Abstract
Mastitis is a serious medical condition of dairy cattle. Here, we evaluated whether the degree of virulence of mastitis pathogens in a mouse model can be linked to the inflammatory response that they provoke. Clinical isolates of Staphylococcus aureus (S. aureus) (strain 556 and 392) and Escherichia coli (E. coli) (676 and 127), and laboratory control strains [8325-4 (S. aureus) and MG1655 (E. coli)], were injected i.p. into mice, followed by the assessment of clinical scores and inflammatory parameters. As judged by clinical scoring, E. coli 127 exhibited the largest degree of virulence among the strains. All bacterial strains induced neutrophil recruitment. However, whereas E. coli 127 induced high peritoneal levels of CXCL1, G-CSF, and CCL2, strikingly lower levels of these were induced by the less virulent bacterial strains. High concentrations of these compounds were also seen in blood samples taken from animals infected with E. coli 127, suggesting systemic inflammation. Moreover, the levels of CXCL1 and G-CSF, both in the peritoneal fluid and in plasma, correlated with clinical score. Together, these findings suggest that highly virulent clinical mastitis isolates produce a distinct cytokine profile that shows a close correlation with the severity of the bacterial infection.
Keywords: mastitis, bovine, inflammation mediators, cytokines, chemokines, bacterial infections
Introduction
Mastitis is the most costly disease in the dairy industry worldwide, incurring economic losses in terms of reduced milk yield, poorer milk quality, and treatment costs (1, 2). Mastitis is defined as an inflammation of the mammary gland, typically the response to an intramammary infection (3, 4). A mastitis case is categorized as either clinical or subclinical. Cows with clinical mastitis present visually recognizable symptoms, such as milk abnormalities and visible changes in the udder. Rapidly developing cases are termed acute clinical mastitis. Subclinical mastitis is marked by the absence of macroscopically visual symptoms, though milk abnormalities or udder changes may occur transiently in a minority of cases. Subclinical cases that persist for at least 2 months are termed chronic. Intramammary infections are typically of bacterial origin and the responsible pathogens are broadly categorized as either contagious (spread between udder quarters or cows) or environmental (opportunistic pathogens) (5, 6). Staphylococci, streptococci, and coliform bacteria constitute the majority of etiological mastitis agents (7, 8).
The immune response toward an intramammary infection in cows is greatly influenced by the species of the invading bacterium. Escherichia coli (E. coli) commonly causes rapid and powerful immune responses (i.e., acute clinical mastitis), whereas Staphylococcus aureus (S. aureus) infections are associated with chronic responses (3). The course of the disease is also influenced by the strain and bacterial virulence factor, as well as by host and environmental factors. In the case of E. coli, it has been shown that strains possessing or having upregulated expression of factors that enable adhesion, invasion, motility (CheA, Tsr, and Tar), and iron acquisition (iroN and sitA) are more likely to cause persistent rather than transient intramammary infections (9–11). Cows infected with more common S. aureus genotypes experienced faster resolution of the inflammatory response than cows infected with less common genotypes (12), possibly because the less common genotypes possessed virulence factors enabling persistent infections. Other studies have found that certain genes are more common in isolates derived from persistent infection. For example, pyrogenic toxin superantigens (PTSAg), enterotoxin-encoding genes (sed and sej), and a penicillin resistance gene (blaZ) were more common in persistent strains (13, 14).
The objective of the current study was to differentiate the virulence of a set of acute clinical mastitis isolates using a mouse infection model. Non-mammary mouse models have previously been used to test the immune response toward mastitis-causing bacterial strains, in particular with the aim of defining novel vaccination strategies for mastitis (15–18). However, such models have not been extensively used to evaluate the inflammatory response toward mastitis pathogens. The current study used an acute intraperitoneal infection model. S. aureus and E. coli were selected by virtue of being common mastitis pathogens (8) and for representing the two mastitis pathogen classes – contagious (S. aureus) and environmental (E. coli). We demonstrate strain-dependent variations in virulence in the mouse model, with highly virulent E. coli inducing a defined set of inflammatory compounds including G-CSF and CXCL1. Moreover, the levels of these compounds correlated significantly with clinical outcome, suggesting the possibility to further evaluate G-CSF and CXCL1 as therapeutic targets, therapeutics, or biomarkers in mastitis.
Materials and Methods
Mice
Female B6JBOM (C57BL/6) mice (11–17 weeks old) were purchased from Taconic. After arrival, the mice were allowed a 1-month period of acclimatization prior to use. The mice were moved to an infection laboratory unit 4 days prior to the start of an experiment. During the entire period, the mice were fed rodent chow and water ad libitum. The animals had access to enrichment in the form of paper and paper houses. All animal experiments were approved by the local ethical committee (Uppsala Djurförsöksetiska Nämnd; no C85/14) and were performed in accordance with relevant guidelines and regulations.
Bacterial Strains
The clinical mastitis isolates 556 (S. aureus), 392 (S. aureus), 676 (E. coli), and 127 (E. coli) were obtained from the collection at the Swedish National Veterinary Institute (SVA). These strains were originally isolated from Swedish dairy cows during a national survey (8). Samples were only taken from lactating cows with acute clinical mastitis that had not undergone treatment with antimicrobials during the previous 30 days, had macroscopic changes in the milk in at least one udder quarter, had no previous record of clinical mastitis during the current lactation period, and a somatic cell count at the latest monthly milking of <200,000 cells/ml.
As controls for the clinical isolates, we used laboratory strains of S. aureus (8325-4) and E. coli (MG1655). The S. aureus strain 8325-4 (19) was obtained from the collection at the Department of Microbiology at the Swedish University of Agricultural Sciences. The E. coli strain MG1655 was a kind gift from Diarmaid Hughes (Department of Medical Biochemistry and Microbiology, Uppsala University).
Genotyping
DNA was extracted from E. coli and S. aureus colony material collected from horse blood agar plates, using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Genotyping was performed using DNA microarrays. The Identibac S. aureus Genotyping System (Alere, Jena, Germany) was used to genotype the S. aureus isolates. The system detects 333 S. aureus genes and gene variants relevant for typing as well as resistance and virulence attributes. E. coli isolates were characterized using the Identibac Ec System (Alere), detecting 124 E. coli virulence genes and gene variants as well as control genes. The assays were performed according to the manufacturer’s instructions. Hybridization results were detected by an ArrayMate Reader (Alere), and signals were registered as positive, ambiguous, or negative for each gene or gene variant.
Bacterial Cultures
Bacterial strains were streaked onto horse blood agar (S. aureus) (SVA, Uppsala, Sweden) or nutrient agar plates (E. coli) (Oxoid, Basingstoke, UK). Irrespective of species, the plates were incubated at 37°C for 24 h, after which colonies were picked and used to inoculate 20 ml of Tryptic Soy Broth (TSB; BD, Franklin Lakes, NJ, USA). Following incubation at 37°C for 16–17 h, 200 μl of overnight culture was transferred to 20 ml fresh TSB and incubated at 37°C until the OD600 reached 0.5. In preparation for injection, bacteria were washed twice in PBS (SVA; taken to room temperature) and finally resuspended in a volume of fresh TSB yielding a 1:1 dilution.
Generation Time
The generation time for each strain was determined from the increase in OD600 during the exponential phase. Briefly, the strains were incubated in TSB at 37°C, and the OD600 was measured every 30th minute until entry into the stationary phase. The assay was performed in triplicates.
In vivo Infection & Clinical Scoring
Mice were injected intraperitoneally with 100 μl TSB medium containing bacteria (~2 × 108 CFU of E. coli or ~5 × 108 CFU of S. aureus). Body weight was monitored prior to injection. After 24 h, the general condition of the mice was assessed by clinical scoring (Table 1), followed by weighing. The mice were subsequently euthanized, and samples were taken in the following order: blood via the eye, peritoneal lavage with 5 ml cold PBS, mesenteric lymph nodes, and spleens. The lymph nodes and spleens were weighed. Presence of E. coli in the blood was determined by streaking a 10 μl aliquot of blood onto MacConkey agar (Oxoid). The colony-forming unit count (CFU) in the peritoneal lavage fluid was determined by plating a 100 μl aliquot onto horse blood agar (S. aureus) or nutrient agar/MacConkey agar (E. coli). The cells in peritoneal lavage fluid were counted. Cytospins were prepared from an aliquot of the peritoneal cells and were stained with May-Grünwald/Giemsa, followed by differential counting. Finally, the remaining cells in the peritoneal lavage fluid were pelleted by centrifugation; the supernatant was collected and stored at −20°C.
Table 1.
Clinical scoring.
| Clinical Scorea | Criteria |
|---|---|
| 1 | Active, responsive, and no signs of any illness |
| 2 | Slower in reaction to stimuli, but otherwise active and healthy |
| 3 | Slow and lethargic, but still active |
| 4 | Inactive but still responsive to stimuli, albeit slowly |
| 5 | Inactive and non-responsive to any stimuli |
| Additional scores of 0.5 were added cumulatively based on the presence of: |
|
aNote that the maximal score is 6.5 (main clinical scores 1–5 + additional scores of 0.5).
Cytokine Arrays & ELISAs
Cytokine levels in the peritoneal lavage fluid were determined using RayBio® Mouse Inflammation Array C1 (RayBiotech, Inc., Norcross, GA, USA), according to the manufacturer’s instructions. Equal volumes of peritoneal lavage fluid from five mice were pooled to give the volume required for one membrane. The integrated density of the upregulated cytokines was quantified using ImageJ software and was normalized using the positive controls on each filter. The density of each spot was then compared with negative controls (peritoneal lavage fluid from non-infected animals); results are given as % density in comparison with the negative control. ELISAs were performed on peritoneal lavage fluid and blood plasma for CCL2 (Ready-SET-Go; eBioscience), CXCL1, and G-CSF (PeproTech), according to the manufacturers’ instructions.
Statistical Analysis
Data are shown as means ± SD. Statistical analyses were performed by using GraphPad Prism 6.0 (GraphPad Software), using one-way ANOVA without matching and Fisher’s LSD post hoc test. Correlation analysis was performed using non-parametric Spearman correlation with a two-tailed P-value.
Results
Genotype and Growth Characteristics of Mastitis Pathogens
The clinical isolates used in this study were all derived from the milk of cows suffering from acute clinical mastitis. The clinical isolates were genotyped (see Tables S1 and S2 in Supplementary Material). As shown in Table S1 in Supplementary Material, the S. aureus strains 556 and 392 clinical isolates shared a high degree of similarity in terms of virulence factors. However, they differed in terms of enterotoxins, with strain 392 harboring genes encoding enterotoxins, whereas S. aureus 556 did not. The E. coli clinical isolate 676 carried genes encoding cytotoxic necrotizing factor (cnf), cytolethal distending toxin (cdt), as well as the F17 fimbriae. This trio of virulence factors encoded on the same plasmid (pVir) is characteristic of type 2 necrotoxic E. coli (NTEC-2), which has been reported to frequently colonize the gastrointestinal tract of healthy ruminants [reviewed by De Rycke et al. (20)], but also been shown to cause both diarrhea and invasion of the blood stream in newborn calves (21), dependent only on factors encoded on the Vir plasmid (22). The E. coli 127 isolate lacked all virulence factors represented in the microarray system (Table S2 in Supplementary Material).
The bacterial growth rate has previously been suggested to influence mastitis strain virulence (3). In order to assess whether differences in growth rate of these bacterial strains could influence their virulence, the generation time for each strain was determined in TSB at 37°C. As shown in Figure 1, all strains of each species had virtually identical generation times, approximately 30 min for S. aureus and 22 min for E. coli.
Figure 1.
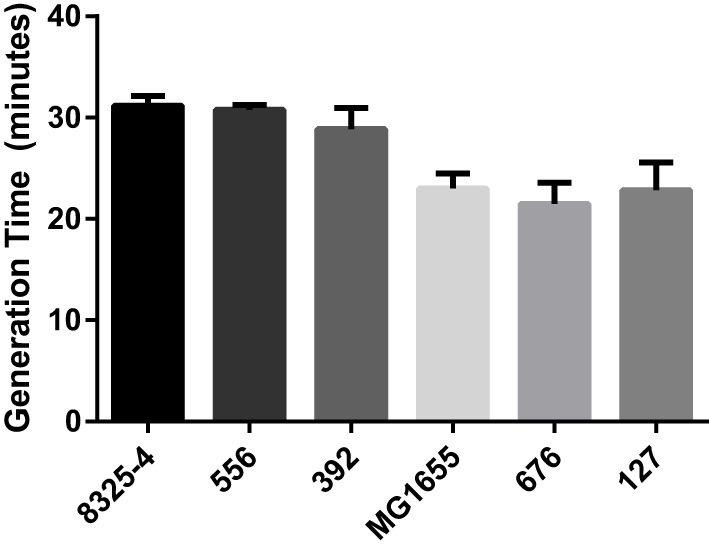
Generation times of bacterial strains. Generation times for the Staphylococcus aureus strains (8325-4, 556, 392) and the Escherichia coli strains (MG1655, 676, 127) were determined from the exponential phase measured in TSB and at OD600.
Mice Infected with E. coli 127 Exhibit Severe Clinical Symptoms
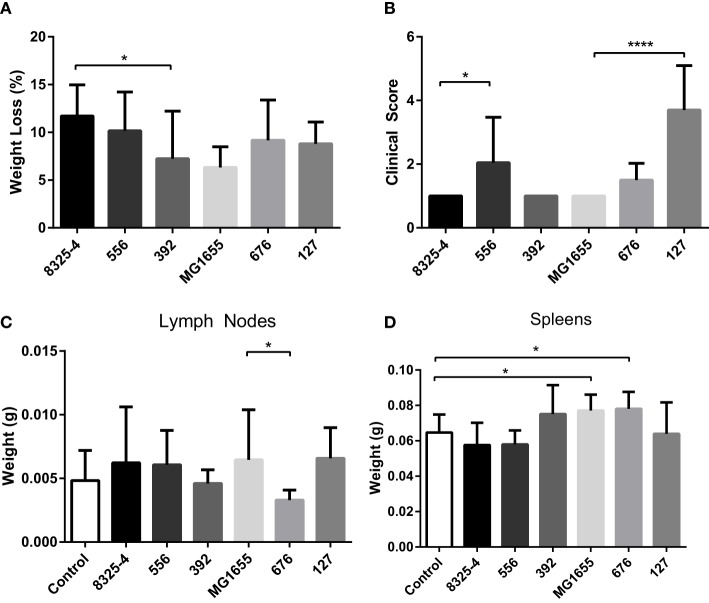
To investigate differences in clinical outcome for the selected strains, a mouse intraperitoneal infection model was used. Clinical outcome was assessed by measuring weight loss and by scoring clinical signs 24 h after infection. The mesenteric lymph nodes and spleens were also collected and weighed. As shown in Figure 2A, the weight loss of the mice in response to the different bacterial strains was relatively similar, with averages ranging from 6–12% over 24 h. In addition to monitoring clinical outcome by weight changes, we also implemented a clinical score system ranging from 1 to 5, where 1 represented animals with normal activity and 5 represented inactive, unresponsive animals (Table 1). Additional scores of 0.5 were added cumulatively for the presence of other clinical signs (ocular changes, intestinal disturbances, and shivering; see Table 1). As displayed in Figure 2B, the E. coli strain 127 caused the highest increase in clinical score out of all the bacterial strains tested. Moreover, mice infected with S. aureus 556 developed higher clinical scores than animals infected with the control S. aureus strain (8325-4).
Figure 2.
Effect of intraperitoneal bacterial infection on weight loss and clinical outcome. Mice were injected intraperitoneally with Staphylococcus aureus (8325-4, 556, 392) or Escherichia coli (MG1655, 676, 127). (A) After 24 h, weight loss of mice was monitored. (B) Severity of infection was monitored using clinical scoring as described in Table 1. (C,D) The weights of mesenteric lymph nodes (C) and spleens (D) were assessed. Data represent mean ± SD. *p < 0.05; ****p < 0.0001 (n = 4–10).
None of the assessed bacterial strains caused a significant increase in the weight of the mesenteric lymph nodes in comparison with non-infected controls (Figure 2C). However, we noted that lymph nodes taken from mice infected with the E. coli strain 676 weighed less than those from mice infected with E. coli MG1655 (Figure 2C). Infection with either of the S. aureus strains did not cause any significant effects on spleen weight (Figure 2D). In contrast, infection with E. coli MG1655 or E. coli 676 caused small, yet significant, increase in the weight of the spleens. Notably though, infection with the E. coli strain 127 did not result in increased spleen weight (Figure 2D).
To monitor any differences in bacterial persistence in vivo, peritoneal lavage was performed on the mice after euthanasia, followed by plating the fluid onto agar plates and quantification of the CFU. Mice infected with the clinical E. coli isolate 127 exhibited a poor bacterial clearance compared with the laboratory strain MG1655 (Figure 3). In particular, the strain 127 persisted at very high numbers after 24 h in the mice. In contrast, the clinical S. aureus isolates 556 and 392 did not yield higher CFU numbers than did the laboratory strain 8325-4.
Figure 3.
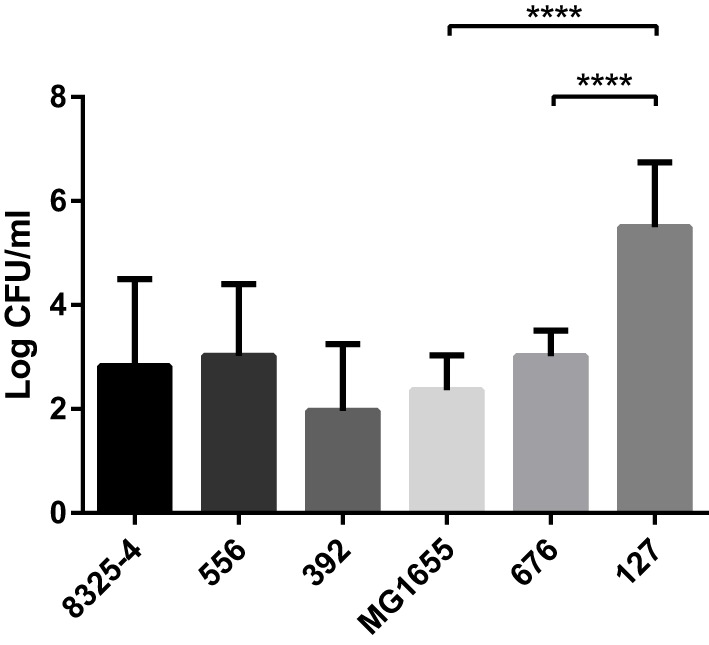
Presence of bacteria in the peritoneal cavities of infected mice. Mice were injected intraperitoneally with Staphylococcus aureus (8325-4, 556, 392) or Escherichia coli (MG1655, 676, 127). After 24 h, the presence of persistent bacteria (colony-forming units; CFU) in the peritoneal lavage fluid was determined. Data represent mean ± SD. ****p < 0.0001 (n = 4–10).
Cows afflicted with acute coliform mastitis have previously been shown to frequently develop bacteremia originating from the mammary infection (23, 24). To determine whether any of the E. coli strains assessed here could spread outside the peritoneal cavity, blood from infected mice was streaked onto MacConkey agar and incubated at 37°C for 24 h. As displayed in Table 2, E. coli strain 127 was present in these blood streaks, while MG1655 and 676 were not.
Table 2.
Bacteremia after Escherichia coli infection.
| Strain | Bacteria detected in blood |
|---|---|
| MG1655 | 0/5 |
| 676 | 0/5 |
| 127 | 5/5 |
Isolate 127 but not 676 or the laboratory strain MG1655 causes bacteremia. Mice were injected intraperitoneally with Escherichia coli (MG1655, 676, 127). After 24 h mice were euthanized. Blood samples were assessed for bacteremia.
Mastitis Pathogens Cause Leukocyte Recruitment into the Peritoneal Cavity
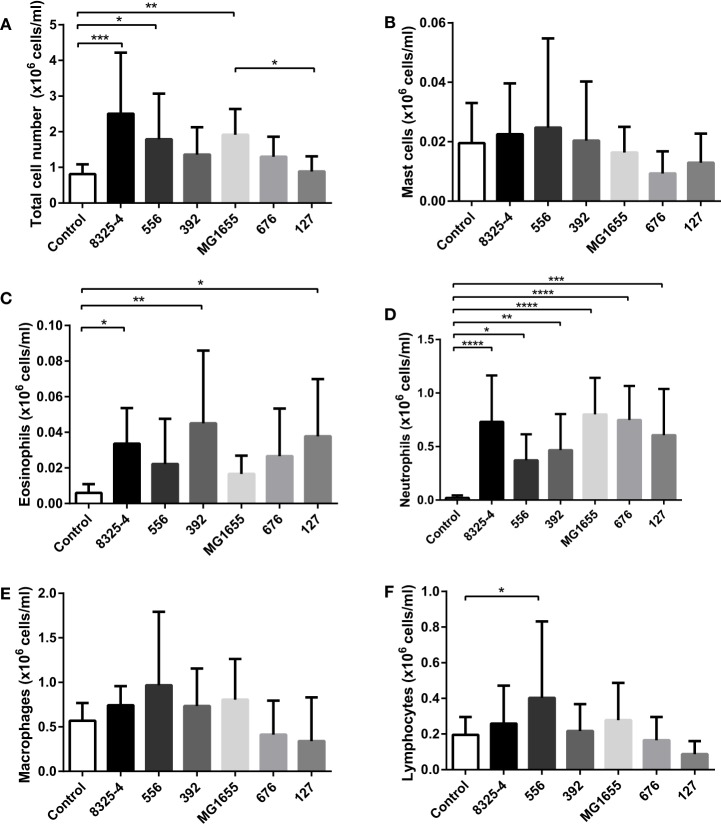
Next, we investigated whether the difference in clinical signs in response to the various bacterial strains tested could be associated with differences in inflammatory parameters. To this end, peritoneal cell populations from the infected mice were collected, followed by the assessment of total cell counts and differential cell counting after May-Grünwald/Giemsa staining of cytospin slides. As seen in Figure 4A, infection with S. aureus 8325-4, S. aureus 556, and E. coli MG1655 caused a significant increase of the total cell counts in the peritoneal cavity. In contrast, there was only a trend of increased total cell counts after infection with S. aureus 392 and E. coli 676, and no effect on total cell counts of the peritoneal cavity were seen after infection with E. coli 127. In response to all of the tested bacterial strains, a significant influx of neutrophils into the peritoneal cavity was seen (Figure 4D). In contrast, the numbers of peritoneal mast cells (Figure 4B) or macrophages (Figure 4E) were not affected by any of the bacteria, and there was no increase in the lymphocyte population in response to any of the bacterial strains, except for a modest increase in lymphocyte numbers in response to S. aureus 556 (Figure 4F). Eosinophil populations were only marginally affected by infection with the bacterial strains used (Figure 4C), although it was noted that infection with S. aureus 392 and E. coli 127 caused a slight, yet significant, increase in peritoneal eosinophil numbers (Figure 4C).
Figure 4.
Effect of bacterial infection on peritoneal leukocyte populations. Mice were injected intraperitoneally with Staphylococcus aureus (8325-4, 556, 392) or Escherichia coli (MG1655, 676, 127). After 24 h, peritoneal lavage was performed and the (A) number of total cells was quantified; differential counting of mast cells (B), eosinophils (C), neutrophils (D), macrophages (E), and lymphocytes (F) was performed after May-Grünwald/Giemsa staining of cytospin slides. Data represent mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 (n = 6–10).
E. coli Strain 127 Causes a Local and a Systemic Cytokine Response
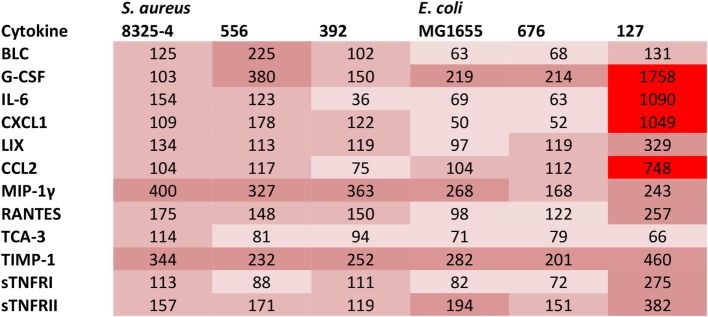
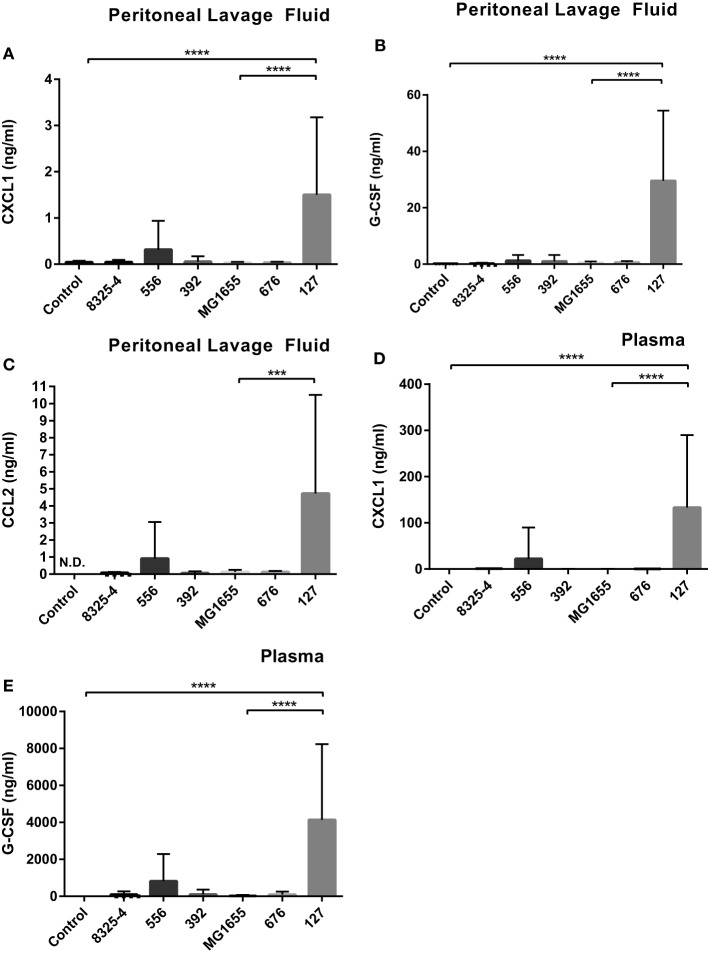
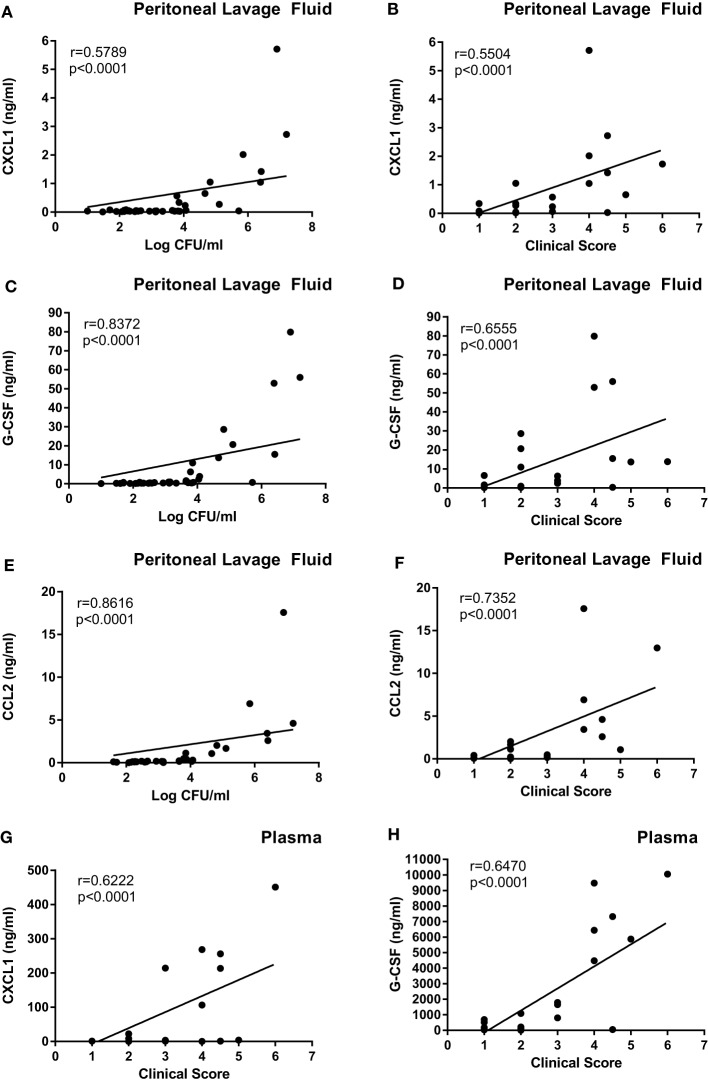
In order to further investigate if the difference in clinical outcome of the different bacterial strains could be related to inflammatory parameters, we assessed differences in the cytokine output in response to the respective bacteria. We used an inflammatory cytokine array as a screening method for this purpose. As shown in Figure S1 in Supplementary Material (shown for the E. coli strains) and Figure 5 (quantification by densitometry of data from all bacteria), several inflammatory cytokines/compounds were induced to varying extents by the various bacterial strains. However, the most striking induction of cytokines was seen after infection with E. coli 127. In particular, there was a dramatic increase in the levels G-CSF, IL-6, CXCL1, and CCL2 in response to this bacterial strain, whereas much lower levels of these compounds were seen in the peritoneal cavity of mice infected with any of the other bacterial strains. To confirm these findings by independent methodology, we used ELISA. As shown in Figures 6A–C, ELISA measurements confirmed a robust induction of CXCL1, G-CSF, and CCL2 in the peritoneal cavity of mice infected with E. coli 127 in comparison with infection with any of the other bacteria. However, a trend of upregulation of these compounds was also seen in response to S. aureus 556. Notably, there was a strong positive correlation between the bacterial counts in the peritoneal lavage fluid (CFU) and the levels of levels of CXCL1, G-CSF, and CCL2 (Figures 7A,C,E). As mice infected with E. coli 127 displayed the most severe clinical signs of infection, we additionally asked whether the levels of these inflammatory compounds showed a correlation with clinical score. Indeed, there was a significant positive correlation between the clinical score and the levels of CXCL1, G-CSF, and CCL2, respectively (Figures 7B,D,F).
Figure 5.
Presence of cytokines in the peritoneal fluids after bacterial infection. Mice were injected intraperitoneally with Staphylococcus aureus (8325-4, 556, 392) or Escherichia coli (MG1655, 676, 127). After 24 h, peritoneal lavage was performed. The presence of pro-inflammatory cytokines in peritoneal lavage fluid was determined by using an antibody-based array. The figure represents quantification of the cytokine array data by densitometry using ImageJ. Data are given as % integrated density in comparison to negative controls (plasma from non-infected animals). Levels of expression correlate with the intensity of red color.
Figure 6.
E. coli 127 induces high levels of G-CSF, CXCL1, and CCL2. Mice were injected intraperitoneally with Staphylococcus aureus (8325-4, 556, 392) or Escherichia coli (MG1655, 676, 127). After 24 h, peritoneal lavage fluid and blood plasma samples were collected. (A–C) The presence of CXCL1 (A), G-CSF (B), and CCL2 (C) in peritoneal lavage fluid was determined by using ELISA. (D,E) The presence of CXCL1 (D) and G-CSF (E) in plasma was determined by using ELISA. Data represent mean ± SD. ***p < 0.001: p < 0.0001 (n = 6–10).
Figure 7.
The levels of G-CSF, CXCL1, and CCL2 correlate with clinical outcome and with bacterial burden. Mice were injected intraperitoneally with Staphylococcus aureus (8325-4, 556, 392) or Escherichia coli (MG1655, 676, 127). (A,C,E) Correlation between bacterial counts (CFU) and CXCL1 (A), G-CSF (C), and CCL2 (E) levels in the peritoneal lavage fluid. (B,D,F) Correlation between clinical score and levels of CXCL1 (B), G-CSF (D), and CCL2 (F) in the peritoneal lavage fluid. (G,H) Correlation between clinical score and levels of CXCL1 (G) and G-CSF (H) in plasma.
Next, we assessed if the induction of inflammatory cytokines in the peritoneal cavity also was reflected by corresponding increases in blood plasma samples, i.e., reflecting a systemic inflammatory response. Indeed, infection with E. coli 127 caused a dramatic increase in the levels of G-CSF and CXCL1 in plasma, whereas none of the other bacterial strains tested caused significant upregulation of these compounds (Figures 6D,E). However, a tendency toward upregulated G-CSF and CXCL1 plasma levels was seen after infection with S. aureus 556 (Figures 6D,E). To further evaluate the association between the levels of G-CSF and CXCL1 with clinical outcome, we assessed whether levels of plasma G-CSF and CXCL1 showed a correlation with clinical score. As seen in Figures 7G,H, the plasma levels of both of these compounds were positively correlated with the clinical score. Together, these findings suggest that the high virulence of E. coli 127 is associated with a dramatic induction of a specific set of inflammatory compounds, such as CXCL1 and G-CSF, and that the levels of these compounds are correlated with clinical outcome and bacterial burden.
Discussion
The outcome of bovine mastitis is determined by host (25, 26) and pathogen factors (27, 28). It is known that the severity of mastitis varies depending on the species of the causative bacteria [reviewed in Ref. (3)]. Additionally, particular genes encoding virulence factors have been frequently isolated from bacterial strains able to cause more severe or persistent intramammary infections. Although there is some knowledge regarding the virulence factors that are expressed by common mastitis pathogens (see Introduction), there is still only limited knowledge of how mastitis pathogens affect the host immune response, a limiting factor for such research efforts being the challenging task of performing relevant studies in the bovine species. As an alternative, it may be possible to address this issue by performing studies in rodent models, and, here, we aimed to elucidate how mastitis pathogens affect the immune response by using a mouse acute intraperitoneal infection model. Although it can be questioned whether such mouse non-mammary models give an adequate picture of the inflammatory events occurring in mastitis, it should be emphasized that non-mammary models have previously been used for this purpose and have been suggested to represent useful tools for studying mastitis (17, 18, 29, 30).
Among the mastitis pathogens evaluated here, we found that the E. coli strain 127 exhibited the highest virulence as judged by clinical scoring. The higher virulence of this strain as opposed to the less virulent E. coli strains tested was most likely not due to a higher growth rate, as shown by analysis of the generation times for the different bacterial strains. This finding is, thus, in some contrast to earlier studies, where it has been suggested that the in vitro growth rate is predictive of in vivo virulence (17, 31). Moreover, E. coli 127 did not induce a more profound effect on lymphoid organ weight as compared with the other strains tested. It was also apparent that all of the bacterial strains tested produced a relatively similar inflammatory response in terms of neutrophil recruitment, whereas no significant recruitment of monocytes/macrophages or other leukocytes populations was observed, indicating that the high virulence of E. coli 127 is not due to a failure of the host to recruit neutrophils to clear the infection. However, E. coli 127 was shown to persist in the peritoneal cavity at markedly higher numbers as compared with the other bacterial strains tested. This suggests that E. coli 127 has a higher capability of evading eradication by the host response than have the other tested strains. Alternatively, although not possessing an intrinsically higher growth rate compared with the other E. coli strains, E. coli 127 may be better adapted than the other strains to maintain growth in the peritoneal milieu. It was also observed that infection with E. coli 127 was associated with bacteremia, whereas the other E. coli strains tested did not penetrate into the blood. This indicates that infection with E. coli 127 results in a systemic infection, well in line with the severe outcome as judged by clinical scoring. In line with these findings, bacteremia is common among cows afflicted with acute coliform mastitis (23, 24, 32). Interestingly, E. coli 127 possessed none of the virulence genes represented in the microarray system, suggesting the involvement of other less obvious virulence or survival factors. In contrast, E. coli 676 belongs to the NTEC-2 pathotype known to be capable of causing bacteremia in cattle (21, 22), but was less virulent as judged by clinical scoring in the present study.
In order to further investigate the reason(s) behind the differential virulence of the bacterial strains tested, we assessed the cytokine output induced by the various bacterial strains. We, thereby, found that the highly virulent E. coli 127 induced a dramatic increase of G-CSF, CXCL1, CCL2, and IL-6, whereas the levels of these were low in peritoneal cavities of mice infected with any of the other bacterial strains. Importantly, we found high levels of such compounds also in the blood plasma, in line with a systemic inflammatory response induced by E. coli 127. G-CSF is a growth factor that promotes the survival, proliferation, and differentiation of neutrophils, as well as enhancing the activity of mature neutrophils (33), and we may, thus, envisage that the high induction of G-CSF reflects an attempt by the host to cope with the bacterial infection by promoting the expansion of the neutrophil population. CXCL1 is a chemokine with neutrophil attracting properties (34), and the induction of this chemokine may, thus, also reflect the need for neutrophil recruitment in order to cope with the severe infection caused by E. coli 127 (34). In agreement with a role of this chemokine in mastitis, CXCL1 is detectable at the protein level in the bovine mammary gland in response to intramammary lipoteichoic acid (LTA) infusion (35). CCL2 is a chemokine that is chemoattractant for macrophages/monocytes (36). The induction of this chemokine may, thus, suggest a role of macrophages/monocytes in the host response. Of note, bovine mammary epithelial cells and mammary tissue express CCL2 mRNA in response to LPS stimuli (37, 38), in agreement with a link between this chemokine and mastitis. IL-6 is a powerful, pro-inflammatory cytokine with wide impact on the host response against various pathogens (39), and it has been shown previously that IL-6 is elevated during mastitis (40). High levels of this cytokine may, thus, be expected to accompany a severe bacterial infection by mastitis pathogens, such as the one caused by E. coli 127.
Altogether, these findings suggest that severe bacterial infection caused by mastitis pathogens results in a distinct profile of induced inflammatory compounds. Most likely, the induction of these compounds may represent a response that is protective for the host, by promoting the recruitment of neutrophils to the site of infection. We may, thus, envisage that treatment of infected subjects with the corresponding compounds may represent a potential therapeutic approach to limit infection. In line with this, attempts to enhance the immune competence of periparturient dairy cows by administrating recombinant G-CSF have been shown to increase the numbers and functional properties of neutrophils (41, 42).
The basis of the high virulence of E. coli 127 is intriguing. As judged by our genotyping for the presence of known virulence factors, E. coli 127 did not harbor genes coding for additional virulence factors in comparison with E. coli 676. The high virulence of E. coli 127 is, therefore, most likely due to non-characterized virulence factors/mechanisms. However, the nature of such factors/mechanisms is at present unknown. Interestingly, these findings are in analogy with the results of an extended survey of E. coli milk isolates from bovine mastitis, where it was observed that many of the pathogens causing acute clinical mastitis lacked well-characterized virulence factor genes (K. Artursson et al., unpublished results). This notion is also supported by a recent study from Kempf et al. who demonstrated by using a genomic comparative approach that mastitis E. coli isolates lacked well-characterized virulence factors (43).
As an alternative to exploiting these findings for therapeutic purposes, we foresee the possibility of using our findings for diagnostic purposes. Clearly, there is a demand for improved tools for diagnosing different grades of mastitis, in particular for detection of subclinical mastitis (44). Based on the findings obtained in this investigation, high levels of G-CSF and CXCL1 may accompany severe, systemic bacterial infection, and it is, thus, possible that levels of G-CSF and/or CXCL1 may be useful diagnostic criteria in mastitis. In agreement with such a notion, we found that the levels of these compounds correlated significantly with severity of infection as judged by clinical scoring. However, further studies are needed to clarify whether induction of these particular pro-inflammatory factors also accompanies bovine mastitis.
Author Contributions
C-FJ planned and performed experiments, interpreted data, and wrote the manuscript; KA collected bacterial strains, performed genotyping, and contributed to the writing of the manuscript; RS performed genotyping and contributed to the writing of the manuscript; BG contributed to experiments and to the writing of the manuscript; ER planned and performed experiments, interpreted data, and contributed to the writing of the manuscript; GP planned the study, interpreted data, and wrote the manuscript. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by grants from Formas, The Swedish Research Council and The Royal Swedish Academy of Agriculture and Forestry. We are grateful to staff at the SVA mastitis laboratory for assistance with handling of bacterial isolates, and to Ulla Johansson and Pedram Effati for assistance with microarray assays. We thank Dr Björn Bengtsson, SVA and the staff at the Antibiotic Laboratory for sharing the strain collection of mastitis isolates with us.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2016.00368
References
- 1.Hogeveen H, Huijps K, Lam TJ. Economic aspects of mastitis: new developments. N Z Vet J (2011) 59:16–23. 10.1080/00480169.2011.547165 [DOI] [PubMed] [Google Scholar]
- 2.Down PM, Green MJ, Hudson CD. Rate of transmission: a major determinant of the cost of clinical mastitis. J Dairy Sci (2013) 96:6301–14. 10.3168/jds.2012-6470 [DOI] [PubMed] [Google Scholar]
- 3.Schukken YH, Gunther J, Fitzpatrick J, Fontaine MC, Goetze L, Holst O, et al. Host-response patterns of intramammary infections in dairy cows. Vet Immunol Immunopathol (2011) 144:270–89. 10.1016/j.vetimm.2011.08.022 [DOI] [PubMed] [Google Scholar]
- 4.De Vliegher S, Fox LK, Piepers S, McDougall S, Barkema HW. Invited review: mastitis in dairy heifers: nature of the disease, potential impact, prevention, and control. J Dairy Sci (2012) 95:1025–40. 10.3168/jds.2010-4074 [DOI] [PubMed] [Google Scholar]
- 5.Bradley A. Bovine mastitis: an evolving disease. Vet J (2002) 164:116–28. 10.1053/tvjl.2002.0724 [DOI] [PubMed] [Google Scholar]
- 6.Thompson-Crispi K, Atalla H, Miglior F, Mallard BA. Bovine mastitis: frontiers in immunogenetics. Front Immunol (2014) 5:493. 10.3389/fimmu.2014.00493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zadoks RN, Middleton JR, McDougall S, Katholm J, Schukken YH. Molecular epidemiology of mastitis pathogens of dairy cattle and comparative relevance to humans. J Mammary Gland Biol Neoplasia (2011) 16:357–72. 10.1007/s10911-011-9236-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ericsson Unnerstad H, Lindberg A, Persson Waller K, Ekman T, Artursson K, Nilsson-Ost M, et al. Microbial aetiology of acute clinical mastitis and agent-specific risk factors. Vet Microbiol (2009) 137:90–7. 10.1016/j.vetmic.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 9.Dogan B, Klaessig S, Rishniw M, Almeida RA, Oliver SP, Simpson K, et al. Adherent and invasive Escherichia coli are associated with persistent bovine mastitis. Vet Microbiol (2006) 116:270–82. 10.1016/j.vetmic.2006.04.023 [DOI] [PubMed] [Google Scholar]
- 10.Lippolis JD, Brunelle BW, Reinhardt TA, Sacco RE, Nonnecke BJ, Dogan B, et al. Proteomic analysis reveals protein expression differences in Escherichia coli strains associated with persistent versus transient mastitis. J Proteomics (2014) 108:373–81. 10.1016/j.jprot.2014.06.008 [DOI] [PubMed] [Google Scholar]
- 11.Fairbrother JH, Dufour S, Fairbrother JM, Francoz D, Nadeau E, Messier S. Characterization of persistent and transient Escherichia coli isolates recovered from clinical mastitis episodes in dairy cows. Vet Microbiol (2015) 176:126–33. 10.1016/j.vetmic.2014.12.025 [DOI] [PubMed] [Google Scholar]
- 12.Lundberg A, Aspan A, Nyman A, Unnerstad HE, Waller KP. Associations between bacterial genotype and outcome of bovine clinical Staphylococcus aureus mastitis. Acta Vet Scand (2014) 56:2. 10.1186/1751-0147-56-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haveri M, Roslof A, Rantala L, Pyorala S. Virulence genes of bovine Staphylococcus aureus from persistent and nonpersistent intramammary infections with different clinical characteristics. J Appl Microbiol (2007) 103:993–1000. 10.1111/j.1365-2672.2007.03356.x [DOI] [PubMed] [Google Scholar]
- 14.Haveri M, Hovinen M, Roslof A, Pyorala S. Molecular types and genetic profiles of Staphylococcus aureus strains isolated from bovine intramammary infections and extramammary sites. J Clin Microbiol (2008) 46:3728–35. 10.1128/JCM.00769-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leitner G, Lubashevsky E, Trainin Z. Staphylococcus aureus vaccine against mastitis in dairy cows, composition and evaluation of its immunogenicity in a mouse model. Vet Immunol Immunopathol (2003) 93:159–67. 10.1016/S0165-2427(03)00062-X [DOI] [PubMed] [Google Scholar]
- 16.Leitner G, Lubashevsky E, Glickman A, Winkler M, Saran A, Trainin Z. Development of a Staphylococcus aureus vaccine against mastitis in dairy cows. I. challenge trials. Vet Immunol Immunopathol (2003) 93:31–8. 10.1016/S0165-2427(03)00051-5 [DOI] [PubMed] [Google Scholar]
- 17.Bogni C, Segura M, Giraudo J, Giraudo A, Calzolari A, Nagel R. Avirulence and immunogenicity in mice of a bovine mastitis Staphylococcus aureus mutant. Can J Vet Res (1998) 62:293–8. [PMC free article] [PubMed] [Google Scholar]
- 18.Pellegrino M, Giraudo J, Raspanti C, Odierno L, Bogni C. Efficacy of immunization against bovine mastitis using a Staphylococcus aureus avirulent mutant vaccine. Vaccine (2010) 28:4523–8. 10.1016/j.vaccine.2010.04.056 [DOI] [PubMed] [Google Scholar]
- 19.Sjostrom JE, Lindberg M, Philipson L. Competence for transfection in Staphylococcus aureus. J Bacteriol (1973) 113:576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Rycke J, Milon A, Oswald E. Necrotoxic Escherichia coli (NTEC): two emerging categories of human and animal pathogens. Vet Res (1999) 30:221–33. [PubMed] [Google Scholar]
- 21.Van Bost S, Roels S, Mainil J. Necrotoxigenic Escherichia coli type-2 invade and cause diarrhoea during experimental infection in colostrum-restricted newborn calves. Vet Microbiol (2001) 81:315–29. 10.1016/S0378-1135(01)00360-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Bost S, Roels S, Oswald E, Mainil J. Putative roles of the CNF2 and CDTIII toxins in experimental infections with necrotoxigenic Escherichia coli type 2 (NTEC2) strains in calves. Microbes Infect (2003) 5:1189–93. 10.1016/j.micinf.2003.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cebra CK, Garry FB, Dinsmore RP. Naturally occurring acute coliform mastitis in Holstein cattle. J Vet Intern Med (1996) 10:252–7. 10.1111/j.1939-1676.1996.tb02058.x [DOI] [PubMed] [Google Scholar]
- 24.Wenz JR, Barrington GM, Garry FB, McSweeney KD, Dinsmore RP, Goodell G, et al. Bacteremia associated with naturally occuring acute coliform mastitis in dairy cows. J Am Vet Med Assoc (2001) 219:976–81. 10.2460/javma.2001.219.976 [DOI] [PubMed] [Google Scholar]
- 25.Kehrli ME, Jr., Shuster DE. Factors affecting milk somatic cells and their role in health of the bovine mammary gland. J Dairy Sci (1994) 77:619–27. 10.3168/jds.S0022-0302(94)76992-7 [DOI] [PubMed] [Google Scholar]
- 26.Steeneveld W, Hogeveen H, Barkema HW, van den Broek J, Huirne RB. The influence of cow factors on the incidence of clinical mastitis in dairy cows. J Dairy Sci (2008) 91:1391–402. 10.3168/jds.2007-0705 [DOI] [PubMed] [Google Scholar]
- 27.Bannerman DD. Pathogen-dependent induction of cytokines and other soluble inflammatory mediators during intramammary infection of dairy cows. J Anim Sci (2009) 87:10–25. 10.2527/jas.2008-1187 [DOI] [PubMed] [Google Scholar]
- 28.Wellnitz O, Arnold ET, Bruckmaier RM. Lipopolysaccharide and lipoteichoic acid induce different immune responses in the bovine mammary gland. J Dairy Sci (2011) 94:5405–12. 10.3168/jds.2010-3931 [DOI] [PubMed] [Google Scholar]
- 29.Leitner G, Krifucks O, Glickman A, Younis A, Saran A. Staphylococcus aureus strains isolated from bovine mastitis: virulence, antibody production and protection from challenge in a mouse model. FEMS Immunol Med Microbiol (2003) 35:99–106. 10.1016/S0928-8244(02)00458-3 [DOI] [PubMed] [Google Scholar]
- 30.Pellegrino M, Giraudo J, Raspanti C, Nagel R, Odierno L, Primo V, et al. Experimental trial in heifers vaccinated with Staphylococcus aureus avirulent mutant against bovine mastitis. Vet Microbiol (2008) 127:186–90. 10.1016/j.vetmic.2007.07.028 [DOI] [PubMed] [Google Scholar]
- 31.Odierno L, Risatti G, Calzolari A, Giraudo JA, Gonzalez Quintana H, Nagel R. Pathogenicity in mice of Staphylococcus aureus mutants deficient in exoprotein synthesis. Vet Microbiol (1994) 41:249–58. 10.1016/0378-1135(94)90105-8 [DOI] [PubMed] [Google Scholar]
- 32.Wenz JR, Barrington GM, Garry FB, Ellis RP, Magnuson RJ. Escherichia coli isolates’ serotypes, genotypes, and virulence genes and clinical coliform mastitis severity. J Dairy Sci (2006) 89:3408–12. 10.3168/jds.S0022-0302(06)72377-3 [DOI] [PubMed] [Google Scholar]
- 33.Roberts AW. G-CSF: a key regulator of neutrophil production, but that’s not all! Growth Factors (2005) 23:33–41. 10.1080/08977190500055836 [DOI] [PubMed] [Google Scholar]
- 34.De Filippo K, Henderson RB, Laschinger M, Hogg N. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol (2008) 180:4308–15. 10.4049/jimmunol.180.6.4308 [DOI] [PubMed] [Google Scholar]
- 35.Rainard P, Fromageau A, Cunha P, Gilbert FB. Staphylococcus aureus lipoteichoic acid triggers inflammation in the lactating bovine mammary gland. Vet Res (2008) 39:52. 10.1051/vetres:2008034 [DOI] [PubMed] [Google Scholar]
- 36.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res (2009) 29:313–26. 10.1089/jir.2008.0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mount JA, Karrow NA, Caswell JL, Boermans HJ, Leslie KE. Assessment of bovine mammary chemokine gene expression in response to lipopolysaccharide, lipotechoic acid + peptidoglycan, and CpG oligodeoxynucleotide 2135. Can J Vet Res (2009) 73:49–57. [PMC free article] [PubMed] [Google Scholar]
- 38.Gilbert FB, Cunha P, Jensen K, Glass EJ, Foucras G, Robert-Granie C, et al. Differential response of bovine mammary epithelial cells to Staphylococcus aureus or Escherichia coli agonists of the innate immune system. Vet Res (2013) 44:40. 10.1186/1297-9716-44-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol (2015) 16:448–57. 10.1038/ni.3153 [DOI] [PubMed] [Google Scholar]
- 40.Sakemi Y, Tamura Y, Hagiwara K. Interleukin-6 in quarter milk as a further prediction marker for bovine subclinical mastitis. J Dairy Res (2011) 78:118–21. 10.1017/S0022029910000828 [DOI] [PubMed] [Google Scholar]
- 41.Kimura K, Goff JP, Canning P, Wang C, Roth JA. Effect of recombinant bovine granulocyte colony-stimulating factor covalently bound to polyethylene glycol injection on neutrophil number and function in periparturient dairy cows. J Dairy Sci (2014) 97:4842–51. 10.3168/jds.2013-7242 [DOI] [PubMed] [Google Scholar]
- 42.Kehrli ME, Jr., Goff JP, Stevens MG, Boone TC. Effects of granulocyte colony-stimulating factor administration to periparturient cows on neutrophils and bacterial shedding. J Dairy Sci (1991) 74:2448–58. 10.3168/jds.S0022-0302(91)78420-8 [DOI] [PubMed] [Google Scholar]
- 43.Kempf F, Slugocki C, Blum SE, Leitner G, Germon P. Genomic comparative study of bovine mastitis Escherichia coli. PLoS One (2016) 11:e0147954. 10.1371/journal.pone.0147954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viguier C, Arora S, Gilmartin N, Welbeck K, O’Kennedy R. Mastitis detection: current trends and future perspectives. Trends Biotechnol (2009) 27:486–93. 10.1016/j.tibtech.2009.05.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.