linear embedding |
| LMCI | Late MCI |
| MA | Mean anisotropy |
| mCCA | Multi-set Canonical Correlation Analysis |
| MCI | Mild Cognitive Impairement |
| MCIC | Multi-site Clinical Imaging Consortium |
| MD | Mean Diffusitivity |
| md-aMCI | Multiple Domains MCI |
| MDD | Major Depressive Disorder |
| MEG | Magnetoencephalography |
| MLSP | Machine Learning for Signal Processing |
| mMLDA | Modified Maximum Uncertainty Linear Discriminant Analysis |
| MMSE | Mini Mental State Examination |
| MPFC | Medial Prefrontal Cortex |
| MRI | Magnetic Resonance Imaging |
| MRMR | Minimum Redundancy Maximum Relevancy |
| MRS | Magnetic Resonance Spectroscopy |
| MTL | Medial Temporal Lobe |
| MTR | Magnetization Transfer Ratio |
| MVPA | Multi voxel pattern analysis |
| N/A | No Answer |
| ncMCI | MCI non-converter |
| NDD | Non-refractory Depressive Disorder |
| NMF | Non-negative Matrix Factorization |
| OCD | Obsessive Compulsive Disorder |
| ODVBA | Optimally-Discriminative Voxel-Based Analysis |
| orPLS | Ordinary Partial Least Square |
| PANSS | Positive and Negative Syndrome Scale |
| PCA | Principal component analysis |
| PCC | Posterior Cingulate Cortex |
| Probability Distribution Functuion | |
| PET | Positron Emission Tomography |
| pMCI | Progressive MCI |
| PPI | Psychophysiological Interaction |
| QDA | Quadratic Discriminant Analysis |
| RAVENS | Regional analysis of brain volumes in normalized space |
| RBF | Radial basis function |
| RDD | Refractory Depressive Disorder |
| ReHo | Regional Homogeneity |
| RMD | Remitted MDD |
| ROC | Receiver Operating Characteristic |
| ROI | Region of interest |
| rsfMRI | Resting-state fMRI |
| RSN | Resting-state Networks |
| RVM | Relevance Vector Machine |
| RVoxM | Relevance Voxel Machine |
| RVR | Relevance Vector Regression |
| sACC | Subgenual Anterior Cingulate Cortex |
| SBM | Surface based morphometry |
| sd-aMCI | Single Domain amnestic MCI |
| sd-fMCI | Single Domain frontal MCI |
| SIFT | Scale-invariant Feature Transform |
| sMCI | Stable MCI |
| sMRI | Structural Magnetic Resonance Imaging |
| SN | Salience Network |
| SNP | Single Nucleotide Polymorphism |
| SSD | Schizophrenia Spectrum Disorders |
| StD | Late-Life Subthreshold Depression |
| SUVr | Standard Uptake Value Ratio |
| SVM | Support Vector Machine |
| SVM-FoBa | Support Vector Machine with a Forward-Backward search strategy |
| SVM-RFE | Support vector machine with recursive feature elimination |
| SZ | Schizophrenia |
| SZA | Schizoaffective |
| TD | Typically Developing |
| TDC | Typically Developing Children |
| uMCI | Unknown MCI |
| VaD | Vascular Dementia |
| VBM | Voxel-based Morphometry |
| VMHC | Voxel-mirrored Homotopic Correlations |
| VOI | Volume of Interest |
| WM | White matter |
| WMD | White Matter Density |
| WMT | Working Memory Task |
2.1 Mild Cognitive Impairment/Alzheimer's Disease
MCI entails cognitive decline more than what is expected for an individual's age and education level, but not to the extent that it interferes notably with activities of daily life (Albert et al., 2011). Unfortunately, more than 50% of the MCI patients progress to dementia within 5 years (Gauthier et al., 2006). So, it is considered a prodromal phase to dementia especially the AD type (Gauthier et al., 2006). The heterogeneous etiology of MCI includes degenerative diseases (AD, fronto-temporal lobe degeneration, dementia with Lewy bodies) as well as vascular and psychiatric disorders (Petersen and Negash, 2008). AD is the most common neurodegenerative disorder, which is increasingly prevalent among adults aged 65 years and older. AD is characterized by the progressive impairment of neurons and their connections, which result in decline and loss of cognitive functions. In 2007, it was estimated that more than 26 million people suffer from AD worldwide (Brookmeyer et al., 2007). In 2001 it was predicted that AD will triple in prevalence by 2050 (Hebert et al., 2001). The detection of AD is based on clinical examinations and an evaluation of the patient's perception and behavior. Considering the prevalence and severity of MCI/AD, the largest number of neuroimaging-based, automatic prediction/classification publications has been devoted to these conditions. Table 2 summarizes the 112 studies that we reviewed in this survey.
Table 2.
Summary of 112 MRI-based AD/MCI classification studies. Overall classification accuracy of sMCI (cMCI) from pMCI (ncMCI) is indicated by MCI-CONV if applicable.
| Modality | Disorder | Features | # Features | Classifier | Number of Subjects | Overall Accuracy | Reference |
|---|---|---|---|---|---|---|---|
| dMRI | AD | FA | 1210 | SVM | HC=25, AD=20, Total=45 |
100% | (Graña et al., 2011) |
| dMRI and sMRI | AD | FA and MD from dMRI and GMD and WMD from sMRI |
26,000 FA, 128,000 MD, 41,000 WMD and 181,000 GMD |
SVM | HC=143, AD=137, Total=280 |
63.6-91.1% | (Dyrba et al., 2013) |
| rsfMRI | AD | Averaged voxel intensities of selected resting-state networks |
4 | Multivariate ROC | HC=16, AD=15, Total=31 |
100% | (Wu et al., 2013) |
| rsfMRI | AD | Graph measures based on FC analysis among ROIs |
454 | SVM | HC=20, AD=20, Total=40 |
100% | (Khazaee et al., 2015) |
| sMRI | AD | Eigen brains of key slices | 10 | SVM | NC=98, AD=28, Total=126 |
92.3% | (Zhang et al., 2015) |
| sMRI | AD | ODVBA of RAVENs maps | N/A | SVM | HC=50, AD=50, Total=100 |
90% | (Zhang and Davatzikos, 2011) |
| sMRI | AD | Hippocampus shape measures using LDDMM and PCA |
20 Principal componen ts (3-4 selected by the classifier) |
Logistic Regression | HC=26, DAT=18, Total=44 |
81.1-84.6% | (Wang et al., 2007) |
| sMRI | AD | GM, WM, and CSF tissue densities along with age, gender and genotype |
237-240 | SVM | HC=190, AD=190, Total=380 |
85.6-89.3% | (Vemuri et al., 2008) |
| sMRI | AD | Cortical thickness measures along mesh vertices |
82000 mesh vertices |
RVoxM | HC= 150, Ad=150, Total=300 |
93.0% (AUC) |
(Sabuncu and Van Leemput, 2012) |
| sMRI | AD | Whole brain and hippocampus VBM measures |
N/A | SVM | EC=31, AD=31, Total=62 |
74-79% | (Polat et al., 2012) |
| sMRI | AD | Volumetric measures | 45 | SVM | HC=20, AD=14, Total=34 |
88.2% | (Oliveira et al., 2010) |
| sMRI | AD | Hippocampus morphometric measures | 9 | LDA | HC=57, AD=38, Semantic dementia=6, Total=101 |
77% | (Miller et al., 2009) |
| sMRI | AD | GM Maps | 10-45 | SVM, ELM, Self- adaptive Resource Allocation Network |
HC=30, AD=30, Total=60 |
97.1-99.7% | (Mahanand et al., 2012) |
| sMRI | AD | GM distribution of ROIs | 90 | SVM | EC=22, AD=16, Total=38 |
94.5% | (Magnin et al., 2009) |
| sMRI | AD | Surface-based measures of hippocampus | N/A | SVM | HC=20, AD=19, Total=39 |
84.6-94.9% | (Li et al., 2007) |
| sMRI | AD | Cortical thickness | N/A | LDA, QDA and Logistic regression |
HC=17, AD=19, Total=36 |
90-100% | (Lerch et al., 2008) |
| sMRI | AD | Cortical thickness data and hippocampus shape |
N/A | LDA | NC=84, AD=33, Total=117 |
87.5% | (Lee et al., 2014) |
| sMRI | AD | GM Probability Maps | Variable | Linear program boosting of voxel- wise weak classifiers with spatial constraints |
Total=183 | 82.0% (AUC) |
(Hinrichs et al., 2009) |
| sMRI | AD | WM and GM voxels selected by SVM-RFE | Variable | SVM | HC=185, AD=185, Total=370 |
94.3-95.1% | (Hidalgo-Muñoz et al., 2014) |
| sMRI | AD | Volumes of hippocampus–amygdala formation |
1 | Thresholding | HC=28, AD=27, Total=55 |
89-96% (Sensitivity) |
(Hampel et al., 2002) |
| sMRI | AD | Linear measurements of several structures | 12 | Linear Discriminant Analysis |
HC=31, AD=46, Total=77 |
81-87% (Sensitivity) |
(Frisoni et al., 1996) |
| sMRI | AD | Texture Features | 260 | Linear Discriminant Function |
HC=40, AD=24, Total=66 |
91% | (Freeborough and Fox, 1998) |
| sMRI | AD | Percentage of brain volume changes | 3 | SVM | NC=30, AD=30, Total=60 |
91.7% | (Farzan et al., 2015) |
| sMRI | AD | GM, WM and CSF volumes and size of hippocampus |
5 | SVM, MLP, and J48 decision tree |
NC=48, AD=37, Total=85 |
93.7% | (Farhan et al., 2014) |
| sMRI | AD | Brain volume, temporal lobe matter and CSF volume |
4 | Discriminant Analysis |
HC=29, DAT=31, Total=60 |
100% | (DeCarli et al., 1995) |
| sMRI | AD | Several voxel-based and cortical thickness- based schemes |
Variable | Regularized SVM | CN=162, AD=137, Total= 299 |
83-91% | (Cuingnet et al., 2013) |
| sMRI | AD | Atrophic patterns of hippocampus and entorhinal cortex |
N/A | QDA | HC=50, AD=50, Total=100 |
93% | (Coupé et al., 2012) |
| sMRI | AD | SIFT Features | 133 | Ensemble of SVMs | HC1=66, AD1=20, HC2=98, AD2=28, Total=212 |
70-87% | (Chen et al., 2014) |
| sMRI | AD | PDF of VOI based on VBM | 100 | SVM | HC=130, AD-130, Total- 260 |
86% | (Beheshti and Demirel, 2015) |
| sMRI | AD | Generative-Discriminative Basis vectors based on RAVEN maps |
30-50 | Logistic Model Trees |
HC=63, AD=54, Total=117 |
87-89% | (Batmanghelich et al., 2012) |
| sMRI | AD | Cortical thickness and volumetric measures | N/A | SVM | HC=25, AD=29, Total=54 |
90.9% (AUC) | (Arimura et al., 2008) |
| sMRI | AD | GM maps based on VBM | 384,065 | SVM | HC=137, AD=108, MCI=203, Total=448 |
63.7-80.3% | (Adaszewski et al., 2013) |
| sMRI | AD | Gray Matter Probability Maps | 2E6 | SVM | HC=226, AD=91, Total=417 |
87% | (Abdulkadir et al., 2011) |
| sMRI and dMRI | AD | FA and GM volumes | 142 | SVM | NC=15, AD=21, Total=36 |
94.3% | (Li et al., 2014b) |
| sMRI and PET | AD | Volumes of interest | 12 | SVM | HC1=28, AD1=28, HC2=13, AD2=21, Total=90 |
86-100% | (Dukart et al., 2013) |
|
sMRI and rsfMRI |
AD | GM Volume from sMRI and ALFF, RcHo and FC from rsfMRI |
Variable | Maximum uncertainty LDA and second level |
HC=22, AD=16, Total=38 |
89.5% | (Z. Dai et al., 2012) |
|
sMRI, rsfMRI and dMRI |
AD | GM volume from sMRI, fiber tract integrity from dMRI and graph-theoretical measures form fMRI |
N/A | SVM | HC=25, AD=28, Total=53 |
74-85% (AUC) |
(Dyrba et al., 2015) |
|
fMRI (confrontation naming task) |
AD (Low and high risk) |
Fractional signal changes ROI | 50 | LDA+orPLS | Low AD Risk HC=11, High AD Risk HC= 13, Total=24 |
83.3% | (Andersen et al., 2012) |
| dMRI | AD/BP | FA Maps | 1500- 14000 |
SVM | HC=25, BP=12, AD=20 | 100% | (Bergouignan et al., 2011) |
| rsfMRI | AD/FTD | ROI-based difference between DMN and SN map |
22 | LDA | HC=12, AD=12, FTD=12, Total=36 |
92% | (Zhou et al., 2010) |
| sMRI | AD/FTD | GM Maps | N/A | SVM | HC=91, AD=85, FTD=19, Total=195 |
87-96% | (Klöppel et al., 2008) |
| sMRI | AD/FTD | GM volume an thickness and complexity estimates |
N/A | LDA | CN=23 AD=24, FTD=19, Total=66 |
81-96% | (Young et al., 2009) |
| sMRI | AD/FTD | Morphometric measures of selected ROIs | 2 | Discriminant Analysis |
EC=12, AD=17, FTD=16, Total=45 |
91% | (Kaufer et al., 1997) |
| dMRI | AD/MCI | FA and MD Values | 12-1080 | SVM | EC=50, AD=37, MCI=113, Total=200 |
68.3-84.9% | (Nir et al., 2015) |
| rsfMRI | AD/MCI | FC among selected AAL regions | 3403 | Bayesian Gaussian process logistic regression |
HC=39,aMCI=50, AD=27, Total=116 |
75-90% | (Challis et al., 2015) |
| sMRI | AD/MCI | 3D hippocampal shape morphology | N/A | SVM | HC=88, MCI=103, AD=71, Total=262 |
MCI-CONV: 80% |
(Costafreda et al., 2011a) |
| sMRI | AD/MCI | GM, WM and CSF volumetric measures and ventricle shape |
18 | Particle swarm optimization SVM |
HC=17, AD=17, MCI=18, Total=52 |
88.9-94.1% | (Yang et al., 2013) |
| sMRI | AD/MCI | Coefficient of ICA on normalized brain images |
N/A | SVM | HC1=316, AD1=98, Total1=416, HC2=200, AD2=200, MCI=400, Total2=800, |
67.5-99% | (Yang et al., 2011) |
| sMRI | AD/MCI | Hippocampal volume, tensor-based morphometry, cortical thickness and Manifold-based learning features |
112-114 | LDA and SVM | HC=231, AD=198, sMCI=238, pMCI=167, Total=834 |
84.0-89.0% MCI- CONV:68.0 % |
(Wolz et al., 2011) |
| sMRI | AD/MCI | ROI-based and correlative features based on cortical and cerebral thickness and WM volumes |
N/A | Multi-kernel SVM | NC=200, AD=198, ncMCI=111, cMCI=89, Total=598 |
79.2-97.4% MCI- CONV:75.1 % |
(Wee et al., 2013) |
| sMRI | AD/MCI | Intensity patches of selected ROIs around hippocampus |
130-150 Patches |
SVM (in multiple instance-Graph Framework) |
CN=231, AD=198, ncMCI=238, cMCI=167, Total=834 |
82.9-89% -MCI- CONV:70% |
(Tong et al., 2014) |
| sMRI | AD/MCI | Diffeomorphometry patterns of subcortical and ventricular structures |
14 | LDA | HC=210, AD=175, cMCI=135, ncMCI=87, Total=607 |
MCI- CONV:77.0 % |
(Tang et al., 2015) |
| sMRI | AD/MCI | Hippocampus, amygdala, and ventricle shape measures |
N/A | LDA | HC=210, AD=175, MCI=369, Total=754 |
86% | (Tang et al., 2014) |
| sMRI | AD/MCI | Whole brain GM and WM maps | 34-127 | SVM | HC=162, AD=137, ncMCI=134, cMCI=76, Total=509 |
72-76%, MCI- CONV:66.0 % |
(Salvatore et al., 2015b) |
| sMRI | AD/MCI | GM Maps | 6000 | SVM | HC=189, AD=144, ncMCI=166, cMCI=136, Total=635 |
80.0% - MCI- CONV:70.7 % |
(Retico et al., 2015) |
| sMRI | AD/MCI | Hippocampal surface deformation measures | 19 | LDA | HC=26, DAT=18, DAT- Converters=9, Total=53 |
77.0-87.0% | (Qiu et al., 2008) |
| sMRI | AD/MCI | GM and WM maps | Variable | SVM, Bayes statistic and voting feature intervals |
HC=18, AD=32, MCI=24, Total=74 |
92%-MCI- CONV:75.0 % |
(Plant et al., 2010) |
| sMRI | AD/MCI | Volumes of the hippocampus and ERC | 2-4 | Discriminant Function Analysis |
HC=59, AD=48, MCI=65, Total=172 |
65.9-90.7% | (Pennanen et al., 2004) |
| sMRI | AD/MCI | GM Density of ROIs | 37 | SVM | ncMCI=38, cMCI=39, Total=77 |
77.7% (MCI Conversion) |
(Ota et al., 2014) |
| sMRI | AD/MCI | Hippocampal volumetric measures | 5 | LDA | HC=53, AD=18, MCI=20, Total=91 |
73.7-77.5% | (Mueller et al., 2010) |
| sMRI | AD/MCI | GM density values and cognitive measures | 309 | Low density separation semi- supervised classifier |
NC=231, AD=200, sMCI=100, pMCI=164, uMCI=130, Total=825 |
MCI-CONV: 76.6-90.0% (AUC) |
(Moradi et al., 2015) |
| sMRI | AD/MCI | Data-driven ROI GM from different templates |
1500 from each template |
SVM | NC=128, AD=97, sMCI=117, pMCI=117, Total=459 |
91.6%, MCI- CONV: 72.4% |
(Min et al., 2014) |
| sMRI | AD/MCI | Longitudinal volumetric MR imaging measures |
N/A | QDA | HC=203 AD=164, MCI=317, Total=684 |
85% | (McEvoy et al., 2011) |
| sMRI | AD/MCI | Volumetric and cortical thickness measures | N/A | LDA | HC=139, AD=84, MCI=175, Total=398 |
89-92% | (McEvoy et al., 2009) |
| sMRI | AD/MCI | GM maps | N/A | Ensemble of SVMs | NC=229, AD=198, MCI=225, Total=652 |
85.3-92.0% | (M. Liu et al., 2014) |
| sMRI | AD/MCI | GM maps registered to multiple templates | 1500 for each template |
Ensemble of SVMs | NC=128, AD=97, sMCI=117, pMCI=117, Total=459 |
93.8% MCI- CONV:80.9 % |
(Liu et al., 2015) |
| sMRI | AD/MCI | Volume and cortical thickness values of ROIs |
162 Original features reduced by LLE |
Logistic regression, SVM and LDA |
CN=137, sMCI=93, cMCI=97, AD=86, Total=413 |
51-89% MCI-CONV: 68% |
(Liu et al., 2013) |
| sMRI | AD/MCI | Surface connectivity and center of mass markers |
N/A | LDA | NC=170, AD=114, MCI=240, Total=524 |
76.6-87.7% (AUC) |
(Lillemark et al., 2014) |
| sMRI | AD/MCI | Proposed local binary pattern features | N/A | SVM | NC=142, AD=80, MCI=141, Total=363 |
61.5-82.8% | (Li et al., 2014a) |
| sMRI | AD/MCI | Cortical thickness measures, cortex thinning dynamics and network features based on longitudinal thickness changes of different ROIs |
262 | SVM | NC=40, sMCI=36, pMCI=39, AD=37, Total=152 |
81.7-96.1% MCI- CONV:80.3 % |
(Li et al., 2012) |
| sMRI | AD/MCI | Hurst's exponents at different scales | N/A | SVM | HC=11, AD=11, MCI=11, Total=33 |
97.1-97.5% | (Lahmiri and Boukadoum, 2014) |
| sMRI | AD/MCI | Volumetric Measures | 120 (115 brain features) |
LDA | CN=125, AD=55, HMCI=114, LMCI=91, Total=385 |
90.8-94.5% | (Goryawala et al., 2015) |
| sMRI | AD/MCI | Spherical harmonics of hippocampi | 2646 | SVM | HC=25, AD=23, aMCI=23, Total=71 |
83-94% | (Gerardin et al., 2009) |
| sMRI | AD/MCI | Three schemes: voxel-based features, cortical thickness features and hippocampus-based features |
Variable | SVM | CN=162, AD=137,cMCI=76, ncMCI=134, Total=509 |
81-95% (for AD vs CN) |
(Cuingnet et al., 2011) |
| sMRI | AD/MCI | Volume, thickness and surface area of selected ROIs |
7 MRI, 2 CSF and 14 neuropsyc hological features |
SVM | HC=111, cMCI=56, ncMCI=111, AD=96, Total=350 |
MCI-CONV: 67.1% |
(Cui et al., 2011) |
| sMRI | AD/MCI | Hippocampus and parahippocampal gyrus GM maps |
11,031 | SVM | HC=188, MCI=260, AD=131 |
70-85% MCI-CONV: 65% |
(Chu et al., 2012) |
| sMRI | AD/MCI | Intensity and texture of selected VOIs in MTL |
>100,000 | Ensemble of SVMs | HC=189, ncMCI=166, cMCI=136, AD=144 |
65-94% MCI- CONV:74.0 % (AUC) |
(Chincarini et al., 2011) |
| sMRI | AD/MCI | GM Map | 50,000- 750,000 |
SVM, Regularized logistic Regression, Linear Regression Classifier |
HC=205, MCI=351, AD= 171, Total= 727 |
80-90% | (Casanova et al., 2012) |
| sMRI | AD/MCI | Volumetric measures of amygdala, hippocampus, and parahippocampal gyrus |
N/A | Discriminant function analysis |
NEC=20, MCI=21, AD=39, Total=60 |
80.5-88.1% | (Bottino et al., 2002) |
| sMRI | AD/MCI | Hippocampal volume and CSF Aβ, t-tau and p-tau levels, and ApoE4 stratification |
N/A | SVM | NC=111, AD=95, MCI=182 |
64-78% | (Apostolova et al., 2014) |
| sMRI | AD/MCI | Cortical thickness and volumetric measures | 57 | SVM | HC=110, AD=116 MCI=119, Total=345 |
88.1% | (Aguilar et al., 2013) |
| sMRI and dMRI | AD/MCI | Network topology, tractography connectivity and flow-based measures |
N/A | SVM | NC=50, AD=38, EMCI=74, LMCI=38, Total=200 |
59.2-78.2% | (Prasad et al., 2015) |
| sMRI and dMRI | AD/MCI | Disease-specific spatial filters | N/A | LDA | NC=22, AD=19, MCI=22, Total=63 |
9.3.0% (AUC) MCI Conversion |
(Oishi et al., 2011) |
| sMRI and dMRI | AD/MCI | Cortical thickness, subcortical volume and white matter integrity |
2-5 | SVM | SMI=27, AD=27, MCI=138 Total=72 |
70.5-96.3% | (Jung et al., 2015) |
| sMRI and PET | AD/MCI | Volumes of GM tissue of seleeted ROIs | 93 | Domain Transfer SVM |
NC=52, AD=51, cMCI=43, ncMCI=56, Total=202 |
MCI-CONV: 79.4% |
(B. Cheng et al., 2015) |
| sMRI and PET | AD/MCI | Functional and structural connectivity measures using sparse inverse covariance estimation |
84 | SVM | HC=68, AD=70, MCI=111, Total=249 |
84-92% | (Ortiz et al., 2015) |
| sMRI and PET | AD/MCI | GM volume of ROIs from sMRI and average intensity of ROIs from PET |
186 (original number of features) |
Multi-kernel SVM | NC=52, AD=51, MCI=99, Total=202 |
78.8-94.8% | (F. Liu et al., 2014) |
| sMRI, FDG-PET | AD/MCI | ROI-based GM, volumes from sMRI and average intensity from PET |
186 | Multi-kernel SVM | NC=52, AD=51, ncMCI=56, cMCI=43, Total=202 |
80.3-95.9%, MCI- CONV:69.8 % |
(Zu et al., 2015) |
|
sMRI, FDG- PET |
AD/MCI | Cortical and volumetric measures and surface based FDG uptakes |
24 | Partial least square LDA |
NC=85, AD=71, MCI=163, Total=319 |
76.5-90.1% | (Yun et al., 2015) |
|
sMRI, FDG- PET |
AD/MCI | GM and WM maps from sMRI and FDG- PET images |
Variable | Multi Kernel Learning |
HC=66, AD=48, MCI=119, Total=233 |
87.6% | (Hinrichs et al., 2011) |
|
sMRI, FDG- PET and Florbetapir PET |
AD/MCI | Mean volume of GM, SUVr value of FDG- PFT and SUVr value of florbetapir PET for selected ROIs |
90 per modalitiy |
Weighted multi- modality sparse representation- based classification |
NC=117, AD=113, sMCI=83, pMCT=27, Total=340 |
74.5-94.8%, MCI- CONV:77.8 % |
(Xu et al., 2015) |
|
sMRI, FDG- PET, and CSF |
AD/MCI | ROI-based GM, WM and CSF volumes from sMRI and average intensity from PET |
189 | SVM | HC=52, AD=51, ncMCI=56, cMCI=43, Total=202 |
76.4-93.2%, MCI- CONV=81.2 % |
(Zhang et al., 2011) |
|
sMRI, FDG- PET, and CSF data |
AD/MCI | Volume of GM from sMRI and average intensity from PET of selected ROIs along with CSF measures |
189 | Graph-guided multi-task learning |
NC=52, AD=50, MCI=97, Total=199 |
80.0-92.6% | (Yu et al., 2015) |
|
sMRI, FDG- PET, CSF, and APOE genotype |
AD/MCI | ROI-based GMD maps, mean activity from PET |
20 ROIs | Gaussian Process Classifier |
HC=73, AD=63, ncMCI=96, cMCI=47, Total=279 |
MCI- CONV:74.1 % |
(Young et al., 2013) |
| sMRI, PET | AD/MCI | Volume of GM from sMRI and average intensity from PET of selected ROIs |
186 | Multi-task Linear Programming Discriminant |
sMCI=226, pMCI=167, Total=393 |
MCI-CONV: 67.2% |
(Yu et al., 2014) |
| sMRI, PET | AD/MCI | GM density relative cerebral metabolic rate for glucose of ROIs |
168-172 | SVM | ncMCI=40, cMCI=40, Total=80 |
MCI-CONV: 74.8-75.0 % (AUC) |
(Ota et al., 2015) |
|
sMRI, PET, CSF and SNP |
AD/MCI | GM volume and average intensity of ROIs along with CSF and SNP features |
93(sMRI, 93(PET), 3 (CSF) and 5677 (SNP) |
SVM | HC=47, AD=49, MCI=93, Total=189 |
71.0-94.8% | (Zhang et al., 2014) |
| sMRI | AD/MCI/Diment ia |
GM and WM maps | N/A | SVM | HC=604, AD=483, FTD=51, LBD=27, ncMCI=290, cMCI=128, Total=1583 |
73-97% (AUC) MCI- CONV:73.0 % (AUC) |
(Klöppel et al., 2015) |
| dMRI, sMRI | AD/VaD | Transcallosal prefrontal FA and Fazckas score |
4 | LDA | HC=22, AD=16, VaD=13, Total=51 |
87.5% | (Zarci et al., 2009) |
| sMRI | Dementia | Hippocampal head and body volumetric measures |
4 | LDA | HC=17, Questionable Deminetia=12, Mild Dimential=10, Total=39 |
76.9% | (Wolf et al., 2001) |
| dMRI | MCI | Clustering coefficient of WM connectivity maps based on fiber count, FA, MD and principal diffusivities |
3 (Most selected ROIs) |
SVM | HC=17, MCI=10, Total=27 |
88.9% | (Wee et al., 2011) |
| dMRI | MCI | FA, DA, DR and MD | 500 | SVM | EC=40, MCI=33, Total=73 |
93.0% | (O'Dwyer et al., 2012) |
| dMRI | MCI | FA and the volume of fiber pathways from selected region |
100-4500 | SVM | NC=45, MCI=39, Total=84 |
100% | (Lee et al., 2013) |
| dMRI | MCI | FA maps | 1000 | SVM | sd-aMCI=18, sd- fMCI=13, ad-aMCI=35, Total=66 |
97% | (Haller et al., 2013) |
| dMRI | MCI | FA, longitudinal, radial, and mean diffusivity features |
N/A | SVM | HC=35, MCI=67, Total=102 |
91.4-97.5% | (Haller et al., 2010) |
| rsfMRI | MCI | local connectivity and global topological properties |
450 | Multiple Kernel Learning |
HC=25, MCI=12, Total=37 |
91.9% | (Jie et al., 2014) |
| rsfMRI | MCI | N/A | 465 | N/A | HC=21, MCI=29, Total=60 |
95.6% | (Beltrachini et al., 2015) |
| sMRI | MCI | Graph properties based on inter-regional co variation of cortical thickness |
Variable | Multiple Kernel Learning |
NC=42,sd-aMCI=38, md- aMCI=32, Total=112 |
56.0-62.0% | (Raamana et al., 2014) |
| sMRI | MCI | Volume, mean T1, MTR and T2* for selected ROIs |
7 ROIs | SVM | HC=77, MCI=42, Total=119 |
75% | (Granziera et al., 2015) |
| sMRI and dMRI | MCI | Subcortical volumetric measures and FA values |
68 | SVM | HC=204, aMCI=79, Total=283 |
71.1% | (Cui et al., 2012) |
|
sMRI, FDG- PET, CSF and Genetics |
MCI | ROI-based volumetric measures from sMRI, voxel-wise intensity measures from PET along with CSF and genetic features |
>1E5 | Random Forest | NC=35, AD=37, sMCI=41, pMCI=34, Total=147 |
74.6-89.0%, MCI- CONV:58.0 % |
(Gray et al., 2013) |
| sMRI, PET | MCI | ROI-based GM, WM and CSF volumes from sMRI and average intensity from PET |
93 ROIs | Multiple Kernel Learning |
ncMCI=50, cMCI=38, Total=88 |
78.4% | (Zhang and Shen, 2012b) |
2.2 Schizophrenia
Schizophrenia is among the most prevalent mental disorders and affects about one percent of the population worldwide (Bhugra, 2005). This devastating, chronic heterogeneous disease is usually characterized by disintegration in perception of reality, cognitive problems, and a chronic course with lasting impairment (Heinrichs and Zakzanis, 1998). Considering the absence of standard clinical test for schizophrenia, there is a growing interest in automatic diagnosis of schizophrenia based on neuroimaging features. We surveyed 65 papers, which are tabulated in Table 3.
Table 3.
Summary of 65 MRI-based schizophrenia classification studies.
| Modality | Disorder | Features | # Features | Classifier | Number of Subjects | Overall Accuracy | Reference |
|---|---|---|---|---|---|---|---|
| dMRI | Schizophrenia | Discriminant PCA of FA Maps | 60 | Fisher's Linear Discriminant |
HC=45, SZ=45 Total=90 |
80% | (Caprihan et al., 2008) |
| dMRI | Schizophrenia | FA Maps | 13 | LDA | HC=24, SZ=34 Total=58 |
75% | (Caan et al., 2006) |
| dMRI | Schizophrenia | Voxels of FA and MA maps reduced by PCA |
11-13 | LDA | HC=50, SZ=50 Total=100 |
96% | (Ardekani et al., 2011) |
|
fMRI (Sensorimotor, AOD, Working Memory tasks) |
Schizophrenia | Mean activation of the largest activation cluster |
1 | Majority vote of 3 decision stumps |
HC=15, SZ=13 Total=28 |
96% | (Honorio et al., 2012) |
|
fMRI (AOD/Sternberg /Sensorimotor tasks) |
Schizophrenia | ICA Spatial Maps | 10:14 | Projection Pursuit | HC=91, SZ=57 Total=138 |
80-90% | (Demirci et al., 2008a) |
|
fMRI (AX-CPT task) |
Schizophrenia (first-episode) |
Voxels of left DLPFC in the contrast map | N/A | LDA | HC=51, SZ=51 Total=102 |
62% | (Yoon et al., 2012) |
|
fMRI- (Monetary Incentive Delay task) |
Schizophrenia | MVPA of task activation pattern (best result for right palladium) |
N/A | Searchlight SVM | HC=44, SZ=44 Total=88 |
93% | (Koch et al., 2015) |
|
fMRI (Sensorimotor task) + SNP |
Schizophrenia | Sparse representation based variable selection |
200 | Sparse representation based classifier |
HC=116, SZ=92 Total=208 |
77% | (Cao et al., 2013) |
|
fMRI (Verbal Fluency task) |
Schizophrenia/bipolar | Thresholded voxels in activation map by ANOVA tests |
N/A | SVM | HC=40, SZ=32, BP=40 Total=104 |
92% | (Costafreda et al., 2011b) |
|
fMRI (Visual task) |
Schizophrenia | Selected active voxels from the contrast map |
346 | Multi voxel pattern analysis (MVPA) |
HC=15, SZ=19 Total=34 |
59-72% | (Yoon et al., 2008) |
|
fMRI (WMT task) |
Schizophrenia with and without OCD |
MVPA on GLM contrast values | 33 | SVM | HC=20, SZ (with OCD)=16, SZ (without OCD)=17, Total=53 |
75-91% | (Bleich-Cohen et al., 2014) |
|
fMRI (AOD task) |
Schizophrenia | ICA Spatial Maps of magnitude and phase data |
135-243 | Multiple kernel learning |
HC=21, SZ=31 Total=52 |
85% | (Castro et al., 2014) |
|
fMRI (AOD task) |
Schizophrenia | ICA (temporal and DMN network) and GLM spatial maps parcellated into AAL atlas |
116 | Recursive composite kernels |
HC=54, SZ=52 Total=106 |
95% | (Castro et al., 2011) |
|
fMRI (AOD task) |
Schizophrenia/bipolar | Distance to mean image for each group build using ICA Spatial Maps (DMN and temporal Lobe) |
3 | Minimum Distance | HC=26, SZ=21, BP=14 Total=61 |
83-95% | (Calhoun et al., 2008) |
|
fMRI (AOD task) |
Schizophrenia/bipolar | ICA Spatial Maps (DMN and temporal Lobe) |
10 | Bayesian Generalized Softmax Perceptron |
HC=25, SZ=21, BP=14 Total=60 |
82-90% (AUC) |
(Arribas et al., 2010) |
|
fMRI (AOD task) and rsfMRI |
Schizophrenia | Kernel PCA on ICA spatial maps | 53 | Fisher's Linear Discriminant |
HC=28, SZ=28 Total=56 |
93-98% | (Du et al., 2012) |
|
fMRI (AOD task) and rsfMRI |
Schizophrenia | FNC scores derived from ICA- based multi- network fusion template for functional normalization |
3 and 100 | LDA and shaplet based classifier |
HC=28, SZ=27 Total=55 |
72% | (Çetin et al., 2015) |
|
fMRI (AOD task) and SNP |
Schizophrenia | Three types of features: selected voxels in fMRI activation map, selected SNPs and ICA components |
261 voxels + 150 SNPs |
Majority voting among 3 SVMs |
HC=20, SZ=20 Total=40 |
87% | (Yang et al., 2010) |
| rsfMRI | Schizophrenia | Functional connectivity among 116 regions in AAL atlas reduced by PCA |
333 | SVM | HC=25, SZ=24, Sibling HC=22 Total=71 |
62% | (Yu et al., 2013b) |
| rsfMRI | Schizophrenia/ MDD |
FC among ROIs | 6670 | SVM | HC=38, SZ=32, MDD=19, Total=89 |
80.9% | (Yu et al., 2013a) |
| rsfMRI | Schizophrenia | Functional connectivity among 347 nodes placed as a grid in the entire brain |
3000 | Fused Lasso, GraphNet |
HC=74, SZ=71 Total=145 |
91% | (Watanabe et al., 2014) |
| rsfMRI | Schizophrenia | fALFF values of the left ITG | N/A | SVM | HC=46, Unaffected Sibling of SCZ patients=46, Total=92 |
75% | (W. Guo et al., 2014) |
| rsfMRI | Schizophrenia | FCamong 90 ROIs | 1096 | Random Forest | HC=18, SZ=18 Total=36 |
75% | (Venkataraman et al., 2012) |
| rsfMRI | Schizophrenia | FC among 90 regions in WFU atlas reduced by PCA |
550 | SVM | HC=22, SZ=22 Total=44 |
93% | (Tang et al., 2012) |
| rsfMRI | Schizophrenia | Functional connectivity (based on extended maximized mutual information) among 116 AAL regions |
6670 | SVM | HC=32, SZ=32 Total=64 |
83% | (Su et al., 2013) |
| rsfMRI | Schizophrenia | Dimension-reduced FC (local linear embedding) among AAL ROIs |
12 | C-Means Clustering |
HC=20, SZ=32 Total=52 |
86% | (Shen et al., 2010) |
| rsfMRI | Schizophrenia | Functional connectivity among 116 regions in AAL atlas |
6670 | Deep Neural Network |
HC=50, SZ=50 Total=100 |
86% | (Kim et al., 2015) |
| rsfMRI | Schizophrenia | Functional connectivity based on ICA decomposition |
46 | Regularized Linear Discriminant classifier |
HC=196, SZ=71 Total=267 |
75-84% | (Kaufmann et al., 2015) |
| rsfMRI | SZ | Graph Measures of Functional connectivity | N/A | SVM | HC=29, SZ=19, Total=48 | 80.0% | (H. Cheng et al., 2015) |
| rsfMRI | Schizophrenia | Local and global Complex network measures |
216 | SVM | HC=10, SZ=8 Total=18 |
100% | (Fekete et al., 2013) |
| rsfMRI | Schizophrenia | Functional connectivity patterns | 6-7 variable |
Ensemble of SVM classifiers |
HC=31, SZ=31 Total=62 |
85-87% | (Fan et al., 2011) |
| rsfMRI | Schizophrenia | Pearson correlation features derived from Regional Homogeneity, ALFF, FALF and Voxel-Mirrored Homotopic Connectivity |
100 | Ensemble of extreme learning machines |
HC=74, SZ=72 Total=146 |
80-91% | (Chyzhyk et al., 2015) |
| rsfMRI | Schizophrenia | Size of connected components in graphs build from correlation among time-courses for 90 AAL regions |
N/A | SVM | HC=29, SZ=29 Total=58 |
75% | (Bassett et al., 2012) |
| rsfMRI | Schizophrenia | Functional network connectivity among 9 ICA time-courses |
45 | SVM (best results) | HC=28, SZ=28 Total=56 |
96% | (Arbabshirani et al., 2013) |
| rsfMRI | Schizophrenia | MVPA based on whole brain thalamic connectivity map |
N/A | SVM | HC=90, SZ=90, Total=180 |
73.9% | (Anticevic et al., 2014) |
| rsfMRI | Schizophrenia | Graph metrics based on FNC computed from ICA |
13 | SVM | HC=74, SZ=72 Total=146 |
65% | (Anderson and Cohen, 2013) |
| sMRI | Schizophrenia | Voxels from five regions based on optimally discriminative voxel-based morphometry |
N/A | SVM | HC=79, SZ=69 Total=148 |
71% | (Zhang and Davatzikos, 2013) |
| sMRI | Schizophrenia (First episode) |
Whole brain volumetric measurements based on RAVENS |
69 | SVM | HC=62, SZ=62 Total=124 |
73% | (Zanetti et al., 2013) |
| sMRI | Schizophrenia (first-episode) |
Volume and mean cortical thickness of selected ROIs |
2.5 | Discriminant Function Analysis |
HC=40, SZ=52 Total=92 |
80% | (Takayanagi et al., 2011) |
| sMRI | Schizophrenia and psychosis |
Cortical GMD | 129 | Sparse multinomial logistic regression classifier |
HC=36, SZ=36 Total=72 |
86% | (Sun et al., 2009) |
| sMRI | Schizophrenia/bipolar | Voxel-wise GM Maps | N/A | SVM | HC1=66, HC2 = 43, SZ1=66, SZ2=46, BP1=66, BP2=47, Total1=198, Total2=136 |
67-90% | (Schnack et al., 2014) |
| sMRI | Schizophrenia | Texture and volumetric measures | N/A | LDA | HC=24, SZ=27, Total=51 | 65.0-72.7% | (Radulescu et al., 2014) |
| sMRI | Schizophrenia | Clinical, neuropsychological, biochemical and volumetric measures |
1050 | SVM | HC=42, SSD=36, Non- SSD=45, Total=123 |
81.0-99.0% | (Pina-Camacho et al., 2015) |
| sMRI | Schizophrenia/bipolar | Volume of 23 ROIs along with 22 neuropsychological test scores |
45 | LDA | HC=8, SZ=10, BP=10 Total=28 |
96% | (Pardo et al., 2006) |
| sMRI | Schizophrenia | GM and CSF volumetric measures of ROIs | 4 | LDA | HC=105, HC2=23, SZ1=38, SZ2=23, Total=189 |
70-76% | (Ota et al., 2012) |
| sMRI | Schizophrenia | Gray matter densities based on voxel-based morphometry of top 10% voxels |
15,700 | SVM | HC1=111, HC2=122, SZ1=128, SZ2=155 Total1=239, Total2=277 |
71% | (Nieuwenhuis et al., 2012) |
| sMRI | Schizophrenia | Volume of several ROIs in the brain | 7 | LDA | HC=47, SZ=57 Total=104 |
78-86% | (Nakamura et al., 2004) |
| sMRI | Schizophrenia/ Mood Disorder |
GM maps of Regional Analysis of brain Volumes in Normalized Space (RAVENS) |
170 | SVM-RFE | Mood Disorder=104, SZ=158 Total=262 |
76% | (Koutsouleris et al., 2015) |
| sMRI | Schizophrenia | The mean expression of Eigen image derived from voxel-based morphometry |
1 | Simple Thresholding |
HC=46, SZ=46 Total=92 |
80-90% | (Kawasaki et al., 2007) |
| sMRI | Schizophrenia (first-episode) |
Whole brain voxel intensity values | N/A(probably thousands) |
maximum- uncertainty LDA |
HC=39, SZ=39 Total=78 |
72% | (Kasparek et al., 2011) |
| sMRI | Recent onset Schizophrenia |
Volumetric measurements of 95 ROIs | 5 | LDA | HC=47, SZ=28 Total=75 |
72% | (Karageorgiou et al., 2011) |
| sMRI | Schizophrenia | MR intensities, gray matter densities and deformation based morphometry |
96 per feature category |
Combination of mMLDA, centroid method, and the average linkage |
HC=49, SZ=49 | 81.6% | (Janousova et al., 2015) |
| sMRI | Schizophrenia | GM and WM maps | N/A | SVM | HC=20, SZ=19, Total=39 | 66.6-77% | (Iwabuchi et al., 2013) |
| sMRI | Schizophrenia (identifying subtypes) |
Multi-edge graphs build from Structural connectivity networks with 78 ROIs |
N/A | Spectral Clustering | HC=29, SZ=23 Total=52 |
78% | (Ingalhalikar et al., 2012) |
| sMRI | Schizophrenia (Childhood onset) |
Cortical Thickness | 74 | Random Forrest | HC=99, SZ=98 Total=197 |
74% | (Greenstein et al., 2012) |
| sMRI | Schizophrenia (cognitive deficit and cognitive spared) |
Whole brain voxel-based morphometry | N/A | SVM | HC=163, SZ=208, SZA=41, Total=412 |
56-72% | (Gould et al., 2014) |
| sMRI | Schizophrenia | Volumetric measurements based on deformation-based morphometry |
39/44 | SVM | HC1=38, HC2=41, SZ1=23, SZ2=46 Total1=61, Total2=87 |
91% | (Fan et al., 2007) |
| sMRI | Schizophrenia | Volumetric measures of all WM, GM and CSF |
69 | SVM-RFE | HC=38, SZ=23 Total=61 |
92% | (Fan et al., 2005) |
| sMRI | Schizophrenia | Whole brain volumetric measurements | N/A | Nonlinear Classifier (not specified) |
HC=79, SZ=69 Total=148 |
81% | (Davatzikos et al., 2005) |
| sMRI | Schizophrenia | Hippocampal and thalamic shape eigenvectors |
25 | Discriminant Function Analysis |
HC=65, SZ=52 Total=117 |
79% | (Csernansky et al., 2004) |
| sMRI | Schizophrenia | Visual words extracted from DLPFC by SIFT and clustered by k-means |
30 | SVM with local kernel |
HC=54, SZ=54 Total=108 |
66-75% | (Castellani et al., 2012) |
| sMRI | Schizophrenia | Surface morpholical Measures | N/A | Semi-supervised (Hierarchical Clustering) |
HC=40, SZ=65, Total=105 |
94.0% | (Bansal et al., 2012) |
| sMRI and dMRI | Schizophrenia/ MDD |
Volume and FA of Insula, thalamus, ACC, Ventricles and corpus callosum |
31 | LDA | MDD=25, SZ=25 Total=50 |
72-88% | (Ota et al., 2013) |
| sMRI, rsfMRI and dMRI | Schizophrenia | Gray matter densities from structural, FA from DTI and ALFF from fMRI |
1863 | SVM | HC=28, SZ=35 Total=63 |
79% | (Sui et al., 2013b) |
2.3 Depressive Disorders
Major depressive disorder (MDD) or unipolar depression characterized by a pervasive low mood, self-esteem and lack of interest in enjoyable activities is a common mental illness affecting adolescents. The lifetime prevalence of MDD is approximately 15–20% (Kessler et al., 2003; Lewinsohn et al., 1986). It is estimated that by the year 2020, depression will account for 15% of the disease burden in the world ranking second after heart disease (Kessler et al., 1994). We reviewed 19 studies that used neuroimaging for automatic diagnose MDD. Those studies are listed in Table 4.
Table 4.
Summary of 19 MRI-based depressive disorder classification studies.
| Modality | Disorder | Features | # Features | Classifier | Number of Subjects | Overall Accuracy | Reference |
|---|---|---|---|---|---|---|---|
| dMRI | MDD | Whole-brain anatomical connectivity patterns |
50 | SVM | HC=26, MDD=22, Total=48 |
91.7% | (Fang et al., 2012) |
|
fMRI (facial affect recognition task) |
MDD | Brain activation maps and ROI-averaged activation features |
N/A | One-class SVM | HC=19, Depressed=19, Total=38 |
63-65.5% (estimated) |
(Mourão-Miranda et al., 2011) |
|
fMRI (gender discrimination and emotional tasks) |
MDD | Sparse network-based features of FC | 9316 | SVM | HC=19, MDD=19, Total=38 |
78.9-85.0% | (Rosa et al., 2015) |
|
fMRI (social concept task) |
MDD | GM maps of PPI analysis | N/A | Maximum Entropy LDA |
HC=21, MDD=25, Total=46 |
78.1% (AUC) |
(Sato et al., 2015) |
|
fMRI (verbal fluency task) |
MDD | Voxel-wise contrast map | 14055 | Regularized Logistic Regression, SVM (best performance) |
HC=31, MDD=31, Total=62 |
90.0-95.0% | (Shimizu et al., 2015) |
| rsfMRI | MDD | FC maps of sACC | N/A | Label Generation Maximum Marging Clustering |
HC=29, MDD=24, Total=53 |
92.5% | (Zeng et al., 2014) |
| rsfMRI | MDI) | Hurst components of resting-state networks | 12 | SVM | HC=20, MDD=20, Total=40 |
90% | (Wei et al., 2013) |
| rsfMRI | MDD | Netowrk-based measures based on FC among ROIs |
2-25 | SVM | HC=22, MDD=21, Total=43 |
99% | (Lord et al., 2012) |
| rsfMRI | MDD | FC among AA1 regions | 31 | SVM | HC=37 MDD=39, Total=76 |
76.6% | (Cao et al., 2014) |
| rsfMRI | First-onset Depressive Disorder |
Graph-theory Measures | 30 | ANN | HC=27, first-onset depression=36, Total=63 |
90.5% | (H. Guo et al., 2014) |
| rsfMRI | Subthreshold Depression |
ReHo features of ROIs | 8 ROIs | Fisher stepwise discriminant analysis |
NC=19, StD=19, Total=37 |
91.9% | (Ma et al., 2013) |
| sMRI | MDD/BP | GM, WM and ventricles volumetric maps (RAVENS) |
53-99 | SVM | HC1=33, HC2=38, MDD=19, BP=23, Total=113 |
54.6-66.1% | (Serpa et al., 2014) |
| sMRI | MDD/BP | Gray matter volumes of Caudate and Ventral Diencephalon |
4 | SVM | HC=61, BP=40, MDD=57, RMD=35, Total=193 |
59.5-62.7% | (Sacchet et al., 2015) |
| sMRI | MDD/BP | Volumetric measurements | 5 | Discriminant function analysis |
HC=22, MDD=32, BP=14, Total=68 |
81.0% | (MacMaster et al., 2014) |
| sMRI | MDD/BP | Cortical thickness and surface area | 18 | SVM | HC=29, MDD=19, BP=16 |
74.3% | (Fung et al., 2015) |
| sMRI | MDD | Feature-based morphometric measures of GM maps |
N/A | SVM and RVM | HC=32, MDD=30, Total=62 |
90.3% | (Mwangi et al., 2012) |
| sMRI | MDD | GM and WM densities | N/A | SVM | HC=42, RDD=23, NDD=23, Total=88 |
58.7-84.6% | (Gong et al., 2011) |
| sMRI | MDD | Cortical thickness of several ROIs | 68 | SVM | HC=15, MDD=18, Total=33 |
70% | (Foland-Ross et al., 2015) |
| sMRI, rsfMRI and dMRI | LLD | Variety of features from each modality | 13 Feature Sets |
Alternating Decision Trees |
EC=35, LLD=33, Total=68 |
87.3% | (Patel et al., 2015) |
2.4 Autism Spectrum Disorder
Autism spectrum disorder (ASD) is a serious neurodevelopmental condition characterized by impaired social communication, deficits in social–emotional reciprocity, deficits in nonverbal communicative behaviors used for social interaction and stereotypic behavior (Association and others, 2003). Although the causation of autism is still largely unknown, it has been suggested that genetic, developmental, and environmental factors could be involved alone or in combination as possible causal or predisposing effects toward developing autism (Minshew and Payton, 1988; Wing, 1997). ASD has an estimated prevalence of 1:68 in the U.S. (Baio, 2012). We surveyed 20 papers in automatic diagnosis of ASD using MRI-based features. Those studies are listed in Table 5.
Table 5.
Summary of 20 MRI-based ASD classification studies.
| Modality | Disorder | Features | # Features | Classifier | Number of Subjects | Overall Accuracy | Reference |
|---|---|---|---|---|---|---|---|
| dMRI | ASD | FA and MD of selected ROIs | 18 | SVM | TDC=30, ASD=45, Total=75 |
80% | (Ingalhalikar et al., 2011) |
|
fMRI (social interaction task) |
ASD | Activation of selected voxels processed by factor analysis |
4 factors | Gaussian Naïve Bayes |
HC=17, TDC=17, Total=34 |
97% | (Just et al., 2014) |
|
fMRI (two language tasks and a Theory- of-Mind task) |
ASD | AG, MPFC and PCC based FC maps | N/A | Logistic Regression | TD=14, ASD=13, Total=27 |
96.0% | (Murdaugh et al., 2012) |
|
fMRI-Task and DMRI |
ASD | Causal connectivity weights, FC values and FA values |
19 | SVM | TDC=15, ASD=15, Total=30 |
95.9% | (Deshpande et al., 2013) |
| rsfMRI | ASD | ICA components of rsfMRI | 10 components |
Logistic Regression | TDC=20, ASD=20, Total=40 |
78.0% | (Uddin et al., 2013) |
| rsfMRI | ASD | FC among ROIs | Variable | Logistic Regression and SVM (best results) |
TD1=59, TD2=89 ASD1=59, ASD2=89, Total=296 |
76.7% | (Plitt et al., 2015) |
| rsfMRI | ASD | FC among 90 ROIs | 4005 | Probabilistic Neural Network |
TDC=328, ASD=312, Total=640 |
90% | (Iidaka, 2015) |
| rsfMRI | ASD | Functional Connectivity among 220 ROIs | 24090 | Random Forest | TDC=126, ASD=126, Total=252 |
91% | (Chen et al., 2015) |
| rsfMRI | ASD | FC among ROIs | 26,393,745 | Threshodling | TD=40, ASD=40, Total=80 |
79.0% | (Anderson et al., 2011) |
| sMRI | ASD | Thickness and volumetric of ROIs along with interregional features |
N/A | Multi-kernel SVM | HC=59, ASD=58, Total=117 |
96.3% | (Wee et al., 2014) |
| sMRI | ASD | Voxel-wise GM and WM maps | N/A | SVM | TD=24, ASD=24, Total=48 |
92.0% | (Uddin et al., 2011) |
| sMRI | ASD | GM volume map | N/A | SVM | HC=40, ASD=52, ASD- Sib=40 |
80.0-85.0% | (Segovia et al., 2014) |
| sMRI | ASD | Regional thickness measurements extracted from SBM |
7 | Logistic model trees |
HC=16, ASD=22, Total=38 |
87% | (Jiao et al., 2010) |
| sMRI | ASD | Morphometric features of selected ROIs | 314 | SVM | HC=20, ASD=21, Total=41 |
74% (AUC) | (Gori et al., 2015) |
| sMRI | ASD | GM and WM maps | >10,000 | SVM | HC=22, ASD=22, Total=44 |
77% | (Ecker et al., 2010b) |
| sMRI | ASD | Volumetric and geometric features of selected cortical locations |
5 features from each ROI |
SVM | HC=20, ASD=20, Total- 40 |
85% | (Ecker et al., 2010a) |
| sMRI | ASD | Gray maps from VBM-DARTEL | 200 | SVM | TDC=38, ASD=30, Total = 76 |
80.0% (AUC) |
(Calderoni et al., 2012) |
| sMRI | ASD | Volumetric measures and cerebellar vermis area |
9 | Discriminant Function Analysis |
TDC=15, ASD=52, Total=67 |
92.3-95.8% | (Akshoomoff et al., 2004) |
|
sMRI, dMRI and MRS |
ASD | Cortical thickness, FA and neurochemical concentration |
3 | Decision Tree | TD=18, ASD=19, Total=37 |
91.9% | (Libero et al., 2015) |
| sMRI, rsfMRI | ASD | Volume of selected subcortical regions, fALFF, number of voxels and Z-values of selected regions and global VMHC voxel number |
22 | Random Tree Classifier |
TDC=153, ASD=127, Total=280 |
70.0% | (Zhou et al., 2014) |
2.5 Attention Deficit Hyperactivity Disorder
Attention Deficit Hyperactivity Disorder (ADHD) is one of the most commonly found functional disorders affecting children. Approximately 3–10% of school aged children are diagnosed with ADHD (Biederman, 2005; Dey et al., 2012). Currently, no biological-based measure exists to detect ADHD and instead behavioral symptoms are investigated to identify it. Despite all the research efforts, the root cause of ADHD is still unknown. In 2011, a global competition called ADHD-200 was held in order to use neuroimaging as well as phonotypic measures to automatically detect ADHD (Consortium and others, 2012). Most of the studies reviewed in this survey were responses to that challenge. The main characteristics of those studies are tabulated in Table 6.
Table 6.
Summary of 22 MRI-based ADHD classification studies.
| Modality | Disorder | Features | # Features | Classifier | Number of Subjects | Overall Accuracy | Reference |
|---|---|---|---|---|---|---|---|
|
fMRI (Stop Task) |
ADHD | Whole brain GLM coefficient map | 21,658 | Gaussian Process Classifier |
HC=30, ADHD=30, Total=60 |
77% | (Hart et al., 2014a) |
|
fMRI (Six Tasks) |
ADHD | Network measures based on FC values | N/A | SVM | ADHD-IA=13, ADHD- C=21, Total=34 |
91.2% | (Park et al., 2015) |
|
fMRI (temporal discrimination task) |
ADHD | Brain Activation Map | N/A | Gaussian Process | HC=20. ADHD=20, Total=40 |
75.0% | (Hart et al., 2014b) |
| rsfMRI | ADHD | ReHo Maps | N/A | PCA-based Fisher discriminative analysis |
HC=12, ADHD=12, Total=24 |
85.0% | (Zhu et al., 2008) |
| rsfMRI | ADHD | ReHo Maps | 6500 | SVM | HC=23, ADHD=23, Total=46 |
80.0% | (Wang et al., 2013) |
| rsfMRI | ADHD | FFT and different varation of PCA on the BOLD signals along with phenotypic measures |
About 7,000 |
SVM | HC=429, ADHD-I=98, ADHD-C=141, Total=668 |
68.86-76% | (Sidhu et al., 2012) |
| rsfMRI | ADHD | ReHO, ALLFand RSN | 400 for each feature type |
Logistic Regression (best performance) |
HC=546, ADHD- IA=122, ADHD-HI=12, ADHD-C=249, Total=929 |
54% ADHD Subtype: 67% |
(Sato et al., 2012) |
| rsfMRI | ADHD | Graph based features based on FC | 150 | SVM-based MVPA | TDC=455, ADHD-I=80, ADHD-C=112 Total=647 |
63.4-82.7% | (Fair et al., 2012) |
| rsfMRI | ADHD | Graph-based measures compressed by Multi-Dimensional Scaling |
2 | SVM | HC=307, ADHD=180, Total=487 |
73.5% | (Dey et al., 2014) |
| rsfMRI | ADHD | Directional connectivity measures | 200 | Artificial Neural Network |
TDC=744, ADHD=433, Total=1177 |
90% | (Deshpande et al., 2015) |
| sMRI | ADHD/Dyslexia | Morphometric measures of ROIs | 6 | Discriminant Function Analysis |
HC=10, ADHD=10, Dyslexia=10, Total=30 |
60.0-87%% | (Semrud-Clikeman et al., 1996) |
| sMRI | ADHD | Cortical thickness measures | 340 | ELM | HC=55, ADHD=55, Total=110 |
90.2% | (Peng et al., 2013) |
| sMRI | ADHD | Voxel-wise GM Volumetric Measures | N/A | Gaussian Process Classifier |
HC=19, ASD=19, ADHD=20 |
68.2-85.2% | (Lim et al., 2013) |
| sMRI | ADHD | WM maps | N/A | SVM | HC=34, ADHD=34, Total=68 |
93% | (Johnston et al., 2014) |
| sMRI | ADHD | Caudate nucleus volumetric measures | N/A | Adaboost and SVM | HC=39 AHDH=39, Total=78 |
72.5% | (Igual et al., 2012) |
| sMRI | ADHD | Texture features based on isotropic local binary patterns on three orthogonal planes |
117- 33630 |
SVM | HC=226, ADHD=210, Total=436 |
69.9% | (Chang et al., 2012) |
| sMRI | ADHD | Surface morphometric measures | N/A | Semi-supervised (Hierarchical Clustering) |
HC=42, ADHD=41, Total=83 |
91.0% | (Bansal et al., 2012) |
|
sMRI + rsfMRI +Phenotypic data |
ADHD | Curvature index, folding index, Gaussian curvature, gray matter volume, mean curvature, surface area, thickness average, and thickness standard deviation along with functional connectivity measures and phenotypic data |
20 | NMF + Decision Tree |
TD=472, ADHD=276, Total=748 |
66.8% | (Anderson et al., 2014) |
| sMRI + rsfMRI | ADHD | Various anatomical, network and non- imaging measures |
5-6000 | SVM | TDC=491, ADHD=285, Total=776 |
80.0% (AUC) |
(Bohland et al., 2012) |
|
sMRI and fMRI- Task(Flanker/NoGo) |
ADHD | Whole brain GLM coefficients and GM maps from VBM |
N/A | SVM | HC=18, ADHD=18, Total=36 |
61.1-77.8% | (Iannaccone et al., 2015) |
| sMRI and rsfMRI | ADHD | Cortical thickness and GM maps from sMRI and ReHo and FC from rsfMRI |
N/A | SVM and Multi- Kernel Learning |
TCD=402, ADHD=222, Total=624 |
61.5% | (D. Dai et al., 2012) |
|
sMRI and rsfMRI |
ADHD | Morphological measures, FC, power spectra and graph measures |
Variable (>100) |
Multiple SVM | TD=491, ADHD=285, Total=776 |
55% | (Colby et al., 2012) |
2.6 Analysis of the Survey
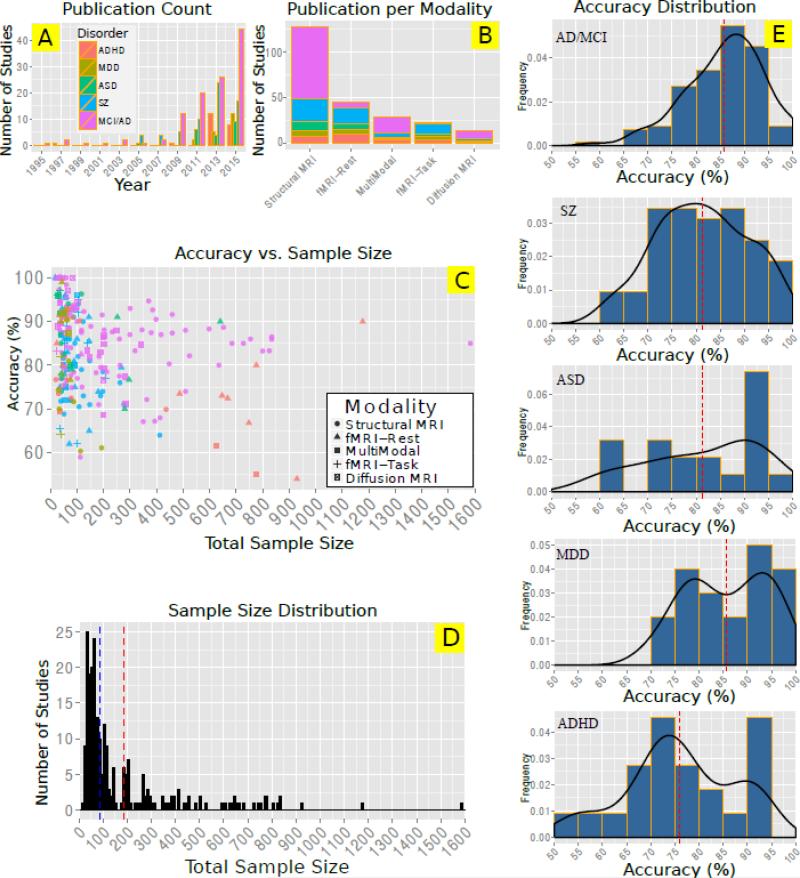
In Figure 3 we illustrate a couple of key aspects of this survey. Figure 3A shows the number of papers published in each year for each disease type. The number of studies has been growing significantly since 2007. There is a peak for ADHD studies in 2012-2013 mainly due to ADHD-200 competition (Consortium and others, 2012) which attracted many scientists. The total number of studies for each modality and each disorder is illustrated in Figure 3B. It is clear that structural MRI is the most popular modality especially for MCI/AD studies thanks to Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset. Combined rest and task fMRI studies are most popular for ADHD and schizophrenia studies. Surprisingly, multimodal studies are more common compared to either task fMRI or diffusion MRI studies. Figure 3C shows the overall accuracy against the total sample size used in the studies. Interestingly, almost all studies that reported very high accuracies, had sample sizes smaller than 100. The reported overall accuracy decreases with sample size in most of disorders such as schizophrenia and ADHD. This pattern raises a serious concern regarding generalizability of many of those studies with small sample sizes. Figure 3D shows the sample size distribution. The dashed lines represent mean (red) and median (blue) sizes, which are 186 and 88 respectively. Finally Figure 3E illustrate the distribution of reported accuracy for each disorder. On average (red dashed lines) MCI/AD and ADHD studies reported the highest and lowest accuracies respectively.
Figure 3.
Visual summary of Table 2-6. A: Total number of papers for two-year intervals for each modality. The inset legend shows the color code for each disorder. This legend also applies to figures in part B and C. B: Number of publications per modality for each disorder C: Scatter plot of overall reported accuracy versus the total sample size. D: Histogram of number of samples used in the surveyed studies. Vertical dashed lines show mean (red) and median (blue) sample size among all studies, which are 186 and 88 respectively. E: Disorder specific histograms of reported accuracies of all surveyed papers. Red dashed line indicates the mean accuracy. Black curves represent the estimated distribution of overall accuracy based on kernel density estimation.
Based on Tables 2-6, the most common extracted features in the surveyed studies are volume and cortical thickness from structural MRI, the activation maps and functional connectivity among ROIs or ICA components from fMRI data and fractional anisotropy from dMRI data. Most common feature reduction methods (not reported in the tables) were based on PCA or univariate statistical tests.
In terms of classification methods, support vector machine (SVM) was by far the most popular method. Different flavors of SVM such as linear, non-linear with different kernel, SVM with recursive feature elimination, SVM with L1 regularization and SVM with L1 and L2 regularization (elastic net) have been used for classification of various disorders. Linear discriminant analysis (under different names) and logistic regression were also popular classification methods among the surveyed studies.
2.7. Predicting Continuous Measures
Most of the studies surveyed above, conducted the diagnosis of a disorder (i.e., assigning a categorical label to each subject) using classification techniques. Pattern regression considers the problem of estimating continuous rather than categorical variables, which can be more challenging as compared to classification. Clinically, pattern regression can be used to estimate the disease stage and progression. Therefore, there is a growing interest in estimating continuous variables such as cognitive scores for brain disorders using neuroimaging measurements. We didn't survey those papers, but we will point out to some of those studies in this section.
Wang et al. proposed a general methodology for estimating continuous clinical variables from high-dimensional imaging data (Wang et al., 2010). Sato et al. used interregional cortical thickness measurements to estimate Autism Diagnostic Observation Schedule (ADOS) score in ASD patients (Sato et al., 2013). Stonnington et al. used relevance vector regression (RVR) to predict number of cognitive scores such as Dementia Rating Scale (DRS) and Alzheimer's Disease Assessment Scale (ADAS) based on structural MRI measures (Stonnington et al., 2010). Tognin et al used RVR to predict Positive and Negative Syndrome Scale (PANSS) scores of subjects at high risk of psychosis based on gray matter volume and cortical thickness measurements (Tognin et al., 2013). Yue et al. showed relationship between functional connectivity and neuropsychological assessment scores such as Rey–Osterrieth Complex Figure Test (CFT) in amnestic MCI patients (Yue et al., 2015). Zahng et al. used MRI, PET and CSF data to predict Mini Mental State Examination (MMSE) and ADAS scores in MCI and AD patients (Zhang and Shen, 2012a).
2.8 Detecting/characterizing at Risk Healthy Subjects
The majority of studies surveyed above tried to automatically diagnose one or more disorders in patients. However, detecting or characterizing healthy individuals who are at high risk of brain disorders could potentially delay or prevent future symptoms. There has been a lot of such studies using genetics information but detecting or characterizing at risk subjects based on neuroimaging data is rare. Mourão-Miranda et al. used functional MRI to detect subjects at high risk of mood disorders (Mourão-Miranda et al., 2012). Guo et al. characterized activity of default-mode network in unaffected siblings of schizophrenia patients using resting-state functional data (W. Guo et al., 2014). In another study, Fan et al. studied structural endophenotypes in unaffected family members of schizophrenia patients using machine learning methods (Fan et al., 2008a).
3. Common Machine-learning Pitfalls in Neuroimaging
In this section, common pitfalls among the surveyed papers are discussed.
3.1 Feature Selection Bias
Most of the papers we surveyed consisted of two consecutive parts: group difference analysis and classification. Usually, statistical tests such as t-tests are used to show group differences on a set of extracted features in the first part of the study, which is followed by a classification approach to assess the discrimination ability of those features on a single subject basis. Unfortunately, it is not rare to see that the results of first part (group differences) are used to select features for the classification part. In general, any use of test samples in any part of the training (such as feature extraction, feature selection and classifier training) poses a bias. Selecting features for classification based on the results of group tests that were conducted on the whole dataset is a form of double dipping and therefore leads to a biased (inflated) result (Bishop, 2006; Demirci et al., 2008b).
This form of feature selection also has another major problem. The significance of group statistical tests, which are the basis of feature selection in some of the studies, is mostly based on p-values. However, the relationship between p-value and discrimination power is not straightforward. Figure 1 shows the p-value of a two-sample t-test as well as overall accuracy based on one or two thresholds in three different scenarios. It is seen that low p-value doesn't necessary mean a strong feature (Figure 1A) and high p-value doesn't mean a weak feature (Figure 1B). However, if the abnormality is one-directional, then a very low p-value might translate to high classification accuracy (Figure 1C). So, by discarding features just based on the result of statistical tests sensitive to group mean, valuable discriminatory information could be lost.
Instead of feature selection based on univariate group-level statistical tests, more common filtering and wrapper methods should be used (Blum and Langley, 1997; Hall and Smith, 1998; Kohavi and John, 1997). Filtering methods assign scores to each feature from which a number of top ones can be selected. A good filtering method should be sensitive to the discriminative power of the features. Most of these methods are univariate and therefore each feature is treated independently from other features. Filtering methods have the advantage of low computational cost, but their main drawback is ignoring the relationship among features.
Wrapper methods, on the other hand, consider selection of a set of feature as a search problem. Different combinations are evaluated and finally the best set of features is selected. A popular wrapper method is the recursive feature elimination (RFE) algorithm (Guyon et al., 2002). Wrapper methods are computationally much more expensive than filtering methods, but can result in superior performance by considering interaction among features.
There are methods that aim at combining both filtering and wrapper methods. Minimum-redundancy maximum relevancy (mRMR) is one the methods popular for genetic feature selection. MRMR tries to select features with maximum mutual information with class labels while minimizing the mutual information among those features (G. Brown et al., 2012)
Finally, there are embedded feature selection methods (Guyon and Elisseeff, 2003). These methods combine classification and feature selection into one unified step. Embedded methods learn the features that contribute the most to the accuracy of the model during the training phase. One of the common categories of the embedded methods is using regularization to enforce the learning algorithm to find more parsimonious models with lower complexity and therefore with fewer parameters. A post training analysis of the model coefficients, determines the selected features. Examples of regularization algorithms used in embedded feature selection methods are LASSO, elastic net and ridge regression (Hastie et al., 2004; Ng, 2004; Park and Hastie, 2007; Zou and Hastie, 2005).
3.2 Overfitting
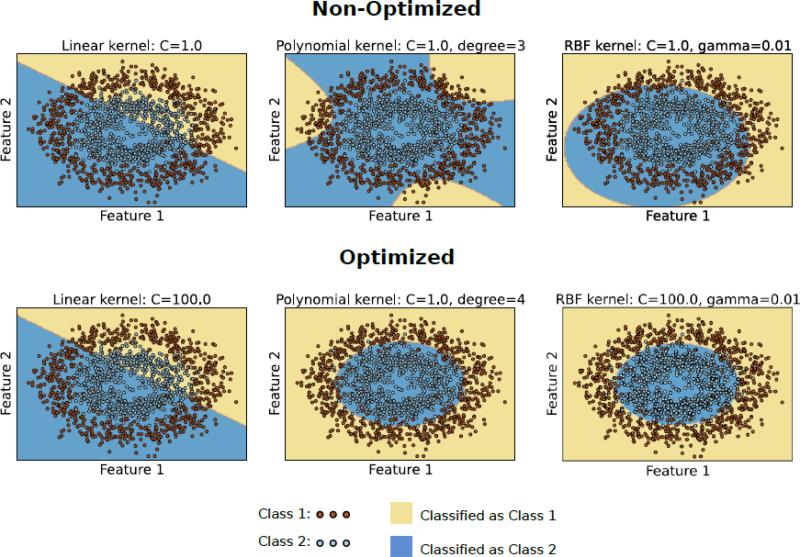
Overfitting happens when a model describes noise in the data rather than the underlying pattern of interest. Overfitting results in very good performance on the observed data and very poor performance on unseen data. Using models that are very complex or have many parameters on datasets with small number of samples and large number of features are more susceptible to overfitting. Neuroimaging datasets have limited number of samples and millions voxels per sample. Based on Figure 3D, the majority of surveyed studies built predictive models based on a very small number of subjects. It is evident from Figure 3C that overall reported accuracy decreases with sample size in our survey. Therefore, it is plausible that many surveyed studies suffer from overfitting problem. It should be noted that by definition, overfitted models work well on the training data and poor on the test data. However, if the process of training and testing is repeated (by varying the model parameters) until a desirable performance on the test data is achieved, the model will likely overfit both the train and test datasets. Cross validation and regularization are common methods to control overfitting. As mentioned earlier, more complex models have a greater chance of overfitting the data. For example, non-linear SVM is more powerful compared to linear SVM but has many more parameters and therefore is also potentially more capable of explaining noise in the data. As discussed in the previous section, proper feature selection can also help to avoiding overfitting.
3.3 Reporting Classification Results
The result of classification is basically a confusion table/matrix also known as a contingency table. The confusion matrix summarizes the results in a table layout where each column represents the predicted class and each row represents the actual class. Confusion matrix is m × m where m represents the number of classes. In the case of binary classification, many statistical measures can be computed from the 2 × 2 confusion matrix, such as sensitivity (or recall), specificity, positive predictive value (or precision), negative predictive value, F1 score, odds ratio, kappa and false negative rate. Confusion matrix and some of the performance measures are shown in Figure 4. In order to understand the performance of a classifier, it is important to report at least sensitivity/specificity or precision/recall along with the overall accuracy. We highly encourage reporting the confusion matrix itself as well. Some of the studies in this review just reported the overall accuracy, which can be very uninformative especially when classes have unequal sample sizes (Alberg et al., 2004). Suppose there are 20 patients and 80 controls in a test dataset. Reporting 80% accuracy is completely uninformative since the classification of all subjects as healthy could result in 80% (one of the scenarios). This problem is easily detectable by looking at the confusion matrix or sensitivity and specificity measures. In unbalanced sample size cases, balanced specificity and sensitivity is more desirable than higher overall accuracy; therefore, measures such as F1 score (harmonic mean of precision and recall) are preferred for evaluating the classifier. The other very common way of reporting results for a binary classifier is by showing “receiver operating characteristic” (ROC) curve. The ROC curve is the plot of sensitivity against “1-specificity” by changing the discrimination threshold and therefore provides a complete picture of classifier's performance. The ROC curve is usually summarized by an area under the curve (AUC), which is a number between 0 and 1 (ideal classifier).
Figure 4.
Confusion matrix and common performance measures for binary classification. Measures such as sensitivity, specificity, precision, accuracy and F1 score are easily computable based on the four elements of the confusion matrix.
The other common reporting issue is unjustified comparison of the achieved overall accuracy with the random chance. This issue is critical in this field due to small sample sizes. For example, an 80% achieved overall accuracy might not be significantly different from a 50% random chance in a statistical sense in a two-class problem if the sample size is too small. Any achieved accuracy in a test sample is just one estimate of the population accuracy. Like any other statistics, a confidence interval can be computed for that measure. In the case of a two-class problem, a binomial confidence interval can be computed for overall accuracy that serves as the basis for comparison with random chance, or any other accuracy. In our example (80% accuracy), if the test sample size is 10, then the 95% exact binomial confidence interval would be [0.444 0.975], which includes the random chance probability (0.50) and therefore is not statistically above chance. Calculating this interval is straightforward using most of statistical and technical computing software such as R and Matlab. This approach should be employed when repeating the classification experiment for number of times is not feasible. However, in most cases, the null distribution of chance is empirically computable by randomly assigning labels to test samples and repeating classification for a number of times. This method, known as a permutation or randomization test, makes it possible to calculate the desired confidence interval of the chance, which consequently can be compared against the achieved classification accuracy using the correct labels (Collingridge, 2013; Fisher et al., 1960; Good, 2006; Mehta et al., 1988) . Recently, for special cases such as SVM, fast analytical estimation of permutation testing has been proposed (Gaonkar and Davatzikos, 2013). Also, it has been shown that p-value for permutation testing can be written in the form of an infinite series whose terms are efficiently computable (Gill, 2007).
3.4 Comparison of Accuracies Across Studies
It was frequently observed that authors claim that their proposed classification framework outperformed some other studies (and sometimes all other studies) just on the basis of overall accuracy. Considering the number of variables in each study—such as sample size, scanner parameters, sample age distribution, patients’ status (e.g., severity, medication), modality, length, type and design of study (for fMRI studies), preprocessing parameters, number and type of extracted features and type of classifier—such a comparison is essentially meaningless. Even in the case of standard neuroimaging datasets, the statistical comparison discussed in the previous section, should be employed to compare the results.
3.5 Hyperparameter Optimization
Hyperparameter optimization or model selection is choosing a set of parameters for the learning algorithm in order to maximize the performance of the algorithm. Hyperparameters should be chosen during training, usually via an inner loop cross validation inside the training data. SVM, which is one of the most popular classifiers in this review and in neuroimaging in general (Orrù et al., 2012), has at least one hyperparameter (linear SVM) called soft margin. In addition to soft margin, non linear SVM has one or more hyperparameters depending on the kernel (e.g. sigma/gamma for RBF kernel and degree for polynomial kernel). Some of the studies that we reviewed just used the default values for these parameters. A lack of parameter optimization can degrade the classification performance significantly. To show this, a toy example is illustrated in Figure 5. SVM with three different kernels is used to classify this simulated two-class problem. In the top row, 1.0 is chosen for soft margin hyperparameter (which is the default of most machine learning software packages) for all kernels, degree of 3 was chosen for the polynomial kernel and gamma of 0.01 was selected for RBF kernel. In the second row, the hyperparameters are optimized. First, it is evident that the linear kernel failed to learn the non linear pattern under both settings. Increasing the polynomial kernel degree by one, dramatically improved the classifier. Also, increased soft margin value, significantly improved SVM with RBF kernel. So, both the choice of kernel and hyperparameters are crucial for building a successful SVM-based classifier. SVM hyperparameters are usually selected based on a grid search over plausible values.
Figure 5.
An example to show the effect of SVM hyperparameter optimization on classification accuracy for linear, polynomial and RBF kernels. Top row: un-optimized, Bottom row: optimized. Since the underlying pattern is non-linear, SVM with linear kernel fails to perform well in both scenarios. Performance of SVM with both polynomial and RBF kernels significantly improve when the parameters are optimized.
4. Machine Learning in Neuroimaging: Shortcomings and Emerging Trends
Machine learning has more than two decades of history in neuroimaging and despite all of the promising results of numerous studies, it is still immature and not ready for integration into clinical healthcare. In this section, we review some of the challenges and emerging solutions.
4.1 Sample Size in Neuroimaging Studies
The most limiting factor in this field is by far the limited sample size issue. As summarized in Figure 3B, the majority of studies in this review and in general have sample size of less than 150. This sample size is miniscule in comparison with other fields in which machine learning is used. As an example, ImageNet2, which is commonly used as standard computer vision dataset, has over one million samples and 1000 classes. As a result of such big datasets, dramatic improvement has been achieved in the field of computer vision in the past few years. However, sample size limitations in neuroimaging pose several problems. First, the classifier performance is directly affected by the sample size. It is shown that large training data sets increase classification accuracy (Franke et al., 2010; Klöppel et al., 2009). Small sample size does not represent the patient population and therefore promising results may not generalize to other patient groups. In a study conducted by Nieuwenhuis et al., it was shown that for small training sample sizes (N<130) the predictive model for classification of schizophrenia patients based on sMRI was not stable (Nieuwenhuis et al., 2012). More than 63% of the studies we reviewed didn't meet this criterion. Large datasets may reduce problems with disease heterogeneity as they can represent the whole spectrum of the disorder. Although there are some machine learning methods that are less sensitive to data, a limited number of data samples can cause model overfitting, resulting in poor generalization of the method to independent data sets (Pereira et al., 2009).
To understand the etiology of complex conditions such as mental health, we must develop a better understanding of the structure of the signals and measurements we make of the brain. Thanks to advances in imaging and assaying technology, we can gather increasingly detailed information about individuals, but the cost and complexity of these techniques means that individual researchers may not have sufficient data to build a compact and informative representation of the data. For example, a single sMRI may have tens of thousands of voxels, but a single site may have only a hundred subjects in their study. With increasingly complex data, the classical “curse of dimensionality” would seem to indicate that there is no way to determine signal from noise in this setting. To address the “small N” problem in other settings, many researchers have proposed open sharing of data to leverage data from multiple sites as well as commercial cloud computing infrastructures to handle the additional computational burden. In the past few years, several multi-site data sharing initiatives such as FBIRN, MCIC and COBRE for schizophrenia, ADNI for Alzheimer's disease, ABIDE for ASD, ADHD-200 for children with ADHD and Functional Connectomes project for healthy have been started.
In neuroscience, measurements often come from human subjects; in some cases legal, ethical, and sociological concerns may preclude or prohibit such open sharing. In particular, local administrative rules, concerns about re-identification of study participants, and a desire to maintain control over data in ongoing research projects may prevent individual research sites from sharing the data (Sarwate et al., 2014). The status quo is a patchwork of institution-to-institution data use agreements whose complexity stymies automated analyses across more than a handful of data sets.
4.2 Operating on Decentralized Data
We believe a more convenient and scalable solution will come from design and implementation of algorithms which learn from data distributed across research groups. These algorithms shall include feature learning as well as classification, prediction and inference. Dropping the requirement of moving the data, these algorithms will better match the current decentralized and efficient organization of research society and substantially lower barriers to entry for collaborative work. The resulting network effect will enable new innovative opportunities for research that we cannot envision today. The need for such approaches to general data computation is realized by some researchers (Bai et al., 2005) but not yet fully appreciated by the neuroimaging field. The field is currently in the state of establishing central repositories of anonymized raw data (Bockholt et al., 2009; Buccigrossi et al., 2007; Di Martino et al., 2014; Jack et al., 2008; Keator et al., 2008; Landis et al., 2015; Marcus et al., 2007; Poldrack et al., 2013; Scott et al., 2011; Turner, 2014; Van Essen et al., 2013). In the past 10 years, release of multi-site neuroimaging datasets such as: FBIRN, MCIC for schizophrenia (Ford et al., 2009; Gollub et al., 2013), ADNI for Alzheimer's disease (Jack et al., 2008), ABIDE for ASD (Di Martino et al., 2014), ADHD-200 for children with ADHD (Consortium and others, 2012) and Functional Connectomes project for healthy subjects (Biswal et al., 2010) have been started.
Certainly, access to raw data is the best way to drill down to the finest details and resolve any inconsistencies due to data handling. However, even in the centralized repositories, it is often more convenient to start analysis from a point in the processing pipeline where less detailed but possibly more informative features are generated. Furthermore, there are three categories of data that pose challenges for public availability for easy access: (1) data that are non-shareable due to obvious re-identification concerns, such as extreme age of the subject or a zip code/disease combination that makes re-identification simple; (2) data that are non-shareable due to more complicated or less obvious concerns, such as genetic data or other data which may be re-identifiable in conjunction with other data not under the investigator's control; and (3) data that are non-shareable due to the local institutional review boards (IRBs) rules or other administrative decisions (e.g., stakeholders in the data collection not allowing sharing). For example, even with broad consent to share the data acquired at the time of data collection, some of the eMERGE sites were required to re-contact the subjects and re-consent prior to sharing within the eMERGE consortium, which can be a permanent show-stopper for some datasets (Ludman et al., 2010). An extensive account of the problems that go along with these concerns is given by Sarwate et al. (Sarwate et al., 2014). An example of how a decentralized data feature learning algorithm could use decentralized data joint ICA in given by Baker et al. (Baker et al., 2015). In short, the algorithm performs a joint ICA on datasets distributed across research sites which enables one to perform temporal ICA on fMRI data as an increasingly large data sample becomes available when many research groups join the collaboration. Importantly, Baker et al. have demonstrated (on synthetic data) that with their approach the estimated components are virtually identical for the pooled data (i.e. a central repository), two sites with data split in half, multiple sites with data evenly split across, and even a very large number of sites with very few subjects at each of them. Once globally consistent features are available they may be used in building classification algorithms.
Nevertheless, decentralized data computation under serious privacy concerns will need additional protection besides simple protection from only sharing summaries and not the raw data samples. A solution for this setting has been offered in the ε-differential privacy model and explained extensively in the neuroimaging context with published examples (Dwork, 2006; Sarwate et al., 2014). This approach defines privacy by quantifying the change in the risk of re-identification as a result of publishing a function of the data. Notably, privacy is a property of an algorithm operating on the data, rather than a property of the sanitized data, which reflects the difference between semantic and syntactic privacy. Importantly for our applications, it can be applied to systems which do not share data itself but instead share data derivatives (functions of the data). Algorithms that guarantee differential privacy are randomized in how they manipulate the data values (e.g., by adding noise) to bound the risk. Enabling individual subject prediction in the classification framework is one of the applications where the above-described approaches can provide the most benefit—especially for rare conditions that are easy to identify by cross referencing when raw data is openly shared and hard to collect enough data at a single site to provide high generalization. The former is perfectly addressed by applying ε-differential privacy approach to classification (Chaudhuri et al., 2011), while the latter can be addressed by running decentralized algorithms over multiple sites. As mentioned already, differentially private algorithms provide guarantees by necessarily lowering the quality of the solution due to the required noise addition. The same happens to differentially private classifiers (Chaudhuri et al., 2011) and the effect is an undesirable increase in prediction error (Sarwate et al., 2014). Fortunately, combining the approaches (differential privacy and decentralized algorithms) can improve the situation considerably by dropping classification error from 25% to 5% while preserving all privacy guarantees (Sarwate et al., 2014).
In these “big data” times, the need for computation on large-scale datasets creates the best climate for software for distributed computation. Many useful and powerful projects came to the scene such as Apache Spark (Zaharia et al., 2010) and H2O (“H2O,” 2015). On closer inspection, these implementations are essentially striving for the efficiency of computation given a big data overload (typically easy to get data stored centrally). They suggest optimization toward an environment that is quite orthogonal to what we have to deal with—hard to get to and expensive to collect data spread across research labs around the nation and the world. The goal of decentralized approaches that we are describing here stands principally as preserving correctness of the computation while minimizing the data passed around and reducing the number of iterations. The tools and methods are not conflicting and decentralized data algorithms can and shall take advantage of what is being developed for large-scale computation in the distributed computing community.
4.3 Differential Diagnosis and Disease Subtype Classification
Using machine learning methods, promising results have been reported for automatic diagnosis of various cognitive and neurodegenerative disorders, usually from healthy controls based on neuroimaging features. However, one of the main challenges in psychiatric and neurology diagnoses is to differentially diagnose a disorder that shares symptoms with multiple other disorders. Examples of such overlapping disorders are schizophrenia, bipolar, schizoaffective, unipolar and mood disorders. Except for differentiating MCI for AD, only a few considered much needed automatic differential diagnosis in the studies we surveyed. Costafreda et al. used fMRI with a verbal fluency task to classify schizophrenia, bipolar and healthy controls (Costafreda et al., 2011b). Calhoun et al., and Arribas et al. both used fMRI with an auditory oddball task and an ICA approach to extract features from the default model network and the temporal lobe of the brain (Arribas et al., 2010; Calhoun et al., 2008). Both of these studies reported high differential accuracy between schizophrenia and bipolar disorder. Pardo et al. used a combination of volumes of 23 ROIs derived from structural MRI along with 22 neurophysiological test scores to automatically classify schizophrenia, bipolar and healthy controls (Pardo et al., 2006). Recently, Schnack et al. proposed using gray matter densities for classification schizophrenia, bipolar and healthy controls (Schnack et al., 2014). Koutsouleris et al., used gray matter maps from structural MRI to classify schizophrenia from mood disorder (Koutsouleris et al., 2015). Ota et al. combined volumetric measures derived from structural MRI with fractional anisotropy from dMRI in selected ROIs to classify schizophrenia from MDD (Ota et al., 2013). Sacchet et al. proposed using gray matter volumes of caudate and ventral diencephalon to differentiate MDD, bipolar and remitted MDD patients (Sacchet et al., 2015).
Pathologies like autism and schizophrenia are spectrum disorders with multiple etiologies under the umbrella of the same diagnostic category. While classification of these disorders using the generic category is commonly used to find diagnostic biomarkers, one of the key issues in mental healthcare is the differential diagnosis of patients across several disease subtypes. Common binary patient-control classification ignores the underlying heterogeneity of the disorder. Usually, the treatment path used for these subtypes differs from each other and therefore the correct subtype diagnosis is very important. For example, several cognitive deficits are observed in schizophrenia patients, but the magnitudes of such symptoms are highly variable among the patients. To reduce this phenotypic heterogeneity two major subtypes named “cognitive deficit” and “cognitively spared” have been defined (Green et al., 2013; Jablensky, 2006). These two subtypes exhibit different genetic and cognitive profiles (Green et al., 2013; Morar et al., 2011). An automatic classification of schizophrenia subtypes has been rarely studied. Ingalhalikar et al. proposed unsupervised spectral clustering of multi-edge graphs built from a structural connectivity network among 78 ROIs be usedto identify subtypes of autism and schizophrenia (Ingalhalikar et al., 2012). Gould et al. proposed using whole brain, voxel-based morphometry to classify schizophrenia patients with cognitive deficit from those that are cognitively spared (Gould et al., 2014).
There are several studies on automatic differentiation of stable MCI from progressive MCI (those that convert to AD within a certain amount of time). Most of these studies reported modest accuracies around 65-80% (Plant et al., 2010; Salvatore et al., 2015a; Tangaro et al., 2015; Tong et al., 2014; Wolz et al., 2011; Zu et al., 2015). ADHD subtype studies are scarce and limited to few studies such as the one by Sato et al. with the intent to automatically differentiate ADHD-IA, ADHD-HI and ADHD-C using resting-state fMRI (Sato et al., 2012).
Again, one major limitation in differential diagnosis and disease subtype classification is the limited sample size. In most of the current datasets, the number of subjects in each diseases subtype is small and therefore provides limited ability to develop robust single-subject predictor to accurately differentiate them.
4.4.1 Multimodal Neuroimaging Studies
Each imaging modality provides a different view of brain function or structure, and data fusion capitalizes on the strengths of each imaging modality/task and their inter-relationships in a joint analysis. This is an important tool to help unravel the pathophysiology of brain disease (Calhoun et al., 2006a; Sui et al., 2012). Recent advances in data fusion include integrating multiple (task) fMRI data sets (Kim et al., 2010; Sui et al., 2015, 2009) from the same participant to specify common versus specific sources of activity to a greater degree than traditional general linear model-based approaches. This can increase confidence in conclusions about the functional significance of brain regions and of activation changes in brain disease. In addition, the combination of function and structure may provide more informative insights into both altered brain patterns and connectivity in brain disorders (McCarley et al., 2008; Michael et al., 2009; Sui et al., 2011). These findings suggest that most studies favor only one data type or do not combine modalities in an integrated manner, and thus miss important changes which are only partially detected by each modality (Calhoun and Adali, 2009). On the other hand, multimodal fusion provides a more comprehensive description of altered brain patterns and connectivity than a single modality, which has shown increasing utility in answering both scientifically interesting and clinically relevant questions.
4.4.2 Single-Subject Prediction using Multimodal Neuroimaging Data
There is increasing evidence from multimodal studies that patients with brain disorders exhibit unique morphological characteristics, connectivity patterns, and functional alterations, which could not have been revealed through separate unimodal analyses as typically performed in the majority of neuroimaging experiments. Hence, applying classification techniques to these characteristics could identify biomarkers for psychiatric diseases. This could expedite differential diagnosis, thus leading to more appropriate treatment and improved outcomes for patients with brain disorders. There has been number of studies showing the benefits of combining both rest and task fMRI data for group differences in functional connectivity between schizophrenia patients and controls (Arbabshirani and Calhoun, 2011; Cetin et al., 2014). The change of functional connectivity from rest to task contains novel information present in neither of the states, which could be beneficial for single subject prediction (Mohammad R Arbabshirani and Calhoun, 2011). Based on these evidences, future studies might benefit from combining resting-state and task-based data for classification of brain disorders.
As another example, MCI is difficult to diagnose due to its rather mild and nearly insignificant symptoms of cognitive impairment. Wee et al. integrated information from DTI and resting fMRI by employing multiple-kemel SVM, yielding statistically significant improvement (>7.4%) in classification accuracy of predicting MCI from HC by using multimodal data (96.3%) compared to using each modality independently (Wee et al., 2012). There are additional studies that demonstrate the potential of the fusion of structural and functional data combined with multi-modal classification techniques to provide more accurate and early detection of brain abnormalities (Fan et al., 2008b). By taking advantage of these two complementary approaches, Sui et al. proposed a framework based on mCCA+jICA, that allows both high and weak connections to be detected and shows excellent source separation performance (Sui et al., 2011). It enables robust identification of correspondence among N diverse data types and enables one to investigate the important question of whether certain disease risk factors are shared or are distinct across multiple modalities, which can also serve as multimodal feature selection method for schizophrenia (Sui et al., 2013a, 2013b). Similarly, Jie et al. adopted SVM-FoBa to classify between bipolar versus unipolar disorders by combining GM and ALFF features, achieving an accuracy of 92% This suggests that using complimentary multimodal biomarkers may be more informative and effective to discriminate brain disorders (Jie et al., 2015).
There are number of recent studies looking at combined biomarkers of sMRI, FDG-PET, and CSF (mostly for ADNI dataset) to discriminate between AD, MCI and HC (Gray et al., 2013; Xu et al., 2015; Yu et al., 2015; Zhang et al., 2011, 2014). Similarly, a few studies combined functional and structural data to build such predictive models (Z. Dai et al., 2012; Dyrba et al., 2015). Most of those studies reported superior performance of models built based on multimodal features compared to those based on a single modality (Calhoun and Sui, 2016).
4.5 Deep Learning in Neuroimaging
In recent years, deep learning methodology has made significant improvement in representation learning and classification in various areas such as speech recognition, natural image classification and text mining. Two main features have made deep learning very attractive to machine learning researchers. First deep learning in contrast with traditional machine learning methods is capable of data-driven automatic feature learning. This important capability removes the subjectivity in selecting the relevant features especially in cases where too many features exist or prior knowledge in selecting features is not conclusive. The second important feature of deep learning is the depth of models. By applying a hierarchy of non-linear layers, deep learning is capable of modeling very complicated data patterns in contrast with traditional shallow models.
Typical approaches in single subject prediction in neuroimaging consist of selecting features sometimes from thousands of voxels. As reviewed in this report, the basis for such a feature selection is usually inefficient univariate statistical tests. Recently, deep belief networks, a class of deep learning, has been applied to both structural and functional MRI data (Plis et al., 2014). Plis et al. showed that deep learning methods could produce physiologically meaningful features and reveal relations from high dimensional neuroimaging data (Plis et al., 2014). Hjelm et al., applied restricted Boltzmann machines (RBM) to identify intrinsic networks in fMRI data (Hjelm et al., 2014). They showed that RBMs could extract spatial networks and their activation with the accuracy of traditional matrix factorization methods such as ICA. Provably, deep models need exponentially smaller number of parameters in order to model the same thing shallow models can model (Bengio, 2013, 2012). Moreover, deep learning structures such as RBM are generative models and therefore it can be sampled from. This way it is easy to access uncertainty in the estimates compared to the point estimates of matrix factorization models. Furthermore, for deep learning RBM could be stacked to obtain deeper models as needed. This cannot be readily done with ICA, NMF, or sparse PCA.
Recently, deep learning is employed in classification of patients using neuroimaging data. Suk et al. used stacked autoencoder (another class of deep learning) to discriminate patients with AD from those with MCI (Suk et al., 2013). Kim et al. used deep learning for classification of schizophrenia patients from healthy controls based on functional connectivity patterns. They showed that their approach outperforms SVM by a significant margin (Kim et al., 2015).
Deep learning is a very promising tool for understanding the neural basis of brain disorders by extracting hidden patterns from high-dimensional neuroimaging data (Kriegeskorte, 2015). In our opinion, this method has the potential to improve brain disorder diagnosis—especially if larger neuroimaging datasets become available and/or improved methods of training based on existing data are developed (Castro et al., 2015).
4.6 Standard Machine Learning Competitions in Neuroimaging
The machine learning field has benefited hugely from standard competitions in many applications. In such competitions, usually the participants are provided with a labeled training dataset and an unlabeled testing dataset. The participants try to develop the best predictive model based on the training dataset, predict the labels of the provided testing dataset and submit the results. Such a setting ensures that the results are not biased. These competitions usually attract many groups, even those with less domain knowledge and expertise. By providing a standard dataset and some initial preprocessing, the participants can primarily concentrate on the machine learning aspect of the analysis.
Due to all of the data sharing problems previously discussed, such competitions are rare in neuroimaging. The ADHD-200 competition was held in 2011 with the goal of predicting ADHD from healthy controls in children and adults, using resting-state fMRI along with anatomical and phonotypical data of 776 subjects (491 TDC and 285 ADHD) for training along with additional 197 subjects for testing (Consortium and others, 2012). The competition was a successful example of large-scale ADHD data sharing among several sites. However, the ‘winning’ team of ADHD-200 competition didn't use the imaging data in their predictive model (just the phenotypical data), which caused discussion in the community about usefulness of brain data in diagnosing a brain disorder (M. R. G. Brown et al., 2012; Consortium and others, 2012).
More recently, The IEEE MLSP workshop held a schizophrenia classification challenge with the goal of automatic classification of schizophrenia patients from healthy controls using just brain imaging features (Silva et al., 2014). Functional network connectivity values of resting-state fMRI along with ICA loadings of source-based morphometry of sMRI were calculated from 144 subjects (75 healthy controls, 69 schizophrenia patients) and shared with participants. Interestingly, 245 teams participated in the competition and the winning team achieved an AUC of around 0.90. Moreover, by combining the top three models, an AUC of around 0.94 was achieved (Silva et al., 2014). In our opinion, sharing ready to use, well-defined features as opposed to imaging data itself, was one of the success factors of the MLSP competition in both attracting numerous groups and also achieving high accuracy results. That experience shows that imaging data has a lot of predictive potential at least in the case of separating schizophrenia patients from healthy controls.
We believe that the field of neuroimaging can benefit a lot from standard machine learning competitions such as the ones discussed above. Such competitions can assess the realistic, unbiased, discriminative power of brain data for detecting brain disorders. Also, by attracting a large number of participants, a variety of machine learning methods will be examined for the specific problem. By providing brain features, machine learning experts with less neuroimaging domain knowledge can participate and develop predictive models.
5. Summary and Conclusions
5.1 Previous Single-subject Prediction Surveys
In this study, we comprehensively reviewed past efforts in neuroimaging-based single subject prediction in several brain disorders such as MCI, AD, ASD, ADHD, schizophrenia and depressive disorders. Previous reviews include disease-specific surveys such as schizophrenia (Calhoun and Arbabshirani, 2012; Dazzan, 2014; Demirci et al., 2008b; Kambeitz et al., 2015; Veronese et al., 2013; Zarogianni et al., 2013), autism spectrum disorder (Retico et al., 2014, 2013), Alzheimer's disease (Falahati et al., 2014; Klöppel et al., 2008) and in general (Klöppel et al., 2012; Orrù et al., 2012) as well as modality-specific reviews such as machine learning based on fMRI (Sundermann et al., 2014). Also, there are few children specific reviews such as a recent one by Levman et al. on multivariate analyses studies in neonatal and pediatric patients (Levman and Takahashi, 2015). Probably the most comprehensive review so far is the recent one by Wolfers et al., where they reviewed about 120 single subject prediction studies in schizophrenia, mood disorders, anxiety disorders, ADHD and ASD (Wolfers et al., 2015). While there is some overlap among the mentioned studies and this survey, to our knowledge, this is by far the largest survey in the field based on the number of papers reviewed (about 240 papers). Moreover, as discussed previously, the majority of single subject prediction studies have been published in recent years; consequently, an updated survey is much needed. In this work, several pitfalls such as feature selection bias, incomplete reporting of results, unfair comparison across studies and improper hyperparameter selection were discussed and suggestions to address those issues were provided. Moreover, emerging trends in this exciting field such as decentralized data sharing, differential diagnosis and disease subtype classification, multimodal neuroimaging, applications of deep learning in neuroimaging and merits of standard machine learning competitions were discussed in detail.
5.2 Limitations
There are several limitations in this work. We limited our search to MRI-based English journal papers in specific disorders. There are other single subject prediction studies that are based on other modalities such as EEG and MEG. Also, other brain disorders such Parkinson disease and anxiety disorders were not reviewed in this work. From the studies we reviewed, we tried to extract the key features as it relates to the machine learning. Many of those studies contained multiple experiments under different scenarios but we just reported one of them (usually the most successful one) here. Also, there are many important details in each study and for that reason interested readers should always refer to each reference for full information on experiment setup and other details.
In terms of common pitfalls, we mostly focused on the potential problems from the machine learning point of view. There are many other important potential issues in topics such as experimental design, effect of head motion and other factors such as the impact of draining veins on fMRI studies (Boubela et al., 2015; Power et al., 2015, 2014, 2012), wakefulness of subjects during rsfMRI (Tagliazucchi and Laufs, 2014) and the selecting of preprocessing steps (Vergara et al., 2015). Effect of those potential issues on single subject prediction deserves a full paper by itself.
In conclusion, we are optimistic about the use of brain imaging for single subject prediction, and many of the issues we recommend are within reach. Larger studies are available and repositories with pooled data across studies are growing rapidly (Eickhoff et al., 2016).
Highlights.
Past efforts on classification of brain disorders are comprehensively reviewed.
The common pitfalls from machine learning point of view are discussed.
Emerging trends related to single-subject prediction are reviewed and discussed.
Acknowledgement
This work was supported by National Institutes of Health grants P20GM103472, R01EB005846 and 1 ROl DA040487 (to V .D. Calhoun); “100 Talents Plan” of Chinese Academy of Sciences (to J. Sui); Chinese National Science Foundation No. 81471367 and the State High-Tech Development Plan (863) No. 2015AA020513; Also, we would like to thank Monica Jaramillo for the initial survey of neuroimaging studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Search Term: (“Machine Learning” OR SVM OR “automatic Classification” OR “discriminant analysis” OR “neural Network” OR “Logistic Regressions” OR “decision tree”) AND (MRI OR “Magnetic Resonance” OR fMRI OR “functional MRI” OR “structural MRI” OR “Diffusion MRI” OR DTI OR DSI) AND (schizophrenia OR bipolar OR Alzheimer's OR “Mild Cognitive Impairment” OR MCI OR autism OR “autism spectrum disorder” OR ASD OR depression OR “depressive disorder” OR ADHD OR “Attention Deficit Hyperactivity Disorder”) concluded on 12/08/2015.
The authors report no financial relationships with commercial interests.
References
- Abdulkadir A, Mortamet B, Vemuri P, Jack CR, Krueger G, Klöppel S. Effects of hardware heterogeneity on the performance of SVM Alzheimer's disease classifier. Neuroimage. 2011;58:785–792. doi: 10.1016/j.neuroimage.2011.06.029. doi:10.1016/j.neuroimage.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adaszewski S, Dukart J, Kherif F, Frackowiak R, Draganski B. How early can we predict Alzheimer's disease using computational anatomy? Neurobiol. Aging. 2013;34:2815–2826. doi: 10.1016/j.neurobiolaging.2013.06.015. doi: 10.1016/j.neurobiolaging.2013.06.015. [DOI] [PubMed] [Google Scholar]
- Aguilar C, Westman E, Muehlboeck J-S, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen FL, Lovestone S, Spenger C, Simmons A, Wahlund L-O. Different multivariate techniques for automated classification of MRI data in Alzheimer's disease and mild cognitive impairment. Psychiatry Res. 2013;212:89–98. doi: 10.1016/j.pscychresns.2012.11.005. doi: 10.1016/j.pscychresns.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Akshoomoff N, Lord C, Lincoln AJ, Courchesne RY, Carper RA, Townsend J, Courchesne E. Outcome classification of preschool children with autism spectrum disorders using MRI brain measures. J Am Acad Child Adolesc Psychiatry. 2004;43:349–357. doi: 10.1097/00004583-200403000-00018. doi:10.1097/00004583-200403000-00018. [DOI] [PubMed] [Google Scholar]
- Alberg AJ, Park JW, Hager BW, Brock MV, Diener-West M. The use of “overall accuracy” to evaluate the validity of screening or diagnostic tests. J. Gen. Intern. Med. 2004;19:460–465. doi: 10.1111/j.1525-1497.2004.30091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, Gamst A, Holtzman DM, Jagust WJ, Petersen RC. The diagnosis of mild cognitive impairment due to Alzheimer's disease: Recommendations from the National Institute on Aging-Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen AH, Rayens WS, Liu Y, Smith CD. Partial least squares for discrimination in fMRI data. Magn Reson Imaging. 2012;30:446–452. doi: 10.1016/j.mri.2011.11.001. doi:10.1016/j.mri.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A, Cohen MS. Decreased small-world functional network connectivity and clustering across resting state networks in schizophrenia: an fMRI classification tutorial. Front. Hum. Neurosci. 2013;7:520. doi: 10.3389/fnhum.2013.00520. doi:10.3389/fnhum.2013.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A, Douglas PK, Kerr WT, Haynes VS, Yuille AL, Xie J, Wu YN, Brown JA, Cohen MS. Non-negative matrix factorization of multimodal MRI, fMRI and phenotypic data reveals differential changes in default mode subnetworks in ADHD. Neuroimage. 2014;102(Pt 1):207–219. doi: 10.1016/j.neuroimage.2013.12.015. doi: 10.1016/j.neuroimage.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Nielsen JA, Froehlich AL, DuBray MB, Druzgal TJ, Cariello AN, Cooperrider JR, Zielinski BA, Ravichandran C, Fletcher PT. Functional connectivity magnetic resonance imaging classification of autism. Brain. 2011;134:3742–3754. doi: 10.1093/brain/awr263. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anticevic A, Cole MW, Repovs G, Murray JD, Brumbaugh MS, Winkler AM, Savic A, Krystal JH, Pearlson GD, Glahn DC. Characterizing thalamo-cortical disturbances in schizophrenia and bipolar illness. Cereb. Cortex. 2014;24:3116–3130. doi: 10.1093/cercor/bht165. doi:10.1093/cercor/bhtl65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolova LG, Hwang KS, Kohannim O, Avila D, Elashoff D, Jack CR, Shaw L, Trojanowski JQ, Weiner MW, Thompson PM. ApoE4 effects on automated diagnostic classifiers for mild cognitive impairment and Alzheimer's disease. Neuroimage Clin. 2014;4:461–472. doi: 10.1016/j.nicl.2013.12.012. doi:10.1016/j.nicl.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbabshirani MR, Calhoun VD. 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE; Boston, MA: 2011. Functional network connectivity during rest and task: Comparison of healthy controls and schizophrenic patients; pp. 4418–4421. [DOI] [PubMed] [Google Scholar]
- Arbabshirani MR, Calhoun VD. Functional network connectivity during rest and task: Comparison of healthy controls and schizophrenic patients. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:4418–4421. doi: 10.1109/IEMBS.2011.6091096. doi:10.1109/IEMBS.2011.6091096. [DOI] [PubMed] [Google Scholar]
- Arbabshirani MR, Kiehl KA, Pearlson GD, Calhoun VD. Classification of schizophrenia patients based on resting-state functional network connectivity. Front. Neurosci. 2013 doi: 10.3389/fnins.2013.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardekani B. a., Tabesh A, Sevy S, Robinson DG, Bilder RM, Szeszko PR. Diffusion tensor imaging reliably differentiates patients with schizophrenia from healthy volunteers. Hum. Brain Mapp. 2011;32:1–9. doi: 10.1002/hbm.20995. doi:10.1002/hbm.20995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura H, Yoshiura T, Kumazawa S, Tanaka K, Koga H, Mihara F, Honda H, Sakai S, Toyofuku F, Higashida Y. Automated method for identification of patients with Alzheimer's disease based on three-dimensional MR images. Acad Radiol. 2008;15:274–284. doi: 10.1016/j.acra.2007.10.020. doi:10.1016/j.acra.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Arribas JI, Calhoun VD, Adali T. Automatic bayesian classification of healthy controls, bipolar disorder, and schizophrenia using intrinsic connectivity maps from fMRI data. IEEE Trans. Biomed. Eng. 2010;57:2850–2860. doi: 10.1109/TBME.2010.2080679. doi:10.1109/TBME.2010.2080679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Association AP. Diagnostic and Statistical Manual of Mental Disorders:: DSM-5. ManMag. 2003 others. [Google Scholar]
- Bai Z-J, Chan RH, Luk FT. Proceedings of the 6th International Conference on Advanced Parallel Processing Technologies, APPT'05. Springer-Verlag; Berlin, Heidelberg: 2005. Principal Component Analysis for Distributed Data Sets with Updating; pp. 471–483. [Google Scholar]
- Baio J. Surveillance Summaries. 3. Vol. 61. Centers Dis. Control Prev; 2012. Prevalence of Autism Spectrum Disorders: Autism and Developmental Disabilities Monitoring Network, 14 Sites, United States, 2008. Morbidity and Mortality Weekly Report. [PubMed] [Google Scholar]
- Baker B, Silva R, Calhoun VD, Sarwate AD, Plis S. IEEE Machine Learning for Signal Processing Workshop. Boston, MA.: 2015. Large scale collabortion with automony: decentralized data ICA. [Google Scholar]
- Bansal R, Staib LH, Laine AF, Hao X, Xu D, Liu J, Weissman M, Peterson BS. Anatomical brain images alone can accurately diagnose chronic neuropsychiatric illnesses. 2012 doi: 10.1371/journal.pone.0050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Nelson BG, Mueller B. a., Camchong J, Lim KO. Altered resting state complexity in schizophrenia. Neuroimage. 2012;59:2196–2207. doi: 10.1016/j.neuroimage.2011.10.002. doi:10.1016/j.neuroimage.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batmanghelich NK, Taskar B, Davatzikos C. Generative-discriminative basis learning for medical imaging. IEEE Trans Med Imaging. 2012;31:51–69. doi: 10.1109/TMI.2011.2162961. doi:10.1109/TMI.2011.2162961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beheshti I, Demirel H. Probability distribution function-based classification of structural MRI for the detection of Alzheimer's disease. Comput. Biol. Med. 2015;64:208–216. doi: 10.1016/j.compbiomed.2015.07.006. doi:10.1016/j.compbiomed.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Beltrachini L, De Marco M, Taylor ZA, Lotjonen J, Frangi AF, Venneri A. Integration of Cognitive Tests and Resting State fMRI for the Individual Identification of Mild Cognitive Impairment. Curr Alzheimer Res. 2015;12:592–603. doi: 10.2174/156720501206150716120332. [DOI] [PubMed] [Google Scholar]
- Bengio Y. Statistical Language and Speech Processing. Springer; 2013. Deep learning of representations: Looking forward; pp. 1–37. [Google Scholar]
- Bengio Y. Deep learning of representations for unsupervised and transfer learning. Unsupervised Transf. Learn. Challenges Mach. Learn. 2012;7:19. [Google Scholar]
- Bergouignan L, Lefranc JP, Chupin M, Morel N, Spano JP, Fossati P. Breast cancer affects both the hippocampus volume and the episodic autobiographical memory retrieval. PLoS One. 2011;6:e25349. doi: 10.1371/journal.pone.0025349. doi:10.1371/journal.pone.0025349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhugra D. The global prevalence of schizophrenia. PLoS Med. 2005;2:372. doi: 10.1371/journal.pmed.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederman J. Attention-deficit/hyperactivity disorder: a selective overview. Biol. Psychiatry. 2005;57:1215–1220. doi: 10.1016/j.biopsych.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Bishop C~M. Pattern Recognition and Machine Learning (Information Science and Statistics) Springer; 2006. [Google Scholar]
- Biswal BB, Mennes M, Zuo X-N, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S. Toward discovery science of human brain function. Proc. Natl. Acad. Sci. 2010;107:4734–4739. doi: 10.1073/pnas.0911855107. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleich-Cohen M, Jamshy S, Sharon H, Weizman R, Intrator N, Poyurovsky M, Hendler T. Machine learning fMRI classifier delineates subgroups of schizophrenia patients. Schizophr. Res. 2014;160:196–200. doi: 10.1016/j.schres.2014.10.033. doi:10.1016/j.schres.2014.10.033. [DOI] [PubMed] [Google Scholar]
- Blum AL, Langley P. Selection of relevant features and examples in machine learning. Artif. Intell. 1997;97:245–271. [Google Scholar]
- Bockholt HJ, Scully M, Courtney W, Rachakonda S, Scott A, Caprihan A, Fries J, Kalyanam R, Segall JM, de la Garza R. Mining the Mind Research Network: {A} Novel framework for exploring large scale, heterogeneous translational neuroscience research data sources. Front. Neuroinform. 2009;3 doi: 10.3389/neuro.11.036.2009. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohland JW, Saperstein S, Pereira F, Rapin J, Grady L. Network, anatomical, and non-imaging measures for the prediction of ADHD diagnosis in individual subjects. Front Syst Neurosci. 2012;6:78. doi: 10.3389/fnsys.2012.00078. doi:10.3389/fnsys.2012.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottino CMC, Castro CC, Gomes RLE, Buchpiguel CA, Marchetti RL, Neto MRL. Volumetric MRI measurements can differentiate Alzheimer's disease, mild cognitive impairment, and normal aging. Int Psychogeriatr. 2002;14:59–72. doi: 10.1017/s1041610202008281. [DOI] [PubMed] [Google Scholar]
- Boubela RN, Kalcher K, Huf W, Seidel E-M, Derntl B, Pezawas L, Nasel C, Moser E. fMRI measurements of amygdala activation are confounded by stimulus correlated signal fluctuation in nearby veins draining distant brain regions. Sci. Rep. 2015;5 doi: 10.1038/srep10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimer's Dement. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- Brown G, Pocock A, Zhao M-J, Lujan M. Conditional Likelihood Maximisation: A Unifying Framework for Mutual Information Feature Selection. J. Mach. Learn. Res. 2012;13:27–66. [Google Scholar]
- Brown MRG, Sidhu GS, Greiner R, Asgarian N, Bastani M, Silverstone PH, Greenshaw AJ, Dursun SM. ADHD-200 Global Competition: diagnosing ADHD using personal characteristic data can outperform resting state fMRI measurements. Front. Syst. Neurosci. 2012;6 doi: 10.3389/fnsys.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccigrossi R, Ellisman M, Grethe J, Haselgrove C, Kennedy DN, Martone M, Preuss N, Reynolds K, Sullivan M, Turner J. The Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC). AMIA... Annual Symposium Proceedings/AMIA Symposium. AMIA Symposium. 2007:1000. others. [PubMed] [Google Scholar]
- Caan M.W. a, Vermeer K. a., van Vliet LJ, Majoie CBLM, Peters BD, den Heeten GJ, Vos FM. Shaving diffusion tensor images in discriminant analysis: A study into schizophrenia. Med. Image Anal. 2006;10:841–849. doi: 10.1016/j.media.2006.07.006. doi: 10.1016/j.media.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Calderoni S, Retico A, Biagi L, Tancredi R, Muratori F, Tosetti M. Female children with autism spectrum disorder: an insight from mass-univariate and pattern classification analyses. Neuroimage. 2012;59:1013–1022. doi: 10.1016/j.neuroimage.2011.08.070. doi: 10.1016/j.neuroimage.2011.08.070. [DOI] [PubMed] [Google Scholar]
- Calhoun V, Arbabshirani MR. Neuroimaging-Based Automatic Classification of Schizophrenia. In: Singh I, Sinnott-Armstrong WP, Savulescu J, editors. Bioprediction, Biomarkers and Bad Behavior. Oxford Univ Press; 2012. pp. 206–230. [Google Scholar]
- Calhoun VD, Adali T, Kiehl KA, Astur R, Pekar JJ, Pearlson GD. A method for multitask f{MRI} data fusion applied to schizophrenia. Hum. Brain Mapp. 2006a;27:598–610. doi: 10.1002/hbm.20204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T. Feature-Based Fusion of Medical Imaging Data. Inf. Technol. Biomed. IEEE Trans. 2009;13:711–720. doi: 10.1109/TITB.2008.923773. doi:10.1109/TITB.2008.923773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Giuliani NR, Pekar JJ, Kiehl KA, Pearlson GD. Method for multimodal analysis of independent source differences in schizophrenia: Combining gray matter structural and auditory oddball functional data. Hum. Brain Mapp. 2006b;27:47–62. doi: 10.1002/hbm.20166. doi:10.1002/hbm.20166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Maciejewski PK, Pearlson GD, Kiehl K. a. Temporal lobe and “default” hemodynamic brain modes discriminate between schizophrenia and bipolar disorder. Hum. Brain Mapp. 2008;29:1265–1275. doi: 10.1002/hbm.20463. doi:10.1002/hbm.20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Sui J. Multimodal fusion of brain imaging data: A key to finding the missing link (s) in complex mental illness. Biol. Psychiatry Cogn. Neurosci. Neuroimaging. 2016 doi: 10.1016/j.bpsc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Duan J, Lin D, Calhoun V, Wang Y-P. Integrating fMRI and SNP data for biomarker identification for schizophrenia with a sparse representation based variable selection method. BMC Med. Genomics 6 Suppl. 2013;3:S2. doi: 10.1186/1755-8794-6-S3-S2. doi:10.1186/1755-8794-6-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Guo S, Xue Z, Hu Y, Liu H, Mwansisya TE, Pu W, Yang B, Liu C, Feng J, Chen EYH, Liu Z. Aberrant functional connectivity for diagnosis of major depressive disorder: a discriminant analysis. Psychiatry Clin. Neurosci. 2014;68:110–119. doi: 10.1111/pcn.12106. doi:10.1111/pcn.l2106. [DOI] [PubMed] [Google Scholar]
- Caprihan a., Pearlson GD, Calhoun VD. Application of principal component analysis to distinguish patients with schizophrenia from healthy controls based on fractional anisotropy measurements. Neuroimage. 2008;42:675–682. doi: 10.1016/j.neuroimage.2008.04.255. doi:10.1016/j.neuroimage.2008.04.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova R, Hsu F-C, Espeland MA. Classification of structural MRI images in Alzheimer's disease from the perspective of ill-posed problems. PLoS One. 2012;7:e44877. doi: 10.1371/journal.pone.0044877. doi:10.1371/journal.pone.0044877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellani U, Rossato E, Murino V, Bellani M, Rambaldelli G, Perlini C, Tomelleri L, Tansella M, Brambilla P. Classification of schizophrenia using feature-based morphometry. J. Neural Transm. 2012;119:395–404. doi: 10.1007/s00702-011-0693-7. doi: 10.1007/s00702-011-0693-7. [DOI] [PubMed] [Google Scholar]
- Castro E, Gómez-Verdejo V, Martínez-Ramon M, Kiehl K. a., Calhoun VD. A multiple kernel learning approach to perform classification of groups from complex-valued fMRI data analysis: Application to schizophrenia. Neuroimage. 2014;87:1–17. doi: 10.1016/j.neuroimage.2013.10.065. doi:10.1016/j.neuroimage.2013.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro E, Martinez-Ramón M, Pearlson G, Sui J, Calhoun VD. Characterization of groups using composite kernels and multi-source fMRI analysis data: Application to schizophrenia. Neuroimage. 2011;58:526–536. doi: 10.1016/j.neuroimage.2011.06.044. doi:10.1016/j.neuroimage.2011.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro E, Ulloa A, Plis SM, Turner JA, Calhoun VD. IEEE International Symposium on Biomedical Imaging. New York, NY, USA.: 2015. Simulation of Structural Magnetic Resonance Images for Deep Learning Pre-Training. [Google Scholar]
- Cetin MS, Christensen F, Abbott CC, Stephen JM, Mayer AR, Cafiive JM, Bustillo JR, Pearlson GD, Calhoun VD. Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia. Neuroimage. 2014;97:117–126. doi: 10.1016/j.neuroimage.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çetin MS, Khullar S, Damaraju E, Michael AM, Baum S. a., Calhoun VD. Enhanced disease characterization through multi network functional normalization in fMRI. Front. Neurosci. 2015;9:1–15. doi: 10.3389/fnins.2015.00095. doi:10.3389/fnins.2015.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis E, Hurley P, Serra L, Bozzali M, Oliver S, Cercignani M. Gaussian process classification of Alzheimer's disease and mild cognitive impairment from resting-state fMRI. Neuroimage. 2015;112:232–243. doi: 10.1016/j.neuroimage.2015.02.037. doi:10.1016/j.neuroimage.2015.02.037. [DOI] [PubMed] [Google Scholar]
- Chang C-W, Ho C-C, Chen J-H. ADHD classification by a texture analysis of anatomical brain MRI data. Front Syst Neurosci. 2012;6:66. doi: 10.3389/fnsys.2012.00066. doi:10.3389/fnsys.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri K, Monteleoni C, Sarwate AD. Differentially private empirical risk minimization. J. Mach. Learn. Res. 2011;12:1069–1109. [PMC free article] [PubMed] [Google Scholar]
- Chen CP, Keown CL, Jahedi A, Nair A, Pflieger ME, Bailey BA, Miiller R-A. Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. Neuroimage Clin. 2015;8:238–245. doi: 10.1016/j.nicl.2015.04.002. doi:10.1016/j.nicl.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Storrs J, Tan L, Mazlack LJ, Lee J-H, Lu LJ. Detecting brain structural changes as biomarker from magnetic resonance images using a local feature based SVM approach. J. Neurosci. Methods. 2014;221:22–31. doi: 10.1016/j.jneumeth.2013.09.001. doi:10.1016/j.jneumeth.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Cheng B, Liu M, Zhang D, Munsell BC, Shen D. Domain Transfer Learning for MCI Conversion Prediction. IEEE Trans Biomed Eng. 2015;62:1805–1817. doi: 10.1109/TBME.2015.2404809. doi:10.1109/TBME.2015.2404809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Newman S, Goni J, Kent JS, Howell J, Bolbecker A, Puce A, O'Donnell BF, Hetrick WP. Nodal centrality of functional network in the differentiation of schizophrenia. Schizophr. Res. 2015;168:345–352. doi: 10.1016/j.schres.2015.08.011. doi:10.1016/j.schres.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chincarini A, Bosco P, Calvini P, Gemme G, Esposito M, Olivieri C, Rei L, Squarcia S, Rodriguez G, Bellotti R, Cerello P, De Mitri I, Retico A, Nobili F. Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer's disease. Neuroimage. 2011;58:469–80. doi: 10.1016/j.neuroimage.2011.05.083. doi:10.1016/j.neuroimage.2011.05.083. [DOI] [PubMed] [Google Scholar]
- Chu C, Hsu A-L, Chou K-H, Bandettini P, Lin C. Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. Neuroimage. 2012;60:59–70. doi: 10.1016/j.neuroimage.2011.11.066. doi: 10.1016/j.neuroimage.2011.11.066. [DOI] [PubMed] [Google Scholar]
- Chyzhyk D, Savio A, Grana M. Computer aided diagnosis of schizophrenia on resting state fMRI data by ensembles of ELM. Neural Networks. 2015;68:23–33. doi: 10.1016/j.neunet.2015.04.002. doi:10.1016/j.neunet.2015.04.002. [DOI] [PubMed] [Google Scholar]
- Colby JB, Rudie JD, Brown JA, Douglas PK, Cohen MS, Shehzad Z. Insights into multimodal imaging classification of ADHD. Front Syst Neurosci. 2012;6:59. doi: 10.3389/fnsys.2012.00059. doi: 10.3389/fnsys.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge DS. A primer on quantitized data analysis and permutation testing. J. Mix. Methods Res. 2013;7:81–97. [Google Scholar]
- Consortium, A.-200 The ADHD-200 consortium: a model to advance the translational potential of neuroimaging in clinical neuroscience. Front. Syst. Neurosci. 2012;6 doi: 10.3389/fnsys.2012.00062. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costafreda SG, Dinov ID, Tu Z, Shi Y, Liu C-Y, Kloszewska I, Mecocci P, Soininen H, Tsolaki M, Vellas B, Wahlund L-O, Spenger C, Toga AW, Lovestone S, Simmons A. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. Neuroimage. 2011a;56:212–219. doi: 10.1016/j.neuroimage.2011.01.050. doi: 10.1016/j.neuroimage.2011.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costafreda SG, Fu CHY, Picchioni M, Toulopoulou T, McDonald C, Kravariti E, Walshe M, Prata D, Murray RM, McGuire PK. Pattern of neural responses to verbal fluency shows diagnostic specificity for schizophrenia and bipolar disorder. BMC Psychiatry. 2011b;11:18. doi: 10.1186/1471-244X-11-18. doi:10.1186/1471-244X-ll-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupé P, Eskildsen SF, Manjon JV, Fonov VS, Collins DL. Simultaneous segmentation and grading of anatomical structures for patient's classification: application to Alzheimer's disease. Neuroimage. 2012;59:3736–3747. doi: 10.1016/j.neuroimage.2011.10.080. doi: 10.1016/j .neuroimage.2011.10.080. [DOI] [PubMed] [Google Scholar]
- Csernansky JG, Schindler MK, Splinter NR, Wang L, Gado M, Selemon LD, Rastogi-Cruz D, Posener J. a., Thompson P. a., Miller MI. Abnormalities of Thalamic Volume and Shape in Schizophrenia. Am. J. Psychiatry. 2004;161:896–902. doi: 10.1176/appi.ajp.161.5.896. doi: 10.1176/appi.ajp.161.5.896. [DOI] [PubMed] [Google Scholar]
- Cui Y, Liu B, Luo S, Zhen X, Fan M, Liu T, Zhu W, Park M, Jiang T, Jin JS. Identification of conversion from mild cognitive impairment to Alzheimer's disease using multivariate predictors. PLoS One. 2011;6:e21896. doi: 10.1371/journal.pone.0021896. doi:10.1371/journal.pone.0021896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Wen W, Lipnicki DM, Beg MF, Jin JS, Luo S, Zhu W, Kochan NA, Reppermund S, Zhuang L, Raamana PR, Liu T, Trollor JN, Wang L, Brodaty H, Sachdev PS. Automated detection of amnestic mild cognitive impairment in community-dwelling elderly adults: a combined spatial atrophy and white matter alteration approach. Neuroimage. 2012;59:1209–1217. doi: 10.1016/j.neuroimage.2011.08.013. doi:10.1016/j.neuroimage.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert M-O, Chupin M, Benali H, Colliot O. Automatic classification of patients with Alzheimer's disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage. 2011;56:766–781. doi: 10.1016/j.neuroimage.2010.06.013. doi:10.1016/j.neuroimage.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Cuingnet R, Glaunés JA, Chupin M, Benali H, Colliot O. Spatial and Anatomical Regularization of SVM: A General Framework for Neuroimaging Data. IEEE Trans Pattern Anal Mach Intell. 2013;35:682–696. doi: 10.1109/TPAMI.2012.142. doi: 10.1109/TPAMI.2012.142. [DOI] [PubMed] [Google Scholar]
- Dai D, Wang J, Hua J, He H. Classification of ADHD children through multimodal magnetic resonance imaging. Front Syst Neurosci. 2012;6:63. doi: 10.3389/fnsys.2012.00063. doi:10.3389/fnsys.2012.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Yan C, Wang Z, Wang J, Xia M, Li K, He Y. Discriminative analysis of early Alzheimer's disease using multi-modal imaging and multi-level characterization with multi-classifier (M3). Neuroimage. 2012;59:2187–2195. doi: 10.1016/j.neuroimage.2011.10.003. doi:10.1016/j.neuroimage.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Davatzikos C, Shen D, Gur RC, Wu X, Liu D, Fan Y, Hughett P, Turetsky BI, Gur RE. Whole-brain morphometric study of schizophrenia revealing a spatially complex set of focal abnormalities. Arch. Gen. Psychiatry. 2005;62:1218–1227. doi: 10.1001/archpsyc.62.11.1218. doi:10.1001/archpsyc.62.11.1218. [DOI] [PubMed] [Google Scholar]
- Dazzan P. Neuroimaging biomarkers to predict treatment response in schizophrenia: the end of 30 years of solitude? Dialogues Clin Neurosci. 2014;16:491–503. doi: 10.31887/DCNS.2014.16.4/pdazzan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCarli C, Murphy DG, Mcintosh AR, Teichberg D, Schapiro MB, Horwitz B. Discriminant analysis of MRI measures as a method to determine the presence of dementia of the Alzheimer type. Psychiatry Res. 1995;57:119–130. doi: 10.1016/0165-1781(95)02651-c. [DOI] [PubMed] [Google Scholar]
- Demirci O, Clark VP, Calhoun VD. A projection pursuit algorithm to classify individuals using fMRI data: Application to schizophrenia. Neuroimage. 2008a;39:1774–82. doi: 10.1016/j.neuroimage.2007.10.012. doi: 10.1016/j.neuroimage.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci O, Clark VP, Magnotta V. a., Andreasen NC, Lauriello J, Kiehl K. a., Pearlson GD, Calhoun VD. A review of challenges in the use of fMRI for disease classification / characterization and A projection pursuit application from A multi-site fMRI schizophrenia study. Brain Imaging Behav. 2008b;2:207–226. doi: 10.1007/s11682-008-9028-1. doi:10.1007/sl 1682-008-9028-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Libera LE, Sreenivasan KR, Deshpande HD, Kana RK. Identification of neural connectivity signatures of autism using machine learning. Front Hum Neurosci. 2013;7:670. doi: 10.3389/fnhum.2013.00670. doi:10.3389/fnhum.2013.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G, Wang P, Rangaprakash D, Wilamowski B. Fully Connected Cascade Artificial Neural Network Architecture for Attention Deficit Hyperactivity Disorder Classification From Functional Magnetic Resonance Imaging Data. IEEE Trans Cybern. 2015;45:2668–2679. doi: 10.1109/TCYB.2014.2379621. doi:10.1109/TCYB.2014.2379621. [DOI] [PubMed] [Google Scholar]
- Dey S, Rao AR, Shah M. Attributed graph distance measure for automatic detection of attention deficit hyperactive disordered subjects. Front Neural Circuits. 2014;8:64. doi: 10.3389/fncir.2014.00064. doi:10.3389/fncir.2014.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Rao AR, Shah M. Exploiting the brain's network structure in identifying ADHD subjects. Front Syst Neurosci. 2012;6:75. doi: 10.3389/fnsys.2012.00075. doi:10.3389/fnsys.2012.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoe EA, Bandettini P, Neitz J, Miller D, Winans P. Functional magnetic resonance imaging (FMRI) of the human brain. J. Neurosci. Methods. 1994;54:171–187. doi: 10.1016/0165-0270(94)90191-0. [DOI] [PubMed] [Google Scholar]
- Di Martino A, Yan C-G, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry. 2014;19:659–667. doi: 10.1038/mp.2013.78. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLuca M, Olesen J. The cost of brain diseases: A burden or a challenge? Neuron. 2014;82:1205–1208. doi: 10.1016/j.neuron.2014.05.044. doi: 10.1016/j .neuron.2014.05.044. [DOI] [PubMed] [Google Scholar]
- Du W, Calhoun VD, Li H, Ma S, Eichele T, Kiehl K. a., Pearlson GD, Adali T. High Classification Accuracy for Schizophrenia with Rest and Task fMRI Data. Front. Hum. Neurosci. 2012;6:1–12. doi: 10.3389/fnhum.2012.00145. doi:10.3389/fnhum.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukart J, Mueller K, Barthel H, Villringer A, Sabri O, Schroeter ML. Meta-analysis based SVM classification enables accurate detection of Alzheimer's disease across different clinical centers using FDG-PET and MRI. Psychiatry Res. 2013;212:230–236. doi: 10.1016/j.pscychresns.2012.04.007. doi:10.1016/j.pscychresns.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Dwork C. Differential privacy. Autom. Lang. Program. 2006:1–12. [Google Scholar]
- Dyrba M, Ewers M, Wegrzyn M, Kilimann I, Plant C, Oswald A, Meindl T, Pievani M, Bokde ALW, Fellgiebel A, Filippi M, Hampel H, Kloppel S, Hauenstein K, Kirste T, Teipel SJ. Robust automated detection of microstructural white matter degeneration in Alzheimer's disease using machine learning classification of multicenter DTI data. PLoS One. 2013;8:e64925. doi: 10.1371/journal.pone.0064925. doi:10.1371/journal.pone.0064925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyrba M, Grothe M, Kirste T, Teipel SJ. Multimodal analysis of functional and structural disconnection in Alzheimer's disease using multiple kernel SVM. Hum Brain Mapp. 2015;36:2118–2131. doi: 10.1002/hbm.22759. doi:10.1002/hbm.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Marquand A, Mourão-Miranda J, Johnston P, Daly EM, Brammer MJ, Maltezos S, Murphy CM, Robertson D, Williams SC, Murphy DGM. Describing the brain in autism in five dimensions--magnetic resonance imaging–assisted diagnosis of autism spectrum disorder using a multiparameter classification approach. J. Neurosci. 2010a;30:10612–10623. doi: 10.1523/JNEUROSCI.5413-09.2010. doi:10.1523/JNEUROSCI.5413-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker C, Rocha-Rego V, Johnston P, Mourao-Miranda J, Marquand A, Daly EM, Brammer MJ, Murphy C, Murphy DG. Investigating the predictive value of whole-brain structural MR scans in autism: a pattern classification approach. Neuroimage. 2010b;49:44–56. doi: 10.1016/j.neuroimage.2009.08.024. doi:10.1016/j.neuroimage.2009.08.024. [DOI] [PubMed] [Google Scholar]
- Eickhoff S, Turner JA, Nichols TE, Van Horn JD. Sharing the wealth: Neuroimaging data repositories. Neuroimage. 2016;124:1065–1068. doi: 10.1016/j.neuroimage.2015.10.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein J, Stern E, Silbersweig D. Neuropsychiatry at the millenium: the potential for mind/brain integration through emerging interdisciplinary research strategies. Clin. Neurosci. Res. 2001;1:10–18. [Google Scholar]
- Ernst RL, Hay JW. The US economic and social costs of Alzheimer's disease revisited. Am. J. Public Health. 1994;84:1261–1264. doi: 10.2105/ajph.84.8.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Nigg JT, Iyer S, Bathula D, Mills KL, Dosenbach NUF, Schlaggar BL, Mennes M, Gutman D, Bangaru S, Buitelaar JK, Dickstein DP, Di Martino A, Kennedy DN, Kelly C, Luna B, Schweitzer JB, Velanova K, Wang Y-F, Mostofsky S, Castellanos FX, Milham MP. Distinct neural signatures detected for ADHD subtypes after controlling for micro-movements in resting state functional connectivity MRI data. Front Syst Neurosci. 2012;6:80. doi: 10.3389/fnsys.2012.00080. doi: 10.3389/fnsys.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falahati F, Westman E, Simmons A. Multivariate data analysis and machine learning in Alzheimer's disease with a focus on structural magnetic resonance imaging. J. Alzheimers Dis. 2014;41:685–708. doi: 10.3233/JAD-131928. doi:10.3233/JAD-131928. [DOI] [PubMed] [Google Scholar]
- Fan Y, Gur RE, Gur RC, Wu X, Shen D, Calkins ME, Davatzikos C. Unaffected family members and schizophrenia patients share brain structure patterns: a high-dimensional pattern classification study. Biol. Psychiatry. 2008a;63:118–124. doi: 10.1016/j.biopsych.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Liu Y, Wu FL, Hao Y, Liu FL, Liu Z, Jiang T. Discriminant analysis of functional connectivity patterns on Grassmann manifold. Neuroimage. 2011;56:2058–2067. doi: 10.1016/j.neuroimage.2011.03.051. doi:10.1016/j.neuroimage.2011.03.051. [DOI] [PubMed] [Google Scholar]
- Fan Y, Resnick SM, Wu X, Davatzikos C. Structural and functional biomarkers of prodromal Alzheimer's disease: a high-dimensional pattern classification study. Neuroimage. 2008b;41:277–285. doi: 10.1016/j.neuroimage.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Shen D, Davatzikos C. Classification of structural images via high-dimensional image warping, robust feature extraction, and SVM. Med. Image Comput. Comput. Assist. Interv. 2005;8:1–8. doi: 10.1007/11566465_1. doi: 10.1007/11566465_1. [DOI] [PubMed] [Google Scholar]
- Fan Y, Shen D, Gur RC, Gur RE, Davatzikos C. COMPARE: Classi cation of Morphological Patterns Using Adaptive Regional Elements. Comp. A J. Comp. Educ. 2007;26:93–105. doi: 10.1109/TMI.2006.886812. [DOI] [PubMed] [Google Scholar]
- Fang P, Zeng L-L, Shen H, Wang L, Li B, Liu L, Hu D. Increased cortical-limbic anatomical network connectivity in major depression revealed by diffusion tensor imaging. PLoS One. 2012;7:e45972. doi: 10.1371/journal.pone.0045972. doi:10.1371/journal.pone.0045972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan S, Fahiem MA, Tauseef H. An ensemble-of-classifiers based approach for early diagnosis of Alzheimer's disease: classification using structural features of brain images. Comput Math Methods Med. 2014;2014:862307. doi: 10.1155/2014/862307. doi:10.1155/2014/862307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan A, Mashohor S, Ramli AR, Mahmud R. Boosting diagnosis accuracy of Alzheimer's disease using high dimensional recognition of longitudinal brain atrophy patterns. Behav. Brain Res. 2015;290:124–130. doi: 10.1016/j.bbr.2015.04.010. doi:10.1016/j.bbr.2015.04.010. [DOI] [PubMed] [Google Scholar]
- Fekete T, Wilf M, Rubin D, Edelman S, Malach R, Mujica-Parodi LR. Combining Classification with fMRI-Derived Complex Network Measures for Potential Neurodiagnostics. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062867. doi:10.1371/journal.pone.0062867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SRA, Genetiker S, Fisher RA, Genetician S, Fisher RA, Geneticien S. The design of experiments. Oliver and Boyd Edinburgh. 1960 [Google Scholar]
- Foland-Ross LC, Sacchet MD, Prasad G, Gilbert B, Thompson PM, Gotlib IH. Cortical thickness predicts the first onset of major depression in adolescence. Int. J. Dev. Neurosci. 2015;46:125–131. doi: 10.1016/j.ijdevneu.2015.07.007. doi: 10.1016/j .ijdevneu.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JM, Roach BJ, Jorgensen KW, Turner JA, Brown GG, Notestine R, Bischoff-Grethe A, Greve D, Wible C, Lauriello J. Tuning in to the voices: a multisite FMRI study of auditory hallucinations. Schizophr. Bull. 2009;35:58–66. doi: 10.1093/schbul/sbn140. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K, Ziegler G, Klöppel S, Gaser C. Estimating the age of healthy subjects from T 1-weighted MRI scans using kernel methods: Exploring the influence of various parameters. Neuroimage. 2010;50:883–892. doi: 10.1016/j.neuroimage.2010.01.005. Initiative, A.D.N., others. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Fox NC. MR image texture analysis applied to the diagnosis and tracking of Alzheimer's disease. IEEE Trans Med Imaging. 1998;17:475–479. doi: 10.1109/42.712137. doi: 10.1109/42.712137. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Beltramello A, Weiss C, Geroldi C, Bianchetti A, Trabucchi M. Linear measures of atrophy in mild Alzheimer disease. AJNR Am J Neuroradiol. 1996;17:913–923. [PMC free article] [PubMed] [Google Scholar]
- Fung G, Deng Y, Zhao Q, Li Z, Qu M, Li K, Zeng Y-W, Jin Z, Ma Y-T, Yu X, Wang Z-R, Shum DHK, Chan RCK. Distinguishing bipolar and major depressive disorders by brain structural morphometry: a pilot study. BMC Psychiatry. 2015;15:298. doi: 10.1186/s12888-015-0685-5. doi:10.1186/sl2888-015-0685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaonkar B, Davatzikos C. Analytic estimation of statistical significance maps for support vector machine based multi-variate image analysis and classification. Neuroimage. 2013;78:270–283. doi: 10.1016/j.neuroimage.2013.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier S, Reisberg B, Zaudig M, Petersen RC, Ritchie K, Broich K, Belleville S, Brodaty FL, Bennett D, Chertkow H. Mild cognitive impairment. Lancet. 2006;367:1262–1270. doi: 10.1016/S0140-6736(06)68542-5. others. [DOI] [PubMed] [Google Scholar]
- Gerardin E, Chételat G, Chupin M, Cuingnet R, Desgranges B, Kim H-S, Niethammer M, Dubois B, Lehericy S, Garnero L, Eustache F, Colliot O. Multidimensional classification of hippocampal shape features discriminates Alzheimer's disease and mild cognitive impairment from normal aging. Neuroimage. 2009;47:1476–1486. doi: 10.1016/j.neuroimage.2009.05.036. doi:10.1016/j.neuroimage.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill PMW. Efficient calculation of p-values in linear-statistic permutation significance tests. J. Stat. Comput. Simul. 2007;77:55–61. [Google Scholar]
- Gollub RL, Shoemaker JM, King MD, White T, Ehrlich S, Sponheim SR, Clark VP, Turner JA, Mueller BA, Magnotta V. The MCIC collection: a shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics. 2013;11:367–388. doi: 10.1007/s12021-013-9184-3. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q, Wu Q, Scarpazza C, Lui S, Jia Z, Marquand A, Huang X, McGuire P, Mechelli A. Prognostic prediction of therapeutic response in depression using high-field MR imaging. Neuroimage. 2011;55:1497–1503. doi: 10.1016/j.neuroimage.2010.11.079. doi:10.1016/j.neuroimage.2010.11.079. [DOI] [PubMed] [Google Scholar]
- Good PI. Permutation, parametric, and bootstrap tests of hypotheses. Springer Science & Business Media; 2006. [Google Scholar]
- Gori I, Giuliano A, Muratori F, Saviozzi I, Oliva P, Tancredi R, Cosenza A, Tosetti M, Calderoni S, Retico A. Gray Matter Alterations in Young Children with Autism Spectrum Disorders: Comparing Morphometry at the Voxel and Regional Level. J Neuroimaging. 2015;25:866–874. doi: 10.1111/jon.12280. doi: 10.Ill 1/jon.12280. [DOI] [PubMed] [Google Scholar]
- Goryawala M, Zhou Q, Barker W, Loewenstein DA, Duara R, Adjouadi M. Inclusion of Neuropsychological Scores in Atrophy Models Improves Diagnostic Classification of Alzheimer's Disease and Mild Cognitive Impairment. Comput Intell Neurosci. 2015;2015:865265. doi: 10.1155/2015/865265. doi:10.1155/2015/865265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould IC, Shepherd AM, Laurens KR, Cairns MJ, Carr VJ, Green MJ. Multivariate neuroanatomical classification of cognitive subtypes in schizophrenia: A support vector machine learning approach. Neuroimage Clin. 2014;6:229–236. doi: 10.1016/j.nicl.2014.09.009. doi:10.1016/j.nicl.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graña M, Termenon M, Savio A, Gonzalez-Pinto A, Echeveste J, Perez JM, Besga A. Computer aided diagnosis system for Alzheimer disease using brain diffusion tensor imaging features selected by Pearson's correlation. Neurosci. Lett. 2011;502:225–229. doi: 10.1016/j.neulet.2011.07.049. doi:10.1016/j.neulet.2011.07.049. [DOI] [PubMed] [Google Scholar]
- Granziera C, Daducci A, Donati A, Bonnier G, Romascano D, Roche A, Bach Cuadra M, Schmitter D, Kloppel S, Meuli R, von Gunten A, Krueger G. A multi-contrast MRI study of microstructural brain damage in patients with mild cognitive impairment. Neuroimage Clin. 2015;8:631–639. doi: 10.1016/j.nicl.2015.06.003. doi:10.1016/j.nicl.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KR, Aljabar P, Heckemann RA, Hammers A, Rueckert D, Initiative ADN. Random forest-based similarity measures for multi-modal classification of Alzheimer's disease. Neuroimage. 2013;65:167–175. doi: 10.1016/j.neuroimage.2012.09.065. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MJ, Cairns MJ, Wu J, Dragovic M, Jablensky A, Tooney PA, Scott RJ, Carr VJ. Genome-wide supported variant MIR 137 and severe negative symptoms predict membership of an impaired cognitive subtype of schizophrenia. Mol. Psychiatry. 2013;18:774–780. doi: 10.1038/mp.2012.84. [DOI] [PubMed] [Google Scholar]
- Greenstein D, Malley JD, Weisinger B, Clasen L, Gogtay N. Using Multivariate Machine Learning Methods and Structural MRI to Classify Childhood Onset Schizophrenia and Healthy Controls. Front. Psychiatry. 2012;3:1–12. doi: 10.3389/fpsyt.2012.00053. doi:10.3389/fpsyt.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Cheng C, Cao X, Xiang J, Chen J, Zhang K. Resting-state functional connectivity abnormalities in first-onset unmedicated depression. Neural Regen Res. 2014;9:153–163. doi: 10.4103/1673-5374.125344. doi:10.4103/1673-5374.125344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Su Q, Yao D, Jiang J, Zhang J, Zhang Z, Yu L, Zhai J, Xiao C. Decreased regional activity of default-mode network in unaffected siblings of schizophrenia patients at rest. Eur Neuropsychopharmacol. 2014;24:545–552. doi: 10.1016/j.euroneuro.2014.01.004. doi:10.1016/j.euroneuro.2014.01.004. [DOI] [PubMed] [Google Scholar]
- Guyon I, Elisseeff A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003;3:1157–1182. [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002;46:389–422. [Google Scholar]
- H20 [WWW Document] 2015 URL http://h2o.ai/
- Hall MA, Smith LA. Practical feature subset selection for machine learning. 1998 [Google Scholar]
- Haller S, Missonnier P, Herrmann FR, Rodriguez C, Deiber M-P, Nguyen D, Gold G, Lovblad K-O, Giannakopoulos P. Individual classification of mild cognitive impairment subtypes by support vector machine analysis of white matter DTI. AJNR Am J Neuroradiol. 2013;34:283–291. doi: 10.3174/ajnr.A3223. doi:10.3174/ajnr.A3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller S, Nguyen D, Rodriguez C, Emch J, Gold G, Bartsch A, Lovblad KO, Giannakopoulos P. Individual prediction of cognitive decline in mild cognitive impairment using support vector machine-based analysis of diffusion tensor imaging data. J. Alzheimers Dis. 2010;22:315–327. doi: 10.3233/JAD-2010-100840. doi:10.3233/JAD-2010-100840. [DOI] [PubMed] [Google Scholar]
- Hampel H, Teipel SJ, Bayer W, Alexander GE, Schwarz R, Schapiro MB, Rapoport SI, Moller H-J. Age transformation of combined hippocampus and amygdala volume improves diagnostic accuracy in Alzheimer's disease. J. Neurol. Sci. 2002;194:15–19. doi: 10.1016/s0022-510x(01)00669-4. [DOI] [PubMed] [Google Scholar]
- Hart H, Chantiluke K, Cubillo AI, Smith AB, Simmons A, Brammer MJ, Marquand AF, Rubia K. Pattern classification of response inhibition in ADHD: toward the development of neurobiological markers for ADHD. Hum Brain Mapp. 2014a;35:3083–3094. doi: 10.1002/hbm.22386. doi:10.1002/hbm.22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart H, Marquand AF, Smith A, Cubillo A, Simmons A, Brammer M, Rubia K. Predictive neurofunctional markers of attention-deficit/hyperactivity disorder based on pattern classification of temporal processing. J. Am. Acad. Child Adolesc. Psychiatry. 2014b;53:569–578. doi: 10.1016/j.jaac.2013.12.024. [DOI] [PubMed] [Google Scholar]
- Hastie T, Rosset S, Tibshirani R, Zhu J. The entire regularization path for the support vector machine. J. Mach. Learn. Res. 2004;5:1391–1415. [Google Scholar]
- Hebert LE, Beckett LA, Scherr PA, Evans DA. Annual incidence of Alzheimer disease in the United States projected to the years 2000 through 2050. Alzheimer Dis. Assoc. Disord. 2001;15:169–173. doi: 10.1097/00002093-200110000-00002. [DOI] [PubMed] [Google Scholar]
- Heinrichs RW, Zakzanis KK. Neurocognitive deficit in schizophrenia: a quantitative review of the evidence. Neuropsychology. 1998;12:426. doi: 10.1037//0894-4105.12.3.426. [DOI] [PubMed] [Google Scholar]
- Hidalgo-Mufioz AR, Ramírez J, Gorriz JM, Padilla P. Regions of interest computed by SVM wrapped method for Alzheimer's disease examination from segmented MRI. Front Aging Neurosci. 2014;6:20. doi: 10.3389/fnagi.2014.00020. doi:10.3389/fnagi.2014.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Mukherjee L, Xu G, Chung MK, Johnson SC. Spatially augmented LPboosting for AD classification with evaluations on the ADNI dataset. Neuroimage. 2009;48:138–149. doi: 10.1016/j.neuroimage.2009.05.056. doi: 10.1016/j.neuroimage.2009.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs C, Singh V, Xu G, Johnson SC. Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage. 2011;55:574–589. doi: 10.1016/j.neuroimage.2010.10.081. doi:10.1016/j.neuroimage.2010.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelm RD, Calhoun VD, Salakhutdinov R, Allen EA, Adali T, Plis SM. Restricted Boltzmann machines for neuroimaging: An application in identifying intrinsic networks. Neuroimage. 2014;96:245–260. doi: 10.1016/j.neuroimage.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honorio J, Tomasi D, Goldstein RZ, Leung HC, Samaras D. Can a single brain region predict a disorder? IEEE Trans. Med. Imaging. 2012;31:2062–2072. doi: 10.1109/TMI.2012.2206047. doi:10.1109/TMI.2012.2206047. [DOI] [PubMed] [Google Scholar]
- Iannaccone R, Hauser TU, Ball J, Brandeis D, Walitza S, Brem S. Classifying adolescent attention-deficit/hyperactivity disorder (ADHD) based on functional and structural imaging. Eur Child Adolesc Psychiatry. 2015;24:1279–1289. doi: 10.1007/s00787-015-0678-4. doi:10.1007/s00787-015-0678-4. [DOI] [PubMed] [Google Scholar]
- Igual L, Soliva JC, Escalera S, Gimeno R, Vilarroya O, Radeva P. Automatic brain caudate nuclei segmentation and classification in diagnostic of Attention-Deficit/Hyperactivity Disorder. Comput Med Imaging Graph. 2012;36:591–600. doi: 10.1016/j.compmedimag.2012.08.002. doi:10.1016/j.compmedimag.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Iidaka T. Resting state functional magnetic resonance imaging and neural network classified autism and control. Cortex. 2015;63:55–67. doi: 10.1016/j.cortex.2014.08.011. doi:10.1016/j.cortex.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Ingalhalikar M, Parker D, Bloy L, Roberts TPL, Verma R. Diffusion based abnormality markers of pathology: toward learned diagnostic prediction of ASD. Neuroimage. 2011;57:918–927. doi: 10.1016/j.neuroimage.2011.05.023. doi:10.1016/j.neuroimage.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingalhalikar M, Smith AR, Bloy L, Gur R, Roberts TPL, Verma R. Identifying sub-populations via unsupervised cluster analysis on multi-edge similarity graphs. Med. Image Comput. Comput. Assist. Interv. 2012;15:254–61. doi: 10.1007/978-3-642-33418-4_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi SJ, Liddle PF, Palaniyappan L. Clinical utility of machine-learning approaches in schizophrenia: improving diagnostic confidence for translational neuroimaging. Front Psychiatry. 2013;4:95. doi: 10.3389/fpsyt.2013.00095. doi:10.3389/fpsyt.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablensky A. Subtyping schizophrenia: implications for genetic research. Mol. Psychiatry. 2006;11:815–836. doi: 10.1038/sj.mp.4001857. [DOI] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, L Whitwell J, Ward C. The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu YC, Waring SC, O'Brien PC, Tangalos EG, Smith GE, Ivnik RJ, Kokmen E. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer's disease. Neurology. 1997;49:786–794. doi: 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri MJ, Pearlson GD, Stevens M, Calhoun VD. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. Neuroimage. 2008;39:1666–1681. doi: 10.1016/j.neuroimage.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janousova E, Schwarz D, Kasparek T. Combining various types of classifiers and features extracted from magnetic resonance imaging data in schizophrenia recognition. Psychiatry Res. Neuroimaging. 2015;232:237–249. doi: 10.1016/j.pscychresns.2015.03.004. doi:10.1016/j.pscychresns.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Chen R, Ke X, Chu K, Lu Z, Herskovits EH. Predictive models of autism spectrum disorder based on brain regional cortical thickness. Neuroimage. 2010;50:589–599. doi: 10.1016/j.neuroimage.2009.12.047. doi: 10.1016/j.neuroimage.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie B, Zhang D, Gao W, Wang Q, Wee C-Y, Shen D. Integration of network topological and connectivity properties for neuroimaging classification. IEEE Trans Biomed Eng. 2014;61:576–589. doi: 10.1109/TBME.2013.2284195. doi:10.1109/TBME.2013.2284195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie N, Zhu M, Xiaoying M, Osuch EA, Wammes M, Theberge J, Li H, Zhang Y, Jiang T, Sui J, Calhoun VD. Discriminating Bipolar Disorder From Major Depression Based on SVM-FoBa: Efficient Feature Selection With Multimodal Brain Imaging Data. IEEE Trans. Auton. Ment. Dev. 2015 doi: 10.1109/TAMD.2015.2440298. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston BA, Mwangi B, Matthews K, Coghill D, Konrad K, Steele JD. Brainstem abnormalities in attention deficit hyperactivity disorder support high accuracy individual diagnostic classification. Hum Brain Mapp. 2014;35:5179–5189. doi: 10.1002/hbm.22542. doi:10.1002/hbm.22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WB, Lee YM, Kim YH, Mun C-W. Automated Classification to Predict the Progression of Alzheimer's Disease Using Whole-Brain Volumetry and DTI. Psychiatry Investig. 2015;12:92–102. doi: 10.4306/pi.2015.12.1.92. doi:10.4306/pi.2015.12.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just MA, Cherkassky VL, Buchweitz A, Keller TA, Mitchell TM. Identifying autism from neural representations of social interactions: neurocognitive markers of autism. PLoS One. 2014;9:e113879. doi: 10.1371/journal.pone.0113879. doi: 10.1371/journal.pone.0113879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambeitz J, Kambeitz-Ilankovic L, Leucht S, Wood S, Davatzikos C, Malchow B, Falkai P, Koutsouleris N. Detecting Neuroimaging Biomarkers for Schizophrenia: A Meta-Analysis of Multivariate Pattern Recognition Studies. Neuropsychopharmacology. 2015 doi: 10.1038/npp.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgiou E, Schulz SC, Gollub RL, Andreasen NC, Ho BC, Lauriello J, Calhoun VD, Bockholt HJ, Sponheim SR, Georgopoulos AP. Neuropsychological testing and structural magnetic resonance imaging as diagnostic biomarkers early in the course of schizophrenia and related psychoses. Neuroinformatics. 2011;9:321–333. doi: 10.1007/s12021-010-9094-6. doi:10.1007/sl2021-010-9094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasparek T, Thomaz CE, Sato JR, Schwarz D, Janousova E, Marecek R, Prikryl R, Vanicek J, Fujita A, Ceskova E. Maximum-uncertainty linear discrimination analysis of first-episode schizophrenia subjects. Psychiatry Res. - Neuroimaging. 2011;191:174–181. doi: 10.1016/j.pscychresns.2010.09.016. doi:10.1016/j.pscychresns.2010.09.016. [DOI] [PubMed] [Google Scholar]
- Kaufer DI, Miller BL, Itti L, Fairbanks LA, Li J, Fishman J, Kushi J, Cummings JL. Midline cerebral morphometry distinguishes frontotemporal dementia and Alzheimer's disease. Neurology. 1997;48:978–985. doi: 10.1212/wnl.48.4.978. [DOI] [PubMed] [Google Scholar]
- Kaufmann T, Skatun KC, Alnaes D, Doan NT, Duff EP, Tonnesen S, Roussos E, Ueland T, Aminoff SR, Lagerberg TV, Agartz I, Melle IS, Smith SM, Andreassen O. a., Westlye LT. Disintegration of Sensorimotor Brain Networks in Schizophrenia. Schizophr. Bull. 2015:1–10. doi: 10.1093/schbul/sbv060. doi:10.1093/schbul/sbv060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Suzuki M, Kherif F, Takahashi T, Zhou SY, Nakamura K, Matsui M, Sumiyoshi T, Seto FL, Kurachi M. Multivariate voxel-based morphometry successfully differentiates schizophrenia patients from healthy controls. Neuroimage. 2007;34:235–242. doi: 10.1016/j.neuroimage.2006.08.018. doi:10.1016/j.neuroimage.2006.08.018. [DOI] [PubMed] [Google Scholar]
- Keator DB, Grethe JS, Marcus D, Ozyurt B, Gadde S, Murphy S, Pieper S, Greve D, Notestine R, Bockholt HJ. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN). Inf. Technol. Biomed. IEEE Trans. 2008;12:162–172. doi: 10.1109/TITB.2008.917893. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, Rush AJ, Walters EE, Wang PS. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). Jama. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- Kessler RC, McGonagle KA, Zhao S, Nelson CB, Hughes M, Eshleman S, Wittchen H-U, Kendler KS. Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States: results from the National Comorbidity Survey. Arch. Gen. Psychiatry. 1994;51:8–19. doi: 10.1001/archpsyc.1994.03950010008002. [DOI] [PubMed] [Google Scholar]
- Khazaee A, Ebrahimzadeh A, Babajani-Feremi A. Identifying patients with Alzheimer's disease using resting-state fMRI and graph theory. Clin Neurophysiol. 2015;126:2132–2141. doi: 10.1016/j.clinph.2015.02.060. doi:10.1016/j.clinph.2015.02.060. [DOI] [PubMed] [Google Scholar]
- Kim DI, Sui J, Rachakonda S, White T, Manoach DS, Clark VP, Ho BC, Schulz SC, Calhoun VD. Identification of Imaging Biomarkers in Schizophrenia: {A} Coefficient-constrained Independent Component Analysis of the Mind Multi-site Schizophrenia Study. Neuroinformatics. 2010:1–17. doi: 10.1007/s12021-010-9077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Calhoun VD, Shim E, Lee J-H. Deep neural network with weight sparsity control and pre-training extracts hierarchical features and enhances classification performance: Evidence from whole-brain resting-state functional connectivity patterns of schizophrenia. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.05.018. doi: 10.1016/j.neuroimage.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, Abdulkadir A, Jack CR, Koutsouleris N, Mourao-Miranda J, Vemuri P. Diagnostic neuroimaging across diseases. Neuroimage. 2012;61:457–463. doi: 10.1016/j.neuroimage.2011.11.002. doi: 10.1016/j.neuroimage.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, Chu C, Tan GC, Draganski B, Johnson H, Paulsen JS, Kienzle W, Tabrizi SJ, Ashburner J, Frackowiak RSJ. Automatic detection of preclinical neurodegeneration Presymptomatic Huntington disease. Neurology. 2009;72:426–431. doi: 10.1212/01.wnl.0000341768.28646.b6. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, Peter J, Ludl A, Pilatus A, Maier S, Mader I, Heimbach B, Frings L, Egger K, Dukart J, Schroeter ML, Perneczky R, Haussermann P, Vach W, Urbach H, Teipel S, Hull M, Abdulkadir A. Applying Automated MR-Based Diagnostic Methods to the Memory Clinic: A Prospective Study. J. Alzheimers Dis. 2015;47:939–954. doi: 10.3233/JAD-150334. doi: 10.3233/JAD-150334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Ashburner J, Frackowiak RSJ. Automatic classification of MR scans in Alzheimer's disease. Brain. 2008;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch SP, Hägele C, Haynes J-D, Heinz A, Schlagenhauf F, Sterzer P. Diagnostic Classification of Schizophrenia Patients on the Basis of Regional Reward-Related fMRI Signal Patterns. PLoS One. 2015;10:eOl19089. doi: 10.1371/journal.pone.0119089. doi: 10.1371/journal.pone.Ol 19089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohavi R, John GH. Wrappers for feature subset selection. Artif. Intell. 1997;97:273–324. [Google Scholar]
- Koutsouleris N, Meisenzahl EM, Borgwardt S, Riecher-Rossler a., Frodl T, Kambeitz J, Kohler Y, Falkai P, Moller H-J, Reiser M, Davatzikos C. Individualized differential diagnosis of schizophrenia and mood disorders using neuroanatomical biomarkers. Brain. 2015 doi: 10.1093/brain/awv111. doi:10.1093/brain/awvlll. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N. Deep neural networks: a new framework for modelling biological vision and brain information processing. bioRxiv. 2015:29876. doi: 10.1146/annurev-vision-082114-035447. [DOI] [PubMed] [Google Scholar]
- Lahmiri S, Boukadoum M. New approach for automatic classification of Alzheimer's disease, mild cognitive impairment and healthy brain magnetic resonance images. Heal. Technol Lett. 2014;1:32–36. doi: 10.1049/htl.2013.0022. doi:10.1049/htl.2013.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis D, Courtney W, Dieringer C, Kelly R, King M, Miller B, Wang R, Wood D, Turner JA, Calhoun VD. An Open Platform for Compiling, Curating, and Disseminating Neuroimaging Data. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.05.049. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan D, Mangin JF, Poupon C, Clark CA, Pappata S, Molko N, Chabriat H. Diffusion tensor imaging: concepts and applications. J. Magn. Reson. imaging. 2001;13:534–546. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- Lee G-Y, Kim J, Kim JH, Kim K, Seong J-K. Online Learning for Classification of Alzheimer Disease based on Cortical Thickness and Hippocampal Shape Analysis. Heal. Inf. Res. 2014;20:61–68. doi: 10.4258/hir.2014.20.1.61. doi:10.4258/hir.2014.20.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Park B, Han K. Classification of diffusion tensor images for the early detection of Alzheimer's disease. Comput. Biol. Med. 2013;43:1313–1320. doi: 10.1016/j.compbiomed.2013.07.004. doi:10.1016/j.compbiomed.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Lerch JP, Pruessner J, Zijdenbos AP, Collins DL, Teipel SJ, Hampel H, Evans AC. Automated cortical thickness measurements from MRI can accurately separate Alzheimer's patients from normal elderly controls. Neurobiol. Aging. 2008;29:23–30. doi: 10.1016/j.neurobiolaging.2006.09.013. doi:10.1016/j.neurobiolaging.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Levman J, Takahashi E. Multivariate analyses applied to fetal, neonatal and pediatric MRI of neurodevelopmental disorders. Neuroimage Clin. 2015;9:532–544. doi: 10.1016/j.nicl.2015.09.017. doi:10.1016/j.nicl.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinsohn PM, Duncan EM, Stanton AK, Hautzinger M. Age at first onset for nonbipolar depression. J. Abnorm. Psychol. 1986;95:378. doi: 10.1037//0021-843x.95.4.378. [DOI] [PubMed] [Google Scholar]
- Li M, Oishi K, He X, Qin Y, Gao F, Mori S. An efficient approach for differentiating Alzheimer's disease from normal elderly based on multicenter MRI using gray-level invariant features. PLoS One. 2014a;9:el05563. doi: 10.1371/journal.pone.0105563. doi:10.1371/journal.pone.0105563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Qin Y, Gao F, Zhu W, He X. Discriminative analysis of multivariate features from structural MRI and diffusion tensor images. Magn Reson Imaging. 2014b;32:1043–1051. doi: 10.1016/j.mri.2014.05.008. doi:10.1016/j.mri.2014.05.008. [DOI] [PubMed] [Google Scholar]
- Li S, Shi F, Pu F, Li X, Jiang T, Xie S, Wang Y. Hippocampal shape analysis of Alzheimer disease based on machine learning methods. AJNR Am J Neuroradiol. 2007;28:1339–1345. doi: 10.3174/ajnr.A0620. doi:10.3174/ajnr.A0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang Y, Wu G, Shi F, Zhou L, Lin W, Shen D. Discriminant analysis of longitudinal cortical thickness changes in Alzheimer's disease using dynamic and network features. Neurobiol. Aging. 2012;33:427, el5–30. doi: 10.1016/j.neurobiolaging.2010.11.008. doi: 10.1016/j.neurobiolaging.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z-P, Lauterbur PC. Principles of magnetic resonance imaging. SPIE Optical Engineering Press; 2000. [Google Scholar]
- Libera LE, DeRamus TP, Lahti AC, Deshpande G, Kana RK. Multimodal neuroimaging based classification of autism spectrum disorder using anatomical, neurochemical, and white matter correlates. Cortex. 2015;66:46–59. doi: 10.1016/j.cortex.2015.02.008. doi:10.1016/j.cortex.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillemark L, Sørensen L, Pai A, Dam EB, Nielsen M. Brain region's relative proximity as marker for Alzheimer's disease based on structural MRI. BMC Med Imaging. 2014;14:21. doi: 10.1186/1471-2342-14-21. doi: 10.1186/1471-2342-14-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Marquand A, Cubillo AA, Smith AB, Chantiluke K, Simmons A, Mehta M, Rubia K. Disorder-specific predictive classification of adolescents with attention deficit hyperactivity disorder (ADHD) relative to autism using structural magnetic resonance imaging. 2013 doi: 10.1371/journal.pone.0063660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wee C-Y, Chen H, Shen D. Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer's Disease and mild cognitive impairment identification. Neuroimage. 2014;84:466–475. doi: 10.1016/j.neuroimage.2013.09.015. doi:10.1016/j.neuroimage.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Adeli-Mosabbeb E, Shen D. Inherent Structure Based Multi-view Learning with Multi-template Feature Representation for Alzheimer's Disease Diagnosis. IEEE Trans Biomed Eng. 2015 doi: 10.1109/TBME.2015.2496233. doi: 10.1109/TBME.2015.2496233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang D, Shen D. Hierarchical fusion of features and classifier decisions for Alzheimer's disease diagnosis. Hum Brain Mapp. 2014;35:1305–1319. doi: 10.1002/hbm.22254. doi:10.1002/hbm.22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Tosun D, Weiner MW, Schuff N. Locally linear embedding (LLE) for MRI based Alzheimer's disease classification. Neuroimage. 2013;83:148–157. doi: 10.1016/j.neuroimage.2013.06.033. doi:10.1016/j.neuroimage.2013.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord A, Horn D, Breakspear M, Walter M. Changes in community structure of resting state functional connectivity in unipolar depression. PLoS One. 2012;7:e41282. doi: 10.1371/journal.pone.0041282. doi:10.1371/journal.pone.0041282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetti V, Allen NB, Fornito A, Yiicel M. Structural brain abnormalities in major depressive disorder: a selective review of recent MRI studies. J. Affect. Disord. 2009;117:1–17. doi: 10.1016/j.jad.2008.11.021. [DOI] [PubMed] [Google Scholar]
- Ludman EJ, Fullerton SM, Spangler L, Trinidad SB, Fujii MM, Jarvik GP, Larson EB, Burke W. Glad you asked: participants' opinions of re-consent for dbGap data submission. J. Empir. Res. Hum. Res. Ethics. 2010;5:9–16. doi: 10.1525/jer.2010.5.3.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Li R, Yu J, He Y, Li J. Alterations in regional homogeneity of spontaneous brain activity in late-life subthreshold depression. PLoS One. 2013;8:e53148. doi: 10.1371/journal.pone.0053148. doi:10.1371/journal.pone.0053148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMaster FP, Carrey N, Langevin LM, Jaworska N, Crawford S. Disorder-specific volumetric brain difference in adolescent major depressive disorder and bipolar depression. Brain Imaging Behav. 2014;8:119–127. doi: 10.1007/s11682-013-9264-x. doi: 10.1007/s 11682-013-9264-x. [DOI] [PubMed] [Google Scholar]
- Magnin B, Mesrob L, Kinkingnehun S, Pelegrini-Issac M, Colliot O, Sarazin M, Dubois B, Lehericy S, Benali H. Support vector machine-based classification of Alzheimer's disease from whole-brain anatomical MRI. Neuroradiology. 2009;51:73–83. doi: 10.1007/s00234-008-0463-x. doi:10.1007/s00234-008-0463-x. [DOI] [PubMed] [Google Scholar]
- Mahanand BS, Suresh S, Sundararajan N, Aswatha Kumar M. Identification of brain regions responsible for Alzheimer's disease using a Self-adaptive Resource Allocation Network. Neural Netw. 2012;32:313–322. doi: 10.1016/j.neunet.2012.02.035. doi:10.1016/j.neunet.2012.02.035. [DOI] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The extensible neuroimaging archive toolkit. Neuroinformatics. 2007;5:11–33. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- McAlonan GM, Cheung V, Cheung C, Suckling J, Lam GY, Tai KS, Yip L, Murphy DGM, Chua SE. Mapping the brain in autism. A voxel-based MRI study of volumetric differences and intercorrelations in autism. Brain. 2005;128:268–276. doi: 10.1093/brain/awh332. [DOI] [PubMed] [Google Scholar]
- McCarley RW, Nakamura M, Shenton ME, Salisbury DF. Combining ERP and structural MRI information in first episode schizophrenia and bipolar disorder. Clin. EEG Neurosci. 2008;39:57–60. doi: 10.1177/155005940803900206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Holland D, Karow DS, Pung CJ, Brewer JB, Dale AM. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251:195–205. doi: 10.1148/radiol.2511080924. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEvoy LK, Holland D, Hagler DJ, Fennema-Notestine C, Brewer JB, Dale AM. Mild cognitive impairment: baseline and longitudinal structural MR imaging measures improve predictive prognosis. Radiology. 2011;259:834–843. doi: 10.1148/radiol.11101975. doi: 10.1148/radiol.11101975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta CR, Patel NR, Senchaudhuri P. Importance sampling for estimating exact probabilities in permutational inference. J. Am. Stat. Assoc. 1988;83:999–1005. [Google Scholar]
- Merboldt K-D, Hanicke W, Frahm J. Self-diffusion NMR imaging using stimulated echoes. J. Magn. Reson. 1985;64:479–486. [Google Scholar]
- Michael AM, Baum SA, Fries JF, Ho BC, Pierson RK, Andreasen NC, Calhoun VD. A method to fuse f{MRI} tasks through spatial correlations: Applied to schizophrenia. Hum. Brain Mapp. 2009;30:25122529. doi: 10.1002/hbm.20691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Priebe CE, Qiu A, Fischl B, Kolasny A, Brown T, Park Y, Ratnanather JT, Busa E, Jovicich J, Yu P, Dickerson BC, Buckner RL. Collaborative computational anatomy: an MRI morphometry study of the human brain via diffeomorphic metric mapping. Hum Brain Mapp. 2009;30:2132–2141. doi: 10.1002/hbm.20655. doi:10.1002/hbm.20655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min R, Wu G, Cheng J, Wang Q, Shen D. Multi-atlas based representations for Alzheimer's disease diagnosis. Hum Brain Mapp. 2014;35:5052–5070. doi: 10.1002/hbm.22531. doi:10.1002/hbm.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshew NJ, Payton JB. New perspectives in autism. Part 2: The differential diagnosis and neurobiology of autism. Curr. Probl. Pediatr. 1988;18:618–694. doi: 10.1016/0045-9380(88)90017-5. [DOI] [PubMed] [Google Scholar]
- Moradi E, Pepe A, Gaser C, Huttunen H, Tohka J. Machine learning framework for early MRI-based Alzheimer's conversion prediction in MCI subjects. Neuroimage. 2015;104:398–412. doi: 10.1016/j.neuroimage.2014.10.002. doi: 10.1016/j .neuroimage.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morar B, Dragović M, Waters FAV, Chandler D, Kalaydjieva L, Jablensky A. Neuregulin 3 (NRG3) as a susceptibility gene in a schizophrenia subtype with florid delusions and relatively spared cognition. Mol. Psychiatry. 2011;16:860–866. doi: 10.1038/mp.2010.70. [DOI] [PubMed] [Google Scholar]
- Mourão-Miranda J, Hardoon DR, Hahn T, Marquand AF, Williams SCR, Shawe-Taylor J, Brammer M. Patient classification as an outlier detection problem: an application of the One-Class Support Vector Machine. Neuroimage. 2011;58:793–804. doi: 10.1016/j.neuroimage.2011.06.042. doi: 10.1016/j.neuroimage.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourão-Miranda J, Oliveira L, Ladouceur CD, Marquand A, Brammer M, Birmaher B, Axelson D, Phillips ML. Pattern recognition and functional neuroimaging help to discriminate healthy adolescents at risk for mood disorders from low risk adolescents. PLoS One. 2012;7:e29482. doi: 10.1371/journal.pone.0029482. doi:10.1371/journal.pone.0029482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Schuff N, Yaffe K, Madison C, Miller B, Weiner MW. Hippocampal atrophy patterns in mild cognitive impairment and Alzheimer's disease. Hum Brain Mapp. 2010;31:1339–1347. doi: 10.1002/hbm.20934. doi:10.1002/hbm.20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdaugh DL, Shinkareva SV, Deshpande HR, Wang J, Pennick MR, Kana RK. Differential deactivation during mentalizing and classification of autism based on default mode network connectivity. 2012 doi: 10.1371/journal.pone.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi B, Ebmeier KP, Matthews K, Steele JD. Multi-centre diagnostic classification of individual structural neuroimaging scans from patients with major depressive disorder. Brain. 2012;135:1508–1521. doi: 10.1093/brain/aws084. doi: 10.1093/brain/aws084. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kawasaki Y, Suzuki M, Hagino H, Kurokawa K, Takahashi T, Niu L, Matsui M, Seto H, Kurachi M. Multiple structural brain measures obtained by three-dimensional magnetic resonance imaging to distinguish between schizophrenia patients and normal subjects. Schizophr. Bull. 2004;30:393–404. doi: 10.1093/oxfordjournals.schbul.a007087. [DOI] [PubMed] [Google Scholar]
- Ng AY. Feature selection, L 1 vs. L 2 regularization, and rotational invariance. Proceedings of the Twenty-First International Conference on Machine Learning. 2004:78. [Google Scholar]
- Nieuwenhuis M, van Haren NEM, Hulshoff Pol HE, Cahn W, Kahn RS, Schnack HG. Classification of schizophrenia patients and healthy controls from structural MRI scans in two large independent samples. Neuroimage. 2012;61:606–612. doi: 10.1016/j.neuroimage.2012.03.079. doi:10.1016/j.neuroimage.2012.03.079. [DOI] [PubMed] [Google Scholar]
- Nir TM, Villalon-Reina JE, Prasad G, Jahanshad N, Joshi SH, Toga AW, Bernstein MA, Jack CR, Weiner MW, Thompson PM. Diffusion weighted imaging-based maximum density path analysis and classification of Alzheimer's disease. Neurobiol. Aging. 2015;36(Suppl 1):SI32–40. doi: 10.1016/j.neurobiolaging.2014.05.037. doi:10.1016/j.neurobiolaging.2014.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dwyer L, Lamberton F, Bokde AL, Ewers M, Faluyi YO, Tanner C, Mazoyer B, O'Neill D, Bartley M, Collins DR. Using support vector machines with multiple indices of diffusion for automated classification of mild cognitive impairment. PLoS One. 2012;7:e32441. doi: 10.1371/journal.pone.0032441. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lee T-M, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. 1990;87:9868–9872. doi: 10.1073/pnas.87.24.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Akhter K, Mielke M, Ceritoglu C, Zhang J, Jiang H, Li X, Younes L, Miller MI, van Zijl PCM, Albert M, Lyketsos CG, Mori S. Multi-modal MRI analysis with disease-specific spatial filtering: initial testing to predict mild cognitive impairment patients who convert to Alzheimer's disease. Front Neurol. 2011;2:54. doi: 10.3389/fneur.2011.00054. doi: 10.3389/fneur.2011.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira P.P. de M., Nitrini R, Busatto G, Buchpiguel C, Sato JR, Amaro E. Use of SVM methods with surface-based cortical and volumetric subcortical measurements to detect Alzheimer's disease. J. Alzheimers Dis. 2010;19:1263–1272. doi: 10.3233/JAD-2010-1322. doi: 10.3233/JAD-2010-1322. [DOI] [PubMed] [Google Scholar]
- Orrú G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A. Using Support Vector Machine to identify imaging biomarkers of neurological and psychiatric disease: A critical review. Neurosci. Biobehav. Rev. 2012;36:1140–1152. doi: 10.1016/j.neubiorev.2012.01.004. doi:10.1016/j.neubiorev.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Ortiz A, Munilla J, Alvarez-Ulan I, Górriz JM, Ramírez J. Exploratory graphical models of functional and structural connectivity patterns for Alzheimer's Disease diagnosis. Front Comput Neurosci. 2015;9:132. doi: 10.3389/fncom.2015.00132. doi:10.3389/fncom.2015.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota K, Oishi N, Ito K, Fukuyama H. Effects of imaging modalities, brain atlases and feature selection on prediction of Alzheimer's disease. J. Neurosci. Methods. 2015;256:168–183. doi: 10.1016/j.jneumeth.2015.08.020. doi:10.1016/j.jneumeth.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Ota K, Oishi N, Ito K, Fukuyama H. A comparison of three brain atlases for MCI prediction. J. Neurosci. Methods. 2014;221:139–150. doi: 10.1016/j.jneumeth.2013.10.003. doi:10.1016/j.jneumeth.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Ota M, Ishikawa M, Sato N, Hori H, Sasayama D, Hattori K, Teraishi T, Noda T, Obu S, Nakata Y, Higuchi T, Kunugi H. Discrimination between schizophrenia and major depressive disorder by magnetic resonance imaging of the female brain. J. Psychiatr. Res. 2013;47:1383–1388. doi: 10.1016/j.jpsychires.2013.06.010. doi:10.1016/j.jpsychires.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Ota M, Sato N, Ishikawa M, Hori H, Sasayama D, Hattori K, Teraishi T, Obu S, Nakata Y, Nemoto K, Moriguchi Y, Hashimoto R, Kunugi FL. Discrimination of female schizophrenia patients from healthy women using multiple structural brain measures obtained with voxel-based morphometry. Psychiatry Clin. Neurosci. 2012;66:611–617. doi: 10.1111/j.1440-1819.2012.02397.x. doi:10.1111/j.l440-1819.2012.02397.x. [DOI] [PubMed] [Google Scholar]
- Pardo PJ, Georgopoulos AP, Kenny JT, Stuve T. a., Findling RL, Schulz SC. Classification of adolescent psychotic disorders using linear discriminant analysis. Schizophr. Res. 2006;87:297–306. doi: 10.1016/j.schres.2006.05.007. doi:10.1016/j.schres.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Park B-Y, Kim M, Seo J, Lee J-M, Park H. Connectivity Analysis and Feature Classification in Attention Deficit Hyperactivity Disorder Sub-Types: A Task Functional Magnetic Resonance Imaging Study. Brain Topogr. 2015 doi: 10.1007/s10548-015-0463-1. doi:10.1007/sl0548-015-0463-l. [DOI] [PubMed] [Google Scholar]
- Park MY, Hastie T. Ll-regularization path algorithm for generalized linear models. J. R. Stat. Soc. Ser. B (Statistical Methodol. 2007;69:659–677. [Google Scholar]
- Patel MJ, Andreescu C, Price JC, Edelman KL, Reynolds CF, Aizenstein HJ. Machine learning approaches for integrating clinical and imaging features in late-life depression classification and response prediction. Int J Geriatr Psychiatry. 2015;30:1056–1067. doi: 10.1002/gps.4262. doi:10.1002/gps.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Lin P, Zhang T, Wang J. Extreme learning machine-based classification of ADHD using brain structural MRI data. PLoS One. 2013;8:e79476. doi: 10.1371/journal.pone.0079476. doi:10.1371/journal.pone.0079476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennanen C, Kivipelto M, Tuomainen S, Hartikainen P, Hanninen T, Laakso MP, Hallikainen M, Vanhanen M, Nissinen A, Helkala E-L, Vainio P, Vanninen R, Partanen K, Soininen H. Hippocampus and entorhinal cortex in mild cognitive impairment and early AD. Neurobiol. Aging. 2004;25:303–310. doi: 10.1016/S0197-4580(03)00084-8. doi: 10.1016/S0197-4580(03)00084-8. [DOI] [PubMed] [Google Scholar]
- Pereira F, Mitchell T, Botvinick M. Machine learning classifiers and fMRI: a tutorial overview. Neuroimage. 2009;45:S199–S209. doi: 10.1016/j.neuroimage.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Negash S. Mild cognitive impairment: an overview. CNS Spectr. 2008;13:45–53. doi: 10.1017/s1092852900016151. [DOI] [PubMed] [Google Scholar]
- Pina-Camacho L, Garcia-Prieto J, Parellada M, Castro-Fornieles J, Gonzalez-Pinto AM, Bombin I, Graell M, Paya B, Rapado-Castro M, Janssen J, Baeza I, Del Pozo F, Desco M, Arango C. Predictors of schizophrenia spectrum disorders in early-onset first episodes of psychosis: a support vector machine model. Eur Child Adolesc Psychiatry. 2015;24:427–440. doi: 10.1007/s00787-014-0593-0. doi:10.1007/s00787-014-0593-0. [DOI] [PubMed] [Google Scholar]
- Plant C, Teipel SJ, Oswald A, Bohm C, Meindl T, Mourao-Miranda J, Bokde AW, Hampel H, Ewers M. Automated detection of brain atrophy patterns based on MRI for the prediction of Alzheimer's disease. Neuroimage. 2010;50:162–174. doi: 10.1016/j.neuroimage.2009.11.046. doi:10.1016/j.neuroimage.2009.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plis SM, Hjelm D, Salakhutdinov R, Allen EA, Bockholt HJ, Long JD, Johnson HJ, Paulsen J, Turner JA, Calhoun VD. Deep learning for neuroimaging: a validation study. Front. Neurosci. 8. 2014 doi: 10.3389/fnins.2014.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plitt M, Barnes KA, Martin A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. Neuroimage Clin. 2015;7:359–366. doi: 10.1016/j.nicl.2014.12.013. doi:10.1016/j.nicl.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polat F, Demirel SO, Kitis O, Simsek F, Haznedaroglu DI, Coburn K, Kumral E, Gonul AS. Computer based classification of MR scans in first time applicant Alzheimer patients. Curr Alzheimer Res. 2012;9:789–794. doi: 10.2174/156720512802455359. [DOI] [PubMed] [Google Scholar]
- Poldrack RA, Barch DM, Mitchell JP, Wager TD, Wagner AD, Devlin JT, Cumba C, Koyejo O, Milham MP. Toward open sharing of task-based {fMRI} data: the {OpenfMRI} project. Front. Neuroinform. 2013;7 doi: 10.3389/fninf.2013.00012. doi:10.3389/fninf.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Petersen SE. Recent progress and outstanding issues in motion correction in resting state fMRI. Neuroimage. 2015;105:536–551. doi: 10.1016/j.neuroimage.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad G, Joshi SH, Nir TM, Toga AW, Thompson PM. Brain connectivity and novel network measures for Alzheimer's disease classification. Neurobiol. Aging 36 Suppl. 2015;1:S121–31. doi: 10.1016/j.neurobiolaging.2014.04.037. doi:10.1016/j.neurobiolaging.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu A, Younes L, Miller MI, Csernansky JG. Parallel transport in diffeomorphisms distinguishes the time-dependent pattern of hippocampal surface deformation due to healthy aging and the dementia of the Alzheimer's type. Neuroimage. 2008;40:68–76. doi: 10.1016/j.neuroimage.2007.11.041. doi:10.1016/j.neuroimage.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raamana PR, Wen W, Kochan NA, Brodaty FL, Sachdev PS, Wang L, Beg MF. Novel ThickNet features for the discrimination of amnestic MCI subtypes. Neuroimage Clin. 2014;6:284–295. doi: 10.1016/j.nicl.2014.09.005. doi:10.1016/j.nicl.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu E, Ganeshan B, Shergill SS, Medford N, Chatwin C, Young RCD, Critchley HD. Grey-matter texture abnormalities and reduced hippocampal volume are distinguishing features of schizophrenia. Psychiatry Res. 2014;223:179–186. doi: 10.1016/j.pscychresns.2014.05.014. doi:10.1016/j.pscychresns.2014.05.014. [DOI] [PubMed] [Google Scholar]
- Retico A, Bosco P, Cerello P, Fiorina E, Chincarini A, Fantacci ME. Predictive Models Based on Support Vector Machines: Whole-Brain versus Regional Analysis of Structural MRI in the Alzheimer's Disease. J Neuroimaging. 2015;25:552–563. doi: 10.1111/jon.12163. doi: 10.1111/jon.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retico A, Tosetti M, Muratori F, Calderoni S. Neuroimaging-based methods for autism identification: a possible translational application? Funct. Neurol. 2014;29:231–239. [PMC free article] [PubMed] [Google Scholar]
- Retico A, Tosetti M, Muratori F, Calderoni S. Neuroimaging-based methods for autism identification: a possible translational application? Funct. Neurol. 2013;29:231–239. [PMC free article] [PubMed] [Google Scholar]
- Rice DP. The economic impact of schizophrenia. J. Clin. Psychiatry. 1999 [PubMed] [Google Scholar]
- Rosa MJ, Portugal L, Hahn T, Fallgatter AJ, Garrido MI, Shawe-Taylor J, Mourao-Miranda J. Sparse network-based models for patient classification using fMRI. Neuroimage. 2015;105:493–506. doi: 10.1016/j.neuroimage.2014.11.021. doi: 10.1016/j.neuroimage.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuncu MR, Van Leemput K. The relevance voxel machine (RVoxM): a self-tuning Bayesian model for informative image-based prediction. IEEE Trans Med Imaging. 2012;31:2290–2306. doi: 10.1109/TMI.2012.2216543. doi: 10.1109/TMI.2012.2216543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchet MD, Livermore EE, Iglesias JE, Glover GH, Gotlib IH. Subcortical volumes differentiate Major Depressive Disorder, Bipolar Disorder, and remitted Major Depressive Disorder. J Psychiatr Res. 2015;68:91–98. doi: 10.1016/j.jpsychires.2015.06.002. doi:10.1016/j.jpsychires.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Salvatore C, Battista P, Castiglioni I. Frontiers for the early diagnosis of AD by means of MRI brain imaging and Support Vector Machines. Curr Alzheimer Res. 2015a doi: 10.2174/1567205013666151116141705. [DOI] [PubMed] [Google Scholar]
- Salvatore C, Cerasa A, Battista P, Gilardi MC, Quattrone A, Castiglioni I. Magnetic resonance imaging biomarkers for the early diagnosis of Alzheimer's disease: a machine learning approach. Front Neurosci. 2015b;9:307. doi: 10.3389/fnins.2015.00307. doi:10.3389/fnins.2015.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarwate AD, Plis SM, Turner JA, Arbabshirani MR, Calhoun VD. Sharing privacy-sensitive access to neuroimaging and genetics data: a review and preliminary validation. Front. Neuroinform. 2014;8:35. doi: 10.3389/fninf.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JR, Hoexter MQ, Fujita A, Rohde LA. Evaluation of pattern recognition and feature extraction methods in ADHD prediction. Front Syst Neurosci. 2012;6:68. doi: 10.3389/fnsys.2012.00068. doi:10.3389/fnsys.2012.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JR, Hoexter MQ, Oliveira P.P. de M., Brammer MJ, Murphy D, Ecker C. Inter-regional cortical thickness correlations are associated with autistic symptoms: a machine-learning approach. J Psychiatr Res. 2013;47:453–459. doi: 10.1016/j.jpsychires.2012.11.017. doi:10.1016/j.jpsychires.2012.11.017. [DOI] [PubMed] [Google Scholar]
- Sato JR, Moll J, Green S, Deakin JFW, Thomaz CE, Zahn R. Machine learning algorithm accurately detects fMRI signature of vulnerability to major depression. Psychiatry Res. 2015;233:289–291. doi: 10.1016/j.pscychresns.2015.07.001. doi:10.1016/j.pscychresns.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack HG, Nieuwenhuis M, van Haren NEM, Abramovic L, Scheewe TW, Brouwer RM, Hulshoff Pol HE, Kahn RS. Can structural MRI aid in clinical classification? A machine learning study in two independent samples of patients with schizophrenia, bipolar disorder and healthy subjects. Neuroimage. 2014;84:299–306. doi: 10.1016/j.neuroimage.2013.08.053. doi:10.1016/j.neuroimage.2013.08.053. [DOI] [PubMed] [Google Scholar]
- Scott A, Courtney W, Wood D, la Garza R, Lane S, King M, Wang R, Roberts J, Turner JA, Calhoun VD. COINS: an innovative informatics and neuroimaging tool suite built for large heterogeneous datasets. Front. Neuroinform. 2011;5 doi: 10.3389/fninf.2011.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segovia F, Holt R, Spencer M, Górriz JM, Ramirez J, Puntonet CG, Phillips C, Chura L, Baron-Cohen S, Suckling J. Identifying endophenotypes of autism: a multivariate approach. Front. Comput. Neurosci. 2014;8 doi: 10.3389/fncom.2014.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrud-Clikeman M, Hooper SR, Hynd GW, Hern K, Presley R, Watson T. Prediction of group membership in developmental dyslexia, attention deficit hyperactivity disorder, and normal controls using brain morphometric analysis of magnetic resonance imaging. Arch Clin Neuropsychol. 1996;11:521–528. [PubMed] [Google Scholar]
- Serpa MH, Ou Y, Schaufelberger MS, Doshi J, Ferreira LK, Machado-Vieira R, Menezes PR, Scazufca M, Davatzikos C, Busatto GF, Zanetti MV. Neuroanatomical classification in a population-based sample of psychotic major depression and bipolar I disorder with 1 year of diagnostic stability. Biomed Res Int2014. 2014:706157. doi: 10.1155/2014/706157. doi: 10.1155/2014/706157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Wang L, Liu Y, Hu D. Discriminative analysis of resting-state functional connectivity patterns of schizophrenia using low dimensional embedding of fMRI. Neuroimage. 2010;49:3110–3121. doi: 10.1016/j.neuroimage.2009.11.011. doi:10.1016/j.neuroimage.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Yoshimoto J, Toki S, Takamura M, Yoshimura S, Okamoto Y, Yamawaki S, Doya K. Toward Probabilistic Diagnosis and Understanding of Depression Based on Functional MRI Data Analysis with Logistic Group LASSO. PLoS One. 2015;10:e0123524. doi: 10.1371/journal.pone.0123524. doi:10.1371/journal.pone.0123524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu GS, Asgarian N, Greiner R, Brown MRG. Kernel Principal Component Analysis for dimensionality reduction in fMRI-based diagnosis of ADHD. Front Syst Neurosci. 2012;6:74. doi: 10.3389/fnsys.2012.00074. doi:10.3389/fnsys.2012.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva RF, Castro E, Gupta CN, Cetin M, Arbabshirani M, Potluru VK, Plis SM, Calhoun VD. THE TENTH ANNUAL MLSP COMPETITION: SCHIZOPHRENIA CLASSIFICATION CHALLENGE The Mind Research Network , 1101 Yale Blvd ., Albuquerque , New Mexico 87106. IEEE INTERNATIONAL WORKSHOP ON MACHINE LEARNING FOR SIGNAL PROCESSING; Remis, France. 2014. [Google Scholar]
- Stonnington CM, Chu C, Kloppel S, Jack CR, Ashburner J, Frackowiak RSJ. Predicting clinical scores from magnetic resonance scans in Alzheimer's disease. Neuroimage. 2010;51:1405–1413. doi: 10.1016/j.neuroimage.2010.03.051. doi:10.1016/j.neuroimage.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L, Wang L, Shen H, Feng G, Hu D. Discriminative analysis of non-linear brain connectivity in schizophrenia: an fMRI Study. Front. Hum. Neurosci. 2013;7:702. doi: 10.3389/fnhum.2013.00702. doi:10.3389/fnhum.2013.00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Adali T, Pearlson GD, Calhoun VD. An {ICA}-based method for the identification of optimal {fMRI} features and components using combined group-discriminative techniques. Neuroimage. 2009;46:73–86. doi: 10.1016/j.neuroimage.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, He H, Liu J, Yu Q, Adali T, Pearlson G, Calhoun V. Three-Way {fMRI}-{DTI}-Methylation Data Fusion Based on {MCCA}+j{ICA} and Its Application to Schizophrenia. Eng. Med. Biol. Soc. 2012. EMBS 2012. 34th Annu. Int. Conf. IEEE. 2012 doi: 10.1109/EMBC.2012.6346519. [DOI] [PubMed] [Google Scholar]
- Sui J, He H, Pearlson GD, Adali T, Kiehl KA, Yu Q, Clark VP, Castro E, White T, Mueller BA. Three-way (N-way) fusion of brain imaging data based on mCCA+ jICA and its application to discriminating schizophrenia. Neuroimage. 2013a;66:119–132. doi: 10.1016/j.neuroimage.2012.10.051. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, He H, Yu Q, Chen J, Rogers J, Pearlson GD, Mayer AR, Bustillo JR, Canive J, Calhoun VD. Combination of Resting State fMRI, DTI, and sMRI Data to Discriminate Schizophrenia by N-way MCCA + jICA. Front. Hum. Neurosci. 2013b;7:235. doi: 10.3389/fnhum.2013.00235. doi:10.3389/fnhum.2013.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Pearlson G, Caprihan A, Adali T, Kiehl KA, Liu J, Yamamoto J, Calhoun VD. Discriminating schizophrenia and bipolar disorder by fusing fMRI and DTI in a multimodal CCA+ joint ICA model. Neuroimage. 2011;57:839–855. doi: 10.1016/j.neuroimage.2011.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Pearlson GD, Du Y, Yu Q, Jones TR, Chen J, Jiang T, Bustillo J, Calhoun VD. In Search of Multimodal Neuroimaging Biomarkers of Cognitive Deficits in Schizophrenia. Biol. Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk H-L, Lee S-W, Shen D, Initiative ADN. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct. Funct. 2013;220:841–859. doi: 10.1007/s00429-013-0687-3. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, van Erp TGM, Thompson PM, Bearden CE, Daley M, Kushan L, Hardt ME, Nuechterlein KH, Toga AW, Cannon TD. Elucidating a Magnetic Resonance Imaging-Based Neuroanatomic Biomarker for Psychosis: Classification Analysis Using Probabilistic Brain Atlas and Machine Learning Algorithms. Biol. Psychiatry. 2009;66:1055–1060. doi: 10.1016/j.biopsych.2009.07.019. doi:10.1016/j.biopsych.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermann B, Herr D, Schwindt W, Pfleiderer B. Multivariate classification of blood oxygen level-dependent fMRI data with diagnostic intention: A clinical perspective. Am. J. Neuroradiol. 2014;39:848–855. doi: 10.3174/ajnr.A3713. doi:10.3174/ajnr.A3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, Laufs H. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron. 2014;82:695–708. doi: 10.1016/j.neuron.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Takayanagi Y, Takahashi T, Orikabe L, Mozue Y, Kawasaki Y, Nakamura K, Sato Y, Itokawa M, Yamasue PL, Kasai K, Kurachi M, Okazaki Y, Suzuki M. Classification of first-episode schizophrenia patients and healthy subjects by automated MRI measures of regional brain volume and cortical thickness. PLoS One. 2011;6:1–10. doi: 10.1371/journal.pone.0021047. doi:10.1371/journal.pone.0021047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Holland D, Dale AM, Younes L, Miller MI. Baseline shape diffeomorphometry patterns of subcortical and ventricular structures in predicting conversion of mild cognitive impairment to Alzheimer's disease. J. Alzheimers Dis. 2015;44:599–611. doi: 10.3233/JAD-141605. doi:10.3233/JAD-141605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Holland D, Dale AM, Younes L, Miller MI. Shape abnormalities of subcortical and ventricular structures in mild cognitive impairment and Alzheimer's disease: detecting, quantifying, and predicting. Hum Brain Mapp. 2014;35:3701–3725. doi: 10.1002/hbm.22431. doi:10.1002/hbm.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Wang L, Cao F, Tan L. Identify schizophrenia using resting-state functional connectivity: an exploratory research and analysis. Biomed. Eng. Online. 2012;11:50. doi: 10.1186/1475-925X-11-50. doi:10.1186/1475-925X-ll-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangaro S, Amoroso N, Brescia M, Cavuoti S, Chincarini A, Errico R, Inglese P, Longo G, Maglietta R, Tateo A, Riccio G, Bellotti R. Feature Selection Based on Machine Learning in MRIs for Hippocampal Segmentation. Comput Math Methods Med. 2015;2015:814104. doi: 10.1155/2015/814104. doi: 10.1155/2015/814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognin S, Pettersson-Yeo W, Valli I, Hutton C, Woolley J, Allen P, McGuire P, Mechelli A. Using structural neuroimaging to make quantitative predictions of symptom progression in individuals at ultra-high risk for psychosis. Front Psychiatry. 2013;4:187. doi: 10.3389/fpsyt.2013.00187. doi:10.3389/fpsyt.2013.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T, Wolz R, Gao Q, Guerrero R, Hajnal JV, Rueckert D. Multiple instance learning for classification of dementia in brain MRI. Med Image Anal. 2014;18:808–818. doi: 10.1016/j.media.2014.04.006. doi:10.1016/j.media.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Turner JA. The rise of large-scale imaging studies in psychiatry. Gigascience. 2014;3:29. doi: 10.1186/2047-217X-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Menon V, Young CB, Ryali S, Chen T, Khouzam A, Minshew NJ, Hardan AY. Multivariate searchlight classification of structural magnetic resonance imaging in children and adolescents with autism. Biol. Psychiatry. 2011;70:833–841. doi: 10.1016/j.biopsych.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin LQ, Supekar K, Lynch CJ, Khouzam A, Phillips J, Feinstein C, Ryali S, Menon V. Salience network–based classification and prediction of symptom severity in children with autism. JAMA psychiatry. 2013;70:869–879. doi: 10.1001/jamapsychiatry.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, Consortium W-MHCP. The WU-Minn human connectome project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri P, Gunter JL, Senjem ML, Whitwell JL, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR. Alzheimer's disease diagnosis in individual subjects using structural MR images: validation studies. Neuroimage. 2008;39:1186–1197. doi: 10.1016/j.neuroimage.2007.09.073. doi:10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman A, Whitford TJ, Westin C-F, Golland P, Kubicki M. Whole brain resting state functional connectivity abnormalities in schizophrenia. Schizophr. Res. 2012;139:7–12. doi: 10.1016/j.schres.2012.04.021. doi:10.1016/j.schres.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara VM, Damaraju E, Mayer AB, Miller R, Cetin MS, Calhoun V. The impact of data preprocessing in traumatic brain injury detection using functional magnetic resonance imaging. Engineering in Medicine and Biology Society (EMBC), 2015 37th Annual International Conference of the IEEE. 2015:5432–5435. doi: 10.1109/EMBC.2015.7319620. [DOI] [PubMed] [Google Scholar]
- Veronese E, Castellani U, Peruzzo D, Bellani M, Brambilla P. Machine learning approaches: from theory to application in schizophrenia. Comput Math Methods Med. 2013;2013:867924. doi: 10.1155/2013/867924. doi:10.1155/2013/867924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Beg F, Ratnanather T, Ceritoglu C, Younes L, Morris JC, Csernansky JG, Miller MI. Large deformation diffeomorphism and momentum based hippocampal shape discrimination in dementia of the Alzheimer type. IEEE Trans Med Imaging. 2007;26:462–470. doi: 10.1109/TMI.2005.853923. doi: 10.1109/TMI.2005.853923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Jiao Y, Tang T, Wang H, Lu Z. Altered regional homogeneity patterns in adults with attention-deficit hyperactivity disorder. Eur J Radiol. 2013;82:1552–1557. doi: 10.1016/j.ejrad.2013.04.009. doi:10.1016/j.ejrad.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Wang Y, Fan Y, Bhatt P, Davatzikos C. High-dimensional pattern regression using machine learning: from medical images to continuous clinical variables. Neuroimage. 2010;50:1519–1535. doi: 10.1016/j.neuroimage.2009.12.092. doi:10.1016/j.neuroimage.2009.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kessler D, Scott C, Angstadt M, Sripada C. Disease prediction based on functional connectomes using a scalable and spatially-informed support vector machine. Neuroimage. 2014;96:183–202. doi: 10.1016/j.neuroimage.2014.03.067. doi:10.1016/j.neuroimage.2014.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee C-Y, Wang L, Shi F, Yap P-T, Shen D. Diagnosis of autism spectrum disorders using regional and interregional morphological features. Hum. Brain Mapp. 2014;35:3414–3430. doi: 10.1002/hbm.22411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee C-Y, Yap P-T, Li W, Denny K, Browndyke JN, Potter GG, Welsh-Bohmer KA, Wang L, Shen D. Enriched white matter connectivity networks for accurate identification of MCI patients. Neuroimage. 2011;54:1812–1822. doi: 10.1016/j.neuroimage.2010.10.026. doi: 10.1016/j.neuroimage.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee C-Y, Yap P-T, Shen D. Prediction of Alzheimer's disease and mild cognitive impairment using cortical morphological patterns. Hum Brain Mapp. 2013;34:3411–3425. doi: 10.1002/hbm.22156. doi:10.1002/hbm.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee C-Y, Yap P-T, Zhang D, Denny K, Browndyke JN, Potter GG, Welsh-Bohmer KA, Wang L, Shen D. Identification of MCI individuals using structural and functional connectivity networks. Neuroimage. 2012;59:2045–2056. doi: 10.1016/j.neuroimage.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M, Qin J, Yan R, Li H, Yao Z, Lu Q. Identifying major depressive disorder using Hurst exponent of resting-state brain networks. Psychiatry Res. 2013;214:306–312. doi: 10.1016/j.pscychresns.2013.09.008. doi:10.1016/j.pscychresns.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Wing L. The autistic spectrum. Lancet. 1997;350:1761–1766. doi: 10.1016/S0140-6736(97)09218-0. [DOI] [PubMed] [Google Scholar]
- Wolf H, Grunwald M, Kruggel F, Riedel-Heller SG, Angerhofer S, Hojjatoleslami A, Hensel A, Arendt T, Gertz H. Hippocampal volume discriminates between normal cognition; questionable and mild dementia in the elderly. Neurobiol. Aging. 2001;22:177–186. doi: 10.1016/s0197-4580(00)00238-4. [DOI] [PubMed] [Google Scholar]
- Wolfers T, Buitelaar JK, Beckmann CF, Franke B, Marquand AF. From estimating activation locality to predicting disorder: A review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neurosci. Biobehav. Rev. 2015;57:328–349. doi: 10.1016/j.neubiorev.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, Soininen H, Lotjonen J. Multi-method analysis of MRI images in early diagnostics of Alzheimer's disease. PLoS One. 2011;6:e25446. doi: 10.1371/journal.pone.0025446. doi:10.1371/journal.pone.0025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Li J, Ayutyanont N, Protas H, Jagust W, Fleisher A, Reiman E, Yao L, Chen K. The receiver operational characteristic for binary classification with multiple indices and its application to the neuroimaging study of Alzheimer's disease. IEEE/ACM Trans Comput Biol Bioinform. 2013;10:173–180. doi: 10.1109/TCBB.2012.141. doi:10.1109/TCBB.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Wu X, Chen K, Yao L. Multi-modality sparse representation-based classification for Alzheimer's disease and mild cognitive impairment. Comput Methods Programs Biomed. 2015;122:182–190. doi: 10.1016/j.cmpb.2015.08.004. doi:10.1016/j.cmpb.2015.08.004. [DOI] [PubMed] [Google Scholar]
- Yang H, Liu J, Sui J, Pearlson G, Calhoun VD. A Hybrid Machine Learning Method for Fusing fMRI and Genetic Data: Combining both Improves Classification of Schizophrenia. Front. Hum. Neurosci. 2010;4:192. doi: 10.3389/fnhum.2010.00192. doi:10.3389/fnhum.2010.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S-T, Lee J-D, Chang T-C, Huang C-H, Wang J-J, Hsu W-C, Chan H-L, Wai Y-Y, Li K-Y. Discrimination between Alzheimer's disease and mild cognitive impairment using SOM and PSO-SVM. Comput Math Methods Med. 2013;2013:253670. doi: 10.1155/2013/253670. doi:10.1155/2013/253670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Lui RLM, Gao J-H, Chan TF, Yau S-T, Sperling RA, Huang X. Independent component analysis-based classification of Alzheimer's disease MRI data. J. Alzheimers Dis. 2011;24:775–783. doi: 10.3233/JAD-2011-101371. doi:10.3233/JAD-2011-101371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Nguyen DV, McVay LM, Deramo P, Minzenberg MJ, Ragland JD, Niendham T, Solomon M, Carter CS. Automated classification of fMRI during cognitive control identifies more severely disorganized subjects with schizophrenia. Schizophr. Res. 2012;135:28–33. doi: 10.1016/j.schres.2012.01.001. doi:10.1016/j.schres.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Tamir D, Minzenberg MJ, Ragland JD, Ursu S, Carter CS. Multivariate Pattern Analysis of Functional Magnetic Resonance Imaging Data Reveals Deficits in Distributed Representations in Schizophrenia. Biol. Psychiatry. 2008;64:1035–1041. doi: 10.1016/j.biopsych.2008.07.025. doi:10.1016/j.biopsych.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Modat M, Cardoso MJ, Mendelson A, Cash D, Ourselin S. Accurate multimodal probabilistic prediction of conversion to Alzheimer's disease in patients with mild cognitive impairment. Neuroimage Clin. 2013;2:735–745. doi: 10.1016/j.nicl.2013.05.004. doi:10.1016/j.nicl.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K, Du A-T, Kramer J, Rosen FL, Miller B, Weiner M, Schuff N. Patterns of structural complexity in Alzheimer's disease and frontotemporal dementia. Hum Brain Mapp. 2009;30:1667–1677. doi: 10.1002/hbm.20632. doi:10.1002/hbm.20632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Liu Y, Shen D. Graph-guided joint prediction of class label and clinical scores for the Alzheimer's disease. Brain Struct Funct. 2015 doi: 10.1007/s00429-015-1132-6. doi:10.1007/s00429-015-l 132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Liu Y, Thung K-H, Shen D. Multi-task linear programming discriminant analysis for the identification of progressive MCI individuals. PLoS One. 2014;9:e96458. doi: 10.1371/journal.pone.0096458. doi:10.1371/journal.pone.0096458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Shen H, Zeng L-L, Ma Q, Hu D. Convergent and divergent functional connectivity patterns in schizophrenia and depression. PLoS One. 2013a;8:e68250. doi: 10.1371/journal.pone.0068250. doi:10.1371/journal.pone.0068250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Shen H, Zhang H, Zeng L-L, Xue Z, Hu D. Functional connectivity-based signatures of schizophrenia revealed by multiclass pattern analysis of resting-state fMRI from schizophrenic patients and their healthy siblings. Biomed. Eng. Online. 2013b;12:10. doi: 10.1186/1475-925X-12-10. doi:10.1186/1475-925X-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue C, Wu D, Bai F, Shi Y, Yu H, Xie C, Zhang Z. State-based functional connectivity changes associate with cognitive decline in amnestic mild cognitive impairment subjects. Behav. Brain Res. 2015;288:94–102. doi: 10.1016/j.bbr.2015.04.013. doi:10.1016/j.bbr.2015.04.013. [DOI] [PubMed] [Google Scholar]
- Yun HJ, Kwak K, Lee J-M. Multimodal Discrimination of Alzheimer's Disease Based on Regional Cortical Atrophy and Hypometabolism. PLoS One. 2015;10:e0129250. doi: 10.1371/journal.pone.0129250. doi:10.1371/journal.pone.0129250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharia M, Chowdhury M, Franklin MJ, Shenker S, Stoica I. Spark: cluster computing with working sets. Proceedings of the 2nd USENIX Conference on Hot Topics in Cloud Computing. 2010:10. [Google Scholar]
- Zanetti MV, Schaufelberger MS, Doshi J, Ou Y, Ferreira LK, Menezes PR, Scazufca M, Davatzikos C, Busatto GF. Neuroanatomical pattern classification in a population-based sample of first-episode schizophrenia. Prog. Neuro-Psychopharmacology Biol. Psychiatry. 2013;43:116–125. doi: 10.1016/j.pnpbp.2012.12.005. doi:10.1016/j.pnpbp.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Zarei M, Damoiseaux JS, Morgese C, Beckmann CF, Smith SM, Matthews PM, Scheltens P, Rombouts SARB, Barkhof F. Regional white matter integrity differentiates between vascular dementia and Alzheimer disease. Stroke. 2009;40:773–779. doi: 10.1161/STROKEAHA.108.530832. doi: 10.1161/STROKEAHA.108.530832. [DOI] [PubMed] [Google Scholar]
- Zarogianni E, Moorhead TWJ, Lawrie SM. Towards the identification of imaging biomarkers in schizophrenia, using multivariate pattern classification at a single-subject level. Neuroimage Clin. 2013;3:279–289. doi: 10.1016/j.nicl.2013.09.003. doi:10.1016/j.nicl.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L-L, Shen H, Liu L, Hu D. Unsupervised classification of major depression using functional connectivity MRI. Hum Brain Mapp. 2014;35:1630–1641. doi: 10.1002/hbm.22278. doi:10.1002/hbm.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D. Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer's disease. Neuroimage. 2012a;59:895–907. doi: 10.1016/j.neuroimage.2011.09.069. doi: 10.1016/j.neuroimage.2011.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Shen D. Predicting future clinical changes of MCI patients using longitudinal and multimodal biomarkers. PLoS One. 2012b;7:e33182. doi: 10.1371/journal.pone.0033182. doi:10.1371/journal.pone.0033182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Wang Y, Zhou L, Yuan H, Shen D, Initiative ADN. Multimodal classification of Alzheimer's disease and mild cognitive impairment. Neuroimage. 2011;55:856–867. doi: 10.1016/j.neuroimage.2011.01.008. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Davatzikos C. Optimally-Discriminative Voxel-Based Morphometry significantly increases the ability to detect group differences in schizophrenia, mild cognitive impairment, and Alzheimer's disease. Neuroimage. 2013;79:94–110. doi: 10.1016/j.neuroimage.2013.04.063. doi:10.1016/j.neuroimage.2013.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Davatzikos C. ODVBA: optimally-discriminative voxel-based analysis. IEEE Trans Med Imaging. 2011;30:1441–1454. doi: 10.1109/TMI.2011.2114362. doi: 10.1109/TMI.2011.2114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Dong Z, Phillips P, Wang S, Ji G, Yang J, Yuan T-F. Detection of subjects and brain regions related to Alzheimer's disease using 3D MRI scans based on eigenbrain and machine learning. Front Comput Neurosci. 2015;9:66. doi: 10.3389/fncom.2015.00066. doi:10.3389/fncom.2015.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Huang H, Shen D. Integrative analysis of multi-dimensional imaging genomics data for Alzheimer's disease prediction. Front Aging Neurosci. 2014;6:260. doi: 10.3389/fnagi.2014.00260. doi: 10.3389/fnagi.2014.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Greicius MD, Gennatas ED, Growdon ME, Jang JY, Rabinovici GD, Kramer JH, Weiner M, Miller BL, Seeley WW. Divergent network connectivity changes in behavioural variant frontotemporal dementia and Alzheimer's disease. Brain. 2010;133:1352–1367. doi: 10.1093/brain/awq075. doi:10.1093/brain/awq075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yu F, Duong T. Multiparametric MRI characterization and prediction in autism spectrum disorder using graph theory and machine learning. PLoS One. 2014;9:e90405. doi: 10.1371/journal.pone.0090405. doi:10.1371/journal.pone.0090405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C-Z, Zang Y-F, Cao Q-J, Yan C-G, He Y, Jiang T-Z, Sui M-Q, Wang Y-F. Fisher discriminative analysis of resting-state brain function for attention-deficit/hyperactivity disorder. Neuroimage. 2008;40:110–120. doi: 10.1016/j.neuroimage.2007.11.029. [DOI] [PubMed] [Google Scholar]
- Zou H, Hastie T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B (Statistical Methodol. 2005;67:301–320. [Google Scholar]
- Zu C, Jie B, Liu M, Chen S, Shen D, Zhang D. Label-aligned multi-task feature learning for multimodal classification of Alzheimer's disease and mild cognitive impairment. Brain Imaging Behav. 2015 doi: 10.1007/s11682-015-9480-7. doi:10.1007/sl 1682-015-9480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]