Abstract
Mass spectrometry (MS) is a mainstream chemical analysis technique in the twenty-first century. It has contributed to numerous discoveries in chemistry, physics and biochemistry. Hundreds of research laboratories scattered all over the world use MS every day to investigate fundamental phenomena on the molecular level. MS is also widely used by industry—especially in drug discovery, quality control and food safety protocols. In some cases, mass spectrometers are indispensable and irreplaceable by any other metrological tools. The uniqueness of MS is due to the fact that it enables direct identification of molecules based on the mass-to-charge ratios as well as fragmentation patterns. Thus, for several decades now, MS has been used in qualitative chemical analysis. To address the pressing need for quantitative molecular measurements, a number of laboratories focused on technological and methodological improvements that could render MS a fully quantitative metrological platform. In this theme issue, the experts working for some of those laboratories share their knowledge and enthusiasm about quantitative MS. I hope this theme issue will benefit readers, and foster fundamental and applied research based on quantitative MS measurements.
This article is part of the themed issue ‘Quantitative mass spectrometry’.
Keywords: bioanalysis, calibration, chemical analysis, mass spectrometry, quantification
Mass spectrometry (MS) is commonly regarded as an instrumental technique for separation of electrically charged species in the gas phase (for general information, see for example [1–11]). The charged species (ions) are produced in the ion source. In some cases, the ion source also assists the transfer of solid-phase or liquid-phase analytes into the gas phase. The gas-phase ions subsequently are transferred into the mass analyser. The mass analyser sorts the ions—in space or time—according to the mass-to-charge ratios (m/z). The separated ions are detected by an ion detector in the space or time domain. Electric signals, produced by the ion detector, are subsequently processed to produce mass spectra. In fact, mass spectra can be viewed as histograms, which provide information on the number of ions at different m/z values. The detected ions may correspond to the original molecules, their fragments or other species formed during the ionization process. MS enables direct identification of molecules based on the mass-to-charge ratio as well as fragmentation patterns. Thus, it fulfils the role of a qualitative analytical technique with high selectivity.
The twentieth century brought a number of milestone developments in the physical and natural sciences, which are qualitative in their nature. Examples include: nuclear fission, discovery of penicillin, elucidation of DNA structure, sequencing genomes or identification of numerous diseases. It also saw remarkable developments in the technologies enabling qualitative studies on the molecular level. Instrumental techniques such as scanning tunnelling microscopy, confocal microscopy, capillary electrophoresis or nuclear resonance spectrometry had enabling roles in the rise of several branches of science—nanotechnology, genetics and bioimaging. In fact, technology is vital for the rapid growth of these disciplines. The employed techniques had to catch up with the demand for the increased performance and outstanding specifications. A mismatch between the capabilities of technology platforms and the requirements of target applications could lead to erroneous results and flawed conclusions, which might subsequently mislead scientists and policy-makers.
While qualitative analysis tools played a huge role in many important discoveries in the past centuries, modern science increasingly relies on quantitative data. Discoveries in fundamental research as well as industrial and clinical applications often require reporting relative or absolute quantities of target molecules. Unfortunately, many of the available MS methods did not perform in quantitative chemical analysis as well as the well-established optical spectroscopic counterparts. It is already clear that in many sub-fields of biochemistry (e.g. proteomics), one cannot move on without quantitative molecular analysis. To address the pressing need for quantitative molecular measurements, several laboratories focused on technological and methodological improvements, which could render MS a fully quantitative metrological platform.
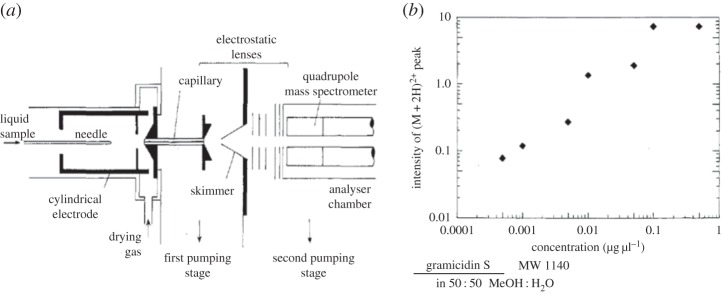
If I were asked to name one single invention that has contributed most to the current state of quantitative MS, I would mention the development of electrospray ionization (ESI) by the team of John Fenn (figure 1a) [12,13], which has now become the first choice for coupling liquid chromatographs with mass spectrometers. While browsing through pages of the early reports on ESI-MS, one can realize how remarkable a transformation has occurred in the design of this popular ion source. For those who are more accustomed to perfectly linear calibration plots obtained by ultraviolet absorption spectroscopy, the fluctuating data points in MS-derived plots (figure 1b) may look somewhat awkward. After more than three decades of intensive research, we still cannot cope with that signal variability, at least in many of the ‘real-word’ applications involving complex samples. The difficulties associated with quantitative MS analysis do not deter the ever growing family of users who find ingenious ways to solve the notorious technical problems. The benefits of MS as a universal chemistry tool—including its superior chemical selectivity—are just too huge to give up using this technique.
Figure 1.
Electrospray interface for mass spectrometry (a), and its application in quantitative analysis of gramicidin S in methanol–water solution (b). Adapted with permission from [13]. Copyright \copyrightlogo (1985) American Chemical Society.
Several factors affect quantitative capabilities of MS (table 1). Most of them fall into one of two categories: (i) instrument-related and (ii) sample-related [11]. The instrument-related factors are setting ion source parameters, transmission efficiency of ions, detection efficiency and contamination of MS parts. The sample-related factors are ionization efficiencies of analytes, concentrations of analytes, degradation of analytes, sample preparation, limited signal stability and repeatability, ionization interferences, detection interferences and spectral interferences [11]. Human operators of mass spectrometers can also be the source of artefacts, degrading quantitative capability of MS workflows. However, thanks to the considerable developments in software and hardware, and the increasing awareness of proper sample handling, nowadays MS analytical workflows are less affected by human errors than before. Among the important factors, sample preparation is the one that requires special attention. Separation techniques are used to decrease interferences caused by sample matrices, and improve quantitative capabilities of mass spectrometric methods.
Table 1.
Challenges associated with different aspects of quantitative mass spectrometry.
| aspect | challenge |
|---|---|
| sample preparation | sample purification or fractionation can reduce sample matrix interferences. Sample concentration and dilution can bring the analytes to the working range of the mass spectrometric method. However, these steps limit sample throughput |
| sample homogeneity | inhomogeneous samples are not amenable to quantitative analysis. Thus, analysis of dry sample/matrix deposits by MALDI-MS proves to be cumbersome |
| labelling | there is a limited range of chemical labels (derivatizing agents) for the target molecules of interest |
| internal standards | in many cases, it is difficult to find the right internal standards with similar ionization efficiencies to those of the analytes. Sometimes, isotopologues of analytes need to be synthesized for use as internal standards |
| interfacing | samples in the solid phase or liquid phase need to be brought to the gas phase, typically before or during ionization of analyte molecules |
| ion suppression | sample matrix components suppress ionization of different analytes to a different extent. The extent of ion suppression may change depending on the operating conditions |
| separation | separation of analytes by liquid or gas chromatography, capillary electrophoresis, ion mobility spectroscopy, or other techniques can reduce sample matrix interferences but it complicates analytical workflows, and (usually) limits sample throughput |
| detection | detection efficiencies for ions with different m/z values are unequal |
| spectral interferences | some sample matrix and/or ionization matrix-derived ions may overlap with the analyte ions |
| concentration/mass calibration | signal–quantity dependencies are not linear over a broad range. Parameters of the calibration equations may drift over time |
| automation | automated sample handling equipment can improve repeatability but it is costly |
| data processing and interpretation | expert knowledge is needed to translate the bare data acquired by a mass spectrometer into useable results, and finally—scientifically sound conclusions |
Arguably, much of the progress in MS methodology, especially regarding quantitative MS, has been motivated by the demands from bioscience fields (e.g. biochemistry, chemical biology). For example, the sub-field of proteomics is aiming to identify and quantify large numbers of proteins present in extracts of cells, tissues and biofluids. Quantification can be absolute or relative. In fact, many of the biochemical applications of MS rely on relative quantification. In these cases, it is sufficient to report a change in the relative levels of biomolecules in the analysed samples. However, in other cases, it is critical to provide the results in absolute units (concentration units such as moles per litre, or amount units such as moles) [14–16]. Although the progress in quantitative MS method development and applications has been enormous, still a lot of effort has to be made to render MS a fully quantitative tool.
Research in the MS field is multi-disciplinary. Quantitative measurements performed with MS build on the achievements in various disciplines of science and engineering. A few available literature positions are particularly relevant to quantitative MS [17–23] (see also related papers published in the Mass Spectrometry Reviews journal). While this literature touched on various aspects of quantitative MS, many of the previous publications focused on specific topics (e.g. proteomics). Arguably, publications on quantitative MS usually do not reach out to those who could benefit from them because they often appear in specialized journals. However, quantitative MS can benefit professionals beyond the narrow fields. Thus, it has been appealing to work on a theme issue containing up-to-date scientific knowledge on many aspects of quantitative MS.
The contents of this theme issue represent the achievements of several disciplines involved in mass spectrometric methods. Consequently, the expertise of the authors extends over several disciplines, including: physics, chemistry, biology, bioinformatics, statistics and engineering. Most of these disciplines heavily rely on the discoveries and developments made during the past two decades. The authors represent academia (universities), research institutes, as well as the private sector. Publishing a theme issue with inputs from experts from various disciplines of science emphasizes the cross-disciplinary character of quantitative MS, and warrants a multi-angle coverage of most quantitative MS approaches. Articles included in this issue encompass original papers, opinion pieces and review articles. They cover topics that can be classified as instrumental developments, sample preparation, data treatment, and applications. Specifically, the invited experts focused on subjects such as proteomics, metabolomics, clinical analysis, pharmaceutical analysis, food science, archaeology, inorganic analysis, hyphenated techniques, mechanistic studies and ionization techniques, as well as instrumentation development. The presented articles are complementary. Minor overlaps between some of the articles were allowed to provide a wider perspective of some of the important topics, taking into account the diverse scientific background of the authors. Apart from discussing their current work, the authors also delineate future research directions in their fields; for instance, addressing the challenges listed in table 1.
Because of the combination of different topics (relevant to various disciplines), this theme issue can serve as a compendium of knowledge for a broad range of readers. It is the guest editor's wish that this theme issue will serve various purposes:
— an up-to-date handbook for students and junior researchers,
— a reference for industrial chemists and engineers,
— a monograph for mass spectrometrists and scientists representing various disciplines,
— a primer for new-comers to the field, and
— a means to popularize one of the most important tools of chemical analysis.
I hope this publication will be accessible for junior and senior scientists from academia and industry, useful for MS professionals as well as students attending courses related to instrumental analysis and bioscience.
Acknowledgements
I would like to thank all the talented co-workers from my research team for many fruitful discussions.
Competing interests
I declare I have no competing interests.
Funding
P.L.U. is supported by Ministry of Science and Technology, Taiwan (MOST 104-2628-M-009-003-MY4).
References
- 1.McLafferty FW, Tureček F. 1993. Interpretation of mass spectra. Sausalito, CA: University Science Books. [Google Scholar]
- 2.Glish GL, Vachet RW. 2003. The basics of mass spectrometry in the twenty-first century. Nat. Rev. Drug Discov. 2, 140–150. ( 10.1038/nrd1011) [DOI] [PubMed] [Google Scholar]
- 3.Gross JH. 2004. Mass spectrometry: a textbook. Berlin, Germany: Springer. [Google Scholar]
- 4.Dass C. 2007. Fundamentals of contemporary mass spectrometry. New York, NY: Wiley. [Google Scholar]
- 5.de Hoffmann E, Stroobant V. 2007. Mass spectrometry: principles and applications. Chichester, UK: Wiley. [Google Scholar]
- 6.Watson JT, Sparkman OD. 2007. Introduction to mass spectrometry: instrumentation , applications, and strategies for data interpretation Chichester, UK: Wiley. [Google Scholar]
- 7.Lee MS. 2012. Mass spectrometry handbook. Hoboken, NJ: Wiley. [Google Scholar]
- 8.Kandiah M, Urban PL. 2013. Advances in ultrasensitive mass spectrometry of organic molecules. Chem. Soc. Rev. 42, 5299–5322. ( 10.1039/C3CS35389C) [DOI] [PubMed] [Google Scholar]
- 9.Greaves J, Roboz J. 2014. Mass spectrometry for the novice. Boca Raton, FL: CRC Press. [Google Scholar]
- 10.Nature Milestones. 2015. Mass spectrometry. New York, NY: Springer Nature. [Google Scholar]
- 11.Urban PL, Chen Y-C, Wang Y-S. 2016. Time-resolved mass spectrometry: from concept to applications. Chichester, UK: Wiley. [Google Scholar]
- 12.Yamashita M, Fenn JB. 1984. Electrospray ion source. Another variation on the free-jet theme. J. Phys. Chem. 88, 4451–4459. ( 10.1021/j150664a002) [DOI] [Google Scholar]
- 13.Whitehouse CM, Dreyer RN, Yamashita M, Fenn JB. 1985. Electrospray interface for liquid chromatographs and mass spectrometers. Anal. Chem. 57, 675–679. ( 10.1021/ac00280a023) [DOI] [PubMed] [Google Scholar]
- 14.Ishihama Y, Schmidt T, Rappsilber J, Mann M, Hartl FU, Kerner MJ, Frishman D. 2008. Protein abundance profiling of the Escherichia coli cytosol. BMC Genomics 9, 102 ( 10.1186/1471-2164-9-102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599. ( 10.1038/nchembio.186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidt A, et al. 2016. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 34, 104–110. ( 10.1038/nbt.3418) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millard BJ. 1977. Quantitative mass spectrometry. Chichester, UK: Wiley. [Google Scholar]
- 18.Duncan MW, Gale PJ, Yergey AL. 2006. The principles of quantitative mass spectrometry. Belfast, UK: Rockpool Productions. [Google Scholar]
- 19.Traldi P, Magno F, Lavagnini I, Seraglia R. 2006. Quantitative applications of mass spectrometry. Chichester, UK: Wiley. [Google Scholar]
- 20.Sechi S (ed.). 2007. Quantitative proteomics by mass spectrometry (methods in molecular biology). Totowa, NJ: Humana Press. [Google Scholar]
- 21.Boyd RK, Basic C, Bethem RA. 2008. Trace quantitative analysis by mass spectrometry. Chichester, UK: Wiley. [Google Scholar]
- 22.Eidhammer I, Barsnes H, Eide GE, Martens L. 2013. Computational and statistical methods for protein quantification by mass spectrometry. Chichester, UK: Wiley. [Google Scholar]
- 23.Eyers CE, Gaskell S (eds). 2014. Quantitative proteomics. Cambridge, UK: RSC. [Google Scholar]