Abstract
Retinoic acid (RA) repression of Fgf8 is required for many different aspects of organogenesis, however relatively little is known about how endogenous RA controls gene repression as opposed to gene activation. Here, we show that nuclear receptor corepressors NCOR1 and NCOR2 (SMRT) redundantly mediate the ability of RA to repress Fgf8. Ncor1;Ncor2 double mutants generated by CRISPR/Cas9 gene editing exhibited a small somite and distended heart phenotype similar to that of RA-deficient Raldh2−/− embryos, associated with increased Fgf8 expression and FGF signaling in caudal progenitors and heart progenitors. Embryo chromatin immunoprecipitation studies revealed that NCOR1/2 but not coactivators are recruited to the Fgf8 RA response element (RARE) in an RA-dependent manner, whereas coactivators but not NCOR1/2 are recruited RA-dependently to a RARE near Rarb that is activated by RA. CRISPR/Cas9-mediated genomic deletion of the Fgf8 RARE in mouse embryos often resulted in a small somite defect with Fgf8 derepression caudally, but no defect was observed in heart development or heart Fgf8 expression. This suggests the existence of another DNA element whose function overlaps with the Fgf8 RARE to mediate Fgf8 repression by RA and NCOR1/2. Our studies support a model in which NCOR1/2 mediates direct RA-dependent repression of Fgf8 in caudal progenitors in order to control somitogenesis.
Keywords: Caudal progenitors, somitogenesis, nuclear receptor corepressor, NCOR, SMRT, Fgf8, Retinoic acid response element, CRISPR/Cas9 gene editing
1. Introduction
Retinoic acid (RA) is a ligand for nuclear RA receptors (RARα, RARβ, RARγ) that form heterodimers with retinoid-X receptor (RXR) and bind to RA response elements (RAREs) near target genes (Cunningham and Duester, 2015). During vertebrate embryogenesis, RA is generated mostly by retinaldehyde dehydrogenase 2 (RALDH2; ALDH1A2) (Duester, 2008; Niederreither and Dolle, 2008). RA synthesis is first observed in the presomitic mesoderm just prior to formation of the first somite when Raldh2 expression initiates (Sirbu et al., 2005). Loss of RA synthesis in Raldh2−/− mouse embryos or vitamin A deficient avian embryos leads to increased Fgf8 expression along the body axis associated with smaller somite size, distended heart tube, loss of forelimb initiation, loss of posterior neurogenesis, and premature termination of body axis extension (Cunningham et al., 2013; Diez del Corral et al., 2003; Mic et al., 2002; Niederreither et al., 1999; Sirbu and Duester, 2006; Vermot et al., 2005; Vermot and Pourquié, 2005).
Recent studies indicate that this early role of RA signaling is mediated at least in part by its effects on a neuromesodermal progenitor (NMP) cell population present in the caudal region of vertebrate embryos that coordinates somitogenesis with spinal cord neurogenesis (Henrique et al., 2015; Tzouanacou et al., 2009; Wilson et al., 2009). Bipotential NMPs expressing the mesodermal marker T (Brachyury) and low levels of the neuroectodermal marker Sox2 are first located in the caudal epiblast lying on each side of the primitive streak and in the tailbud at later developmental stages (Martin and Kimelman, 2012; Olivera-Martinez et al., 2012; Tsakiridis et al., 2014). As the body axis extends, NMPs entering the primitive streak undergo epithelial-to-mesenchymal transition to differentiate into presomitic mesodermal cells expressing Tbx6 that functions to repress Sox2, while NMPs that stay in the caudal epiblast epithelial layer differentiate to neural plate cells expressing high levels of Sox2 (Martin and Kimelman, 2009; Takemoto et al., 2011). This model is supported by Tbx6 loss-of-function studies showing conversion of somites into ectopic neural tubes (Chapman and Papaioannou, 1998; Takemoto et al., 2011). Caudal FGF and Wnt signaling is required to maintain NMPs to promote body axis extension (Aulehla et al., 2003; Boulet and Capecchi, 2012; Ciruna and Rossant, 2001; Dunty et al., 2008; Martin and Kimelman, 2012; Naiche et al., 2011; Olivera-Martinez et al., 2012). Treatment of chick embryos with an RALDH inhibitor, or loss of RA synthesis in Raldh2−/− mouse embryos, results in expanded caudal Fgf8 expression responsible for a small somite phenotype as well as loss of Sox2 expression coupled with increased Tbx6 expression in the NMP niche (Cunningham et al., 2015; Olivera-Martinez et al., 2012). Loss of RA synthesis in Raldh2−/− embryos results not only in ectopic anterior expansion of the caudal Fgf8 domain, but also ectopic posterior expansion of the heart Fgf8 domain that controls early second heart field progenitors, resulting in ectopic FGF signaling throughout the developing trunk that leads to a distended heart tube and inhibition of forelimb initiation (Cunningham et al., 2013; Ryckebusch et al., 2008; Sirbu et al., 2008; Zhao et al., 2009). Thus, RA antagonism of Fgf8 represents a key developmental pathway controlling progenitor cell differentiation that is required for many different aspects of organogenesis.
The mechanism through which RA antagonizes Fgf8 is currently being explored. As caudal progenitor cells migrate to the developing trunk during body axis extension, their Fgf8 chromosomal locus becomes located more peripherally in the nucleus (a location associated with repression), plus this shift to the nuclear periphery requires RA generated by Raldh2 (Patel et al., 2013). Analysis of wild-type and mutant bacterial artificial chromosome transgenes carrying the mouse Fgf8 locus fused to lacZ demonstrated that a conserved RARE was able to restrict Fgf8-lacZ expression to the caudal progenitors of mouse embryos; also, chromatin immunoprecipitation studies on wild-type and Raldh2−/− embryos showed that Polycomb repressive complex 2 (PRC2) and the repressive H3K27me3 mark are recruited near the Fgf8 RARE in an RA-dependent manner (Kumar and Duester, 2014). These studies suggest that RA directly represses Fgf8 transcription.
While control of gene activation by RA has been widely studied, relatively little is known about how RA controls gene repression (Cunningham and Duester, 2015). Signal-induced repression has been suggested to play a major role in developmental signaling (Affolter et al., 2008). Therefore, an increased understanding of the epigenetic mechanism used by RA to control gene repression will allow us to more fully understand how RA controls developmental processes, thus providing insight into how RA may be best used for differentiation of stem/progenitor cells in vitro. Early studies on nuclear receptor function performed mostly in transfected cell lines suggested that liganded receptors (such as RAR bound to RA) recruit nuclear receptor coactivators (NCOA1, NCOA2, NCOA3; also known as SRC1/2/3) that stimulate transcriptional activation, whereas unliganded receptors recruit the nuclear receptor corepressors NCOR1 and NCOR2 (also known as silencing mediator of retinoic acid and thyroid hormone receptors; SMRT) that mediate transcriptional repression (Chen et al., 1996; Hörlein et al., 1995; Xu et al., 1999). Nuclear receptor coactivators and corepressors continually bind and release to allow quick changes in gene activation (Perissi et al., 2004). However, no clear example of gene repression by ligand-dependent recruitment of NCOR1/2 to RA receptors has been found (Glass and Rosenfeld, 2000; Jepsen et al., 2000; Perissi et al., 2010). Here, analysis of mouse Ncor1 and Ncor2 double mutants generated by CRISPR/Cas9-mediated gene editing combined with other studies demonstrates that NCOR1/2 mediates RA-dependent repression of Fgf8 in order to control somitogenesis. Our in vivo studies provide new mechanistic insight into NCOR1/2 function that support a model for direct RA repression of Fgf8 via RA-dependent recruitment of NCOR1/2 to the Fgf8 RARE.
2. Results
2.1. Loss of both Ncor1 and Ncor2 results in a small somite phenotype and ectopic FGF signaling
The Ncor1 knockout dies at E15.5 with defects in erythropoiesis and brain neurogenesis, whereas the Ncor2 knockout dies at E16.5 with reduced heart growth and forebrain defects, but no early embryonic defects were reported; also, double mutants have not been reported (Mottis et al., 2013). In order to determine whether Ncor1 and Ncor2 have redundant functions in early development, double knockout mutations were generated using CRISPR/Cas9 gene editing (Wang et al., 2013). We performed CRISPR/Cas9 gene editing using two sgRNAs each for Ncor1 and Ncor2 (targeting both the ATG start codon and first splice donor sites for each gene). Of 24 embryos dissected at E8.5, yolk sac DNA sequencing of exon 1 identified 7 embryos that are Ncor1;Ncor2 double biallelic null mutants based on deletions/insertions in all four alleles that affect translation initiation or RNA splicing, or that create frameshifts, with 10 other embryos harboring these types of mutations in two or three alleles, and 1 embryo with no mutations; 6 embryos possessed more than two detectable alleles (mosaicism) at the Ncor1 and/or Ncor2 loci (Table 1). Thus, 29% of the embryos exhibited non-mosaic double biallelic mutations. DNA sequencing of embryos after in situ hybridization revealed the same biallelic deletions in Ncor1 and Ncor2 as observed in the original yolk sac DNA sequence; we obtained successful post-hybridization PCR for embryos #9 and #23, and their Ncor1 and Ncor2 DNA sequences exhibited the same mutations observed in yolk sac DNA sequences.
Table 1.
Embryos obtained from CRISPR/Cas9 gene editing of Ncor1/Ncor2.
| Embryo | Number of alleles with deletions/ Genotype |
Somites (Uncx) |
Heart Tube | FGF signaling (Fgf8 or Spry2) |
|---|---|---|---|---|
| 10 | (0) Ncor1+/+;Ncor2+/+ | normal | normal | # |
| 2 | (2) Ncor1+/−;Ncor2+/− | normal | normal | normal Fgf8 |
| 7 | (2) Ncor1+/−;Ncor2+/− | normal | normal | # |
| 8 | (2) Ncor1+/−;Ncor2+/− | normal | normal | # |
| 4 | (2) Ncor1+/−;Ncor2+/− | normal | normal | normal Fgf8 |
| *19 | (3) Ncor1−/−;Ncor2+/− | small | distended | # |
| *20 | (3) Ncor1−/−;Ncor2+/− | small | distended | # |
| *5 | (3) Ncor1+/−;Ncor2−/− | small | distended | # |
| *6 | (3) Ncor1+/−;Ncor2−/− | small | distended | # |
| *11 | (3) Ncor1−/−;Ncor2+/− | # | distended | Ectopic Spry2 |
| *16 | (3) Ncor1−/+;Ncor2−/− | small | distended | # |
| *12 | (4) Ncor1−/−;Ncor2−/− | # | distended | Ectopic Fgf8 |
| *14 | (4) Ncor1−/−;Ncor2−/− | small | distended | # |
| *3 | (4) Ncor1−/−;Ncor2−/− | # | distended | Ectopic Spry2 |
| *23 | (4) Ncor1−/−;Ncor2−/− | small | distended | # |
| *9 | (4) Ncor1−/−;Ncor2−/− | small | distended | # |
| *13 | (4) Ncor1−/−;Ncor2−/− | # | distended | Ectopic Spry2 |
| *17 | (4) Ncor1−/−;Ncor2−/− | # | distended | Ectopic Fgf8 |
Total 24 embryos with 6 embryos (numbers 1, 15, 18, 21, 22, 24) not examined as they were mosaic for DNA sequence near exon 1 for Ncor1 and/or Ncor2 (more than two alleles were obtained often including wild-type sequence).
, embryos with mutant phenotype;
, not analyzed.
All embryos harboring null mutations in three or four alleles exhibited distended heart tubes, whereas embryos carrying only two null mutant alleles had normal heart tubes (Fig. 1, Table 1). Embryos carrying three or four null mutant alleles that were examined for Uncx expression by in situ hybridization all displayed small somites, whereas embryos carrying two null mutant alleles did not exhibit small somites (Fig. 1, Table 1). Measurements of somite height along the anteroposterior axis demonstrated that Ncor1;Ncor2 mutants carrying three or four null mutant alleles possess a somite height that is reduced to approximately 55% that observed in wild-type.
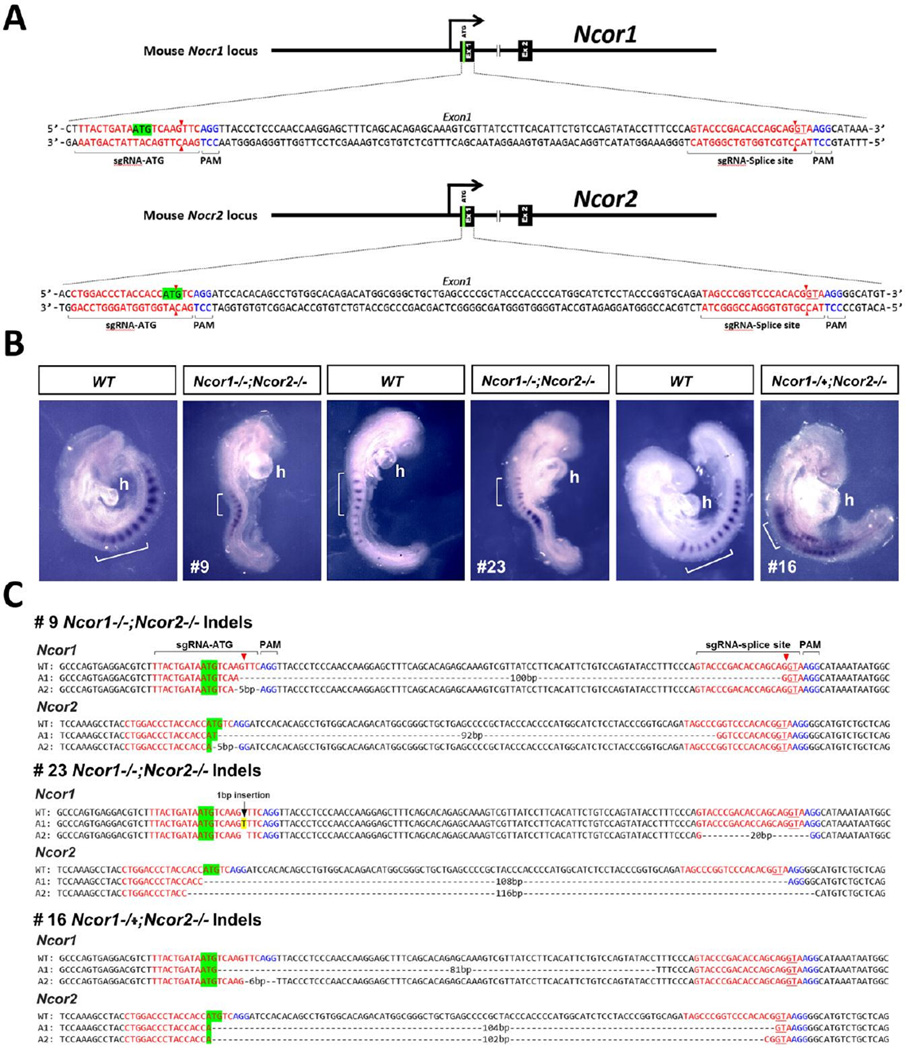
Fig. 1.
CRISPR/Cas9 gene editing of both Ncor1 and Ncor2 results in disruption of somitogenesis and heart development. (A) Strategy for generating mutations in the first exons of Ncor1 and Ncor2 using sgRNAs targeting the ATG start codon and GT splice donor site. (B) Shown is expression of the somite marker Uncx for wild-type (WT) or CRISPR mutants; brackets mark 6-somite regions for each embryo; h, heart (mutants exhibit distended heart tube). (C) DNA sequences for WT and both alleles (A1 and A2; sense strand) of Ncor1 and Ncor2 for the three CRISPR mutants shown above demonstrate that each mutant carries biallelic insertion or deletion mutations in both Ncor1 and Ncor2 that disrupt the ATG start codon, GT splice donor site, or reading frame. As Ncor1 A2 for embryo #16 carries an in-frame 6 bp deletion expected to delete only amino acids 4 and 5, this embryo is likely to have three null alleles rather than four.
Most Ncor1 and Ncor2 alleles with deletions between the ATG start codon and GT spice donor are considered null mutations as either the ATG or GT is lost or the deletion creates a translational frameshift since it is not multiple of 3 bp. Some embryos contain alleles with deletions between the ATG and GT that are a multiple of 3 bp, thus resulting in shorter proteins that may still retain some function. For example, embryo #16 contains one Ncor1 allele with an in-frame 6 bp deletion after the ATG start codon expected to remove only amino acids 4 and 5 that may not affect function, and another Ncor1 allele with an in-frame 81 bp deletion expected to remove 27 amino acids that may affect function as the N-terminal region harbors a repressive domain (Perissi et al., 2010). Thus, embryo #16 is considered to have 3 null alleles (Ncor1−/+; Ncor2−/−), a designation that is consistent with the observation of smaller somites although perhaps not as severe a somite defect as for embryos containing 4 null alleles for Ncor1 and Ncor2 (Fig. 1).
Embryos carrying four mutant alleles also exhibited ectopic expression of Fgf8 or Spry2 (induced by FGF signaling) in both the heart domain (expanded posteriorly) and the caudal domain (expanded anteriorly) (Fig. 2; Table 1). DNA sequences near exon 1 of Ncor1 and Ncor2 demonstrated a wide variety of mutations, most expected to be null mutations due to loss of the ATG start codon, loss of the GT splice donor, or frame shift (Figs.1–2; Fig. S1). These genetic loss-of-function studies provide evidence that Ncor1 and Ncor2 are both required for the epigenetic mechanism that represses Fgf8 along the body axis during early development in order to reduce FGF signaling in the developing trunk needed for normal somitogenesis and heart tube formation.
Fig. 2.
Double knockout mutations of Ncor1 and Ncor2 generated using CRISPR/Cas9 gene editing result in ectopic Fgf8 expression and FGF signaling in both heart and caudal domains. (A–B) Shown is expression of Fgf8 (panel A) or the FGF target gene Spry2 (panel B) for wild-type (WT) or CRISPR mutants; arrows mark the posterior border of the heart expression domain and the anterior border of the caudal expression domain; h, heart (mutants exhibit distended heart tube); the caudal end of embryo #12 was cut off in order to align with a WT embryo; embryo #3 lateral view (left panels) has the heads aligned with WT to best compare the heart domains, whereas the dorsal view (middle panels) has the tails aligned with WT to best compare the caudal domains. (C) DNA sequences of the three embryos shown above demonstrate biallelic insertion or deletion mutations in both Ncor1 and Ncor2 that are expected to be null mutations.
2.2. RA-dependent recruitment of NCOR1/2 to the Fgf8 RARE
RA response elements consist of two direct repeats, with each repeat binding either the RA receptor (RAR) or its heterodimer partner retinoid X receptor (RXR); most function in activation of transcription, but a few repressive RAREs have been reported (Cunningham and Duester, 2015). Previous studies demonstrated that the repressive Fgf8 RARE recruits the RA receptor (RAR) (Kumar and Duester, 2014), but recruitment of its heterodimer partner RXR had not been analyzed. Here, we used chromatin immunoprecipitation (ChIP) on E8.25 mouse embryo trunk tissue to show that the Fgf8 RARE recruits RXRa as does the Rarb RARE (Fig. 3A–B). Thus, the repressive Fgf8 RARE appears to function by recruitment of RAR/RXR heterodimers similar to activating RAREs such as that near the Rarb gene (Mendelsohn et al., 1991).
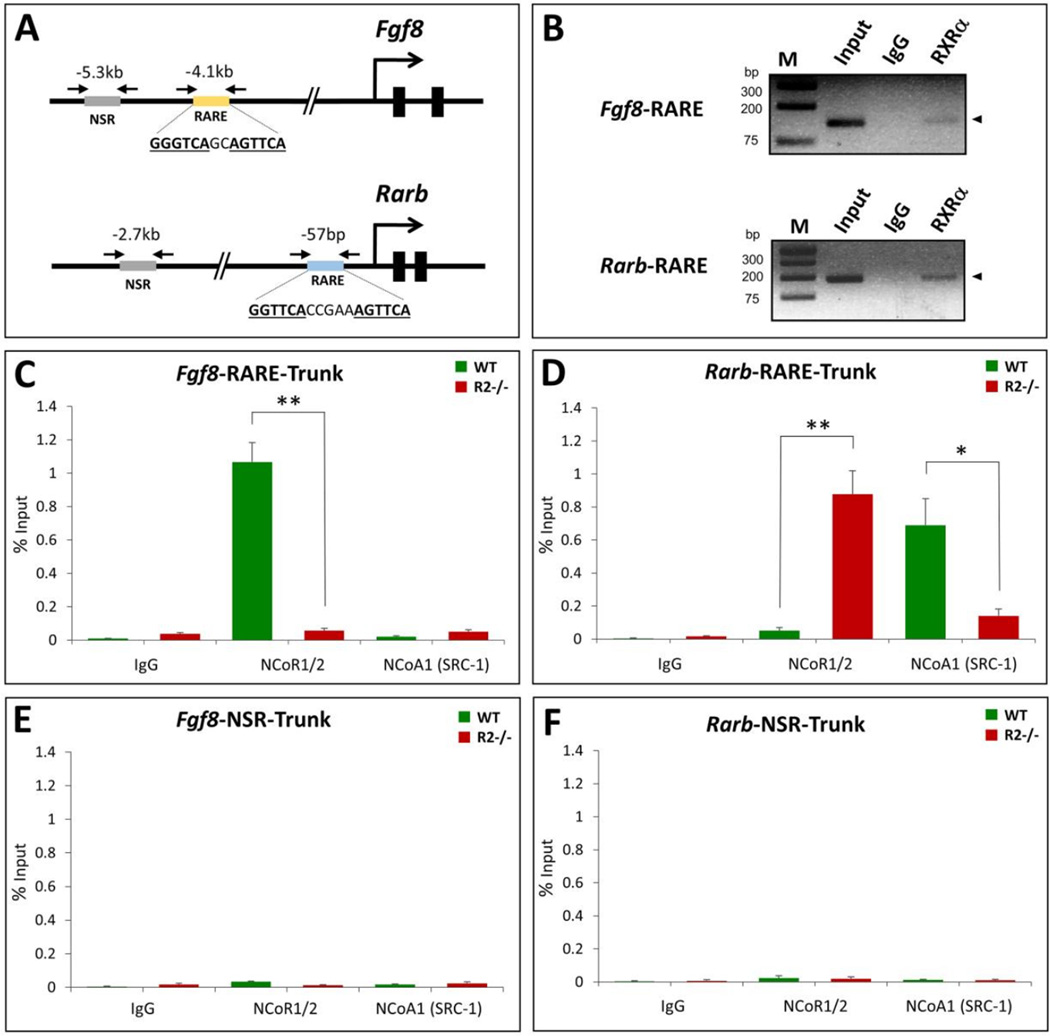
Fig. 3.
RA-dependent recruitment of NCOR1/2 to the Fgf8 RARE. Shown are ChIP studies on chromatin from pooled embryonic trunk tissues of wild-type (WT) or Raldh2−/− (R2−/−) E8.25 embryos that were cut just posterior to the heart and just posterior to the most recently formed somite to release the trunk. (A) Schematic showing locations of RAREs near Fgf8 and Rarb genes; arrows point to location of primers used for ChIP PCR to examine recruitment of proteins to the RARE or a non-specific region (NSR). (B) ChIP was performed using antibodies against RXRa or IgG (control); input DNA (diluted 100-fold) and immunoprecipitated DNA was analyzed by PCR using primers flanking Fgf8 and Rarb RAREs; M, molecular size markers. (C–F) ChIP assays using trunk chromatin from pooled wild-type (WT) or Raldh2−/− (KO) embryos showing effect of loss of RA activity (R2−/− mutants) on recruitment of NCOR1/2 (pooled antibodies for NCOR1 and NCOR2) or NCOA1 (SRC-1) near the Fgf8 RARE (panel C), the Rarb RARE (panel D), an NSR near Fgf8 as a negative control (panel E), or an NSR near Rarb as a negative control (panel F). Controls included assays with non-specific IgG antibody. Data shown as % input, mean ± SEM; *p < 0.05, **p < 0.01 (t test).
As NCOR1 and NCOR2 are known to be recruited to RAR/RXR heterodimers bound to RAREs, we performed studies to determine if these nuclear receptor corepressors are recruited to the repressive Fgf8 RARE. Previous studies using embryo ChIP demonstrated that the Fgf8 RARE recruits Polycomb Repressive Complex 2 (PRC2) and histone deacetylase 1 (HDAC1), plus it stimulates introduction of the repressive histone mark H3K27me3 in an RA-dependent fashion; interestingly, the coregulator RERE was recruited in the absence of RA but removed by RA suggesting that it does not function as a corepressor but could be a coactivator for Fgf8 in tissues that lack RA (Kumar and Duester, 2014). Those studies were performed using wild-type embryos and RA-deficient Raldh2−/− embryos lacking RA synthesis collected by traditional timed matings of Raldh2+/− adult mice, a time-consuming process if many embryos are needed as is the case for embryo ChIP studies. Here, in order to more efficiently perform embryo ChIP on RA-deficient mouse embryos, CRISPR/Cas9 gene editing was used to generate a large number of Raldh2−/− embryos. Using this method we obtained 52 embryos out of a total of 60 (86%) at E8.25-E8.5 that all exhibited the characteristic distended heart and small trunk phenotype of the previously published conventional Raldh2−/− knockout (Ryckebusch et al., 2008; Sirbu and Duester, 2006). DNA sequences from three embryos with this phenotype revealed deletions or insertions in Raldh2 exon 1 that are expected to be null mutations due to loss of the ATG start codon, loss of the GT splice donor, or frame shift; staining of one embryo for Uncx expression revealed the characteristic small somite phenotype of conventional Raldh2−/− embryos (Fig. 4). Even though the CRISPR-generated embryos exhibiting the morphological defects of Raldh2 mutants each carry a different mutation in Raldh2, each is very likely to be a null mutant lacking RA synthesis by RALDH2 and pooling of these embryos for ChIP studies should not pose a problem.
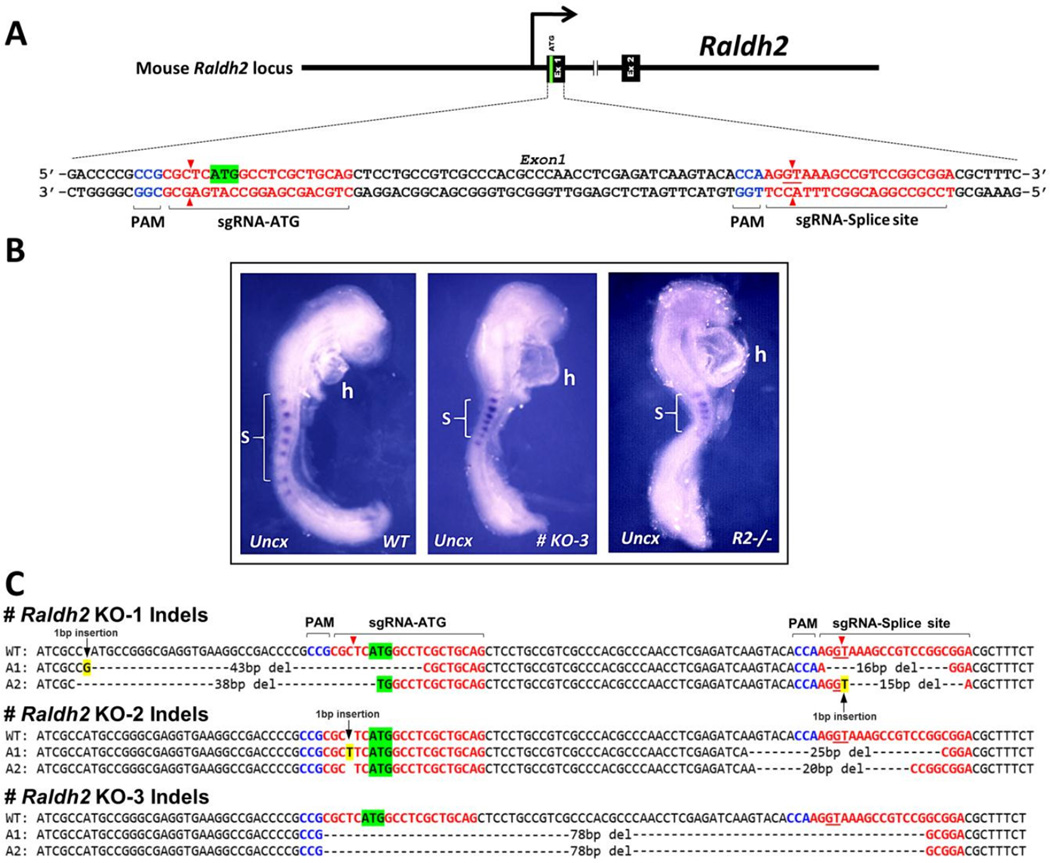
Fig. 4.
Raldh2 mutants generated by CRISPR/Cas9 gene editing phenocopy Raldh2−/− embryos generated by the traditional knockout strategy. (A) Strategy for generating mutations in the first exon of Raldh2 using sgRNAs targeting the ATG start codon and GT splice donor site. (B) Shown is Uncx somite expression for wild-type (WT), CRISPR mutant KO-3, and a conventional Raldh2−/− embryo; brackets mark 7-somite regions for each embryo. (C) DNA sequences for WT and both alleles (A1 and A2) of three CRISPR mutants show that each mutant carries biallelic insertion or deletion mutations in Raldh2 that disrupt the ATG start codon, GT splice donor site, or reading frame.
Embryo ChIP on E8.25-E8.5 mouse trunk tissue was performed to examine recruitment of NCOR1/2 to either the Fgf8 repressive RARE or the Rarb activating RARE using a mixture of antibodies against NCOR1 and NCOR2 (NCOR1/2). We found that NCOR1/2 was recruited to the vicinity of the Fgf8 RARE in wild-type tissue, but that the signal was greatly reduced in Raldh2−/− mutants that are deficient for RA synthesis (Fig. 3C). In contrast, NCOR1/2 exhibited very little recruitment to the Rarb RARE in wild-type tissue, but the signal was greatly increased in Raldh2−/− mutants (Fig. 3D). We also used embryo ChIP to examine recruitment of the nuclear receptor coactivator NCOA1 (SRC-1) previously shown to be recruited to RAREs that mediate gene activation in RA-treated cells in vitro. We found very little recruitment of NCOA1 to the Fgf8 RARE in either wild-type or Raldh2−/− tissue, whereas NCOA1 was recruited to the Rarb RARE in wild-type tissue and the signal was greatly reduced in Raldh2−/− mutants (Fig. 3C–D). Neither NCOR1/2 nor NCOA1 were recruited to non-specific regions (NSRs) located 1.2 kb upstream of the Fgf8 RARE or 2.6 kb upstream from the Rarb RARE in wild-type or Raldh2−/− embryos, suggesting the RAREs are involved in recruitment (Fig. 3E–F). These findings indicate that the Fgf8 RARE recruits NCOR1/2 in an RA-dependent fashion and that it does not recruit NCOA1 in the presence of RA. Thus, regarding recruitment of NCOR1/2 in vivo, the repressive Fgf8 RARE functions oppositely to the well-established activating RARE for Rarb.
2.3. Genomic deletion of the Fgf8 RARE results in somite defects and ectopic Fgf8 expression
Previous studies using mouse embryos carrying Fgf8-lacZ transgenes demonstrated that deletion of a conserved RARE upstream of Fgf8 resulted in anterior expansion of caudal Fgf8-lacZ expression, thus showing that this RARE can limit the anterior extent of caudal Fgf8 expression in vivo (Kumar and Duester, 2014). This observation together with our studies above suggest that RA may directly repress Fgf8 transcription. However, as transgenes are randomly integrated in the genome outside of their normal chromosomal location, it remains unclear if the Fgf8 RARE is required in its normal chromosomal context to restrict caudal Fgf8 expression. Here, we performed CRISPR/Cas9 gene editing to delete the endogenous Fgf8 RARE in the mouse genome and thus determine how this would affect axial development and caudal Fgf8 expression in developing mouse embryos.
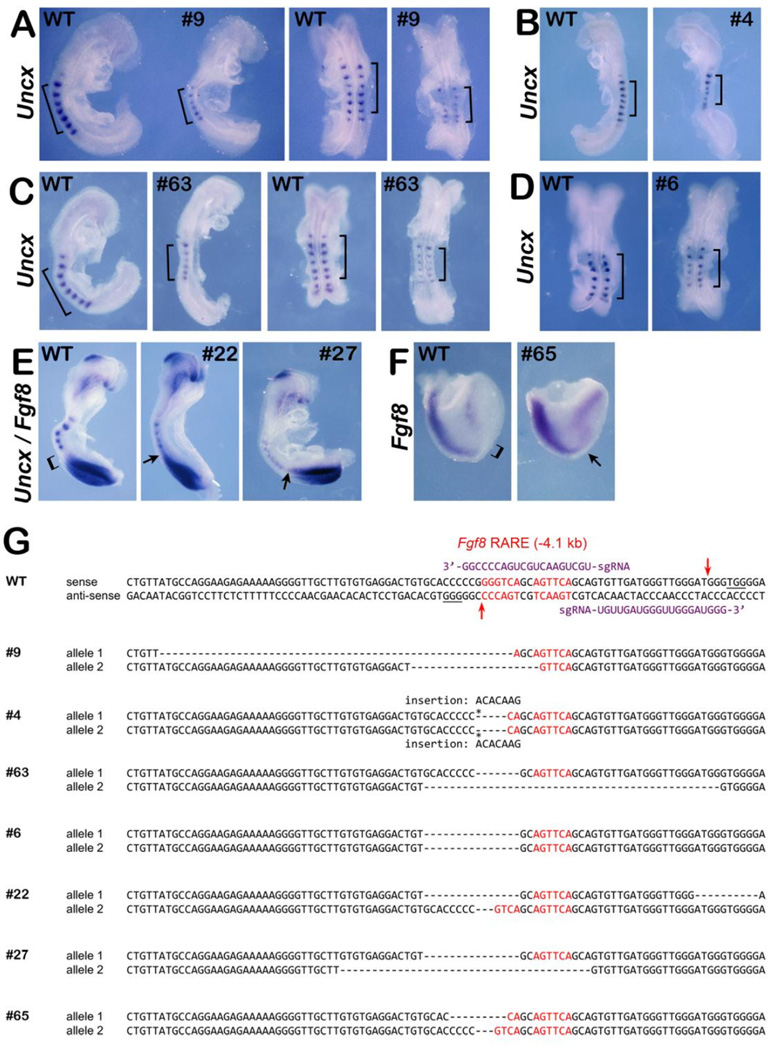
Mouse zygotes were injected with Cas9 mRNA and a pair of sgRNAs flanking the Fgf8 RARE genomic sequence to induce indel mutations (Fig. 5H). We harvested embryos at E8.5 and obtained 70 live embryos that were subjected to DNA sequencing and categorized by the severity of mutations in the Fgf8 RARE (Table 2); 16 embryos exhibited mosaic sequence (more than three alleles often including wild-type) and were not further examined. The remaining 54 embryos were examined for defects. In contrast to the results obtained above for Ncor1;Ncor2 double mutants, none of the Fgf8 RARE mutants exhibited a distended heart tube although 9 embryos exhibited a small somite phenotype and/or ectopic Fgf8 expression (Table 2). Seven mutant embryos stained for Uncx or Fgf8 expression by in situ hybridization are shown with their DNA sequences at the Fgf8 RARE locus (Fig. 5). Embryos #9, #4, #63, and #6 stained for Uncx expression show smaller somites than wild-type; somite height along the anteroposterior axis for mutant embryos is approximately 70% that observed in WT embryos. (Fig. 5A–D). Embryos #22 and #27 simultaneously stained for Fgf8 and Uncx mRNA exhibit smaller somites and ectopic caudal Fgf8 expression extending anteriorly and abutting the last somite formed rather than showing a clear open space between the Fgf8 and Uncx mRNA domains as seen in wild-type (Fig. 5E). Embryo #65 examined only for Fgf8 mRNA exhibits ectopic Fgf8 expression in the developing trunk between the heart and caudal Fgf8 domains (Fig. 5F). We note that the observed ectopic caudal Fgf8 expression is not very extensive, and this may be why the small somite phenotype observed in Fgf8 RARE mutants is less severe than that observed in Ncor1;Ncor2 double mutants. However, our observation of double-stained Fgf8 RARE mutants exhibiting both small somites and ectopic caudal Fgf8 expression suggests that the somitic phenotype we see is caused by Fgf8 overexpression. DNA sequences of these mutants, that exhibit defects in somitogenesis or ectopic caudal Fgf8 expression, show that they all contain biallelic deletions and/or insertions in the Fgf8 RARE that would be expected to impact binding of RAR/RXR (Fig. 5G).
Fig. 5.
Deletion of Fgf8 RARE in mouse embryos results in small somite phenotype and ectopic caudal Fgf8 expression. Shown are the results obtained from mutant embryos generated by CRISPR/Cas9 gene editing near the Fgf8 RARE. (A–D) Wild-type (WT) and mutant embryos with the same number of somites (or differing by 1 somite) analyzed for Uncx mRNA (a somite marker); brackets mark 6-somite regions for each embryo. (E) Embryos analyzed for expression of both Fgf8 and Uncx and (F) embryo analyzed for expression of only Fgf8; brackets indicate regions normally devoid of Fgf8 expression in WT whereas arrows point out regions of ectopic Fgf8 expression in mutants. (G) dsDNA sequence of wild-type Fgf8 RARE region (Kumar and Duester, 2014); sequence in red indicates the two 6 bp half sites of the Fgf8 RARE; PAM sequences are underlined; sgRNA guide sequences and their respective binding positions are shown in purple; arrows represent targeted Cas9 cleavage sites. Also, shown are the DNA sequences of CRISPR/Cas9-generated mutants (sense strand of both alleles shown); sequences in red indicate the Fgf8 RARE half-sites; dashes indicate deletions and asterisks indicate insertions.
Table 2.
Embryos obtained from CRISPR/Cas9 gene editing of Fgf8 RARE.
| Embryo | bp deleted from RARE for each allele |
Somites (Uncx) |
Heart Tube | Fgf8 expression |
|---|---|---|---|---|
| 1 | 0/0 | # | normal | normal |
| 2 | 0/0 | normal | normal | # |
| 7 | 0/0 | normal | normal | # |
| 14 | 0/0 | normal | normal | # |
| 28 | 0/0 | normal | normal | # |
| 30 | 0/0 | normal | normal | normal |
| 34 | 0/0 | normal | normal | # |
| 16 | 0/1 | normal | normal | normal |
| 17 | 0/0 | normal | normal | # |
| 24 | 0/0 | normal | normal | normal |
| 49 | 0/0 | normal | normal | # |
| 10 | 0/2 | normal | normal | # |
| 18 | 1/1 | normal | normal | # |
| 25 | 0/2 | normal | normal | normal |
| 45 | 0/1 | normal | normal | # |
| 12 | 0/3 | normal | normal | # |
| 60 | 0/3 | # | normal | normal |
| 48 | 1/2 | normal | normal | # |
| 11 | 2/2 | normal | normal | # |
| 42 | 2/2 | normal | normal | normal |
| 43 | 2/2 | normal | normal | # |
| 20 | 0/6 | normal | normal | normal |
| 33 | 0/6 | normal | normal | # |
| 40 | 0/6 | normal | normal | normal |
| 47 | 0/6 | normal | normal | # |
| 70 | 0/6 | # | normal | normal |
| 23 | 0/7 | normal | normal | normal |
| 29 | 0/8 | normal | normal | normal |
| 31 | 0/9 | normal | normal | normal |
| 19 | 0/10 | # | normal | |
| 50 | 0/10 | normal | normal | # |
| 21 | 0/12 | normal | normal | normal |
| 26 | 0/12 | normal | normal | normal |
| 58 | 0/8 | # | normal | |
| 5 | 1/11 | normal | normal | # |
| *65 | 2/4 | # | ectopic | |
| 55 | 2/5 | normal | normal | normal |
| *38 | 2/6 | small | normal | # |
| *22 | 2/6 | small | normal | ectopic |
| 41 | 2/6 | normal | normal | normal |
| 32 | 4/6 | normal | normal | # |
| 64 | 5/5 | normal | normal | # |
| *3 | 6/6 | small | normal | ectopic |
| *4 | 6/6 | small | normal | # |
| *6 | 6/6 | small | normal | # |
| *9 | 5/7 | small | normal | # |
| 44 | 6/6 | normal | normal | # |
| 46 | 6/6 | normal | normal | # |
| 54 | 6/7 | normal | normal | # |
| *27 | 6/12 | small | normal | ectopic |
| 61 | 6/12 | # | normal | normal |
| *63 | 6/12 | small | normal | # |
| 52 | 12/12 | normal | normal | # |
| 56 | 12/12 | # | normal | normal |
Total 70 embryos with 16 embryos (numbers 8, 13, 15, 35, 36, 37, 39, 51, 53, 57, 59, 62, 66, 67, 68, 69) not examined as they were mosaic for DNA sequence near the Fgf8 RARE (more than two alleles were obtained often including wild-type sequence.
, embryos with mutant phenotype;
, too young for determination;
, not analyzed.
The DNA sequences of the remaining embryos, most of which did not have defects, reveal changes often only on one allele (monoallelic), or biallelic mutations mostly outside of the RARE (Fig. S2). However, 10 embryos that did not display small somites or ectopic Fgf8 expression carry Fgf8 RARE mutations that could be considered as severe as those for the 9 embryos that did have defects; i.e. if embryo #65 is considered to be the least affected mutant with 2 bp deleted from one RARE allele and 4 bp deleted from the other RARE allele, all embryos listed below embryo #65 have more severe RARE mutations but 10 have no phenotype (Table 1). This observation is not due to chimerism between yolk sac and embryo proper, as DNA sequencing of several embryos after in situ hybridization revealed the same biallelic deletions in the RARE as observed in the original yolk sac DNA sequence. Among mutant embryos that did not show defects in somite size or Fgf8 expression, we obtained successful post-hybridization PCR for embryos #52, #61, and #55 carrying biallelic RARE deletions of 12/12, 6/12, and 2/5, respectively, and their RARE mutant DNA sequences were identical to the yolk sac sequences; successful PCR for embryos #3, #4, and #9 that do show defects in somite size and/or Fgf8 expression also exhibited RARE mutant DNA sequences identical to that seen for yolk sac DNA. These results demonstrate that our CRISPR/Cas9 methodology often generated biallelic mutations at an early stage in mouse embryos (likely at the 1- or 2-cell stage), thus resulting in identical mutations in both yolk sac and embryo.
These findings demonstrate that the repressive Fgf8 RARE is not needed to restrict heart Fgf8 expression in order to obtain a normal heart tube, plus it is not always needed to control caudal Fgf8 expression, suggesting the existence of at least one additional repressive control element that mediates RA restriction of Fgf8 expression in the early heart and/or caudal domains. Based on these findings we conclude that the Fgf8 RARE contributes in vivo to repression of caudal Fgf8 transcription in order to control somitogenesis, but its function is not always essential or could be modulated by additional DNA elements.
3. Discussion
Although RA signaling through RARs has mostly been reported to directly activate genes during development, recent studies suggest that RA can directly repress Fgf8 in embryos (Kumar and Duester, 2014; Patel et al., 2013). Here, we report that the mechanism through which RA represses Fgf8 requires nuclear receptor corepressors NCOR1 and NCOR2 (SMRT). Previous studies suggested that NCOR1 and NCOR2 are recruited to RAR only in the absence of RA ligand, thus turning off genes that would normally be activated if RA is present (Chen et al., 1996; Hörlein et al., 1995). These in vitro studies were performed on RAREs from genes known to be activated by RA using transfected cell lines, leaving it unknown how RAREs associated with repression may function, particularly under in vivo conditions. With the advent of CRISPR/Cas9 gene editing technology it is now feasible to analyze mechanisms of gene activation and repression in vivo. Ncor1 and Ncor2 single mutants are not associated with early embryonic defects caused by excessive expression of Fgf8, perhaps due to functional redundancy. Here, Ncor1;Ncor2 double mutant embryos generated by CRISPR/Cas9 gene editing exhibited excessive Fgf8 expression along the body axis associated with defects in somitogenesis and heart development similar to those observed in Raldh2−/− embryos lacking RA signaling. We found that NCOR1/2 is recruited to the Fgf8 RARE in an RA-dependent manner. Thus, we provide in vivo evidence that NCOR1/2 can directly repress gene transcription via recruitment to a RARE in a RA-dependent manner.
Interestingly, whereas Ncor1 and Ncor2 are required to restrict Fgf8 expression in both the caudal progenitors and heart progenitors, loss of the Fgf8 RARE results in Fgf8 derepression only in the caudal domain and does not result in a heart defect. Also, whereas the small somite phenotype and ectopic Fgf8 expression observed in Ncor1;Ncor2 double mutants is consistent for all mutants carrying three or four mutant alleles, only about half of the embryos carrying Fgf8 RARE mutations in both alleles exhibit these defects, plus the reduction in somite height along the anteroposterior axis is not as severe for Fgf8 RARE mutants compared to Ncor1;Ncor2 double mutants which are similar to Raldh2−/− embryos. This result is unlikely due to mosaicism since DNA sequences of yolk sac and embryo proper for many embryos show the same mutations. Also, this result is very unlikely due to off-target effects of the sgRNAs used for CRISPR/Cas9 gene editing. Our method of using sgRNAs having no more than 17 out of 20 matches with any other site in the mouse genome has been shown by others to greatly minimize off-targeting events (Wang et al., 2013). Also, previous studies showed that even when an off-target site is mutated it occurs with much less frequency than mutation of the intended target (Tan et al., 2015). So, by analyzing several embryos carrying the desired mutation we can be assured that many embryos do not also carry an off-site mutation that is responsible for the phenotype (or the lack of a phenotype). Instead, our findings suggest the existence of another DNA control element that recruits NCOR1/2 to mediate RA repression of Fgf8 in the heart progenitor domain, and in some embryos this additional control element may also be sufficient to repress caudal Fgf8 to minimize or prevent the small somite phenotype.
Our studies provide more clues for understanding the mechanism through which RA represses Fgf8. Fgf8 expression in the presomitic mesoderm is reported to be mainly a reflection of Fgf8 mRNA stability, with transcription occurring only in progenitors in the caudal epiblast that give rise to both presomitic mesoderm and neural plate (Dubrulle and Pourquié, 2004). This transcriptional load in the caudal epiblast then forms a caudal-high gradient of Fgf8 mRNA along the anteroposterior axis of the presomitic mesoderm as epiblast cells ingress to form the presomitic mesoderm (Mallo, 2016). Thus, RA-mediated repression of Fgf8 could be due to either a reduction in the transcriptional load in the caudal epiblast or an increase in Fgf8 mRNA decay in the presomitic mesoderm. Recent studies demonstrated that repression of caudal Fgf8 operates through RA action in the neural plate where it joins the caudal epiblast, not RA activity in presomitic mesoderm, consistent with RA repressing Fgf8 via a transcriptional mechanism in caudal progenitors (Cunningham et al., 2015). Here, our observation that loss of the transcriptional corepressors NCOR1 and NCOR2 results in up-regulation of caudal Fgf8 expression provides more evidence that RA represses Fgf8 through a transcriptional mechanism in both caudal and heart domains.
RA receptor signaling through RAREs is commonly associated with gene activation, but there are a few examples of repressive RAREs (Cunningham and Duester, 2015). For instance, activation of Hoxb1 in the mouse hindbrain during the early stages of body axis extension requires RAR and a 3'-RARE (Marshall et al., 1994) that recruits the histone acetyltransferase coactivator MOZ in an RA-dependent manner (Voss et al., 2009). However, Hoxb1 also possesses a 5'-RARE that represses Hoxb1 in rhombomeres 3 and 5 to limit expression to only rhombomere 4 (Studer et al., 1994); the Hoxb1 5'-RARE was found to recruit PRC2 in an RA-dependent manner (Kumar and Duester, 2014). Here, our Ncor1;Ncor2 and Fgf8 RARE gene editing studies in mouse embryos combined with our previous transgenic mouse Fgf8 RARE deletion studies and epigenetic analyses (Kumar and Duester, 2014) support a model in which the Fgf8 RARE bound to RAR/RXR functions repressively in vivo through RA-dependent recruitment of NCOR1/2 and PRC2, both of which are associated with transcriptional repression. As the repressive Fgf8 and Hoxb1 RAREs are composed of sequences very similar to activating RAREs (Cunningham and Duester, 2015), we conclude that the ability of some RAREs to function repressively is not due to a different variant RARE sequence. Likewise, genome-wide analyses from embryonic stem cells suggest that various RAREs of similar sequence identified in genome-wide RAR ChIP analyses may be associated with either upregulation and downregulation of nearby genes in RA-treated cells (Moutier et al., 2012). Thus, the ability of particular RAREs to function either in gene activation or repression may be decided by their locations relative to other DNA control elements near target genes.
It is also important to note that ligand-dependent repression mediated by RA-liganded RAR that we discuss here (specifically for RARE-mediated repression of Fgf8 in trunk tissue anterior to the caudal progenitors), is the opposite of ligand-independent repression mediated by RARs bound to RAREs that recruit corepressors in the absence of RA. In the latter case, ligand-independent repression by RAR has been suggested to occur in Xenopus embryos in the RA-free region caudal to the neural plate where it functions to maintain caudal progenitors (Janesick et al., 2014). Thus, RA-independent RAR-mediated repression occurring in regions devoid of RA is likely a mechanism designed to keep genes that are normally activated by RA turned off until RA is present. In contrast, RA-dependent RAR-mediated repression that we report here is designed to turn off genes that are already expressed, but only when RA accumulates to a high enough level. Derepression of genes other than Fgf8 in our Ncor1/Ncor2 double mutants may contribute to the severe phenotype we see, plus this may explain why our Fgf8 RARE mutants have a less severe defect.
It is interesting that Ncor1;Ncor2 double mutants display a phenotype that is very similar to Raldh2−/− embryos. In particular, the distended heart tube, small somites, and ectopic Fgf8 expression in both the heart and caudal domains are very similar between the two mutants, suggesting that ectopic Fgf8 expression is a major contributor to the severe defects observed in both mutants. Also, we note that dissection at E10.5 revealed no Ncor1;Ncor2 double mutants (data not shown), suggesting that they exhibit lethality between E8.5-E10.5 similar to Raldh2−/− embryos (Mic et al., 2002; Niederreither et al., 1999). However, a full comparison of the differences between Ncor1;Ncor2 double mutants and Raldh2−/− mutants will be needed to determine how similar the phenotypes are across multiple organs. Also, further studies on Ncor1;Ncor2 double mutants are needed to determine if alterations in expression of other genes occurs that may contribute to the mesodermal defects we report, and whether loss of NCOR1/2 affects the mechanism originally proposed for its function based on in vitro studies, i.e. ligand-independent repression of RA-activated genes in order to prevent their expression when RA is not present.
Our studies also demonstrate how CRISPR/Cas9 gene editing can greatly accelerate genetic loss-of-function studies in mouse embryos by obviating the need to first create heterozygous adult mice and then perform timed matings to generate homozygous mutant embryos. This is particularly important when analysis of double mutants is needed. Such methodology has the potential to greatly advance our understanding of developmental mechanisms.
In order to fully understand the function of RA and realize its full potential as a differentiation agent for stem/progenitor cells in regenerative medicine, it will be important to further explore the genetic and epigenetic mechanisms that determine whether RA acts in a repressive or activating manner for specific genes. Our experience here indicates that CRISPR/Cas9 gene editing of mouse embryos will be an important tool for quickly determining in vivo functions of DNA control elements and transcriptional coregulators.
4. Materials and methods
4.1. Generation of conventional Raldh2−/− knockout mouse embryos
Raldh2−/− mice have been previously described (Sirbu and Duester, 2006); genotyping was performed by PCR analysis of yolk sac DNA. All mouse studies conformed to the regulatory standards adopted by the Institutional Animal Care and Use Committee at the Sanford Burnham Prebys Medical Discovery Institute which approved this study under Animal Welfare Assurance Number A3053-01 (permit #15-104).
4.2. CRISPR/Cas9 gene editing of mouse embryos
CRISPR/Cas9 gene editing was performed using methods similar to those described previously (Tan et al., 2015; Wang et al., 2013). Single-guide RNAs (sgRNAs) were generated that target sites near the ATG translation initiation site and the first GT splice donor sites for Ncor1, Ncor2, and Raldh2 plus sites flanking the Fgf8 RARE; sgRNAs were designed with maximum specificity using the tool at crispr.mit.edu to ensure that each sgRNA had no more than 17 out of 20 matches with any other site in the mouse genome. DNA templates for sgRNAs were generated by PCR amplification (Phusion DNA Polymerase; New England Biolabs) of ssDNA oligonucleotides (purchased from Integrated DNA Technologies) containing on the 5' end a minimal T7 promoter, then a 20 nucleotide sgRNA target sequence (underlined below), and finally the tracrRNA sequence utilized by Cas9 on the 3' end, shown as follows: 5'-GCGTAATACGACTCACTATAGGNNNNNNNNNNNNNNNNNNNNGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT-3' The 20 nucleotide target sequences used were as follows: Ncor1 ATG (TTACTGATAATGTCAAGTTC), Ncor1 1st splice donor site (GTACCCGACACCAGCAGGTA), Ncor2 ATG (CTGGACCCTACCACCATGTC), Ncor2 1st splice donor site (TAGCCCGGTCCCACACGGTA), Raldh2 ATG (CTGCAGCGAGGCCATGAGCG), Raldh2 1st splice donor site (TCCGCCGGACGGCTTTACCT), Fgf8 RARE upstream (TGCTGAACTGCTGACCCCGG), Fgf8 RARE downstream (TGTTGATGGGTTGGGATGGG).
sgRNAs were then transcribed from templates using HiScribe T7 High Yield RNA Synthesis Kit (New England Biolabs) and purified using Megaclear Kit (Life Technologies). sgRNAs were tested in vitro for their cleavage ability in combination with Cas9 nuclease (New England Biolabs); briefly, genomic regions flanking the target sites were PCR amplified, then 100 ng was incubated with 30 nM Cas9 nuclease and 30 ng sgRNA in 30 µl for 1 hour at 37°C, followed by analysis for cleavage by gel electrophoresis.
For injection into mouse embryos, a solution containing 50 ng/µl Cas9 mRNA (Life Technologies) and 20 ng/µl for each sgRNA used was prepared in nuclease free water. Fertilized oocytes were collected from 3–4 week-old superovulated C57Bl6 females prepared by injecting 5 IU each of pregnant mare serum gonadotrophin (PMSG) (Sigma Aldrich) and human chorionic gonadotropin (hCG) (Sigma Aldrich). Fertilized oocytes were then transferred into M2 medium (Millipore) and injected with the Cas9 mRNA/sgRNA solution into the cytoplasm. Injected embryos were cultured in KSOMaa medium (Zenith) in a humidified atmosphere with 5% CO2 at 37°C overnight to maximize the time for CRISPR/Cas9 gene editing to occur at the 1-cell stage, then re-implanted at the 2-cell stage into recipient pseudo-pregnant ICR female mice. Implanted females were sacrificed 8 days after re-implantation, For genotyping, yolk sac DNA was collected and PCR products generated using primers flanking the target sites were subjected to DNA sequence analysis from both directions using either upstream or downstream primers. In selected cases, embryos were genotyped after in situ hybridization to confirm that the embryo proper carried the same biallelic mutations as the yolk sac.
4.3. Gene expression analysis
Embryos were fixed in paraformaldehyde at 4°C overnight, dehydrated into methanol, and stored at −20°C. Detection of mRNA was performed by whole mount in situ hybridization as previously described (Sirbu and Duester, 2006).
4.4. Embryo chromatin immunoprecipitation (ChIP)
Embryo ChIP was performed according to the manufacturer’s protocol (Active Motif) as described (Frank et al., 2001). For ChIP assays, trunk tissues from embryos at stages E8.25-E8.5 were pooled from 78 wild-type embryos or 73 Raldh2−/− embryos (51 generated from CRISPR/Cas9 gene editing and 22 conventional mutants); trunk tissue was separated from the rest of the embryo by cutting posterior to the heart and posterior to the last somite were used as previously described (Kumar and Duester, 2014). ChIP-grade antibodies used include Anti-Nuclear Receptor Corepressor NCoR1 antibody, (Abcam, ab24552), SMRT (NCOR2) (Santa Cruz Biotechnology, sc-32298), SRC-1 (NCOA1) (Thermo Fisher, MA1-840), RXRa (Santa Cruz Biotechnology, sc-553) or control IgG antibody (Cell Signaling Technology, 2729). For all ChIP reactions, 3 µg antibody was used for each immunoprecipitation reaction; for NCOR1 and NCOR2 (NCOR1/2) studies, 3 µg of each antibody was mixed together. Analysis of immunoprecipitated DNA was performed by PCR amplification using primers flanking the mouse Fgf8 RARE and RARb RARE. Oligonucleotide sequences used in ChIP studies; Fgf8-RARE-Fwd 5'-CAG CAC TCT GCC ATA CTG TCT TA-3’, Fgf8-RARE-Rev 5'-TCT GTC AGT CTT CAG CTT GTC TG-3’, Fgf8-NSR-Fwd 5'- TAG CAG CTG AAT GAG TGG CTC TA -3’, Fgf8-NSR-Rev 5'- GTA GCA AGC AGT TAC CTG ATC TG -3’, RARb-RARE-Fwd 5’-TGG CAT TGT TTG CAC GCT GA-3’, RARb-RARE-Rev 5’-CCC CCC TTT GGC AAA GAA TAG A-3’, RARb-NSR-Fwd 5′-AGTACAGACCTTCCAAGAGTGCCT-3’, RARb-NSR-Rev 5′ GTCATGGGAAAGAGAGGTTGAGC-3′. RXRa ChIP results were obtained by gel electrophoretic analysis of PCR products. For quantitation of other ChIP results, enrichment of specific DNA fragments was measured by real-time qPCR using LightCycler® 96 System (Roche Diagnostics) and EXPRESS SYBR® GreenER™ qPCR Supermix (Thermo Fisher). Each ChIP analysis was repeated in at least in three independent experiments and results are reported as ± SEM; using the t test.
Supplementary Material
Highlights.
Genetic loss of Ncor1 and Ncor2 results in ectopic Fgf8 expression and small somites.
NCOR1/2 corepressors, but not coactivators, are recruited to the Fgf8 RARE by RA.
Genomic deletion of Fgf8 RARE with CRISPR/Cas9 often results in small somite defect.
Nuclear receptor corepressors NCOR1 and NCOR2 mediate RA-dependent Fgf8 repression.
Acknowledgments
We thank the Sanford Burnham Prebys Medical Discovery Institute Animal Resources Core Facility for conducting timed-matings to generate mouse embryos and the UCSD Transgenic Mouse Facility for carrying out fertilized oocyte injections to perform CRISPR/Cas9 gene editing of mouse embryos. This work was funded by National Institutes of Health grant GM062848 (G.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
S.K. and T.C. carried out experiments. S.K., T.C., and G.D. designed the study, analyzed data, and wrote the final manuscript.
Appendix A and Appendix B. Supplementary material
Supplementary data associated with this article can be found in the online version at ?.
References
- Affolter M, Pyrowolakis G, Weiss A, Basler K. Signal-induced repression: the exception or the rule in developmental signaling. Dev. Cell. 2008;15:11–22. doi: 10.1016/j.devcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wehrle C, Brand-Saberi B, Kemler R, Gossler A, Kanzler B, Herrmann BG. Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell. 2003;4:395–406. doi: 10.1016/s1534-5807(03)00055-8. [DOI] [PubMed] [Google Scholar]
- Boulet AM, Capecchi MR. Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev. Biol. 2012;371:235–245. doi: 10.1016/j.ydbio.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DL, Papaioannou VE. Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature. 1998;391:695–697. doi: 10.1038/35624. [DOI] [PubMed] [Google Scholar]
- Chen JD, Umesono K, Evans RM. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc. Natl. Acad. Sci. USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev. Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- Cunningham TJ, Brade T, Sandell LL, Lewandoski M, Trainor PA, Colas A, Mercola M, Duester G. Retinoic Acid Activity in Undifferentiated Neural Progenitors Is Sufficient to Fulfill Its Role in Restricting Fgf8 Expression for Somitogenesis. PLoS ONE [Electronic Resource] 2015;10:e0137894. doi: 10.1371/journal.pone.0137894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nature Rev. Mol. Cell Bio. 2015;16:110–123. doi: 10.1038/nrm3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TJ, Zhao X, Sandell LL, Evans SM, Trainor PA, Duester G. Antagonism between retinoic acid and fibroblast growth factor signaling during limb development. Cell Reports. 2013;3:1503–1511. doi: 10.1016/j.celrep.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40:65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- Dubrulle J, Pourquié O. fgf8 mRNA decay establishes a gradient that couples axial elongation to patterning in the vertebrate embryo. Nature. 2004;427:419–422. doi: 10.1038/nature02216. [DOI] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunty WC, Jr, Biris KK, Chalamalasetty RB, Taketo MM, Lewandoski M, Yamaguchi TP. Wnt3a/beta-catenin signaling controls posterior body development by coordinating mesoderm formation and segmentation. Development. 2008;135:85–94. doi: 10.1242/dev.009266. [DOI] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- Henrique D, Abranches E, Verrier L, Storey KG. Neuromesodermal progenitors and the making of the spinal cord. Development. 2015;142:2864–2875. doi: 10.1242/dev.119768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörlein AJ, Näär AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamel Y, Söderström M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Janesick A, Nguyen TT, Aisaki K, Igarashi K, Kitajima S, Chandraratna RA, Kanno J, Blumberg B. Active repression by RAR signaling is required for vertebrate axial elongation. Development. 2014;141:2260–2270. doi: 10.1242/dev.103705. [DOI] [PubMed] [Google Scholar]
- Jepsen K, Hermanson O, Onami TM, Gleiberman AS, Lunyak V, McEvilly RJ, Kurokawa R, Kumar V, Liu F, Seto E, Hedrick SM, Mandel G, Glass CK, Rose DW, Rosenfeld MG. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Kumar S, Duester G. Retinoic acid controls body axis extension by directly repressing Fgf8 transcription. Development. 2014;141:2972–2977. doi: 10.1242/dev.112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo M. Revisiting the involvement of signaling gradients in somitogenesis. FEBS Journal. 2016;283:1430–1437. doi: 10.1111/febs.13622. [DOI] [PubMed] [Google Scholar]
- Marshall H, Studer M, Pöpperl H, Aparicio S, Kuroiwa A, Brenner S, Krumlauf R. A conserved retinoic acid response element required for early expression of the homeobox gene Hoxb-1. Nature. 1994;370:567–571. doi: 10.1038/370567a0. [DOI] [PubMed] [Google Scholar]
- Martin BL, Kimelman D. Wnt signaling and the evolution of embryonic posterior development. Curr. Biol. 2009;19:R215–R219. doi: 10.1016/j.cub.2009.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BL, Kimelman D. Canonical Wnt Signaling Dynamically Controls Multiple Stem Cell Fate Decisions during Vertebrate Body Formation. Dev. Cell. 2012;22:223–232. doi: 10.1016/j.devcel.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn C, Ruberte E, LeMeur M, Morriss-Kay G, Chambon P. Developmental analysis of the retinoic acid-inducible RAR-b2 promoter in transgenic animals. Development. 1991;113:723–734. doi: 10.1242/dev.113.3.723. [DOI] [PubMed] [Google Scholar]
- Mic FA, Haselbeck RJ, Cuenca AE, Duester G. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development. 2002;129:2271–2282. doi: 10.1242/dev.129.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev. 2013;27:819–835. doi: 10.1101/gad.214023.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutier E, Ye T, Choukrallah MA, Urban S, Osz J, Chatagnon A, Delacroix L, Langer D, Rochel N, Moras D, Benoit G, Davidson I. Retinoic Acid Receptors Recognize the Mouse Genome through Binding Elements with Diverse Spacing and Topology. J. Biol. Chem. 2012;287:26328–26341. doi: 10.1074/jbc.M112.361790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiche LA, Holder N, Lewandoski M. FGF4 and FGF8 comprise the wavefront activity that controls somitogenesis. Proc. Natl. Acad. Sci. USA. 2011;108:4018–4023. doi: 10.1073/pnas.1007417108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nature Rev. Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dollé P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nature Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- Olivera-Martinez I, Harada H, Halley PA, Storey KG. Loss of FGF-Dependent Mesoderm Identity and Rise of Endogenous Retinoid Signalling Determine Cessation of Body Axis Elongation. Plos Biology. 2012;10:e1001415. doi: 10.1371/journal.pbio.1001415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NS, Rhinn M, Semprich CI, Halley PA, Dolle P, Bickmore WA, Storey KG. FGF Signalling Regulates Chromatin Organisation during Neural Differentiation via Mechanisms that Can Be Uncoupled from Transcription. PLoS Genetics. 2013;9:e1003614. doi: 10.1371/journal.pgen.1003614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nature Rev. Genet. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Ryckebusch L, Wang Z, Bertrand N, Lin S-C, Chi X, Schwartz R, Zaffran S, Niederreither K. Retinoic acid deficiency alters second heart field formation. Proc. Natl. Acad. Sci. USA. 2008;105:2913–2918. doi: 10.1073/pnas.0712344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu IO, Duester G. Retinoic acid signaling in node ectoderm and posterior neural plate directs left-right patterning of somitic mesoderm. Nature Cell Biol. 2006;8:271–277. doi: 10.1038/ncb1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu IO, Gresh L, Barra J, Duester G. Shifting boundaries of retinoic acid activity control hindbrain segmental gene expression. Development. 2005;132:2611–2622. doi: 10.1242/dev.01845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu IO, Zhao X, Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev. Dyn. 2008;237:1627–1635. doi: 10.1002/dvdy.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M, Pöpperl H, Marshall H, Kuroiwa A, Krumlauf R. Role of a conserved retinoic acid response element in rhombomere restriction of Hoxb-1. Science. 1994;265:1728–1732. doi: 10.1126/science.7916164. [DOI] [PubMed] [Google Scholar]
- Takemoto T, Uchikawa M, Yoshida M, Bell DM, Lovell-Badge R, Papaioannou VE, Kondoh H. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature. 2011;470:394–398. doi: 10.1038/nature09729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan EP, Li Y, Del Castillo Velasco-Herrera M, Yusa K, Bradley A. Off-target assessment of CRISPR-Cas9 guiding RNAs in human iPS and mouse ES cells. Genesis: the Journal of Genetics & Development. 2015;53:225–236. doi: 10.1002/dvg.22835. [DOI] [PubMed] [Google Scholar]
- Tsakiridis A, Huang Y, Blin G, Skylaki S, Wymeersch F, Osorno R, Economou C, Karagianni E, Zhao S, Lowell S, Wilson V. Distinct Wnt-driven primitive streak-like populations reflect in vivo lineage precursors. Development. 2014;141:1209–1221. doi: 10.1242/dev.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V, Nicolas JF. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell. 2009;17:365–376. doi: 10.1016/j.devcel.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Vermot J, Llamas JG, Fraulob V, Niederreither K, Chambon P, Dollé P. Retinoic acid controls the bilateral symmetry of somite formation in the mouse embryo. Science. 2005;308:563–566. doi: 10.1126/science.1108363. [DOI] [PubMed] [Google Scholar]
- Vermot J, Pourquié O. Retinoic acid coordinates somitogenesis and left-right patterning in vertebrate embryos. Nature. 2005;435:215–220. doi: 10.1038/nature03488. [DOI] [PubMed] [Google Scholar]
- Voss AK, Collin C, Dixon MP, Thomas T. Moz and retinoic acid coordinately regulate H3K9 acetylation, Hox gene expression, and segment identity. Dev. Cell. 2009;17:674–686. doi: 10.1016/j.devcel.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V, Olivera-Martinez I, Storey KG. Stem cells, signals and vertebrate body axis extension. Development. 2009;136:1591–1604. doi: 10.1242/dev.021246. [DOI] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Zhao X, Sirbu IO, Mic FA, Molotkova N, Molotkov A, Kumar S, Duester G. Retinoic acid promotes limb induction through effects on body axis extension but is unnecessary for limb patterning. Curr. Biol. 2009;19:1050–1057. doi: 10.1016/j.cub.2009.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.