Abstract
Intraspecific coevolution between selfish elements and suppressors may promote interspecific hybrid incompatibility, but evidence of this process is rare. Here, we use genomic data to test alternative models for the evolution of cytonuclear hybrid male sterility in Mimulus. In hybrids between Iron Mountain (IM) Mimulus guttatus × Mimulus nasutus, two tightly linked M. guttatus alleles (Rf1/Rf2) each restore male fertility by suppressing a local mitochondrial male-sterility gene (IM-CMS). Unlike neutral models for the evolution of hybrid incompatibility loci, selfish evolution predicts that the Rf alleles experienced strong selection in the presence of IM-CMS. Using whole-genome sequences, we compared patterns of population-genetic variation in Rf at IM to a neighbouring population that lacks IM-CMS. Consistent with local selection in the presence of IM-CMS, the Rf region shows elevated FST, high local linkage disequilibrium and a distinct haplotype structure at IM, but not at Cone Peak (CP), suggesting a recent sweep in the presence of IM-CMS. In both populations, Rf2 exhibited lower polymorphism than other regions, but the low-diversity outliers were different between CP and IM. Our results confirm theoretical predictions of ubiquitous cytonuclear conflict in plants and provide a population-genetic mechanism for the evolution of a common form of hybrid incompatibility.
Keywords: cytoplasmic male sterility, cytonuclear coevolution, hybrid incompatibility, Mimulus guttatus, selective sweep, speciation
1. Introduction
The sterility and inviability of interspecific hybrids has been a puzzle since before Darwin, and the origins of hybrid incompatibilities remain a central problem in evolutionary biology. How can natural selection allow, or even favour, alleles causing low fitness? The Bateson–Dobzhansky–Muller (BDM) model [1–3] resolves this paradox by invoking multi-locus epistasis: incompatible alleles at two or more loci interact to reduce fitness only in hybrids. Abundant empirical evidence (reviewed in [4–6]) supports BDM as a major source of hybrid post-zygotic barriers in diverse taxa. Alleles contributing to hybrid breakdown via epistatic BDM interactions are generally modelled to accumulate independently in allopatric populations by drift or ordinary natural selection (i.e. gradually, and without local effects on fertility or viability). By contrast, molecular-genetic evidence suggests that the loci involved in BDM incompatibilities are a non-random subset of genes (e.g. heterochromatin proteins; reviewed in [4,7]), and the few BDM genes characterized exhibit signatures of strong natural selection [8–10]. These patterns are not consistent with the gradual accumulation of chance incompatibilities between loci with no strong fitness effects. Thus, it has been argued that BDM incompatibilities may often derive from the evolution of costly selfish elements and complementary coevolution of their suppressors within lineages [9,11–16].
Selfish genetic elements, which distort genetic transmission to their own advantage, are an appealing source of hybrid incompatibilities for three reasons. First, selfish elements (by definition) incur fitness costs, including sterility and inviability. Second, because of their costs, the rest of the genome experiences selection for their suppression, and the resultant interactions between selfish elements and suppressors are inherently epistatic. Third, selfish evolution can cause rapid genetic divergence between populations, contributing to the development of post-zygotic barriers relatively early during speciation. Although selfish genetic elements are impressively diverse [17–19], a general scenario for the selfish evolution of hybrid incompatibility is easy to imagine. A selfish genetic element with fitness costs spreads within a population, prompting the evolution of a suppressor that restores both equal transmission and individual fitness. Both selfish element and suppressor are locally driven to fixation by strong positive selection (a ‘selective sweep’). Finally, hybridization generates mismatch between selfish elements and suppressor loci, revealing fitness costs of the selfish element as hybrid breakdown. Such scenarios are compelling, and circumstantial evidence suggests that selfish elements may play a disproportionate role in the evolution of post-zygotic isolation [9,20]. However, direct evidence for selfish evolution of a known pair of interacting loci is rare. In this study, we show that the loci underlying a cytonuclear hybrid incompatibility in yellow monkeyflowers (Mimulus) fit a selfish evolution/coevolution scenario, providing a population-genetic mechanism for the evolution of a BDM incompatibility on a local scale.
In flowering plants, a common form of hybrid incompatibility is cytonuclear male sterility (reviewed in [4,21,22]). Cytoplasmic male sterility (CMS) loci and their nuclear restorers of fertility (Rf) not only cause asymmetric reproductive incompatibility between hermaphroditic species [23–25], but they are also classic examples of selfish genetic elements and suppressors [16,19]. Maternally transmitted cytoplasmic mutations that prevent pollen production in otherwise hermaphroditic plants can spread rapidly through populations (resulting in gynodioecy) with even small positive effects on female fertility [26,27]. However, loss of male fertility eventually exerts strong selection favouring alleles that suppress selfish CMS and restore male fertility. Under broad theoretical conditions, both nuclear restorers and their target CMS are expected to spread to fixation within populations [18,28], reinstating monomorphic hermaphroditism. The now-cryptic CMS can be revealed in crosses with individuals that do not carry appropriate Rf [29], creating a cytonuclear BDM incompatibility. Because the same fitness effects that allow selfish CMS to spread within species may also drive them across species boundaries, they may not often serve as ‘speciation genes’ [4]. However, the impact of cytonuclear conflict on speciation may be enhanced by differences in mating system (as in our study system; see below), repeated CMS–Rf coevolution, and effects on linked organellar and nuclear loci. Regardless of their direct role as species barriers, cytonuclear incompatibilities manifest fundamental and theoretically widespread processes of nuclear and organellar coevolution. Cytonuclear hybrid male sterility is extremely common in angiosperms [21], and cross-species patterns of molecular evolution suggest a long-term history of conflict between CMS and the widespread class of nuclear Rf genes (pentatricopeptide repeat genes, PPRs; [30,31]). Selfish models of CMS–Rf coevolution are widely accepted, but the history of natural selection on Rf or CMS loci has not been demonstrated in any system. Thus, direct evidence of selfish CMS–Rf coevolution as the source of cytonuclear male sterility in plant hybrids is key to validating population-genetic theory for both mating-system evolution and the evolution of hybrid incompatibilities.
Here, we test alternative scenarios for the evolution of cytonuclear incompatibility in Mimulus (=Erythranthe sect. Simiola, Phrymaceae). Hybrids between the outcrossing species Mimulus guttatus (specifically, the IM62 inbred line from Iron Mountain (IM), OR, USA) and closely related selfer Mimulus nasutus (multiple lines) exhibit asymmetric hybrid sterility. Hybrids carrying the M. guttatus IM62 cytoplasm and M. nasutus alleles at a nuclear Rf locus make deformed anthers that produce no pollen; this phenotype is absent in the reciprocal hybrids [29]. The candidate CMS gene is a rearranged version of the upstream region of mitochondrial NADH dehydrogenase subunit 6 (nad6; [32]), and restoration has been mapped to an approximately 1.3 cm region of Linkage Group 7 (LG7; [33]). Fine-mapping of the Rf region revealed that the M. guttatus parent carries dominant alleles at two tightly linked loci—Rf1 and Rf2—each able to completely restore male fertility [33]. Both Rf loci occur within a cluster of tandemly repeated PPR genes, and LG7 contains the highest concentration of restorer-like PPRs in the M. guttatus genome [33]. Thus, the molecular basis of the CMS–Rf system in Mimulus resembles the cryptic CMS seen in diverse crop hybrids [30], and is probably representative of evolutionary processes commonly occurring throughout flowering plants [31,34].
The IM M. guttatus population provides a particularly fertile context for investigating the evolutionary history of hybrid incompatibilities, as its selection regimes and phenotypic variation have been well characterized (e.g. [10,35,36]). Despite intensive research documenting standing variation for both male [35,37] and female fertility [36], there is no evidence of segregating anther sterility (i.e. gynodioecy) within the IM population. Nonetheless, both molecular screens [32] and marker-based surveys [38] suggest that nearly all (more than 97%) IM plants carry the CMS mitochondrial type. The high frequency of the IM-CMS gene in the absence of the CMS phenotype indicates that at least one of the Rf alleles must also be present in all individuals at IM. Importantly, while ubiquitous at IM, the IM-CMS is absent or at extremely low frequency in neighbouring (less than 2 km) populations [32,38]. This geographical localization of the IM-CMS sets the stage for our assessment of its evolutionary history. The local fixation of a given mitochondrial haplotype cannot itself be used as evidence of selection on a particular gene; organellar genes are inherited together and are subject to elevated drift/founder effects [39,40]. However, because the IM-CMS appears local (and probably recent) in origin, we can uniquely infer local selection on the IM-Rf loci under a selfish CMS–Rf coevolution scenario. Thus, we can use population-genetic variation in the nuclear Rf region within M. guttatus to test alternative scenarios for the evolutionary history of this interspecific cytonuclear hybrid incompatibility (figure 1).
Figure 1.
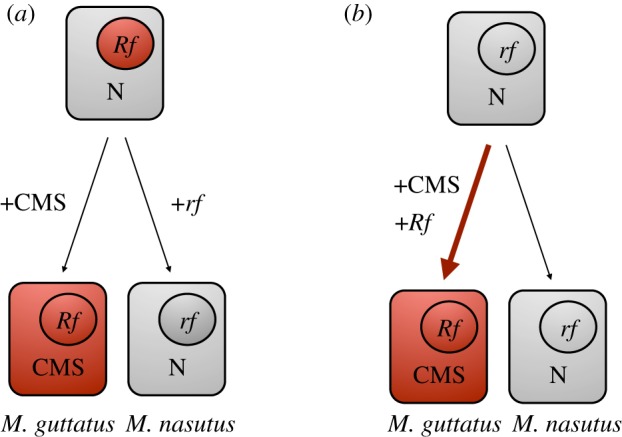
Models for the evolution of cytonuclear hybrid incompatibility in Mimulus make alternative predictions about the history of selection on Rf alleles. Each box depicts a plant cell with nuclear (inside circles) and cytoplasmic (outside circles) genotypes: CMS = male-sterilizing cytoplasm; N = non-CMS cytoplasm; Rf = restorer allele; rf = non-restorer allele. All genotypes shown are phenotypically male-fertile (i.e. CMS is cryptic at endpoints). (a) Non-selfish Bateson–Dobzhansky–Muller model: incompatible genotypes (CMS and rf) evolved independently in Iron Mountain (IM) M. guttatus and in M. nasutus, respectively; the IM-CMS was never expressed, and thus there should be no evidence of recent selection at Rf loci. (b) Selfish CMS–Rf coevolution model: the vulnerable rf (M. nasutus-like) allele is ancestral, so the selfish spread of CMS within IM M. guttatus generated strong directional selection for Rf alleles (red arrow).
In this study, we took advantage of a rare opportunity to examine patterns of divergence, polymorphism, and haplotype structure in the Rf region in neighbouring populations with and without the IM-CMS. We expect to find evidence of recent Rf selection unique to the IM population, where the CMS mitotype is prevalent. Under a non-selfish scenario for the evolution of cytonuclear hybrids incompatibility (figure 1a), the CMS mitochondrial type arose and spread in the IM population, but Rf alleles suppressing the IM-CMS were already fixed. Thus, the CMS phenotype was never expressed, and the new mitochondrial type had neither a selfish transmission advantage nor individual fitness costs. In this ‘traditional’ model for the origin of BDM incompatibility, non-restoring rf alleles would arise and fix (e.g. by drift) independently in M. nasutus or other populations incompatible with IM-CMS. This scenario, plausible given potentially strong effects of genetic drift under the highly selfing mating system of M. nasutus, predicts no recent or local history of selection on the Rf region in the IM population. By contrast, if the IM-Rf alleles evolved in response to the selfish (i.e. male-sterilizing) local spread of the novel IM-CMS, then they should exhibit signatures of population-specific selection (figure 1b).
To distinguish selfish versus non-selfish scenarios, we conducted population-genetic analyses of IM and a nearby population lacking the IM-CMS (Cone Peak: CP). First, we characterized population differentiation (FST) between IM and CP, genome-wide and across the Rf region, using Illumina re-sequence data from inbred lines, as well as genetic markers in wild-collected samples. Under the selfish CMS–Rf model, nuclear genes in the Rf region should exhibit elevated between-population differentiation (FST) matching the local distribution of the IM-CMS. Second, we analysed patterns of linkage disequilibrium (LD, haplotype structure) and polymorphism (π, Tajima's D) across the Rf region within each population. Under the selfish scenario, we expect elevated LD, reduced polymorphism, and a skewed site-frequency spectrum characteristic of a recent selective sweep at IM. Our results are consistent with recent, CMS-driven selection on the Rf region at IM, directly confirming theoretical models of cytonuclear conflict in plants, and revealing the selfish origins of an interspecific hybrid incompatibility.
2. Material and methods
(a). Study system
The yellow monkey flowers of the M. guttatus (=Erythranthe guttata) species complex are a morphologically diverse but largely inter-fertile group of wildflowers with their centre of diversity in Western North America [41,42]. Mimulus guttatus, the most common species, has large, insect-pollinated flowers and is predominantly outcrossing [43]. A large annual M. guttatus population in the Oregon Cascades (IM; 44°24′15″ N, 122°8′35″ W) has been intensively studied. A fully annotated reference genome derived from a single IM inbred line (IM62) is publicly available (Mimulus guttatus v. 2.0; http://phytozome.jgi.doe.gov/pz/p2ortal.html), as are whole-genome re-sequence data from the 10 IM inbred lines used in this study [44]. The very large census size at IM (more than 100 000 individuals most years) is reflected in migration- and selection-dominated patterns of genetic variation, including very high levels of genic sequence diversity and mean positive Tajima's D [45]. In addition, local balancing selection has been shown to maintain at least two chromosomal polymorphisms at IM [35,36,46], and one locus involved in a distinct nuclear–nuclear hybrid incompatibility with M. nasutus also appears to be under strong selection [10].
Importantly, the cytoplasmic component of the incompatibility (IM-CMS) is nearly fixed at IM: more than 97% of IM individuals (n = 545) carry the IM-CMS gene [32,38]. It is absent or at extremely low frequency in nearby populations (n = 9 sites in the Oregon Cascades, 544 plants haplotyped outside of IM). Most notably, IM-CMS has not been found at CP (n = 65; 44°24′7″ N, 122°7′59″ W), which is less than 2 km from IM, similar in mating system [47], population size, and overall ecology. Thus, we can use analyses of variation within and between the two populations to test the hypothesis of IM-specific, CMS-driven Rf evolution. To complement the IM re-sequence data, we generated a set of inbred lines (three generations of inbreeding) from wild-collected CP plants, and analysed re-sequence data from eight lines (see the electronic supplementary material).
(b). Delineation of Rf loci in the Mimulus guttatus genome
The nuclear component of the incompatibility comprises a single Mendelian locus on LG7 [29] with two tightly linked loci on LG7—Rf1 and Rf2—spanning approximately 1 cm in a region containing a large number of restorer-like PPR genes [33]. Inconsistencies in the location of restorer-linked marker loci in multiple M. guttatus IM62 genome assemblies, and between physical and genetic maps, suggest that the Rf region is probably misassembled in the v. 2.0 reference. The Rf region is centred on two gene-based markers (191_27 and 191_45), which were on a single contig (less than 110 kb apart) in an initial draft genome assembly, and approximately 0.2 cm apart in M. nasutus × M. guttatus linkage maps [33]. However, these markers were split between two scaffolds (sc97 and sc14) of the v. 1.0 assembly, and more recently placed approximately 8 Mb apart on LG7 in the v. 2.0 pseudochromosome assembly. Within M. guttatus linkage maps (L. Fishman and J. K. Kelly 2016, unpublished data) confirm that these markers are less than 1 cm apart, and that the placement and orientation of sc97 (Migut.G00884–G00829) and sc14 (Migut.G01156–G01178) in our previous fine-mapping experiments is more representative of the IM62 M. guttatus genome. Therefore, we present our analyses of divergence and polymorphism across the Rf region using linkage-defined landmarks to scaffold sections of the v. 2.0 physical map, with the caveat that Rf2 may contain some additional genomic sequence. However, this re-orientation does not materially affect our conclusions.
(c). Population-genomic analyses
We used whole-genome Illumina re-sequence data from 10 inbred IM lines [44] and 8 inbred CP lines to examine local (Rf region) versus larger-scale patterns of population-genetic variation. In addition, we analysed a Pool-Seq dataset (n = 200 wild individuals) representing a broader sampling of variation from IM. Read mapping, filtering and handling of missing data are described in the electronic supplementary material.
Under the selfish CMS–Rf coevolution model (figure 1b), we expect elevated differentiation (FST) between IM and CP within one or both Rf loci, reflecting local selection on one or both restorer alleles only in the presence of the IM-CMS. We used the CP and IM line datasets to compare FST in the Rf region to the genome-wide distribution and conducted chromosome-level outlier analyses to identify unusually differentiated single nucleotide polymorphisms (SNPs). A history of recent selection on Rf at IM should also generate locally elevated LD and reduced diversity, classic signatures of a selective sweep. We explored patterns of LD by examining pairwise r2 in the Rf region and elsewhere, and genewise identity to the IM62 reference haplotype (known to carry functional Rf1 and Rf2 alleles). For analyses of polymorphism across the Rf region and elsewhere, we calculated nucleotide diversity (π) and Tajima's D per gene for the IM and CP lines. Two genomic regions outside of Rf were chosen for comparison: (i) the gene-rich, PPR-poor and Rf-lacking, distal arm of LG7 (LG7NR; Migut.G00001–Migut.G00600) and (ii) linkage group 6 (LG6; Migut.F00001–Migut.F02142), which contains the second highest density of restorer-like PPRs [33]. We used the larger Pool-Seq dataset from IM to examine the allele frequency spectrum and patterns of nucleotide diversity at Rf genes. We corroborated the genomic findings with Sanger sequencing (nucleotide diversity; n = 26 IM lines) of loci within and outside Rf and genotyping of gene-based markers in wild plants (FST; IM n = 48; CP n = 39). See the electronic supplementary material for details of all genetic and genomic analyses.
3. Results
As predicted by the CMS–Rf coevolution model, the Rf region exhibits unusual differentiation between IM and the mitochondrially distinct CP population (figure 2). FST is significantly elevated in the Rf region (mean = 0.141; s.e. = 0.0171; n = 47 genes) compared with the rest of the genome (mean = 0.0376; s.e. = 0.0007; n = 18 669 genes). An excess of genes in Rf2 (9 of 38; p < 0.0001) had FST values above the genome-wide 95th percentile, and eight SNPs in Rf2 were also individual FST outliers in a bootstrapping analysis (electronic supplementary material). Genes in Rf1 also show elevated FST (mean FST = 0.232; s.e. = 0.055) relative to the genomic background. Several PPR genes, which are functional candidates for CMS restoration, were among the FST outliers in both Rf1 and Rf2 (red points, figure 2). Locally elevated FST was also seen in a larger field-collected sample genotyped at PCR-based markers, where the two Rf-linked genetic markers were the only significant FST outliers (electronic supplementary material, figure S1 and table S1). These results are consistent with strong selection in one or both populations driving differentiation in the Rf region, matching the distribution of IM-CMS.
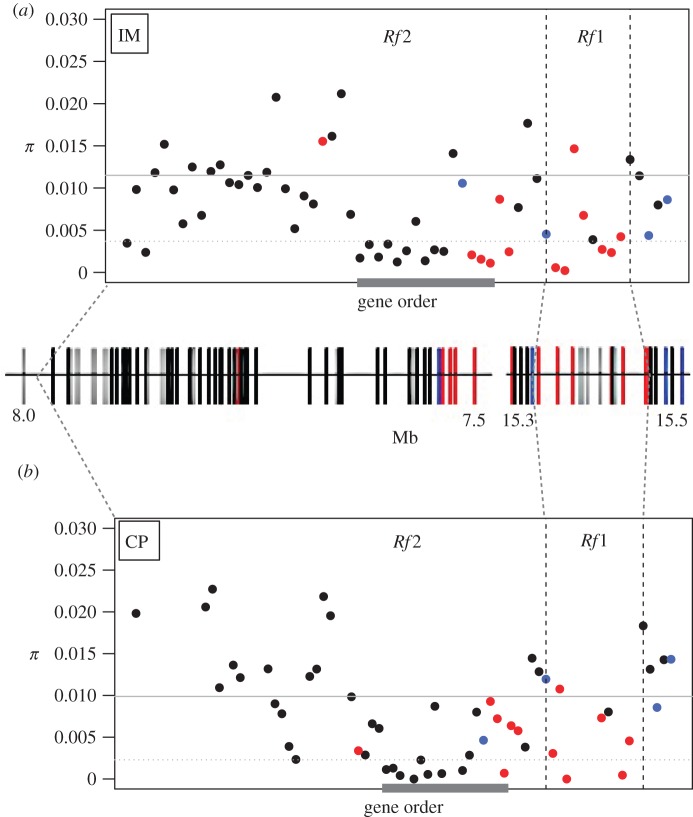
Figure 2.

Elevated population-genetic differentiation across the Rf region between M. guttatus populations at Iron Mountain (with IM-CMS) and nearby Cone Peak (without IM-CMS). Mean FST per gene (n = 47) was calculated based on genome re-sequence data from 10 IM lines and 8 CP lines. Genome-wide mean FST is shown by the white line, and 95% confidence limits in grey shading. Gene order follows re-orientation of genome segments to match the genetic map in [33], anchored by four fine-mapped genetic markers (blue points). Red denotes pentatricopeptide repeat genes.
Consistent with a recent partial selective sweep at IM, we find elevated LD and distinct extended haplotype structure across the Rf2 region (figure 3a and electronic supplementary material, figures S2 and S3). Pairwise LD values across Rf2 at IM (mean r2 = 0.459; s.e. = 0.013) are much higher than LD across similarly sized blocks across the chromosome (mean = 0.201; s.e. = 0.001). At more than 20 genes across the Rf2 region, 5/9 non-reference IM lines and 0/8 CP lines are identical or nearly identical (purple and blue in figure 3) to the IM62 reference, which is known to carry the functional Rf2 allele [33]. By contrast, we see no evidence of elevated LD or extended haplotypes in Rf1, and no significant LD between the two Rf regions at IM. At CP, the Rf2 region does not stand out as a block of elevated LD (mean r2 = 0.272; s.e. = 0.0171) within the chromosome (mean r2 = 0.271; s.e. = 0.0017), and there was no evidence of near-fixation of the reference haplotype at CP (figure 3b and electronic supplementary material, figures S4 and S5).
Figure 3.
Haplotype structure across the Rf region based on genome re-sequence data from (a) 10 IM lines and (b) 8 CP lines. Vertical bars represent genes (n = 159), horizontal bands represent individual lines, and colours show the proportion of variant sites per gene matching the M. guttatus v. 2.0 reference. Genes with less than seven polymorphic sites genotyped for an individual are white. For each population, lines are ordered according to average genotype match score across Rf2. IM62, the reference genome, is the top band in (a). Within Rf2 and Rf1, genes are ordered as in figure 2. Outside the Rf regions, gene order follows the v. 2.0 map, with the positions of v. 1.0 scaffolds noted.
Nucleotide diversity is notably low across portions of Rf2 and Rf1 in both populations (figure 4). At IM, 15 contiguous genes nearest to Rf1 (v. 1.0 sc97; Migut.G00830–Migut.G00847, with three genes dropped for excess missing data) span a 240-kb region of low diversity (mean π = 0.004). Twelve of the 15 genes, including three PPRs, were in the lower fifth percentile of nucleotide diversity (dotted line in figure 4a). Post hoc permutation tests confirm that this portion of Rf2 is significantly less diverse than similar-sized regions on LG6 and LG7NR (electronic supplementary material, figure S6). In the larger pooled sample of IM, 29 10-kb windows within Rf2 are also negative outliers (electronic supplementary material, figure S7). At CP, Rf2 exhibited a partially overlapping region of greatly reduced polymorphism (figure 4b), and the 15-gene block analysed at IM was also an outlier in permutation analyses (mean π = 0.0032, p < 0.0001; electronic supplementary material, figure S8). In contrast with IM, however, only one Rf2 PPR is in the lower fifth percentile of the overall distribution at CP.
Figure 4.
Pairwise nucleotide diversity (π, averaged per gene) in (a) 10 IM lines and (b) 8 CP lines. Each point represents a gene annotated in the M. guttatus v. 2.0 reference genome. The heavy line denotes a 240-kb block of 15 contiguous genes in Rf2 with unusually low diversity (mean π = 0.004) in IM. Physical positions (Mb, v. 2.0 genome) are shown in the hatched bars between the data panels, but genes are ordered as in figure 2. The left segment corresponds to sc97 (inverted; Migut.G00884–G00829) and the right segment to sc14 (Migut.G01156–G001178) of the M. guttatus v. 1.0 genome. Blue denotes fine-mapped genetic markers; red denotes pentatricopeptide repeat genes; grey denotes genes excluded from analysis (15/60 Rf2 genes; 4/14 Rf1 genes).
In both populations, polymorphism across Rf1 (figure 4) is also significantly reduced relative to LG7NR and LG6 (p < 0.005; electronic supplementary material, figures S6 and S8). Again, however, the particular low-diversity loci are not the same, suggesting distinct directional selection between populations, rather than shared purifying or directional selection. Consistent with positive selection in the Rf region, rather than low polymorphism of PPRs in general, nucleotide diversity of PPRs inside the Rf locus (n = 13) is significantly lower than for PPRs on LG6 and LG7NR at IM (n = 18; F1,29 = 5.17, p = 0.0306; electronic supplementary material, figure S9). The sharp reduction in sequence diversity in the Rf region at IM was confirmed with Sanger sequencing of gene-based markers (n = 26 IM inbred lines; electronic supplementary material, figure S10 and table S4).
The genic site-frequency spectrum in Rf2 was also skewed in both populations, with the 240-kb block of low-diversity genes exhibiting strongly negative values of Tajima's D (electronic supplementary material, figures S11–S13). At IM, mean Tajima's D of the entire block (−0.75) is significantly reduced relative to the LG6 and LG7NR means (−0.11 and −0.21, respectively), and was an outlier in permutations of similar-sized blocks (p < 0.026; electronic supplementary material, figure S12a,b). This regional signal at IM is largely driven by very negative values (less than −1.0) at four PPRs near Rf1, the most extreme of which (Migut.G00831) is predicted to be mitochondrially targeted [33]. Accordingly, in the IM Pool-Seq sample, seven of the 11 SNPs across LG7 with Tajima's D values below the first percentile are found in Rf2. At CP, this region is an even more extreme outlier (p < 0.0001; mean Tajima's D = −0.91 versus 0.054 and 0.026 across LG6 and LG7NR respectively; electronic supplementary material, figure S12c,d). However, evidence of selection at CP is strongest at the end of the block away from Rf1. Eleven contiguous genes (eight individual outliers, including one PPR) have Tajima's D < −1 at CP, but none of these loci are individual outliers at IM. Notably, the single PPR that appears swept at CP has a highly positive value of Tajima's D at IM (Migut.G00851), and is not predicted to be mitochondrially targeted [33].
Unlike Rf2, mean Tajima's D values for genes in Rf1 region are not significantly lower than LG6 and LG7NR (p > 0.70; IM mean = 0.20, n = 8 genes; CP mean = 0.23, n = 6 genes; electronic supplementary material, figures S11 and S13). Interestingly, two loci (the gene containing marker 191_45 and a nearby PPR) were individual negative outliers at IM, but had high positive values at CP. These data support a history of selection in both populations, but invoke a scenario more complex than a simple local sweep at IM. Such complexity is expected both from the theory of constant CMS–Rf conflict, and the existence of multiple functional Rf alleles at IM.
4. Discussion
Selfish genetic elements and coevolved suppressors are often invoked as sources of hybrid incompatibility [9,16,48], but direct evidence for a specific role of genomic conflict in the evolution of BDM incompatibilities is rare. Cryptic CMS in plants, where mismatch between organellar and nuclear genes results in hybrid male sterility, epitomizes this gap. Despite abundant evidence that cryptic CMS is common [21,23,24] and robust theory that it should evolve selfishly [28], the links between pattern and process have been circumstantial to date [31]. Here, we present, to our knowledge, the first direct evidence that mitochondrial CMS loci and associated nuclear restorers have evolved under the positive selection predicted by the conflict model. Our findings strongly point to selfish evolution/coevolution within one parental species, rather than negative epistasis limited only to hybrids, as the source of cytonuclear incompatibilites in crosses between hermaphroditic plant species.
Population-genomic data, guided by our previous characterization of Rf and CMS loci [29,32,33], support geographically and genetically localized selection in the Rf region—a pattern uniquely predicted by conflict models for the evolution of hybrid incompatibility. Genes in the Rf region (particularly Rf2) are highly differentiated from a nearby population without the IM-CMS (figure 2), and exhibit a distinct extended haplotype structure (figure 3), reduced polymorphism (figure 4), and skewed site-frequency spectrum relative to flanking and unlinked regions. Within the Rf region, we see particularly strong evidence of population-specific selection on PPR genes that are functional candidates to suppress the locally restricted CMS [32,33]; this argues against other causes of cytonuclear coevolution [49,50] at IM. Furthermore, these molecular population-genetic patterns are not an artefact of the high density of PPR genes in the Rf region, as the signatures of a selective sweep are not seen in PPR clusters elsewhere. Thus, we conclude that the IM-CMS has a history of selfish spread, during which the IM M. guttatus population must have been transiently gynodioecious (i.e. containing female as well as hermaphroditic individuals). Our data also suggest distinct selection at the nearby CP population, which does not carry IM-CMS but is demographically similar, and may have its own history of CMS–Rf coevolution. It is likely that similar dynamics can account for the cryptic CMS present in many hermaphroditic plant species [4,11,18,21–23,30], confirming the conventional wisdom that cytonuclear conflict is an important source of hybrid incompatibilities in plants.
In addition to revealing a history of mitochondrial–nuclear coevolution at IM, our results suggest that these dynamics may be more widespread. Although we observed strong evidence of selection at IM, and the signal appears to be centred on a distinct set of candidate PPRs within the Rf region, we also see non-neutral patterns in the Rf region at CP (in the absence of IM-CMS). Because the populations are highly differentiated across the Rf region (figure 2), and different loci appear to be foci of strong selection (figures 3 and 4), we conclude that the populations are not responding to shared selection. For Rf2, at least, IM exhibits a strong haplotype structure consistent with a selective sweep: across an extended (more than 20 gene) region, a majority of lines match the reference, which is known to carry both functional restorer alleles. The extremely low polymorphism and high LD we observed in the Rf region is atypical for the IM genome [45], and the lack of similarly high local LD at CP implies recent, local selection on Rf2 only at IM. However, we cannot rule out selection on Rf2 at CP; in particular, we see a small (11-gene) region of low diversity and very negative Tajima's D, but not at the genes showing strongest signatures of selection at IM. The low-diversity genes at CP include one extremely swept PPR (arrow in electronic supplementary material, figure S11b), but this PPR is never an outlier at IM and not predicted to be mitochondrially targeted [33]. Given the theoretical prediction that selfish CMS should be constantly turning over both mitochondrial genomes and corresponding Rf alleles, it is plausible that CP has an independent history of CMS–restorer coevolution, or some other gene in this small segment of Rf2 has experienced recent, parallel positive selection at both IM and CP.
Understanding the history of the IM-Rf region is complicated by the presence of two dominant, redundant and tightly linked Rf alleles in at least some IM plants (including the IM62 reference line). At IM, a distinct, extended haplotype (figure 3) is consistent with a recent partial selective sweep of the reference haplotype across Rf2. By contrast, the reference haplotype is a minor component of the variation in Rf1, and there is no evidence of elevated LD in this region. The Rf1 allele that was genetically mapped in interspecific crosses may have little history of strong selection, perhaps arriving at IM once Rf2 was already at high frequency. In that case, the weaker signatures of selection in Rf1 at IM (a few individual loci with extremely negative values of Tajima's D and low nucleotide diversity) could represent a scenario in which recombination during a much slower sweep in a population already containing the restoring Rf2 alleles broke up associations with flanking markers and limited the signal to the causal PPR(s) alone. Notably, the two apparently swept PPRs at IM (arrows in electronic supplementary material, figure S11a) are significant FST outliers, show opposite patterns of nucleotide diversity and Tajima's D between IM and CP, and are both predicted to be mitochondrially targeted [33]. Together, these data point to two PPRs as strong candidate IM restorer loci, and further investigation of their functional effects and local population genetics promise an even clearer picture of the evolutionary dynamics of CMS–Rf systems.
Given that cryptic CMS in Mimulus guttatus (and probably other species) appears to have a local history of selfish coevolution, how do such dynamics play out species-wide in this and other systems? In Mimulus, cryptic CMS occurs in crosses of M. nasutus to several M. guttatus populations with distinct mitochondrial haplotypes (C. M. Barr, L. Fishman and A. L. Case 2016, unpublished data) and also in a few within-guttatus hybrids [51], but each CMS type appears restricted to a small geographical area. Similar patterns are seen in other taxa (e.g. Helianthus, reviewed in [4]), suggesting that relatively high levels of nuclear Rf gene flow (via seeds and pollen) often inoculate surrounding populations against the invasion of novel CMS (via seeds only). Conversely, any CMS that can spread beyond the bounds of a single population may also be able introgress into related species, making it a weak reproductive barrier (though this may depend on genetic background effects [4]). For the above reasons, we do not argue that the selfish IM-CMS is a major contributor to speciation of M. guttatus and M. nasutus, or a substantial barrier to introgression (although selfing M. nasutus—in which male and female fitness are tightly coupled—should be resistant to CMS; [29]). Indeed, it is precisely the highly localized nature of IM-CMS that has allowed us to infer its history from the molecular population genetics of the Rf region. However, if selfish CMS–restorer coevolution is as common as theory and our data suggest, even infrequent cases of non-local effects would make CMS–Rf systems an important contributor to species divergence in absolute terms.
Across flowering plants, selfish CMS–Rf coevolution can plausibly contribute to the evolution of population differentiation and species barriers via multiple mechanisms. In particular, situations in which seed flow is high regionally, but restricted between geographical isolates, (e.g. glacial refugia; [52]) might allow local CMS–restorer dynamics to generate species-wide incompatibility. Furthermore, incipient species of flowering plants frequently evolve through local habitat specialization [53]. Thus, even when CMS–Rf dynamics remain local, they may have long-term effects on the establishment of species-level post-zygotic barriers. In addition to such direct effects on hybrid breakdown, selfish CMS–Rf coevolution will also indirectly contribute to genomic divergence among populations and species. For example, sweeps by CMS mutations may carry associated mitochondrial and chloroplast variation to high frequency locally, with consequences for both organelle function and the (co-)evolution of nuclear genes that interact with organelles. Similarly, sweeps by Rf alleles responding to CMS will alter the dynamics of linked variation (as with the M. guttatus IM-Rf2 haplotype), having potentially important consequences for large regions of the genome. Thus, through either direct or indirect effects on genomic variation, the selfish CMS–Rf interactions we have documented within M. guttatus may frequently drive population and species divergence in flowering plants.
Supplementary Material
Acknowledgements
We thank: A. Saunders for assistance with Sanger sequencing; Y-W. Lee and J. Willis for sharing DNA from IM inbred lines; J. K. Kelly for access to the IM Pool-Seq data; S. Flannagan for help with fhetboot; A. Sweigart, J. Stinchcombe, and three anonymous referees for constructive comments on the manuscript.
Data accessibility
BAM files of Illumina resequence data are available via NCBI Sequence Read Archive (SRP082265 and SRP082275); Sanger sequences via GenBank (accessions KX707450–KX707631); marker genotypes via Dryad [54]: http://dx.doi.org/10.5061/dryad.h966b.
Authors' contributions
A.L.C. analysed the marker data and contributed to manuscript preparation and revision; F.R.F. assembled and analysed the genomic datasets and contributed to manuscript preparation and revision; C.M.B. designed the project, generated and analysed the marker data; L.F designed the project, contributed to all data analyses and to manuscript preparation and revision. All living authors have approved the manuscript for publication.
Competing interests
We have no competing interests.
Funding
L.F. and C.M.B. were supported by NSF EF-0328326; L.F. and F.R.F. by NSF DEB-0846089; A.L.C. by sabbatical support from Kent State University. Funding for the IM Pool-Seq re-sequencing was provided by NIH R01 GM073990-02 to J. K. Kelly.
References
- 1.Bateson W. 1909. Heredity and variation in modern lights. In Darwin and modern science (ed. Seward AC.), pp. 85–101. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 2.Dobzhansky T. 1937. Genetics and the origin of species. New York, NY: Columbia University Press. [Google Scholar]
- 3.Muller HJ. 1942. Isolating mechanisms, evolution, and temperature. Biol. Symp. 6, 71–125. [Google Scholar]
- 4.Rieseberg LH, Blackman BK. 2010. Speciation genes in plants. Ann. Bot. 106, 439–455. ( 10.1093/aob/mcq126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutter AD. 2012. The polymorphic prelude to Bateson–Dobzhansky–Muller incompatibilities. Trends Ecol. Evol. 27, 210–219. ( 10.1016/j.tree.2011.11.004) [DOI] [PubMed] [Google Scholar]
- 6.Sweigart AL, Willis JH. 2012. Molecular evolution and genetics of postzygotic reproductive isolation in plants. F1000 Biol. Rep. 4, 23 ( 10.3410/B4-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maheshwari S, Barbash DA. 2011. The genetics of hybrid incompatibilities. Annu. Rev. Genet. 45, 331–355. ( 10.1146/annurev-genet-110410-132514) [DOI] [PubMed] [Google Scholar]
- 8.Barbash DA, Siino DF, Tarone AM, Roote J. 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl Acad. Sci. USA 100, 5302–5307. ( 10.1073/pnas.0836927100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Presgraves DC. 2010. The molecular evolutionary basis of species formation. Nat. Rev. Gen. 11, 175–180. ( 10.1038/nrg2718) [DOI] [PubMed] [Google Scholar]
- 10.Sweigart AL, Flagel LE. 2015. Evidence of natural selection acting on a polymorphic hybrid incompatibility locus in Mimulus. Genetics 199, 543–554. ( 10.1534/genetics.114.171819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank SA. 1991. Divergence of meiotic drive-suppression systems as an explanation for sex- biased hybrid sterility and inviability. Evolution 45, 262–267. ( 10.2307/2409661) [DOI] [PubMed] [Google Scholar]
- 12.Hurst LD, Pomiankowski A. 1991. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's rule and related phenomena. Genetics 128, 841–858. ( 10.1534/genetics.114.167536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orr HA. 2005. The genetic basis of reproductive isolation: insights from Drosophila. Proc. Natl Acad. Sci. USA 102(Suppl 1), 6522–6526. ( 10.1073/pnas.0501893102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314, 1292–1295. ( 10.1126/science.1133953) [DOI] [PubMed] [Google Scholar]
- 15.Presgraves DC. 2007. Does genetic conflict drive rapid molecular evolution of nuclear transport genes in Drosophila? Bioessays 29, 386–391. ( 10.1002/bies.20555) [DOI] [PubMed] [Google Scholar]
- 16.Ågren JA. 2013. Selfish genes and plant speciation. Evol. Biol. 40, 439–449. ( 10.1007/s11692-012-9216-1) [DOI] [Google Scholar]
- 17.Hurst GD, Werren JH. 2001. The role of selfish genetic elements in eukaryotic evolution. Nat. Rev. Gen. 2, 597–606. ( 10.1038/35084545) [DOI] [PubMed] [Google Scholar]
- 18.Burt A, Trivers R. 2006. Genes in conflict: the biology of selfish genetic elements. Cambridge, MA: Harvard University Press. [Google Scholar]
- 19.Fishman L, Jaenike J. 2013. Selfish genetic elements and genetic conflict. In The Princeton guide to evolution (eds Baum DA, Futuyma DJ, Hoekstra HE, Lenski RE, Moore AJ, Peichel CL, Schluter D, Whitlock MC), pp. 347–355. Princeton, NJ: Princeton University Press. [Google Scholar]
- 20.Crespi B, Nosil P. 2013. Conflictual speciation: species formation via genomic conflict. Trends Ecol. Evol. 28, 48–57. ( 10.1016/j.tree.2012.08.015) [DOI] [PubMed] [Google Scholar]
- 21.Kaul MLH. 1988. Male sterility in higher plants. Berlin, Germany: Springer Science & Business Media. [Google Scholar]
- 22.Bomblies K. 2010. Doomed lovers: mechanisms of isolation and incompatibility in plants. Annu. Rev. Plant Biol. 61, 109–124. ( 10.1146/annurev-arplant-042809-112146) [DOI] [PubMed] [Google Scholar]
- 23.Tiffin P, Olson M, Moyle LC. 2001. Asymmetrical crossing barriers in angiosperms. Proc. R. Soc. Lond. B 268, 861–867. ( 10.1098/rspb.2000.1578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turelli M, Moyle LC. 2007. Asymmetric postmating isolation: Darwin‘s corollary to Haldane's rule. Genetics 176, 1059–1088. ( 10.1534/genetics.106.065979) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aalto EA, Koelewijn H-P, Savolainen O. 2013. Cytoplasmic male sterility contributes to hybrid incompatibility between subspecies of Arabidopsis lyrata. G3 3, 1727–1740. ( 10.1534/g3.113.007815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charlesworth B, Charlesworth D. 1978. A model for the evolution of dioecy and gynodioecy. Am. Nat. 112, 975–997. ( 10.2307/2460344) [DOI] [Google Scholar]
- 27.Frank SA. 1989. The evolutionary dynamics of cytoplasmic male sterility. Am. Nat. 133, 345–376. ( 10.1086/284923) [DOI] [Google Scholar]
- 28.Frank SA, Barr CM. 2001. Spatial dynamics of cytoplasmic male sterility. In Integrating ecology and evolution in a spatial context (eds Silvertown J, Antonovics J), pp. 219–243. Oxford, UK: Blackwell Science. [Google Scholar]
- 29.Fishman L, Willis JH. 2006. A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evolution 60, 1372–1381. ( 10.1111/j.0014-3820.2006.tb01216.x) [DOI] [PubMed] [Google Scholar]
- 30.Hanson MR, Bentolila S. 2004. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16, S154–S169. ( 10.1105/tpc.015966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujii S, Bond CS, Small ID. 2011. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl Acad. Sci. USA 108, 1723–1728. ( 10.1073/pnas.1007667108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Case AL, Willis JH. 2008. Hybrid male sterility in Mimulus (Phrymaceae) is associated with a geographically restricted mitochondrial rearrangement. Evolution 62, 1026–1039. ( 10.1111/j.1558-5646.2008.00360.x) [DOI] [PubMed] [Google Scholar]
- 33.Barr CM, Fishman L. 2010. The nuclear component of a cytonuclear hybrid incompatibility in Mimulus maps to a cluster of pentatricopeptide repeat genes. Genetics 184, 455–465. ( 10.1534/genetics.109.108175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahan J, Mireau H. 2013. The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 10, 1469–1476. ( 10.4161/rna.25568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fishman L, Saunders A. 2008. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science 322, 1559–1562. ( 10.1126/science.1161406) [DOI] [PubMed] [Google Scholar]
- 36.Fishman L, Kelly JK. 2015. Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution 69, 1208–1218. ( 10.1111/evo.12661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee YW. 2009. Genetic analysis of standing variation for floral morphology and fitness components in a natural population of Mimulus guttatus (common monkeyflower). Dissertation, Duke University, Durham, USA.
- 38.Floro ER, McCauley DE, Fishman L, Case AL. In preparation. Distribution of a male-sterility inducing mitochondrial haplotype within and among populations of Mimulus guttatus.
- 39.Birky CW, Fuerst P, Maruyama T. 1989. Organelle gene diversity under migration, mutation, and drift: equilibrium expectations, approach to equilibrium, effects of heteroplasmic cells, and comparison to nuclear genes. Genetics 121, 613–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG. 2005. Invited review: comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 14, 689–701. ( 10.1111/j.1365-294X.2004.02410.x) [DOI] [PubMed] [Google Scholar]
- 41.Vickery RK. 1978. Case studies in the evolution of species complexes in Mimulus. In Evolutionary biology (eds Hecht MK, Steere WC, Wallace B), pp. 405–507. Boston, MA: Springer. [Google Scholar]
- 42.Wu CA, Lowry DB, Cooley AM, Wright KM, Lee YW, Willis JH. 2008. Mimulus is an emerging model system for the integration of ecological and genomic studies. Heredity 100, 220–230. ( 10.1038/sj.hdy.6801018) [DOI] [PubMed] [Google Scholar]
- 43.Willis JH. 1993. Effects of different levels of inbreeding on fitness components in Mimulus guttatus. Evolution 47, 864 ( 10.2307/2410190) [DOI] [PubMed] [Google Scholar]
- 44.Flagel LE, Willis JH, Vision TJ. 2014. The standing pool of genomic structural variation in a natural population of Mimulus guttatus. Genome Biol. Evol. 6, 53–64. ( 10.1093/gbe/evt199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puzey JR, Wills JH, Kelly JK. In press. Whole genome sequencing of 56 Mimulus individuals illustrates population structure and local selection. Biorxiv ( 10.1101/031575) [DOI] [Google Scholar]
- 46.Lee YW, Fishman LF, Kelly JK, Willis JH. 2016. Fitness variation is generated by a segregating inversion in yellow monkeyflower (Mimulus guttatus). Genetics 202, 1473–1484. ( 10.1534/genetics.115.183566) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willis JH. 1996. Measures of phenotypic selection are biased by partial inbreeding. Evolution 50, 1501–1511. ( 10.2307/2410887) [DOI] [PubMed] [Google Scholar]
- 48.Presgraves DC. 2007. Speciation genetics: epistasis, conflict and the origin of species. Curr. Biol. 17, R125–R127. ( 10.1016/j.cub.2006.12.030) [DOI] [PubMed] [Google Scholar]
- 49.Burton RS, Pereira RJ, Barreto FS. 2013. Cytonuclear genomic interactions and hybrid breakdown. Annu. Rev. Ecol. Evol. Syst. 44, 281–302. ( 10.1146/annurev-ecolsys-110512-135758) [DOI] [Google Scholar]
- 50.Bock DG, Andrew RL, Rieseberg LH. 2014. On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol. 23, 4899–4911. ( 10.1111/mec.12920) [DOI] [PubMed] [Google Scholar]
- 51.Martin NH, Willis JH. 2010. Geographical variation in postzygotic isolation and its genetic basis within and between two Mimulus species. Phil. Trans. R. Soc. B 365, 2469–2478. ( 10.1098/rstb.2010.0030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gérardi S, Jaramillo-Correa JP, Beaulieu J, Bousquet J. 2010. From glacial refugia to modern populations: new assemblages of organelle genomes generated by differential cytoplasmic gene flow in transcontinental black spruce. Mol. Ecol. 19, 5265–5280. ( 10.1111/j.1365-294X.2010.04881.x) [DOI] [PubMed] [Google Scholar]
- 53.Levin DA. 2000. The origin, expansion, and demise of plant species. Oxford, UK: Oxford University Press. [Google Scholar]
- 54.Case AL, Finseth FR, Barr CM, Fishman LF. 2016. Data from: selfish evolution of cytonuclear hybrid incompatibility in Mimulus. Dryad digital repository, 10.5061/dryad.h966b. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BAM files of Illumina resequence data are available via NCBI Sequence Read Archive (SRP082265 and SRP082275); Sanger sequences via GenBank (accessions KX707450–KX707631); marker genotypes via Dryad [54]: http://dx.doi.org/10.5061/dryad.h966b.