Abstract
Darwin's naturalization hypothesis (DNH), which predicts that alien species more distantly related to native communities are more likely to naturalize, has received much recent attention. The mixed findings from empirical studies that have tested DNH, however, seem to defy generalizations. Using meta-analysis to synthesize results of existing studies, we show that the predictive power of DNH depends on both the invasion stage and the spatial scale of the studies. Alien species more closely related to natives tended to be less successful at the local scale, supporting DNH; invasion success, however, was unaffected by alien–native relatedness at the regional scale. On the other hand, alien species with stronger impacts on native communities tended to be more closely related to natives at the local scale, but less closely related to natives at the regional scale. These patterns are generally consistent across different ecosystems, taxa and investigation methods. Our results revealed the different effects of invader–native relatedness on invader success and impact, suggesting the operation of different mechanisms across invasion stages and spatial scales.
Keywords: biological invasions, Darwin's naturalization conundrum, invasion stage, meta-analysis, pre-adaptation hypothesis, spatial scale
1. Introduction
Increasing human activities, such as transportation, agriculture and aquaculture, have significantly promoted the invasion of alien species into their non-native habitats [1,2]. Such biological invasions, which are occurring worldwide, have brought about widespread ecological, economic and social consequences [3,4]. A central goal of invasion biology has been to gain a better understanding of why some, but not all, introduced species successfully establish in their recipient ecosystems, and even fewer of these species become problematic [5–7]. Both environmental filtering and biotic interactions have been identified as potentially important factors influencing outcomes of biological invasions [8,9]. Notably, their roles in regulating biological invasions were recognized as early as Darwin [10], who proposed two seemingly contradictory hypotheses emphasizing the importance of one factor over the other. On the one hand, Darwin [10] postulated that alien species more closely related to native communities are less likely to invade, based on the premise that native species more closely related to alien invaders tend to share more similar niches (i.e. phylogenetic niche conservatism [11]) with them, and thus offer stronger biotic resistance (Darwin's naturalization hypothesis, DNH [12]). On the other hand, Darwin also hypothesized that alien species closely related to native species may be favoured during invasion, reasoning that closely related species tend to share similar traits that allow them to adapt to the same environments (the pre-adaptation hypothesis, PAH [13]). Empirical tests of Darwin's hypotheses have been quickly accumulating in recent years (summarized by Proches et al. [14], Thuiller et al. [15] and Jones et al. [16]), coinciding with the emergence of phylogenetic community ecology that links species evolutionary relationships with species interactions [17]. These studies have produced mixed findings that echo Darwin's diametric views (termed ‘Darwin's naturalization conundrum’ by Diez et al. [18]), raising the question of whether considering invader–native evolutionary relationships could help understand mechanisms regulating biological invasions.
Searching for simple solutions to Darwin's naturalization conundrum is not straightforward, given the multitude of factors that need to be considered for properly testing the hypotheses [14,15,18]. For example, the relationship between invader–native relatedness and invasion may depend on the spatial scale considered. While Darwin [10] implicitly assumes local spatial scales at which biotic interactions are important in developing DNH, patterns predicted by PAH may be more likely to emerge at regional scales where related species may pass the same environmental filters and co-occur without necessarily driving one another to extinction [14,15,18,19]. Moreover, the effect of invader–native relatedness on invasion could vary among different invasion stages. The invasion process goes through a series of stages such as transport, naturalization/establishment, spread and impact [20], and the relative importance of mechanisms influencing alien species invasion may change across these stages [5,21,22], with potential consequences for native–alien relatedness patterns. For example, if environmental filtering and competition are, respectively, more important at two different invasion stages [23], we might expect to find PAH more applicable to the stage associated with environmental filtering and DNH more applicable to the stage associated with strong competition. The two hypotheses of Darwin, originally proposed with the establishment stage in mind, have been applied to other invasion stages in recent empirical studies (e.g. the stage of invasion spread [23] and the stage of invasion impact [24]). In addition, the effect of invader–native relatedness on invasion may also vary among taxa and ecosystems [25]. Taxon-specific life-history, physiological and behavioural characteristics could influence the strength of invader–native interactions, and, in turn, the outcome of invasion [26]. The differences among ecosystems (e.g. aquatic versus terrestrial) may also carry similar consequences, although across-ecosystem-type studies of Darwin's hypotheses have not been conducted.
During the past decade, an appreciable number of empirical studies have tested the relationships between invader–native relatedness and invasion, making it possible for a quantitative synthesis on this topic. There have been several recent reviews of relevant empirical studies [14–16], but these reviews were based on vote counting that does not account for potential differences in effect sizes among studies. Here, we build on these reviews by including more recent empirical studies and by using quantitative meta-analysis to more rigorously synthesize the data from the existing observational and experimental studies that have examined the relationship between invader–native relatedness and invasion. Our goal was to identify potentially general relationships between invader–native relatedness and invasion, and the sources of variation (if any) in the relationships across studies, in an effort to provide a possible solution to Darwin's naturalization conundrum. Nevertheless, only a modest number of studies satisfied our selection criteria and were included in our meta-analysis, and the number of studies for some subcategories was limited. Therefore, the trends detected in our analyses are best viewed as hypotheses, which can be reexamined later when substantially more studies are available.
2. Material and methods
We followed the PRISMA guidelines (http://www.prisma-statement.org/) in our meta-analysis for data collection, selection, analysis and reporting (see the electronic supplementary material, table S1).
(a). Literature search and data selection
We searched for the list of relevant empirical studies testing DNH and/or PAH, using the ISI Web of Science database. The following key words were used: ‘Darwin's* AND (naturalization hypothesis OR pre-adaptation hypothesis OR conundrum)’ and ‘phylogenetic AND (relatedness OR distance) AND invasi*’. In addition, more studies were identified by examining the reference lists of the electronically retrieved studies. From all these studies, we further selected those that reported the relationships between invader–native relatedness and invasion success/impact, or reported data that can be used to calculate these relationships, for meta-analysis. Studies were excluded if they (i) address issues other than the relationship between invader–native relatedness and invasion (e.g. studying the influence of invader–native relatedness on enemy release), (ii) are reviews, commentaries or mathematical models that contained no empirical data, or (iii) contained duplicate data from other studies (electronic supplementary material, figure S1 and table S2). Our screening yielded 33 articles retained for meta-analysis, contributing 87 data entries (electronic supplementary material, tables S3 and S4).
We classified studies included in our meta-analysis according to the spatial scale of the investigation (local versus regional), stage of invasion (invasion success versus impact), method of investigation (observational versus experimental), study taxa (plants versus animals versus microorganisms) and study ecosystems (terrestrial versus aquatic). We classified studies into local and regional studies according to their grain size (i.e. the size of the individual units of observation): local scale where species interactions probably occur (e.g. the scale of 10 × 10 m plot), and regional scale where species may co-occur but not directly interact with one another (e.g. the scale of California). We followed Lockwood et al. [20] in their classification of invasion stages (transport, establishment, spread and impact) and focused on the three post-transport stages in our analyses. Further, because the majority of studies in our analysis examined establishment, and because alien naturalization and spread may be similarly influenced by competition from native communities [27], we pooled studies of alien establishment and spread together. Considering studies of establishment and spread separately yielded similar results on how native–invader relatedness influences invasion success (electronic supplementary material, figure S2), but resulted in the difficulty of further analysing spread studies given the paucity of relevant data. We did not equate invader spread with impact as in some previous invasion studies, because the ability of alien invaders to spread in their introduced habitats may not necessarily predict their impacts on native species and ecosystems [9,27]. Some of the studies included in our meta-analysis viewed problematic alien species either as aliens that spread beyond the initial area of their introduction or those with demonstrated ecological and economic impacts (i.e. invasive species); we classified these studies into invasion success and impact studies, respectively. Therefore, studies included in our meta-analysis were grouped into studies of invasion success and studies of invasion impact. Invasion success was typically measured by whether the alien species could successfully establish or spread, or in a few cases the abundance that the alien species attain, in the recipient communities; invasion impact was generally measured by whether the alien species are invasive (that is, those that resulted in significant ecological and/or economic damages) or not. Note that, ideally, quantitative measures of invasion success and impacts should be used when studying relatedness–invasion relationships; such data, however, were unavailable for the majority of invasion success studies and all invasion impact studies (see the electronic supplementary material, table S4). Our analyses are thus mainly based on discrete measures of invasion success and impact. For both invasion success and impact, we considered cases where multiple native communities were invaded by the same alien species as well as cases where the same native communities were invaded by multiple alien species.
(b). Data extraction
Correlation coefficients (in the form of Pearson's r) between invader–native relatedness and invader success/impact, after Fisher's z-transformation, were used as effect sizes in our meta-analysis. The use of correlation coefficients as effect sizes maximized the number of studies that can be included in the meta-analysis, as a variety of other statistics, such as the values of the χ2-test [18] and Student's t-test [24], can be readily converted to correlation coefficients [28]. The individual effect size was calculated as
| 2.1 |
where r is the correlation coefficient. The asymptotic variance of z (i.e. the variance of its limit distribution) to be used for summary statistics estimation and publication bias testing was calculated as
| 2.2 |
where n is the sample size [28]. For each data entry, we obtained from the corresponding article either the correlation coefficient or other statistics that could be converted to correlation coefficient. Some of the correlation coefficient values were obtained by extracting raw data from graphs using GetData Graph Digitizer 2.2.6 (http://www.getdata-graph-digitizer.com) and running correlative analyses with the extracted data. For data that could not be obtained from the articles, we requested them from authors and included those that were provided by the authors.
To minimize the inclusion of non-independent data, we further adopted the following criteria to screen data from the assembled studies: (i) when the studies quantified taxonomy-based metrics of species relatedness (e.g. congeneric or coordinal species occurring in the same habitat), we retained the data based on the lowest classification level only; (ii) when the studies reported both the mean phylogenetic relatedness (MPR) of the invader to resident species and the phylogenetic relatedness of the invader to its nearest native relatives (nearest phylogenetic relatedness, NPR), we chose to use NPR as it showed similar patterns to MPR (electronic supplementary material, figure S3) but was more frequently reported; (iii) when the same studies quantified invader performance with multiple metrics (e.g. density, body size and biomass), we used the metrics representing invader overall performance (e.g. biomass) only; (iv) when the studies were conducted with multiple grain sizes at the local spatial scales, we selected only the smallest grain data for the local-scale analysis; and (v) when the studies considered resident communities that differ in their species diversity (e.g. monocultures versus polycultures), we pooled all communities together for a single comprehensive analysis even if data at different diversity levels were reported.
(c). Meta-analysis
Given the inherent random component of effect size variations among studies [29], we chose mixed-effects models that contain both fixed and random components to assess the overall relationships between invader–native relatedness and invasion success/impact. Mixed-effects models were also used to assess the difference in the relatedness–invasion relationship between different types of studies (i.e. regional versus local, experimental versus observational, animals versus plants versus microorganisms, terrestrial versus aquatic). The significance of mean effect sizes was assessed by comparing the bias-corrected 95% bootstrap confidence intervals, based on 9999 iterations, to zero. Significant negative and positive z-values would indicate patterns consistent with DNH and PAH, respectively. The significance of the difference in mean effect sizes between categories was tested using Cochran's Q-test statistic.
We checked for potential publication bias in our meta-analysis using the regression test for funnel plot asymmetry, as suggested by Egger et al. [30], and the trim and fill method, as suggested by Duval & Tweedie [31]. We did not evaluate the risk of bias in individual studies, as the information on the quality of each individual study cannot be obtained. Testing for publication bias was done using the metafor package in R [32], and all other analyses were performed in MetaWin 2.0 [28].
3. Results
Our dataset consisted of 87 data entries, with 65 and 22 entries associated with invasion success and impact, respectively (electronic supplementary material, table S4). Neither the regression test for funnel plot asymmetry (t = −0.482, d.f. = 85, p = 0.631) nor the trim and fill method revealed significant publication bias.
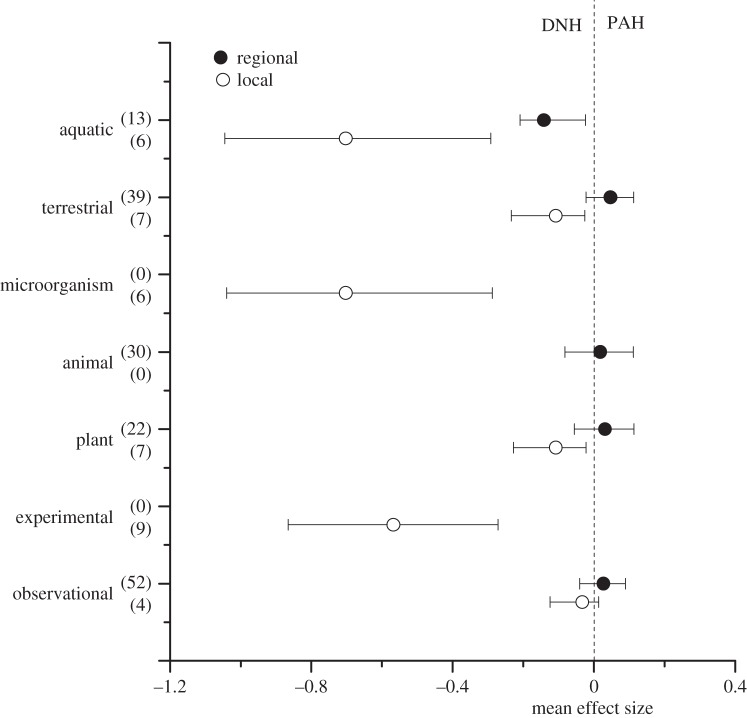
Meta-analysis of studies of invader–native relatedness and invasion success showed that the overall mean effect size did not differ from zero (figure 1). Different patterns, however, emerged for local and regional studies, as indicated by the significant Cochran's Q-test for the two types of studies (figure 1 and table 1). While mean effect size at the local scale was significantly negative, indicating that alien species more closely related to native are less successful at this scale, mean effect size at the regional scale did not differ from zero. This trend largely persisted when studies were further classified according to the method of investigation, taxa or ecosystem type, although the smaller sample sizes rendered most of the between-scale comparisons non-significant (table 1 and figure 2).
Figure 1.
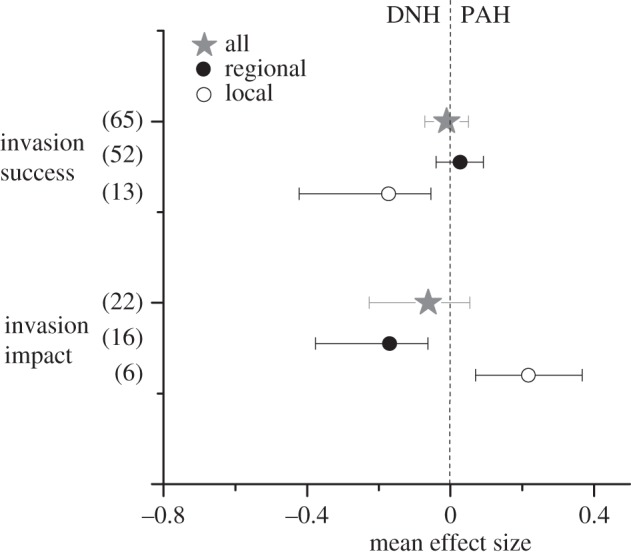
Different effects of invader–native phylogenetic relatedness on invader success and impact for both local and regional scales. Shown are the mean effect sizes (±bias-corrected 95% bootstrap confidence intervals) of the relationships between invader–native phylogenetic relatedness and invader success/impact. Studies were classified into local and regional studies. Mean effect sizes were calculated as Fisher's z-transformations of correlation coefficients between relatedness and invasion success/impact. Values in parentheses represent the sample sizes. Positive mean effect sizes are consistent with the PAH, and negative mean effect sizes are consistent with DNH.
Table 1.
Summary of mixed-effects models estimating invasion success and impact effects sizes and heterogeneity between regional and local studies. The analyses show consistently different effects of invader–native phylogenetic relatedness on invader success and impact for both local and regional scales. Significance values of Q are based on randomization tests. n.a. indicates no data due to low sample size.
| analysis | spatial scale | n | raw data model mean effect size | bias-corrected 95% bootstrap CI | p-value (Cochran's Q-test) |
|---|---|---|---|---|---|
| invasion success | |||||
| all | regional | 52 | 0.0269 | −0.03679 to 0.086 | 0.02 |
| local | 13 | −0.1729 | −0.3944 to −0.0558 | ||
| observational | regional | 52 | 0.0269 | −0.0401 to 0.0893 | 0.593 |
| local | 4 | −0.0116 | −0.0753 to −0.0135 | ||
| experimental | regional | 0 | n.a. | n.a. | n.a. |
| local | 9 | −0.5877 | −0.5877 to −0.5877 | ||
| plant | regional | 22 | 0.0315 | −0.0662 to 0.1082 | 0.361 |
| local | 7 | −0.0528 | −0.1447 to −0.0041 | ||
| animal | regional | 30 | 0.0012 | 0.0012 to 0.0839 | n.a. |
| local | 0 | n.a. | n.a. | ||
| microorganism | regional | 0 | n.a. | n.a. | n.a. |
| local | 6 | −0.7447 | −1.0531 to −0.3333 | ||
| terrestrial | regional | 39 | 0.047 | −0.0223 to 0.1076 | 0.222 |
| local | 7 | −0.0528 | −0.1893 to −0.005 | ||
| aquatic | regional | 13 | −0.1045 | −0.1951 to −0.0045 | 0.003 |
| local | 6 | −0.7313 | −1.0622 to −0.346 | ||
| invasion impact | |||||
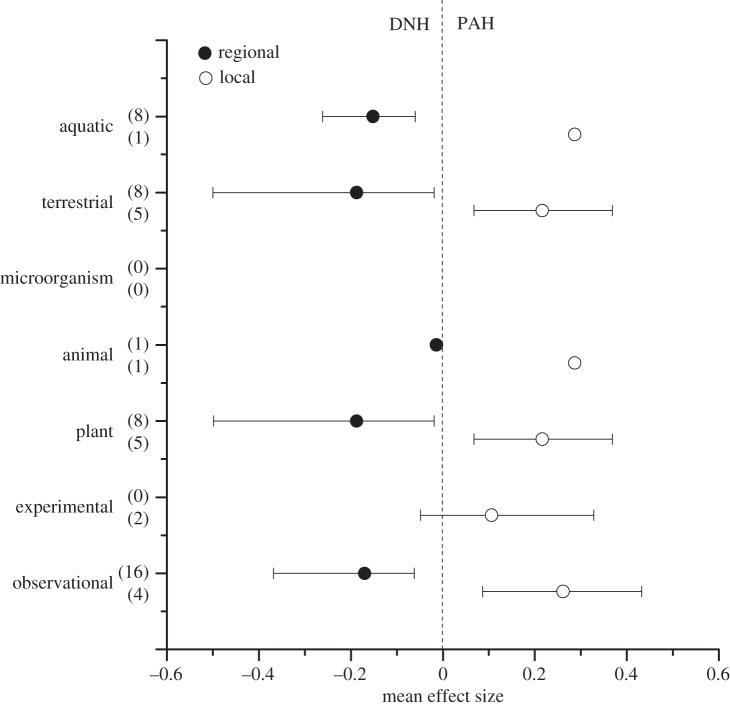
| all | regional | 16 | −0.1701 | −0.3697 to −0.062 | 0.002 |
| local | 6 | 0.2077 | 0.057 to 0.3659 | ||
| observational | regional | 16 | −0.1701 | −0.371 to −0.0644 | 0.007 |
| local | 4 | 0.2483 | 0.0475 to 0.4323 | ||
| experimental | regional | 0 | n.a. | n.a. | n.a. |
| local | 2 | 0.1151 | −0.049 to 0.3283 | ||
| plant | regional | 8 | −0.1874 | −0.5007 to −0.0227 | 0.026 |
| local | 5 | 0.1929 | 0.018–0.362 | ||
| animal | regional | 1 | −0.014 | n.a. | n.a. |
| local | 1 | 0.2866 | n.a. | ||
| microorganism | regional | 0 | n.a. | n.a. | n.a. |
| local | 0 | n.a. | n.a. | ||
| terrestrial | regional | 8 | −0.1874 | −0.5032 to −0.0222 | 0.033 |
| local | 5 | 0.1929 | 0.018–0.3826 | ||
| aquatic | regional | 8 | −0.0987 | −0.2366 to −0.2366 | n.a. |
| local | 1 | 0.2866 | n.a. | ||
Figure 2.
The significant negative relationship between invader–native phylogenetic relatedness and invader success at the local scale, but not the regional scale. Shown are the mean effect sizes (±bias-corrected 95% bootstrap confidence intervals) of the relationships between invader–native phylogenetic relatedness and invader success. Local and regional studies were sub-classified according to ecosystem type (aquatic and terrestrial), taxa (microorganisms, animals and plants) and method of investigation (experimental and observational). For other details see figure 1.
When considering all studies of invader–native relatedness and invasion impact together, the overall mean effect size again did not differ from zero (figure 1). Spatial scale again altered the effect of invader–native relatedness on invasion impact. In contrast to invasion success, mean effect size associated with invader impact was significantly positive at the local scale and significantly negative at the regional scale (figure 1), indicating that alien species with stronger impacts tended to be more closely related to natives at the local scale, but less closely related to natives at the regional scale. This pattern was also largely consistent across different investigation methods, taxa and ecosystems (table 1 and figure 3).
Figure 3.
The positive and negative relationship between invader–native phylogenetic relatedness and invader impact at the local and regional scales, respectively. Shown are the mean effect sizes (±bias-corrected 95% bootstrap confidence intervals) of the relationships between invader–native phylogenetic relatedness and invader impact. Local and regional studies were sub-classified according to ecosystem type (aquatic and terrestrial), taxa (microorganisms, animals and plants) and method of investigation (experimental and observational). For other details see figure 1.
4. Discussion
Our meta-analysis, which represents the first quantitative synthesis of the effects of invader–native relatedness on biological invasions, produced interesting findings. At the local scale, we found that close evolutionary relatedness of invaders and native species made it less likely for the invaders to be successful, supporting DNH. This contrasts with the lack of relationships between invader–native relatedness and invasion success at the regional scale. At the local scale, we also found that close relatedness of invaders and natives was associated with increased invader impact. This pattern was reversed at the regional scale. These results highlight the difference in the relatedness–invasion relationships across different invasion stages and spatial scales.
Perhaps not surprisingly, the results of studies of invasion success at small scales provided strong support for DNH. Biotic interactions, known to be important for regulating invasions at the neighbourhood scale [33], probably contributed to this pattern. In developing DNH, Darwin reasoned that greater niche overlap exists between invaders and their closer native relatives under phylogenetic niche conservatism, resulting in stronger competition from the latter on the former [12]. Few studies in our analyses have tested for phylogenetic niche conservatism (but see [34]), whose strength is known to vary among taxonomic groups [35]. An alternative mechanism leading to patterns consistent with DNH is that aliens may be more likely to share natural enemies with their close native relatives (i.e. phylogenetic niche conservatism with regard to natural enemies [36,37]), resulting in stronger biotic resistance from the latter through apparent competition. Future studies of DNH could benefit from quantifying resident and native species niches (including natural enemies) to gain a more mechanistic understanding of the phylogenetic signature of biological invasions, similar to studies that have quantified species niches to better understand the phylogenetic signature of species coexistence [38–40].
In contrast to the local-scale pattern, invasion success at the regional scale was unrelated to invader–native relatedness. This result is a bit surprising, as we had expected environmental filtering to become more important at the regional scale, potentially leading to patterns predicted by PAH (e.g. [41]). This result, however, could be explained by the fact that a host of other factors, such as propagule pressure and residence time [19], could strongly influence invasion patterns at the regional scale. It is likely that the effects of those other factors overwhelmed those of environmental filtering and biotic interactions, resulting in non-significant relatedness–invasion success patterns. Consistent with this idea, Lambdon & Hulme [19] studied plant invasions on Mediterranean islands and found that the effect of invader–native relatedness on invasion success was visible only after taking the frequency of invader introduction into account. Alternatively, the effects of environmental filtering and biotic interactions could have counteracted each other at the regional scale [42], resulting in the lack of relatedness–invasion success patterns at this scale.
It has been suggested that mechanisms regulating invasion success and impact may be different, such that the two are not necessarily positively correlated [27]. Consistent with this idea, our results on invader impact differ drastically from those on invader success. Specifically, alien species with stronger impacts on native communities tend to be more closely related to natives at the local scale, a pattern consistent with PAH, but tend to be less closely related to natives at the regional scale, a pattern consistent with DNH. While the positive relatedness–impact relationship at the local scale may seem puzzling at first, it is nevertheless consistent with contemporary species coexistence theory, which differentiates species niche and fitness differences [43]. According to this theory, although niche differences may allow invaders to establish in the presence of natives, which was also predicted by Darwin [10], it is invaders' fitness differences with natives that determine their impacts [44]. In particular, high-impact invaders are expected to be those that share similar niches with natives but have fitness advantage over them [44]. Under phylogenetic niche conservatism, this prediction translates into the scenario where invaders that are more closely related to natives, if they do manage to establish by virtue of greater fitness, would impose stronger competitive effect on the natives, resulting in the positive relatedness–impact associations we found at the local scale. Note that this scenario would not occur if species fitnesses are also phylognetically conserved (i.e. little fitness diffence between invaders and their closely related natives), but may emerge if species fitness differences are instead determined by their hierarchical trait distances, a pattern increasingly recognized for plant communities [45,46]. However, although a recent meta-analysis has revealed the general fitness advantage of invasive plants over their native competitors [6], how invader–native fitness difference changes across phylogeny remains an open question.
Prior to our meta-analysis, several studies have reported that invaders with larger impacts tend to be more distantly related to native species at regional scales [24,47]. The results of our meta-analysis strengthen their findings. One hypothesis invoked to explain this negative relatedness–impact relationship at the regional scale is the vulnerability of native communities to the invasion of ecologically novel invaders, to which native species had little exposure [48]. An alternative explanation is that the invaders with close native relatives may share common natural enemies that limit their impacts, whereas those without can impose their forces freely. Note that these hypotheses alone cannot explain the local-scale invasion impact pattern (i.e. alien species with stronger impacts tend to be more closely related to natives), suggesting that the role of the aforementioned processes could vary across spatial scales. Identifying mechanisms underlying the observed phylogenetic patterns of invasion impacts at multiple scales should be an important component of future research.
One issue of note is that the number of studies included in some of our analyses was limited (figures 2 and 3). While this issue is somwhat allieviated by the consistency of our findings across categories of studies (e.g. taxa, ecosystems and investigation methods), it can only be fully addressed with the availability of more data in the future. Another issue is that our meta-analyses were based mainly on discrete measures of invasion success and impact, owing to the lack of quantatitive data. Future invasion studies should aim to collect quantitative data on invasion success and impact, which would allow stronger tests of Darwin's hypotheses and more rigorous syntheses via meta-analysis. A third issue is the lack of studies that have examined invasion success and impact together. In our meta-analysis, all but four studies examined invasion success or impact, but not both (see the electronic supplementary material, table S4). More studies are therefore needed to investigate the success and impact of the same set of invader species within the same ecosystems, which constitute the most rigorous empirical tests of Darwin's hypotheses across invasion stages. With these caveats in mind, our meta-analysis revealed striking differences in how evolutionary relationships between alien and native species influence biological invasions across invasion stages and at different spatial scales. These different patterns clearly indicate situations where predictions of DNH and PAH are expected, providing a possible solution to Darwin's naturalization conundrum. More importantly, these different patterns suggest that mechanisms regulating invader performance differ across invasion stages and spatial scales (e.g. niche differences important for invader success and fitness differences important for invader impact at local spatial scales), which can be specifically explored by future studies.
Supplementary Material
Supplementary Material
Acknowledgements
We thank authors of studies included in our meta-analysis for sharing their data and/or providing data-related information, and Erin Ga-Eun Park and Qixin He for their contributions at the early stage of this project. We are grateful for Xian Yang and two anonymous reviewers for valuable suggestions that significantly improved this manuscript.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
L.J., M.L., C.M. and SP.L. designed research. C.M. and J.T. prepared the database. C.M., SP.L. and Z.P. analysed data. C.M., L.J., SP.L., M.L., H.L. and J.Z. wrote the article.
Competing interests
The authors declare no competing interests.
Funding
C.M. and M.L. were supported by the National Key R&D Program (2016YFD0200305), the National Natural Science Foundation of China (41371263), the PADA (Priority Academic Program Development of Jiangsu Higher Education Institutions) and the Fundamental Research Funds for the Central University of China (KYTZ201404). J.T. was supported by a British Ecological Society Research Grant (5174-6216). This project was funded by the National Science Foundation of USA (DEB-1257858 and DEB-1342754) and National Natural Science Foundation of China (NSFC31361123001).
References
- 1.D'Antonio CM, Vitousek PM. 1992. Biological invasions by exotic grasses, the grass/fire cycle, and global change. Annu. Rev. Ecol. Evol. Syst. 23, 63–87. ( 10.2307/2097282) [DOI] [Google Scholar]
- 2.Vitousek PM, Mooney HA, Lubchenco J, Melillo JM. 1997. Human domination of Earth's ecosystems. Science 277, 494–499. ( 10.1126/science.277.5325.494) [DOI] [Google Scholar]
- 3.Pyšek P, Richardson DM. 2010. Invasive species, environmental change and management, and health. Annu. Rev. Environ. Resour. 35, 25–55. ( 10.1146/annurev-environ-033009-095548) [DOI] [Google Scholar]
- 4.Sala OE, et al. 2000. Global biodiversity scenarios for the year 2100. Science 287, 1770–1774. ( 10.1126/science.287.5459.1770) [DOI] [PubMed] [Google Scholar]
- 5.Kolar CS, Lodge DM. 2001. Progress in invasion biology: predicting invaders. Trends Ecol. Evol. 16, 199–204. ( 10.1016/S0169-5347(01)02101-2) [DOI] [PubMed] [Google Scholar]
- 6.van Kleunen M, Dawson W, Schlaepfer D, Jeschke JM, Fischer M. 2010. Are invaders different? A conceptual framework of comparative approaches for assessing determinants of invasiveness. Ecol. Lett. 13, 947–958. ( 10.1111/j.1461-0248.2010.01503.x) [DOI] [PubMed] [Google Scholar]
- 7.Vila M et al. 2011. Ecological impacts of invasive alien plants: a meta-analysis of their effects on species, communities and ecosystems. Ecol. Lett. 14, 702–708. ( 10.1111/j.1461-0248.2011.01628.x) [DOI] [PubMed] [Google Scholar]
- 8.Rejmánek M, Richardson DM, Pyšek P. 2013. Plant invasions and invasibility of plant communities. In Vegetation ecology, 2nd edn (eds E van der Maarel, J Franklin), pp. 387–424. Oxford, UK: Wiley-Blackwell. [Google Scholar]
- 9.Gallien L, Carboni M, Munkemuller T. 2014. Identifying the signal of environmental filtering and competition in invasion patterns: a contest of approaches from community ecology. Methods Ecol. Evol. 5, 1002–1011. ( 10.1111/2041-210x.12257) [DOI] [Google Scholar]
- 10.Darwin C. 1859. The origin of species. London, UK: Murray. [Google Scholar]
- 11.Wiens JJ, et al. 2010. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol. Lett. 13, 1310–1324. ( 10.1111/j.1461-0248.2010.01515.x) [DOI] [PubMed] [Google Scholar]
- 12.Daehler CC. 2001. Darwin's naturalization hypothesis revisited. Am. Nat. 158, 324–330. ( 10.1086/321316) [DOI] [PubMed] [Google Scholar]
- 13.Ricciardi A, Mottiar M. 2006. Does Darwin's naturalization hypothesis explain fish invasions? Biol. Invasions 8, 1403–1407. ( 10.1007/s10530-006-0005-6) [DOI] [Google Scholar]
- 14.Proches S, Wilson JRU, Richardson DM, Rejmanek M. 2008. Searching for phylogenetic pattern in biological invasions. Glob. Ecol. Biogeogr. 17, 5–10. ( 10.1111/j.1466-8238.2007.00333.x) [DOI] [Google Scholar]
- 15.Thuiller W, Gallien L, Boulangeat I, de Bello F, Munkemuller T, Roquet C, Lavergne S. 2010. Resolving Darwin's naturalization conundrum: a quest for evidence. Divers. Distrib. 16, 461–475. ( 10.1111/j.1472-4642.2010.00645.x) [DOI] [Google Scholar]
- 16.Jones EI, Nuismer SL, Gomulkiewicz R. 2013. Revisiting Darwin's conundrum reveals a twist on the relationship between phylogenetic distance and invasibility. Proc. Natl Acad. Sci. USA 110, 20 627–20 632. ( 10.1073/pnas.1310247110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Webb CO, Ackerly DD, Mcpeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Evol. Syst. 8, 475–505. ( 10.2307/3069271) [DOI] [Google Scholar]
- 18.Diez JM, Sullivan JJ, Hulme PE, Edwards G, Duncan RP. 2008. Darwin's naturalization conundrum: dissecting taxonomic patterns of species invasions. Ecol. Lett. 11, 674–681. ( 10.1111/j.1461-0248.2008.01178.x) [DOI] [PubMed] [Google Scholar]
- 19.Lambdon PW, Hulme PE. 2006. How strongly do interactions with closely-related native species influence plant invasions? Darwin's naturalization hypothesis assessed on Mediterranean islands. J. Biogeogr. 33, 1116–1125. ( 10.1111/j.1365-2699.2006.01486.x) [DOI] [Google Scholar]
- 20.Lockwood JL, Hoopes MF, Marchetti MP. 2013. Invasion ecology. New York, NY: John Wiley & Sons. [Google Scholar]
- 21.Theoharides KA, Dukes JS. 2007. Plant invasion across space and time: factors affecting nonindigenous species success during four stages of invasion. New Phytol. 176, 256–273. ( 10.1111/j.1469-8137.2007.02207.x) [DOI] [PubMed] [Google Scholar]
- 22.Dawson W, Burslem DFRP, Hulme PE. 2009. Factors explaining alien plant invasion success in a tropical ecosystem differ at each stage of invasion. J. Ecol. 97, 657–665. ( 10.1111/j.1365-2745.2009.01519.x) [DOI] [Google Scholar]
- 23.Schaefer H, Hardy OJ, Silva L, Barraclough TG, Savolainen V. 2011. Testing Darwin's naturalization hypothesis in the Azores. Ecol. Lett. 14, 389–396. ( 10.1111/j.1461-0248.2011.01600.x) [DOI] [PubMed] [Google Scholar]
- 24.Strauss SY, Webb CO, Salamin N. 2006. Exotic taxa less related to native species are more invasive. Proc. Natl Acad. Sci. USA 103, 5841–5845. ( 10.1073/pnas.0508073103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen CR, et al. 2013. Predictors of regional establishment success and spread of introduced non-indigenous vertebrates. Glob. Ecol. Biogeogr. 22, 889–899. ( 10.1111/geb.12054) [DOI] [Google Scholar]
- 26.Ferreira RB, Beard KH, Peterson SL, Poessel SA, Callahan CM. 2012. Establishment of introduced reptiles increases with the presence and richness of native congeners. Amphib-Reptil. 33, 387–392. ( 10.1163/15685381-00002841) [DOI] [Google Scholar]
- 27.Ricciardi A, Cohen J. 2007. The invasiveness of an introduced species does not predict its impact. Biol. Invasions 9, 309–315. ( 10.1007/s10530-006-9034-4) [DOI] [Google Scholar]
- 28.Rosenberg MS, Adams DC, Gurevitch J. 2000. MetaWin: statistical software for meta-analysis. Sunderland, MA: Sinauer Associates Sunderland. [Google Scholar]
- 29.Gurevitch J, Hedges LV. 1999. Statistical issues in ecological meta-analyses. Ecology 80, 1142–1149. ( 10.1890/0012-9658(1999)080%5B1142:Siiema;2.0.Co;2) [DOI] [Google Scholar]
- 30.Egger M, Smith GD, Schneider M, Minder C. 1997. Bias in meta-analysis detected by a simple, graphical test. Brit. Med. J. 315, 629–634. ( 10.1136/bmj.315.7109.629) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duval SJ, Tweedie RL. 2000. Trim and fill: a simple funnel-plot–based method of testing and adjusting for publication bias in meta-analysis. Biometrics 56, p455–463(459). ( 10.1111/j.0006-341X.2000.00455.x) [DOI] [PubMed] [Google Scholar]
- 32.Viechtbauer W. 2010. Conducting meta-analyses in R with the metafor Package. J. Stat. Softw. 36, 1–48. ( 10.18637/jss.v036.i03) [DOI] [Google Scholar]
- 33.Kennedy TA, Naeem S, Howe KM, Knops JMH, Tilman D, Reich P. 2002. Biodiversity as a barrier to ecological invasion. Nature 417, 636–638. ( 10.1038/Nature00776) [DOI] [PubMed] [Google Scholar]
- 34.Jiang L, Tan JQ, Pu ZC. 2010. An experimental test of Darwin's naturalization hypothesis. Am. Nat. 175, 415–423. ( 10.1086/650720) [DOI] [PubMed] [Google Scholar]
- 35.Losos JB. 2008. Phylogenetic niche conservatism, phylogenetic signal and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol. Lett. 11, 995–1003. ( 10.1111/j.1461-0248.2008.01229.x) [DOI] [PubMed] [Google Scholar]
- 36.Liu XB, Liang MX, Etienne RS, Wang YF, Staehelin C, Yu SX. 2012. Experimental evidence for a phylogenetic Janzen–Connell effect in a subtropical forest. Ecol. Lett. 15, 111–118. ( 10.1111/j.1461-0248.2011.01715.x) [DOI] [PubMed] [Google Scholar]
- 37.Parker IM, Saunders M, Bontrager M, Weitz AP, Hendricks R, Magarey R, Suiter K, Gilbert GS. 2015. Phylogenetic structure and host abundance drive disease pressure in communities. Nature 520, 542–544. ( 10.1038/nature14372) [DOI] [PubMed] [Google Scholar]
- 38.Narwani A, Alexandrou MA, Oakley TH, Carroll IT, Cardinale BJ. 2013. Experimental evidence that evolutionary relatedness does not affect the ecological mechanisms of coexistence in freshwater green algae. Ecol. Lett. 16, 1373–1381. ( 10.1111/ele.12182) [DOI] [PubMed] [Google Scholar]
- 39.Godoy O, Kraft NJB, Levine JM. 2014. Phylogenetic relatedness and the determinants of competitive outcomes. Ecol. Lett. 17, 836–844. ( 10.1111/ele.12289) [DOI] [PubMed] [Google Scholar]
- 40.Tan J, Slattery MR, Yang X, Jiang L. 2016. Phylogenetic context determines the role of competition in adaptive radiation. Proc. R. Soc. B 283, 1–8. ( 10.1098/rspb.2016.0241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carboni M, Munkemuller T, Gallien L, Lavergne S, Acosta A, Thuiller W. 2013. Darwin's naturalization hypothesis: scale matters in coastal plant communities. Ecography 36, 560–568. ( 10.1111/j.1600-0587.2012.07479.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helmus MR, Savage K, Diebel MW, Maxted JT, Ives AR. 2007. Separating the determinants of phylogenetic community structure. Ecol. Lett. 10, 917–925. ( 10.1111/j.1461-0248.2007.01083.x) [DOI] [PubMed] [Google Scholar]
- 43.Chesson P. 2000. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. ( 10.1146/annurev.ecolsys.31.1.343) [DOI] [Google Scholar]
- 44.MacDougall AS, Gilbert B, Levine JM. 2009. Plant invasions and the niche. J. Ecol. 97, 609–615. ( 10.1111/j.1365-2745.2009.01514.x) [DOI] [Google Scholar]
- 45.Kunstler G, Lavergne S, Courbaud B, Thuiller W, Vieilledent G, Zimmermann NE, Kattge J, Coomes DA. 2012. Competitive interactions between forest trees are driven by species’ trait hierarchy, not phylogenetic or functional similarity: implications for forest community assembly. Ecol. Lett. 15, 831–840. ( 10.1111/j.1461-0248.2012.01803.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kraft NJB, Crutsinger GM, Forrestel EJ, Emery NC. 2014. Functional trait differences and the outcome of community assembly: an experimental test with vernal pool annual plants. Oikos 123, 1391–1399. ( 10.1111/oik.01311) [DOI] [Google Scholar]
- 47.Ricciardi A, Atkinson SK. 2004. Distinctiveness magnifies the impact of biological invaders in aquatic ecosystems. Ecol. Lett. 7, 781–784. ( 10.1111/j.1461-0248.2004.00642.x) [DOI] [Google Scholar]
- 48.Ricciardi A, Hoopes MF, Marchetti MP, Lockwood JL. 2013. Progress toward understanding the ecological impacts of nonnative species. Ecol. Monogr. 83, 263–282. ( 10.1890/13-0183.1) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.