Abstract
During last decades canine health and well being is becoming an important issue for human owners. In dogs, several factors including diet, pathogenic bacterial and stress conditions can affect the composition of the gut microbiota. In this study, we evaluated the effect of dietary chabazitic zeolitite (CZ) supplementation on the contribution of bifidobacteria to the fecal microbiota in training hunting dogs. Fecal microbiota cataloging based on 16S rRNA microbial profiling analyses highlighted an increase of Lactobacillus and Bifidobacterium in animals treated with CZ, with a simultaneous decrease of pathogens associated with dog gastrointestinal infections, such as Klebsiella and Enterobacter. A detailed profiling of the bifidobacterial population of dogs receiving CZ based on the ITS-based sequencing approach, revealed an enhancement bifidobacterial of species typical of animals such as Bifidobacterium animalis and B. pseudolongum. Moreover, these analyses identified the occurrence of putative new bifidobacterial taxa in both treated and untreated samples.
Keywords: dogs, gut microbiota, chabazitic zeolitite, Bifidobacterium, adsorptive capacity
Introduction
Pet population is increasing in western countries, and dogs are the major human companions. Mutual interest has evolved into companion animals being a stable part of human life and therefore, the health and wellbeing of pets have increasingly raised interest during last decades. During history, the dog diet has changed, starting from a carnivorous behavior and a high protein diet (Clauss et al., 2010) to a carbohydrate rich diet and an urban life-style.
Despite the long span history of human-dog co-evolution, the knowledge of canine intestinal microbiota composition is much less complete than for humans. The dog gastro-intestinal tract (GIT) represents a rich ecosystem, composed of a wide range of metabolically active microorganisms (Simpson et al., 2002; Suchodolski et al., 2008; Kerr et al., 2013a). The predominant bacterial phyla in the colon and faeces of dogs are represented by Firmicutes (40–60%), Bacteroidetes (5–10%), Proteobacteria (15–20%), and Fusobacteria (5%) (Kerr et al., 2013b; Deng and Swanson, 2015), representing approximately 99% of the gut microbiota in dogs. However, very little is known about the occurrence of healthy promoting microorganisms such as bifidobacteria in the gut especially using metagenomics based approaches (Gavini et al., 2006; Jia et al., 2010).
Bifidobacteria are Gram positive bacteria that colonize different ecological niches, but represents one of the dominant colonizers of mammals at the very early first stages of life (Milani et al., 2015). The analyses of the gut microbiota of different mammals indicate that some bifidobacterial species, usually detected in the human GIT, were also identified in many other animals (Lamendella et al., 2008). For example, Bifidobacterium bifidum, B. adolescentis, B. catenulatum, and B. dentium are human-type bifidobacteria (Duranti et al., 2015, 2016), but these taxa displayed a cosmopolitan ecological behavior among different mammals (Lamendella et al., 2008).
In hunting dogs, emotional stress to which they are submitted during the training, can alter the habitat of the GIT (Rutgers et al., 1996). Therefore, to keep a suitable function of the GIT through appropriate feeding strategies is interesting, to avoid the intestinal colonization by enteropathogens (e.g., Escherichia coli, Salmonella ssp., Clostridium perfringens, C. difficile) (McKenzie et al., 2010; Kerr et al., 2013a).
To avoid antibiotic therapies, alternative products are under investigation. Zeolitites are aluminosilicates characterized by an open structure, which can accommodate a wide variety of ions. The particle size, crystallite size, and the degree of aggregation of the zeolitic material, as well as the porosity of individual particles, determine the access of ingesta fluids to the zeolitic surface during the passage across the GIT, and strongly affect its ion exchange, adsorption and catalytic properties (Papaioannou et al., 2005). The mechanism of action of zeolite is likely to be multifunctional. Different health and performance promoting properties were highlighted for zeolite in animal diet. These include ammonia binding effect, fecal elimination of p-cresol, retarding effect on digesta transit, enhanced pancreatic ezymes activity, and aflatoxin sequestering effect (Papaioannou et al., 2005). Moreover, recently it was reported the application of zeolite in reducing pathogens counts in broiler chicken (Prasai et al., 2016). Among zeolitites, the chabazitic zeolitite (CZ) has a high cation-exchange capacity and bulk density (Pabalan and Bertetti, 2001). Dietary inclusion of zeolitites has been effective in animals (e.g., pigs, calves) and humans suffering from gastrointestinal disturbances (RodriguezFuentes et al., 1997; Papaioannou et al., 2005). To date, no data exist about the evaluation of the effects of zeolitites on dog intestinal microbiota. The aim of the present study was to assess the effect of dietary CZ supplementation on the fecal microbiota with particular emphasis on bifidobacterial populations in training hunting dogs through culture-dependent methods and 16S rRNA/ITS (internal transcribed spacer) microbial profiling approach.
Materials and Methods
Ethics Statement
This study was carried out in accordance with the recommendations of the ethical committee of the University of Parma. The protocol was approved by the “Comitato di Etica Università degli Studi di Parma”, Italy. All animal procedures were performed according to national guidelines (Decreto legislativo 26/2014) on the protection of animals used for scientific purposes.
Animals and Experimental Procedure
Twenty adult English Setter dogs, reared in the same kennel, were selected to be homogeneous with reference to age (mean age ± SD: 3.50 ± 1.9 years), body weight (mean weight ± SD: 18.83 ± 2.96 kg) and gender (10 males, 10 not pregnant females). Based on age, weight, and sex animals were equally divided into two groups (10 dogs group-1), individually penned with a rest area inside (2.70 m× 1.40 m) and a paddock outside (4.50 m× 1.40 m). Animals were free of any clinical symptoms indicating gastrointestinal disease and they did not receive medications that are expected to alter the gut microbiota such as antibiotics. Dogs were wormed one month before the start of study. The characteristics of the groups are reported in Table 1. During a period of 28 days, both groups received a diet, based on raw poultry meat (25% crude protein, 24% ether extract, 5% ash, 2% crude fiber, and 18.4 MJ kg-1 ME, on dry matter). The individual ration, administered at about 25 g dry matter kg-1 of body weight0.75, once a day, was supplemented (group Tr) or not (group NTr) with CZ powder at the dose of 5 g day-1. For each dog, zeolitite was weighed and added to the ration at each meal. Free access to water was provided. During the study, all dogs were daily subjected to an aerobic physical activity characterized by gallop for 20 min, according to the trainer’s practices. Training was performed in two outdoor next areas, at a mean temperature and relative humidity of 24 ± 3°C and of 67 ± 10%, respectively. Inside each group, five pairs of dogs were identified and each of them assigned alternatively to one or to the other of the training areas.
Table 1.
Characteristics of the experimental groups (mean ± SD).
| Parameter | Groups∗ |
|
|---|---|---|
| NTr | Tr | |
| Animals (No.) | 10 | 10 |
| Age (years) | 3.41 ± 1.59 | 3.50 ± 1.60 |
| Body weight (kg) | 19.59 ± 2.85 | 18.08 ± 2.74 |
∗NTr, untreated group; Tr, treated group.
Chabazitic Zeolitite Source
The powder of CZ, was obtained after sterilization at 200°C for 20 min (Chabasite 70® Verdi S.p.A, Italy). The total zeolitic content was 70 ± 5%, of which 65 ± 3% due to chabazite (Na0.14K1.03Ca1.00Mg0.17) [Al3.46Si8.53O24] × 9.7H2O and 5 ± 3% to phillipsite (Na0.9Ca0.5K0.6) [Si5.2A12.8O16] × 6H2O. No traces of clinoptilolite were found. The composition of zeolitic powder was determined by Rietveld-RIR method (Gualtieri, 2000). The cation-exchange capacity and bulk density in relation to particles size were 2.2 ± 0.1 mEq g-1 and 0.70-0.90 g (cm3)-1, respectively (Gualtieri, 2000; Cresswell and Hamilton, 2002). Water retention in relation to particles size was about 30-40% (w/w). The granulometry of the powder was less than 100 μm.
Collection of Fecal Samples
Feces consistency was scored using a scale of 1 (hard) to 5 (watery) (Grieshop et al., 2002) at days 0 (Time point 0, T0), 16 (Time point 1, T1) from the beginning of the dietary treatment, and at the end of experimental period (day 29, Time point 2, T2). During the same days, individual fecal samples were collected directly from the rectum, using a sterile glove lubricated with water. The feces were placed in sterile polyethylene bags, immediately transported to the laboratory on ice packs and frozen at -20°C until analysis.
16S rRNA/ITS Microbial Profiling
Upon arrival at the laboratory, individual fecal samples were aliquoted and combined with other individual samples from the same treatment to form pooled samples. In fact, in animal health it has been shown recently that pooling stool samples allows a rapid assessment of infection intensity and drug efficacy (Mekonnen et al., 2013). Each individual dog sample was equally represented in the respective pooled sample. DNA was extracted from pooled fecal samples using the QIAamp DNA Stool Mini kit following the manufacturer’s instructions (Qiagen Ltd., Strasse, Germany).
Partial 16S rRNA gene sequences were amplified from extracted DNA using primer pair Probio_Uni and /Probio_Rev, which target the V3 region of the 16S rRNA gene sequence, as previously reported (Milani et al., 2013). Partial ITS sequences were amplified from extracted DNA using the primer pair Probio-bif_Uni/Probiobif_Rev as described by Milani et al. (2014b). The PCR conditions used were 5 min at 95°C and 35 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C, followed by 10 min at 72°C. Amplification was carried out using a Veriti Thermocycler (Applied Bio-systems).
16S rRNA gene and ITS sequencing were performed using a MiSeq (Illumina) according to the protocols previously published (Milani et al., 2013, 2014b).
16S rRNA Gene-Based Microbiota Analysis
The achieved individual sequence reads were filtered by the Illumina software to remove low quality and polyclonal sequences. All Illumina quality-approved, trimmed, and filtered data were exported as.fastq files. The.fastq files were processed using a custom script based on the QIIME software suite (Caporaso et al., 2010). Paired-end reads pairs were assembled to reconstruct the complete Probio_Uni/Probio_Rev amplicons. Quality control retained sequences with a length between 140 and 400 bp and mean sequence quality score >20 while sequences with homopolymers >7 bp and mismatched primers were omitted. In order to calculate downstream diversity measures (alpha diversity indices, Unifrac analysis), 16S rRNA Operational Taxonomic Units (OTUs) were defined at ≥97 % sequence homology using uclust (Edgar, 2010) and OTUs with less than 10 sequences were filtered. All reads were classified to the lowest possible taxonomic rank using QIIME (Caporaso et al., 2010) and a reference dataset from the SILVA database (Quast et al., 2013). Biodiversity of the samples (alpha-diversity) were calculated with Chao1 index.
ITS-Based Microbiota Analysis
For ITS-based microbiota analysis Fastq files obtained by sequencing of the ITS amplicons were analyzed using a custom script, named bif_ITS_analysis.sh script 1. This script requires QIIME (Caporaso et al., 2010) to be installed (or works in a QIIME virtual machine) and accepts.bam or.fastq input files containing sequencing reads. Input data were processed as previously described (Milani et al., 2014b).
Bacterial Counts
The homogenates fecal specimens were serially diluted with both half-strength Wilkins-Chalgren Anaerobe Broth (WCAB) and Buffered Peptone Water (ThermoScientific-Oxoid, UK). Dilutions in duplicate were plated on MacConkey agar (Merck, Germany) for Enterobacteriaceae, Perfringens agar Base (OPSP) (Oxoid, UK) for C. perfringens, vancomycin and bromocresol green (LAMVAB) agar (Hartemink and Rombouts, 1999) for lactobacilli, and Azide maltose agar (Biolife, Italy) for enterococci counts. MacConkey agar and Azide maltose agar plates were incubated aerobically at 37°C for 24 and 48 h, respectively. Other media were incubated anaerobically at 37°C for 48-72 h. The taxonomy of colonies isolated random on selective media were determined at genus and species level by API System (Bio-Merieux, Italy) to verify the reliability of the media utilized.
In vitro CZ Adsorptive Capacity
The ability of CZ to bind to enteropathogens bacteria was evaluated in pooled feces using two reference strains, i.e., E. coli ATCC 35218 and C. perfringens ATCC 13124. Strains were grown in Mueller-Hinton Broth (Difco, MI, USA) at 37°C for 24 h, then transferred to 10 ml of broth and grown for another 8 h to reach the final exponential phase.
Adsorptive capacity of CZ was evaluated, measuring spectrophotometrically the OD of the samples (An and Friedman, 1997). Twenty-five grams of pooled feces obtained by NTr groups and collected on days 0, 16, and 29 were placed, in triplicate, into flasks containing 225 ml of Buffered Sodium Chloride-Peptone Solution pH 7.0 (Oxoid, UK). CZ was added in different quantities (0, 0.25, 0.5, 1 g). Lastly, C. perfringens ATCC 13124 or E. coli ATCC 35218 strains were added to medium and incubated at 37°C. At 0, 2, 4, 6, and 24 h, 150 μl of the suspension were transferred into a microtiter plate in four replicates and the absorbance was immediately evaluated (VICTOR3, 1420 multilabel counter, PerkinElmer, Italy) at 620 nm.
Statistical Analysis
Data for fecal score and fecal bacteria counts were checked for normality and then analyzed by ANOVA using the GLM procedure in SAS (Version 9.4, SAS Institute Inc., USA). The mixed model included the fixed effects of group (two levels), of sampling time (three levels), the interaction between group and sampling time and the random effect of animal. Values of colony forming units (CFU) have been expressed as log10 g-1 of feces.
Statistical significance was reached for P ≤ 0.05 as a P-value >0.05 and ≤0.10 was considered as a trend.
Data Deposition
Raw sequences of 16S rRNA gene profiling are accessible through SRA study accession number SRP075756. Raw sequences of ITS profiling are accessible through SRA study accession number SRP080281.
Results
16S rRNA Profiling of CZ Treated Dog
Pooled fecal samples from CZ treated (Tr) and no-treated (NTr) dogs were obtained in order to assess the microbiota composition based on 16S rRNA-sequencing analysis as described previously (Milani et al., 2013). The sequencing produced a total of 589784 reads with an average of 98297 reads per sample (Supplementary Table S1).
Assessment of rarefaction curves, based on the Chao1 biodiversity indexes calculated for 10 subsampling of sequenced read pools, indicated that both curves tend to reach a plateau. Therefore, in all cases the obtained sequencing data was deemed adequate to cover the vast majority of the biodiversity contained within the samples (Figure 1A). Moreover, the two curves did not show relevant differences, thus indicating that the analyzed samples have similar biodiversity.
FIGURE 1.
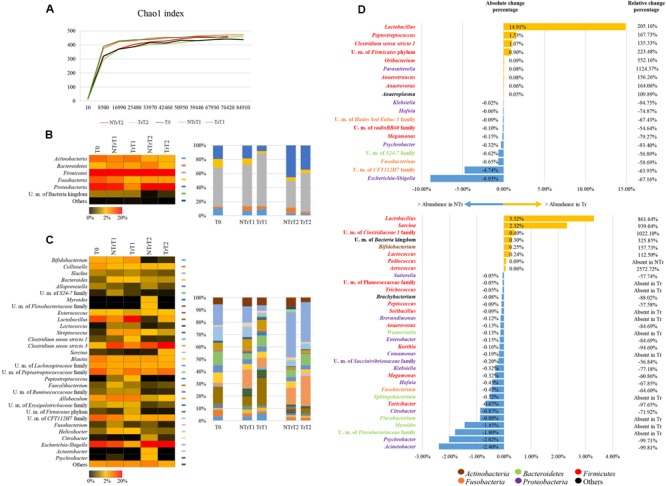
Exploration of the taxonomic profile of NTr and Tr groups. (A) Shows the rarefaction curves representing variation of the Chao1 and the Shannon diversity indexes at increasing sequencing depth of NTr and Tr fecal samples. (B) Displays bar plots and heat map of the identified bacterial phyla in the pooled CZ treated or untreated samples. (C) Represents bar plots and heat map of the identified bacterial genera in the pooled Tr or NTr samples. (D) exhibits the variation of taxa at time point T1 (upward) and T2 (below). We reported the bacterial genera with absolute change percentage >0.05% and showing increase >100% or decrease <-50% of relative change percentage in Tr data sets as compared to those obtained from NTr samples. In all panels the term unclassified member is abbreviated to U. m..
Gut Microbiota Composition of CZ Treated Dogs
During the study, diarrhea events were not observed in the CZ treated dogs. CZ did not affect the palatability of the feed, which was eaten completely within 30 min after dosing. Fecal scores were not affected by the factors in the statistical model (P > 0.05; Table 2). Fecal microbiota differences were observed in relation to group and sampling time (P < 0.05).
Table 2.
Effects of chabazitic zeolitite (CZ) supplementation on fecal score and fecal microbial concentration (least squares means of log10 CFU g-1 of feces).
| Parameter | Groups∗ |
Sampling time |
SEM† |
P-values |
|||||
|---|---|---|---|---|---|---|---|---|---|
| NT | T | 0 | T1 | T2 | G‡ | St‡ | GxSt‡ | ||
| Faecal score§ | 3.15 | 3.35 | 3.07 | 3.33 | 3.37 | 1.18 | NS | NS | NS |
| Lactobacillus ssp. | 7.22 | 7.59 | 7.18 | 7.33 | 7.72 | 0.08 | <0.001 | <0.001 | <0.001 |
| Enterococcus ssp. | 7.19 | 7.51 | 7.15 | 7.22 | 7.68 | 0.05 | <0.001 | <0.001 | <0.001 |
| Enterobacteriaceae | 7.18 | 6.85 | 7.17 | 7.19 | 6.69 | 0.06 | <0.001 | <0.001 | <0.001 |
| Clostridium perfringens | 7.36 | 6.99 | 8.18 | 6.64 | 6.71 | 1.27 | NS | NS | NS |
∗NTr, untreated group; Tr, treated group. ‡SEM, standard error of the difference of means. ‡G, group effect; St, sampling time effect; G × St, interaction. § On a scale of 1 (hard) to 5 (watery).
Inspection of predicted taxonomic profiles at phylum level for all NTr samples (T0, NTrT1, NTrT2) highlighted that Firmicutes (average 51.15% ± 11.46%) represented the dominant phylum of the cecal community in dogs, outnumbering the Proteobacteria (average 27.06% ± 15.75%), the Fusobacteria (average 8.54% ± 3.46%) and the Bacteroidetes (average 5.49% ± 2.62%) phyla (Figures 1B,C).
The comparison of the average relative abundance of NTr and Tr samples at time point T1 revealed a decrease of members of the Enterobacteriaceae family (-66.99 %), such as Escherichia (-67.16%), Klebsiella (-94.75%), and Hafnia (-74.87%), in Tr samples (Figure 1D) and an increase of Lactobacillus (205.16%) and Bifidobacterium (75.35%) in CZ treated animals (Figure 1D). At time point T2 in CZ treated animals (Figure 1D), the decrease in Enterobacteriaceae (-15.34%), includes a reduction of the genera Hafnia (-67.85 %), Klebsiella (-77.18%), and Enterobacter (-84.69%), along with an increase in relative abundance of Lactobacillus (861.64%) and Bifidobacterium (157.73%) (Figure 1D).
Notably, data achieved with culture-dependent approaches largely confirmed results obtained with 16S rRNA microbial profiling. In fact, Lactobacillus ssp. and Enterococcus ssp. counts were higher, while Enterobacteriaceae counts were lower in Tr than in NTr group (P < 0.05). Lactobacillus ssp. counts tended to be higher in Tr than in NTr group on day 16 (T1; 7.43 vs. 7.24; P < 0.10) and were higher on day 29 (T2; 8.18 vs. 7.25; P < 0.05). An increase of Enterococcus ssp. concentration (8.10 vs. 7.27) and a decrease of Enterobacteriaceae counts (6.24 vs. 7.14) were found in Tr compared to NTr group on day 29 (T2; P < 0.05). Besides, no change on the fecal C. perfringens counts was reported in relation both to the sampling time and to the treatment (P > 0.05).
Bifidobacterial Community Modulation by CZ
Focusing on the contribution of bifidobacteria to the overall dog microbiota, it is worth noticing that at day 0 (T0) and in NTr animals at days 16 and 29 (T1 and T2, respectively), this genus represents 2.32% ± 1.88% of the gut microbiota of hunting dogs. In treated animals (Tr) the presence of the Bifidobacterium genus showed an increase of about 157.73% compared with Tr animals at T2, after 29 days of CZ diet (Figure 1D).
In order to precisely catalog the effects on the bifidobacterial population of dogs after CZ treatment, we performed an ITS profiling of bifidobacterial communities in stool samples of Tr and NTr dogs.
Quality filtering of the sequenced ITS amplicons produced an average of 52468 high-quality and full-length reads per sample (Supplementary Table S2) that were taxonomically attributed reaching the minimal taxonomical rank of species.
The composition of bifidobacterial populations of dogs included in the analysis showed the presence of peculiar species, such as B. pseudolongum (average of 60.70% ± 24.61% for T0 and NTr samples) and B. animalis (average of 7.84% ± 7.50% in T0 and NTr dogs) (Figure 2A), which have been previously described to be typical of the animal GIT (Milani et al., 2014a) and especially of the dog GIT (Gavini et al., 2006). Notably, other bifidobacterial species previously described to be typical of the human gut such B. catenulatum and B. bifidum were detected at a lower extend (Figure 2B).
FIGURE 2.
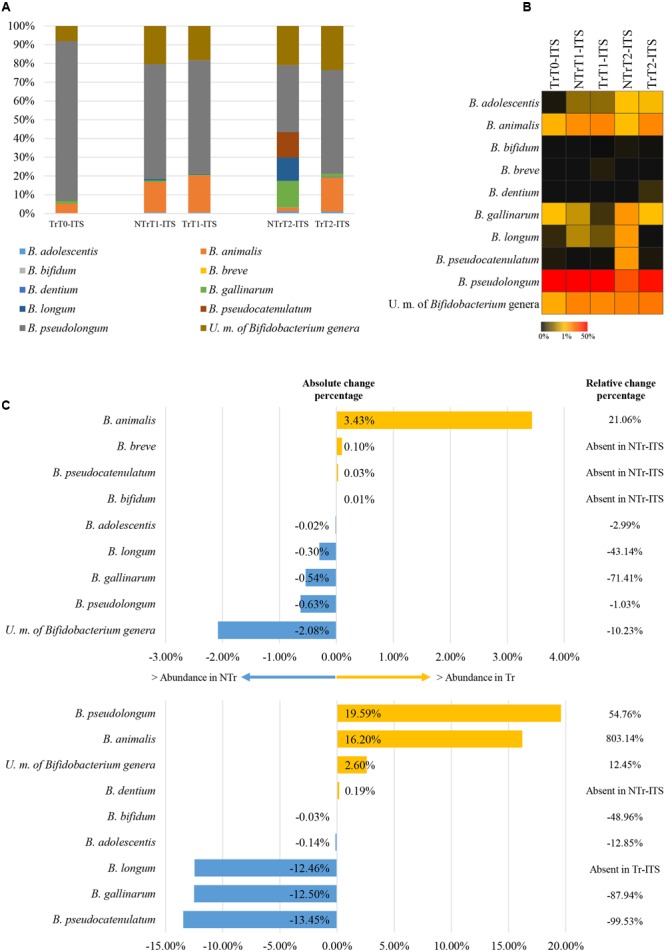
Exploration of the bifidobacterial population of NTr and Tr groups. (A) Represents the bar plots of the identified bifidobacteria in the pooled CZ treated or untreated samples through the ITS analysis. (B) Shows heat map of the identified bifidobacteria in the pooled Tr-ITS or NTr-ITS samples. (C) Displays the variation of the bifidobacterial population at time point T1 (upward) and T2 (below). We reported the Bifidobacterium species with absolute change percentage >0.05 % and showing increase >100% or decrease <-50% of relative change percentage in Tr-ITS data sets as compared to those obtained from NTr-ITS samples. In all panels the term unclassified member is abbreviated to U. m..
Furthermore, in untreated animal samples (NTr1 and NTr2), ITS analysis revealed the occurrence of B. longum, B. gallinarum, and B. pseudocatenulatum species, typical human bifidobacterial taxa (Milani et al., 2014a). One possible explanation of the presence of these species in the canine gut microbiota could be a bacterial transmission between animals and trainers as previously reported in literature (Song et al., 2013). However, further investigations will be needed. Notably, a large proportion of the OTUs defined as ‘unclassified’ in T0 dog samples (Figures 2A,B) clusters separately from any current known bifidobacterial taxon, thus putatively representing novel Bifidobacterium taxa. These putative new unclassified bifidobacterial species represents the second most present bifidobacterial taxa in the dog microbiota, in both Tr and NTr animals (Figures 2A,B).
As reported above, at time point T2 in CZ treated animals, there was an increase in relative abundance of the genus Bifidobacterium (Figure 1D). ITS profiling experiments revealed an increase of 803.14 and 54.76% of B. animalis species and B. pseudolongum species, respectively, after the addition of CZ. Moreover, a slight increase was detected also for the here identified putative new bifidobacterial taxa in TrT2 (12.45 %) compared to NTrT2 (Figure 2C).
In Vitro Bacterial Adsorptive Test
CZ showed an adsorptive capacity toward E. coli (Figure 3A) and C. perfringens (Figure 3B) strains in a dose- and time-dependent trial. Differences among CZ levels were registered for both strains after 2, 4, 6, and 24 h of incubation (P < 0.05). In particular, higher adsorptive capability against E. coli strain, was observed when CZ was added to the medium at a dose of 0.5 and 1 g rather than of 0 and 0.25 g (P < 0.05). When CZ was added at a dose of 1 g, negative values of OD starting from 0 h of incubation was observed for E. coli. During the first six hours of incubation, the adsorptive effect of CZ on C. perfringens strain was higher for levels of 0.5 and 1 g, than of 0 and 0.25 g (P < 0.05; Figure 3).
FIGURE 3.
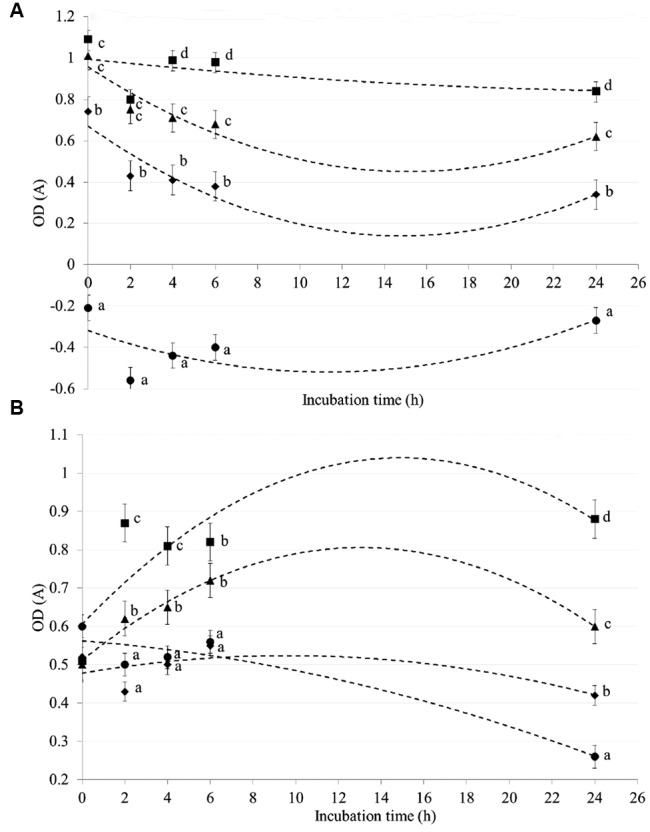
In vitro adsorptive capacity of chabazitic zeolitite (CZ) toward Escherichia coli(A) and Clostridium perfringens(B). CZ levels: (▪) 0 g, (▲) 0.25 g, (◆) 0.5 g, (●) 1 g. Error bars indicate standard errors; a, b, c, d, P < 0.05 (differences among CZ levels).
Discussion
In hunting dogs, emotional factors, such as those to which they are submitted during the training, can affect the GIT permeability, motility, secretion and mucin production. Thus, ultimately altering the habitat of resident gut bacteria and promoting changes in the gut microbiota composition (Gagne et al., 2013). Therefore, various feeding strategies have been developed in order to keep a suitable function of the GIT tract. Zeolite and in particular CZ have shown efficacy in animals (such as pigs, calves) and humans suffering from gastrointestinal disturbances (RodriguezFuentes et al., 1997; Papaioannou et al., 2005).
In this study, 20 adult English Setter dogs were trained and fed with a diet supplemented with CZ to evaluate how the microbiota and in particular bifidobacterial population as well as specific gut pathogens, could be modulated.
The results obtained after 29 days of CZ diet, showed that CZ affects the fecal microbial concentration but not the fecal score, which remained in a desirable range (well-formed, soft stools) for healthy dogs (Gagne et al., 2013). Notably, we observed an increase in relative abundance of Lactobacillus ssp. as well as Bifidobacterium ssp. phylotypes, accompanied by a decrease in phylotypes belonging to Enterobacteriaceae family in CZ fecal samples. This could be supported by the adsorptive capacity exploited by CZ toward E. coli and C. perfringens. Furthermore, E. coli and Enterobacter are common causes of extra-intestinal opportunistic infections in in dogs (Ogeer-Gyles et al., 2006), while C. perfringens is strongly related to hemorrhagic gastroenteritis (Schlegel et al., 2012).
Moreover, the major presence of lactobacilli and bifidobacteria could be very interesting since these bacterial taxa are considered to exploit beneficial roles on the health of their hosts (Gibson et al., 2005). In this context, various members of Lactobacillus and Bifidobacterium species are the most exploited probiotic bacteria utilized for pet (Kelley et al., 2010; Strompfova et al., 2014) and some of them have been suggested to improve the health and brain function of dogs (Biagi et al., 2007; Bravo et al., 2011). Increased concentrations of these microorganisms have been associated with decreased fecal concentrations of potentially pathogenic bacteria and decreased levels of carcinogenic and putrefactive compounds in digesta (Grieshop et al., 2002).
This is the first study where the bifidobacterial community of healthy dog was explored through a Next Generation Sequencing approach involving bifidobacterial ITS profiling. The obtained results allowed the identification of a bifidobacterial profile in English setter hunting dogs and revealed the presence of typical animal bifidobacteria such as B. animalis and B. pseudolongum and many putative new taxa. CZ treatment led to an increase of the abundance of B. animalis and B. pseudolongum species, which are characterized by the presence of genes encoding for exopolysaccharides structures that could lead to a special cell protection (Ferrario et al., 2016; Hidalgo-Cantabrana et al., 2016). Increase of the bifidobacterial strains coupled with the adsorptive capacity of CZ could bring to a reduction of species belonging to the Enterobacteriaceae family, such as Klebsiella and Enterobacter, typical dog pathogens (Gibson et al., 2008). Combined CZ treatment with probiotic supplementation, such as bifidobacterial strains, might enhance the reduction of canine pathogens as well as strength the beneficial effects on the animal health.
Conclusion
Dietary CZ supplementation can help to maintain a balanced intestinal microbial ecosystem and to prevent stress-related GIT upsets in healthy dogs, with a decrease of gut pathogens and a remarkable increase of bifidobacteria. This is particularly relevant in training hunting dogs where the mental and physical stress, to which they are subjected during training periods, can affect GI permeability and motility. Further studies are needed to confirm the beneficially effect by CZ also in diseased dogs.
Author Contributions
AS, PS, VB, and MO designed and performed experiments. MO, LM, AS, PS, and CF wrote the manuscript. LM and CM performed bioinformatic analyses. AS, CF, PS, MO, and VB performed experiments. CM, LM, ER, and FDI commented the manuscript. PS, AS, and MO conceived the study, revised and approved the manuscript. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer FB and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
The authors express sincere thanks to Verdi S.p.A, Italy for generously providing the Chabasite 70® product, Della Bassana kennel, Pegognaga (MN), Italy for providing necessary facilities and dogs and Ms Mirella Masini for her operating contribution in the present study.
Funding. LM is supported by Fondazione Cariparma, Parma, Italy.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01491
References
- An Y. H., Friedman R. J. (1997). Laboratory methods for studies of bacterial adhesion. J. Microbiol. Methods 30 141–152. 10.1016/S0167-7012(97)00058-4 [DOI] [Google Scholar]
- Biagi G., Cipollini I., Pompei A., Zaghini G., Matteuzzi D. (2007). Effect of a Lactobacillus animalis strain on composition and metabolism of the intestinal microflora in adult dogs. Vet. Microbiol. 124 160–165. 10.1016/j.vetmic.2007.03.013 [DOI] [PubMed] [Google Scholar]
- Bravo J. A., Forsythe P., Chew M. V., Escaravage E., Savignac H. M., Dinan T. G., et al. (2011). Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc. Natl. Acad. Sci. U.S.A. 108 16050–16055. 10.1073/pnas.1102999108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss M., Kleffner H., Kienzle E. (2010). Carnivorous mammals: nutrient digestibility and energy evaluation. Zoo Biol. 29 687–704. 10.1002/zoo.20302 [DOI] [PubMed] [Google Scholar]
- Cresswell H. P., Hamilton G. J. (2002). “Particle size analysis,” in Soil Physical Measurement and Interpretation for Land Evaluation eds McKenzie N. J., Cresswell H. P., Coughlan K. J. (Collingwood, VIC: CSIRO Publishing; ) 224–239. [Google Scholar]
- Deng P., Swanson K. S. (2015). Gut microbiota of humans, dogs and cats: current knowledge and future opportunities and challenges. Br. J. Nutr. 113 S6–S17. 10.1017/S0007114514002943 [DOI] [PubMed] [Google Scholar]
- Duranti S., Milani C., Lugli G. A., Mancabelli L., Turroni F., Ferrario C., et al. (2016). Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci. Rep. 6 23971 10.1038/srep23971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duranti S., Milani C., Lugli G. A., Turroni F., Mancabelli L., Sanchez B., et al. (2015). Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ. Microbiol. 17 2515–2531. 10.1111/1462-2920.12743 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Ferrario C., Milani C., Mancabelli L., Lugli G. A., Duranti S., Mangifesta M., et al. (2016). Modulation of the eps-ome transcription of bifidobacteria through simulation of human intestinal environment. FEMS Microbiol. Ecol. 92 fiw056 10.1093/femsec/fiw056 [DOI] [PubMed] [Google Scholar]
- Gagne J. W., Wakshlag J. J., Simpson K. W., Dowd S. E., Latchman S., Brown D. A., et al. (2013). Effects of a synbiotic on fecal quality, short-chain fatty acid concentrations, and the microbiome of healthy sled dogs. BMC Vet. Res. 9:246 10.1186/1746-6148-9-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavini F., Delcenserie V., Kopeinig K., Pollinger S., Beerens H., Bonaparte C., et al. (2006). Bifidobacterium species isolated from animal feces and from beef and pork meat. J. Food Prot. 69 871–877. [DOI] [PubMed] [Google Scholar]
- Gibson G. R., McCartney A. L., Rastall R. A. (2005). Prebiotics and resistance to gastrointestinal infections. Br. J. Nutr. 93 S31–S34. 10.1079/Bjn20041343 [DOI] [PubMed] [Google Scholar]
- Gibson J. S., Morton J. M., Cobbold R. N., Sidjabat H. E., Filippich L. J., Trott D. J. (2008). Multidrug-resistant E. coli and enterobacter extraintestinal infection in 37 dogs. J. Vet. Intern. Med. 22 844–850. 10.1111/j.1939-1676.2008.00124.x [DOI] [PubMed] [Google Scholar]
- Grieshop C. M., Flickinger E. A., Fahey G. C., Jr. (2002). Oral administration of arabinogalactan affects immune status and fecal microbial populations in dogs. J. Nutr. 132 478–482. [DOI] [PubMed] [Google Scholar]
- Gualtieri A. F. (2000). Accuracy of XRPD QPA using the combined Rietveld-RIR method. J. Appl. Crystallogr. 33 267–278. 10.1107/S002188989901643x [DOI] [Google Scholar]
- Hartemink R., Rombouts F. M. (1999). Comparison of media for the detection of bifidobacteria, lactobacilli and total anaerobes from faecal samples. J. Microbiol. Methods 36 181–192. 10.1016/S0167-7012(99)00031-7 [DOI] [PubMed] [Google Scholar]
- Hidalgo-Cantabrana C., Algieri F., Rodriguez-Nogales A., Vezza T., Martinez-Camblor P., Margolles A., et al. (2016). Effect of a Ropy Exopolysaccharide-Producing Bifidobacterium animalis subsp. lactis strain orally administered on DSS-induced colitis mice model. Front. Microbiol. 7:868 10.3389/fmicb.2016.00868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J., Frantz N., Khoo C., Gibson G. R., Rastall R. A., McCartney A. L. (2010). Investigation of the faecal microbiota associated with canine chronic diarrhoea. FEMS Microbiol. Ecol. 71 304–312. 10.1111/j.1574-6941.2009.00812.x [DOI] [PubMed] [Google Scholar]
- Kelley R. L., Park J. S., O’Mahony L., Minikhiem D., Fix A. (2010). Safety and tolerance of dietary supplementation with a canine-derived probiotic (Bifidobacterium animalis strain AHC7) fed to growing dogs. Vet. Ther. 11 E1–E14. [PubMed] [Google Scholar]
- Kerr K. R., Beloshapka A. N., Swanson K. S. (2013a). 2011 and 2012 early careers achievement awards: use of genomic biology to study companion animal intestinal microbiota. J. Anim. Sci. 91 2504–2511. 10.2527/jas.2012-6225 [DOI] [PubMed] [Google Scholar]
- Kerr K. R., Forster G., Dowd S. E., Ryan E. P., Swanson K. S. (2013b). Effects of dietary cooked navy bean on the fecal microbiome of healthy companion dogs. PLoS ONE 8:e74998 10.1371/journal.pone.0074998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamendella R., Santo Domingo J. W., Kelty C., Oerther D. B. (2008). Bifidobacteria in feces and environmental waters. Appl. Environ. Microbiol. 74 575–584. 10.1128/AEM.01221-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie E., Riehl J., Banse H., Kass P. H., Nelson S., Marks S. L. (2010). Prevalence of diarrhea and enteropathogens in racing sled dogs. J. Vet. Intern. Med. 24 97–103. 10.1111/j.1939-1676.2009.0418.x [DOI] [PubMed] [Google Scholar]
- Mekonnen Z., Meka S., Ayana M., Bogers J., Vercruysse J., Levecke B. (2013). Comparison of individual and pooled stool samples for the assessment of soil-transmitted helminth infection intensity and drug efficacy. PLoS Negl. Trop. Dis. 7:e2189 10.1371/journal.pntd.0002189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C., Hevia A., Foroni E., Duranti S., Turroni F., Lugli G. A., et al. (2013). Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS ONE 8:e68739 10.1371/journal.pone.0068739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C., Lugli G. A., Duranti S., Turroni F., Bottacini F., Mangifesta M., et al. (2014a). Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl. Environ. Microbiol. 80 6290–6302. 10.1128/AEM.02308-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani C., Lugli G. A., Turroni F., Mancabelli L., Duranti S., Viappiani A., et al. (2014b). Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol. Ecol. 90 493–503. 10.1111/1574-6941.12410 [DOI] [PubMed] [Google Scholar]
- Milani C., Mancabelli L., Lugli G. A., Duranti S., Turroni F., Ferrario C., et al. (2015). Exploring vertical transmission of Bifidobacteria from mother to child. Appl. Environ. Microbiol. 81 7078–7087. 10.1128/AEM.02037-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogeer-Gyles J., Mathews K., Weese J. S., Prescott J. F., Boerlin P. (2006). Evaluation of catheter-associated urinary tract infections and multi-drug-resistant Escherichia coli isolates from the urine of dogs with indwelling urinary catheters. J. Am. Vet. Med. Assoc. 229 1584–1590. 10.2460/javma.229.10.1584 [DOI] [PubMed] [Google Scholar]
- Pabalan R. T., Bertetti F. P. (2001). Cation-exchange properties of natural zeolites. Nat. Zeolites 45 453–518. 10.2138/rmg.2001.45.14 [DOI] [Google Scholar]
- Papaioannou D., Katsoulos P. D., Panousis N., Karatzias H. (2005). The role of natural and synthetic zeolites as feed additives on the prevention and/or the treatment of certain farm animal diseases: a review. Microporous Mesoporous Mater. 84 161–170. 10.1016/j.micromeso.2005.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasai T. P., Walsh K. B., Bhattarai S. P., Midmore D. J., Van T. T., Moore R. J., et al. (2016). Biochar, bentonite and zeolite supplemented feeding of layer chickens alters intestinal microbiota and reduces campylobacter load. PLoS ONE 11: e0154061 10.1371/journal.pone.0154061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RodriguezFuentes G., Barrios M. A., Iraizoz A., Perdomo I., Cedre B. (1997). Enterex: anti-diarrheic drug based on purified natural clinoptilolite. Zeolites 19 441–448. 10.1016/S0144-2449(97)00087-0 [DOI] [Google Scholar]
- Rutgers H. C., Batt R. M., Proud F. J., Sorensen S. H., Elwood C. M., Petrie G., et al. (1996). Intestinal permeability and function in dogs with small intestinal bacterial overgrowth. J. Small Anim. Pract. 37 428–434. 10.1111/j.1748-5827.1996.tb02443.x [DOI] [PubMed] [Google Scholar]
- Schlegel B. J., Van Dreumel T., Slavic D., Prescott J. F. (2012). Clostridium perfringens type A fatal acute hemorrhagic gastroenteritis in a dog. Can. Vet. J. 53 555–557. [PMC free article] [PubMed] [Google Scholar]
- Simpson J. M., Martineau B., Jones W. E., Ballam J. M., Mackie R. I. (2002). Characterization of fecal bacterial populations in canines: effects of age, breed and dietary fiber. Microb. Ecol. 44 186–197. 10.1007/s00248-002-0001-z [DOI] [PubMed] [Google Scholar]
- Song S. J., Lauber C., Costello E. K., Lozupone C. A., Humphrey G., Berg-Lyons D., et al. (2013). Cohabiting family members share microbiota with one another and with their dogs. Elife 2 e00458 10.7554/eLife.00458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompfova V., Pogany Simonova M., Gancarcikova S., Mudronova D., Farbakova J., Mad’ari A., et al. (2014). Effect of Bifidobacterium animalis B/12 administration in healthy dogs. Anaerobe 28 37–43. 10.1016/j.anaerobe.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Suchodolski J. S., Camacho J., Steiner J. M. (2008). Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol. Ecol. 66 567–578. 10.1111/j.1574-6941.2008.00521.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


