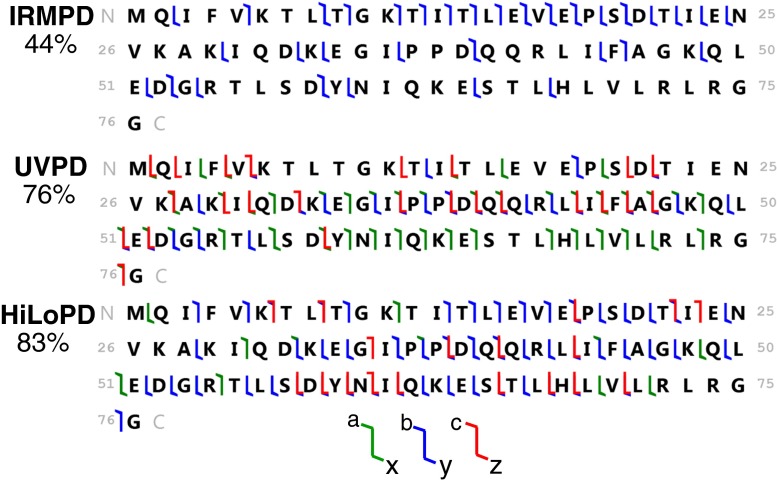Abstract
Herein we report the successful implementation of the consecutive and simultaneous photodissociation with high (213 nm) and low (10.6 μm) energy photons (HiLoPD, high-low photodissociation) on ubiquitin in a quadrupole-Orbitrap mass spectrometer. Absorption of high-energy UV photon is dispersed over the whole protein and stimulates extensive C–Cα backbone fragmentation, whereas low-energy IR photon gradually increases the internal energy and thus preferentially dissociates the most labile amide (C–N) bonds. We noticed that simultaneous irradiation of UV and IR lasers on intact ubiquitin in a single MS/MS experiment provides a rich and well-balanced fragmentation array of a/x, b/y, and z ions. Moreover, secondary fragmentation from a/x and z ions leads to the formation of satellite side-chain ions (d, v, and w) and can help to distinguish isomeric residues in a protein. Implementation of high-low photodissociation in a high-resolution mass spectrometer may offer considerable benefits to promote a comprehensive portrait of protein characterization.
Graphical Abstract.
ᅟ
Electronic supplementary material
The online version of this article (doi:10.1007/s13361-016-1419-8) contains supplementary material, which is available to authorized users.
Keywords: Photodissociation, UVPD, IRMPD, Ubiquitin, Top-down proteomics
Introduction
Photon-based activation methods, including ultraviolet photodissociation (UVPD) [1–3] and infrared multiphoton dissociation (IRMPD) [4–6], have received great attention as alternative to electron-driven methods [7–11]. In recent years, UVPD has been implemented in high resolution mass spectrometry and employed for peptide and whole protein characterizations [12–19]. High energy UV photons preferentially cleave Cα–C bond in peptides and proteins producing abundant a/x ions. Other fragment ions, such as c/z and y ions, are also detected in UVPD providing nearly complete sequence coverage [12, 20].
Contrasting to UVPD and electron transfer dissociation (ETD), multiple low energy IR photon excitation selectively breaks the most labile amide (C–N) bonds and generates b and y ions similar to the traditional slow-heating collision activation dissociation (CAD) method [21]. IRMPD has been implemented in different instruments, including quadrupole ion traps [22] and dual pressure linear ion traps [4, 6, 23]. Vasicek et al. reported the execution of IRMPD in the high collision dissociation (HCD) cell of a modified hybrid linear ion trap-Orbitrap mass spectrometer [24].
The dissociation mechanisms involved after high and low energy photon excitations are quite different. Absorption of a single high energy photon (in the UV) is sufficient to induce dissociation of a peptide and protein in the gas phase. On the other hand, multiple absorption of low energy photons (in the IR) are required before fragmentation. Excitation is followed by fast internal vibrational redistribution (IVR) and causes a slow and steady rise of the internal energy until it exceeds the dissociation threshold and thus induces cleavage of the labile bonds [2].
Despite some analytical challenges, coupling of high and low energy activation pathways in a single MS/MS event is expected to offer diverse fragmentation arrays and thus deliver improved, efficient, and well-balanced fragmentation for whole protein characterization. Tsybin et al. reported the implementation of IRMPD with electron capture dissociation (ECD) in a FT-ICR mass spectrometer [25]. Electron and photon irradiation significantly improved the formation of sequence ions for peptides and proteins. Simultaneous IR photoactivation with ETD, known as activated ion electron transfer dissociation (AI-ETD), is also implemented in an ion trap-Orbitrap Elite system [26]. Moreover, tandem ETD spectra exhibited abundant peaks related to unreacted and charge reduced precursors. Hybrid AI-ETD showed better performance for lower charge states and production of specific fragment ions. The combination of UVPD with ETD (known as ETUVPD) in an ion trap-Orbitrap has also been reported [27]. The combined ETUVPD method showed balanced fragment ions with increased number of c and z ions. The fragmentation efficiency of ETD can also be enhanced by other means, such as additional activation with CID and HCD, known as ETciD and EThcd [28, 29]. These hybrid methods showed rich fragmentation spectra compared with CID, HCD, and ETD alone.
Although a few studies report on coupling electron and photon based methods, integrating electron-driven technique with low or high collision activation approaches, so far there is no study reporting the combination of high and low energy photons for characterizing protein within a single MS/MS framework. Here, we report the implementation of a method combining solid-state fifth harmonic 213 nm laser excitation with 10.6 μm CO2 laser excitation in hybrid quadrupole-Orbitrap mass spectrometer using different excitation schemes (consecutive IR + UV, UV + IR, and simultaneous UV/IR) for top-down characterization of ubiquitin. This high-low energy photon-based method (HiLoPD) improves the fragmentation pattern, providing well-proportioned a/x, b/y, and z-ions with richness of secondary fragment ions, including d, v, and w.
Materials and Methods
Laser Setup and Experiments
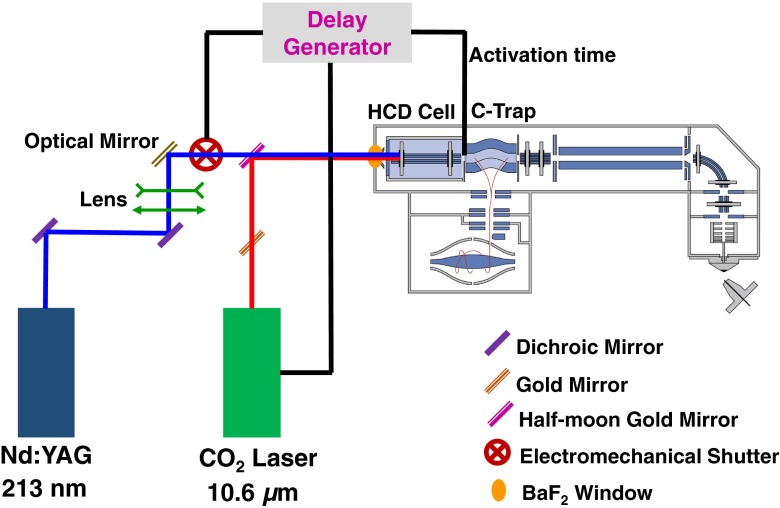
A simple schematic presentation of the coupling of IRMPD and UVPD irradiation in the HCD cell of a hybrid quadrupole-Orbitrap is presented in Figure 1. IRMPD experiments were performed using a 50 W cw-CO2 laser (ULR-50; Universal Laser System, Scottsdale, AZ, USA). The wavelength of the CO2 laser is 10.6 μm with a beam diameter and divergence (full angle) of 4 ± 1 mm and 5 ± 1 mrad, respectively. Sixty percent of the nominal laser power was used. The IR beam was directed to the HCD cell using gold mirrors. The IR beam was gated on an external TTL signal. Irradiation times from 0.1 to 1 s were tested. The N2 pressure in the HCD cell was adjusted to optimize the IR fragmentation while avoiding significant loss of signal (pressure controller set to ~0.09 MPa). For the UVPD experiments, the fifth harmonic (λ = 213 nm, ~1 mJ/pulse) of a 20 Hz BrillantB solid-state Nd:YAG laser (Quantel, Les Ulis, France) was used. A mechanical shutter (SH05/TSC001; Thorslab) was used to allow, on demand, the beam in the HCD cell. For UVPD, the optimal shutter open time was determined to be 0.2 s (four laser shots). In order to combine both CO2 and UV laser beams, a half-moon gold mirror was used on the IR beam path to the HCD cell. Also, a BaF2 window (wavelength range 0.2–12 μm, Ø 25.4 mm, thickness 5 mm) was placed at the rear of the HCD cell, which transmits both IR (10.6 μm) and UV (213 nm) beams with 90% and 85% efficiency, respectively.
Figure 1.
Schematic representation of the execution of combined IRMPD and UVPD in the HCD cell of a hybrid quadrupole-Orbitrap mass spectrometer
In order to irradiate ions only when they are in the HCD cell, the voltage on test-point 18 (TP18), located on Q-Exactive electronic board, was monitored. In our experimental conditions, the falling edge (–10 V → –350 V) on the TP18 is used to determine the moment when ions are ejected from the C-trap to the HCD-cell (see Supplementary Figure S1). Two independent TTL pulses are then generated, with width and delay adjustable with regards to the TP18 trigger. The TTL pulses are used to lift the gate on the CO2 laser and open the shutter on the UV beam path.
Three different coupling schemes between IR and UV were implemented (Supplementary Figure S1). In Scheme I, CO2 laser was ON for 1 s and then followed by 4 UV pulses (0.2 s). In Scheme II, four pulses of UV were admitted in the HCD cell first, and followed by 1 s of CO2 laser. In the first two schemes, IR and UV were used consecutively: when CO2 laser was ON, the UV laser was OFF and vice versa. In Scheme III, the CO2 laser was turned ON and the UV shutter was open concomitantly. As in previous schemes, IR was left ON for 1 s while the UV shutter was left open for 0.2 s (four pulses). In each scheme, the coupled IR/UV irradiation takes place during single HCD events in MS2 sequences.
Mass Spectrometry
All experiments were performed on a hybrid quadrupole-Orbitrap Q-Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) equipped with a HESI ion source. Ubiquitin (76 residues, 8.6 kDa) from bovine erythrocytes was obtained from Sigma-Aldrich and used without any further purification. Ubiquitin samples were prepared at 10 μM concentration in 50/49/1 (v/v/v) methanol/water/acetic acid and directly infused to MS at a flow rate of 5 μL/min. All mass spectra were acquired using a mass range of 200–2000 m/z and resolving power of 140,000 at m/z 400. The automatic gain control (AGC) target was set to 5 × 106 and the maximum injection time was set at 250 ms. The isolation width was 8–10 Th. To avoid collisions and CID contamination, HCD collision energy was set to the minimum 2 eV. All experiments were performed for three microscans and averaging for 50 scans.
Data Analysis
Raw files were deconvoluted and de-isotoped to the neutral monoisotopic masses using Xtract algorithm provided by Thermo Scientific Inc. Manual analysis of IRMPD, UVPD, and combined UVPD and IRMPD data was performed with the aid of ProSight Light software [30] and Protein Prospector v5.14.4. (http://prospector.ucsf.edu/prospector/mshome.htm). All major ion types (a, a + 1, a + 2, b – 1, b, b + 1, b + 2, c – 1, c, c + 1, x – 1, x, x + 1, x + 2, y, y – 1, y – 2, z – 1, z, z + 1) were considered. We observed a substantial number of secondary fragment ions, including d, v, and w, which were analyzed by Protein Prospector. H2O and NH3 losses from the fragment ions were also considered. Single protein mode with a fragment mass tolerance set to 15 ppm was used for all methods.
Results and Discussion
Optimization of IRMPD on Intact Protein
The overall performance of IRMPD is hindered by the failure to provide adequate fragmentation of peptide or proteins at the standard pressure in the HCD cell. Although relatively high pressure is desirable for collision cooling during the ion accumulation to obtain maximum trapping efficiency, it is disadvantageous to ion activation and dissociation [31]. In the Q-Exactive mass spectrometer, the HCD cell and C-trap are filled with N2 gas with chamber pressure of ~10–5 mbar known as High Vacuum (HV) region whereas Orbitrap kept the low pressure at ~10–10 mbar designated as Ultra High Vacuum (UHV) region. A pressure regulator allows control of the collision gas valve and, hence, the pressure in the HCD cell. The position of the pressure controller also has an effect on the HV pressure value. Here, where we discuss high and low pressure, it is the HCD cell pressure governed by the pressure controller position and estimated via HV gauge that is being considered. A previous study on hybrid QLT-Orbitrap indicated that the level of the collision gas (N2) must be lowered [32]. At high pressure in the chamber (HV ~4.6 × 10–5 mbar, pressure controller 0.5 MPa), there is no noticeable photodissociation observed for +12 charge state ion of ubiquitin even at longer (1 s) irradiation time (Figure 2). The collision frequency, which is associated with the collision cross-section [33] of the protein, of +12 charge state ion of ubiquitin is typically around 6700 s–1 at high pressure of 10-4 mbar (Supplementary Figure S2). This high collision rate promotes collision deactivation and cooling of the protein before it can undergo fragmentation, resulting in the limited photodissociation observed. The fragmentation efficiency improves as the pressure is reduced. At low pressure (HV ~9.3 × 10–6 mbar, pressure controller ~0.09 MPa), the dissociation efficiency is augmented significantly for +12 charged precursor ion of ubiquitin. Indeed, at ~9.3 × 10–6 mbar pressure, the collision frequency of +12 ion of ubiquitin is reduced. It is noticed that pressure lower than ~0.09 MPa on the pressure controller can lead to more fragmented ions; however, the signal is not very stable at this range and, moreover, sensitivity and resolution are also decreased. The irradiation time also has a major impact on the photodissociation yield of ubiquitin (Supplementary Figure S3). At lower pressure, when ubiquitin is irradiated for 0.1 s, the fragmentation efficiency is only about 25%, which is considerably improved to 68% for 1 s irradiation time (Supplementary Figure S4). Most of the previous studies related to IRMPD used a laser irradiation time lower than 0.1 s in LIT [6, 26, 34, 35].
Figure 2.
Impact of pressure on the fragmentation of the +12 charge state precursor ion (m/z = 714.7279) of ubiquitin at 1 s irradiation time by IRMPD
The IRMPD on the +12 charge state ion of ubiquitin induces a total of 141 fragment ions of which 41 are b-type and 98 are y-type ions. Exact masses and assignments of the ions detected in the IRMPD of the 12+ precursor ion (m/z = 714.7279) of ubiquitin are summarized in Supplementary Table S1. For this charge state, twice as many y-type ions as b-type ions are identified. The sequence coverage for the +12 ion is 59% (44 bonds break), which is significantly higher than the coverage of 24% (18 bonds break) reported earlier when IRMPD was first implemented in high resolution Orbitrap mass spectrometer (Supplementary Figure S5) [24]. We found that a combination of lower pressure (HV ~9.3 × 10–6 mbar) and longer irradiation time (~1 s) are optimal for characterization of intact protein by IRMPD in a quadrupole-Orbitrap system.
UVPD, IRMPD, and HiLoPD on Ubiquitin
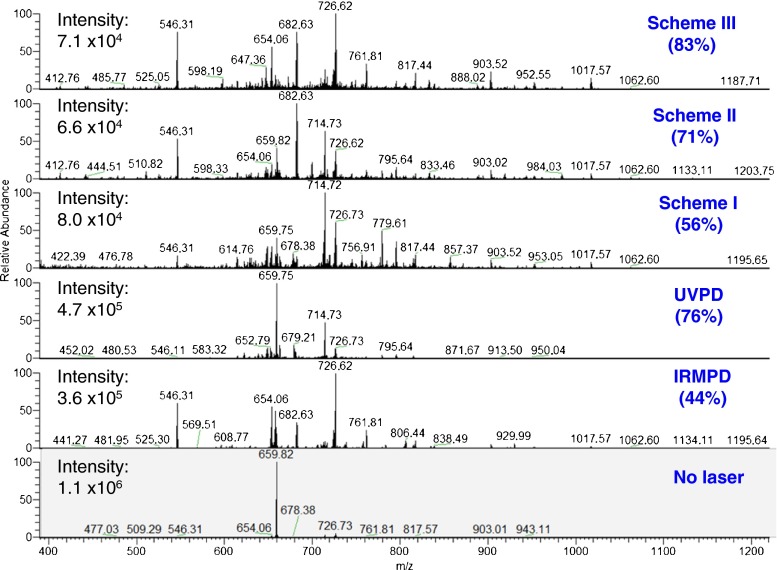
The photodissociation mass spectra using IRMPD, UVPD, and combined IR and UV (Scheme I, II, III) of the +13 precursor ion of ubiquitin are presented in Figure 3.
Figure 3.
Combined IR and UV (Schemes I, II, III), IRMPD, and UVPD spectra of the +13 charge state precursor ion (m/z = 659.8249) of ubiquitin. Isolation spectrum with no activation is also presented. Sequence coverages are indicated in brackets
First the +13 precursor ion of ubiquitin was subjected to UVPD only. All 213 nm UVPD experiments were performed in the low pressure regime (~9.3 × 10–6 mbar) to make unbiased comparison with consecutive or simultaneous irradiation of IRMPD and UVPD. Even at low pressure, the 213 nm UVPD on the +13 charge state ion produces a total of 209 fragment ions (Figure 4a), including 68 a-type, 5 b-type, and 10 c-type ions as well as 38 x-type, 59 y-type, and 28 z-type ions (Supplementary Table S2). Along with traditional a/x, y, and c/z ions, a + 1/x + 1, x + 2, y – 1, y – 2, c – 1, and c + 1 fragment ions of ubiquitin are also detected. Recently, we reported that the radical-driven backbone fragmentation provides 22 distinctive fragment ion types for peptide anions at 213 nm UVPD [15]. High energy UVPD (157 and 193 nm) yields to abundant formation of radical a + 1 and x + 1 ions [36–38]. Here, we observed the similar feature at 213 nm (5.8 eV) UVPD, which produces a significant number of a + 1/x + 1 ions. The mechanism of the homolytic cleavage of the Cα–C(O) bond, which produces a + 1/x + 1 ions, has been proposed elsewhere [15]. Moreover, the formation of y – 1 and y – 2 occurs from the secondary dissociation of the x + 1 radical and is associated with presence of proline residues [18, 37]. Neutral losses of NH3 are also detected from a and y ions. The UVPD sequence coverage achieved for the +13 precursor ion is 76%.
Figure 4.
(a) Number of fragment ions detected by IRMPD, UVPD, and combined IR and UV (Schemes I, II, and III) of the +13 charge state precursor ion (m/z = 659.8249) of ubiquitin. (b) Number of fragment ions detected by IRMPD, UVPD, and HiLoPD (Scheme III) of the +8 charge states precursor ion (m/z = 1071.5864) of ubiquitin
The IRMPD experiment on the +13 ion of ubiquitin allows detecting a total of 121 fragment ions (Figure 4a). Exact masses and assignments of ions detected in the IRMPD of the +13 ion (m/z = 659.8249) of ubiquitin are summarized in Supplementary Table S3. Among them, 49 ions are b-type and 67 ions are y-type fragments. The formation of only b- and y-type ions is expected from cleavages of C–N bonds proceeding via vibrationally-excited ground state dissociation. H2O and NH3 losses from the b and y ions are also observed, with H2O losses being more widespread than NH3 loss. The loss of water is energetically favorable from the protonated acidic group [39]. Ubiquitin has seven threonine (T), six glutamic acid (E), five aspartic acid (D), and three serine (S) residues, which may promote the extensive water loss. Low (z = +1) to high charge states (z = +12) of the b and y ions are detected. The same fragment ion is often observed in many different charge states. For example, +2, +3, and +4 charge states b17 ions are detected at m/z 952.5491, 635.3695, and 476.7778, respectively. The IRMPD sequence coverage of this charge state precursor ion is 44%.
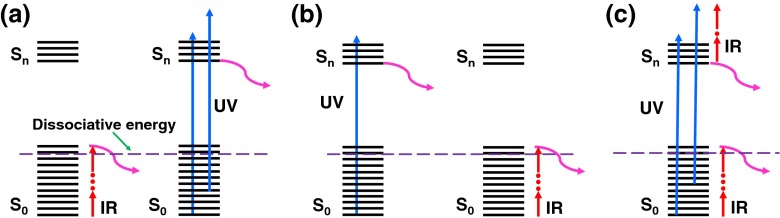
The same precursor ion (z = +13) was then fragmented with UVPD in combination with IRMPD. In the consecutive Scheme I, in which first IR then UV irradiation was performed, the total number of detected fragment ions declined remarkably compared with UVPD (Figure 4a) alone. Despite this decrease, the number of b-type ions detected increased significantly. The y-type ions remain the same as the one with UVPD alone. The a/x and c/z ions are also remarkably suppressed. Overall, the sequence coverage using this scheme is only 56% (Figure 3). In Scheme I, IR laser pulses produce ubiquitin in its vibrationally hot electronic ground state (Figure 5a). Excitation promotes formation of hot ions and eventually ground state dissociation. Thus, less parent ions are then available for UV fragmentation, although UV excitation of hot ions is also possible.
Figure 5.
Ground and excited state dissociation channels in Scheme I (a), Scheme II (b), and Scheme III (c)
In the consecutive Scheme II, when irradiation with UV laser pulses is followed by IR irradiation, the overall number of detected fragment ions is considerably higher compared with Scheme I (Figure 4a). The number of b- and y-type ions increased sharply compared with both UVPD and Scheme I. The relaxation following electronic excitation either by light emission, internal conversion through a conical intersection or via fragmentation is expected to be fast (typically ranging from fs to ns timescales). In Scheme II, IR excitation occurs after electronic excitation and relaxation has occurred. The UV laser promotes excited states dissociation, whereas IR laser subsequently leads to the ground state dissociation (Figure 5b). The combination of the two dissociation mechanisms explains the large amount of detected fragment ions in Scheme II. The sequence coverage for this charge state precursor ion is 71% and is comparable to the one observed in UVPD.
The simultaneous introduction of UV and IR lasers (HiLoPD, high-low photodissociation, Scheme III), on the +13 ion of ubiquitin produces a more diverse range of fragment ions than any of IRMPD, UVPD, Scheme I, and Scheme II (Figure 4a). Exact masses and assignments of the ions detected in the combined UVPD and IRMPD of the +13 ion (m/z = 659.8249) of ubiquitin are summarized in Supplementary Table S4. Compared with UVPD, a substantial increase in b and y ions is observed in Scheme III. The number of b-type ions increases from 5 to 48 while the number of y-type ions rises from 59 to 106 ions. The number of c-type ions increases slightly from 10 to 18. The number of x- and z-type of ions remains nearly the same as the one in UVPD, whereas the number of a-type ions decreases noticeably from 68 to 24 ions compared with UVPD. Secondary fragmentation, which leads to the formation of d, v, and w ions, also increases significantly in Scheme III compared with UVPD, Scheme I, and Scheme II (Supplementary Figure S6). Due to the excess energy in Scheme III, eliminations of other groups such as R, CO, and CONH are observed near the position of primary cleavage. The side-chain losses from the a + 1/x + 1 ions, allowing formation of d, v, and w ions, were also reported with high energy 157 and 193 nm UVPD [38]. Kjeldsen et al. observed the formation of d and w ions from Leu and Ile containing peptides using hot electron capture dissociation [40]. Zhang and Reilly also observed the formation of v, wa, and wb ions from x + 1 ion of Leu and Ile containing peptides by UVPD at 157 nm [41]. It is interesting to note that ubiquitin has a total number of 16 Leu and Ile residues, and thus formation of these secondary ions allows distinguishing isomeric residues. In addition, HiLoPD can be applied for de novo sequencing of peptides [42]. The sequence coverage of the +13 ion of ubiquitin obtained with Scheme III is 83%. The photophysical interpretation of this increase in fragmentation yield is that IR irradiation concomitant to UV irradiation can lead to vibrational excitation as well as excitation of higher electronic states and thus produce a rich fragmentation array (Figure 5c). Figure 6 shows the sequence maps obtained for the +13 charge state precursor ion in IRMPD, UVPD, and HiLoPD (Scheme III). The sequence coverage is improved in HiLoPD thanks to the combination of IR and UV irradiation. While IRMPD yields more fragment ions from the N-terminal, UVPD produces ions from the mid- and C-terminal regions. Interestingly, HiLoPD is able to produce fragment ions from all regions.
Figure 6.
Sequence coverage of the +13 charge state precursor ion (m/z = 659.8249) of ubiquitin observed by IRMPD, UVPD, and HiLoPD (Scheme III)
For a lower charge state (z = +8), the simultaneous irradiation in Scheme III (HiLoPD) also showed a balanced fragmentation pattern. The total number of detected fragment ions for this charge state is higher than both the ones observed with IRMPD and UVPD (Figure 4b) alone. Large numbers of b and y ions are observed for this lower charge state, and c-type ions are also detected. Only z-type ions remain essentially the same as in UVPD. The sequence coverage of the +8 ion of ubiquitin obtained with HiLoPD (Scheme III) is 85%. Formation of d, v, and w ions is also observed for this charge state similar to +13 precursor ion.
Overall, IRMPD selectively produces b/y and b-H2O/y-H2O ions, whereas UVPD preferentially yields to a + 1/x + 1, a/x, y – 1, y – NH3, z, v, and w ions (Supplementary Figure S7a). The hybrid HiLoPD (Scheme III) method generates b/y, b – H2O/y – H2O, x, x + 1, y – 1, y – 2, y – NH3, z, v, and w ions. Bond breaking and sequence coverage of high (z = +13) and low (z = +8) charge state ions of ubiquitin obtained by IRMPD, UVPD, and HiLoPD (Scheme III) are shown in Supplementary Figure S7b and c. HiLoPD allows to improve the efficiency of structural characterization of ubiquitin compared with IRMPD and UVPD. Moreover, sequence coverages obtained with HiLoPD are similar to those theoretically expected by combining UVPD and IRMPD (calculated IR + UV, see Supplementary Figure S7c).
Conclusion
We report IRMPD, 213 nm UVPD, and HiLoPD patterns of ubiquitin in a hybrid quadrupole-Orbitrap mass spectrometer. Improved performance of IRMPD is observed when a combination of very low pressure and longer irradiation time in the HCD cell are used. Significant numbers of b/y ions and neutral losses of NH3 and H2O are detected by IRMPD. Similar to excimer 193 nm UVPD, solid-state 213 nm UVPD can promote Cα–C cleavage generating abundant a/x, y, and z fragment ions for ubiquitin.
The coupling of low-energy IRMPD and high-energy UVPD was implemented using three different irradiation schemes. In Scheme I, where IR irradiation is followed by UV, the number of detected fragment ions is decreased compared with the one obtained by UVPD only, which is mainly due to intense IR fragmentation prior to UV excitation. When UV irradiation was followed by IR (Scheme II), the total number of detected fragment ions is slightly increased. In Scheme III, while UV and IR lasers irradiation are simultaneous, the total number of detected fragment ions is maximal. Excited and ground state dissociation channels promote widespread fragmentation of ubiquitin precursor ions. Compared with UVPD, b/y-type ions are increased. We observed that while a/x fragment ions are decreasing, nearly equal number of d, v, and w ions emerge, which can lead to identifying the isomeric residues in a protein.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOCX 1.72 mb)
Acknowledgments
The research leading to these results has received funding from the European Research Council under the European Union’s Seventh Framework Program (FP7/2007-2013 Grant agreement No. 320659). The authors thank Christian Clavier for his invaluable technical assistance. They also thank Dr. Steven Daly (ILM, CNRS et Université Lyon 1, France) for improving the English of this manuscript.
References
- 1.Reilly JP. Ultraviolet photofragmentation of biomolecular ions. Mass Spectrom. Rev. 2009;28:425–447. doi: 10.1002/mas.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodbelt JS. Photodissociation mass spectrometry: new tools for characterization of biological molecules. Chem. Soc. Rev. 2014;43:2757–2783. doi: 10.1039/c3cs60444f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoine R, Lemoine J, Dugourd P. Electron photodetachment dissociation for structural characterization of synthetic and biopolymer anions. Mass Spectrom. Rev. 2014;33:501–522. doi: 10.1002/mas.21402. [DOI] [PubMed] [Google Scholar]
- 4.Gardner MW, Smith SI, Ledvina AR, Madsen JA, Coon JJ, Schwartz JC, Stafford GC, Jr, Brodbelt JS. Infrared multiphoton dissociation of peptide cations in a dual pressure linear ion trap mass spectrometer. Anal. Chem. 2009;81:8109–8118. doi: 10.1021/ac901313m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raspopov SA, El-Faramawy A, Thomson BA, Siu KWM. Infrared multiphoton dissociation in quadrupole time-of-flight mass spectrometry: top-down characterization of proteins. Anal. Chem. 2006;78:4572–4577. doi: 10.1021/ac052248i. [DOI] [PubMed] [Google Scholar]
- 6.Ledvina AR, Lee MV, McAlister GC, Westphall MS, Coon JJ. Infrared multiphoton dissociation for quantitative shotgun proteomics. Anal. Chem. 2012;84:4513–4519. doi: 10.1021/ac300367p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhurov KO, Fornelli L, Wodrich MD, Laskay UA, Tsybin YO. Principles of electron capture and transfer dissociation mass spectrometry applied to peptide and protein structure analysis. Chem. Soc. Rev. 2013;42:5014–5030. doi: 10.1039/c3cs35477f. [DOI] [PubMed] [Google Scholar]
- 8.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 1998;120:3265–3266. doi: 10.1021/ja973478k. [DOI] [Google Scholar]
- 9.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breuker K, Oh H, Lin C, Carpenter BK, McLafferty FW. Nonergodic and conformational control of the electron capture dissociation of protein cations. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14011–14016. doi: 10.1073/pnas.0406095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breuker K, Oh H, Horn DM, Cerda BA, McLafferty FW. Detailed unfolding and folding of gaseous ubiquitin ions characterized by electron capture dissociation. J. Am. Chem. Soc. 2002;124:6407–6420. doi: 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- 12.Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation. J. Am. Chem. Soc. 2013;135:12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannon JR, Cammarata MB, Robotham SA, Cotham VC, Shaw JB, Fellers RT, Homas PM, Kelleher NL, Brodbelt JS. Ultraviolet photodissociation for characterization of whole proteins on a chromatographic time scale. Anal. Chem. 2014;86:2185–2192. doi: 10.1021/ac403859a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cammarata MB, Brodbelt JS. Structural characterization of holo- and apo-myoglobin in the gas phase by ultraviolet photodissociation mass spectrometry. Chem. Sci. 2015;6:1324–1333. doi: 10.1039/C4SC03200D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halim MA, Girod M, MacAleese L, Lemoine J, Antoine R, Dugourd P. 213 nm Ultraviolet photodissociation on peptide anions: radical-directed fragmentation patterns. J. Am. Soc. Mass Spectrom. 2016;27:474–486. doi: 10.1007/s13361-015-1297-5. [DOI] [PubMed] [Google Scholar]
- 16.Cammarata MB, Thyer R, Rosenberg J, Ellington A, Brodbelt JS. Structural characterization of dihydrofolate reductase complexes by top-down ultraviolet photodissociation mass spectrometry. J. Am. Chem. Soc. 2015;137:9128–9135. doi: 10.1021/jacs.5b04628. [DOI] [PubMed] [Google Scholar]
- 17.Cannon JR, Martinez-Fonts K, Robotham S, Matouschek A, Brodbelt JS. Top-Down 193-nm ultraviolet photodissociation mass spectrometry for simultaneous determination of polyubiquitin chain length and topology. Anal. Chem. 2015;87:1812–1820. doi: 10.1021/ac5038363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Girod M, Sanader Z, Vojkovic M, Antoine R, MacAleese L, Lemoine J, Bonacic-Koutecky V, Dugourd P. UV photodissociation of proline-containing peptide ions: insights from molecular dynamics. J. Am. Soc. Mass Spectrom. 2014;26:432–443. doi: 10.1007/s13361-014-1038-1. [DOI] [PubMed] [Google Scholar]
- 19.Fort KL, Dyachenko A, Potel CM, Corradini E, Marino F, Barendregt A, Makarov AA, Scheltema RA, Heck AJR. Implementation of ultraviolet photodissociation on a benchtop Q Exactive mass spectrometer and its application to phosphoproteomics. Anal. Chem. 2016;88:2303–2310. doi: 10.1021/acs.analchem.5b04162. [DOI] [PubMed] [Google Scholar]
- 20.Brodbelt JS. Ion activation methods for peptides and proteins. Anal. Chem. 2016;88:30–51. doi: 10.1021/acs.analchem.5b04563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLuckey SA. Principles of collisional activation in analytical mass spectrometry. J. Am. Soc. Mass Spectrom. 1992;3:599–614. doi: 10.1016/1044-0305(92)85001-Z. [DOI] [PubMed] [Google Scholar]
- 22.Crowe MC, Brodbelt JS. Infrared multiphoton dissociation (IRMPD) and collisionally activated dissociation of peptides in a quadrupole ion trap with selective IRMPD of phosphopeptides. J. Am. Chem. Soc. 2004;15:1581–1592. doi: 10.1016/j.jasms.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Madsen JA, Gardner MW, Smith SI, Ledvina AR, Coon JJ, Schwartz JC, Stafford GC, Brodbelt JS. Top-down protein fragmentation by infrared multiphoton dissociation in a dual pressure linear ion trap. Anal. Chem. 2009;81:8677–8686. doi: 10.1021/ac901554z. [DOI] [PubMed] [Google Scholar]
- 24.Vasicek LA, Ledvina AR, Shaw J, Griep-Raming J, Westphall MS, Coon JJ, Brodbelt JS. Implementing photodissociation in an orbitrap Mass Spectrometer. J. Am. Soc. Mass Spectrom. 2011;22:1105–1108. doi: 10.1007/s13361-011-0119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsybin YO, Witt M, Baykut G, Kjeldsen F, Håkansson P. Combined infrared multiphoton dissociation and electron capture dissociation with a hollow electron beam in Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 2003;17:1759–1768. doi: 10.1002/rcm.1118. [DOI] [PubMed] [Google Scholar]
- 26.Riley NM, Westphall MS, Coon JJ. Activated ion electron transfer dissociation for improved fragmentation of intact proteins. Anal. Chem. 2015;87:7109–7116. doi: 10.1021/acs.analchem.5b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cannon JR, Holden DD, Brodbelt JS. Hybridizing ultraviolet photodissociation with electron transfer dissociation for intact protein characterization. Anal. Chem. 2014;86:10970–10977. doi: 10.1021/ac5036082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swaney DL, McAlister GC, Wirtala M, Schwartz JC, Syka JEP, Coon JJ. Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 2007;79:477–485. doi: 10.1021/ac061457f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frese CK, Altelaar AFM, van den Toorn H, Nolting D, Griep-Raming J, Heck AJR, Mohammed S. Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 2012;84:9668–9673. doi: 10.1021/ac3025366. [DOI] [PubMed] [Google Scholar]
- 30.Fellers RT, Greer JB, Early BP, Yu X, LeDuc RD, Kelleher NL, Thomas PM. ProSight lite: graphical software to analyze top-down mass spectrometry data. Proteomics. 2014;15:1235–1238. doi: 10.1002/pmic.201400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashimoto Y, Hasegawa H, Waki I. High sensitivity and broad dynamic range infrared multiphoton dissociation for a quadrupole ion trap. Rapid Commun. Mass Spectrom. 2004;18:2255–2259. doi: 10.1002/rcm.1619. [DOI] [PubMed] [Google Scholar]
- 32.Ledvina, A.R., Rose, C.M., Mcalister, G.C., Syka, J.E.P., Westphall, M.S., Griep-Raming, J., Schwartz, J.C., Coon, J.J.: Activated ion ETD performed in a modified collision cell on a hybrid QLT-Oribtrap mass spectrometer. J. Am. Soc. Mass Spectrom. 24, 1623–1633 (2013) [DOI] [PMC free article] [PubMed]
- 33.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal. Chem. 2010;82:9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 34.Brodbelt JS, Wilson JJ. Infrared multiphoton dissociation in quadrupole ion traps. Mass Spectrom. Rev. 2009;28:390–424. doi: 10.1002/mas.20216. [DOI] [PubMed] [Google Scholar]
- 35.Ledvina AR, Beauchene NA, McAlister GC, Syka JEP, Schwartz JC, Griep-Raming J, Westphall MS, Coon JJ. Activated-ion electron transfer dissociation improves the ability of electron transfer dissociation to identify peptides in a complex mixture. Anal. Chem. 2010;82:10068–10074. doi: 10.1021/ac1020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Cui W, Thompson MS, Reilly JP. Structures of alpha-type ions formed in the 157 nm photodissociation of singly-charged peptide ions. J. Am. Soc. Mass Spectrom. 2006;17:1315–1321. doi: 10.1016/j.jasms.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 37.Madsen J, Cheng RR, Kaoud TS, Dalby K, Makarov DE, Brodbelt J. Charge-site-dependent dissociation of hydrogen-rich radical peptide cations upon vacuum UV photoexcitation. Chem. Eur. J. 2012;18:5374–5383. doi: 10.1002/chem.201103534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui W, Thompson MS, Reilly JP. Pathways of peptide ion fragmentation induced by vacuum ultraviolet light. J. Am. Soc. Mass Spectrom. 2005;16:1384–1398. doi: 10.1016/j.jasms.2005.03.050. [DOI] [PubMed] [Google Scholar]
- 39.Van Stipdonk M, Kullman M, Berden G, Oomens J. IRMPD and DFT study of the loss of water from protonated 2-hydroxynicotinic acid. Int. J. Mass Spectrom. 2012;330/332:134–143. doi: 10.1016/j.ijms.2012.06.026. [DOI] [Google Scholar]
- 40.Kjeldsen F, Haselmann KF, Budnik BA, Jensen F, Zubarev RA. Dissociative capture of hot (3–13 eV) electrons by polypeptide polycations: an efficient process accompanied by secondary fragmentation. Chem. Phys. Lett. 2002;356:201–206. doi: 10.1016/S0009-2614(02)00149-5. [DOI] [Google Scholar]
- 41.Zhang L, Reilly JP. Peptide photodissociation with 157 nm light in a commercial tandem time-of-flight mass spectrometer. Anal. Chem. 2009;81:7829–7838. doi: 10.1021/ac9012557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Reilly JP. De novo sequencing of tryptic peptides derived from deinococcus radiodurans ribosomal proteins using 157 nm photodissociation MALDI TOF/TOF mass spectrometry. J. Proteome Res. 2010;9:3025–3034. doi: 10.1021/pr901206j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1.72 mb)