Abstract
Detection of somatic mutations in non–small cell lung cancers (NSCLCs), especially adenocarcinomas, is important for directing patient care when targeted therapy is available. Here, we present our experience with genotyping NSCLC using the Ion Torrent Personal Genome Machine (PGM) and the AmpliSeq Cancer Hotspot Panel v2. We tested 453 NSCLC samples from 407 individual patients using the 50 gene AmpliSeq Cancer Hotspot Panel v2 from May 2013 to July 2015. Using 10 ng of DNA, up to 11 samples were simultaneously sequenced on the Ion Torrent PGM (316 and 318 chips). We identified variants with the Ion Torrent Variant Caller Plugin, and Golden Helix's SVS software was used for annotation and prediction of the significance of the variants. Three hundred ninety-eight samples were successfully sequenced (12.1% failure rate). In all, 633 variants in 41 genes were detected with a median of 2 (range of 0 to 7) variants per sample. Mutations detected in BRAF, EGFR, ERBB2, KRAS, NRAS, and PIK3CA were considered potentially actionable and were identified in 237 samples, most commonly in KRAS (37.9%), EGFR (11.1%), BRAF (4.8%), and PIK3CA (4.3%). In our patient population, all mutations in EGFR, KRAS, and BRAF were mutually exclusive. The Ion Torrent Ampliseq technology can be utilized on small biopsy and cytology specimens, requires very little input DNA, and can be applied in clinical laboratories for genotyping of NSCLC. This targeted next-generation sequencing approach allows for detection of common and also rare mutations that are clinically actionable in multiple patients simultaneously.
Introduction
Lung cancers are broadly classified as small cell or non–small cell cancers (NSCLCs), with NSCLCs further subtyped largely on the basis of histologic features and immunohistochemistry profile. NSCLCs include adenocarcinoma (ADC), squamous cell carcinoma (SqCC), large cell carcinoma, and other less common subtypes (e.g., adenosquamous carcinoma and sarcomatoid carcinoma) [1]. The genomic profile of NSCLC is highly variable both across and within histologic subtypes [2], [3].
Incorporation of molecular analysis in the pathologic evaluation of nonsquamous NSCLC is now considered the standard of care in clinical practice [4], [5], [6]. Once the molecular profile of a tumor is known, the appropriate use of targeted clinical therapies or eligibility for clinical trials can be determined. It is desirable to have the ability to analyze several genes simultaneously to assess for the presence of a known clinically actionable variant in a tumor. In cases without clinically actionably mutations, it is also beneficial to document the genomic profile of a tumor should a targeted therapy be discovered. In addition, immunotherapies may be an alternative therapeutic option for patients who lack known actionable mutations, forming another pathway to targeted therapy.
Next-generation sequencing (NGS) is one testing modality that can detect multiple gene variants simultaneously, allowing for the precise diagnosis of a tumor at the genetic level. The Ion Torrent platform can be used in the clinical laboratory for sequencing of NSCLC, among other cancer types, in an efficient and cost-effective manner. In many instances, only a small biopsy or cytology specimen is available for molecular testing; therefore, the ability to detect known targetable driver mutations from a small amount of input DNA is often required. Here, we present our experience with NGS using the Ion Torrent Personal Genome Machine (PGM) to detect somatic mutations in NSCLC; this assay covers 2855 COSMIC-cited mutations in 50 cancer-related genes.
Methods
All NSCLCs with a diagnosis of ADC or poorly differentiated NSCLC, favor ADC (small biopsy and cytology samples), and adenosquamous carcinoma or those in which adenosquamous carcinoma cannot be excluded are reflexively genotyped at our institution. In May 2013, our laboratory introduced a targeted NGS panel, the Ion AmpliSeq 50-gene Cancer Hotspot Panel v2, for this purpose followed by reflex ALK fluorescence in situ hybridization testing for tumors that are negative for EGFR, KRAS, and BRAF [4]. Rarely, an NSCLC of other histology (sarcomatoid, SqCC, or large cell neuroendocrine carcinoma) was also tested on a per-request basis. Institutional review board approval, including a waiver of consent, was granted to review our genotyping experience in lung cancer.
DNA Extraction
Hematoxylin-eosin–stained slides of resection specimens, biopsy, or fine needle aspiration (FNA) cell blocks were reviewed to determine the area of tumor for extraction and the percent tumor content within that area; a minimum of 10% tumor content was considered acceptable for further processing. DNA was extracted from 8 unstained sections (4 μm) using a manual extraction, the Gentra Puregene Kit (Qiagen Inc., Valencia, CA), or the QIAamp DNA FFPE Tissue Kit automated on the Qiacube Instrument (after August 2015) (Qiagen Inc., Valencia, CA) according to the manufacturer's instructions following deparaffinization with xylene. DNA quantification was performed using the Quant-iT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific Inc.) according to the manufacturer's directions on a TECAN microplate reader (Tecan Group Ltd., Männedorf, Switzerland).
Sequencing
The validation of this targeted panel was previously reported [7]. Briefly, bar-coded libraries were prepared using 10 ng of DNA. Up to 11 samples were simultaneously sequenced on the Ion Torrent PGM (Life Technologies, Rockville, MD) (316 and 318 chips) using the manufacturer's recommended protocol. The Ion Torrent Variant Caller Plugin v4.0 was used to align reads to the reference genome hg19. All variant calls were initially filtered to remove benign polymorphisms and noncoding and synonymous.
Variants
Golden Helix SNP and Variation Suite software v.8.2.1 was used for annotation and prediction of the significance of the variants. A report detailing the detected variants and resultant amino acid changes was included in each patient's medical record.
Quality Assurance Metrics
At various steps in the sequencing process, all samples were subject to six quality assurance measures (Table 1): minimum tumor cellularity, DNA quantification, DNA quality, library quantification, ISP quantification, and data analysis metrics [8].
Table 1.
Quality Assurance Metrics Applied during the Preanalytic and Analytic Processing and Analysis of the Samples
| QA Measure | Method | Acceptable Criteria |
|---|---|---|
| Minimum tumor cellularity | Pathologist review | >10% |
| DNA quantification | PicoGreen | 1.7 ng/μl of DNA⁎ |
| DNA quality | Kapa Library Quantification Kit (Kapa Biosystems, Wilmington, MA) | Q129/Q41 > 0.4⁎ |
| Library quantification | qPCR (7500 Fast Real-Time PCR System) | (100 pM each) and Pooled (100 pM) |
| ISP quantification | qPCR (Qubit 2.0 Fluorometer, Life Technologies) | 10% TO 30% (<10%: FAILED E-PCR >30%: POLYCLONAL AMPLIFICATION) |
| Data analysis metrics | Ion Variant Caller Plug-in Golden Helix SVS | Postsequencing metrics were established at the run, sample, and variant levels.† |
Samples must fail both DNA concentration and KAPA to be deemed quality not sufficient.
For each run, the following sequencing metrics were verified: chip loading (>70.0%), usable sequences (>55.0%), polyclonality (<35.0%), and low-quality reads (<20.0%). For each individual sample, the metrics assessed were on-target reads (>90.0%), coverage uniformity (>90.0%), and ≥95% amplicons with 500× coverage (to avoid amplicon dropouts and false negatives). And finally, for each variant, the metrics assessed were coverage ≥500×, allelic frequency of ≥5%, and strand bias between 0.40 and 0.59.
Results
Four hundred fifty-three (453) samples from 407 individual patients were submitted for sequencing from May 2013 to July 2015. There were a total of 204 females and 203 males. All tumors were diagnosed as NSCLC; the vast majority were ADC or poorly differentiated carcinoma, favor ADC (n= 437). Additional histologic types included cases where adenosquamous carcinoma could not be excluded (n= 8), squamous cell carcinoma (n= 4), sarcomatoid carcinoma (n= 3), or large cell neuroendocrine carcinoma (n= 1).
Quantity Not Sufficient (QNS) Cases
Overall, 55 of 453 samples tested (12.1%) from 48 individual patients had an insufficient amount of material for sequencing; all of these were of the ADC subtype. Of these, the lung core biopsies had the highest failure rate comprising 42% (23/55) of all QNS specimens (Figure 1A). Twenty-two of the patients whose sample originally failed processing underwent sequencing with a different sample in which 19 (86.4%) were successfully sequenced on the second attempt. Twenty-six of the patients with QNS samples did not undergo additional testing during the study period. In total, 378 of 407 patients (93%) had sequencing results.
Figure 1.
Distribution of sample types that were successfully sequenced (n= 398). LN – lymph node; FNA – Fine needle aspiration.
Sequencing Results
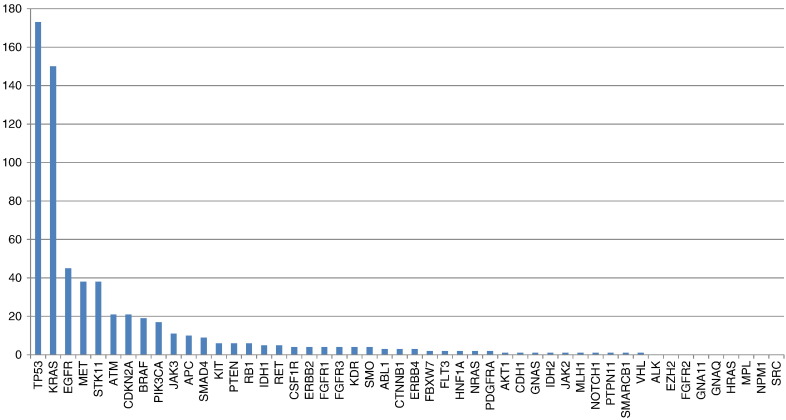
Sequenced samples (n= 398) were from primary and metastatic sites and consisted of the following: lung core biopsies (n= 110, 28%), regional lymph node (LN) FNA (n= 92, 23%), excision of the primary lung tumor (n= 59, 15%), biopsies of metastatic sites (other than regional LN) (n= 51, 13%), lung FNAs (n= 38, 9%), cell block of a fluid (pleural/pericardial) (n= 21, 5%), outside consult cases sent for molecular testing (n= 18, 4%), and regional LN biopsies or excisions (n= 9, 2%) (Figure 1B). Metastatic sites included the following: abdominal metastasis (1), adrenal (4), bone (2,) brain (22,) epidural tumor (1), paraspinal tumor (2), gastroesophageal junction metastasis (1), inguinal LN (2), liver (4), neck LN (3), kidney (3), pleural biopsy (3), skin metastasis (2), and spleen (1). Among the successfully sequenced samples, 633 variants in 41 genes were detected (Figure 2) with a median of 2 variants and a range of 0 to 7 variants per case (45 samples with 0 variant [wild type]; 145 samples with 1 variant; 132 samples with 2 variants; 58 samples with 3 variants; 15 samples with 4 variants; 2 samples with 5 variants; and 1 sample [the LCNEC case] with 7 variants).
Figure 2.
Number of cases with variants detected in the respective 50 genes included in the NGS assay.
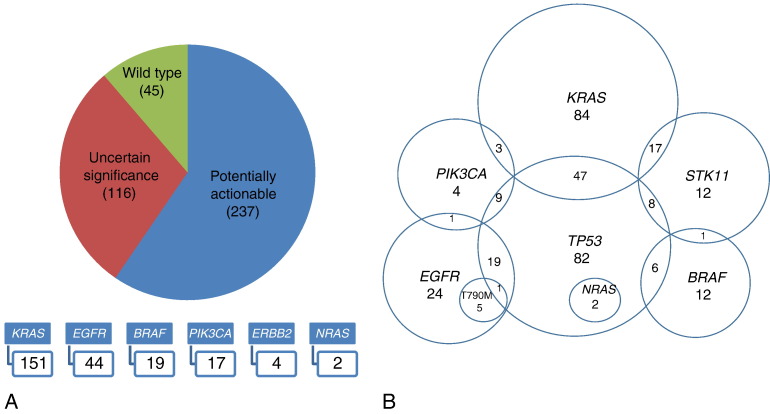
Mutations detected in BRAF, EGFR, ERBB2, KRAS, NRAS, and PIK3CA were considered potentially actionable. For the purpose of this manuscript, we defined actionable as any variant that either has an FDA-approved therapy assigned to it or for which there is a clinical trial indication. Such actionable mutations were identified in 237 samples, most commonly in KRAS (n= 151, 37.9%), EGFR (n= 44, 11.1%; excluding 6 secondary p. T790 M mutations), BRAF (n= 19, 4.8%) and PIK3CA (n= 17, 4.3%). Potentially actionable mutations were detected in 60% of samples (237/398). The breakdown of the specific mutations identified in the genes considered actionable is shown in Figure 3 and Table S1. One hundred sixteen samples had variants detected in other genes that are currently not clinically actionable, and 45 samples had no mutations detected with the 50 gene panel.
Figure 3.
Clinically actionable mutations identified.
The vast majority of identified potentially actionable mutations were single nucleotide variants (205) followed by insertions and deletions (31) and only 1 frame shift mutation. Within EGFR, common and rare mutations were detected in exons 18 to 21 including known activating mutations p. G719C, exon 19 deletions, and p. L858R as well as resistance mutations including p. L747S, p. L761I, exon 20 insertions, p. T790M, and p. L861Q. The 6 samples that had p. T790M mutations also had a coexisting exon 19 deletion, consistent with acquired resistance. Interestingly, within the 1 sample that harbored EGFR p. L747S, a described acquired resistance mutation, no other mutations were identified; it is currently not known if this patient was tyrosine kinase inhibitor (EGFR TKI) naive. Four ERBB2 exon 20 mutations and 2 NRAS codon 61 mutations were identified. And in BRAF, 19 mutations were identified, 7 of which were p. V600E (37%) with 10 (53%) occurring in exon 11.
We also identified co-occurrence of some of the most frequently altered and clinically significant genes (Figure 4). Not surprisingly, TP53 mutations co-occurred with mutations in KRAS, EGFR, BRAF, PIK3CA, NRAS, and STK11. STK11 mutations were most commonly seen in association with KRAS mutations. PIK3CA mutations were only rarely identified co-occurring with other driver mutations in KRAS or EGFR. Similar to prior reports in lung ADC, all mutations in EGFR, KRAS, and BRAF were mutually exclusive in our patient population. Interestingly, we also noticed a mutually exclusive pattern among some additional genes: SMO, SMARCB1, SMAD4, RET, RB1, PTPN11, PTEN, PIK3CA, and PDGFRA (which is currently of uncertain significance).
Figure 4.
Co-occurrence of clinically actionable mutations.
Patients with Multiple Tumors Tested
Although most of the patients who had testing performed on multiple samples were due to an insufficient quantity of material on the first sample, we did have a cohort of patients who had multiple tumors tested. Four patients had multiple synchronous lung tumors tested: three bilateral and one ipsilateral. In all four cases, divergent mutational profiles were identified (Table 2). Eight patients either had the primary lung tumor and metastatic sites (four patients) or multiple metastatic sites tested. Metastatic sites included LN metastases, pleural fluid or biopsy, adrenal and liver metastases, brain metastases, and a skin metastasis. Among these tumors, there were a consistent mutational profile among primary tumors and metastatic sites and identification of EGFR p. T790M resistance mutations in the metastatic tumors of two patients (Table 2).
Table 2.
In a Select Number of Patients, Multiple Tumors Were Tested: Mutations Identified in Four Cases with Synchronous Tumors and Eight Cases with Metastatic Tumors
| Synchronous Tumors | Site | Mutations Identified | Site | Mutations Identified | Site | Mutations Identified |
|---|---|---|---|---|---|---|
| Patient S1 | Lung, left | EGFR L858R | Lung, right | TP53 H61R | ||
| Patient S2 | Lung, left | KRAS G12 V; ATM G2869R | Lung, right | KRAS G12C; CCNTB1 S37C | ||
| Patient S3 | Lung, left | KRAS G12 V; CDKN2A c.151-2 A > T | Lung, right | STK11 D194Y | ||
| Patient S4 | Lung, right | KRAS G12C; TP53 R158L | Lung, right | PIK3CA R335S; TP53 G245C | ||
| Metastatic tumors | ||||||
| Patient M1 | LN | EGFR ex19 del | Pleural fluid | EGFR ex 19 del; EGFR T790 M | Pleural fluid | EGFR ex 19 del; EGFR T790 M |
| Patient M2 | Lung, right | EGFR ex 19 del | Pleural fluid | EGFR ex 19 del | ||
| Patient M3 | Pleural fluid | EGFR ex 19 del; TP53 H193Y | Adrenal metastasis | EGFR ex 19 del; TP53 H193Y | Liver metastasis | EGFR ex 19 del; EGFR T790 M; TP53 H193Y |
| Patient M4 | Lung | None detected | Subcutaneous metastasis | None detected | ||
| Patient M5 | Lung | ERBB2 ex 20 ins; TP53 Y88C | Brain metastasis | ERBB2 ex 20 ins; TP53 Y88C | ||
| Patient M6 | Lung | FGFR1 A268P; TP53 A159P | LN | FGFR1 A268P; TP53 A159P | ||
| Patient M7 | Pleural biopsy | TP53 P190L | LN | TP53 P190L; PTEN Y16* | ||
| Patient M8 | LN | KRAS G12C; IDH1 R132L | Brain metastasis | KRAS G12C; IDH1 R132L | ||
S, synchronous tumors; M, metastatic tumors; LN, lymph node.
Discussion
Next-generation sequencing is a powerful tool that allows for testing multiple targets in multiple genes in multiple patients simultaneously. NGS assays are rapidly being adopted in clinical laboratories for this purpose, replacing individual single gene assays and expanding the testing capacity that can be performed on relatively small amounts of tissue. Here, we report from our experience using the AmpliSeq 50 gene Cancer Hotspot Panel v2 in the clinical testing of a large number of lung adenocarcinomas. The Ion Torrent platform requires very little input DNA (10 ng), thus allowing for successful testing of small biopsy and FNA specimens. As we showed, we had a very high success rate with theses samples. Surprisingly, our highest failure rate was with lung core needle biopsies. We speculate that this may in part be due to challenges of getting the fixed tumor cells into solution when associated with a fibrous background/scar as opposed to FNA cell blocks where the cells are already in solution prior to fixation. Additionally, many of the lung core biopsy procedures at our institution have immediate onsite cytologic assessment performed via touch imprint preparations. Transfer of tumor cells to the cytology slides may result in some decreased tumor cellularity in the formalin-fixed paraffin tissue block. Requesting additional passes, minimizing tissue “rolling,” and preparing unstained slides upfront are measures that we have utilized to increase molecular testing yield.
In our population, we identified a higher percentage of KRAS mutations (37.9% vs 28% and 26%) and fewer EGFR mutations (11.1% vs 14% and 20%) than reported by Hageman et al. and Dogan et al., respectively [9], [10]. This may be due to different prevalence of smoking and ethnicity in the different sample sets or selection bias of the various populations or testing methods (for instance, we tested multiple samples from the same patient in several instances).
Using this targeted NGS panel, we identified clinically actionable rare mutations that have specific therapeutic significance that would have gone undetected using single gene assays previously used in our laboratory. Examples include detection of EGFR and ERBB2 exon 20 insertions, EGFR p. T790M secondary resistance mutations, as well as additional variants within EGFR, KRAS, and BRAF (Table S1 and Figure 3) [11]. Recently, the efficacy of afatinib, a second-generation irreversible TKI, was reviewed in 75 patients with uncommon EGFR mutations that had been enrolled in the LUX-lung trials 2, 3, and 6. Whereas afatinib was active in patients with uncommon mutations including p. L861Q (which is typically resistant to first-generation TKIs) and p. G719X, the response rate was low in tumors with exon 20 insertions [12]. EGFR exon 20 insertions are biologically different than common EGFR mutations because they do not affect the ATP-binding pocket of the kinase domain but rather the c-helix domain. This results in increased kinase activity but decreased affinity to TKIs [12], [13]. Alternative therapies for these patients are needed. Currently, a phase 2 clinical trial of a heat shock protein 90 inhibitor (AUY922) is under way in patients with EGFR exon 20 insertion mutations (NCT01854034). Another EGFR mutation we detected, an exon 7 p. A289V, was classified as a variant of uncertain significance; this mutation in the EGFR extracellular domain is commonly seen in glioblastomas, but the significance is uncertain in NSCLC [14].
ERBB2 exon 20 insertion mutations are identified in a small proportion of ADCs (approximately 2%) and also define a distinct subset of tumors. The most common mutation is a 12-bp insertion (p. A775 G776insYVMA) found in 50% to 80% of ERBB2 mutant lung cancers [15], [16]. ERBB2 exon 20 mutations are mutually exclusive with common EGFR and KRAS mutations and are distinct from tumors showing ERBB2 amplification [17]. ERBB2 targeted therapies for this indication are available including lapatinib, dacomitinib, and afatinib (NCT02369484) [17].
In BRAF, we identified 19 mutations; however, only 7 of these (37%) were p. V600E and 10 (53%) occurred in exon 11. These findings are consistent with those in a recent report by Carter et al. that demonstrated a broad spectrum of unique BRAF mutations in lung cancers with a high prevalence of mutations located in exon 11 (41%) [18]. The response to selective BRAF inhibitors in patients with non–codon 600 mutations is unclear, although some report that tumors with exon 11 or mutations impairing the kinase activity are predicted to be unresponsive to current BRAF inhibitors [18], [19]. However, preclinical studies suggest that dasatinib and MEK inhibitors with or without a BRAF inhibitor may have efficacy in such patients [18], [20]. Presently, patients with non-V600 mutations are also eligible for an NCI-Match trial with the MEK inhibitor trametinib (NCT02465060). We also detected mutations in KIT, IDH1, IDH2, PTEN, CDKN2A, and JAK2 that are well characterized in other tumor types and may allow patients to meet eligibility criteria for novel clinical trials. Patients with mutations that not known to be actionable are referred to the Norris Cotton Cancer Center Molecular Tumor Board [21].
The detection of multiple, co-occurring, potentially actionable mutations in an individual tumor represents an advance in molecular pathology with possible significant clinical, therapeutic, and research implications based on the different combinations of mutations. Eng et al. recently published their data on the impact of concurrent PIK3CA mutations with other oncogenic driver mutations on response to therapy. Overall, they found that a concurrent PIK3CA mutation was a poor prognostic factor in EGFR or KRAS mutant lung ADC, although it did not significantly alter the benefit of EGFR TKI therapy in the EGFR mutant patients [22]. However, co-occurrence with KRAS, as was identified in three cases in our study, may in fact be a contraindication for targeted therapies as mutations in both of these genes effect the PI3K-AKT pathway and alternative therapies such as immunotherapies may be more effective. Overall, among EGFR and KRAS mutant lung cancers, those with a concurrent PIK3CA mutation are unique with prognostic and predictive implications.
STK11 mutations, commonly identified in lung ADC, often coexist with KRAS mutations and likely also have a confounding prognostic significance. Pécuchet et al., who examined a cohort of 567 resected nonsquamous NSCLC patients and validated their findings in 2 publically available datasets, found that patients with STK11 mutations, specifically in exons 1 and 2, had a significantly worse prognosis than wild-type tumors or tumors with exon 3 to 9 mutations [23]. Recent reports also suggest that co-occurring genomic alterations can define heterogeneous subsets of KRAS-mutant lung ADC with distinct clinical implications [24]. Skoulidis et al. describe three robust subsets of KRAS-mutant ADC dominated by co-occurring mutations in STK11, TP53, and CDKN2A/B inactivation. They identified differences in drug sensitivity patterns including expression of PD-L1 and susceptibility to HSP90-inhibitor therapy, significant findings given the challenges to date of therapeutically targeting KRAS-mutant lung ADC [24].
In our study, we have shown the utility of the Ion Torrent Ampliseq technology for clinical genotyping of NSCLC which requires very little input DNA and can successfully be performed on small biopsy and cytology specimens. This targeted NGS approach allows for detection of common and also rare clinically actionable mutations and profiles of co-mutations in multiple patients simultaneously.
The following is the supplementary data related to this article.
Potentially Actionable Mutations Detected by Gene and Tumor Type
Acknowledgements
The authors thank the staff of the Laboratory for Clinical Genomics and Advanced Technology (CGAT). The data presented in this manuscript were in part generated through CGAT in the Department of Pathology and Laboratory Medicine of the Geisel School of Medicine at Dartmouth, the Dartmouth Hitchcock Medical Center, and the Norris Cotton Cancer Center.
Footnotes
Disclosure/Conflict of Interest: The authors of this paper have no conflicts of interest to report.
References
- 1.Travis W.D., Brambilla E., Burke A.P., Marx A., Nicholson A.G., editors. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. 4th ed. IAEC; Lyon: 2015. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013;137:828–860. doi: 10.5858/arpa.2012-0720-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Comprehensive Cancer Network . 2015. NCCN Guidelines Version 4. [Google Scholar]
- 6.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba II. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsongalis GJ, Peterson JD, de Abreu FB, Tunkey CD, Gallagher TL, Strausbaugh LD. Routine use of the Ion Torrent Ampliseq(™) Cancer Hotspot Panel for identification of clinically actionable somatic mutations. Clin Chem Lab Med. 2014;52:707–714. doi: 10.1515/cclm-2013-0883. [DOI] [PubMed] [Google Scholar]
- 8.De Abreu FB, Peterson JD, Amos CI, Wells WA, Tsongalis GJ. Effective quality management practices in routine clinical next generation sequencing. Clin Chem Lab Med. 2016 doi: 10.1515/cclm-2015-1190. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagemann IS, O'Neill PK, Erill I, Pfeifer JD. Diagnostic yield of targeted next generation sequencing in various cancer types: an information-theoretic approach. Cancer Genet. 2015;208:441–447. doi: 10.1016/j.cancergen.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 10.Dogan S, Shen R, Ang DC, Johnson ML, D'Angelo SP, Paik PK. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massarelli E, Johnson FM, Erickson HS, Wistuba II, Papadimitrakopoulou V. Uncommon epidermal growth factor receptor mutations in non–small cell lung cancer and their mechanisms of EGFR tyrosine kinase inhibitors sensitivity and resistance. Lung Cancer. 2013;80:235–241. doi: 10.1016/j.lungcan.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Yang JC, Sequist LV, Geater SL, Tsai CM, Mok TS, Schuler M, Yamamoto N, Yu CJ, Ou SH, Zhou C. Clinical activity of afatinib in patients with advanced non–small-cell lung cancer harbouring uncommon EGFR mutations: a combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015;16:830–838. doi: 10.1016/S1470-2045(15)00026-1. [DOI] [PubMed] [Google Scholar]
- 13.Tafe LJ, Tsongalis GJ. A 78-year-old woman with brain metastases. Clin Chem. 2015;61:584–586. doi: 10.1373/clinchem.2014.229864. [DOI] [PubMed] [Google Scholar]
- 14.Vivanco I, Robins HI, Rohle D, Campos C, Grommes C, Nghiemphu PL. Differential sensitivity of glioma- versus lung cancer–specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li BT, Lee A, O'Toole S, Cooper W, Yu B, Chaft JE, Arcila ME, Kris MG, Pavlakis N. HER2 insertion YVMA mutant lung cancer: long natural history and response to afatinib. Lung Cancer. 2015;90:617–619. doi: 10.1016/j.lungcan.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, Zakowski MF, Kris MG, Ladanyi M. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12:220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li BT, Ross DS, Aisner DL, Chaft JE, Hsu M, Kako SL, Kris MG, Varella-Garcia M, Arcila ME. HER2 amplification and HER2 mutation are distinct molecular targets in lung cancers. J Thorac Oncol. 2016;11:414–419. doi: 10.1016/j.jtho.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carter J, Tseng LH, Zheng G, Dudley J, Illei P, Gocke CD. Non-p.V600E BRAF mutations are common using a more sensitive and broad detection tool. Am J Clin Pathol. 2015;144:620–628. doi: 10.1309/AJCP85ATMJOZOUDJ. [DOI] [PubMed] [Google Scholar]
- 19.Gautschi O, Peters S, Zoete V, Aebersold-Keller F, Strobel K, Schwizer B. Lung adenocarcinoma with BRAF G469L mutation refractory to vemurafenib. Lung Cancer. 2013;82:365–367. doi: 10.1016/j.lungcan.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Sen B, Peng S, Tang X, Erickson HS, Galindo H, Mazumdar T. Kinase-impaired BRAF mutations in lung cancer confer sensitivity to dasatinib. Sci Transl Med. 2012;4:136ra70. doi: 10.1126/scitranslmed.3003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tafe LJ, Gorlov IP, de Abreu FB, Lefferts JA, Liu X, Pettus JR. Implementation of a molecular tumor board: the impact on treatment decisions for 35 patients evaluated at Dartmouth-Hitchcock Medical Center. Oncologist. 2015;20:1011–1018. doi: 10.1634/theoncologist.2015-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eng J, Woo KM, Sima CS, Plodkowski A, Hellmann MD, Chaft JE. Impact of concurrent PIK3CA mutations on response to EGFR tyrosine kinase inhibition in EGFR-mutant lung cancers and on prognosis in oncogene-driven lung adenocarcinomas. J Thorac Oncol. 2015;10:1713–1719. doi: 10.1097/JTO.0000000000000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pécuchet N, Laurent-Puig P, Mansuet-Lupo A, Legras A, Alifano M, Pallier K. Different prognostic impact of STK11 mutations in non-squamous non–small-cell lung cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.6379. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skoulidis F, Byers LA, Diao L, Papadimitrakopoulou VA, Tong P, Izzo J. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov. 2015;5:860–877. doi: 10.1158/2159-8290.CD-14-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Potentially Actionable Mutations Detected by Gene and Tumor Type