Abstract
Deficiency of the TCA cycle enzyme Succinyl-CoA Synthetase/Ligase (SCS), due to pathogenic variants in subunits encoded by SUCLG1 and SUCLA2 causes mitochondrial encephalomyopathy, methylmalonic acidemia, and mitochondrial DNA (mtDNA) depletion. In this study, we report an 11 year old patient who presented with truncal ataxia, chorea, hypotonia, bilateral sensorineural hearing loss and preserved cognition. Whole exome sequencing identified a heterozygous known pathogenic variant and a heterozygous novel missense variant of uncertain clinical significance (VUS) in SUCLG1. To validate the suspected pathogenicity of the novel VUS, molecular and biochemical analyses were performed using primary skin fibroblasts from the patient. The patient’s cells lack the SUCLG1 protein, with significantly reduced levels of SUCLA2 and SUCLG2 protein. This leads to essentially undetectable SCS enzyme activity, mtDNA depletion, and cellular respiration defects. These abnormal phenotypes are rescued upon ectopic expression of wild-type SUCLG1 in the patient’s fibroblasts, thus functionally confirming the pathogenic nature of the SUCLG1 VUS identified in this patient and expanding the phenotypic spectrum for SUCLG1 deficiency.
Keywords: Mitochondria, Mitochondrial DNA depletion, TCA cycle, Succinyl-CoA Synthetase, Methylmalonic aciduria
1. Introduction
Succinyl-Co-A ligase/synthetase (SCS) is a component of citric acid cycle that converts succinyl-Co-A to succinate, coupled to the phosphorylation of ADP/GDP. In eukaryotes and gram-positive bacteria, SCS exists as a heterodimer consisting of an α-subunit, SUCLG1, and one of two β-subunit isoforms. The βisoforms, SUCLA2 and SUCLG2, determine the specificity of the SCS complex for ADP or GDP, respectively [1]. The expression pattern of SUCLG1 is ubiquitous, while SUCLA2 is predominantly expressed in brain, heart, and muscle and SUCLG2 is primarily restricted to liver and kidney [2]. In humans, mutations in SUCLG1 and SUCLA2 are reported to cause mitochondrial encephalomyopathy with mitochondrial DNA (mtDNA) depletion [3].
Over the last few years a number of genes responsible for mtDNA depletion syndromes have been identified and the spectrum of phenotypes related to mtDNA depletion further delineated [3]. Mitochondrial DNA depletion syndromes are defined by a global or tissue-specific reduction in mtDNA copy number. These disorders are clinically heterogeneous and exhibit autosomal recessive inheritance, with a broad spectrum of clinical features including combinations of infantile/childhood encephalopathy with severe intellectual disability, myopathy, cardiomyopathy and hepatopathy [4]. Nuclear encoded genes associated with mtDNA depletion syndromes include genes required for mtDNA replication (e.g., POLG, TWINKLE), regulation of mitochondrial nucleotide pools (e.g., DGUOK, TP, TK2, RRM2B), and genes with poorly defined functions related to mtDNA maintenance, including, MPV17 and, the subunits of SCS (SUCLG1, SUCLA2) [5]. Recently two more genes, FBXL4 [6, 7] and ABAT have been associated with mtDNA depletion syndromes, with ABAT (GABA transaminase) demonstrated to be required for normal mitochondrial nucleoside metabolism [8].
Multiple patients with mutations in either SUCLG1 or SUCLA2 have been previously reported (21 and 50, respectively) [9, 10], but no patients with pathogenic variants in SUCLG2 have been reported. The clinical phenotypes of SUCLG1 patients are more severe when compared to the SUCLA2 patients [9]. Patients with SUCLA2 deficiency typically present with encephalopathy, hearing loss, hypotonia with mtDNA depletion, and mild methylmalonic acidemia [9]. Patients with mutations in SUCLG1 often share clinical problems with SUCLA2 patients, but also can exhibit hepatopathy and/or hypertrophic cardiomyopathy [9]. In a recently reported case series, mean survival for 50 SUCLA2 patients was 20 years. In contrast, mean survival was 20 months for 21 SUCLG1 patients [9]. In this report, we present an 11-year-old patient with a history of hearing loss, chorea, hypotonia, mild MMA and preserved cognition, for which two heterozygous missense variants in SUCLG1, one known pathogenic variant and one variant of uncertain clinical significance (VUS) were identified by whole exome sequencing. Molecular, biochemical, and cellular respiration analyses of the patient’s fibroblasts demonstrate abnormalities that are fully rescued by ectopic expression of wild type SUCLG1 cDNA, validating the pathogenicity of the SUCLG1 variants. This patient’s features are compared to previously reported SUCLG1 cases, expanding the phenotypic spectrum of SUCLG1-related disease.
2. Materials and methods
2.1. Participants
The proband and mother provided informed consent to be enrolled in the study. The study was approved by Institutional Review Board (IRB) of BCM.
2.2. Cell culture
Control and patient fibroblasts were grown in culture medium (DMEM with 25 mM Glucose, 5 mM Pyruvate, 10% FBS, 1% Antibiotic-Antimycotic and 50 µg/mL Uridine). SUCLG1 cDNA was cloned into pLenti6.3 destination vector and fibroblasts were transduced with the SUCLG1-lentivirus as previously described [11]. The control fibroblasts lines were established from five different healthy individuals as previously described [11]. Transduced cells were put under Blasticidin selection (5 µg/mL) for a period of 10 days before using them for analysis.
2.3. mtDNA quantification
mtDNA content of the fibroblasts was analyzed using real-time qPCR method as described before [11]. The beta 2 microglobulin gene (B2M) was used as the nuclear gene (nDNA) normalizer for the calculation of mtDNA/nDNA ratio. The ND1 region of human mtDNA was amplified using forward primer 5’ GTCAACCTCGCTTCCCCACCCT 3’ and reverse primer 5’ TCCTGCGAATAGGCTTCCGGCT’, giving a fragment of 108bp; A fragment of human beta 2 microglobulin gene was amplified using forward primer 5’ CGACGGGAGGGTCGGGACAA 3’ and reverse primer 5’ GCCCCGCGAAAGAGCGGAAG 3’ giving a fragment of 118bp. The real time PCR reaction was performed in triplicate for both mtDNA and B2M gene for each DNA sample. The 15 µl PCR reaction contained 1× iTaq™ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA), 500nM of each primer, and 10ng of total genomic DNA extract. Real-time PCR conditions were 2 min at 50°C, 10 min at 95°C, followed by 45 cycles of 15 seconds of denaturation at 95°C, and 60 seconds of annealing/extension at 60°C. Real-time qPCR analysis was performed and fluorescent signal intensity was recorded and analyzed on The Bio-Rad iCycler Sequence Detection System. Dissociation curves for the amplicons were generated after each run to confirm that the fluorescent signals were not attributable to nonspecific signals (primer-dimers). The mtDNA content (mtDNA/B2M ratio) was calculated using the formula: mtDNA content = 1/2ΔCt, where ΔCt=CtmtDNA−CtB2M.
2.4. Cellular respiration assay
XF24 extracellular flux analyzer from Seahorse Biosciences was used to measure the rates of fibroblasts oxygen consumption, as previously described [11]. Cells were plated the previous day of experiment on the XF24 cell culture microplates at a density of 60,000 cells per well. XF24 cartridge was equilibrated with the calibration solution overnight at 37°C. XF assay media (5 mM glucose, 2 mM Pyruvate, in DMEM (Seahorse Biosciences) was prepared and pH adjusted to 7.0 on the day of the experiment. XF assay media was used to prepare cellular stress reagents, 500 nM Oligomycin, 500 nM FCCP, 100 nM Antimycin and 100 nM Rotenone (final concentration). All the reagents were loaded in the ports as suggested by Seahorse biosciences. Oxygen consumption rates were measured for 3 min with 3 min of mixing and 2 min of waiting period. After the assay was completed, cells in each well were counted using ViCell cell counter and the counts were used to normalize the OCR rates. OCR was expressed as nmoles of oxygen/min/1000 cells.
2.5. Western blotting
Whole cell lysates from fibroblasts were prepared in RIPA buffer (50 mM TrisHCl pH7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.1% SDS). After centrifugation of the lysate (10000 g, 4 min), soluble proteins were isolated in the supernatant, and protein concentration was determined according to the Bradford-Lowry method. Samples were mixed with equal volume of 2× SDS-PAGE sample loading buffer (0.5 M Tris-Cl, pH6.8, 10%SDS, 50% glycerol, 2% β-mercaptoethanol, and 5% bromophenol blue). Protein samples were separated on a 10% SDS-polyacrylamide gradient mini-gel. Proteins were transferred electrophoretically to 0.45 "m of polyvinylidine difluoride membranes for 1 h 15 min at 100 V. Membranes were blocked for 3h in 5% milk-PBS and incubated overnight in 1:10000 rabbit SUCLA2 polyclonal antibody, 1:10000 rabbit SUCLG1 and SUCLG2 polyclonal antibody, 1:500 rabbit ND5 antibody ((Gene Tex), 1:1000 mouse monoclonal SDHA antibody (Mitosciences), 1:3000 Citrate synthase antibody (AbCam), and 1:100000 mouse monoclonal GAPDH antibody for loading control. After three washes with PBS −0.05% Tween 20, the membranes were incubated for 2 h with horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse (Bio-Rad) diluted in 5% milk-PBS. This secondary antibody was detected using the chemiluminescent ECL Plus reagent (Millipore) and exposing the membrane to x-ray film.
2.6. Succinyl-CoA synthetase activity assay
Succinyl-CoA synthetase (SCS) activity was measured in whole cell lysates from fibroblasts in the direction of succinate to succinyl-CoA reaction. Assays of A-SCS and G-SCS activities were performed at 30°C using a modification of the procedure of Alarcon et al [13]. The complete assay mixture in a volume of 175 µl contained 50 mM potassium phosphate, pH 7.2, 10 mM MgCl2, 0.2 mM succinyl-CoA, 2 mM ADP (for A-SCS) or 1 mM GDP (for G-SCS), and 0.2 mM DTNB. The reactions were initiated by adding succinyl-CoA and DTNB in quick succession to the above mixture along with cell lysates containing 5µg of protein. Rates were corrected by subtracting the rate observed in the absence of ADP or GDP. The formation of thionitrobenzoate was followed at 412 nm. Controls demonstrated that the observed succinyl-CoA synthetase activities were dependent on magnesium, succinyl-CoA, and nucleoside diphosphate. All activities were calculated as nmoles/min/mg protein, and expressed as a percentage of control activity.
3. Results
3.1. Case Report
The subject of this study was an 11-year-old female who exhibited normal development during the first 12 months of life, but was observed to have truncal ataxia and chorea upon first walking at 14 months of age. Subsequently, bilateral sensorineural hearing loss was detected in the presence of speech delay during the second year of life, and generalized hypotonia in the context of gross motor delay was appreciated on physical exam. At 11 years of age, she has a cochlear implant and demonstrates average academic skills with appropriate accommodations for hearing impairment. She is able to ambulate for short distances using a walker and orthotics, but requires a wheelchair for longer distances. She has had a normal MRI of the brain and normal echocardiogram. Notable laboratory evaluations include sustained elevations of plasma methylmalonic acid (MMA) (0.9–2.8 mmol/L with normal range 0.09–0.32 mmol/L) and urine MMA (15–16.9 mmol/mol creatinine with normal range 0.3–1.9 mmol/mol creatinine). Plasma lactate levels were normal on three separate occasions. Examination of skeletal muscle biopsy revealed histopathological features mildly suggestive of mitochondrial myopathy: 1) a few fibers with mildly increased subsarcolemmal oxidative activity; 2) evidence of abnormal substrate utilization, with mild fat accumulation by histochemistry and electron microscopy; 3) type I fiber predominance; 4) occasional subsarcolemmal mitochondrial aggregates on electron microscopy. Analysis of electron transport chain activities from muscle biopsy revealed no reductions that meet modified Walker criteria (Complex I-277 nmoles/min/mg protein which is 116% of control mean and 86% normalized to citrate synthase (CS) with normal range 239+82.5, complex II-7.18 nmoles/min/mg protein which is 99% of control mean and 74% normalized to CS with normal range 7.22+1.32, complex I+III – 46.8 nmoles/min/mg protein which is 57% of control mean and 42% normalized to CS with normal range 82.2+16.2, Complex I+III+R – 21.64 nmoles/min/mg protein which is 58% of control mean and 43% normalized to CS with normal range 37.0+7.6, Complex II+III- 4.62 nmoles/min/mg protein which is 89% of control mean and 66% normalized to CS with normal range 5.21+2.63, Complex IV – 33.9 nmoles/min/mg protein which is 139% of control mean and 103% normalized to CS with normal range 24.4+9.9, and citrate synthase – 326 nmoles/min/mg protein which is 135% of control mean). Quantitation of mtDNA of two separate samples from muscle biopsy revealed mtDNA content level of 103% and 72% of mean age-matched control level, respectively. Clinical whole exome sequencing identified two heterozygous variants in SUCLG1: c.40A>T (p.M14L) and c.635A>G (p.Q212R). Analysis of DNA from the patient’s mother identified the c.40A>T (p.M14L) variant but not the c.635A>G (p.Q212R) variant (the patient’s father is deceased and no sample was available for analysis), suggesting that these two variants are in trans configuration. While a de novo mutation in cis with the identified mutation is not excluded, the absence of enzyme activity is most consistent with a trans configuration.
3.2. Loss of SCS activity, mtDNA depletion, and cellular respiration defects in fibroblasts
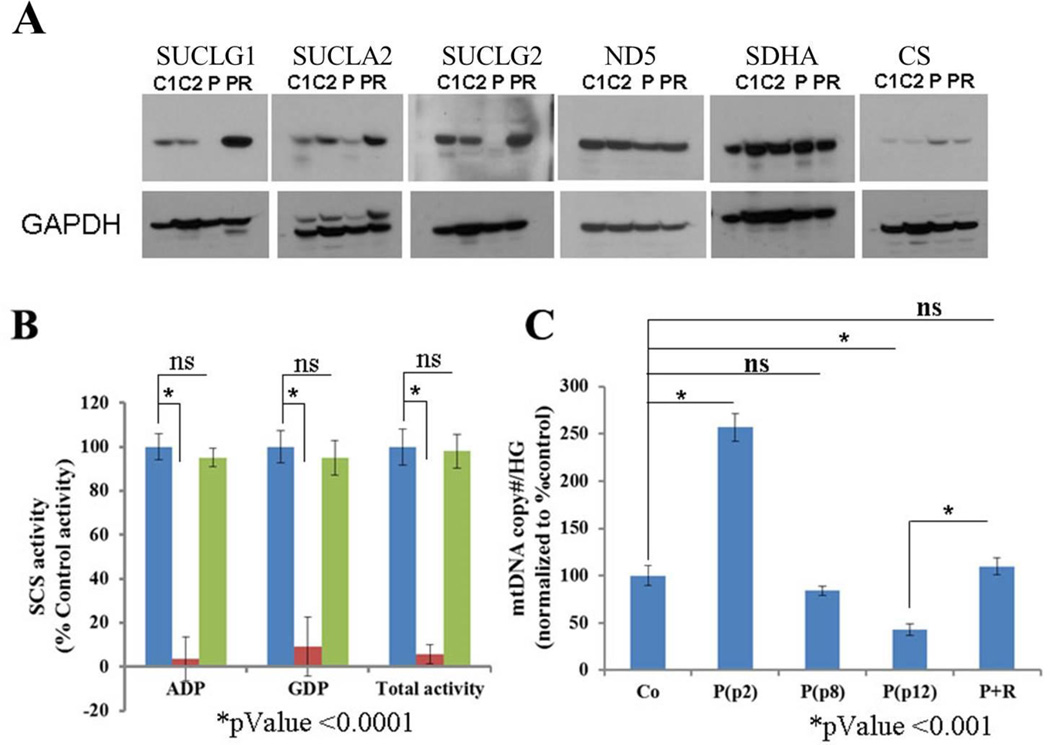
Western blot analysis of the patient’s fibroblasts demonstrates a lack of detectable SUCLG1 protein, suggesting that the identified missense variants may cause protein misfolding and instability (Figure 1A). Additionally, stable expression of both β subunits of SCS complex, SUCLA2 and SUCLG2, is absent or significantly reduced in the patient fibroblasts (Figure 1A). Levels of representative subunits from ETC complex I and II, (ND5 and SDHA, respectively) are not significantly altered, while Citrate synthase (CS) appears to be slightly elevated in the patient’s fibroblasts (Figure 1A). SCS enzyme activity was measured in control and patient fibroblast lysates. The patient’s cells exhibit profound enzymatic deficiency for total SCS activity, including both ADP-specific and GDP-specific SCS activity (Figure 1B, Table S1). Analysis of relative mtDNA copy number by qPCR in the patient’s fibroblasts show significantly elevated mtDNA (~2.5 fold compared to controls) at early cell passage (P2) that progresses to mtDNA depletion (~45%) with later passage (P12) (Figure 1C, Table S2).
Figure 1. Loss of SUCLG1 results in loss of SCS components, SCS activity and mtDNA depletion.
(A) Western blot analysis of fibroblasts cell lysates for SCS enzyme complex components (SUCLG1, SUCLA2, & SUCLG2), selected ETC subunits (ND5, SDHA) and TCA cycle enzyme, citrate synthase (CS). C1, C2 = controls; P = patient; PR = patient fibroblast cell line rescued by ectopic expression of wild type SUCLG1 cDNA. (B) ADP-specific, GDP-specific, and Total SCS (ADP-specific + GDP-specific) activities were measured for control (blue), patient (red), and patient + rescue (green) fibroblasts. (C) mtDNA relative copy number analysis by qPCR. Patient fibroblasts exhibit mtDNA depletion with multiple passages when compared to control (Co) fibroblasts and patient fibroblasts rescued (P+R) by ectopic expression of wild type SUCLG1 cDNA.
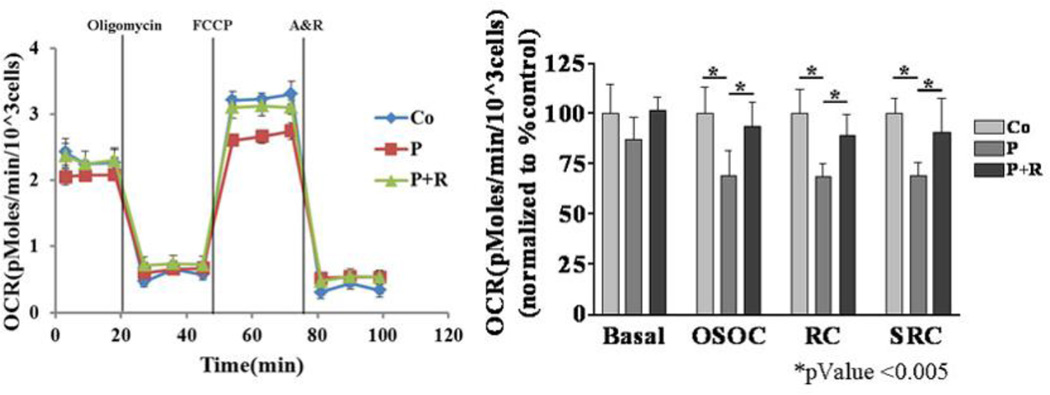
Analysis of cellular respiration revealed evidence of a respiratory deficiency (Figure 2, Table S3). Patient and control cells exhibit similar basal respiration rates; while there is a trend of the patient’s cells having a lower basal respiration rate, it is not statistically different from controls. The patient’s cells show a decreased response to the ATP inhibitor oligomycin, indicating altered coupling efficiency [14]. Addition of FCCP functionally uncouples mitochondrial respiration from ADP phosphorylation, allowing cells to reach full respiratory capacity. Compared to the control cells, the patient’s fibroblasts demonstrate significantly lower coupled respiratory capacity and spare respiratory capacity. These analyses demonstrate that the SUCLG1-deficient fibroblasts have a SCS deficiency, mtDNA depletion, and a significant defect in mitochondrial respiration.
Figure 2. Cellular respiration defect in SUCLG1 patient fibroblasts is dependent on SUCLG1 deficiency.
Cellular respiration analysis demonstrates reduced respiratory capacity in patient fibroblasts (P) compared to controls (Co) (n = 5) that is completely restored with ectopic expression of wild type SUCLG1 in patient cells (“P+R”). Experiment was done in triplicate. Error bars indicate standard deviation. “OCR” = oxygen consumption rate. Oligomycin = complex V (ATP synthase) inhibitor; FCCP = proton ionophore uncoupler; Antimycin & Rotenone (A&R) = inhibitors of ETC complex III and complex I, respectively.
3.3. Restoration of SCS activity, mtDNA content and cellular respiration with ectopic expression of wild type SUCLG1
SCS deficient fibroblasts transduced with lentivirus packaged with a wild type cDNA clone of SUCLG1 show restoration of SUCLG1 protein, as well as SUCLA2 and SUCLG2 protein levels similar to control cells (Figure 1A). Both ADP and GDP-specific SCS activities are completely restored in the patient’s cells with ectopic expression of SUCLG1 (Figure 1B). In fibroblasts with rescue construct there was restoration of mtDNA content comparable to control cells (Figure 1C). Finally, the cellular respiration defects observed in the SCS deficient fibroblasts were completely reversed with the ectopic expression of SUCLG1 (Figure 2). The rescue of the abnormal cellular phenotypes with ectopic SUCLG1 expression functionally validates the SUCLG1 missense VUS identified in this patient as a loss of function allele.
4. Discussion
To date, twenty five patients with mutations in SUCLG1, including the patient in this study, have been reported [9, 10, 15–23]. The age of onset for many of these patients (12/25) is at birth. Only two patients, P20 [8] and P25 (our patient) became symptomatic beyond one year of age. Out of twenty five patients, thirteen are female and twelve are male, indicating there is no sex bias. Only eleven patients were alive at the time of the reports, with the mortality rate being 56%. Some signs and symptoms that are consistent among the patients include hypotonia (25/25), elevated blood lactate (21/24), mildly elevated methylmalonic acid (21/21), and EEG abnormalities (17/19) (Table 1). The majority of the patients required respiratory support (14/23) and some had hearing loss (8/22) and multiple congenital abnormalities (4/25) (Table 1). Patient 5 had renal fusion abnormalities and hypospadias [20]. Patient 6 had a persistent ductus arteriosus with a permanent left to right shut, right ventricular hypertrophy and an interrupted aortic arch [20]. Patient 13 has asymmetrical IUGR, craniofacial dysmorphisms, and a right single palmar crease [19], and patient 16 had a cleft lip with clefting of the anterior hard palate and a coarctation of aorta [15]. The methylmalonic acid elevation seen in SUCLG1 patients is considerably lower than that typically observed in case of methylmalonic acidemia, propionic acidemia, or cobalamin deficiency [24]. Also, patients with methylmalonic acidemia, propionic acidemia, and cobalamin deficiency typically do not experience lactic acidosis. When a diagnostic laboratory observes methylmalonic elevatation in urine of patients who are negative for most of the metabolic disorders, it may be misinterpreted to be a result of gut bacterial activity. The patient in this report clearly has a phenotype that extends the mild end of the SUCLG1-deficiency clinical spectrum.
Table 1.
Clinical phenotype of twenty five patients with mutations in SUCLG1.
| Sex | Age at onset | Status | Hypotonia | Hearing loss |
MCAs | EEG/MRI | RS | Lactate | MMA | Genotype | Amino acid change |
Allele Phase | Source | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | F | Day of life 1 | Died on day 3 | Yes | No | No | Abnormal | Yes | E | E(M) | c.112_113del | p.Ile38Serfs*15 | Homozygous | Ostergaard et al, 2007 |
| P2 | M | Day of life 1 | Died on day 4 | Yes | No | No | Abnormal | Yes | E | E(M) | c.112_113del | p.Ile38Serfs*15 | Homozygous | Ostergaard et al, 2007 |
| P3 | F | Day of life 1 | Died on day 2 | Yes | No | No | NA | Yes | E | E(M) | c.112_113del | p.Ile38Serfs*15 | Homozygous | Ostergaard et al, 2007 |
| P4 | M | 3mo | Died at 3yrs | Yes | Yes | No | Abnormal | Yes | E(M) | E(M) | c.215G>C | p.Gly72Ala | Homozygous | Ostergaard et al, 2010 |
| P5 | M | Day of life 1 | Died on day 3.5 | Yes | No | Yes | NA | Yes | E | E(M) | c.626C > A | p.Ala209Glu | Homozygous | Rivera et al.2010 |
| P6 | F | Day of life 1 | Died on day 3 | Yes | No | Yes | NA | Yes | E | E(M) | c.626C > A | p.Ala209Glu | Homozygous | Rivera et al.2010 |
| P7 | M | Day of life 1 | Died at 1yr | Yes | No | No | Abnormal | Yes | E | E(M) | c.509C>G & c.97+3G>C |
p.Pro170Arg & Aberrant splicing |
Compound heterozygous |
Rouzier et el., 2010 |
| P8 | M | 3mo | Alive at 12yrs | Yes | No | No | Abnormal | Yes | E | E(M) | c.448C>T | p.Gln150× | Heterozygous(2nd allele unidentified) |
Rouzier et el., 2010 |
| P9 | M | 2mo | Alive at 9yrs | Yes | No | No | Abnormal | No | E | E(M) | c.626C > A (p.Ala209Glu) |
p.Ala209Glu | Heterozygous(2nd allele unidentified) |
Valayannopoulos et al., 2010 |
| P10 | M | 3mo | Alive at 5.5yrs | Yes | No | No | Abnormal | No | E | E(M) | c.309_3 10delTG & c.428T > G |
p.Thr103fs & p.Ile143Ser |
Compound heterozygous |
Valayannopoulos et al., 2010 |
| P11 | F | 5mo | Died at 18mo | Yes | No | No | Abnormal | Yes | E | NA | c.137C > T | p.Ser46Phe | Homozygous | Valayannopoulos et al., 2010 |
| P12 | F | Day of life 1 | Died at 17mo | Yes | Yes | No | Abnormal | Yes | E | E(M) | c.40A>T | p.Met14Leu | Homozygous | Van Hove, et al, 2010 |
| P13 | F | At birth | Died at 10mo | Yes | No | Yes | NA | NA | E | E(M) | c.280-1G>A | Aberrant splicing | Homozygous | Randolph et al, 2011 |
| P14 | F | Day of life 2 | Died at 20mo | Yes | Yes | No | NA | Yes | E | E(M) | c.41T > C & c.599C > T |
p.Met14Leu & p.Ser200Phe |
Compound heterozygous |
Sakamoto et al., 2011 |
| P15 | F | 1mo | Alive at 20yrs | Yes | Yes | No | Abnormal | No | E(M) | E(M) | c.626C>A & c.531+4A>T |
p.Ala209Glu & Aberrant splicing |
Compound heterozygous |
Navarro-Sastre et al., 2012 |
| P16 | M | Day of life 1 | Died at 1mo | Yes | NA | Yes | Abnormal | Yes | E | E(M) | c.749ANG | p.Glu263Gly | Homozygous | Landsverk et. al., 2014 |
| P17 | M | 6 mo | Alive at 2yrs 4mo |
Yes | No | No | Normal | No | E | E(M) | c.809A>C & c.826-2A>G |
p.Leu270Trp & Aberrant splicing |
Compound heterozygous |
Liu Y. et al., 2015 |
| P18 | M | 4 mo | Alive at 3yrs 6mo |
Yes | No | No | Abnormal | No | E | E(M) | c.550G>A & c.826-2A>G |
p.Gly184Ser & Aberrant splicing |
Compound heterozygous |
Liu Y. et al., 2015 |
| P19 | F | Neonate | Alive at 2yrs 9mo |
Yes | No | No | Abnormal | No | E | E(M) | c.751C>T & c.961C>G |
p.Gly251Ser & p.Ala321Pro |
Compound heterozygous |
Liu Y. et al., 2015 |
| P20 | F | 1–2yrs | Alive at 21 yrs | Yes | Yes | No | NA | No | Normal | E(M) | c.212A>G | p.His71Arg | Homozygous | Carrozzo et al., 2016 |
| P21 | F | Day of life 1 | Alive at 6 yrs | Yes | Yes | No | Abnormal | No | Normal | E(M) | c.215G>C | p.Gly72Ala | Homozygous | Carrozzo et al., 2016 |
| P22 | M | 4 mo | Died at 21 mo | Yes | NA | No | Abnormal | NA | NA | NA | c.215G>C | p.Gly72Ala | Homozygous | Carrozzo et al., 2016 |
| P23 | M | Day of life 1 | Died at 8 mo | Yes | NA | Yes | Abnormal | Yes | E | NA | c.787G>A & c.626C>A |
p.Glu263Lys & p.Ala209Glu |
Compound heterozygous |
Carrozzo et al., 2016 |
| P24 | F | 2mo | Alive at 10 mo | Yes | Yes | No | Abnormal | Yes | E(M) | NA | c.137C > T | p.Ser46Phe | Homozygous | Carrozzo et al., 2016 |
| P25 | F | 14 mo | Alive at 10 yrs | Yes | Yes | No | Normal | No | Normal | E(M) | c.40A>T & c.635A>G |
p.Met14Leu & p.Gln212Arg |
Compound heterozygous |
Current study |
Abbreviations are as follows: F, female; M, male; mo, months; E, elevated; E(M), moderately elevated; NA, not available; MMA, methylmalonic acid; SCS, succinylcoA-synthetase; RS, respiratory support; ETC, electron transport chain; MCAs, Multiple congenital abnormalities
Out of the twenty five patients, only two had SCS enzyme activity tested and in both cases SCS activity was reduced (Table 3). The majority of the patients had electron transport chain (ETC) activity tested (19/25), and most (17/19) showed reduced ETC activities. The mtDNA copy number in samples from all the patients tested (18/18) was abnormal. Two patients had interesting mtDNA copy number results, one with 128% mtDNA content in muscle compared to controls [23] and another patient with normal results in fibroblasts but a doubling of mtDNA content in muscle [22]. In our patient’s fibroblasts we observed a significant increase in mtDNA content in very early passage before it declined to ~50% at a later passage (Figure 1C). A similar phenomenon was observed in a Sucla2 deficient mouse model, where mtDNA content was significantly increased in early passage fibroblasts as well as in the tissues of earlier stage (e15.5) embryos [25]. This may reflect a compensatory response to loss of SCS activity by increased mtDNA synthesis and/or turnover. All the patients that were tested (4/4), either using mitochondria from muscle or fibroblasts, showed reduced oxygen consumption. Samples from all the patients that have had western blot analysis for SUCLG1 (7/25) showed reduced or absent protein content, while SUCLA2 protein levels were reduced in all the patients tested (3/25). The patient reported in this study is the only one who has had all the SCS components tested by western blot analysis, demonstrating an absence of SUCLG1 and SUCLG2 and a significantly reduced amount of SUCLA2. The deficiency of beta subunits in the patient’s fibroblasts was corrected upon ectopic expression of SUCLG1, indicating that the abnormal cellular phenotypes were the result of SUCLG1 deficiency (Figure 1A).
With the significant advances made in the field of whole exome sequencing (WES), the technology is increasingly employed for the diagnosis of rare disorders. WES studies that report novel variants of unknown significance present a specific challenge to clinicians trying to associate molecular findings with a clinical diagnosis [26]. Ultimately, confirming the clinical significance of these variants by appropriate functional assays will facilitate to overcome uncertainty and may allow a clear diagnosis. In the patient reported in this study, the two heterozygous variants identified in SUCLG1 appear to be in trans by Sanger sequencing (Table 1). The c.40A>T (p.M14L) variant is not reported previously while the c.635A>G (p.Q212R) variant is known. The c.40A>T (p.M14L) variant is not found in the ExAC database and is predicted to be deleterious by both SIFT and PolyPhen programs. Biochemical analyses with genetic rescue functionally validated the loss of function nature of these alleles, confirming the diagnosis of SUCLG1-dependent mtDNA Depletion Syndrome, providing another example of the utility of cellular functional analyses in metabolic and mitochondrial disorders.
5. Conclusion
Mitochondrial encephalomyopathy with mitochondrial DNA (mtDNA) depletion is a rare, genetically heterogenous disorder with a broad phenotypic spectrum. In the clinical setting, when a mtDNA depletion syndrome is suspected, one should consider WES or a mtDNA depletion gene panel. When sequencing results report variants of unknown significance in a candidate gene, it is important to confirm the clinical significance of these variants by biochemical functional analysis and/or by ectopic expression of the gene whenever possible.
Supplementary Material
Table 2.
Lab findings of twenty five patients with SUCLG1 mutations
| SCS activity | ETC activity | mtDNA copy# | Respiration analysis |
SUCLG1 levels by WB |
SUCLG2 levels by WB |
SUCLA2 levels by WB |
|
|---|---|---|---|---|---|---|---|
| P1 | NA | Abnormal in mu | 15% of controls(mu) | NA | Abscent(fb) | NA | NA |
| P2 | NA | Abnormal in Liver | 15% of controls in Liver | NA | Abscent(fb) | NA | NA |
| P3 | NA | NA | NA | NA | NA | NA | NA |
| P4 | NA | Very mildly reduced | Decreased(mu) by southern blot | Mildly reduced | Severely reduced(fb) | NA | NA |
| P5 | NA | Reduced(mu) | 7–10% of controls(mu) | NA | NA | NA | NA |
| P6 | NA | Reduced(mu) | 7–10% of controls(mu) | NA | NA | NA | NA |
| P7 | NA | Reduced (mu & fb) | 11% of controls(mu) | NA | Abscent(fb) | NA | Abscent(fb) |
| P8 | NA | Reduced (mu & fb) | 18% of controls(mu) | NA | Reduced(fb) | NA | Reduced(fb) |
| P9 | NA | Reduced(mu) | 48% of controls(mu) | Reduced(mu) | NA | NA | NA |
| P10 | NA | Reduced(mu) | 25% of controls(mu) | Reduced(mu) | NA | NA | NA |
| P11 | NA | NA | 128% of controls(mu) | NA | NA | NA | NA |
| P12 | Deficient | NA | 50% of controls in Liver | NA | Severely reduced | NA | NA |
| P13 | NA | severe C IV and II+III deficiencies | 18% of controls(mu) | NA | NA | NA | NA |
| P14 | NA | Reduced(fb & mu) | No change (fb) & doubled (mu) | NA | NA | NA | NA |
| P15 | NA | NA | NA | NA | NA | NA | NA |
| P16 | NA | Severely reduced(mu) | 27% of controls(mu) | NA | NA | NA | NA |
| P17 | NA | Reduced CV (leu) | 90% of controls(leu) | NA | NA | NA | NA |
| P18 | NA | Reduced CI (leu) | 75% of controls(leu) | NA | NA | NA | NA |
| P19 | NA | Reduced CI (leu) | 68% of controls(leu) | NA | NA | NA | NA |
| P20 | NA | Normal | NA | NA | NA | NA | NA |
| P21 | NA | Mildly reduced CI &CIV | NA | NA | NA | NA | NA |
| P22 | NA | NA | NA | NA | NA | NA | NA |
| P23 | NA | Severely reduced CI &CIV | NA | NA | NA | NA | NA |
| P24 | NA | Normal | NA | NA | NA | NA | NA |
| P25 | Severly reduced | NA | 72% of control(mu) & 50% of control (fb) |
Reduced(fb) | Absent(fb) | Abscent(fb) | Reduced(fb) |
Abbreviations are as follows: NA, not available; SCS, succinylcoA-synthetase; ETC, electron transport chain; mu, muscle; fb, fibroblasts; leu, leukocytes
Highlights.
Pathogenicity of the novel variant of unknown significance, c.40A>T (p.M14L), is confirmed by molecular and biochemical functional analyses using primary skin fibroblasts from the patient.
A mild phenotype for SUCLG1 deficiency is reported
This study expands the phenotypic spectrum of patients with SUCLG1 deficiency
This study shows that in the absence of Suclg1 protein, Sucla2 and Suclg2 are also absent or severely reduced.
Acknowledgments
We would like to thank the patient and the patient’s family for their participation in the study. We acknowledge the expert assistance of Joel M. Sederstrom of the Cytometry and Cell Sorting Core at Baylor College of Medicine.
Funding sources
These studies were supported, in part, by NIHR01GM098387 (BHG) and R21GM110190(BHG). This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574). The project was also supported in part by IDDRC grant number 1U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development, Clinical Translation Core – Biobanking Unit. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare that no conflict of interest exists.
References
- 1.Johnson JD, Mehus JG, Tews K, Milavetz BI, Lambeth DO. Genetic evidence for the expression of ATP- and GTP-specific succinyl-CoA synthetases in multicellular eucaryotes. J Biol Chem. 1998;273:27580–27586. doi: 10.1074/jbc.273.42.27580. [DOI] [PubMed] [Google Scholar]
- 2.Lambeth DO, Tews KN, Adkins S, Frohlich D, Milavetz BI. Expression of two succinyl-CoA synthetases with different nucleotide specificities in mammalian tissues. J Biol Chem. 2004;279:36621–36624. doi: 10.1074/jbc.M406884200. [DOI] [PubMed] [Google Scholar]
- 3.El-Hattab AW, Scaglia F. Mitochondrial DNA depletion syndromes: review and updates of genetic basis, manifestations, and therapeutic options. Neurotherapeutics. 2013;10:186–198. doi: 10.1007/s13311-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graham BH. Diagnostic challenges of mitochondrial disorders: complexities of two genomes. Methods Mol Biol. 2012;837:35–46. doi: 10.1007/978-1-61779-504-6_3. [DOI] [PubMed] [Google Scholar]
- 5.Suomalainen A, Isohanni P. Mitochondrial DNA depletion syndromes--many genes, common mechanisms. Neuromuscul Disord. 2010;20:429–437. doi: 10.1016/j.nmd.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 6.Bonnen PE, Yarham JW, Besse A, Wu P, Faqeih EA, Al-Asmari AM, Saleh MA, Eyaid W, Hadeel A, He L, Smith F, Yau S, Simcox EM, Miwa S, Donti T, Abu-Amero KK, Wong LJ, Craigen WJ, Graham BH, Scott KL, McFarland R, Taylor RW. Mutations in FBXL4 cause mitochondrial encephalopathy and a disorder of mitochondrial DNA maintenance. Am J Hum Genet. 2013;93:471–481. doi: 10.1016/j.ajhg.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gai X, Ghezzi D, Johnson MA, Biagosch CA, Shamseldin HE, Haack TB, Reyes A, Tsukikawa M, Sheldon CA, Srinivasan S, Gorza M, Kremer LS, Wieland T, Strom TM, Polyak E, Place E, Consugar M, Ostrovsky J, Vidoni S, Robinson AJ, Wong LJ, Sondheimer N, Salih MA, Al-Jishi E, Raab CP, Bean C, Furlan F, Parini R, Lamperti C, Mayr JA, Konstantopoulou V, Huemer M, Pierce EA, Meitinger T, Freisinger P, Sperl W, Prokisch H, Alkuraya FS, Falk MJ, Zeviani M. Mutations in FBXL4, encoding a mitochondrial protein, cause early-onset mitochondrial encephalomyopathy. Am J Hum Genet. 2013;93:482–495. doi: 10.1016/j.ajhg.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besse A, Wu P, Bruni F, Donti T, Graham BH, Craigen WJ, McFarland R, Moretti P, Lalani S, Scott KL, Taylor RW, Bonnen PE. The GABA transaminase, ABAT, is essential for mitochondrial nucleoside metabolism. Cell Metab. 2015;21:417–427. doi: 10.1016/j.cmet.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrozzo R, Verrigni D, Rasmussen M, de Coo R, Amartino H, Bianchi M, Buhas D, Mesli S, Naess K, Born AP, Woldseth B, Prontera P, Batbayli M, Ravn K, Joensen F, Cordelli DM, Santorelli FM, Tulinius M, Darin N, Duno M, Jouvencel P, Burlina A, Stangoni G, Bertini E, Redonnet-Vernhet I, Wibrand F, Dionisi-Vici C, Uusimaa J, Vieira P, Osorio AN, McFarland R, Taylor RW, Holme E, Ostergaard E. Succinate-CoA ligase deficiency due to mutations in SUCLA2 and SUCLG1: phenotype and genotype correlations in 71 patients. J Inherit Metab Dis. 2016;39:243–252. doi: 10.1007/s10545-015-9894-9. [DOI] [PubMed] [Google Scholar]
- 10.Van Hove JL, Saenz MS, Thomas JA, Gallagher RC, Lovell MA, Fenton LZ, Shanske S, Myers SM, Wanders RJ, Ruiter J, Turkenburg M, Waterham HR. Succinyl-CoA ligase deficiency: a mitochondrial hepatoencephalomyopathy. Pediatr Res. 2010;68:159–164. doi: 10.1203/PDR.0b013e3181e5c3a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadat R, Barca E, Masand R, Donti TR, Naini A, De Vivo DC, DiMauro S, Hanchard NA, Graham BH. Functional cellular analyses reveal energy metabolism defect and mitochondrial DNA depletion in a case of mitochondrial aconitase deficiency. Mol Genet Metab. 2016;118:28–34. doi: 10.1016/j.ymgme.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai RK, Perng CL, Hsu CH, Wong LJ. Quantitative PCR analysis of mitochondrial DNA content in patients with mitochondrial disease. Ann N Y Acad Sci. 2004;1011:304–309. doi: 10.1007/978-3-662-41088-2_29. [DOI] [PubMed] [Google Scholar]
- 13.Alarcon C, Wicksteed B, Prentki M, Corkey BE, Rhodes CJ. Succinate is a preferential metabolic stimulus-coupling signal for glucose-induced proinsulin biosynthesis translation. Diabetes. 2002;51:2496–2504. doi: 10.2337/diabetes.51.8.2496. [DOI] [PubMed] [Google Scholar]
- 14.Divakaruni AS, Paradyse A, Ferrick DA, Murphy AN, Jastroch M. Analysis and interpretation of microplate-based oxygen consumption and pH data. Methods Enzymol. 2014;547:309–354. doi: 10.1016/B978-0-12-801415-8.00016-3. [DOI] [PubMed] [Google Scholar]
- 15.Landsverk ML, Zhang VW, Wong L-JC, Andersson HC. A SUCLG1 mutation in a patient with mitochondrial DNA depletion and congenital anomalies. Molecular Genetics and Metabolism Reports. 2014;1:451–454. doi: 10.1016/j.ymgmr.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Li X, Wang Q, Ding Y, Song J, Yang Y. Five novel SUCLG1 mutations in three Chinese patients with succinate-CoA ligase deficiency noticed by mild methylmalonic aciduria. Brain Dev. 2015 doi: 10.1016/j.braindev.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Ostergaard E, Christensen E, Kristensen E, Mogensen B, Duno M, Shoubridge EA, Wibrand F. Deficiency of the alpha subunit of succinate-coenzyme A ligase causes fatal infantile lactic acidosis with mitochondrial DNA depletion. Am J Hum Genet. 2007;81:383–387. doi: 10.1086/519222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostergaard E, Schwartz M, Batbayli M, Christensen E, Hjalmarson O, Kollberg G, Holme E. A novel missense mutation in SUCLG1 associated with mitochondrial DNA depletion, encephalomyopathic form, with methylmalonic aciduria. Eur J Pediatr. 2010:201–205. doi: 10.1007/s00431-009-1007-z. [DOI] [PubMed] [Google Scholar]
- 19.Randolph LM, Jackson HA, Wang J, Shimada H, Sanchez-Lara PA, Wong DA, Wong LJ, Boles RG. Fatal infantile lactic acidosis and a novel homozygous mutation in the SUCLG1 gene: a mitochondrial DNA depletion disorder. Mol Genet Metab. 2011;102:149–152. doi: 10.1016/j.ymgme.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 20.Rivera H, Merinero B, Martinez-Pardo M, Arroyo I, Ruiz-Sala P, Bornstein B, Serra-Suhe C, Gallardo E, Marti R, Moran MJ, Ugalde C, Perez-Jurado LA, Andreu AL, Garesse R, Ugarte M, Arenas J, Martin MA. Marked mitochondrial DNA depletion associated with a novel SUCLG1 gene mutation resulting in lethal neonatal acidosis, multi-organ failure, and interrupted aortic arch. Mitochondrion. 2010;10:362–368. doi: 10.1016/j.mito.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 21.Rouzier C, Le Guedard-Mereuze S, Fragaki K, Serre V, Miro J, Tuffery-Giraud S, Chaussenot A, Bannwarth S, Caruba C, Ostergaard E, Pellissier JF, Richelme C, Espil C, Chabrol B, Paquis-Flucklinger V. The severity of phenotype linked to SUCLG1 mutations could be correlated with residual amount of SUCLG1 protein. J Med Genet. 2010;47:670–676. doi: 10.1136/jmg.2009.073445. [DOI] [PubMed] [Google Scholar]
- 22.Sakamoto O, Ohura T, Murayama K, Ohtake A, Harashima H, Abukawa D, Takeyama J, Haginoya K, Miyabayashi S, Kure S. Neonatal lactic acidosis with methylmalonic aciduria due to novel mutations in the SUCLG1 gene. Pediatr Int. 2011;53:921–925. doi: 10.1111/j.1442-200X.2011.03412.x. [DOI] [PubMed] [Google Scholar]
- 23.Valayannopoulos V, Haudry C, Serre V, Barth M, Boddaert N, Arnoux JB, Cormier-Daire V, Rio M, Rabier D, Vassault A, Munnich A, Bonnefont JP, de Lonlay P, Rotig A, Lebre AS. New SUCLG1 patients expanding the phenotypic spectrum of this rare cause of mild methylmalonic aciduria. Mitochondrion. 2010;10:335–341. doi: 10.1016/j.mito.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Schmedes A, Brandslund I. Analysis of methylmalonic acid in plasma by liquid chromatography-tandem mass spectrometry. Clin Chem. 2006;52:754–757. doi: 10.1373/clinchem.2005.058586. [DOI] [PubMed] [Google Scholar]
- 25.Donti TR, Stromberger C, Ge M, Eldin KW, Craigen WJ, Graham BH. Screen for abnormal mitochondrial phenotypes in mouse embryonic stem cells identifies a model for succinyl-CoA ligase deficiency and mtDNA depletion. Dis Model Mech. 2014;7:271–280. doi: 10.1242/dmm.013466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindor NM, Goldgar DE, Tavtigian SV, Plon SE, Couch FJ. BRCA1/2 sequence variants of uncertain significance: a primer for providers to assist in discussions and in medical management. The oncologist. 2013;18:518–524. doi: 10.1634/theoncologist.2012-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.