Abstract
Fibronectin is a multidomain glycoprotein found ubiquitously in human body fluids and extracellular matrices of a variety of cell types from all human tissues and organs, including intestinal epithelial cells. Fibronectin plays a major role in the regulation of cell migration, tissue repair, and cell adhesion. Importantly, fibronectin also serves as a common target for bacterial adhesins in the gastrointestinal tract. Fibronectin-binding proteins (FnBPs) have been identified and characterized in a wide variety of host-associated bacteria. Single bacterial species can contain multiple, diverse FnBPs. In pathogens, some FnBPs contribute to virulence via host cell attachment, invasion, and interference with signaling pathways. Although FnBPs in commensal and probiotic strains are not sufficient to confer virulence, they are essential for attachment to their ecological niches. Here we describe the interaction between human fibronectin and bacterial adhesins by highlighting the FnBPs of Gram-positive pathogens and commensals. We provide an overview of the occurrence and diversity of FnBPs with a focus on the model pathogenic organisms in which FnBPs are most characterized. Continued investigation of FnBPs is needed to fully understand their divergence and specificity in both pathogens and commensals.
Keywords: fibronectin, Lactobacillus, Acidophilus, lactobacilli, streptococci
Introduction
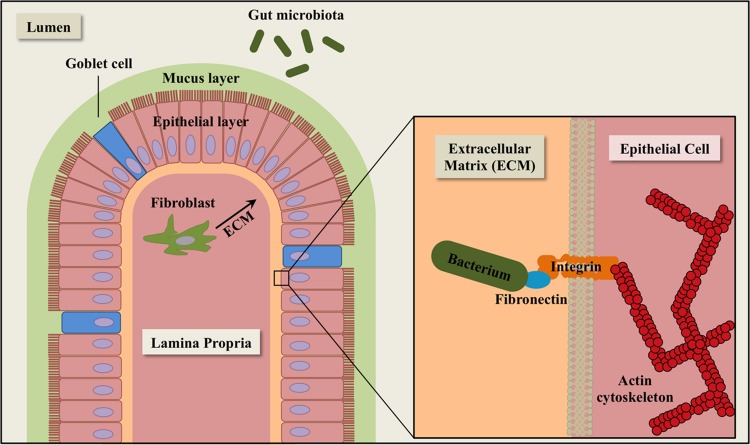
Fibronectin is a multidomain glycoprotein found ubiquitously in human body fluids and extracellular matrices (ECM) of a variety of human tissues and organs, including intestinal epithelial cells (Hynes, 1973; Frantz et al., 2010) (Figure 1). After secretion, fibronectin molecules bind to transmembrane integrins, which facilitate dimerization and cytoskeletal coupling (Schmidt and Friedl, 2010). The integrin-bound fibronectin is capable of binding to ECM components such as collagen and laminin. Human fibronectin plays a major role in the regulation of cell migration, tissue repair, and adhesion. Fibronectin is also a common target for bacterial adhesins in the gastrointestinal tract.
FIGURE 1.
Schematic diagram of extracellular matrix (ECM) components in the intestinal epithelium. The epithelial layer is comprised of simple columnar epithelial cells (pink). Goblet cells (blue) secrete mucin for cell lubrication and protection. Fibroblasts (dark green) synthesize components of the ECM, including fibronectin. The gut microbiota (green ovals) consists of a complex community of microorganisms that inhabit the gastrointestinal tract of animals.
After its discovery in the mid-1970s, fibronectin was described as a non-integral glycoprotein that mediates attachment to fibroblasts and hepatocytes (Hynes, 1973; Klebe, 1974). Researchers first showed that Staphylococcus aureus binds to fibronectin in vitro (Kuusela, 1978). In the nearly 40 years since the discovery of fibronectin-bacterial interactions, fibronectin-binding proteins (FnBPs) have been identified in both Gram-positive and Gram-negative bacteria, including pathogens and commensals. Notably, no common sequence features have been identified among the large collection of known FnBPs. To further complicate the classification of bacterial FnBPs, single bacterial species often contain multiple, diverse FnBPs. In this review, we describe the interaction between human fibronectin structures and bacterial adhesins by highlighting the FnBPs of Gram-positive pathogens and commensals. We provide an overview of the multiplicity and diversity of FnBPs, with a focus on the model pathogenic organisms in which FnBPs are best characterized.
Fibronectin Structure
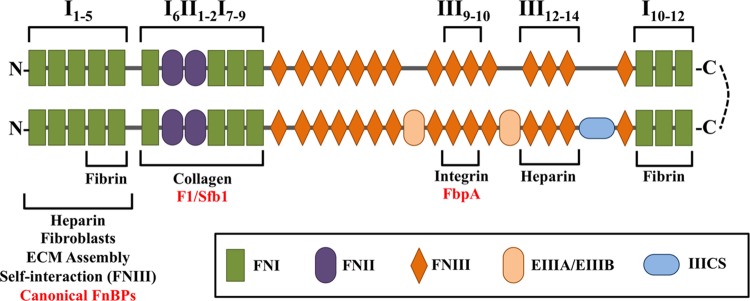
The mature form of fibronectin exists as a heterodimer linked by two C-terminal disulfide bonds (Keski-Oja et al., 1977) (Figure 2). There are two distinct forms of mature fibronectin: soluble and insoluble. Soluble fibronectin is produced by liver cells and secreted into the bloodstream. Meanwhile, fibroblasts and endothelial cells synthesize insoluble, cellular fibronectin. Cellular fibronectin is involved in cell adhesion, migration, and the deposition of other ECM proteins (Knox et al., 1986; Sottile and Hocking, 2002). In general, fibronectin consists of 12 FN type I repeats (FNI), 2 FN type II repeats (FNII), and 15 FN type III repeats (FNIII). The modular structure of insoluble fibronectin can include two alternatively spliced FNIII domains (EIIIA/EIIIB) and one FNIII connecting segment (IIICS). Notably, soluble fibronectin does not contain the EIIIA and EIIIB domains (Tressel et al., 1991; Wilson and Schwarzbauer, 1992). Though both forms of fibronectin are encoded by a single gene, they contain different arrangements of domains due to alternative splicing (Schwarzbauer et al., 1983). In fact, 20 isoforms of insoluble fibronectin have been identified in humans (Ffrench-Constant, 1995). Specific domain organizations are responsible for interaction with other host proteins, including collagen, laminin, integrin, and fibrin (Engvall and Ruoslahti, 1977; McDonald et al., 1982; Hayashi and Yamada, 1983; Tamkun et al., 1986; Potts and Campbell, 1994). Modifications to subdomain structure have been shown to affect structural conformation of fibronectin, thus affecting the presentation of domains (Pickford and Campbell, 2004). Changes in loop structures and domain availability can alter the intricate and specific interactions of fibronectin with its surroundings (Spitzfaden et al., 1997).
FIGURE 2.
Schematic diagram of the multidomain architecture of a cellular fibronectin heterodimer, consisting of 12 FN type I repeats (FNI), 2 FN type II repeats (FNII), and 15 FN type III repeats (FNIII). The lower branch contains splice variants, which can include two alternatively spliced FNIII domains (EIIIA/EIIIB) and one FNIII connecting segment (IIICS). The presence and arrangement of these domains are responsible for interaction with bacterial FnBPs (red) and host proteins (black).
The N-terminal FNI1–FNI5 modules were the first domains in fibronectin shown to interact specifically with bacteria (Mosher and Proctor, 1980). As many FnBPs have since been shown to bind to this region, the FNI1–FNI5 modules represent the canonical bacterial binding site on fibronectin. These domains are also required for binding to heparin, fibroblasts, and fibrin (Sottile et al., 1991; Potts and Campbell, 1994). However, the FNI4–FNI5 modules alone are sufficient to bind fibrin (Matsuka et al., 1994). The FNI1–FNI5 modules are required for proper assembly of the ECM, as well as self-interaction with FNIII domains (Schwarzbauer, 1991; Vakonakis et al., 2009).
The region immediately downstream of the FNI1–FNI5 modules, consisting of the domains FNI6FNII1-2FNI7-9, is necessary for binding collagen (Owens and Baralle, 1986a,b; Banyai et al., 1990). This region is also a non-canonical bacterial binding site for select FnBPs in Streptococcus pyogenes (Sela et al., 1993). Additional non-canonical bacterial binding sites are located at the FNIII12 module and FNIII9–FNIII10 modules, which have been shown to bind FnBPs from Staphylococcus epidermidis and Clostridium perfringens (Christner et al., 2010; Katayama et al., 2015). The FNIII12–FNIII14 modules are necessary for heparin binding, although FNIII13 has been identified as the primary binding site (Novokhatny et al., 1992; Ingham et al., 1993). A second fibrin-binding site is located at the C-terminal FNI10–FNI12 modules (Rostagno et al., 1994; Williams et al., 1994).
Fibronectin attaches to the host cell surface via membrane-spanning α5β1 integrin receptor molecules (Hynes et al., 1987). Integrins bind fibronectin at the RGD loop of the FNIII10 module and the adjacent PHSRN sequence of the FNIII9 module (Tamkun et al., 1986; Aota et al., 1994). By this mechanism, fibronectin, integrin, and FnBPs form a three-component bridge between host cells and bacterial cells (Sinha et al., 1999).
Fibronectin-Binding Proteins
In 1978, researchers showed that S. aureus binds to fibronectin in vitro (Kuusela, 1978; Espersen and Clemmensen, 1982; Froman et al., 1987). The proteins FnBPA and FnBPB were initially identified as FnBPs in S. aureus (Flock et al., 1987; Jonsson et al., 1991). The two proteins contain N-terminal signal peptides with the YSIRK/GS motif that direct the proteins to localize at the cell surface, while a C-terminal region with the LPXTG motif anchors them to the cell wall (Signas et al., 1989; Bae and Schneewind, 2003; DeDent et al., 2008). Once anchored to the cell wall, an array of fibronectin-binding repeats (FnBRs) mediates direct interactions with fibronectin (Schwarz-Linek et al., 2003). Originally, a series of 38-amino acid C-terminal repeats were thought to constitute the FnBPA binding site (Signas et al., 1989). However, the binding site has since expanded to contain 11 tandem repeats in FnBPA and 10 tandem repeats in FnBPB, with each repeat consisting of 30–40 amino acids (Massey et al., 2001; Schwarz-Linek et al., 2003). These domains bind fibronectin with differing affinities at the N-terminal five-module region (FNI1–FNI5) by a tandem β-zipper model (Joh et al., 1994; Meenan et al., 2007). Recent studies examine the structure of FnBPA in complex with fibronectin and reveal the role of each domain in fibronectin attachment (Bingham et al., 2008; Casillas-Ituarte et al., 2012). These findings suggest multivalent binding between a single copy of FnBPA/B and multiple fibronectin molecules.
Studies on FnBPA and FnBPB of S. aureus are guided by an interest in virulence factors of model pathogenic organisms. However, S. aureus expresses many other FnBPs that contribute to the complexity of bacterial adherence to host ligands. The largest of these is 1.1-MDa Ebh (>10,000 amino acids), a surface protein with 44 imperfect repeats of 126 amino acids (Clarke et al., 2002). Ebh is tightly associated with the bacterial cell surface despite the absence of an LPXTG motif. A region within the central repeat sequence has been identified as the binding site for fibronectin (Clarke et al., 2002). Recent studies on S. aureus show that inactivation of Ebh leads to a drastic increase in cell volume with irregular shape and thickness, suggesting Ebh plays a major role in cell growth and envelope assembly (Cheng et al., 2014). An additional FnBP in S. aureus, the 15-kDa cell wall-attached protein Eap, mediates fibronectin binding using an alternative cell wall-anchoring mechanism in which externally added protein can bind cells of S. aureus in addition to a variety of ECM proteins (Braun et al., 1997; Palma et al., 1999). Eap contains a central MAP domain that is presumed to bind fibronectin, S. aureus cells, and a variety of extracellular proteins (Jonsson et al., 1995; Harraghy et al., 2003; Geisbrecht et al., 2005). The ECM-binding protein (Emp) also mediates fibronectin-binding in S. aureus (Hussain et al., 2001). Like Ebh, Emp is tightly associated with the bacterial cell surface despite the absence of an LPXTG motif. Notably, Emp exhibits broad affinity for ECM components, including fibronectin, fibrinogen, collagen, and vitronectin. This highlights an important problem inherent in the study of FnBPs: though they have long been studied with the assumption of single ligand-specificity, a multifunctional model of bacterial adhesins is emerging (Hartleib et al., 2000; Foster et al., 2014). For example, FnBPA binds to fibrinogen and elastin (Wann et al., 2000; Keane et al., 2007); Eap binds vitronectin, fibrinogen, and prothrombin (Jonsson et al., 1995; Harraghy et al., 2003); Aaa binds to vitronectin and fibrinogen (Heilmann et al., 2005; Hirschhausen et al., 2012). Given the limited number of cell wall-associated adhesion proteins and their importance in evasion of host immune responses, cell invasion and biofilm formation, it is expected that FnBPs have evolved to bind multiple ligands (Foster et al., 2014). Furthermore, the apparent functional redundancy of FnBPs makes it difficult to attribute definitive adhesion phenotypes.
While many of the FnBPs in S. aureus are conserved across staphylococci, other Gram-positive bacteria possess an entirely different collection of FnBPs. The human pathogen S. pyogenes, for example, expresses at least 11 additional distinct FnBPs (Henderson et al., 2011). Perhaps the most studied of these is a set of homologous proteins, F1 and Sfb1 (Talay et al., 1991; Hanski and Caparon, 1992). As with many of the S. aureus FnBPs, both F1 and Sfb1 are cell wall-anchored. Another shared feature between S. aureus FnBPs and F1/Sfb1 is a series of central FnBRs similar to those observed in FnBPA/FnBPB (Ozeri et al., 1998). Like FnBPA/B, the FnBRs of F1/Sfb1 bind to fibronectin at the N-terminal FNI1-FNI5 region (Schwarz-Linek et al., 2004). In F1/Sfb1, a 43-amino acid N-terminal region also binds fibronectin, but at modules FNI6–FNI9 (Sela et al., 1993) (Figure 2).
Fibronectin-binding repeats with sequence similarity to those in S. aureus have been found in other FnBPs from S. pyogenes, including F2, FbaB, Sof, SfbX, and FbaA (Henderson et al., 2011). F2 is similar to F1, though it lacks the domain for binding modules FNI6–FNI9 (Kreikemeyer et al., 2004). FbaB shows homology to the C-terminal domain of protein F2 (Terao et al., 2002). Although serum opacity factor (Sof) contains functional FnBRs, an additional N-terminal opacity domain is necessary for cell binding (Rakonjac et al., 1995). SfbX features a C-terminal array of four FnBRs. The sfbX gene, which occurs immediately downstream of sof, is found only in sof-positive streptococci (Jeng et al., 2003). The dominant theme in this set of FnBPs is the role of FnBRs in binding the N-terminal domain of fibronectin (FNI1–FNI5). Furthermore, these proteins contain C-terminal LPXTG cell wall anchors.
A second subset of FnBPs in S. pyogenes and other streptococci do not possess the canonical FnBRs. These include the M1 protein, GAPDH, protein H, Shr, and Scl1. Protein M1 anchors to the cell wall by an LPXTG motif binds fibronectin with two N-terminal domains (Cue et al., 2001). Unlike the other FnBPs discussed, protein H binds to FNIII modules instead of FNI modules (Frick et al., 1995). Glyceraldehyde-3-phosphate-dehydrogenase (GAPDH) also shows fibronectin-binding activity (Pancholi and Fischetti, 1992). Shr and Scl1 are relatively new additions to the non-FnBR subset of S. pyogenes FnBPs (Fisher et al., 2008; Caswell et al., 2010). The streptococcal surface enolase, a glycolytic pathway enzyme with plasminogen-binding capability, has been identified as a FnBP in S. suis (Pancholi and Fischetti, 1998; Esgleas et al., 2008). More recently a putative peptidase (Ssa) in S. suis and an endopeptidase (PepO) in S. pneumoniae have been implicated in fibronectin-binding (Agarwal et al., 2013; Li et al., 2013). The discovery of these novel FnBPs represents a new paradigm in which bacterial proteins with other known functions double as FnBPs.
A 54-kDa protein was originally identified in streptococci and termed Fbp54 after it was shown to bind to fibronectin and fibrinogen, despite a lack of typical fibronectin-binding sequences (Courtney et al., 1994). Since the initial characterization of Fbp54, distant homologs have been found among a variety of host-associated bacteria including streptococci, lactococci, lactobacilli, clostridia, listeria, pneumococci, enterococci, and bacilli. There has been inconsistency in the naming of Fbp54 homologs, such as PavA in S. pneumoniae, FbpA in S. gordonii, and FbpS is S. suis (Holmes et al., 2001; Christie et al., 2002; de Greeff et al., 2002). This has led to confusion about the prevalence and identity of this FnBP. The Gram-positive pathogen C. perfringens, a common cause of wound-associated infections and food poisoning, also expresses an Fbp54 homolog (FbpA). FbpA recognizes a non-canonical FNIII9-FNIII10 region of fibronectin (Katayama et al., 2009, Katayama et al., 2015). Despite its ubiquity, little is known about the binding mechanism of the Fbp54 family of FnBPs in other organisms.
Host Interactions
The ability to attach to the surface of host cells, followed by entry and proliferation, can lead to severe host diseases specifically mediated by FnBPs (Joh et al., 1999; Lammers et al., 1999; Henderson et al., 2011; Ribet and Cossart, 2015; Stones and Krachler, 2015). Pathogenic strains of staphylococci are one of the most common causes of skin and bloodstream infections in the United States (Lowy, 1998; Wisplinghoff et al., 2004; Moran et al., 2005; Tong et al., 2015). Bacterial cells use FnBPs to form a three-component bridge between themselves and the host cell through attachment to fibronectin molecules, which are further attached to α5β1 integrins (Tamkun et al., 1986; Hynes et al., 1987). The linkage between integrins and the bacteria-fibronectin complex brings about the recruitment of cell signaling molecules and a rearrangement of the cytoskeleton that facilitates host cell invasion (Hoffmann et al., 2011). The absence of FnBPA/B in S. aureus leads to a nearly 500-fold reduction in the internalization of bacteria (Sinha et al., 2000). Importantly, expression of S. aureus FnBPA in non-invasive Lactococcus lactis bacteria confers the ability to invade human endothelial cells (Heying et al., 2009).
The same mechanism of host cell invasion via integrin-binding is observed in streptococci (LaPenta et al., 1994; Molinari et al., 1997). Protein F1 and Sfb1 of S. pyogenes interact with fibronectin on the surface of non-phagocytic cells to trigger bacterial internalization (Molinari et al., 1997; Jadoun et al., 1998; Ozeri et al., 1998). Though not as essential as protein F1 and Sfb1, other FnBPs such as FbaA, FbaB, Ssa, and protein M1 promote cell invasion (Henderson et al., 2011; Li et al., 2013). Because fibronectin interacts with integrin by means of its RGD peptide, it has been proposed that FnBPs with the RGD integrin attachment domain, such as FbaB, interact directly with integrin (Lamont, 2004).
Arguably the most prevalent FnBP, Fbp54 and its homologs (FbpA, FbpS, and PavA) play an important role in virulence-associated internalization (Holmes et al., 2001). An fbpA-deficient mutant of Listeria monocytogenes exhibited a reduced ability to invade hepatocytes (Dramsi et al., 2004; Osanai et al., 2013). A pavA-deficient mutant of S. pneumoniae exhibited a similar decrease in adherence and internalization ability (Pracht et al., 2005). Recent evidence suggests that staphylococcal FnBPs are also required to form biofilms. A homolog of the 1.1-MDa S. aureus FnBP (Ebh) was identified in S. epidermidis and found to be sufficient and necessary for biofilm formation (Christner et al., 2010). The introduction of mutations into fnbpA and fnbpB, encoding FnBPA and FnBPB, reduced biofilm formation in multiple methicillin-resistant strains of S. aureus (O’Neill et al., 2008). A full deletion of fnbpA and fnbpB from S. aureus also reduced biofilm formation, highlighting reduced initial bacterial aggregation as the underlying mechanism (McCourt et al., 2014). Further evidence suggests that low-affinity homophilic interactions between FnBPA domains on adjacent cells promote cell accumulation and contribute to biofilm formation (Herman-Bausier et al., 2015).
In addition to exploiting fibronectin as a method of host cell attachment and invasion, bacterial FnBPs can modify the signaling activity of human fibronectin. Fragments of fibronectin are often found in the blood after injury or infection (Clark et al., 1982). These fragments are important for host cell signaling and have been linked to essential biological functions (Woods et al., 1986; Hanenberg et al., 1996). Fibronectin fragments of 110 kDa stimulate human macrophages in vitro, significantly increasing output of TNF-alpha, FGF-1, IGF-1, and LIF (Trial et al., 2004a). Fibronectin fragments can also influence monocyte behavior in HIV-1-infected patients (Trial et al., 2004b). The role of fibronectin fragments in biological processes appears to be shaped by the domains present on the fibronectin fragment. For example, the alternatively spliced EIIIA domain is associated with cell motility and fibrosis. However, the EIIIA domain is non-essential for differentiation of hepatic stellate cells and portal fibroblasts to myofibroblasts (Olsen et al., 2012).
Smaller sequences within fibronectin domains have also been linked with specific biological functions. A 13-residue stretch of fibronectin (FN13) is responsible for inducing matrix assembly in cultured cells. In the absence of this peptide, migration of tumorigenic cells is inhibited (Colombi et al., 2003). An N-terminal 29-kDa fragment of fibronectin increases phosphorylation of ERK1/2, p38 and JNK1/2 protein kinases, leading to enhanced cartilage matrix damage (Ding et al., 2009). Larger fibronectin fragments of 50 and 140-kDa show less kinase activation, though all three fragments show significantly more activity than native fibronectin, which is inactive in terms of cartilage degradation (Ding et al., 2008). In binding these fragments, FnBPs may interfere with host cell signaling. A 49-residue sequence of the F1 protein in S. pyogenes binds the N-terminal 70-kDa region of fibronectin and inhibits matrix assembly (Tomasini-Johansson et al., 2001). This interaction illustrates the ability of FnBPs to block the activity of fibronectin fragments.
It is important to note that because fibronectin is produced at basolateral surfaces, bacteria must bypass the epithelial barrier to gain access. However, adenosine, a proinflammatory signaling molecule, induces transport of fibronectin to the apical surface where it is accessible to bacteria (Walia et al., 2004). Adenosine-induced apical display was shown to facilitate the adherence and consequent invasion of Salmonella enterica. By this mechanism, other signaling molecules could induce apical display of fibronectin, providing an ecological advantage to species with FnBPs.
Non-Pathogenic FnBPs
In both pathogenic and commensal bacteria, host attachment allows access to nutrients, suitable environmental conditions, and interaction with the host immune system by promoting retention in a particular niche. The diverse array of FnBPs identified in pathogens is unparalleled in commensals, though some FnBPs are expressed in both pathogens and commensal species. The clearest example is Fbp54, which is found across a variety of host-associated commensals, as well as the probiotic species Lactobacillus acidophilus, L. casei, L. plantarum, L. brevis, L. rhamnosus, and Bacillus subtilis (Altermann et al., 2005; Boekhorst et al., 2006; Velez et al., 2007; Munoz-Provencio et al., 2010). Purified FbpA from L. casei exhibits a stronger affinity for immobilized fibronectin than soluble fibronectin — a trend also seen in the FbpA homolog of S. pneumoniae (Holmes et al., 2001; Munoz-Provencio et al., 2010). In L. acidophilus, a mutant with inactivated fbpA exhibited a significant decrease in adhesion to epithelial cells in vitro (Buck et al., 2005).
A subset of lactobacilli forms surface layers (S-layers) that are crystalline arrays self-assembling, proteinaceous subunits called S-layer proteins (Boot and Pouwels, 1996; Sara and Sleytr, 2000). S-layer proteins are important for protection, cell shape, immunomodulation, and adhesion (Sara and Sleytr, 2000; Buck et al., 2005; Hynönen and Palva, 2013; Lightfoot et al., 2015). The S-layer protein in L. brevis (SlpA) binds fibronectin, while inactivation of the S-layer protein in L. acidophilus (SlpA) reduced binding to epithelial cells (Hynonen et al., 2002; Buck et al., 2005). Although SlpA has not been further investigated for specific fibronectin-binding, the recent identification of S-layer associated proteins (SLAPs) in L. acidophilus has led to the implication of an additional FnBP, termed FbpB (Johnson et al., 2013; Hymes et al., 2016). FbpB contains an FNIII domain, which bears homology to the FNIII domain of human fibronectin. This suggests that FbpB may interact with the self-binding region of fibronectin (FNI1–FNI5) known to target the FNIII domain (Vakonakis et al., 2009). Strikingly, homologs of FbpB are found only within the S-layer-forming subset of gut-associated lactobacilli. The unique FnBPs of lactobacilli and other non-pathogens may possess distinctive mechanisms to bind fibronectin in competition with pathogens.
Concluding Remarks
There appears to be fewer FnBPs in commensals than pathogens, but this is likely due to sampling bias: pathogen “virulence factors” have been studied more often than commensal adhesins. Consequently, commensal and probiotic FnBPs are less understood than the FnBPs in pathogenic bacteria. Due to the presence of so-called “virulence factors” in commensals, it may be more accurate to refer to bacterial adhesins as “niche factors,” as suggested in Hill (2012). It is proposed that attachment proteins be categorized as niche factors because they are found in both pathogens and commensals that occupy an identical niche. However, proteins unique to pathogens that play a significant role in pathogenesis, such as exotoxins or coagulases, would remain classified as virulence factors. Addressing these concerns will be important from a regulatory perspective, as the probiotic potential of gut microbes is being increasingly investigated.
Bacteria employ adhesins as a means of attachment to their ecological niches. Adhesins play an important role in competition between organisms on host cell surfaces. The evolution of diverse FnBPs that interact with distinct regions of human fibronectin would likely provide an advantage to a bacterial species. Advances in genome sequencing technologies will enable extensive characterization of FnBPs in a growing number of microorganisms. The continued investigation of FnBPs will enhance our understanding of their diversity and specificity.
Author Contributions
TK, project design and management. JH, research scientist and review author.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewers MS and AH and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Footnotes
Funding. This research and the graduate stipend of JH was supported by the North Carolina Agricultural Foundation, and Danisco/Dupont Nutrition and Health.
References
- Agarwal V., Kuchipudi A., Fulde M., Riesbeck K., Bergmann S., Blom A. M. (2013). Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J. Biol. Chem. 288 6849–6863. 10.1074/jbc.M112.405530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altermann E., Russell W. M., Azcarate-Peril M. A., Barrangou R., Buck B. L., McAuliffe O., et al. (2005). Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U.S.A. 102 3906–3912. 10.1073/pnas.0409188102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aota S., Nomizu M., Yamada K. M. (1994). The short amino acid sequence Pro-His-Ser-Arg-Asn in human fibronectin enhances cell-adhesive function. J. Biol. Chem. 269 24756–24761. [PubMed] [Google Scholar]
- Bae T., Schneewind O. (2003). The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J. Bacteriol. 185 2910–2919. 10.1128/JB.185.9.2910-2919.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyai L., Trexler M., Koncz S., Gyenes M., Sipos G., Patthy L. (1990). The collagen-binding site of type-II units of bovine seminal fluid protein PDC-109 and fibronectin. Eur. J. Biochem. 193 801–806. 10.1111/j.1432-1033.1990.tb19403.x [DOI] [PubMed] [Google Scholar]
- Bingham R. J., Rudino-Pinera E., Meenan N. A., Schwarz-Linek U., Turkenburg J. P., Hook M., et al. (2008). Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc. Natl. Acad. Sci. U.S.A. 105 12254–12258. 10.1073/pnas.0803556105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhorst J., Wels M., Kleerebezem M., Siezen R. J. (2006). The predicted secretome of Lactobacillus plantarum WCFS1 sheds light on interactions with its environment. Microbiology 152(Pt 11) 3175–3183. 10.1099/mic.0.29217-0 [DOI] [PubMed] [Google Scholar]
- Boot H. J., Pouwels P. H. (1996). Expression, secretion and antigenic variation of bacterial S-layer proteins. Mol. Microbiol. 21 1117–1123. 10.1046/j.1365-2958.1996.711442.x [DOI] [PubMed] [Google Scholar]
- Braun L., Dramsi S., Dehoux P., Bierne H., Lindahl G., Cossart P. (1997). InlB: an invasion protein of Listeria monocytogenes with a novel type of surface association. Mol. Microbiol. 25 285–294. 10.1046/j.1365-2958.1997.4621825.x [DOI] [PubMed] [Google Scholar]
- Buck B. L., Altermann E., Svingerud T., Klaenhammer T. R. (2005). Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71 8344–8351. 10.1128/AEM.71.12.8344-8351.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casillas-Ituarte N. N., Lower B. H., Lamlertthon S., Fowler V. G., Jr., Lower S. K. (2012). Dissociation rate constants of human fibronectin binding to fibronectin-binding proteins on living Staphylococcus aureus isolated from clinical patients. J. Biol. Chem. 287 6693–6701. 10.1074/jbc.M111.285692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell C. C., Oliver-Kozup H., Han R., Lukomska E., Lukomski S. (2010). Scl1 the multifunctional adhesin of group A Streptococcus, selectively binds cellular fibronectin and laminin, and mediates pathogen internalization by human cells. FEMS Microbiol. Lett. 303 61–68. 10.1111/j.1574-6968.2009.01864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. G., Missiakas D., Schneewind O. (2014). The giant protein Ebh is a determinant of Staphylococcus aureus cell size and complement resistance. J. Bacteriol. 196 971–981. 10.1128/JB.01366-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie J., McNab R., Jenkinson H. F. (2002). Expression of fibronectin-binding protein FbpA modulates adhesion in Streptococcus gordonii. Microbiology 148(Pt 6) 1615–1625. 10.1099/00221287-148-6-1615 [DOI] [PubMed] [Google Scholar]
- Christner M., Franke G. C., Schommer N. N., Wendt U., Wegert K., Pehle P., et al. (2010). The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol. Microbiol. 75 187–207. 10.1111/j.1365-2958.2009.06981.x [DOI] [PubMed] [Google Scholar]
- Clark R. A., Quinn J. H., Winn H. J., Lanigan J. M., Dellepella P., Colvin R. B. (1982). Fibronectin is produced by blood vessels in response to injury. J. Exp. Med. 156 646–651. 10.1084/jem.156.2.646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. R., Harris L. G., Richards R. G., Foster S. J. (2002). Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect. Immun. 70 6680–6687. 10.1128/IAI.70.12.6680-6687.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombi M., Zoppi N., De Petro G., Marchina E., Gardella R., Tavian D., et al. (2003). Matrix assembly induction and cell migration and invasion inhibition by a 13-amino acid fibronectin peptide. J. Biol. Chem. 278 14346–14355. 10.1074/jbc.M211997200 [DOI] [PubMed] [Google Scholar]
- Courtney H. S., Li Y., Dale J. B., Hasty D. L. (1994). Cloning, sequencing, and expression of a fibronectin/fibrinogen-binding protein from group A streptococci. Infect. Immun. 62 3937–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Lam H., Cleary P. P. (2001). Genetic dissection of the Streptococcus pyogenes M1 protein: regions involved in fibronectin binding and intracellular invasion. Microb. Pathog. 31 231–242. 10.1006/mpat.2001.0467 [DOI] [PubMed] [Google Scholar]
- de Greeff A., Buys H., Verhaar R., Dijkstra J., van Alphen L., Smith H. E. (2002). Contribution of fibronectin-binding protein to pathogenesis of Streptococcus suis serotype 2. Infect. Immun. 70 1319–1325. 10.1128/IAI.70.3.1319-1325.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeDent A., Bae T., Missiakas D. M., Schneewind O. (2008). Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 27 2656–2668. 10.1038/emboj.2008.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Guo D., Homandberg G. A. (2008). The cartilage chondrolytic mechanism of fibronectin fragments involves MAP kinases: comparison of three fragments and native fibronectin. Osteoarthritis Cartilage 16 1253–1262. 10.1016/j.joca.2008.02.015 [DOI] [PubMed] [Google Scholar]
- Ding L., Guo D., Homandberg G. A. (2009). Fibronectin fragments mediate matrix metalloproteinase upregulation and cartilage damage through proline rich tyrosine kinase 2 c-src, NF-kappaB and protein kinase Cdelta. Osteoarthritis Cartilage 17 1385–1392. 10.1016/j.joca.2009.03.024 [DOI] [PubMed] [Google Scholar]
- Dramsi S., Bourdichon F., Cabanes D., Lecuit M., Fsihi H., Cossart P. (2004). FbpA, a novel multifunctional Listeria monocytogenes virulence factor. Mol. Microbiol. 53 639–649. 10.1111/j.1365-2958.2004.04138.x [DOI] [PubMed] [Google Scholar]
- Engvall E., Ruoslahti E. (1977). Binding of soluble form of fibroblast surface protein, fibronectin, to collagen. Int. J. Cancer 20 1–5. 10.1002/ijc.2910200102 [DOI] [PubMed] [Google Scholar]
- Esgleas M., Li Y., Hancock M. A., Harel J., Dubreuil J. D., Gottschalk M. (2008). Isolation and characterization of alpha-enolase, a novel fibronectin-binding protein from Streptococcus suis. Microbiology 154(Pt 9) 2668–2679. 10.1099/mic.0.2008/017145-0 [DOI] [PubMed] [Google Scholar]
- Espersen F., Clemmensen I. (1982). Isolation of a fibronectin-binding protein from Staphylococcus aureus. Infect. Immun. 37 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant C. (1995). Alternative splicing of fibronectin—many different proteins but few different functions. Exp. Cell Res. 221 261–271. 10.1006/excr.1995.1374 [DOI] [PubMed] [Google Scholar]
- Fisher M., Huang Y. S., Li X., McIver K. S., Toukoki C., Eichenbaum Z. (2008). Shr is a broad-spectrum surface receptor that contributes to adherence and virulence in group A streptococcus. Infect. Immun. 76 5006–5015. 10.1128/IAI.00300-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock J. I., Froman G., Jonsson K., Guss B., Signas C., Nilsson B., et al. (1987). Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J. 6 2351–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Geoghegan J. A., Ganesh V. K., Hook M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12 49–62. 10.1038/nrmicro3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz C., Stewart K. M., Weaver V. M. (2010). The extracellular matrix at a glance. J. Cell Sci. 123(Pt 24) 4195–4200. 10.1242/jcs.023820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick I. M., Crossin K. L., Edelman G. M., Bjorck L. (1995). Protein H–a bacterial surface protein with affinity for both immunoglobulin and fibronectin type III domains. EMBO J. 14 1674–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froman G., Switalski L. M., Speziale P., Hook M. (1987). Isolation and characterization of a fibronectin receptor from Staphylococcus aureus. J. Biol. Chem. 262 6564–6571. [PubMed] [Google Scholar]
- Geisbrecht B. V., Hamaoka B. Y., Perman B., Zemla A., Leahy D. J. (2005). The crystal structures of EAP domains from Staphylococcus aureus reveal an unexpected homology to bacterial superantigens. J. Biol. Chem. 280 17243–17250. 10.1074/jbc.M412311200 [DOI] [PubMed] [Google Scholar]
- Hanenberg H., Xiao X. L., Dilloo D., Hashino K., Kato I., Williams D. A. (1996). Colocalization of retrovirus and target cells on specific fibronectin fragments increases genetic transduction of mammalian cells. Nat. Med. 2 876–882. 10.1038/nm0896-876 [DOI] [PubMed] [Google Scholar]
- Hanski E., Caparon M. (1992). Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. U.S.A. 89 6172–6176. 10.1073/pnas.89.13.6172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harraghy N., Hussain M., Haggar A., Chavakis T., Sinha B., Herrmann M., et al. (2003). The adhesive and immunomodulating properties of the multifunctional Staphylococcus aureus protein Eap. Microbiology 149(Pt 10) 2701–2707. 10.1099/mic.0.26465-0 [DOI] [PubMed] [Google Scholar]
- Hartleib J., Kohler N., Dickinson R. B., Chhatwal G. S., Sixma J. J., Hartford O. M., et al. (2000). Protein A is the von Willebrand factor binding protein on Staphylococcus aureus. Blood 96 2149–2156. [PubMed] [Google Scholar]
- Hayashi M., Yamada K. M. (1983). Domain structure of the carboxyl-terminal half of human plasma fibronectin. J. Biol. Chem. 258 3332–3340. [PubMed] [Google Scholar]
- Heilmann C., Hartleib J., Hussain M. S., Peters G. (2005). The multifunctional Staphylococcus aureus autolysin aaa mediates adherence to immobilized fibrinogen and fibronectin. Infect. Immun. 73 4793–4802. 10.1128/IAI.73.8.4793-4802.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson B., Nair S., Pallas J., Williams M. A. (2011). Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol. Rev. 35 147–200. 10.1111/j.1574-6976.2010.00243.x [DOI] [PubMed] [Google Scholar]
- Herman-Bausier P., El-Kirat-Chatel S., Foster T. J., Geoghegan J. A., Dufrene Y. F. (2015). Staphylococcus aureus fibronectin-binding protein A mediates cell-cell adhesion through low-affinity homophilic bonds. MBio 6 e00413-15. 10.1128/mBio.00413-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heying R., van de Gevel J., Que Y. A., Piroth L., Moreillon P., Beekhuizen H. (2009). Contribution of (sub)domains of Staphylococcus aureus fibronectin-binding protein to the proinflammatory and procoagulant response of human vascular endothelial cells. Thromb. Haemost. 101 495–504. [PubMed] [Google Scholar]
- Hill C. (2012). Virulence or niche factors: what’s in a name? J. Bacteriol. 194 5725–5727. 10.1128/JB.00980-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhausen N., Schlesier T., Peters G., Heilmann C. (2012). Characterization of the modular design of the autolysin/adhesin Aaa from Staphylococcus aureus. PLoS ONE 7:e40353 10.1371/journal.pone.0040353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann C., Ohlsen K., Hauck C. R. (2011). Integrin-mediated uptake of fibronectin-binding bacteria. Eur. J. Cell Biol. 90 891–896. 10.1016/j.ejcb.2011.03.001 [DOI] [PubMed] [Google Scholar]
- Holmes A. R., McNab R., Millsap K. W., Rohde M., Hammerschmidt S., Mawdsley J. L., et al. (2001). The pavA gene of Streptococcus pneumoniae encodes a fibronectin-binding protein that is essential for virulence. Mol. Microbiol. 41 1395–1408. 10.1046/j.1365-2958.2001.02610.x [DOI] [PubMed] [Google Scholar]
- Hussain M., Becker K., von Eiff C., Schrenzel J., Peters G., Herrmann M. (2001). Identification and characterization of a novel 38.5-kilodalton cell surface protein of Staphylococcus aureus with extended-spectrum binding activity for extracellular matrix and plasma proteins. J. Bacteriol. 183 6778–6786. 10.1128/JB.183.23.6778-6786.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymes J. P., Johnson B. R., Barrangou R., Klaenhammer T. R. (2016). Functional analysis of an S-layer-associated fibronectin-binding protein in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 82 2676–2685. 10.1128/AEM.00024-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O. (1973). Alteration of cell-surface proteins by viral transformation and by proteolysis. Proc. Natl. Acad. Sci. U.S.A. 70 3170–3174. 10.1073/pnas.70.11.3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R. O., Schwarzbauer J. E., Tamkun J. W. (1987). Isolation and analysis of cDNA and genomic clones of fibronectin and its receptor. Methods Enzymol. 144 447–463. 10.1016/0076-6879(87)44194-3 [DOI] [PubMed] [Google Scholar]
- Hynönen U., Palva A. (2013). Lactobacillus surface layer proteins: structure, function and applications. Appl. Microbiol. Biotechnol. 97 5225–5243. 10.1007/s00253-013-4962-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynonen U., Westerlund-Wikstrom B., Palva A., Korhonen T. K. (2002). Identification by flagellum display of an epithelial cell- and fibronectin-binding function in the SlpA surface protein of Lactobacillus brevis. J. Bacteriol. 184 3360–3367. 10.1128/JB.184.12.3360-3367.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham K. C., Brew S. A., Migliorini M. M., Busby T. F. (1993). Binding of heparin by type III domains and peptides from the carboxy terminal hep-2 region of fibronectin. Biochemistry 32 12548–12553. 10.1021/bi00097a035 [DOI] [PubMed] [Google Scholar]
- Jadoun J., Ozeri V., Burstein E., Skutelsky E., Hanski E., Sela S. (1998). Protein F1 is required for efficient entry of Streptococcus pyogenes into epithelial cells. J. Infect. Dis. 178 147–158. 10.1086/515589 [DOI] [PubMed] [Google Scholar]
- Jeng A., Sakota V., Li Z., Datta V., Beall B., Nizet V. (2003). Molecular genetic analysis of a group A Streptococcus operon encoding serum opacity factor and a novel fibronectin-binding protein, SfbX. J. Bacteriol. 185 1208–1217. 10.1128/JB.185.4.1208-1217.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joh D., Wann E. R., Kreikemeyer B., Speziale P., Hook M. (1999). Role of fibronectin-binding MSCRAMMs in bacterial adherence and entry into mammalian cells. Matrix Biol. 18 211–223. 10.1016/S0945-053X(99)00025-6 [DOI] [PubMed] [Google Scholar]
- Joh H. J., House-Pompeo K., Patti J. M., Gurusiddappa S., Hook M. (1994). Fibronectin receptors from gram-positive bacteria: comparison of active sites. Biochemistry 33 6086–6092. 10.1021/bi00186a007 [DOI] [PubMed] [Google Scholar]
- Johnson B., Selle K., O’Flaherty S., Goh Y. J., Klaenhammer T. (2013). Identification of extracellular surface-layer associated proteins in Lactobacillus acidophilus NCFM. Microbiology 159(Pt 11) 2269–2282. 10.1099/mic.0.070755-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson K., McDevitt D., McGavin M. H., Patti J. M., Hook M. (1995). Staphylococcus aureus expresses a major histocompatibility complex class II analog. J. Biol. Chem. 270 21457–21460. 10.1074/jbc.270.37.21457 [DOI] [PubMed] [Google Scholar]
- Jonsson K., Signas C., Muller H. P., Lindberg M. (1991). Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202 1041–1048. 10.1111/j.1432-1033.1991.tb16468.x [DOI] [PubMed] [Google Scholar]
- Katayama S., Nozu N., Okuda M., Hirota S., Yamasaki T., Hitsumoto Y. (2009). Characterization of two putative fibronectin-binding proteins of Clostridium perfringens. Anaerobe 15 155–159. 10.1016/j.anaerobe.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Katayama S., Tagomori M., Morita N., Yamasaki T., Nariya H., Okada M., et al. (2015). Determination of the Clostridium perfringens-binding site on fibronectin. Anaerobe 34 174–181. 10.1016/j.anaerobe.2014.11.007 [DOI] [PubMed] [Google Scholar]
- Keane F. M., Loughman A., Valtulina V., Brennan M., Speziale P., Foster T. J. (2007). Fibrinogen and elastin bind to the same region within the A domain of fibronectin binding protein A, an MSCRAMM of Staphylococcus aureus. Mol. Microbiol. 63 711–723. 10.1111/j.1365-2958.2006.05552.x [DOI] [PubMed] [Google Scholar]
- Keski-Oja J., Mosher D. F., Vaheri A. (1977). Dimeric character of fibronectin, a major cell surface-associated glycoprotein. Biochem. Biophys. Res. Commun. 74 699–706. 10.1016/0006-291X(77)90359-X [DOI] [PubMed] [Google Scholar]
- Klebe R. J. (1974). Isolation of a collagen-dependent cell attachment factor. Nature 250 248–251. 10.1038/250248a0 [DOI] [PubMed] [Google Scholar]
- Knox P., Crooks S., Rimmer C. S. (1986). Role of fibronectin in the migration of fibroblasts into plasma clots. J. Cell Biol. 102 2318–2323. 10.1083/jcb.102.6.2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B., Oehmcke S., Nakata M., Hoffrogge R., Podbielski A. (2004). Streptococcus pyogenes fibronectin-binding protein F2: expression profile, binding characteristics, and impact on eukaryotic cell interactions. J. Biol. Chem. 279 15850–15859. 10.1074/jbc.M313613200 [DOI] [PubMed] [Google Scholar]
- Kuusela P. (1978). Fibronectin binds to Staphylococcus aureus. Nature 276 718–720. 10.1038/276718a0 [DOI] [PubMed] [Google Scholar]
- Lammers A., Nuijten P. J., Smith H. E. (1999). The fibronectin binding proteins of Staphylococcus aureus are required for adhesion to and invasion of bovine mammary gland cells. FEMS Microbiol. Lett. 180 103–109. 10.1111/j.1574-6968.1999.tb08783.x [DOI] [PubMed] [Google Scholar]
- Lamont R. J. (2004). Bacterial Invasion of Host Cells. New York, NY: Cambridge University Press. [Google Scholar]
- LaPenta D., Rubens C., Chi E., Cleary P. P. (1994). Group A streptococci efficiently invade human respiratory epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 91 12115–12119. 10.1073/pnas.91.25.12115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wan Y., Tao Z., Chen H., Zhou R. (2013). A novel fibronectin-binding protein of Streptococcus suis serotype 2 contributes to epithelial cell invasion and in vivo dissemination. Vet. Microbiol. 162 186–194. 10.1016/j.vetmic.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Lightfoot Y. L., Selle K., Yang T., Goh Y. J., Sahay B., Zadeh M., et al. (2015). SIGNR3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J. 34 881–895. 10.15252/embj.201490296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339 520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Massey R. C., Kantzanou M. N., Fowler T., Day N. P., Schofield K., Wann E. R., et al. (2001). Fibronectin-binding protein A of Staphylococcus aureus has multiple, substituting, binding regions that mediate adherence to fibronectin and invasion of endothelial cells. Cell. Microbiol. 3 839–851. 10.1046/j.1462-5822.2001.00157.x [DOI] [PubMed] [Google Scholar]
- Matsuka Y. V., Medved L. V., Brew S. A., Ingham K. C. (1994). The NH2-terminal fibrin-binding site of fibronectin is formed by interacting fourth and fifth finger domains. Studies with recombinant finger fragments expressed in Escherichia coli. J. Biol. Chem. 269 9539–9546. [PubMed] [Google Scholar]
- McCourt J., O’Halloran D. P., McCarthy H., O’Gara J. P., Geoghegan J. A. (2014). Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol. Lett. 353 157–164. 10.1111/1574-6968.12424 [DOI] [PubMed] [Google Scholar]
- McDonald J. A., Kelley D. G., Broekelmann T. J. (1982). Role of fibronectin in collagen deposition: Fab’ to the gelatin-binding domain of fibronectin inhibits both fibronectin and collagen organization in fibroblast extracellular matrix. J. Cell Biol. 92 485–492. 10.1083/jcb.92.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meenan N. A., Visai L., Valtulina V., Schwarz-Linek U., Norris N. C., Gurusiddappa S., et al. (2007). The tandem beta-zipper model defines high affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J. Biol. Chem. 282 25893–25902. 10.1074/jbc.M703063200 [DOI] [PubMed] [Google Scholar]
- Molinari G., Talay S. R., Valentin-Weigand P., Rohde M., Chhatwal G. S. (1997). The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect. Immun. 65 1357–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran G. J., Amii R. N., Abrahamian F. M., Talan D. A. (2005). Methicillin-resistant Staphylococcus aureus in community-acquired skin infections. Emerg. Infect. Dis. 11 928–930. 10.3201/eid1106.040641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher D. F., Proctor R. A. (1980). Binding and factor XIIIa-mediated cross-linking of a 27-kilodalton fragment of fibronectin to Staphylococcus aureus. Science 209 927–929. 10.1126/science.7403857 [DOI] [PubMed] [Google Scholar]
- Munoz-Provencio D., Perez-Martinez G., Monedero V. (2010). Characterization of a fibronectin-binding protein from Lactobacillus casei BL23. J. Appl. Microbiol. 108 1050–1059. 10.1111/j.1365-2672.2009.04508.x [DOI] [PubMed] [Google Scholar]
- Novokhatny V., Schwarz F., Atha D., Ingham K. (1992). Domain structure and domain-domain interactions in the carboxy-terminal heparin binding region of fibronectin. J. Mol. Biol. 227 1182–1191. 10.1016/0022-2836(92)90530-W [DOI] [PubMed] [Google Scholar]
- Olsen A. L., Sackey B. K., Marcinkiewicz C., Boettiger D., Wells R. G. (2012). Fibronectin extra domain-A promotes hepatic stellate cell motility but not differentiation into myofibroblasts. Gastroenterology 142 928.e3–937.e3. 10.1053/j.gastro.2011.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E., Pozzi C., Houston P., Humphreys H., Robinson D. A., Loughman A., et al. (2008). A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190 3835–3850. 10.1128/JB.00167-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osanai A., Li S. J., Asano K., Sashinami H., Hu D. L., Nakane A. (2013). Fibronectin-binding protein, FbpA, is the adhesin responsible for pathogenesis of Listeria monocytogenes infection. Microbiol. Immunol. 57 253–262. 10.1111/1348-0421.12030 [DOI] [PubMed] [Google Scholar]
- Owens R. J., Baralle F. E. (1986a). Exon structure of the collagen-binding domain of human fibronectin. FEBS Lett. 204 318–322. 10.1016/0014-5793(86)80836-5 [DOI] [PubMed] [Google Scholar]
- Owens R. J., Baralle F. E. (1986b). Mapping the collagen-binding site of human fibronectin by expression in Escherichia coli. EMBO J. 5 2825–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeri V., Rosenshine I., Mosher D. F., Fassler R., Hanski E. (1998). Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol. Microbiol. 30 625–637. 10.1046/j.1365-2958.1998.01097.x [DOI] [PubMed] [Google Scholar]
- Palma M., Haggar A., Flock J. I. (1999). Adherence of Staphylococcus aureus is enhanced by an endogenous secreted protein with broad binding activity. J. Bacteriol. 181 2840–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V., Fischetti V. A. (1992). A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176 415–426. 10.1084/jem.176.2.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V., Fischetti V. A. (1998). alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273 14503–14515. 10.1074/jbc.273.23.14503 [DOI] [PubMed] [Google Scholar]
- Pickford A. R., Campbell I. D. (2004). NMR studies of modular protein structures and their interactions. Chem. Rev. 104 3557–3566. 10.1021/cr0304018 [DOI] [PubMed] [Google Scholar]
- Potts J. R., Campbell I. D. (1994). Fibronectin structure and assembly. Curr. Opin. Cell Biol. 6 648–655. 10.1016/0955-0674(94)90090-6 [DOI] [PubMed] [Google Scholar]
- Pracht D., Elm C., Gerber J., Bergmann S., Rohde M., Seiler M., et al. (2005). PavA of Streptococcus pneumoniae modulates adherence, invasion, and meningeal inflammation. Infect. Immun. 73 2680–2689. 10.1128/IAI.73.5.2680-2689.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakonjac J. V., Robbins J. C., Fischetti V. A. (1995). DNA sequence of the serum opacity factor of group A streptococci: identification of a fibronectin-binding repeat domain. Infect. Immun. 63 622–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D., Cossart P. (2015). How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 17 173–183. 10.1016/j.micinf.2015.01.004 [DOI] [PubMed] [Google Scholar]
- Rostagno A., Williams M. J., Baron M., Campbell I. D., Gold L. I. (1994). Further characterization of the NH2-terminal fibrin-binding site on fibronectin. J. Biol. Chem. 269 31938–31945. [PubMed] [Google Scholar]
- Sara M., Sleytr U. B. (2000). S-Layer proteins. J. Bacteriol. 182 859–868. 10.1128/JB.182.4.859-868.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S., Friedl P. (2010). Interstitial cell migration: integrin-dependent and alternative adhesion mechanisms. Cell Tissue Res. 339 83–92. 10.1007/s00441-009-0892-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E. (1991). Identification of the fibronectin sequences required for assembly of a fibrillar matrix. J. Cell Biol. 113 1463–1473. 10.1083/jcb.113.6.1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzbauer J. E., Tamkun J. W., Lemischka I. R., Hynes R. O. (1983). Three different fibronectin mRNAs arise by alternative splicing within the coding region. Cell 35(2 Pt 1) 421–431. 10.1016/0092-8674(83)90175-7 [DOI] [PubMed] [Google Scholar]
- Schwarz-Linek U., Pilka E. S., Pickford A. R., Kim J. H., Hook M., Campbell I. D., et al. (2004). High affinity streptococcal binding to human fibronectin requires specific recognition of sequential F1 modules. J. Biol. Chem. 279 39017–39025. 10.1074/jbc.M405083200 [DOI] [PubMed] [Google Scholar]
- Schwarz-Linek U., Werner J. M., Pickford A. R., Gurusiddappa S., Kim J. H., Pilka E. S., et al. (2003). Pathogenic bacteria attach to human fibronectin through a tandem beta-zipper. Nature 423 177–181. 10.1038/nature0189 [DOI] [PubMed] [Google Scholar]
- Sela S., Aviv A., Tovi A., Burstein I., Caparon M. G., Hanski E. (1993). Protein F: an adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10 1049–1055. 10.1111/j.1365-2958.1993.tb00975.x [DOI] [PubMed] [Google Scholar]
- Signas C., Raucci G., Jonsson K., Lindgren P. E., Anantharamaiah G. M., Hook M., et al. (1989). Nucleotide sequence of the gene for a fibronectin-binding protein from Staphylococcus aureus: use of this peptide sequence in the synthesis of biologically active peptides. Proc. Natl. Acad. Sci. U.S.A. 86 699–703. 10.1073/pnas.86.2.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha B., Francois P., Que Y. A., Hussain M., Heilmann C., Moreillon P., et al. (2000). Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect. Immun. 68 6871–6878. 10.1128/IAI.68.12.6871-6878.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha B., Francois P. P., Nusse O., Foti M., Hartford O. M., Vaudaux P., et al. (1999). Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin alpha5beta1. Cell. Microbiol. 1 101–117. 10.1046/j.1462-5822.1999.00011.x [DOI] [PubMed] [Google Scholar]
- Sottile J., Hocking D. C. (2002). Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Mol. Biol. Cell 13 3546–3559. 10.1091/mbc.E02-01-0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottile J., Schwarzbauer J., Selegue J., Mosher D. F. (1991). Five type I modules of fibronectin form a functional unit that binds to fibroblasts and Staphylococcus aureus. J. Biol. Chem. 266 12840–12843. [PubMed] [Google Scholar]
- Spitzfaden C., Grant R. P., Mardon H. J., Campbell I. D. (1997). Module-module interactions in the cell binding region of fibronectin: stability, flexibility and specificity. J. Mol. Biol. 265 565–579. 10.1006/jmbi.1996.0736 [DOI] [PubMed] [Google Scholar]
- Stones D. H., Krachler A. M. (2015). Fatal attraction: how bacterial adhesins affect host signaling and what we can learn from them. Int. J. Mol. Sci. 16 2626–2640. 10.3390/ijms16022626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talay S. R., Ehrenfeld E., Chhatwal G. S., Timmis K. N. (1991). Expression of the fibronectin-binding components of Streptococcus pyogenes in Escherichia coli demonstrates that they are proteins. Mol. Microbiol. 5 1727–1734. 10.1111/j.1365-2958.1991.tb01921.x [DOI] [PubMed] [Google Scholar]
- Tamkun J. W., DeSimone D. W., Fonda D., Patel R. S., Buck C., Horwitz A. F., et al. (1986). Structure of integrin, a glycoprotein involved in the transmembrane linkage between fibronectin and actin. Cell 46 271–282. 10.1016/0092-8674(86)90744-0 [DOI] [PubMed] [Google Scholar]
- Terao Y., Kawabata S., Nakata M., Nakagawa I., Hamada S. (2002). Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes strains isolated from toxic shock-like syndrome patients. J. Biol. Chem. 277 47428–47435. 10.1074/jbc.M209133200 [DOI] [PubMed] [Google Scholar]
- Tomasini-Johansson B. R., Kaufman N. R., Ensenberger M. G., Ozeri V., Hanski E., Mosher D. F. (2001). A 49-residue peptide from adhesin F1 of Streptococcus pyogenes inhibits fibronectin matrix assembly. J. Biol. Chem. 276 23430–23439. 10.1074/jbc.M103467200 [DOI] [PubMed] [Google Scholar]
- Tong S. Y., Davis J. S., Eichenberger E., Holland T. L., Fowler V. G., Jr. (2015). Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28 603–661. 10.1128/CMR.00134-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tressel T., McCarthy J. B., Calaycay J., Lee T. D., Legesse K., Shively J. E., et al. (1991). Human plasma fibronectin. Demonstration of structural differences between the A- and B-chains in the III CS region. Biochem. J. 274(Pt 3) 731–738. 10.1042/bj2740731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trial J., Rossen R. D., Rubio J., Knowlton A. A. (2004a). Inflammation and ischemia: macrophages activated by fibronectin fragments enhance the survival of injured cardiac myocytes. Exp. Biol. Med. (Maywood) 229 538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trial J., Rubio J. A., Birdsall H. H., Rodriguez-Barradas M., Rossen R. D. (2004b). Monocyte activation by circulating fibronectin fragments in HIV-1-infected patients. J. Immunol. 173 2190–2198. 10.4049/jimmunol.173.3.2190 [DOI] [PubMed] [Google Scholar]
- Vakonakis I., Staunton D., Ellis I. R., Sarkies P., Flanagan A., Schor A. M., et al. (2009). Motogenic sites in human fibronectin are masked by long range interactions. J. Biol. Chem. 284 15668–15675. 10.1074/jbc.M109.003673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velez M. P., De Keersmaecker S. C., Vanderleyden J. (2007). Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol. Lett. 276 140–148. 10.1111/j.1574-6968.2007.00908.x [DOI] [PubMed] [Google Scholar]
- Walia B., Castaneda F. E., Wang L., Kolachala V. L., Bajaj R., Roman J., et al. (2004). Polarized fibronectin secretion induced by adenosine regulates bacterial-epithelial interaction in human intestinal epithelial cells. Biochem. J. 382(Pt 2) 589–596. 10.1042/BJ20040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wann E. R., Gurusiddappa S., Hook M. (2000). The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J. Biol. Chem. 275 13863–13871. 10.1074/jbc.275.18.13863 [DOI] [PubMed] [Google Scholar]
- Williams M. J., Phan I., Harvey T. S., Rostagno A., Gold L. I., Campbell I. D. (1994). Solution structure of a pair of fibronectin type 1 modules with fibrin binding activity. J. Mol. Biol. 235 1302–1311. 10.1006/jmbi.1994.1083 [DOI] [PubMed] [Google Scholar]
- Wilson C. L., Schwarzbauer J. E. (1992). The alternatively spliced V region contributes to the differential incorporation of plasma and cellular fibronectins into fibrin clots. J. Cell Biol. 119 923–933. 10.1083/jcb.119.4.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H., Bischoff T., Tallent S. M., Seifert H., Wenzel R. P., Edmond M. B. (2004). Nosocomial bloodstream infections in US hospitals: analysis of 24179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39 309–317. 10.1086/421946 [DOI] [PubMed] [Google Scholar]
- Woods A., Couchman J. R., Johansson S., Hook M. (1986). Adhesion and cytoskeletal organisation of fibroblasts in response to fibronectin fragments. EMBO J. 5 665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]