Abstract
Environmental stresses experienced by individual parents can influence offspring phenotypes in ways that enhance survival under similar conditions. Although such adaptive transgenerational plasticity is well documented, its transmission mechanisms are generally unknown. One possible mechanism is environmentally induced DNA methylation changes. We tested this hypothesis in the annual plant Polygonum persicaria, a species known to express adaptive transgenerational plasticity in response to parental drought stress. Replicate plants of 12 genetic lines (sampled from natural populations) were grown in dry versus moist soil. Their offspring were exposed to the demethylating agent zebularine or to control conditions during germination and then grown in dry soil. Under control germination conditions, the offspring of drought-stressed parents grew longer root systems and attained greater biomass compared with offspring of well-watered parents of the same genetic lines. Demethylation removed these adaptive developmental effects of parental drought, but did not significantly alter phenotypic expression in offspring of well-watered parents. The effect of demethylation on the expression of the parental drought effect varied among genetic lines. Differential seed provisioning did not contribute to the effect of parental drought on offspring phenotypes. These results demonstrate that DNA methylation can mediate adaptive, genotype-specific effects of parental stress on offspring phenotypes.
Keywords: ecological epigenetics, non-genetic inheritance, maternal effects, phenotypic plasticity, drought stress
1. Introduction
Stressful parental environments can influence the phenotypes of offspring in many plant and animal taxa [1–4]. Such inherited environmental effects on development (transgenerational plasticity or parental environmental effects) were initially understood to directly reflect resource levels, with resource-deprived parents producing low-quality offspring due to reduced provisioning (e.g. to seeds or eggs). While these direct effects of provisioning are indeed widespread [3,4], more specialized developmental effects of parental environments have increasingly come to light. In some cases, stressed parents produce offspring with specific developmental alterations that mitigate that particular type of stress. When these offspring encounter similar conditions, such alterations result in heritable, environmentally induced adaptation [5–10]. As with other aspects of phenotypic plasticity [11,12], genotypes from natural populations differ in the precise pattern and degree of these inherited environmental effects [8,13–15].
Despite increasing awareness of the potential adaptive value of transgenerational plasticity, in most cases, the mechanisms responsible for the inheritance of these effects remain unclear [16–18]. In some systems, adaptive transgenerational effects are known to stem from increased provisioning of nutritive resources to seeds or eggs (e.g. [19,20]). However, as provisioning-based effects are mediated directly by maternal individuals, environmental effects that persist for multiple generations (e.g. [21–24]) must be mediated by mechanisms capable of longer-term stability.
One potential mechanism is heritable, environmentally induced changes to DNA methylation [25,26]. In many taxa, the addition or removal of methyl groups on cytosine residues can influence transcriptional activity at specific loci. In plants, numerous environmental stimuli can induce changes to DNA methylation throughout the genome, ranging from biotic stresses such as pathogen infection [27–29] to abiotic stresses such as drought [30–33]. Such environmentally induced methylation changes vary by genotype [34] and have been shown to be heritable in a variety of species, in some cases for multiple generations [35,36]. Furthermore, heritable methylation variation can have substantial impacts on ecologically important traits. For instance, studies of epigenetic recombinant inbred lines in Arabidopsis thaliana showed that DNA methylation variants can cause substantial heritable variation in key traits such as primary root length and flowering time in the absence of DNA sequence diversity [37].
This combination of genotype-specific environmental sensitivity, transgenerational stability, and phenotypic impact makes DNA methylation a primary candidate mechanism for adaptive transgenerational plasticity, particularly in plants [16,17]. Yet a central question remains unresolved: to what extent do genotypes in natural populations vary in their epigenetic responses to stressful parental environments [38]? The prevalence of genotype × parental environment interactions on offspring phenotypes suggests that genotype-specific patterns of epigenetic response may underlie this common form of genetic variation. Such G × parental E variation provides the substrate for the adaptive evolution of transgenerational plasticity, just as genetic variation for responses to the immediate environment (G × E) fuels the adaptive evolution of within-generation plasticity [11,39,40]. Hence, demonstrating a role for DNA methylation in genotype × parental environment interactions would add a new dimension of evolutionary relevance to our understanding of environmentally induced epigenetic variation.
Experimental demethylation using pharmacological agents is a well-established method for investigating whether DNA methylation mediates phenotypic expression in diverse animal, plant, and fungal systems [41–46]. Several such agents are known to interfere with the DNA methyltransferase enzymes that establish and maintain methylation of cytosine residues, leading to genome-wide reductions in DNA methylation levels. Zebularine is particularly useful as an experimental demethylation agent because its methyltransferase-inhibiting effects are transient and dose-dependent [47,48]. Alternative demethylation agents such as 5-azacytidine produce more side effects [49], and mutational approaches to demethylation cause long-lasting and drastic methylation changes [37].
We used zebularine to investigate the functional role of DNA methylation in the adaptive, drought-induced transgenerational plasticity that is differentially expressed by genotypes of the annual plant Polygonum persicaria (=Persicaria maculosa [8,24,50]). In this common species, seedling offspring of drought-stressed parents or grandparents develop more extensive, deeper root systems, and have enhanced growth and survival, in dry soil [24,51]. This Eurasian plant has successfully spread throughout most of North America, occupying ecologically diverse sites that vary both spatially and temporally in soil-moisture content [52]. As P. persicaria has a mixed breeding system with a high natural rate of self-fertilization [53], highly inbred lines can be generated without causing inbreeding depression. Such experimental lines provide replicate parent plants of each genotype that can be raised in contrasting environments and allowed to produce progeny, to compare genotypic patterns of transgenerational plasticity [8,24].
We raised replicate parent plants in both dry and moist soil from 12 inbred genetic lines that had been sampled initially from natural P. persicaria populations. We then examined the effects of these parental environments on drought-stressed offspring germinated either in control conditions, or with a zebularine concentration known to cause moderate genome-wide demethylation. We raised these offspring under drought stress to test the adaptive value of the drought-induced parental effects [24]. We also estimated seed provisioning for each offspring individual in the study. We tested the following predictions. (1) If enhanced seed provisioning mediates transgenerational response to drought, then offspring of drought-stressed parents should have greater provisioning than offspring of well-watered parents. (2) If DNA methylation regulates drought-induced transgenerational plasticity, demethylating offspring should reduce or remove the specific effects of parental drought on offspring growth without significantly altering growth in offspring of well-watered parents. (Note that (1) and (2) need not be exclusive alternatives.) (3) If environmentally induced DNA methylation patterns are genotype specific, then the effect of demethylation on offspring expression of parental drought effects should vary among genetic lines.
2. Material and methods
(a). Parental generation
Twelve genetic lines drawn from five ecologically distinct natural populations (field environmental data in [52]) were propagated by self-fertilization and single-seed descent for five generations under uniform, favourable glasshouse conditions. In the parental generation, we stratified achenes (1 seeded propagules) from each inbred line for 28 days in distilled water at 4°C to break dormancy, and sowed them in vermiculite-filled flats positioned randomly on a glasshouse bench in full sun (March 2014). Individual seedlings were transplanted (approx. 20 d after emergence) into 1 l clay pots filled with a 2 : 2 : 1 mix of sterilized topsoil: horticultural sand: fritted clay (Turface™, Profile Products, Buffalo Grove, IL, USA). Soil moisture was maintained at 100% of field capacity (approx. 31% moisture by weight) for all plants for one week. We then assigned one seedling from each highly inbred genetic line to a dry soil environment (approx. 42% of soil field capacity), and another seedling from the same line to a moist-soil environment (approx. 84% of soil field capacity). Parental moisture environments were maintained using an automatic watering system, with pot-specific manual watering as needed. The 24 parental plants (12 genetic lines × 1 parent per treatment × 2 moisture treatments) were grown for 53 d, with bench positions re-randomized weekly. Self-fertilized achenes (offspring) were harvested from each parent plant, air-dried, and stored with desiccant at 4°C [54].
(b). Offspring generation
We individually weighed 24 achenes from each parental plant on a Cahn C-33 microbalance (Cahn Instruments, Cerritos, CA, USA), placed them into 96 well plates, and submerged them in distilled water at 4°C for five weeks to break dormancy. Demethylation treatment was imposed during seed germination. Achenes were sown in Petri plates (8 August 2014) on solidified 0.8% agar containing either 0 or 45 µM zebularine (hereafter referred to as Control and Zebularine germination treatments, respectively); this zebularine concentration did not disrupt plant development (electronic supplementary material, Appendix S1). (Note that 40 µM zebularine reduced global 5-methyldeoxycytidine levels by 15–18% in Medicago truncatula and A. thaliana; in these plants methylation levels returned to normal after several weeks' growth in the absence of zebularine [47].) Petri plates were placed on a glasshouse bench and their positions were re-randomized daily.
Six days after germination, we transplanted four replicate seedlings from each genetic line × parental environment × germination treatment combination into individual growth containers filled with a 2 : 2 : 1 mix of sterilized topsoil: horticultural sand: fritted clay pre-moistened lightly with 40 ml of water per litre of soil mix (details in [51]). The total experimental sample was 192 offspring (12 genetic lines × 2 parental moisture treatments × 2 germination treatments × 4 replicate offspring). The seedlings were placed in a randomized complete-block design in a dual Conviron growth chamber (Controlled Environments, Winnipeg, Manitoba, Canada) at a 25°C : 18°C 14 : 10 h day : night cycle with approximately 500 µmol m−2 s−1 'photosynthetically active radiation (PAR) daytime illumination. A constant low soil-moisture level (approx. 40% of field capacity; comparable with the 42% in the parental generation) was manually maintained for all seedlings. Seedling positions were re-randomized within blocks weekly; seedlings were grown for 21 days.
(c). Data collection
To estimate seed provisioning for each seedling offspring, we subtracted the mass of the pericarp (retrieved after germination) from the initial achene mass. This measure thus includes only embryo and nutritive tissue to most precisely capture provisioning [8]. On day 21, aboveground tissues from each seedling were separated and dried at 100°C for 1 h and then at 65°C for greater than or equal to 48 h before weighing. The following data were collected for each seedling. The first three true leaves were scanned on a LI-3100 leaf area meter (LICOR, Inc., Lincoln, NE, USA), dried, and weighed to estimate specific leaf area (leaf surface area per unit mass; cm2 leaf/g leaf). Total leaf area was estimated by multiplying this ratio by the total biomass of leaves from that plant. We washed root systems free of all soil mix before measuring total root system length for each seedling with a Comair optical scanner (Hasker de Havilland, Melbourne, Australia). Root systems were dried at 65°C for greater than or equal to 48 h and weighed. We calculated seedling biomass as the sum of shoot and root biomass. Two plants were not included in the final sample due to insufficient germination, and one control-germinated plant was removed from the experiment due to abnormal development. Owing to missing data and the removal of two outliers from the analysis, final sample sizes for total root length, leaf area, and seedling biomass were 177, 177, and 180, respectively.
(d). Data analysis
We used a linear mixed-effects model to analyse the effects of parental environment (dry versus moist soil), genetic line, and their interaction on seed provisioning, treating genetic line and its interaction with parental environment as random effects (variance components estimated by restricted maximum likelihood (REML); [55]). This approach was also used to analyse the effects on offspring (seedling) phenotypes of parental environment (dry versus moist soil), germination treatment (zebularine versus control), genetic line, and all two- and three-way interactions among these factors, again treating genetic line and its interactions as random effects. A significant parental environment × germination treatment interaction would indicate that the demethylation treatment altered the expression of transgenerational plasticity (i.e. that the effect of dry versus moist parental environment differed if offspring were germinated in zebularine rather than control conditions). Seed provisioning was included as a covariate in the analyses, and spatial block was included in the model as a fixed effect.
To assess the effect size of each random effect, we expressed the variance for each random effect as a percentage of the remaining variance that was not explained by fixed effects: (random effect variance/[sum of all random effect variances + residual variance]) × 100 [56] (see electronic supplementary material Appendix S1 for the rationale for forgoing significance testing of random effects in complex linear mixed-effects models). If the three-way interaction of genetic line × parental environment × germination treatment explained a substantial percentage of remaining variance that would indicate that the effect of demethylation on the expression of transgenerational plasticity varied among genetic lines. We set a threshold of greater than or equal to 10% of variance explained to indicate a biologically meaningful source of variation, but we report the actual per cent of variance explained for each effect so that the reader may set this threshold however s/he sees fit. We verified (qualitatively) the random effects results obtained from linear mixed-effects models by running similar analyses in a mixed ANOVA framework. (Linear mixed models estimate variance components via maximum-likelihood procedures, in contrast with the least-squares approach of mixed ANOVA. Note that linear mixed models provide more robust estimates of random effects than mixed ANOVA, especially when there are missing data [55,57].) We also ran linear mixed models within each germination treatment to calculate the percentage of variance explained by the genetic line × parental environment interaction.
We used one-way ANOVA to test a priori hypotheses regarding the effect of parental drought versus moist-soil environments on offspring within each germination treatment. Visual inspection of the results suggested that genetic lines that most strongly increased seedling biomass in response to parental drought under control germination conditions were also the most inhibited in their growth when demethylated. We used Spearman's rank correlation coefficient to test the significance of this apparent negative correlation [58]. Mixed-model ANOVAs were performed in JMP 11 (SAS Institute, Cary, NC, USA). All other analyses were conducted with R v. 3.1.2 [59]. The nlme package was used to perform linear mixed-effects models [60].
3. Results
(a). Parental drought did not increase seed provisioning
Offspring of drought-stressed and well-watered parents had equivalent seed provisioning (electronic supplementary material, figure S1, parental environment, F1,172 = 0.547, p = 0.461). Offspring provisioning did not significantly contribute to variation in any seedling trait (table 1).
Table 1.
Effects of parental environment (PE; drought versus moist soil), germination treatment (GT; demethylation versus control), genetic line, and their interactions on total root system length, leaf area, and seedling biomass from linear mixed-effects models. An estimate of seed provisioning was included as a covariate. The variance for each random effect is expressed as the percentage of the variance that was unexplained by fixed effects (%Var. = (random effect variance/(sum of all random effect variances + residual variance)) × 100). Significance levels for fixed effects indicated as †p < 0.10, **p < 0.01, ***p < 0.001.
| fixed effects | total root length (N = 177) |
leaf area (N = 177) |
seedling biomass (N = 180) |
|||
|---|---|---|---|---|---|---|
| F | p-value | F | p-value | F | p-value | |
| parent env. (PE) | 0.108 | 0.743 | 0.016 | 0.899 | 1.390 | 0.240 |
| germination trt. (GT) | 8.634 | 0.004** | 3.135 | 0.079† | 13.485 | <0.001*** |
| PE × GT | 6.934 | 0.009** | 7.562 | 0.007** | 7.594 | 0.007** |
| seed provisioning | 1.041 | 0.309 | 0.127 | 0.721 | 1.125 | 0.291 |
| block | 5.274 | 0.002** | 23.398 | <0.001*** | 8.677 | <0.001*** |
| random effects | %Var. | %Var. | %Var. | |||
| genetic line | 28.323 | 23.925 | 19.771 | |||
| PE × Line | 14.193 | 13.822 | 14.822 | |||
| GT × Line | 10.913 | 18.002 | 17.262 | |||
| PE × GT × Line | 17.076 | 23.990 | 30.566 | |||
(b). Demethylation removed the adaptive effect of parental drought stress
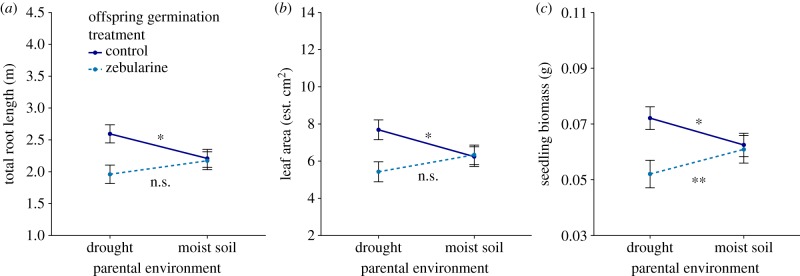
On average, the zebularine (demethylating) germination treatment did not alter any seedling traits in offspring of well-watered parents (figure 1), but did alter the development of seedlings of drought-stressed parents (cf. significant interaction between parental environment and germination treatment for total seedling root length, leaf area, and biomass, table 1). Under control germination conditions, as found in previous studies, the offspring of drought-stressed parents had on average approximately 20% longer root systems (figure 1a, F1,90 = 5.218, p = 0.025), approximately 23% greater leaf area (figure 1b, F1,89 = 5.04, p = 0.027), and approximately 16% greater biomass (figure 1c, F1,91 = 4.749, p = 0.032) compared with offspring of the same genetic lines whose parents had been well watered. By contrast, when germinated in the presence of zebularine, offspring of drought-stressed parents had approximately 17% lower biomass (figure 1c, F1,91 = 8.294, p = 0.005) as well as non-significantly shorter roots and lower leaf area (11% and 13% reduction, figure 1a,b, respectively) compared with either control-germinated or zebularine-treated offspring of well-watered parents.
Figure 1.
Demethylation of offspring DNA removes the adaptive effect of parental drought on total root length (a), leaf area (b), and total biomass (c) of individual seedling offspring grown in dry soil. Means ± s.e. are shown for offspring exposed to 0 µM (control) or 45 µM zebularine during germination. Asterisks indicate significance of the parental drought effect (one-way ANOVA separately testing the effect of parental environment on control-germinated and zebularine-germinated seedlings, *p < 0.05, **p < 0.01, n.s., non-significant, see Results section for details). (Online version in colour.)
Demethylation reduced the positive growth effects of parental drought stress on seedling traits more strongly than expected, resulting in reduced rather than equivalent root system length, leaf area, and seedling biomass compared with offspring of non-stressed parents as described above (significant main effect of germination treatment, table 1). We examined germination dynamics to test a possible explanation for this unexpected effect: that offspring of drought-stressed parents may have experienced a more extreme zebularine treatment than those of well-watered parents by virtue of germinating later and consequently remaining longer in the Petri plate. This was not the case: there was no difference in germination timing (i.e. number of days between sowing and germination) between offspring of drought-stressed versus well-watered parents in the zebularine germination treatment (F1,177 = 0.398, p = 0.529). In fact, 114 of the 190 transplanted seedlings germinated on the same day, and this subsample of seedlings displayed the same pattern of response differences observed in the full dataset (electronic supplementary material, figure S2), indicating that offspring from both parental environments received equivalent demethylation treatments.
(c). Genetic lines varied in both the effect of parental drought and its alteration by demethylation
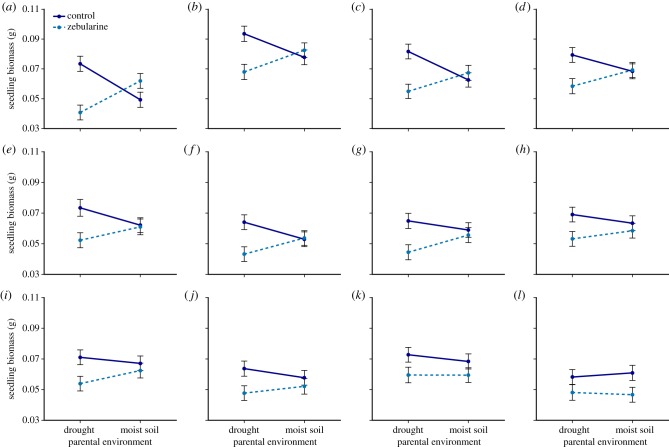
The response patterns of most genetic lines were qualitatively similar to the general pattern explained above: in most cases, parental drought increased offspring biomass, total root length, and leaf area, and demethylation inhibited this effect (figure 2; electronic supplementary material, figures S3 and S4). However, there was substantial variation among genetic lines in the magnitude of the parental drought effect and its alteration by demethylation. Under control germination conditions, parental drought increased seedling biomass between 5.8% and 48.8% in 11 of 12 lines (figure 2a–j), while slightly decreasing biomass in one line (figure 2l) by 4.7% (within the control germination treatment, genetic line × parental environment interaction accounted for 23.6% of variance in biomass after accounting for fixed effects). Lines in which control-germinated offspring most sharply increased biomass in response to parental drought stress were also the most growth inhibited by parental drought stress when demethylated (figure 3; Spearman's rank correlation ρ = −0.776, p = 0.002). In offspring of well-watered parents, demethylation caused either no reduction or a slight growth reduction in most lines but increased biomass substantially in one genetic line and very slightly in two lines (figure 2a–c). In two other lines, demethylation had similar effects on offspring from both drought-stressed and well-watered parents (figure 2k,l). This complex pattern of genetic variation for the effects of parental soil-moisture treatment and DNA methylation status is reflected in the three-way interaction between genetic line, parental environment, and germination treatment, which explained approximately 30.5% of the remaining variance in seedling biomass after accounting for fixed effects (table 1). Variation in total seedling leaf area revealed very similar patterns of genotype-specific differences in parental environment × germination treatment effects, explaining approximately 24% of the remaining variance in this trait after accounting for fixed effects (electronic supplementary material, figure S4; table 1). Effects of parental drought and demethylation on total root length varied less among genetic lines (genetic line × parental environment × germination treatment explained 17.1% of remaining root length variance; electronic supplementary material, figure S3; table 1). Variance components for the three-way interactions estimated by mixed-model ANOVAs were qualitatively similar (though slightly smaller) to those estimated by REML in linear mixed models: after accounting for fixed effects, the remaining variance explained by the three-way interaction was 28.59% for seedling biomass (p = 0.007), 15.19% for leaf area (p = 0.064), and 10.47% for total root length (p = 0.143).
Figure 2.
Genetic variation for the effect of parental drought on seedling biomass and its alteration by demethylation, reflecting the interaction between genetic line, parental environment, and zebularine versus control germination treatment in the linear mixed-effects model (table 1). Each plot displays means ± s.e. for one genetic line. (Online version in colour.)
Figure 3.
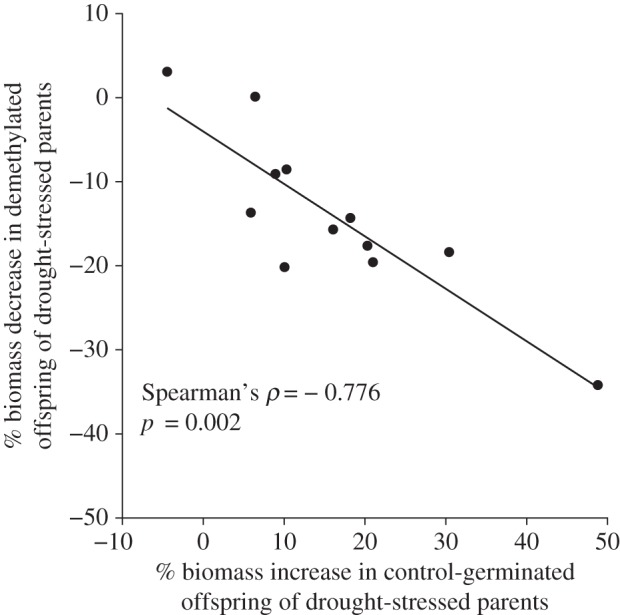
Genetic lines that most strongly increased seedling biomass in response to parental drought when germinated under control conditions were also the most inhibited in their growth when germinated in the presence of zebularine. The x-axis represents the per cent increase in biomass of control-germinated offspring of drought-stressed parents relative to the biomass of control-germinated offspring of well-watered parents. The y-axis represents the per cent decrease in biomass of zebularine-germinated offspring of drought-stressed parents relative to the biomass of zebularine-germinated offspring of well-watered parents. One point represents one genetic line (based on three to four replicates in each germination treatment). Significance test is shown based on Spearman's rank correlation coefficient (see Material and methods and Data analysis sections).
4. Discussion
(a). DNA methylation mediates adaptive transgenerational plasticity in Polygonum persicaria
Results from the control germination treatment showed that drought-stressed P. persicaria parents produced drought-adapted offspring, compared with parent plants of the same genetic lines that had been given ample moisture. This finding confirmed previous studies documenting adaptive transgenerational plasticity to drought stress in this system. Earlier work showed that offspring of drought-stressed parents developed longer root systems in dry soil, and extended roots at a faster rate, compared with offspring of the same inbred genetic lines whose parents were well watered [51]. Consequently, the former seedlings had better access to the limited amount of moisture available in their own dry soil environments, and produced more biomass than did offspring of well-watered parents. Note that seedling biomass is a reliable proxy for survival in this species [24] and is generally a robust seedling-stage indicator of fitness since early growth differentials generally increase with age under natural conditions [61]. These effects of drought stress were found to persist for two generations, increasing the survival of grandoffspring grown under severe drought conditions [24]. Such multigenerational inheritance confirms that these developmental effects are truly transgenerational, rather than direct influences of the parental plant's environment on its developing seeds.
In this study, control-germinated offspring of drought-stressed parents likewise produced extensive root systems in dry soil, allowing them to support greater leaf areas and produce more biomass than offspring of well-watered parents grown in the same conditions. These results add to a growing number of cases in which environmental stress induces inherited phenotypic effects that enhance offspring performance under the same stress (e.g. [5–7,62,63]).
Experimental demethylation of offspring DNA removed these adaptive effects of parental drought, indicating that DNA methylation is required for the expression of these inherited environmental effects on offspring development. Adaptive transgenerational plasticity in this system derives much of its benefit from the initial advantage that parental exposure to drought confers on offspring in dry soil at the outset of their growth and development [51]. These inherited effects cause seedlings to enhance root extension immediately after emergence without experiencing a lag time between sensing dry conditions and initiating a response. We removed this initial advantage by briefly demethylating offspring with zebularine during seed germination. It is possible that zebularine could have had effects on the Polygonum genome in addition to demethylation, such as mobilization of transposable elements [47], resulting in generalized changes to seedling development. However, demethylation-induced transposition or other systemic effects would have occurred in offspring produced in both parental environments, so the specific effect of zebularine on offspring of drought-stressed parents suggests that such accessory treatment effects were not appreciable. Rather, the results indicate that it is specifically the inherited effect of parental drought stress that is removed when DNA methylation levels are experimentally reduced.
DNA methylation is but one process among a suite of mechanisms that can independently or jointly transmit environmental effects across generations. Such mechanisms also include environmentally induced changes in seed or egg provisions such as starches and proteins, changes to cytoplasmic factors including hormones, defensive chemicals, and other secondary metabolites, and heritable changes to epigenetic regulatory molecules such as small RNAs that work in concert with DNA methylation to regulate gene expression [16,17,64–66]. In plants, altered provisioning to nutritive seed tissues is often expected to be the primary mechanism of both beneficial and maladaptive effects of parental environment, and indeed is a major source of variation in many cases [3]. We found no evidence that changes to seed provisioning accounted for the inherited effects of parental drought in this study. Previous studies of P. persicaria that used smaller genotypic samples found some evidence for drought-induced changes in provisioning [8,24], suggesting that this effect occurs in some genetic lines but is not predominant in the species. In one such study, adaptive transgenerational effects of drought remained after effects of provisioning were removed via covariate analysis, confirming that the expression of transgenerational plasticity in this system relies on additional mechanisms [24]. Studies in other plant systems have also identified effects of parental environment that are independent of seed provisioning [67–69], suggesting that other regulatory mechanisms may be involved in many cases of transgenerational plasticity in plants.
Despite a surge of new findings about the role of DNA methylation in regulating environment-specific gene expression in plants and animals [35,70,71] only a few studies have tested whether methylation is involved in mediating adaptive transgenerational plasticity. In a study of the perennial plant Boechera stricta, Alsdurf & colleagues [33] found that parental drought caused changes in DNA methylation that correlated with increased drought-tolerance in offspring. These transgenerational changes also correlated with decreased production of defensive chemicals in offspring, thus implicating epigenetic inheritance in an ecologically meaningful trade-off between offspring tolerances to different stresses. A recent study of simulated herbivory in Mimulus guttatus found that demethylation of offspring DNA removed potentially adaptive maternal, but not paternal, effects on leaf trichome density in offspring, suggesting that distinct epigenetic mechanisms regulate inheritance of these effects [45]. DNA methylation has also been implicated in transgenerationally induced tolerance of salt stress in A. thaliana [44,72], nutrient deficiency in rice [23], and resistance to pathogen attack in Nicotiana tabacum [73]. Similarly, a recent study of the aquatic invertebrate Artemia found that heritable, heat-shock-induced resistance to both temperature stress and pathogen infection correlated with heritable changes in DNA methylation and histone modifications [74]. These initial studies suggest that adaptive transgenerational plasticity that is regulated by DNA methylation may be phylogenetically widespread.
(b). Genetic variation for methylation-mediated transgenerational plasticity
A sample of 12 naturally evolved and then inbred P. persicaria genetic lines altered offspring to varying degrees in response to parental drought stress. These genotype-specific environmental responses were mirrored by genotypic differences in the degree to which demethylation changed the expression of the parental drought effect. These results suggest that parental drought stress induces genotype-specific changes to DNA methylation in offspring, and that it is these methylation changes that underlie the adaptive transgenerational phenotypic effects of the stress.
Studies of transgenerational environmental effects commonly reveal variation among genotypes for those effects (i.e. genotype × environment variation for transgenerational plasticity; e.g. [8,13,14]). Our results show that, in some cases, such transgenerational genotype × environment variation may result from genotype-specific differences in environmentally induced, inherited DNA methylation changes. As DNA methylation occurs primarily at cytosine bases in eukaryotes, variation among genetic lines in cytosine content in or near either key regulatory sequences or protein-coding genes constitutes genetic differences in the potential for methylation changes at those loci [66,75,76]. Gene expression can also be modulated by environmentally induced methylation changes at nearby transposable elements [29]. Differences in transposon insertions among genetic lines are, therefore, an additional avenue for genotype-specific changes in methylation. The genetic lines in our sample may have varied in their susceptibility to environmentally induced methylation changes due to these types of local, sequence-based constraints. The recent finding that, in ants, quantitative differences in DNA methylation levels are associated with quantitative phenotypic variation suggests that the genotype-specific frequency of methylated sites may influence development [46].
Environmentally induced methylation patterns at specific loci can also be influenced by allelic variation at distant loci throughout the genome. For instance, a recent study of within-generation environmental effects in 150 Swedish A. thaliana accessions found that both cis and trans genetic variants substantially influenced temperature-induced changes in DNA methylation at hundreds of transposable elements [34] (see [77] for similar findings in humans). The sequence-specific nature of those changes points to DNA methylation as a source of genotype × environment interaction: in other words, induced methylation changes may translate specific environmental signals into genotype-specific adjustments in trait expression. When such environmentally induced methylation changes are heritable (which appears to be especially common in plants, [35,36]), they may also underlie the expression of genotype × parental environment variation.
Our results indicate that DNA methylation plays an integral role in the expression of genotype-specific effects of parental drought. Yet it remains unclear precisely what changes are induced by parental drought and inherited by offspring. It is possible that drought induces targeted modifications in DNA methylation in parents that are transmitted through meiosis to shape the expression of adaptive phenotypes in offspring [71]. In this case, genetic variation for transgenerational plasticity could reflect either (a) differences among genetic lines in their potential for methylation changes (as described above), (b) the transgenerational stability of those changes, or (c) genotypic differences in their phenotypic impact (or some combination of these effects). In support of possibility (b), a recent study of DNA methylation transmission through male gametes in the perennial plant Helleborus foetidus documented genetic variation for the ability of methylation marks to persist unchanged through meiosis [78]. However, changes in DNA methylation may not themselves be inherited, but instead may be reconstituted by inheritance of other directive factors such as small RNAs or hormones. These factors may also be subject to genetic variation in their inducibility, transmissibility, and/or influence on phenotypes.
The distinct pattern of genotype × parental environment × demethylation effects in our study may offer additional clues to the nature of these interactions: genetic lines that most strongly increased biomass in response to parental drought were also most strongly inhibited in their growth when offspring of droughted parents were demethylated. This growth reduction resulted in 17% lower rather than equivalent biomass (on average) compared with offspring of well-watered parents. Demethylation may have revealed a maladaptive developmental effect of parental drought that is normally overcompensated by the effects of drought-induced DNA methylation. Alternatively, the genetic lines that most strongly increased offspring biomass in response to parental drought stress may have had the fastest root extension rates at the outset of growth. By initially taking up more water, these seedlings would have acquired more zebularine immediately after germination compared with seedlings with slower root extension rates, potentially reducing seedling growth due to more extreme demethylation effects than those experienced by smaller rooted seedlings. However, greater-than-expected reductions in growth did not occur in all cases, indicating that the removal of the parental drought effect by demethylation did not stem simply from enhanced uptake of zebularine.
The genetic differences that we observed in the degree of adaptive response to parental drought may also stem from (epi)genetic constraints that limit the ability of some genotypes to express adaptive transgenerational responses. As a result of genotype-specific constraints, genotypes may differ in the specific combinations of plastic trait adjustments they express in response to a given stress [79]. Genetic lines that did not express adaptive transgenerational responses to drought (and those that did so weakly) may express particularly effective within-generation plastic responses to drought stress, while genetic lines that strongly expressed the adaptive parental drought effect may be less efficient at mounting an immediate response to moisture limitation. More broadly, genetic differences in the expression of within- versus transgenerational plasticity may derive from differences among source populations in historical exposure to particular regimes of environmental change [80]. Recent theory predicts that adaptive within-generation responses will evolve when there is high temporal environmental variability, whereas adaptive transgenerational plasticity is likely to evolve when environments are stable over generations such that parents and offspring experience similar conditions [81,82] (for an empirical example, see [10]).
By demonstrating a role for DNA methylation in mediating genotype × environment interactions for adaptive transgenerational plasticity, this research contributes to the emerging picture that genetic variation, epigenetic variation, and environmental cues interactively contribute to adaptive diversity. Further exploring the interactions between these factors in naturally evolved systems represents a promising new direction in evolutionary biology.
Supplementary Material
Acknowledgements
We thank Hea-Ream Lee, Rachel Budker, Lars Berg, Emma Broder, and Eva Steinberg for their assistance. We also thank Frederick Cohan, Christina Richards, Michael Singer, and Hamish Spencer for valuable discussion, Jennifer Rose for statistical consultation and two anonymous reviewers for constructive comments. Koen Verhoeven kindly shared the demethylation protocol with us.
Data accessibility
Data for all analyses are available in the Dryad repository: http://dx.doi.org/10.5061/dryad.6824t [83].
Authors' contributions
J.J.H. and S.E.S. designed the experiment; J.J.H. conducted the experiment and analyses; J.J.H. and S.E.S. interpreted results and wrote the manuscript.
Competing interests
We have no competing interests.
Funding
This research was funded by Wesleyan University and the New Phytologist Trust (S.E.S.).
References
- 1.Bateson P, et al. 2004. Developmental plasticity and human health. Nature 430, 419–421. ( 10.1038/nature02725) [DOI] [PubMed] [Google Scholar]
- 2.Gluckman P, Hanson M. 2004. The fetal matrix: evolution, development and disease. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 3.Roach D, Wulff R. 1987. Maternal effects in plants. Annu. Rev. Ecol. Syst. 18, 209–235. ( 10.1146/annurev.es.18.110187.001233) [DOI] [Google Scholar]
- 4.Falconer DS, Mackay TFC. 1996. Introduction to quantitative genetics, 4th edn, p. 480 San Francisco, CA: Benjamin Cummings. [Google Scholar]
- 5.Galloway LF, Etterson JR. 2007. Transgenerational plasticity is adaptive in the wild. Science 318, 1134–1136. ( 10.1126/science.1148766) [DOI] [PubMed] [Google Scholar]
- 6.Holeski LM. 2007. Within and between generation phenotypic plasticity in trichome density of Mimulus guttatus. J. Evol. Biol. 20, 2092–2100. ( 10.1111/j.1420-9101.2007.01434.x) [DOI] [PubMed] [Google Scholar]
- 7.Salinas S, Munch SB. 2012. Thermal legacies: transgenerational effects of temperature on growth in a vertebrate. Ecol. Lett. 15, 159–163. ( 10.1111/J.1461-0248.2011.01721.X) [DOI] [PubMed] [Google Scholar]
- 8.Sultan SE. 1996. Phenotypic plasticity for offspring traits in Polygonum persicaria. Ecology 77, 1791–1807. ( 10.2307/2265784) [DOI] [Google Scholar]
- 9.Mousseau TA, Fox CW. 1998. The adaptive significance of maternal effects. Trends Ecol. Evol. 13, 403–407. ( 10.1016/S0169-5347(98)01472-4) [DOI] [PubMed] [Google Scholar]
- 10.Walsh MR, Castoe T, Holmes J, Packer M, Biles K, Walsh M, Munch SB, Post DM. 2016. Local adaptation in transgenerational responses to predators. Proc. R. Soc. B 283, 20152271 ( 10.1098/rspb.2015.2271) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheiner SM. 1993. Genetics and evolution of phenotypic plasticity. Annu. Rev. Ecol. Syst. 24, 35–68. ( 10.1146/Annurev.Ecolsys.24.1.35) [DOI] [Google Scholar]
- 12.Des Marais DL, Hernandez KM, Juenger TE. 2013. Genotype-by-environment interaction and plasticity: exploring genomic responses of plants to the abiotic environment. Annu. Rev. Ecol. Evol. Syst. 44, 5–29. ( 10.1146/annurev-ecolsys-110512-135806) [DOI] [Google Scholar]
- 13.Schmitt J, Niles J, Wulff R. 1992. Norms of reaction of seed traits to maternal environments in Plantago lanceolata. Am. Nat. 139, 451–466. ( 10.1086/285338) [DOI] [Google Scholar]
- 14.Stjernman M, Little TJ. 2011. Genetic variation for maternal effects on parasite susceptibility. J. Evol. Biol. 24, 2357–2363. ( 10.1111/J.1420-9101.2011.02363.X) [DOI] [PubMed] [Google Scholar]
- 15.Alsdurf JD, Ripley TJ, Matzner SL, Siemens DH. 2013. Drought-induced trans-generational tradeoff between stress tolerance and defence: consequences for range limits? AoB Plants 5, plt038. ( 10.1093/aobpla/plt038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman JJ, Sultan SE. 2011. Adaptive transgenerational plasticity in plants: case studies, mechanisms, and implications for natural populations. Front. Plant Sci. 2, 1–10. ( 10.3389/fpls.2011.00102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holeski LM, Jander G, Agrawal AA. 2012. Transgenerational defense induction and epigenetic inheritance in plants. Trends Ecol. Evol. 27, 618–626. ( 10.1016/J.Tree.2012.07.011) [DOI] [PubMed] [Google Scholar]
- 18.Salinas S, Brown SC, Mangel M, Munch SB. 2013. Non-genetic inheritance and changing environments. Non-Genetic Inheritance 1, 38–50. ( 10.2478/ngi-2013-0005) [DOI] [Google Scholar]
- 19.Fox CW, Thakar MS, Mousseau TA. 1997. Egg size plasticity in a seed beetle: an adaptive maternal effect. Am. Nat. 149, 149–163. ( 10.1086/285983) [DOI] [PubMed] [Google Scholar]
- 20.Donohue K, Schmitt J. 1998. Maternal environmental effects in plants: adaptive plasticity? In Maternal effects as adaptations (eds Mousseau T, Fox CW), pp. 137–158. New York, NY: Oxford University Press. [Google Scholar]
- 21.Shama LNS, Wegner KM. 2014. Grandparental effects in marine sticklebacks: transgenerational plasticity across multiple generations. J. Evol. Biol. 27, 2297–2307. ( 10.1111/jeb.12490) [DOI] [PubMed] [Google Scholar]
- 22.Whittle C, Otto S, Johnston M, Krochko J. 2009. Adaptive epigenetic memory of ancestral temperature regime in Arabidopsis thaliana. Botany 87, 650–657. ( 10.1139/B09-030) [DOI] [Google Scholar]
- 23.Kou HP, Li Y, Song XX, Ou XF, Xing SC, Ma J, Liu B, Von Wettstein D. 2011. Heritable alteration in DNA methylation induced by nitrogen-deficiency stress accompanies enhanced tolerance by progenies to the stress in rice (Oryza sativa L.). J. Plant Physiol. 168, 1685–1693. ( 10.1016/J.Jplph.2011.03.017) [DOI] [PubMed] [Google Scholar]
- 24.Herman JJ, Sultan SE, Horgan-Kobelski T, Riggs C. 2012. Adaptive transgenerational plasticity in an annual plant: grandparental and parental drought stress enhance performance of seedlings in dry soil. Integr. Comp. Biol. 52, 77–88. ( 10.1093/icb/ics041) [DOI] [PubMed] [Google Scholar]
- 25.Bossdorf O, Richards CL, Pigliucci M. 2008. Epigenetics for ecologists. Ecol. Lett. 11, 106–115. ( 10.1111/j.1461-0248.2007.01130.x) [DOI] [PubMed] [Google Scholar]
- 26.Kappeler L, Meaney MJ. 2010. Epigenetics and parental effects. Bioessays 32, 818–827. ( 10.002/bies.201000015) [DOI] [PubMed] [Google Scholar]
- 27.Pastor V, Luna E, Mauch-Mani B, Ton J, Flors V. 2013. Primed plants do not forget. Environ. Exp. Bot. 94, 46–56. ( 10.1016/j.envexpbot.2012.02.013) [DOI] [Google Scholar]
- 28.Yu A, et al. 2013. Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc. Natl Acad. Sci. USA 110, 2389–2394. ( 10.1073/pnas.1211757110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowen RH, Pelizzola M, Schmitz RJ, Lister R, Dowen JM, Nery JR, Dixon JE, Ecker JR. 2012. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl Acad. Sci. USA 109, E2183–E2191. ( 10.1073/pnas.1209329109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W-S, Pan Y-J, Zhao X-Q, Dwivedi D, Zhu L-H, Ali J, Fu B-Y, Li Z-K. 2011. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J. Exp. Bot. 62, 1951–1960. ( 10.1093/jxb/erq391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labra M, Ghiani A, Citterio S, Sgorbati S, Sala F, Vannini C, Ruffini-Castiglione M, Bracale M. 2002. Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol. 4, 694–699. ( 10.1055/s-2002-37398) [DOI] [Google Scholar]
- 32.Zheng X, Chen L, Li M, Lou Q, Xia H, Wang P, Li T, Liu H, Luo L. 2013. Transgenerational variations in DNA methylation induced by drought stress in two rice varieties with distinguished difference to drought resistance. PLoS ONE 8, e80253 ( 10.1371/journal.pone.0080253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alsdurf J, Anderson C, Siemens DH. 2015. Epigenetics of drought-induced trans-generational plasticity: consequences for range limit development. AoB Plants 8, plv146. ( 10.1093/aobpla/plv146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubin MJ, et al. 2015. DNA methylation in Arabidopsis has a genetic basis and shows evidence of local adaptation. eLife 4, e05255 ( 10.7554/eLife.05255) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feil R, Fraga MF. 2012. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109. ( 10.1038/nrg3142) [DOI] [PubMed] [Google Scholar]
- 36.Jablonka E, Raz G. 2009. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q. Rev. Biol. 84, 131–176. ( 10.1086/598822) [DOI] [PubMed] [Google Scholar]
- 37.Cortijo S, et al. 2014. Mapping the epigenetic basis of complex traits. Science 343, 1145–1148. ( 10.1126/science.1248127) [DOI] [PubMed] [Google Scholar]
- 38.Schaefer S, Nadeau JH. 2015. The genetics of epigenetic inheritance: modes, molecules, and mechanisms. Q. Rev. Biol. 90, 381–415. ( 10.1086/683699) [DOI] [PubMed] [Google Scholar]
- 39.Via S, Lande R. 1985. Genotype-environment interaction and the evolution of phenotypic plasticity. Evolution 39, 505–522. ( 10.2307/2408649) [DOI] [PubMed] [Google Scholar]
- 40.Sultan SE. 2015. Organism and environment: ecological development, niche construction, and adaptation. New York, NY: Oxford University Press. [Google Scholar]
- 41.Bossdorf O, Arcuri D, Richards CL, Pigliucci M. 2010. Experimental alteration of DNA methylation affects the phenotypic plasticity of ecologically relevant traits in Arabidopsis thaliana. Evol. Ecol. 24, 541–553. ( 10.1007/s10682-010-9372-7) [DOI] [Google Scholar]
- 42.Herrera CM, Pozo MI, Bazaga P. 2012. Jack of all nectars, master of most: DNA methylation and the epigenetic basis of niche width in a flower-living yeast. Mol. Ecol. 21, 2602–2616. ( 10.1111/j.1365-294X.2011.05402.x) [DOI] [PubMed] [Google Scholar]
- 43.Verhoeven KJF, van Gurp TP. 2012. Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS ONE 7, e38605 ( 10.1371/journal.pone.0038605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyko A, Blevins T, Yao YL, Golubov A, Bilichak A, Ilnytskyy Y, Hollander J, Meins F, Kovalchuk I. 2010. Transgenerational adaptation of Arabidopsis to stress requires DNA methylation and the function of Dicer-like proteins. PLoS ONE 5, e0009514 ( 10.1371/journal.pone.0009514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Akkerman K, Sattirin A, Kelly JK, Scoville AG. 2016. Transgenerational plasticity is sex-dependent and persistent in yellow monkeyflower (Mimulus guttatus). Environ. Epigenet. 2, dvw003. ( 10.1093/eep/dvw003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alvarado S, Rajakumar R, Abouheif E, Szyf M. 2015. Epigenetic variation in the Egfr gene generates quantitative variation in a complex trait in ants. Nat. Commun. 6, 6513 ( 10.1038/ncomms7513) [DOI] [PubMed] [Google Scholar]
- 47.Baubec T, Pecinka A, Rozhon W, Mittelsten Scheid O. 2009. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 57, 542–554. ( 10.1111/j.1365-313x.2008.03699.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. 2002. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J. Mol. Biol. 321, 591–599. ( 10.1016/S0022-2836(02)00676-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, Jones PA, Selker EU. 2003. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J. Natl Cancer Inst. 95, 399–409. ( 10.1093/jnci/95.5.399) [DOI] [PubMed] [Google Scholar]
- 50.Kim S, Sultan SE, Donoghue M. 2008. Allopolyploid speciation in Persicaria (Polygonaceae): insights from a low-copy nuclear region. Proc. Natl Acad. Sci. USA 105, 12 370–12 375. ( 10.1073/pnas.0805141105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sultan SE, Barton K, Wilczek AM. 2009. Contrasting patterns of transgenerational plasticity in ecologically distinct congeners. Ecology 90, 1831–1839. ( 10.1890/08-1064.1) [DOI] [PubMed] [Google Scholar]
- 52.Sultan SE, Wilczek A, Hann S, Brosi B. 1998. Contrasting ecological breadth of co-occurring annual Polygonum species. J. Ecol. 86, 363–383. ( 10.1046/j.1365-2745.1998.00265.x) [DOI] [Google Scholar]
- 53.Mulligan GA, Findlay JN. 1970. Reproductive systems and colonization in Canadian weeds. Can. J. Bot. 48, 859–860. ( 10.1139/b70-119) [DOI] [Google Scholar]
- 54.Sultan SE, Horgan-Kobelski T, Nichols LM, Riggs CE, Waples RK. 2012. A resurrection study reveals rapid adaptive evolution within populations of an invasive plant. Evol. Appl. 6, 266–278. ( 10.1111/j.1752-4571.2012.00287.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinheiro J, Bates D. 2000. Mixed-effects models in S and S-plus. Berlin, Germany: Springer. [Google Scholar]
- 56.Martin RA, Pfennig DW. 2010. Maternal investment influences expression of resource polymorphism in amphibians: implications for the evolution of novel resource-use phenotypes. PLoS ONE 5, e9117 ( 10.1371/journal.pone.0009117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaw RG. 1987. Maximum-likelihood approaches applied to quantitative genetics of natural populations. Evolution 41, 812–826. ( 10.2307/2408890) [DOI] [PubMed] [Google Scholar]
- 58.Zar JH. 1999. Biostatistical analysis, 4th edn Upper Saddle River, NJ: Prentice Hall. [Google Scholar]
- 59.R Core Team. 2015. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 60.Pinheiro J, Bates D, DebRoy S, Sarkar D. 2012. nlme: Linear and nonlinear mixed effects models. R package version 3.1–120.
- 61.Moles AT, Leishman MR. 2008. The seedling as part of a plant's life history strategy. In Seedling ecology and evolution (eds Leck MA, Parker VT, Simpson RL), pp. 217–238. New York, NY: Cambridge University Press. [Google Scholar]
- 62.Agrawal A, Laforsch C, Tollrian R. 1999. Transgenerational induction of defences in animals and plants. Nature 401, 60–63. ( 10.1038/43425) [DOI] [Google Scholar]
- 63.Scoville A, Barnett L, Bodbyl-Roels S, Kelly JK, Hileman LC. 2011. Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 191, 251–263. ( 10.1111/j.1469-8137.2011.03656.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyko A, Kovalchuk I. 2011. Genome instability and epigenetic modification—heritable responses to environmental stress? Curr. Opin. Plant Biol. 14, 260–266. ( 10.1016/j.pbi.2011.03.003) [DOI] [PubMed] [Google Scholar]
- 65.Bonduriansky R, Day T. 2009. Nongenetic inheritance and its evolutionary implications. Annu. Rev. Ecol. Evol. Syst. 40, 103–125. ( 10.1146/annurev.ecolsys.39.110707.173441) [DOI] [Google Scholar]
- 66.Duncan EJ, Gluckman PD, Dearden PK. 2014. Epigenetics, plasticity, and evolution: How do we link epigenetic change to phenotype? J. Exp. Zool. B 322, 208–220. ( 10.1002/jez.b.22571) [DOI] [PubMed] [Google Scholar]
- 67.Agrawal A. 2002. Herbivory and maternal effects: mechanisms and consequences of transgenerational induced plant resistance. Ecology 83, 3408–3415. ( 10.1890/0012-9658(2002)083%5B3408:HAMEMA%5D2.0.CO;2) [DOI] [Google Scholar]
- 68.Dyer AR, Brown CS, Espeland EK, Mckay JK, Meimberg H, Rice KJ. 2010. The role of adaptive trans-generational plasticity in biological invasions of plants. Evol. Appl. 3, 179–192. ( 10.1111/j.1752-4571.2010.00118.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Case A, Lacey E, Hopkins R. 1996. Parental effects in Plantago lanceolata L. II. Manipulation of grandparental temperature and parental flowering time. Heredity 76, 287–295. ( 10.1038/hdy.1996.42) [DOI] [Google Scholar]
- 70.Meyer P. 2015. Epigenetic variation and environmental change. J. Exp. Bot. 66, 3541–3548. ( 10.1093/jxb/eru502) [DOI] [PubMed] [Google Scholar]
- 71.Jablonka E. 2013. Epigenetic inheritance and plasticity: the responsive germline. Prog. Biophys. Mol. Biol. 111, 99–107. ( 10.1016/j.pbiomolbio.2012.08.014) [DOI] [PubMed] [Google Scholar]
- 72.Bilichak A, Ilnystkyy Y, Hollunder J, Kovalchuk I. 2012. The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS ONE 7, e30515 ( 10.1371/journal.pone.0030515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kathiria P, Sidler C, Golubov A, Kalischuk M, Kawchuk LM, Kovalchuk I. 2010. Tobacco mosaic virus infection results in an increase in recombination frequency and resistance to viral, bacterial, and fungal pathogens in the progeny of infected tobacco plants. Plant Physiol. 153, 1859–1870. ( 10.1104/pp.110.157263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Norouzitallab P, Baruah K, Vandegehuchte M, Van Stappen G, Catania F, Bussche JV, Vanhaecke L, Sorgeloos P, Bossier P. 2014. Environmental heat stress induces epigenetic transgenerational inheritance of robustness in parthenogenetic Artemia model. FASEB J. 8, 3552–3563. ( 10.1096/fj.14-252049) [DOI] [PubMed] [Google Scholar]
- 75.Gutierrez-Arcelus M, et al. 2013. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife 2, e00523 ( 10.7554/eLife.00523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ladd-Acosta C, Fallin MD. 2016. The role of epigenetics in genetic and environmental epidemiology. Epigenomics 8, 271–283. ( 10.2217/epi.15.102) [DOI] [PubMed] [Google Scholar]
- 77.Teh AL, et al. 2014. The effect of genotype and in utero environment on inter-individual variation in neonate DNA methylomes. Genome Res 24, 1064–1074. ( 10.1101/gr.171439.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Herrera C, Medrano M, Bazaga P. 2014. Variation in DNA methylation transmissibility, genetic heterogeneity and fecundity-related traits in natural populations of the perennial herb Helleborus foetidus. Mol. Ecol. 23, 1085–1095. ( 10.1111/mec.12679) [DOI] [PubMed] [Google Scholar]
- 79.Heschel M, Sultan SE, Glover S, Sloan D. 2004. Population differentiation and plastic responses to drought stress in the generalist annual Polygonum persicaria. Int. J. Plant Sci. 165, 817–824. ( 10.1086/421477) [DOI] [Google Scholar]
- 80.Herman JJ, Spencer HG, Donohue K, Sultan SE. 2014. How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 68, 632–643. ( 10.1111/evo.12324) [DOI] [PubMed] [Google Scholar]
- 81.Leimar O, McNamara JM. 2015. The evolution of transgenerational integration of information in heterogeneous environments. Am. Nat. 185, E55–E69. ( 10.1086/679575) [DOI] [PubMed] [Google Scholar]
- 82.Uller T, English S, Pen I. 2015. When is incomplete epigenetic resetting in germ cells favoured by natural selection?. Proc. R. Soc. B 282, 20150682 ( 10.1098/rspb.2015.0682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Herman JJ, Sultan SE. 2016. Data from: DNA methylation mediates genetic variation for adaptive transgenerational plasticity. Dryad Digital Repository ( 10.5061/dryad.6824t) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for all analyses are available in the Dryad repository: http://dx.doi.org/10.5061/dryad.6824t [83].