Abstract
Sphingosine 1-phosphate (S1P) is a bioactive lipid that acts via G protein-coupled receptors. The S1P receptor S1P1, encoded by S1pr1, is expressed in developing heart but its roles there remain largely unexplored. Analysis of S1pr1 LacZ knockin embryos revealed β-galactosidase staining in cardiomyocytes in the septum and in the trabecular layer of hearts collected at 12.5 days post coitus (dpc) and weak staining in the inner aspect of the compact layer at 15.5 dpc and later. Nkx2-5-Cre− and Mlc2a-Cre−mediated conditional knockout of S1pr1 led to ventricular noncompaction and ventricular septal defects at 18.5 dpc and to perinatal lethality in the majority of mutants. Further analysis of Mlc2a-Cre conditional mutants revealed no gross phenotype at 12.5 dpc but absence of the normal increase in the number of cardiomyocytes and the thickness of the compact layer at 13.5 dpc and after. Consistent with relative lack of a compact layer, in situ hybridization at 13.5 dpc revealed expression of trabecular markers extending almost to the epicardium in mutants. Mutant hearts also showed decreased myofibril organization in the compact but not trabecular myocardium at 12.5 dpc. These results suggest that S1P signaling via S1P1 in cardiomyocytes plays a previously unknown and necessary role in heart development in mice.
Keywords: Sphingosine 1-phosphate, Sphingosine 1-phosphate receptor, S1P1, S1pr1, GPCR, cardiomyocyte, noncompaction, heart development
INTRODUCTION
Sphingosine 1-phosphate (S1P) is a bioactive lipid that acts via five G protein-coupled receptors (GPCRs) called S1P1-S1P5 (Blaho and Hla, 2011; Mutoh et al., 2012). The S1P1 receptor, encoded by S1pr1, contributes to a variety of different cellular processes including actin polymerization, cell migration, survival, proliferation and other responses in endothelial cells, lymphocytes and other cell types (Cyster and Schwab, 2012; Hla et al., 2001). Global and “endothelial-specific” knockout of S1pr1 in mouse leads to embryonic lethality by approximately 12.5–14.5 days post coitus (dpc) due to abnormal blood vessel development (Allende et al., 2003; Gaengel et al., 2012; Liu et al., 2000). In cultured human endothelial monolayers, S1P1 promotes barrier integrity, and removal of S1P from the plasma compartment or pharmacological inhibition of S1P1 in postnatal mice impairs endothelial barrier function (Donati and Bruni, 2006; Garcia et al., 2001; Singleton et al., 2005) (Camerer et al., 2009; Rosen et al., 2009). These and other data suggest a vital role for S1P signaling via S1P1 in endothelial cells in blood vessel development and maintenance of vascular integrity. S1pr1 is also highly expressed in mouse heart during embryonic development (Liu et al., 2000; Poulsen et al., 2011), but the role of S1P1 in cardiomyocytes or other cardiac cells and whether it contributes to cardiac development is unknown.
As the mammalian heart develops, the ventricular walls undergo significant morphological changes through processes known as trabeculation and compaction. From ~9.5 dpc, cardiomyocytes proliferate asymmetrically from the subepicardial myocardium and project into the ventricular space to form an endocardium-covered meshwork of sheets and cords called the trabecular layer (Li et al., 2016). Cardiomyocytes in the subepicardial region are morphologically and molecularly distinct from trabecular cardiomyocytes and organize in a dense arrangement known as the compact layer. Between ~9.5 and ~12.5 dpc, the compact layer of cardiomyocytes remains very thin, with the majority of the ventricular mass in the trabecular layer. Beginning at ~12.5 dpc and extending until birth, the compact layer expands as cardiomyocytes proliferate and adopt a compact morphology; trabecular layer growth is rapidly outpaced by the compact myocardium, which eventually composes the majority of left ventricular mass (Ieda et al., 2009; Lee et al., 2000; Luxán et al., 2013). Disruption of either trabeculation or compaction has important consequences; failure to undergo trabeculation results in growth arrest and embryonic lethality, while defective compaction results in left ventricular non-compaction cardiomyopathy. To date the molecular mechanisms that regulate the proliferation and differentiation required for trabeculation and compaction remain incompletely understood (Paige et al., 2015; Towbin et al., 2015; Wilsbacher and McNally, 2016).
To detect potential roles for S1P1 in heart development, we generated mice in which S1pr1 was excised in cardiomyocytes. We report evidence that S1P1 in cardiomyocytes is required for heart development, specifically for normal ventricular compaction as well as septation and normal postnatal survival.
METHODS
Mouse strains and genotyping
All procedures were approved by the UCSF Institutional Animal Care and Use Committee. Richard Proia (National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, USA) provided S1pr1 β-galactosidase knockin mice (S1pr1tm2Rlp, here called S1pr1+/Lac−) (Liu et al., 2000). Dr. Jerold Chun provided mice carrying a floxed S1pr1 allele (S1pr1tm1Jch, ; here called S1pr1f/f in the text) (Choi et al., 2011). Dr. Robert Schwartz provided Nkx2-5-Cre knockin mice (Nkx2-5tm1(cre)Rjs) (Moses et al., 2001). Dr. Xu Peng (Scott & White Memorial Hospital, Temple, USA) provided Mlc2a-Cre knockin mice (Myl7tm1(cre)Krc) (Wettschureck et al., 2001). Mice carrying Cre knockin or wild-type alleles at Nkx2-5 or Mlc2a are called Cre + or – herein. "Mutants" refers to mice/embryos with genotypes predicted to have no functional S1pr1 alleles in cardiomyocytes (Cre+;S1pr1f/−). "Controls", usually littermates, refers to mice/embryos predicted to have at least one functional S1pr1 allele (Cre−;S1pr1f/+, Cre− ;S1pr1f/−, and Cre+;S1pr1f/+ as indicated). Genotyping primer sequences are listed in Supplemental Table 1.
Mouse embryo collection, histology, antibody staining, and myocardial wall measurements
Noon of the day a vaginal plug was observed was defined as 0.5 dpc. Embryonic hearts were hematoxylin and eosin stained using standard protocols. S1pr1 expression was assessed in S1pr1+/LacZ embryonic hearts using whole mount X-gal staining followed by 4% PFA post-fixation overnight and either paraffin or cryosectioning. For antibody staining, whole embryos collected up to 13.5 dpc and excised embryonic hearts collected from 14.5 to 18.5 dpc were snap frozen and immunostained as previously described (Wilsbacher and Coughlin, 2015) using sarcomeric α-actinin (Sigma A7811) and fibronectin (Sigma F3648) primary antibodies. Measurements of compact and trabecular myocardial wall thickness were obtained from sections of embryonic hearts collected at 12.5, 13.5, 15.5, and 16.5 dpc from controls and mutants using Fiji image analysis software (Schindelin et al., 2012). Measurements were taken at two to six evenly spaced intervals along the left ventricular free wall of each section. For one control and one mutant heart at both 15.5 dpc and 16.5 dpc, only high power images of the compact wall were obtained; trabecular wall measurements were not made in these hearts. The average of measurements from each heart was used for statistical analysis (see Statistics).
5-ethynyl-2’-deoxyuridine (Edu) proliferation assay and TUNEL assay
Pregnant females received 30 mg/kg Edu i.p. at 12.5 and 13.5 dpc. Embryos were collected after two hours and fixed overnight in 4% PFA in PBS at 4°C prior to cryoprotection and cryosectioning. Transverse embryo sections were collected and the sections with maximal LV diameter were analyzed for each heart. Cardiomyocytes were identified by s-α-actinin staining. TUNEL detection, Edu detection, and Hoechst nuclear staining were performed per the manufacturer’s protocol (Roche 11684795910, Life Technologies C10337). In each section, contiguous, non-overlapping boxes were tiled over the myocardium along the left ventricular free wall and cardiomyocytes were classified as compact or trabecular based on position and morphology. Total and Edu-positive cardiomyocytes per high power field, were counted blind to genotype using manual detection and Fiji image analysis software (Schindelin et al., 2012). The average number per high power field for individual ventricles was used for statistical analyses (see Statistics).
In situ hybridization
Embryos were collected at 13.5 dpc, and fixed overnight in 4% PFA in PBS at 4°C prior to cryoprotection and cryosectioning. The plasmids for Nppa and Bmp10 were those previously described (Koibuchi and Chin, 2007; Morikawa and Cserjesi, 2008). Each plasmid was linearized with XhoI and transcribed with T3 polymerase to generate the digoxigenin-labeled probe, which was hybridized at 70°C overnight at a concentration of 1–1.5 microgram/ml. Digoxigenin detection was performed using anti-digoxigenin labeling and chromogenic alkaline phosphatase development with Roche BM Purple (Roche 11442074001).
Electron microscopy
Electron microscopy was performed as described (Kurrasch et al., 2009). Briefly, 12.5 dpc embryonic hearts (n =1 each for Nkx2-5-Cre−; S1pr1f/− control, Nkx2-5-Cre+; S1pr1f/−mutant, Mlc2a-Cre−; S1pr1f/− control, and Mlc2a-Cre+; S1pr1f/− mutant) were dissected in ice-cold PBS and fixed in 0.1 mol/L sodium cacodylate buffer (pH 7.4) containing 2% glutaraldehyde and 1% PFA. Hearts were post-fixed in the same buffer containing 2% osmium tetroxide, then block-stained in 2% aqueous uranyl acetate, dehydrated, and embedded in LX-112 resin (Ladd Research Industries). Semi-thin sections were stained with toluidine blue to identify areas of interest. Ultrathin sections were taken on a Reichert-Jung Ultracut S ultramicrotome (Leica Microsystems) and stained with 0.8% lead citrate. Grids were visualized on a JEOL JEM-1230 transmission electron microscope and images were captured using a Gatan Ultrascan 1000 digital camera. Z-disc length measurement was made across the electron-dense portion of the Z-disc perpendicular to myofibrils. For the Nkx2-5 animals, 117 total sarcomere units in 13 fields were analyzed for Cre− and 59 units in 11 fields for Cre+. For Mlc2a animals, 83 sarcomere units in 8 fields were analyzed for Cre− and 49 units in 8 fields for Cre+. Each field was 8.3 µm by 8.3 µm.
Statistics
All measurements were done blind to genotype. The Chi-square test was used to analyze viability data at 18.5 dpc and weaning. Differences in ventricular wall thickness and cell number and Edu labeling per high-power field were assessed using two-way ANOVA by genotype (control, mutant) and embryo age (12.5 dpc, 13.5 dpc, 15.5 dpc, 16.5 dpc). When permitted, follow-on individual comparisons were performed using the Tukey test. All statistical analyses were performed using GraphPad software (La Jolla, CA).
RESULTS
Pattern of S1pr1 expression in the myocardium of the developing mouse heart
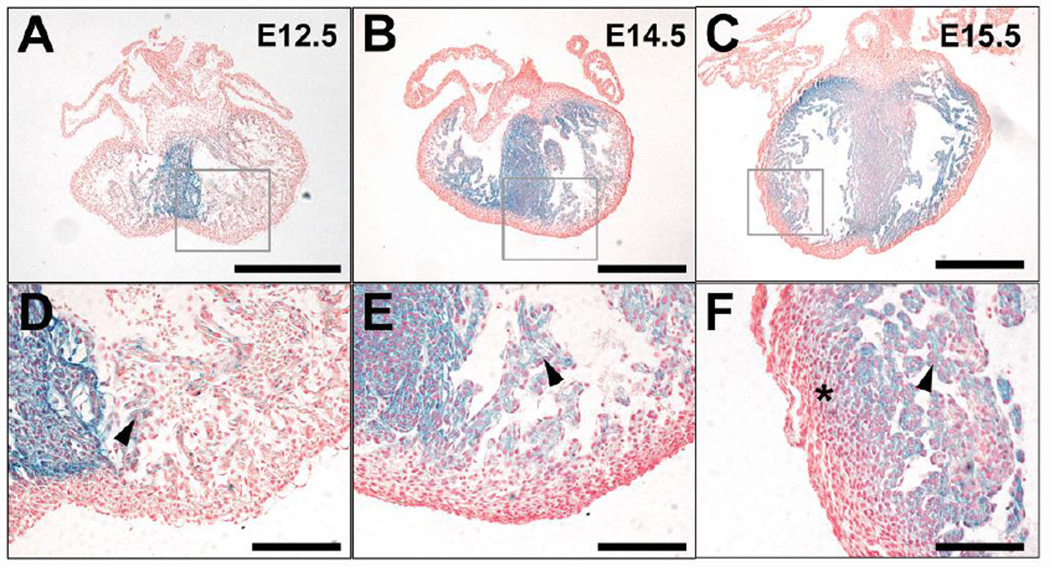
To determine the pattern of S1pr1 expression in the developing heart, we examined paraffin (Figure 1) and frozen sections (Supplemental Figure 1) of hearts collected from S1pr1+/LacZ embryos in which one S1pr1 allele is a β-galactosidase reporter knock-in (Liu et al., 2000). β-galactosidase staining was detected in ventricular myocardium as early as 9.5 dpc (Supplemental Figure 1 and Liu et al., 2000). At 12.5, 14.5, 15.5 and 18.5 dpc, β-galactosidase staining was detected in cardiomyocytes in the septum and trabeculae (Figure 1 and Supplemental Figure 1). At 15.5 dpc, lower-intensity staining could be detected in compact layer cells adjacent to the trabecular layer (Figure 1C, F, I). At 18.5 dpc, staining extended farther into the ventricular wall but usually spared the immediate subepicardial myocardium (Supplemental Figure 1). No staining was detected in atrial cardiomyocytes (Figure 1A–C). While differences in tissue penetration of Xgal could contribute to the observed pattern, this seems unlikely as the thicker septum showed more staining than the thinner ventricular free walls. Extraction of stain during tissue processing might also influence the staining pattern, but the pattern was similar in hearts processed for paraffin and frozen sections (Figure 1 and Supplemental Figure 1); the latter undergo minimal processing with no exposure to organic solvents between staining and sectioning. Moreover, staining spanned the thin ventricular wall in hearts from 9.5 dpc embryos (Supplemental Figure 1G). Thus, at face value, the temporal and spatial pattern of β-galactosidase staining seen in hearts from S1pr1+/LacZ embryos suggests that S1pr1 is expressed in most ventricular cardiomyocytes beginning at 9.5 dpc, is localized in the septum and trabecular myocardium at 12.5 and 14.5 dpc, and extends at lower levels into the inner compact myocardium as development proceeds.
Figure 1. S1pr1 reporter expression in embryonic mouse heart.
Hearts were collected from S1pr1+/LazZ embryos in which one S1pr1 allele was a β-galactosidase reporter knock-in at 12.5 dpc (A, D), 14.5 dpc (B, E), and 15.5 dpc (C, F). Hearts were Xgal-stained as whole mounts, paraffin embedded, sectioned and counterstained with Nuclear Fast Red. Panels D–F are high-power views of the areas indicated in A–C. Note prominent staining for β-galactosidase activity in cells that appear to be cardiomyocytes by morphology and location in the septum at all three time points, in trabeculae increasing in intensity and distribution from 12.5 to 15.5 dpc, and in the inner aspect of the compact layer at 15.5 dpc (asterisk). Scale bar 500 µm for A–C; 100 µm for D–F. Similar results were obtained with 3–5 hearts analyzed at each time point.
S1pr1 in cardiomyocytes is necessary for normal cardiac structure and postnatal survival
We probed for possible roles of S1P1 in cardiomyocytes during heart development by deleting S1pr1 using Nkx2-5-Cre and Mlc2a-Cre knockin mice in a flox/null S1pr1 background (Choi et al., 2011). Nkx2-5-Cre excises floxed sequences in cardiomyocytes as well as in endocardium and epicardium, while Mlc2a-Cre excises specifically in cardiomyocytes (Peng et al., 2008). At weaning, Nkx2-5-Cre+; S1pr1f/− and Mlc2a-Cre+; S1pr1f/− mice were present at only 17% and 32% of the expected Mendelian rate, respectively (Table 1). By contrast, at 18.5 dpc, Nkx2-5-Cre+; S1pr1f/− and Mlc2a-Cre+; S1pr1f/− embryos were present at the expected Mendelian rate (Table 1) and were indistinguishable from control littermates by external examination. Thus, mutant embryos died between 18.5 dpc and weaning. Three Nkx2-5-Cre+; S1pr1f/− and Mlc2a-Cre+; S1pr1f/− pups were found dead within 24 hours of birth and none were found thereafter, suggesting that those mutants that died did so shortly after birth (with the "missing" dead pups presumably cannibalized).
Table 1. Genotypes of live 18.5 dpc embryos and pups from S1pr1f/f X S1pr1+/−; Cre+/0 crosses.
Numbers of live, grossly normal embryos of each genotype present at 18.5 dpc and of live pups at weaning are shown. Numbers in parentheses indicate number expected assuming equal survival of the control genotypes. The Cre driver is indicated at left.
| S1pr1f/+, Cre− |
S1pr1f/+, Cre+ |
S1pr1f/−, Cre− | S1pr1f/−, Cre+ | |
|---|---|---|---|---|
| Nkx2-5, 18.5 dpc | 19 (13.3) | 8 (13.3) | 13 (13.3) | 17 (13.3) |
| Mlc2a, 18.5 dpc | 8 (6.3) | 4 (6.3) | 7 (6.3) | 6 (6.3) |
| Nkx2-5, weaning | 16 (17.7) | 21 (17.7) | 16 (17.7) | 3* (17.7) |
| Mlc2a, weaning | 28 (25) | 24 (25) | 23 (25) | 8* (25) |
Note that live Cre+;S1pr1f/− mutant embryos were present at the expected Mendelian frequency at 18.5 dpc when either “cardiac-specific” Nkx2-5-Cre or cardiomyocyte-specific Mlc2a-Cre was used to generate conditional knockouts, but at weaning both conditional knockouts were markedly underrepresented
p=0.005 and 0.007 by Chi-squared analysis, respectively).
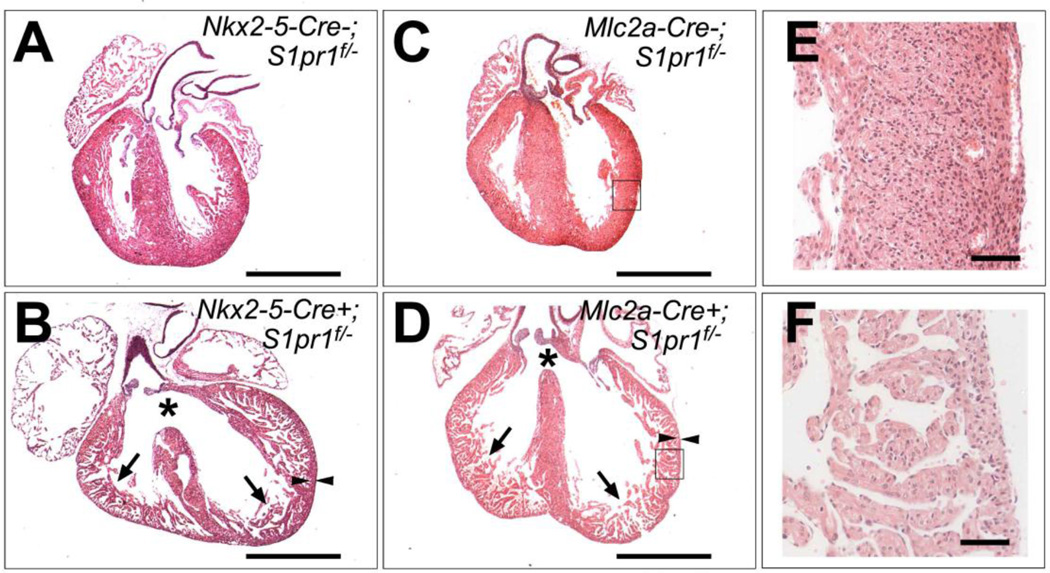
All Nkx2-5-Cre+; S1pr1f/− (4/4) and Mlc2a-Cre+; S1pr1f/− (3/3) hearts collected at 18.5 dpc all showed a markedly thin compact layer and a thick trabecular layer, a phenotype known as ventricular noncompaction, as well as a membranous ventricular septal defects (VSD) (Figure 2). The three dead pups collected at P0 also showed ventricular noncompaction and VSD, but hearts collected from the four mutants that survived beyond weaning showed ventricular noncompaction with complete septation. We did not observe other endocardial cushion defects such as atrial septal defects, double outlet right ventricle, or truncus arteriosus (Figure 2). These observations suggest that deletion of S1pr1 in cardiomyocytes is associated with ventricular noncompaction and VSD and that postnatal lethality in mutants was likely due to the latter.
Figure 2. Cardiac abnormalities in cardiomyocyte-specific S1pr1 knockouts.
(A–D) Hematoxylin and eosin stained sections of hearts from (A) Nkx2-5-Cre−; S1Pr1f/− control, (B) Nkx2-5-Cre+; S1Pr1f/− mutant, (C) Mlc2a-Cre−; S1Pr1f/− control and (D) Mlc2a-Cre+; S1Pr1f/− mutant embryos collected at 18.5 dpc. Note ventricular noncompaction with persistence of trabeculae (arrows) and thin compact layer (arrowheads) as well as ventricular septal defects (asterisks) in mutant hearts. Scale bar 1mm. (E and F) Higher magnification view of areas boxed in C and D, respectively. Scale bar 100 µm. VSD and noncompaction were noted in 4/4 Nkx2-5-Cre+; S1Pr1f/− mutant and 3/3 Mlc2a-Cre+; S1Pr1f/− mutant hearts analyzed at 18.5 dpc.
As Nkx2.5-Cre excises floxed alleles in multiple lineages in the developing heart, and given the similarity of the Nkx2-5-Cre+; S1pr1f/− and Mlc2a-Cre+; S1pr1f/− phenotypes and our goal of defining the role of S1P1 specifically in cardiomyocytes, we focused our subsequent studies on analysis of Mlc2a-Cre+; S1pr1f/− mutants and what appeared to be a fully penetrant ventricular noncompaction phenotype.
S1pr1 in cardiomyocytes is necessary for ventricular compaction
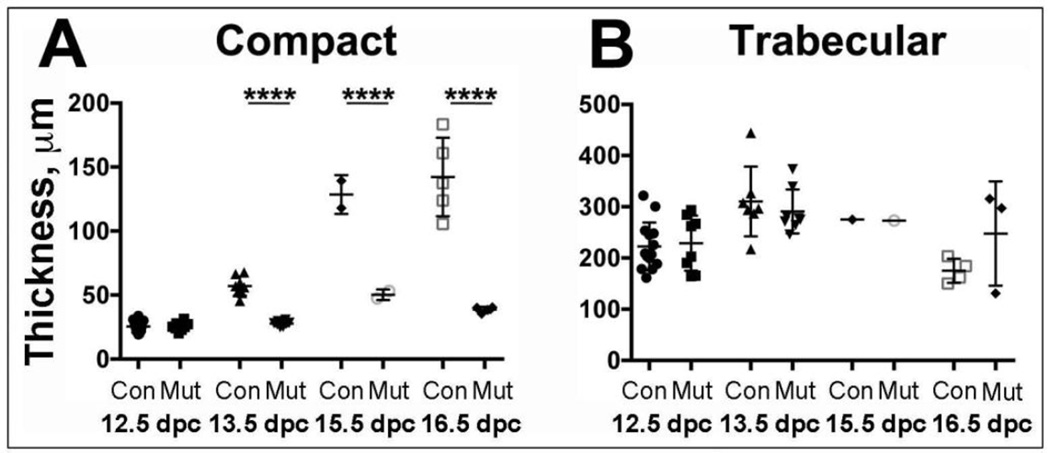
To quantitate the noncompaction phenotype, we measured the thickness of the compact and trabecular layers at different developmental time points in mutants and littermate controls (Figure 3). At 12.5 dpc, the thickness of the compact layer was not different in control and mutant hearts. However, while in control hearts compact layer thickness increased approximately 5-fold from 12.5 to 16.6 dpc, compact layer thickness did not increase after 12.5 dpc in mutant hearts, such that at 13.5, 15.5, and 16.5 dpc, the compact layer was significantly thinner in mutant vs. control hearts (Figure 3A). Trabecular layer thickness was not different between mutant and control hearts at any time point examined (Figure 3B).
Figure 3. Quantitation of compact and trabecular layer thickness.
Quantitation of (A) compact layer thickness and (B) trabecular layer thickness in Mlc2a-Cre−; S1Pr1f/− control (Con) and Mlc2a-Cre+; S1Pr1f/− mutant (Mut) hearts collected at the indicated gestational ages. By two-way ANOVA, groups were significantly different by gestational age and genotype with a significant interaction term. Follow-on individual comparisons by Tukey test revealed compact layer thickness to be different in controls and mutants to be different at 13.5, 15.5 and 16.5 dpc (**** p < 0.0001). Trabecular layer thickness did not differ between control and mutants at these time points.
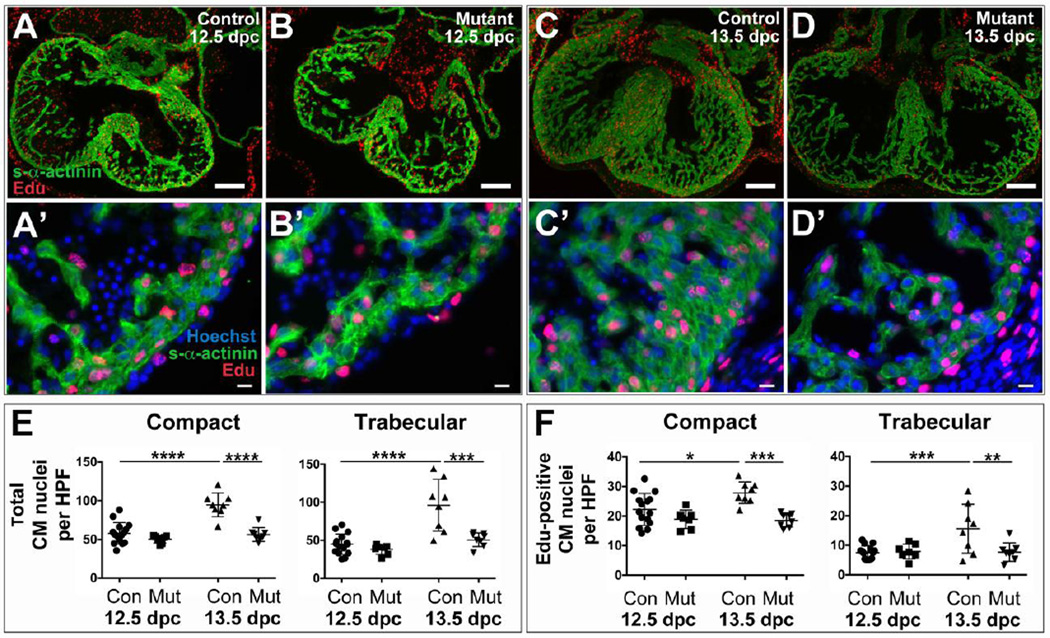
An analysis of the number of cardiomyocytes per high-power field confirmed that the number of compact and trabecular cardiomyocytes increased in control but not mutant hearts between 12.5 and 13.5 dpc; likewise, Edu labeling revealed that the total number of compact and trabecular cardiomyocytes in S-phase was different in control vs. mutant hearts at 13.5 dpc (Figure 4). As noted in global S1pr1 knockouts (Poulsen et al., 2011), virtually no apoptosis was detected in cardiomyocytes in mutants or controls at either time point: the fraction of cardiomyocytes that was TUNEL-positive was less than 0.1% in all samples tested and was not different between controls and mutants (e.g., 0–2 TUNEL-positive cardiomyocytes per heart section, with an average of 0.105% cardiomyocytes TUNEL-positive in controls (n=13) and 0.127% in mutants (n=7) at 12.5 dpc). These results strongly suggest that S1P1 function in cardiomyocytes is necessary for normal expansion of cardiomyocyte number during a time period critical for normal ventricular compaction. Interestingly, the fraction of cardiomyocytes that became Edu-positive in the windows sampled at 12.5 and 13.5 dpc was not different in controls and mutants (Supplemental Figure 5). Thus, while the presence of fewer total cardiomyocytes with fewer cardiomyocytes in S-phase in mutant hearts at 13.5 dpc suggests that a decrease in the number of cardiomyocytes available to proliferate may contribute to failed expansion of the compact layer thereafter, the basis for the initial failure to increase cardiomyocyte number between 12.5 and 13.5 dpc remains to be determined (see Discussion).
Figure 4. Cardiomyocyte (CM) density and proliferation at 12.5 dpc and 13.5 dpc in cardiomyocytespecific S1pr1 knockouts.
To quantitate the number of CM and those in S-phase, Edu-positive and s-α-actinin-positive cells were detected in sections of hearts collected from Mlc2a-Cre−;S1pr1f/+ Control (Con) or Mlc2a-Cre+;S1pr1f/− Mutant embryos at 12.5 or 13.5 dpc as indicated. (A–D) Representatitive images with s-α-actinin staining in green and Edu in red. Scale bar 100 µm. (A’-D’) High-power images from AD, respectively. Note Edu-positive nuclei stained with Hoechst in s-α-actinin-positive CM and non-CM. Scale bar 25 µm. Note also the increased thickness of the s-α-actinin-positive CM in the compact layer of Control (C,C') vs. Mutant (D,D') hearts at 13.5 dpc. (E) CM number per high-power field (HPF) in the compact and trabecular layers from control or mutant hearts collected at 12.5 or 13.5 dpc. By two-way ANOVA, CM per HPF in both the compact and the trabecular layers was different by gestational age and genotype with a significant interaction term. Follow-on individual comparisons by Tukey test revealed that the total number of compact and trabecular cardiomyocytes increased in controls but not mutants from 12.5 to 13.5 dpc such that CM per HPF was different in control and mutant hearts at 13.5 dpc. (F) Edu-positive CM per HPF in the hearts analyzed in (E). For the compact layer, two-way ANOVA revealed a significant difference by genotype with a significant interaction term; individual comparisons by Tukey test demonstrated that the number of Edu-positive compact CM per HPF in controls at 13.5 dpc was different from mutant at 13.5 dpc and control at 12.5 dpc. For the trabecular layer, two-way ANOVA detected a significant difference by gestational age and genotype with a positive interaction effect, and individual comparisons by Tukey test showed that the number of Edu-positive trabecular CM per HPF in controls at 13.5 dpc was different from mutant at 13.5 and control at 12.5 dpc. *, p < 0.05; **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001.
Expanded distribution of trabecular cardiomyocyte markers in cardiomyocyte-specific S1pr1 knockout hearts
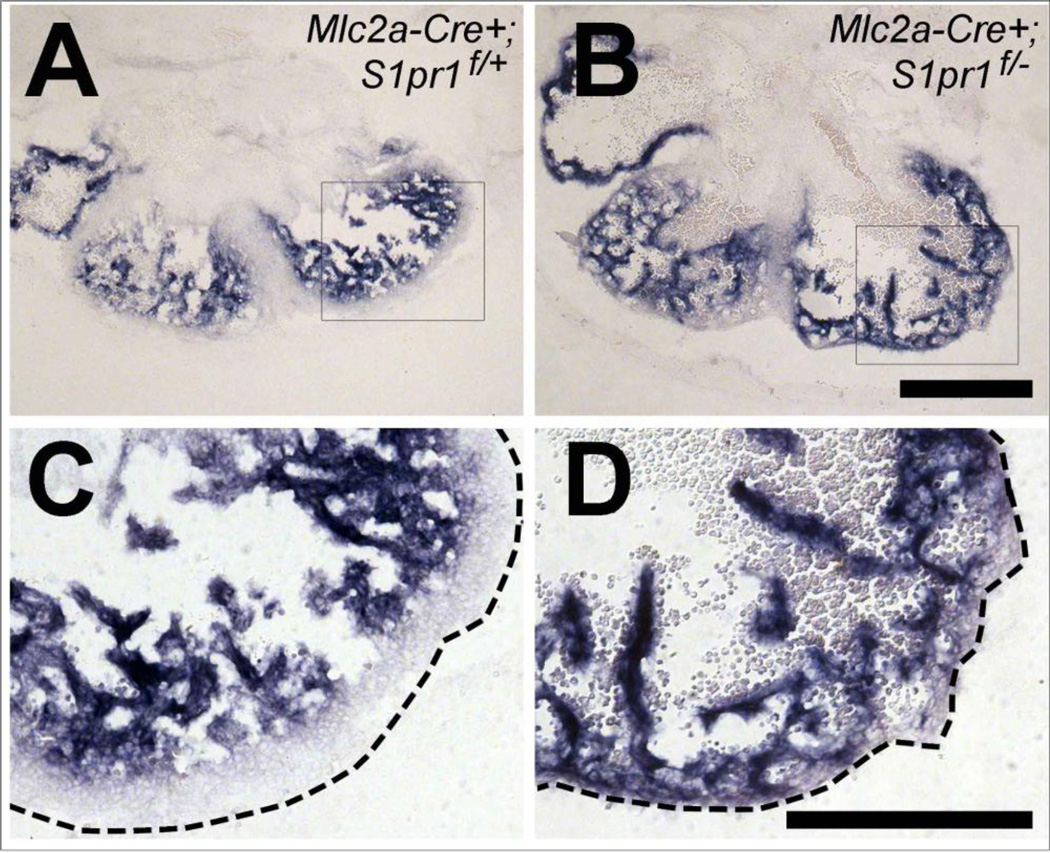
In addition to morphological differences, trabecular and compact myocardium have differential expression and distribution of markers including Nppa, Bmp10, and fibronectin. We next examined whether loss of S1pr1 affected the distribution of these markers using in situ hybridization and immunostaining. In hearts from control embryos collected at 13.5 dpc, Nppa and Bmp10 expression was detected in the trabecular layer and absent from the compact layer. In hearts from littermate Mlc2a-Cre+; S1pr1f/− mutants, Nppa and Bmp10 expression was detected in trabecular structures that extended to or almost to the epicardium (Figure 5 and Supplemental Figure 2). Similarly, fibronectin distribution was expanded into the compact layer in mutant hearts (Supplemental Figure 3). In controls collected at 16.5 dpc, fibronectin staining was most abundant in basement membrane just beneath endocardium, with high staining in endocardiumcovered trabeculae and low staining in the compact layer. In littermate Mlc2a-Cre+; S1pr1f/− mutant hearts, fibronectin staining extended almost to the epicardium (Supplemental Figure 3). Overall, the presence of trabecular markers close to the subepicardial region is consistent with a relative lack of a compact layer in the mutants and suggests that S1P1 signaling in cardiomyocytes is necessary for normal compaction.
Figure 5. Trabecular marker expression differences in cardiomyocyte-specific S1pr1 mutant hearts.
In situ hybridization for the trabecular marker Nppa was performed on littermate 13.5 dpc Mlc2a-Cre+;S1pr1f/+ control (A, C) and Mlc2a-Cre+;S1pr1f/− mutant (B, D) embryos. C and D are higher magnification views of the boxed areas of A and B, respectively. The dotted lines indicate the position of the epicardium. Note the presence of a zone free of blue Nppa staining (the compact layer) between the trabecular layer and epicardium in control but not mutant heart. Similar results were obtained with two embryos of each genotype. Scale bar 500 µm for low-power and 300 µm for high-power magnification.
Disorganization of the thin compact layer in cardiomyocyte-specific S1pr1 knockout hearts
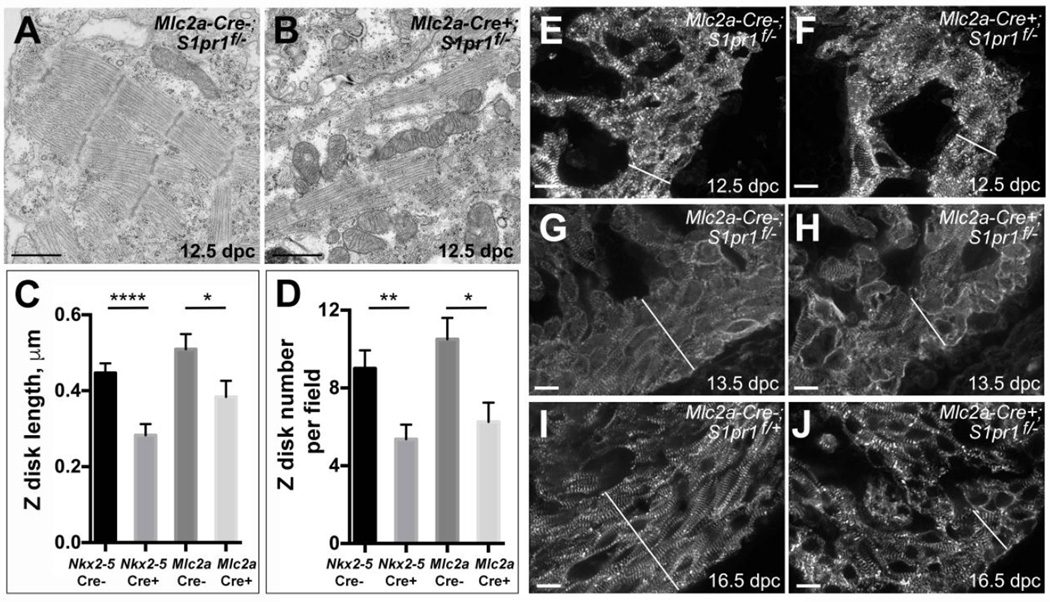
To determine whether the thin outer layer of cardiomyocytes in mutant hearts has features of normal compact myocardium, we examined cardiomyocyte and myofibril organization by transmission electron microscopy (TEM). TEM revealed myofibrils with Z discs and repeated sarcomeric units in compact layer cardiomyocytes in control hearts collected at 12.5 dpc. (Figure 6A and Supplemental Figure 4). In compact layer cardiomyocytes in mutant hearts, myofibrils appeared to be smaller with less overall organization than in controls (Figure 6B and Supplemental Figure 3) despite the absence of gross morphological changes at this time point (Figures 3 and 4). Quantitation of Z-disc size and number per field in TEM images revealed smaller and fewer Z-discs in mutant hearts than in control hearts (Figure 6C,D).
Figure 6. Structural changes in compact layer cardiomyocytes in cardiomyocyte-specific S1pr1 knockouts.
(A, B) Representative transmission electron microscopy in the left ventricle compact layer of hearts from (A) Mlc2aCre−;S1pr1f/− control and (B) Mlc2aCre+;S1pr1f/− mutant embryos collected at 12.5 dpc. Note narrower and less continuous myofibrils in the mutant heart. Scale bars 1 µm. (C, D) Mean Zdisc length (C) and number of Z-discs per field (D) in EM images of hearts from S1pr1f/− 12.5 dpc embryos negative and positive for Mlc2a-Cre or Nkx2-5-Cre transgenes. Error bars represent SEM. *, p < 0.05; **, p <0.01, and ****, p < 0.05 by Student’s unpaired t-test. (E-J) Representative s-α-actinin immunostaining of the left ventricle in control (E, G, H) or mutant (F, H, J) embryos collected at 12.5 dpc (E, F), 13.5 (G, H) and 16.5 dpc (I, J). Specific genotypes were as indicated. The epicardium is to the lower right of the field in each image, and the white diagonal line indicates the compact layer. At 12.5 dpc, no differences in s-α-actinin immunostaining were apparent between control and mutant hearts. At 13.5 dpc and 16.5 dpc, compact layer cardiomyocytes in control hearts appeared to align parallel to the epicardium, while cardiomyocytes in the thin compact layer of mutant hearts failed to align with each other or the epicardium. Similar results were seen in 8–14 hearts of each genotype at 12.5 dpc, 3–5 hearts per genotype at 13.5 dpc, and 4 hearts of each genotype at 16.5 dpc. Scale bars 10 µm.
Despite the ultrastructural differences noted in TEM images but in keeping with our other histological observations, immunostaining for the Z-disc component s-α-actinin did not reveal differences in pattern or intensity of immunofluorescence in compact layer cardiomyocytes in control and mutant hearts at 12.5 dpc (Figure 6E, F). However, at 13.5 dpc and 16.5 dpc, s-α-actinin of cardiomyocytes in the thin compact layer of mutant hearts displayed less regular myofibril alignment both within cardiomyocytes and between adjacent cardiomyocytes compared to the normal compact region of control hearts (Figure 6G–J and Supplemental Figure 4). In contrast to the compact layer, no differences in cell or myofibril alignment or sarcomere structure were observed in trabecular cardiomyocytes between control and mutant hearts: well-formed myofibrils aligned from cell to cell were readily detected by immunofluorescence and TEM, including within the trabecular cardiomyocytes adjacent to the thin mutant compact layer (Supplemental Figure 4). Taken together, the persistence of trabecular myocardium, the thin outer layer of cardiomyocytes where the compact myocardium would normally reside, and the decreased cellular and myofibril organization in this outer layer in the mutants suggests a necessary role for S1P1 in formation of a normal compact layer.
DISCUSSION
Our results demonstrate an unexpected role for S1pr1 in cardiomyocytes in heart development in mice. Specifically, loss of S1P1 in cardiomyocytes led to postnatal lethality, VSD, and ventricular noncompaction. To our knowledge, this necessary role for S1P1 in cardiomyocytes in heart development represents a previously unknown role for S1P signaling and a new role for cardiomyocyte GPCRs in the process of ventricular compaction.
Loss of S1P1 appears to affect the process of compaction by an overall reduction in trabecular and compact layer cardiomyocyte number after 12.5 dpc. Trabeculation and compaction are largely temporally distinct processes, with trabeculation beginning at ~9.5 dpc and ending by ~13.5 dpc, during which time the trabecular myocardium appears to be more differentiated than outer ventricular wall myocardium and provides the bulk of the contractile force of the heart (Samsa et al., 2013). Compaction begins at ~12.5 dpc, with compact layer cardiomyocytes maintaining a higher proliferative rate than trabecular myocardium until the compact layer comprises the majority of ventricular mass. Trabecular and compact cardiomyocyte numbers failed to increase normally after 12.5 in mutants lacking S1P1 in cardiomyocytes. The time of onset of this phenotype, after the majority of trabeculation has occurred but at the onset of compaction, probably accounts for the failed expansion of the compact layer but relative preservation of the trabecular layer in the mutants as seen by both cardiac morphology and trabecular marker expression in our study. The decreased myofibrillar organization in cardiomyocytes in the compact layer in the mutants at 12.5 dpc, before expansion of the compact layer begins and when mutant hearts were still morphologically indistinguishable from controls, raises the possibility that S1P1 may also contribute directly to differentiation of compact layer cardiomyocytes.
The number of total and Edu-positive (S-phase) trabecular and compact cardiomyocytes increased in control hearts but not cardiomyocyte S1pr1 mutant hearts between 12.5 dpc and 13.5 dpc; however, the ratio of Edu-positive cardiomyocytes to total cardiomyocytes was not different between control and mutant hearts in the windows sampled at either 12.5 dpc or 13.5 dpc (Supplemental Figure 5). Rates of apoptosis at these time points were insignificant. Thus, while a decreased number of cardiomyocytes available for proliferation might contribute to subsequent failure of compact layer expansion, how the difference in cardiomyocyte number between control and S1pr1 mutants arose during the period from 12.5 to 13.5 dpc is not clear. Precedent for alterations in cardiomyocyte number without detectable changes in the fraction of S-phase cells exists in other systems. In chick hearts lacking epicardium, the loss of one round of cell division over four days was associated with a 50% reduction in the number of cardiomyocytes compared to control without a detectable change in the rate of Brdu positivity (Pennisi et al., 2003). Lack of a detectable difference in cardiomyocyte proliferation or apoptosis in association with ventricular noncompaction was also noted in RXRalpha-deficient mouse embryos (Kastner et al., 1994). Whether slowing of cell cycle progression outside of S-phase, decreased DNA replication in a window not sampled in our Edu study (see below), or other explanations account for decreased cardiomyocyte number without a decreased fraction of Edupositive cardiomyocytes in the window examined remains to be determined.
S1pr1 expression, as assessed by β-galactosidase staining of hearts from S1pr1+/LacZ embryos, appeared confined to trabecular cardiomyocytes at and after of onset of the noncompaction phenotype. How might loss of S1P1 function in cardiomyocytes alter compaction if S1pr1 expression is limited to the trabecular layer? S1pr1 appears to be widely expressed in ventricular myocardium at 9.5 dpc in this (Supplemental Figure 1) and other studies (Liu et al., 2000; Poulsen et al., 2011). Thus, loss of S1P1 function at an early stage of myocardial development, before separation of the myocardium into trabecular and nascent compact layers has occurred, might alter the properties of cardiomyocytes in a manner that leads to a later decrease in expansion of cardiomyocyte numbers. The outsized effect on expansion of the compact layer may be for reasons of timing, as outlined above. Alternatively, altered differentiation with decreased responsiveness to epicardial growth/trophic factors in cardiomyocytes destined to form the compact layer might also contribute to noncompaction and perhaps to the decreased myofibril organization noted in compact layer cardiomyocytes at 12.5 dpc (Pennisi et al., 2003). Lastly, S1pr1 might be expressed in compact layer cardiomyocytes during compaction at levels that are functionally important but below the limits of detection of the β-galactosidase reporter, or S1P1 function in trabecular myocardium might somehow indirectly influence the behavior of compact layer cardiomyocytes.
Multiple complex and essential interactions between the myocardium and endocardium, epicardium, and cardiac fibroblasts contribute to ventricular compaction (Kastner et al., 1994; Lavine et al., 2005; Li et al., 2011; Pennisi et al., 2003). In accord, more than 60 genes have been linked to ventricular noncompaction in human and mouse studies (Paige et al., 2015; Towbin et al., 2015; Wilsbacher and McNally, 2016). Disruption of endocardial and myocardial Notch signaling pathways after the onset of compaction leads to ventricular noncompaction, but in contrast to cardiomyocyte-specific S1pr1 deletion, these mutants display increased proliferation especially within the trabecular myocardium (D'Amato et al., 2016; Luxán et al., 2013). Disruption of epicardial signals, such as retinoic acid, fibroblast growth factors, and insulin-like growth factor lead to noncompaction with reduced proliferation (Kastner et al., 1994; Lavine et al., 2005; Li et al., 2011); whether S1P1 signaling intersects with signals from these factors and their receptors remains unknown. Finally, myocardial excision of FOG2 (Zfpm2 gene) and focal adhesion kinase (FAK; Ptk2 gene), as well as cardiomyocyte-specific overexpression of the FAK inhibitory protein FRNK, all result in a noncompaction phenotype with decreased cardiomyocyte proliferation similar to that seen in cardiomyocyte S1pr1 mutants; these provide attractive candidate pathways for further study.
The source of S1P driving S1P1 signaling in cardiomyocytes during cardiac development remains unknown. S1P, primarily generated by erythrocytes (Pappu et al., 2007), is abundant in plasma, and it is plausible that plasma S1P might traverse endocardium in the developing heart to reach cardiomyocytes. Alternatively, S1P generated by the epicardium, endocardium, fibroblasts or cardiomyocytes themselves might contribute to activation of cardiomyocyte S1P1. Finally, S1P1 in cardiomyocytes may participate in a mechanosensing program in response to stretch or strain, as has been suggested in endothelial cells in response to flow (Jung et al., 2012). In such a scenario, S1P1 might sense wall stress as a milestone in cardiac development and trigger compaction as a result. Future studies will address potential sources of S1P via conditional deletion of sphingosine kinases 1 and 2, the enzymes that generate S1P (Camerer et al., 2009; Gaengel et al., 2012; Wang and Dudek, 2009).
In conclusion, our results demonstrate an unexpected role for S1P1 signaling in cardiomyocytes in the developing heart in expansion of cardiomyocyte number and ventricular compaction. Exploration of gene expression and signaling pathways altered by loss of cardiomyocyte S1P1 may provide further insight into the mechanisms by which ventricular compaction is regulated. Future studies will identify the cellular and molecular mechanisms by which S1P1 signaling in cardiomyocytes contributes to compaction, the source of the S1P ligand important for cardiomyocyte S1P1 function, and whether those mutant mice that survive to adulthood might be a useful model of cardiomyopathy associated with noncompaction.
Supplementary Material
Highlights.
Mouse embryo cardiomyocytes (CM) express S1P1, a sphingosine 1-phosphate receptor.
Normal expansion of cardiomyocyte number in mouse embryos requires S1P1.
Loss of S1P1 in CM results in ventricular noncompaction.
These results suggest a new role for S1P signaling.
Acknowledgments
The authors thank Drs. Brian Black, Ken Chien, and Xu Peng for sharing mouse lines and genotyping protocols, Dr. Lisa Hua for sharing the Nppa and Bmp10 plasmids for in situ probes, Dr. Konstantin Gaengel for S1pr1 genotyping protocols, Jinny Wong for Gladstone Electron Microscopy Core services, Caroline Miller for Gladstone Histology Core services, and Cherry Concengco for animal husbandry. Confocal microscopy was performed at the Cardiovascular Research Institute Imaging Core at the University of California, San Francisco. The authors also thank Drs. Kathleen Ruppel, Brian Black, and Takashi Mikawa for helpful discussions.
Funding sources
This work was supported in part by NIH T32HL007731 and American Heart Association 10POST3020068 (HC), NIH K08HL105657 and the Gilead Research Scholars Program in Cardiovascular Disease (LDW), NIH NS048478 and DA019674 (JC), and NIH HL65590 and HL054737 (SRC). The funding sources had no role in the design, collection and analysis of data, writing, or submission of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors have no competing or financial interests.
Nomenclature
Sphingosine 1-phosphate receptor-1 protein is designated S1P1 per IUPHAR nomenclature. S1pr1 designates the mouse gene encoding S1P1.
REFERENCES
- Allende ML, Yamashita T, Proia RL. G-protein-coupled receptor S1P1 acts within endothelial cells to regulate vascular maturation. Blood. 2003;102:3665–3667. doi: 10.1182/blood-2003-02-0460. [DOI] [PubMed] [Google Scholar]
- Blaho VA, Hla T. Regulation of mammalian physiology, development, and disease by the sphingosine 1-phosphate and lysophosphatidic acid receptors. Chemical reviews. 2011;111:6299–6320. doi: 10.1021/cr200273u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerer E, Regard JB, Cornelissen I, Srinivasan Y, Duong DN, Palmer D, Pham TH, Wong JS, Pappu R, Coughlin SR. Sphingosine-1-phosphate in the plasma compartment regulates basal and inflammation-induced vascular leak in mice. J Clin Invest. 2009;119:1871–1879. doi: 10.1172/JCI38575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JW, Gardell SE, Herr DR, Rivera R, Lee C-W, Noguchi K, Teo ST, Yung YC, Lu M, Kennedy G, Chun J. FTY720 (fingolimod) efficacy in an animal model of multiple sclerosis requires astrocyte sphingosine 1-phosphate receptor 1 (S1P1) modulation. Proc Natl Acad Sci USA. 2011;108:751–756. doi: 10.1073/pnas.1014154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster JG, Schwab SR. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annual review of immunology. 2012;30:69–94. doi: 10.1146/annurev-immunol-020711-075011. [DOI] [PubMed] [Google Scholar]
- D'Amato G, Luxan G, Del Monte-Nieto G, Martinez-Poveda B, Torroja C, Walter W, Bochter MS, Benedito R, Cole S, Martinez F, Hadjantonakis AK, Uemura A, Jimenez-Borreguero LJ, de la Pompa JL. Sequential Notch activation regulates ventricular chamber development. Nat Cell Biol. 2016;18:7–20. doi: 10.1038/ncb3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati C, Bruni P. Sphingosine 1-phosphate regulates cytoskeleton dynamics: implications in its biological response. Biochim Biophys Acta. 2006;1758:2037–2048. doi: 10.1016/j.bbamem.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Gaengel K, Niaudet C, Hagikura K, Siemsen BL, Muhl L, Hofmann JJ, Ebarasi L, Nyström S, Rymo S, Chen LL, Pang M-F, Jin Y, Raschperger E, Roswall P, Schulte D, Benedito R, Larsson J, Hellstrom M, Fuxe J, Uhlén P, Adams R, Jakobsson L, Majumdar A, Vestweber D, Uv A, Betsholtz C. The Sphingosine-1-Phosphate Receptor S1PR1 Restricts Sprouting Angiogenesis by Regulating the Interplay between VE-Cadherin and VEGFR2. Dev. Cell. 2012;23:587–599. doi: 10.1016/j.devcel.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Garcia JG, Liu F, Verin AD, Birukova A, Dechert MA, Gerthoffer WT, Bamberg JR, English D. Sphingosine 1-phosphate promotes endothelial cell barrier integrity by Edg-dependent cytoskeletal rearrangement. J Clin Invest. 2001;108:689–701. doi: 10.1172/JCI12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hla T, Lee MJ, Ancellin N, Paik JH, Kluk MJ. Lysophospholipids--receptor revelations. Science. 2001;294:1875–1878. doi: 10.1126/science.1065323. [DOI] [PubMed] [Google Scholar]
- Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong T-T, Shaw RM, Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev. Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B, Obinata H, Galvani S, Mendelson K, Ding B-s, Skoura A, Kinzel B, Brinkmann V, Rafii S, Evans T, Hla T. Flow-Regulated Endothelial S1P Receptor-1 Signaling Sustains Vascular Development. Dev. Cell. 2012;23:600–610. doi: 10.1016/j.devcel.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastner P, Grondona JM, Mark M, Gansmuller A, LeMeur M, Decimo D, Vonesch JL, Dollé P, Chambon P. Genetic analysis of RXR alpha developmental function: convergence of RXR and RAR signaling pathways in heart and eye morphogenesis. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- Koibuchi N, Chin MT. CHF1/Hey2 plays a pivotal role in left ventricular maturation through suppression of ectopic atrial gene expression. Circ Res. 2007;100:850–855. doi: 10.1161/01.RES.0000261693.13269.bf. [DOI] [PubMed] [Google Scholar]
- Kurrasch DM, Nevin LM, Wong JS, Baier H, Ingraham HA. Neuroendocrine transcriptional programs adapt dynamically to the supply and demand for neuropeptides as revealed in NSF mutant zebrafish. Neural development. 2009;4:22. doi: 10.1186/1749-8104-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev. Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lee Y, Song AJ, Baker R, Micales B, Conway SJ, Lyons GE. Jumonji, a nuclear protein that is necessary for normal heart development. Circulation Research. 2000;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- Li J, Miao L, Shieh D, Spiotto E, Li J, Zhou B, Paul A, Schwartz RJ, Firulli AB, Singer HA, Huang G, Wu M. Single-Cell Lineage Tracing Reveals that Oriented Cell Division Contributes to Trabecular Morphogenesis and Regional Specification. Cell reports. 2016;15:158–170. doi: 10.1016/j.celrep.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Cavallero S, Gu Y, Chen THP, Hughes J, Hassan AB, Brüning JC, Pashmforoush M, Sucov HM. IGF signaling directs ventricular cardiomyocyte proliferation during embryonic heart development. Development. 2011;138:1795–1805. doi: 10.1242/dev.054338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wada R, Yamashita T, Mi Y, Deng CX, Hobson JP, Rosenfeldt HM, Nava VE, Chae SS, Lee MJ, Liu CH, Hla T, Spiegel S, Proia RL. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J Clin Invest. 2000;106:951–961. doi: 10.1172/JCI10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxán G, Casanova JC, Martínez-Poveda B, Prados B, D'Amato G, Macgrogan D, Gonzalez-Rajal A, Dobarro D, Torroja C, Martinez F, Izquierdo-García JL, Fernández-Friera L, Sabater-Molina M, Kong Y-Y, Pizarro G, Ibañez B, Medrano C, García-Pavía P, Gimeno JR, Monserrat L, Jiménez-Borreguero LJ, de la Pompa JL. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat. Med. 2013;19:193–201. doi: 10.1038/nm.3046. [DOI] [PubMed] [Google Scholar]
- Morikawa Y, Cserjesi P. Cardiac neural crest expression of Hand2 regulates outflow and second heart field development. Circ Res. 2008;103:1422–1429. doi: 10.1161/CIRCRESAHA.108.180083. [DOI] [PubMed] [Google Scholar]
- Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- Mutoh T, Rivera R, Chun J. Insights into the pharmacological relevance of lysophospholipid receptors. Br J Pharmacol. 2012;165:829–844. doi: 10.1111/j.1476-5381.2011.01622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Plonowska K, Xu A, Wu SM. Molecular regulation of cardiomyocyte differentiation. Circ Res. 2015;116:341–353. doi: 10.1161/CIRCRESAHA.116.302752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappu R, Schwab SR, Cornelissen I, Pereira JP, Regard JB, Xu Y, Camerer E, Zheng YW, Huang Y, Cyster JG, Coughlin SR. Promotion of lymphocyte egress into blood and lymph by distinct sources of sphingosine-1-phosphate. Science. 2007;316:295–298. doi: 10.1126/science.1139221. [DOI] [PubMed] [Google Scholar]
- Peng X, Wu X, Druso JE, Wei H, Park AY-J, Kraus MS, Alcaraz A, Chen J, Chien S, Cerione RA, Guan J-L. Cardiac developmental defects and eccentric right ventricular hypertrophy in cardiomyocyte focal adhesion kinase (FAK) conditional knockout mice. Proc Natl Acad Sci USA. 2008;105:6638–6643. doi: 10.1073/pnas.0802319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi DJ, Ballard VLT, Mikawa T. Epicardium is required for the full rate of myocyte proliferation and levels of expression of myocyte mitogenic factors FGF2 and its receptor, FGFR-1, but not for transmural myocardial patterning in the embryonic chick heart. Dev. Dyn. 2003;228:161–172. doi: 10.1002/dvdy.10360. [DOI] [PubMed] [Google Scholar]
- Poulsen RR, McClaskey CM, Rivkees SA, Wendler CC. The Sphingosine-1-phospate receptor 1 mediates S1P action during cardiac development. BMC Dev. Biol. 2011;11:37. doi: 10.1186/1471-213X-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen H, Gonzalez-Cabrera PJ, Sanna MG, Brown S. Sphingosine 1-phosphate receptor signaling. Annual review of biochemistry. 2009;78:743–768. doi: 10.1146/annurev.biochem.78.072407.103733. [DOI] [PubMed] [Google Scholar]
- Samsa LA, Yang B, Liu J. Embryonic cardiac chamber maturation: Trabeculation, conduction, and cardiomyocyte proliferation. American journal of medical genetics. Part C, Seminars in medical genetics. 2013;163C:157–168. doi: 10.1002/ajmg.c.31366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton PA, Dudek SM, Chiang ET, Garcia JG. Regulation of sphingosine 1-phosphate-induced endothelial cytoskeletal rearrangement and barrier enhancement by S1P1 receptor, PI3 kinase, Tiam1/Rac1, and alpha-actinin. FASEB J. 2005;19:1646–1656. doi: 10.1096/fj.05-3928com. [DOI] [PubMed] [Google Scholar]
- Towbin JA, Lorts A, Jefferies JL. Left ventricular non-compaction cardiomyopathy. Lancet. 2015;386:813–825. doi: 10.1016/S0140-6736(14)61282-4. [DOI] [PubMed] [Google Scholar]
- Wang L, Dudek SM. Regulation of vascular permeability by sphingosine 1-phosphate. Microvasc Res. 2009;77:39–45. doi: 10.1016/j.mvr.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettschureck N, Rütten H, Zywietz A, Gehring D, Wilkie TM, Chen J, Chien KR, Offermanns S. Absence of pressure overload induced myocardial hypertrophy after conditional inactivation of Galphaq/Galpha11 in cardiomyocytes. Nat. Med. 2001;7:1236–1240. doi: 10.1038/nm1101-1236. [DOI] [PubMed] [Google Scholar]
- Wilsbacher L, McNally EM. Genetics of Cardiac Developmental Disorders: Cardiomyocyte Proliferation and Growth and Relevance to Heart Failure. Annu Rev Pathol. 2016;11 doi: 10.1146/annurev-pathol-012615-044336. Epub ahead of Print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbacher LD, Coughlin SR. Analysis of Cardiomyocyte Development using Immunofluorescence in Embryonic Mouse Heart. Journal of visualized experiments : JoVE. 2015 doi: 10.3791/52644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.