Abstract
Cancer metastasis is a multistep process associated with the induction of an epithelial-mesenchymal transition (EMT) and cancer stem cells (CSCs). Although significant progress has been made in understanding the molecular mechanisms regulating EMT and the CSC phenotype, little is known of how these processes are regulated by epigenetics. Here we demonstrate that reduced expression of DNA methyltransferase 1 (DNMT1) plays an important role in the induction of EMT and the CSC phenotype by prostate cancer (PCa) cells, with enhanced tumorigenesis and metastasis. First, we observed that reduction of DNMT1 by 5-azacitidine (5-Aza) promotes EMT induction as well as CSCs and sphere formation in vitro. Reduced expression of DNMT1 significantly increased PCa migratory potential. We showed that the increase of EMT and CSC activities by reduction of DNMT1 is associated with the increase of protein kinase C. Furthermore, we confirmed that silencing DNMT1 is correlated with enhancement of the induction of EMT and the CSC phenotype in PCa cells. Additionally, chromatin immunoprecipitation assay reveals that reduction of DNMT1 promotes the suppression of H3K9me3 and H3K27me3 on the Zeb2 and KLF4 promoter region in PCa cells. Critically, we found in an animal model that significant tumor growth and more disseminated tumor cells in most osseous tissues were observed following injection of 5-Aza pretreated–PCa cells compared with vehicle-pretreated PCa cells. Our results suggest that epigenetic alteration of histone demethylation regulated by reduction of DNMT1 may control induction of EMT and the CSC phenotype, which facilitates tumorigenesis in PCa cells and has important therapeutic implications in targeting epigenetic regulation.
Introduction
Prostate cancer (PCa) is the second leading cause of cancer deaths in American males [1]. Death by PCa in most patients is accompanied by bone metastasis [2]. Therefore, understanding the mechanisms underlying the generation of metastasis is crucial for developing future therapies for cancer treatment. Increasing evidence demonstrates that cytokines and growth factors from the stromal tumor microenvironment are essential in neoplastic progression, which results in deregulation of normal growth-control mechanisms in the primary tumor sites [3]. In the prostate, many stromal signals from the tumor microenvironment contribute to the development of epithelial-mesenchymal transition (EMT) and cancer stem cell (CSC) phenotypes initiating the metastatic cascade [3], [4], [5], [6]. Recent studies also demonstrate that the induction of EMT creates a self-renewing state (CSCs) that enables redifferentiation to allow growth and colonization at distant metastatic sites [5], [6]. Although many investigations provide important clues as to which stromal elements and factors lead to tumor progression and metastasis, the molecular mechanisms underlying EMT and stemness remain unclear.
Recent studies suggest that epigenetic regulation by DNA methyltransferases (DNMTs) associates with EMT and CSC populations within tumors [7], [8], [9], [10], [11], [12]. DNA methyltransferase 1 (DNMT1) is a member of the DNA methyltransferase family that is responsible for maintaining methylation patterns located in CG dinucleotide-rich regions as well as for transcriptional repression in cells [13], [14], [15]. Aberrant expression of DNMT1 during the G0/G1 phase in the cell cycle may silence tumor suppressors and eliminate normal arrest signals, leading to the uncontrolled growth in cancer cells [15], [16]. It has been shown that cell cycle regulation of DNMT1 is disrupted in colorectal cancer cells in vivo [17], [18] and in other cancer cells [19], [20]. It has also been demonstrated that murine DNMT1 mRNA is undetectable in growth-arrested BALB/c 3T3 cells but is highly induced at the G1-S boundary of the cell cycle [21]. In addition, both DNMT1 and p21 have been shown to bind to the proliferating cell nuclear antigen (PCNA) at the same site in human acute lymphoblastic leukemia cells, acting as competitive inhibitors [22]. Elevated p21 competes with DNMT1 for PCNA binding in G0/G1 phase, whereas during S phase, elevated DNMT1 competes with p21 for binding to PCNA, allowing replication to occur [22]. In PCa, the studies have shown that high DNMT1 expression is correlated with advanced-stage cancers [23], [24], [25], [26]. Conversely, there is also some evidence that decreased expression of DNMT1 promotes progression of early-stage cancers [26]. Importantly, growing evidence suggests that loss of DNA methylation induces EMT and cancer stem-like phenotypes, resulting in tumorigenesis and migratory potential and ultimately increasing tumorigenesis and metastatic capacities [7], [8], [9], [10].
In the present study, we explored whether DNMT1 facilitates EMT induction and the transition of CSCs in PCa cells, which results in tumorigenesis and metastasis. We demonstrated that reduction of DNMT1 enhances EMT and CSC phenotypes in association with protein kinase C (PKC). We also observed that loss of DNMT1 increases migration of PCa cells. Moreover, we demonstrated in an animal model that reduction of DNMT1 by pretreatment of 5-azacitidine (5-Aza) in vitro significantly increases primary tumor growth and bone metastasis potential of PCa cells. Our results suggest that DNMT1 regulates PCa metastasis by associating with EMT and CSC properties and has important implications in targeting of epigenetic therapies.
Materials and Methods
Cell Culture
The human PCa cell lines PC3 and DU145 were obtained from the American Type Culture Collection (Rockville, MD). The green fluorescent protein (GFP)–expressing PCa cell lines (PC3GFP and DU145GFP cells) were established by lentiviral transduction. PCa cells were grown in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin.
Quantitative Reverse Transcriptase Polymerase Chain Reaction (QRT-PCR)
Total RNA was extracted from cells using the RNeasy mini kit (cat. 74104, Qiagen, Valencia, CA) and converted into cDNA using a First-Strand Synthesis Kit (Invitrogen). Quantitative PCR (real-time PCR) was performed on an ABI 7700 sequence detector using TaqMan Universal PCR Master Mix according to the directions of manufacturer (Applied Biosystems, Foster City, CA). TaqMan MGB probes (Applied Biosystems) were as follows: DNMT1 (Hs00154749_m1), E-Cadherin (Hs01013952_m1), N-Cadherin (Hs00169953_m1), Vimentin (Hs00185584_m1), CD44 (Hs01075861_m1), NFkB1 (HS00765730_m1), Snail1 (Hs00195591_m1), Snail2 (Hs00950344_m1), Zeb1 (Hs00232783_m1), Zeb2 (Hs00207691_m1), TGFBR2 (Hs00234253_m1), KLF4 (Hs00358836_m1), and PKCα (Hs00925193_m1). β-Actin (Hs01060665_g1) was used as internal control for the normalization of target gene expression.
RNA Interference
PC3 or DU145 cells at 60% confluence were seeded onto 6-well culture plates. After 24 hours, negative control siRNA (cat. 4390843; Ambion, Foster City, CA) or DNMT1 siRNA (cat. 439082, Ambion) with OPTI-MEM (cat. 31985-062; Life Technologies, Carlsbad, CA) was transfected into PCa cells using Lipofectamine RNAiMAX (cat. 56532, Life Technologies) according to the manufacturer's instructions. Transfected cells were incubated at 37°C for 72 hours, and the cells were used in various cell assays. Silencing was verified by real-time PCR and Western blots.
CSC (CD133+/CD44+ Phenotype) Analysis
PCa cells (PC3 or DU145) (1 × 105) were seeded onto 12-well culture plates. The cells were treated with vehicle or a demethylating agent, 5-Aza (0-20 μM) for 4 days. PCa cells were incubated with PE-anti-CD133 antibody (cat. 130-080-901; Miltenyi Biotec, San Diego, CA) and APC-anti-CD44 antibody (cat. 559942; BD Biosciences, San Jose, CA) for 20 minutes at 4°C. The CD133+/CD44+ fraction was analyzed with a FACS Aria High-Speed Cell Sorter (BD Biosciences).
Prostatosphere Culture and Assay
The PCa cells (PC3 or DU145) were dissociated to single cells by standard trypsinization and washed three times with PBS. The cells were plated in stem cell culture medium (Dulbecco’s modified Eagle’s medium: F12 plus 10 ng/ml bFGF, 20 ng/ml EGF, 5 mg/ml insulin, and 0.4% BSA) supplemented with 1% KO serum replacement (cat. 10828-028, Invitrogen) [27] at a density of 1000 cells/ml in low-attachment 6-well culture plates. Seven-day-old spheres were enumerated as cell clusters comprised of >30 cells.
Transwell Chemotaxis Assay
PC3 or DU145 cells at 60% confluence (1 × 105) were seeded onto 24-well culture plates. The cells were treated with vehicle or 5-Aza (5 μM) for 4 days prior to use in Transwell chemotaxis assays. For these studies, the cells were labeled with 2.5 μg/ml of carboxyfluorescein diacetate (cat. V12883; Molecular Probes, Eugene, OR), which facilitated enumeration. The vehicle- or 5-Aza–treated PCa cells were resuspended in serum-free RPMI 1640 and equilibrated for 10 minutes at 37°C. Carboxyfluorescein diacetate–labeled PCa cells were loaded into the top chambers of 8-μm Transwell microporous membrane 24-well plates (cat. 3422; Costar Corp, Cambridge, MA). A total of 650 μl of conditioned medium with chemoattractant CXCL12 (200 ng/ml) (cat. 350-NS-050; R& D System, Minneapolis, MN) was added in the bottom. The plates were incubated at 37°C. At 3 hours, the number of cells which had migrated into the lower chambers was determined by plate reader (Molecular Probes, Eugene, OR).
Immunostaining
Cells, tumor sections, and long-bone sections were used for immunostaining. Cells were fixed with 10% neutral-buffered formalin (cat. HT501320, Sigma) and permeabilized with Perm/Wash Buffer (cat. 554723; BD Biosciences, San Jose, CA). Tumor sections and long bone sections were blocked, deparaffinized, hydrated, then blocked with Image-iT FX signal enhancer for 30 minutes, and incubated for 2 hours at room temperature with primary antibodies combined with reagents of Zenon Alexa Fluor 488 (green) or 555 (red) labeling kit (Invitrogen). Human E-Cadherin (1:25 dilution, cat. 610181, BD Biosciences), N-Cadherin (1:25 dilution, cat. 610921, BD Biosciences), CD133 (1:50 dilution, cat. 130-090-423; Miltenyi Biotec, San Diego, CA), CD44 (1:100 dilution, cat. ab6124; Abcam, Burlingame, CA), DNMT1 (1:50 dilution, cat. ab19905; Abcam, Cambridge, MA), Zeb2 (1:10 dilution, cat. NBP1-82991; Novus Biologicals, Littleton, CO), KLF4 (1:50 dilution, cat. ab75486, Abcam), GFP (1:200 dilution, cat. ab290, Abcam), and (1:100 dilution, cat. ab25117, Abcam) primary antibodies were used. After washing with PBS, the slides were mounted with ProLong Gold antifade reagent with DAPI (Invitrogen). Images were taken with a Olympus FV-500 confocal microscope. Human prostate tissue microarrays (TMAs) were purchased from US Biomax, Inc. (Rockville, MD). Tumors were graded using stage progressing system. Co-staining of anti-human DNMT1 and Zeb2, DNMT1 and N-Cadherin, or DNMT1 and KLF4 antibodies was applied to TMAs.
Western Blot
PCa cells were cultured in RPMI 1640 with 10% fetal bovine serum and 1% penicillin/streptomycin. The cells were treated with vehicle or 5-Aza (5 μM) for 4 days at 37°C. Whole cell lysates were prepared from cells, separated on 4% to 20% Tris-Glycine gel, and transferred to PVDF membranes. The membranes were incubated with 5% milk for 1 hour and incubated with primary antibodies overnight at 4°C. Primary antibodies used were as follows: monoclonal anti-DNMT1 (1:1000 dilution, cat. 5032; Cell Signaling, Danvers, MA), polyclonal anti–E-Cadherin (1:1000 dilution, cat. sc-7870; Santa Cruz, Santa Cruz, CA), monoclonal anti–N-Cadherin (1:1000 dilution, cat. 610921; BD Transduction Laboratory, San Jose, CA), polyclonal anti-Zeb2 (1:1000 dilution, cat. PA5-20980; Thermo Fisher, Waltham, MA), polyclonal anti-Vimentin (1:1000 dilution, cat. 3932, Cell Signaling), monoclonal anti-PKCα (1:1000 dilution, cat. 2056, Cell Signaling), monoclonal anti-KLF4 (1:1000 dilution, cat. 12173, Cell Signaling), monoclonal anti-OCT4 (1:1000 dilution, cat. 2750, Cell Signaling), and monoclonal anti-SOX2 (1:1000 dilution, cat. 3579, Cell Signaling). Blots were incubated with peroxidase-coupled anti-mouse IgG secondary antibody (cat. 7076, 1:2000 dilution, Cell Signaling) or peroxidase-coupled anti-rabbit IgG secondary antibody (cat. 7074, 1:2000 dilution, Cell Signaling) for 1 hour, and protein expression was detected with SuperSignal West Dura Chemiluminescent Substrate (cat. prod. 34075; Thermo Scientific, Rockford, IL). Membranes were reprobed with monoclonal anti–β-Actin antibody (1:1000 dilution, cat. 4970, Cell Signaling) to control for equal loading.
Protein Kinase C (PKC) ELISA
PCa cells (PC3 or DU145) (1 × 105) were seeded onto 12-well culture plates. The cells were treated with vehicle or 5-Aza (5 μM) for 4 days. Whole cell lysates were prepared to evaluate PKC kinase activity by following the directions of the manufacturer (cat. ADI-EKS-420A; Enzo Life Sciences, Farmingdale, NY). The levels of PKC kinase activity were normalized to total cell numbers.
In Vitro Chromatin Immunoprecipitation (CHIP)
CHIP assays were performed in vehicle- or 5-Aza (5 μM)–treated PCa cells (PC3 or DU145) (4 × 106) and siControl- or siDNMT1-treated PCa cells (PC3 or DU145) (4 × 106) by following the directions of the manufacturer (cat. 334471, EpiTech CHIP OneDay Kit, Qiagen). PCa cells were treated with 1% formaldehyde to cross-link histones to DNA. The cross-linking was stopped by treating the samples with stop buffer for 5 minutes. The chromatin was extracted and fragmented by sonication, and the lysate was used to immunoprecipitation using Protein A + G beads and the following antibodies; H3K9me3 (cat. GAH-6204, Qiagen), H3K27me3 (cat. GAH-9205, Qiagen), or rabbit control IgG (cat. GAH-9205, Qiagen). Immunocomplexes were pulled down and washed, and DNA was isolated to run SYBR qPCR with Zeb2 (assay tile: −0.4 kb; assay position: −3502) (cat. GPH1021580(−)04A, Qiagen) and KLF4 (assay tile: −0.4Kb; assay position: −3110) (cat. GPH1026871(−)04A, Qiagen) specific primers on promoter sites.
Subcutaneous Tumor Growth
All experimental animal procedures were performed in compliance with the institutional ethical requirements and approved by the University of Michigan Committee for the Use and Care of Animals. To evaluate tumor growth, subcutaneous tumors were established. PCa cells (PC3GFP or DU145GFP) were treated with vehicle or 5 Aza (5 µM) for four days and then 2 x 105 cells were loaded with Collagen I (cat. 354236; BD Bioscience, Bedford, MA) and were injected subcutaneously (s.c.) into 5- to 7-week-old male SCID mice (n = 8/group). The animals were monitored daily. After 4 weeks, the animals were sacrificed. The tumors and tissue samples were collected. The tumor volumes and tumor weights were evaluated. Tumor volumes were calculated using the formula V = (the shortest diameter) × (the longest diameter) × height. The tumors and femorae were prepared for histology, and tissue samples were used for tumor metastasis assays.
In Vivo Metastasis Assay
To evaluate metastasis, PCa cells labeled with GFP were treated in vitro for 4 days with 5-Aza (5 µM) or vehicle, and were subsequently implanted into 5-7 week old male SCID mice. Following tissue harvest, cancer cell engraftment was assessed by real-time PCR using human Alu TaqMan probes (F: 5′-CAT GGT GAA ACC CCG TCT CTA-3′, R: 5′-GCC TCA GCC TCC CGA GTA G-3′, TaqMan probe: 5′-FAM-ATT AGC CGG GCG TGG TGG CG-TAMRA-3′, Applied Biosystems). The DNA levels were expressed as relative copies (% control) normalized against murine β-actin (Mm00607939_s1, Applied Biosystems), and a standard curve constructed from serial dilutions of a purified Alu cDNA fragment was cloned by classic PCR. Numerical data were determined against a standard curve established using murine bone marrow containing log-fold dilutions of human PCa cells. Positive and negative controls include tissues obtained from non–PCa-injected mice or DNA derived directly from PCa cells [28]. Immunohistochemistry for PCa cells in the marrow was also used for metastasis assays. The numbers of GFP-positive PCa cells were quantified in the endosteal region of the long bones defined as 10 cell diameters from the bone surfaces.
Statistical Analyses
Results are presented as mean ± standard deviation. Distribution of the data was determined by skewness [29]. Values within a 1.95 (5%) confidence interval were accepted as a normal distribution and evaluated further using parametric tests for significance. Unpaired Student's t tests were conducted on data sets of two comparisons, and significance of data with multiple comparisons was evaluated using an analysis of variance. Values of P < .05 were considered significant.
Results
Reduction of DNMT1 Promotes EMT Induction in PCa Cells
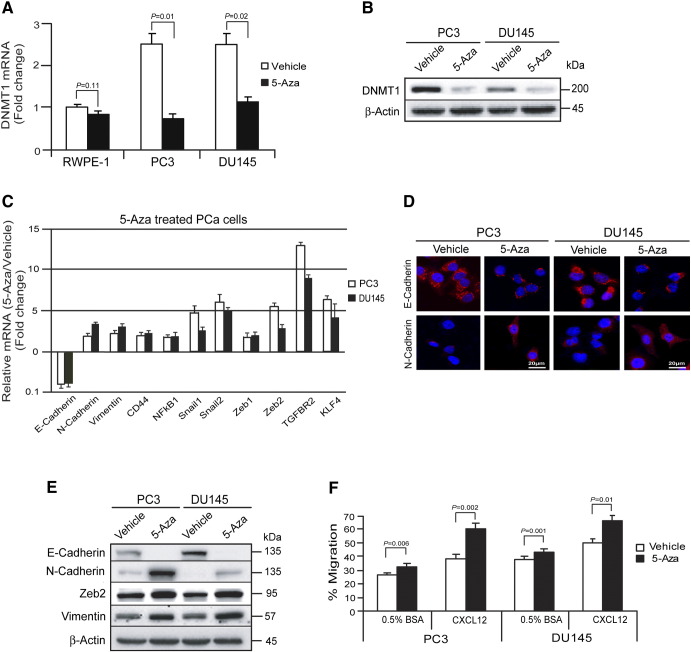
To explore the role that the epigenetic alteration of DNMT1 plays in tumor progression, we examined the expression levels of DNMT1 in PCa cells following treatment with 5-Aza. At baseline, high levels of DNMT1were expressed in human PCa cells. After 4 days of 5-Aza treatment, DNMT1 expression was significantly decreased in PCa cells (Figure 1, A and B). These findings suggest that 5-Aza alters the level of DNMT1 in PCa cells. To further determine if reduction of DNMT1 expression regulates EMT induction, we first examined the expression of epithelial and mesenchymal phenotypes in vehicle- or 5-Aza–treated PCa cells by real-time PCR, immunofluorescence staining, and Western blots. Expression of N-Cadherin and Vimentin were significantly increased by 5-Aza treatment in PCa cells versus vehicle-treated PCa cells, indicating the induction of a mesenchymal phenotype. In sharp contrast, more E-Cadherin was expressed in vehicle-treated PCa cells versus 5-Aza–treated PCa cells (Figure 1, C–E). Moreover, expression of mRNA for mesenchymal associated transcription factors Snail1, Snail2, Zeb1, Zeb2, and KLF4 was increased in 5-Aza–treated PCa cells (Figure 1C). At the protein level, a significant increase in N-Cadherin, Vimentin, and the Zeb2 transcription factor expression was observed in response to 5-Aza treatment, whereas E-Cadherin levels decreased (Figure 1E). These data suggest that reduction of DNMT1 by 5-Aza is associated with an EMT in PCa cells. To explore the functional role that 5-Aza treatment has on PCa cells, a migration assay was performed. We observed that more 5-Aza–treated PCa cells migrated toward the chemoattractant CXCL12 compared with vehicle-treated PCa cells (Figure 1F). Taken together, these findings suggest that reduction of DNMT1 expression by 5-Aza may play a role in PCa migration.
Figure 1.
Reduction of DNMT1 promotes EMT induction in PCa cells.
PCa cells (PC3 or DU145) (1 × 105) were seeded in 6-well culture plates. The cells were treated by vehicle or 5-Aza (0-20 μM) for 4 days.
(A) mRNA levels of DNMT1 expression in vehicle- or 5-Aza (5 μM)–treated PCa cells (PC3 or DU145) as quantified by real-time PCR. The human prostate epithelial cell RWPE-1 was used as a control. (B) Protein levels of DNMT1 following vehicle or 5-Aza (5 μM) treatment in PCa cells (PC3 or DU145) as quantified by Western blots. (C) mRNA levels of EMT markers in vehicle- or 5-Aza (5 μM)–treated PCa cells (PC3 or DU145) as quantified by real-time PCR. (D) E-Cadherin or N-Cadherin expression in vehicle- or 5-Aza (5 μM)–treated PCa cells (PC3 or DU145) as detected by immunofluorescence staining. Blue, DAPI nuclear stain. Bar = 20 μm. (E) EMT markers in vehicle- or 5-Aza (5 μM)–treated PCa cells (PC3 or DU145) as quantified by Western blots. (F) Migration of vehicle- or 5-Aza (5 μM)–treated PCa cells toward the chemoattractant CXCL12 was performed in Transwell plates. Percent migration was set at 100% for the initial cell numbers in top chamber. The data in A, C, and F are representative of mean ± SD (n = 6). P values were calculated by Student's t test.
Reduction of DNMT1 Stimulates Transition of CSCs in PCa Cells
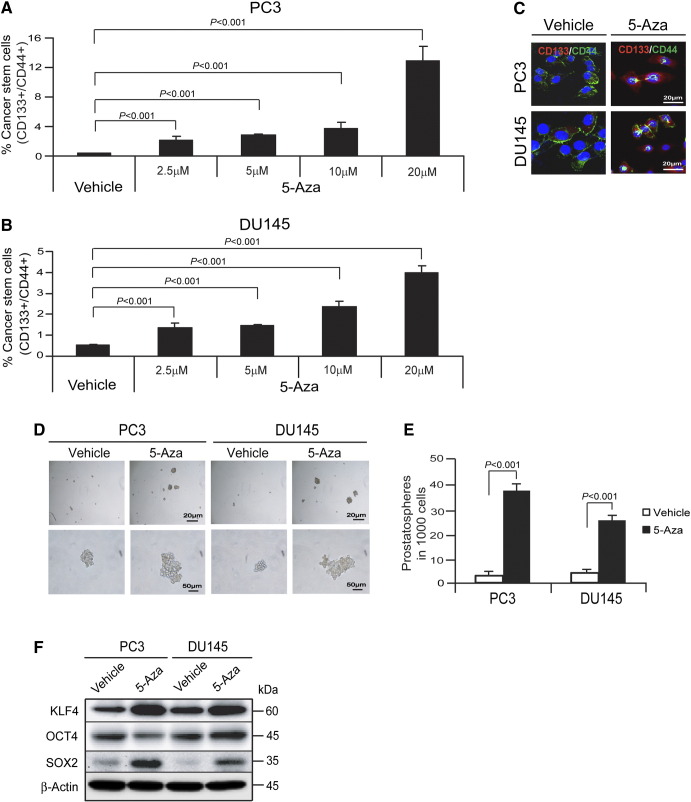
The transition to a CSC phenotype is a critical event that enables redifferentiation to allow growth and colonization at distant metastatic sites. To expand our understanding of the interplay between the EMT and CSC activities, we examined the impact of 5-Aza treatment on CSCs. To determine if reduction of DNMT1 expression regulates the CSC phenotype (CD133+/CD44+), PCa cells were treated with 5-Aza for 4 days, and CSCs were examined by FACS analysis. A dose-dependent increase in CSCs was identified in 5-Aza–treated PC3 cells (Figure 2A) and DU145 cells (Figure 2B). Immunofluorescence staining was used to confirm the results obtained by FACS analysis. As shown in Figure 2C, the CD133/CD44 phenotype was significantly enhanced in PCa cells treated with 5-Aza compared with PCa cells treated with vehicle. Next, we evaluated the impact of DNMT1 expression on prostatosphere formation. When DNMT1 expression is reduced following 5-Aza treatment, a greater number of prostatospheres formed in culture, and the spheres had a significantly greater range of sizes (Figure 2, D and E). We further examined whether reduction of DNMT1 by 5-Aza treatment alters their expression of stem cell–related transcription factors KLF4, OCT4, and SOX2 in PCa cells. Western blot analysis demonstrates that the levels of KLF4 and SOX2 were significantly enhanced in the PC3 and DU145 cells treated with 5-Aza compared with PCa cells treated with vehicle. OCT4 was decreased in PC3 cells, whereas it was increased in DU145 cells treated with 5-Aza compared with the cells treated with vehicle (Figure 2F). Collectively, these results suggest that reduction of DNMT1 expression plays a crucial role in transition to the CSC phenotype in PCa cells.
Figure 2.
Reduction of DNMT1 stimulates transition of CSCs in PCa cells.
CSCs (CD133+/CD44+ phenotype) from vehicle- or 5-Aza (5 μM)–treated (A) PC3 cells or (B) DU145 cells were evaluated by FACS analysis. (C) Coexpression of CD133 (red) and CD44 (green) in vehicle- or 5-Aza (5 μM)–treated PCa cells as detected by immunofluorescence staining. Blue, DAPI nuclear stain. Bar = 20 μm. (D) In vitro prostatosphere formation of vehicle or 5-Aza–treated PCa cells (bar = 20-50 μm) and (E) quantification of Figure 2D. (F) Stem cell–related transcription factors as quantified by Western blots. The data in A, B, and E are representative of mean ± SD (n = 6).
P values were calculated by Student's t test.
PKCα Associates with DNMT1-Mediated EMT and CSCs in PCa Cells
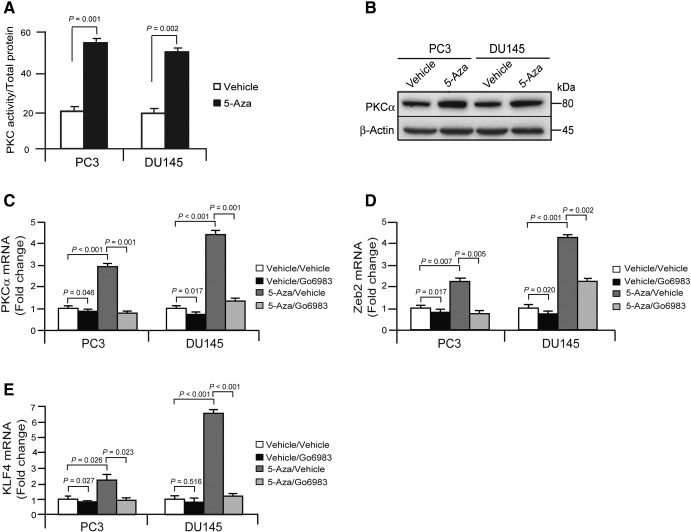
The PKC protein family plays a central role in cellular signal processing. Growing evidence suggests that various isoforms of PKC participate in the regulation of cell proliferation, differentiation, survival, and death in normal epithelial cells as well as cancers [30], [31], [32], [33]. It has been demonstrated that the PKCα signaling network is activated specifically in CSC-enriched populations [31] and that PKCα suppresses keratinocyte proliferation by increasing p21Cip1 levels by a KLF4 transcription factor–dependent mechanism [32]. Therefore, we evaluated the involvement of PKCα in EMT induction and transition to a CSC phenotype in 5-Aza–treated PCa cells with reduced DNMT1 expression. The protein of PKCα was significantly enhanced in the PCa cells treated with 5-Aza compared with PCa cells treated with vehicle (Figure 3, A and B). Moreover, we examined whether the enhancement of PKCα by the reduction of DNMT1 links to inductions of EMT and CSCs by PCa cells. First, we found that mRNA levels of PKCα were significantly increased in 5-Aza–treated PCa cells compared with vehicle-treated PCa cells, and critically, mRNA levels of PKCα were reduced following treatment with a pan-PKC inhibitor, Go6983 (Figure 3C). Likewise, we also observed that mRNA levels of Zeb2 and KLF4 transcriptional factors were correlated to PKCα expression levels in 5-Aza treatment with the presence or absence of the pan-PKC inhibitor (Figure 3, D and E). These data indicate that the reduction of DNMT1 by 5-Aza regulates the EMT and CSC phenotypes in the presence of PKCα expression.
Figure 3.
PKCα associates with DNMT1-mediated EMT and CSCs in PCa cells.
PCa cells (PC3 or DU145) (1 × 105) were seeded in 6-well culture plates, and the cells were treated with vehicle or 5-Aza (5 μM) for 4 days.
PKC kinase activities as quantified by (A) ELISA (the data are representative of mean ± SD; Student's t test) and (B) PKCα protein expression as quantified by Western blots. mRNA levels of (C) PKCα, (D) Zeb2, and (E) KLF4 expression in vehicle or 5-Aza (5 μM) with the presence or absence of a pan-PKC inhibitor, Go6983 (cat. A8343; ApexBio, Houston, TX 77054) treated PCa cells (PC3 or DU145) for 4 days as quantified by real-time PCR. The data in A, C, D, and E are representative of mean ± SD (n = 3). P values were calculated by Student's t test.
Silencing DNMT1 Is Correlated with EMT Induction and Stemness in PCa Cells
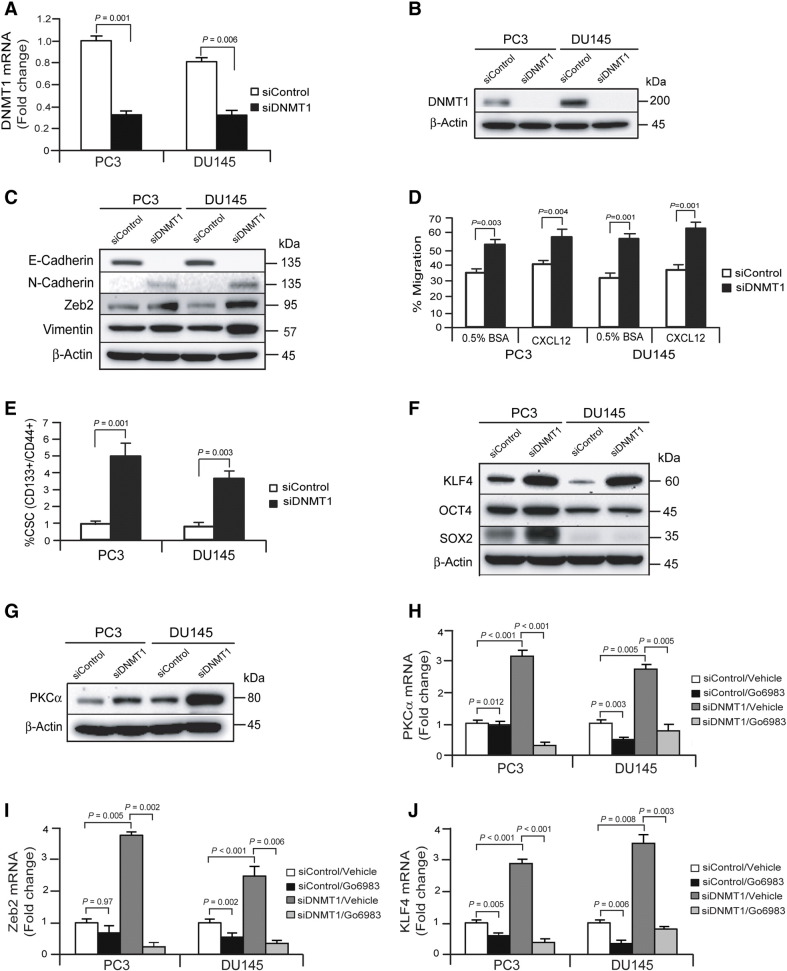
To further explore whether knockdown of endogenous DNMT1 regulates induction of EMT and the CSC phenotype, DNMT1 siRNA was used to silence DNMT1 expression in PCa cells (PC3siDNMT1 or DU145siDNMT1). mRNA (Figure 4A) and protein (Figure 4B) levels of DNMT1 were significantly decreased in PC3siDNMT1 or DU145siDNMT1 cells compared with control PCa cells (PC3siContol or DU145siControl). To test the correlation of DNMT1 expression with the EMT induction, EMT markers were first examined in DNMT1 silenced PCa cells by Western blots. E-Cadherin was decreased, whereas N-Cadherin and Vimentin were significantly increased, in PCasiDNMT1 cells compared with PCasiControl cells (Figure 4C). Moreover, Zeb2 transcription factors were significantly increased in PCasiDNMT1 cells compared with PCasiControl cells (Figure 4C). As expected, the silencing DNMT1 in PCa cells increased the migration (Figure 4D). Regarding CSC transition, more CSCs were recovered from PCasiDNMT1 cells compared with PCasiControl cells (Figure 4E). We also examined whether PCasiDNMT1 cells had increased expression of stem cell–related transcription factors KLF4, OCT4, and SOX2. The levels of KLF4, OCT4, and SOX2 were significantly enhanced in PC3siDNMT1 cells compared with PC3siControl cells, and KLF4 expression was significantly increased in DU145 siDNMT1 cells compared with DU145siControl cells (Figure 4F). Furthermore, we explored how silencing DMMT1 expression is linked to PKC expression in PCa cells. There were significantly higher levels of expression of PKCα in PCa cells (PC3siDNMT1 and DU145siDNMT1) compared with PCasiControl cells (PC3siControl and DU145siControl) (Figure 4G). We also found that mRNA levels of PKCα were significantly increased in PCasiDNMT1 cells compared with PC3siControl cells, and critically, mRNA levels of PKCα were reduced following treatment with a pan-PKC inhibitor, Go6983 (Figure 4H). We also observed that mRNA levels of Zeb2 and KLF4 transcriptional factors were correlated to PKCa expression levels in PCasiDNMT1 cells in the presence or absence of the pan-PKC inhibitor (Figure 4, I and J). These data indicate that the reduction of DNMT1 by RNA silencing regulates EMT and CSC phenotypes in the presence of PKCα expression. Taken together, these findings suggest that silencing DNMT1 is strongly correlated with enhancement of EMT and CSC phenotypes in the presence of PKCα expression in PCa cells.
Figure 4.
Silencing DNMT1 is correlated with EMT induction and stemness in PCa cells.
PCa cells (PC3 or DU145) were transiently transfected with siRNA targeting DNMT1 and cultured for 72 hours. (A) mRNA levels of DNMT1 in control or DNMT1-silenced PCa cells (PC3 or DU145) as quantified by real-time PCR. (B) Protein levels of DNMT1 in control or DNMT1-silenced PCa cells (PC3 or DU145) as quantified by Western blots. (C) EMT markers in control or DNMT1-silenced PCa cells (PC3 or DU145) as quantified by Western blots. (D) Migration of DNMT1-silenced PCa cells was performed by chemotaxis assays. (E) CSC populations (CD133+/CD44+ phenotype) from control or DNMT1-silenced PCa cells (PC3 or DU145) as evaluated by FACS analysis. The data in A, D, and E are representative of mean ± SD (n = 6). P values were calculated by Student's t test. (F) Stem cell–related transcription factors in control or DNMT1-silenced PCa cells (PC3 or DU145) as quantified by Western blots. (G) PKCα in control or DNMT1-silenced PCa cells (PC3 or DU145) as quantified by Western blots. mRNA levels of (H) PKCα, (I) Zeb2, and (J) KLF4 expression in control or DNMT1-silenced PCa cells (PC3 or DU145) with the presence or absence of a pan-PKC inhibitor for 3 days as quantified by real-time PCR. The data in H, I, and J are representative of mean ± SD (n = 3). P values were calculated by Student's t test.
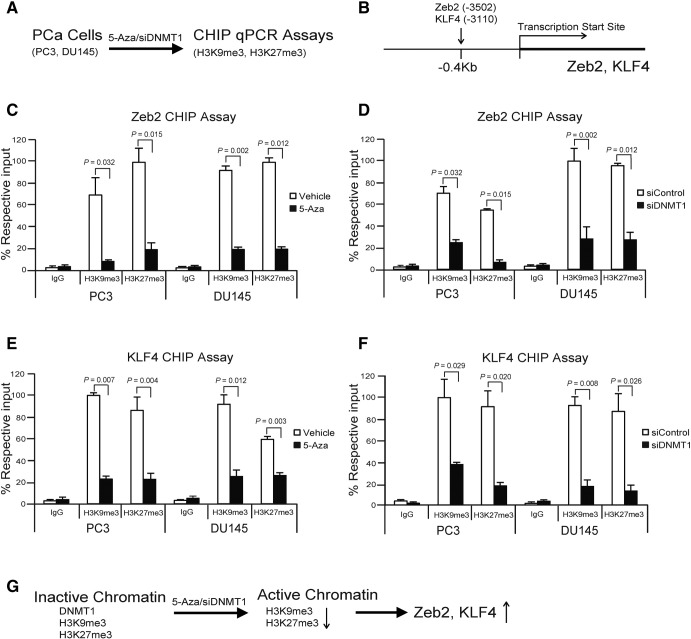
Reduction of DNMT1 Induces Histone Demethylation on the Zeb2 or KLF4 Promoter in PCa Cells
DNMT1 is necessary to maintain trimethylation of lysine 9 at histone H3 in pericentromeric regions [34], and loss of DNMT1 by 5-Aza regulates global reduction of di- and trimethylation of histone H3 lysine 9 (H3K9me2 and H3K9me3) in the many human cancer cells [34], [35]. We examined whether reduction of DNMT1 induces Zeb2 or KLF4 expression through promoter histone demethylation of H3K9me3 and H3K27me3 in the 5-Aza– or siDNMT1-treated PCa cells by CHIP qPCR assays (Figure 5, A and B). We found that the levels of both silent marks H3K9me3 and H3K27me3 were significantly suppressed in the promotor sites of Zeb2 (−0.4 kb [−3502] sites) or KLF4 (−04 kb [−3110] sites) in DNMT1 downregulated PCa cells, which promote the activation of chromatin, resulting in the increase of transcription of both Zeb2 and KLF4 (Figure 5, C–G). The data suggest that reduction of DNMT1-induced histone demethylation on the Zeb2 and KLF4 promoter is important in the regulation of Zeb2 or KLF4 transcription in EMT or CSC induction.
Figure 5.
Reduction of DNMT1 induces histone demethylation of Zeb2 or KLF4 promoter in PCa cells.
(A) Experimental design of CHIP qPCR assays in 5-Aza– or siDNMT1-treated PCa cells. (B) A scheme shows the primer sites at the promoter region from transcription start site of the human Zeb2 or KLF4 genomic locus. Percent respective input of H3K9me3 and H3K27me3 levels at the Zeb2 genomic locus by CHIP qPCR assays (C) in vehicle- or 5-Aza (5 μM)–treated PCa cells (PC3, DU145) and (D) in siControl- or siDNMT1-treated PCa cells (PC3, DU145). Percent respective input of H3K9me3 and H3K27me3 levels at the KLF4 genomic locus by CHIP qPCR assays (E) in vehicle- or 5-Aza–treated PCa cells (PC3, DU145) and (F) in siControl- or siDNMT1-treated PCa cells (PC3, DU145). The data in C–F are representative of mean ± SD (n = 3). P values are calculated by Student's t test. (G) A summary diagram shows the activation of chromatin by histone demethylation of H3K9me3 and H3K27me3 following 5-Aza or siDNMT1 treatment, resulting in the increase of Zeb2 and KLF4 transcription.
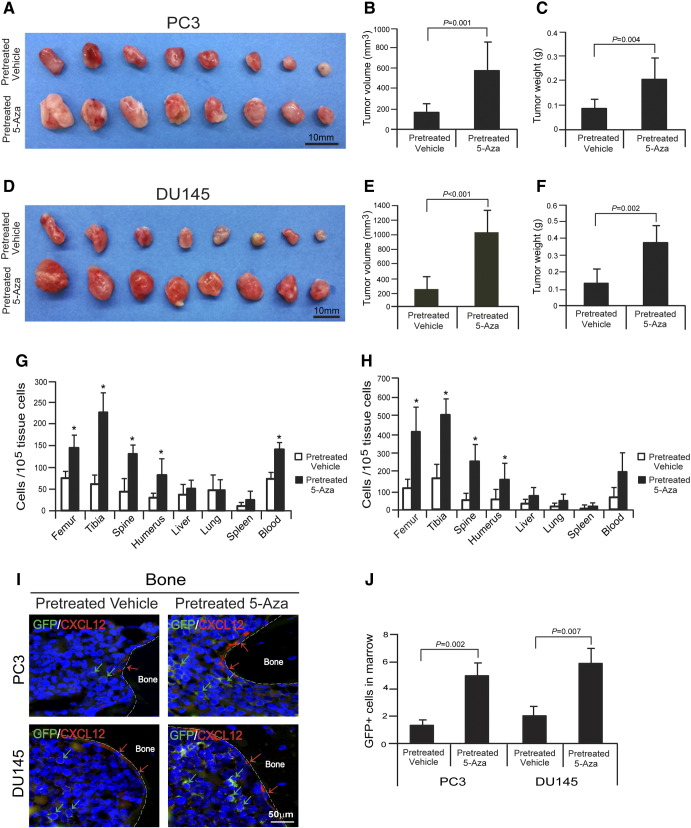
Reduction of DNMT1 Stimulates PCa Tumor Growth and Metastasis
To determine whether reduction of DNMT1 by 5-Aza regulates tumor growth or bone metastasis by PCa cells, GFP-expressing PCa cells (PC3GFP or DU145GFP) were pretreated with vehicle or 5-Aza and injected s.c. into 5- to 7-week-old male SCID mice. After 4 weeks, the mice were sacrificed. Significantly greater tumor volumes were observed in animals injected with PCa cells pretreated with 5-Aza in vitro compared with vehicle-pretreated PCa cells (Figure 6, A–F). To evaluate the metastatic capabilities of DNMT1 inhibited cells, the numbers of disseminated tumor cells (DTCs) were evaluated in the animals. All animals had significant numbers of DTCs in their bones following injection of vehicle- or 5-Aza–pretreated PCa cells. Strikingly, animals receiving 5-Aza–pretreated PCa cells showed a significant increase of the total DTC load in most osseous tissues compared with animals injected with vehicle-pretreated PCa cells (Figure 6, G and H). Critically, significant numbers of DTCs were observed in the bone marrow near osteoblasts on the bone surface following injection of 5-Aza–pretreated PCa cells (Figure 6, I and J). Together, these data suggest that reduction of DNMT1 promotes induction of EMT and CSCs, which plays critical role in PCa growth, cell dissemination, and metastasis.
Figure 6.
Reduction of DNMT1 stimulates PCa metastasis.
GFP-expressing PCa cells (PC3GFP or DU145GFP) pretreated with vehicle or 5-Aza (5 μM) for 4 days were injected s.c. into 5- to 7-week-old male SCID mice (n = 8/group). After 4 weeks, mice were sacrificed. Tumor growth was shown in tumors generated with (A–C) PC3 cells or (D–F) DU145 cells. Bar = 10 mm. (G and H) Metastasis was assessed by real-time PCR for Alu in a number of tissues. * Denotes P < .05 between vehicle- versus 5-Aza–pretreated PCa cells by Student's t test (n = 5). (I) The GFP-expressing PCa cells were identified in the femorae of SCID mice following s.c. injection. Green arrows identify PCa cells. Red arrows identify osteoblasts on the bone surface staining positive for CXCL12 expression. Bar = 50 μm. (J) Quantification of I. The numbers of PCa cells were quantified on the endosteal region of the long bones. Endosteal regions were defined as 10 cell diameters or less from the bone surfaces. Data are representative of mean ± SD (n = 8/group) on the 5 images per tumor. P values were calculated by Student's t test.
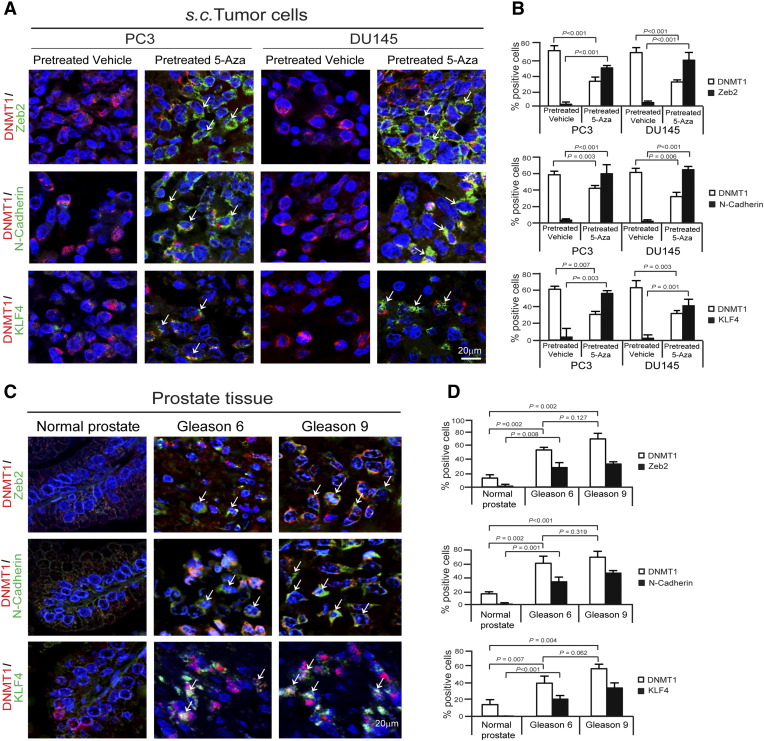
Reduction of DNMT1 Associates with Zeb2, N-Cadherin, or KLF4 in S.C. PCa Tumors and Human Primary PCa Tumors
To further determine whether EMT or CSC phenotypic changes by reduction of DNMT1 in in vitro model reflect to s.c. PCa tumors, DNMT1 and Zeb2, DNMT1 and N-Cadherin, or DNMT1 and KLF4 were co-stained in tumors generated by vehicle- or 5-Aza–pretreated PCa cells. High levels of DNMT1 expression were detected in vehicle-treated PCa tumor cells, whereas more Zeb2, N-Cadherin, or KLF4 expression was detected in the subset of DNMT1-reduced cell populations in 5-Aza–pretreated PCa tumor cells (Figure 7, A and B). To further examine whether these in vitro and animal results are correlated with human PCa tumors, DNMT1 and Zeb2, DNMT1 and N-Cadherin, or DNMT1 and KLF4 were also co-stained in TMA samples. TMAs from PCa patients demonstrated that significant levels of DNMT1 expression are detected in tumor cells compared with normal prostate epithelial tissues, and that DNMT1 expression is increased in advanced tumor cells (Figure 7, C and D). We further found that Zeb2, N-Cadherin, or KLF4 expression was increased in DNMT1-reduced tumor cells in advanced stages, which correlated with our findings in s.c. animal models. The data suggest that reduction of DNMT1 is associated with Zeb2, N-Cadherin, or KLF4, which plays critical role in PCa tumor growth and metastasis.
Figure 7.
Association of DNMT1 reduction with Zeb2, N-Cadherin, or KLF4 in s.c. PCa tumors and human primary PCa tumors.
(A) Zeb2 (green), N-Cadherin (green), or KLF4 (green) expression (white arrow) in DNMT1 (red)-expressing cells in vehicle- or 5-Aza–pretreated s.c. PCa tumors as detected by immunofluorescence co-staining. Blue, DAPI nuclear stain. Bar = 20 μm. (B) Quantification of A. Data represent mean ± SD (n = 8/group) (Student's t test). (C) Zeb2 (green), N-Cadherin (green), or KLF4 (green) expression (white arrow) in DNMT1 (red)-expressing cells in TMA samples from PCa patients as detected by immunofluorescence co-staining. Blue, DAPI nuclear stain. Bar = 20 μm. TMAs are normal prostate tissue (n = 8), Gleason 6 PCa tissue (n = 9), and Gleason 9 PCa tissue (n = 5). (D) Quantification of C. Data represent mean ± SD (Student's t test).
Discussion
Epigenetic regulation of genetic programs by DNMTs is associated with EMT and CSC induction within tumors [7], [8], [9], [10], [11], [12]. In this study, we demonstrate that reduction of DNMT1 stimulates EMT induction and the transition to a CSC phenotype by PCa cells in association with PKC expression. Importantly, these changes led to increased primary tumorigenesis and dissemination in vivo. Together, these data suggest that reduction of DNMT1 facilitates tumorigenesis in PCa cells and has important therapeutic implications in targeting epigenetic regulation.
In the prostate, tumor stroma is known to secrete cytokines and growth factors, which lay the foundations for self-renewal of CSCs that lead to tumor progression and EMT for distant metastasis [5], [6]. Cytokines and growth factors play an important role in EMT induction for the development of bone metastasis [3], [4], [36], [37], [38]. For example, HGF, CXCL12, and TGF-β produced by cancer-associated fibroblasts (CAFs) have been reported as stimulators of EMT in cancer cells [3], [4], [36], [37], [38]. CAF-derived CXCL12 triggering CXCR4 expression in cells having undergone an EMT is consistent with the essential role of CXCL12-CXCR4 signaling during metastasis, as the level of CXCL12 is elevated in the bone [39]. Recently, we demonstrated that enhanced levels of stromal CXCL12 by CAFs promote EMT that supports PCa bone metastasis [3], [4]. Furthermore, transient induction of EMT creates a self-renewing state (CSCs) that enables redifferentiation to allow growth and colonization at distant metastatic sites [5], [6]. Interestingly, a recent report shows that CXCL12α stimulates the increase of cardiac stem cell progenitor cells with c-kit expression through the inhibition of DNMT1 and DNMT3b expression as well as demethylation of the c-kit gene [40]. In this study, we demonstrated that reduction of DNMT1 induces EMT and CSCs phenotypes by PCa cells within primary tumors, resulting in tumorigenesis and metastasis. Elucidating stromal signals in the tumor microenvironment that are responsible for epigenetic regulation of EMT and CSC phenotypes may increase the success of targeted therapies.
Understanding the epigenetic mechanisms of altering DNA methylation underlying the functional balance of quiescent and proliferating cancer cells is one of the key pieces of data which is essential for our ability to better target cancer therapy [9], [10], [11], [12]. Deregulated expression of DNMT1 during the cell cycle may be associated with cellular transformation [15]. In many cancers, high levels of DNMT1 expression are observed in advanced and metastatic progression [16], [17], [18], [19], [20], [23], [24], [25], [26]. We also found that high levels of DNMT1 are expressed in PCa cell lines. It has also been demonstrated that aberrant expression of DNMT1 eliminates normal arrest signals, leading to the uncontrolled growth in cancer cells [13], [14]. The DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-Aza-CdR), a potential anticancer agent, functions in part to reduce DNMT1 expression by the activation of normal arresting signals, resulting in antiproliferative effects and antitumorigenic activities. Yet these actions also lead to the generation of cell cycle arrest signals, which induce CSCs and EMT phenotypes and ultimately increase tumorigenesis, and metastatic capacities [9], [10].
Many CSCs are not likely cycling and stay in G0, regulated by arresting signals with the expression of cell cycle inhibitors (e.g., p21 and p27) [41], [42], [43], [44], [45], [46]. The epigenetic regulation by DNMTs associates with CSC populations within tumors [9], [10], [11], [12]. CSCs may increase tumorigenesis through the stem cell processes of self-renewal and differentiation [9], [10], [11], [12]. Further evidence demonstrates that the loss of pericentromeric DNA methylation with altered DNMT expression is associated with the stem cell compartment in human glioblastomas [9], and DNMT1 inhibitor zebularine induces significant levels of liver CSCs resulting in the increase of highly tumorigenesis [10]. In addition, 5-Aza treatment increased invasiveness, tumorigenesis, and metastatic capacities with the upregulation of proinvasive EMT-associated genes in MCF-7 breast cancer cells [8]. The loss of DNMT1 dramatically increases migration and invasive potential and mRNA expression of CDKN3, Claudin-3, and PKC in PCa cells [23]. Growing evidence shows that the association of DNMT1 with histone methylation marks. DNMT1 is necessary to maintain trimethylation of lysine 9 at histone H3 in pericentromeric regions [34], and loss of DNMT1 by 5-Aza regulates global reduction of di- and trimethylation of histone H3 lysine 9 (H3K9me2 and H3K9me3) in the many human cancer cells [34], [35]. Here, we demonstrated that reduction of DNMT1 induced histone demethylation of H3K9me3 and H3K27me3 on the Zeb2 or KLF4 promoter region, resulting in the increase of Zeb2 or KLF4 transcription in EMT or CSC induction of PCa cells. Collectively, reduction of DNMT1 increases migration in concert with enhanced expression of EMT and CSC phenotypes in PCa cells. Therefore, careful clarification of the activity of DNA methylation inhibitors is important to develop anticancer therapies.
Several mechanisms have been proposed for tumor inhibition by the DNA methylation inhibitor 5-Aza-CdR, most notably the demethylation of tumor suppressor genes which results in antitumor activities. 5-Azacitidine (5-Aza-CdR, decitabine) is currently used for the treatment of hematologic malignancies, myelodysplastic syndrome, and acute myeloid leukemia [47]. Preclinical studies show that low-dose 5-Aza-CdR is an effective treatment for tumors with reduced toxicity [47]. There is evidence that estimated plasma levels (0.04-15 μM) in various doses and schedules of 5-Aza-CdR are correlated with the clinical responses and remission in hematologic malignancies [47]. In solid tumors, the clinical response to low-dose 5-Aza-CdR is less clear. One pilot study reported the use of high-dose 5-Aza-CdR (660 mg/m2 as an 8-hour infusion) in stage III/IV non–small cell lung cancer, which produced an estimated plasma level in the range of 3 μM [48]. Patients with metastatic malignancy and poor prognosis are currently recommended for the higher intensive dose of 5-Aza-CdR (60 mg/m2 per hour administered as an 18-hour infusion/total dose of 1080 mg/m2) with an estimated plasma level in the range of 4 μM [47]. In the patients with refractory metastatic PCa, treatment with 1-hour infusions of 75 mg/m2 5-Aza-CdR every 8 hours for 3 doses was reported to effectively treat the disease in 2 of 12 patients with an evaluable response [49]. 5-Aza-CdR continues to be an effective agent of growth inhibition and induction of apoptosis in part by the upregulation of growth arrest signals during the chemotherapy. Importantly, the developing effective cancer therapeutics is required to block primary tumor growth and tumor metastases to distant organs in solid tumors. Our data showed that reduction of DNMT1 by 5-Aza treatment increased EMT and CSC induction, resulting in tumor growth and metastasis which are likely to be counterproductive when treating PCa. Thus, careful evaluations and monitoring of patients receiving DNA methylation inhibitors will be required. Taken together, novel optimization of dose and schedules for 5-Aza-CdR is needed to establish effective cancer treatments.
Growing attention has focused on the various PKC isoforms involved in regulation of cell proliferation, differentiation, survival, and death [30], [31], [32], [33]. PKC isoforms are activated by extracellular signals, which regulate the activities of cellular proteins including receptors, enzymes, cytoskeletal proteins, and transcription factors [30], [31], [32], [33]. A recent report shows that PKC activation is associated with the initiation of squamous cell carcinoma formation and progression to a malignant phenotype [30]. In addition, inhibition of PKCα reduces tumor-initiating activity and tumor growth of the CSC enriched populations in vivo [31]. Furthermore, PKCδ suppresses keratinocyte proliferation by increasing p21Cip1 levels through a KLF4 transcription factor–dependent mechanism [32], and KLF4 is required for maintenance of breast CSCs and for cell migration and invasion [33]. Collectively, these studies strongly support our data that PKC signals are involved in the development of EMT and CSC phenotypes.
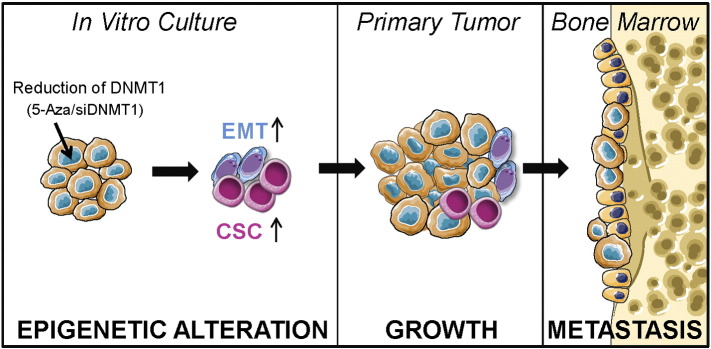
In summary, this study lays the foundation for a greater understanding of molecular mechanisms pertaining to DNMT1 function and how it regulates EMT induction and a CSC phenotype, which facilitates tumorigenesis in PCa cells. Thus, these studies have important implications for preclinical studies of epigenetic targeted therapies (Figure 8).
Figure 8.
Experimental model.
Reduction of DNMT1 expression by 5-Aza treatment or silencing of DNMT1 induces EMT and CSC phenotypes by PCa cells in vitro resulting in tumorigenesis within primary tumor and bone metastasis.
Author Contributions
E. L. and R. S. T. designed experiments. E. L., J. W., K. Y., Y. J., F. C. C., A. M. D., and Y. L. performed experiments and analyzed the data. R. T. F. and K. J. P. discussed the results and gave valuable critique on the paper. E. L. and R. S. T. wrote the manuscript.
Acknowledgements
This work is directly supported by the National Cancer Institute (CA093900 to K. J. Pienta and R. S. Taichman; CA163124 to Y. Shiozawa, K. J. Pienta, and R. S. Taichman; U54CA143803 to K. J. Pienta; CA143055 to K. J. Pienta), the Department of Defense (W81XWH-11-1-0636 to K. J. Pienta and R. S. Taichman, W81XWH-15-1-0413 and W81XWH-15-1-0637 to R. S. Taichman), and the Prostate Cancer Foundation (Y. Shiozawa, K. J. Pienta, and R. S. Taichman). K.J. Pienta receives support as an American Cancer Society Clinical Research Professor.
Footnotes
This work is directly supported by the National Cancer Institute (CA093900 to K. J. Pienta and R. S. Taichman; CA163124 to Y. Shiozawa, K. J. Pienta, and R. S. Taichman; U54CA143803 to K. J. Pienta; CA143055 to K. J. Pienta), the Department of Defense (W81XWH-11-1-0636 to K. J. Pienta and R. S. Taichman, W81XWH-15-1-0413, and W81XWH-15-1-0637), and the Prostate Cancer Foundation (K. J. Pienta, and R. S. Taichman). K.J. Pienta receives support as an American Cancer Society Clinical Research Professor.
References
- 1.Pienta KJ, Esper PS. Risk factors for prostate cancer. Ann Intern Med. 1993;118:793–803. doi: 10.7326/0003-4819-118-10-199305150-00007. [DOI] [PubMed] [Google Scholar]
- 2.Koutsilieris M. Osteoblastic metastasis in advanced prostate cancer. Anticancer Res. 1993;13:443–449. [PubMed] [Google Scholar]
- 3.Wang J, Ying G, Wang J, Jung Y, Lu J, Zhu J, Pienta KJ, Taichman RS. Characterization of phosphoglycerate kinase-1 expression of stromal cells derived from tumor microenvironment in prostate cancer progression. Cancer Res. 2010;70:471–480. doi: 10.1158/0008-5472.CAN-09-2863. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Jung Y, Kim JK, Shiozawa Y, Wang J, Mishra A, Joseph J, Berry JE, McGee S, Lee E, Sun H. Recruitment of mesenchymal stem cells into prostate tumors promotes metastasis. Nat Commun. 2013;4 doi: 10.1038/ncomms2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell. 2012;22:699–701. doi: 10.1016/j.ccr.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Shang Y. Epigenetic control of epithelial-to-mesenchymal transition and cancer metastasis. Exp Cell Res. 2013;319:160–169. doi: 10.1016/j.yexcr.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 8.Ateeq B, Unterberger A, Szyf M, Rabbani SA. Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia. 2008;10:266–278. doi: 10.1593/neo.07947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fanelli M, Caprodossi S, Ricci-Vitiani L, Porcellini A, Tomassoni-Ardori F, Amatori S, Andreoni F, Magnani M, De Maria R, Santoni A. Loss of pericentromeric DNA methylation pattern in human glioblastoma is associated with altered DNA methyltransferases expression and involves the stem cell compartment. Oncogene. 2008;27:358–365. doi: 10.1038/sj.onc.1210642. [DOI] [PubMed] [Google Scholar]
- 10.Marquardt JU, Factor VN, Thorgeirsson SS. Epigenetic regulation of cancer stem cells in liver cancer: current concepts and clinical implications. J Hepatol. 2010;53(3):568–577. doi: 10.1016/j.jhep.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 13.Szyf M, Detich N. Regulation of the DNA methylation machinery and its role in cellular transformation. Prog Nucleic Acid Res Mol Biol. 2001;69:47–79. doi: 10.1016/s0079-6603(01)69044-5. [DOI] [PubMed] [Google Scholar]
- 14.Bigey P, Ramchandani S, Theberge J, Araujo FD, Szyf M. Transcriptional regulation of the human DNA methyltransferase (dnmt1) gene. Gene. 2000;242:407–418. doi: 10.1016/s0378-1119(99)00501-6. [DOI] [PubMed] [Google Scholar]
- 15.Szyf M. The role of DNA methyltransferase 1 in growth control. Front Biosci. 2001;6:D599–D609. doi: 10.2741/szyf. [DOI] [PubMed] [Google Scholar]
- 16.Robertson KD, Keyomarsi K, Gonzales FA, Velicescu M, Jones PA. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G(0)/G(1) to S phase transition in normal and tumor cells. Nucleic Acids Res. 2000;28:2108–2113. doi: 10.1093/nar/28.10.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.el-Deiry WS, Nelkin BD, Celano P, Yen RW, Falco JP, Hamilton SR, Baylin SB. High expression of the DNA methyltransferase gene characterizes human neoplastic cells and progression stages of colon cancer. Proc Natl Acad Sci U S A. 1991;88:3470–3474. doi: 10.1073/pnas.88.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Issa JP, Vertino PM, Wu J, Sazawal S, Celano P, Nelkin BD, Hamilton SR, Baylin SB. Increased cytosine DNA-methyltransferase activity during colon cancer progression. J Natl Cancer Inst. 2000;85:1235–1240. doi: 10.1093/jnci/85.15.1235. [DOI] [PubMed] [Google Scholar]
- 19.Nass SJ, Ferguson AT, El-Ashry D, Nelson WG, Davidson NE. Expression of DNA methyl-transferase (DMT) and the cell cycle in human breast cancer cells. Oncogene. 1999;18:7453–7461. doi: 10.1038/sj.onc.1203138. [DOI] [PubMed] [Google Scholar]
- 20.Wu CT, Wu CF, Lu CH, Lin CC, Chen WC, Lin PY, Chen MF. Expression and function role of DNA methyltransferase 1 in human bladder cancer. Cancer. 2011;117:5221–5233. doi: 10.1002/cncr.26150. [DOI] [PubMed] [Google Scholar]
- 21.Szyf M, Bozovic V, Tanigawa G. Growth regulation of mouse DNA methyltransferase gene expression. J Biol Chem. 1991;266:10027–10030. [PubMed] [Google Scholar]
- 22.Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 23.Yaqinuddin A, Qureshi SA, Qazi R, Farooq S, Abbas F. DNMT1 silencing affects locus specific DNA methylation and increases prostate cancer derived PC3 cell invasiveness. J Urol. 2009;182:756–761. doi: 10.1016/j.juro.2009.03.082. [DOI] [PubMed] [Google Scholar]
- 24.Valdez CD, Davis JN, Odeh HM, Layfield TL, Cousineau CS, Berton TR, Johnson DG, Wojno KJ, Day ML. Repression of androgen receptor transcription through the E2F1/DNMT1 axis. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valdez CD, Kunju L, Daignault S, Wojno KJ, Day ML. The E2F1/DNMT1axis is associated with the development of AR negative castration resistant prostate cancer. Prostate. 2013;73:1776–1785. doi: 10.1002/pros.22715. [DOI] [PubMed] [Google Scholar]
- 26.Kinney SR, Moser MT, Pascual M, Greally JM, Foster BA, Karpf AR. Opposing roles of Dnmt1 in early- and late-stage murine prostate cancer. Mol Cell Biol. 2010;30:4159–4174. doi: 10.1128/MCB.00235-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duhagon MA, Hurt EM, Sotelo-Silveira JR, Zhang X, Farrar WL. Genomic profiling of tumor initiating prostatospheres. BMC Genomics. 2010;11:324. doi: 10.1186/1471-2164-11-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Havens AM, Pedersen EA, Shiozawa Y, Ying C, Jung Y, Sun Y, Neeley C, Wang J, Mehra R, Keller ET. In vivo mouse model for human prostate cancer metastasis. Neoplasia. 2008;10(4):371–380. doi: 10.1593/neo.08154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HY. Statistical notes for clinical researchers: assessing normal distribution (2) using skewness and kurtosis. Restor Dent Endod. 2013;38(1):52–54. doi: 10.5395/rde.2013.38.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breitkreutz D, Braiman-Wiksman L, Daum N, Denning MF, Tennenbaum T. Protein kinase C family: on the crossroads of cell signaling in skin and tumor epithelium. J Cancer Res Clin Oncol. 2007;133:793–808. doi: 10.1007/s00432-007-0280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, Wu ZJ, Krall JA, Bierie B, Guo W. Protein kinase Cα is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24:347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chew YC, Adhikary G, Wilson GM, Reece EA, Eckert RL. Protein kinase C (PKC) delta suppresses keratinocyte proliferation by increasing p21(Cip1) level by a KLF4 transcription factor–dependent mechanism. J Biol Chem. 2011;286:28772–28782. doi: 10.1074/jbc.M110.205245. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Yu F, Li J, Chen H, Fu J, Ray S, Huang S, Zheng H, Ai W. Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene. 2011;30:2161–2172. doi: 10.1038/onc.2010.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espada J, Ballestar E, Fraga MF, Villar-Garea A, Juarranz A, Stockert JC, Robertson KD, Fuks F, Esteller M. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J Biol Chem. 2004;279(35):37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- 35.Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2(6):607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 37.Kojima Y, Acar A, Eaton EN, Mellody KT, Scheel C, Ben-Porath I, Onder TT, Wang ZC, Richardson AL, Weinberg RA. Autocrine TGF-beta and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc Natl Acad Sci U S A. 2010;107:20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daly AJ, McIlreavey L, Irwin CR. Regulation of HGF and SDF-1 expression by oral fibroblasts—implications for invasion of oral cancer. Oral Oncol. 2008;44:646–651. doi: 10.1016/j.oraloncology.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Taichman RS, Cooper C, Keller ET, Pienta KJ, Taichman NS, McCauley LK. Use of the stromal cell–derived factor-1/CXCR4 pathway in prostate cancer metastasis to bone. Cancer Res. 2002;62:1832–1837. [PubMed] [Google Scholar]
- 40.Chen Z, Pan X, Yao Y, Yan F, Chen L, Huang R, Ma G. Epigenetic regulation of cardiac progenitor cells marker c-kit by stromal cell derived factor-1α. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069134. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Cheng T. Cell cycle inhibitors in normal and tumor stem cells. Oncogene. 2004;23:7256–7266. doi: 10.1038/sj.onc.1207945. [DOI] [PubMed] [Google Scholar]
- 42.Moore N, Lyle S. Quiescent, slow-cycling stem cell populations in cancer: a review of the evidence and discussion of significance. J Oncol. 2011 doi: 10.1155/2011/396076. [pii: 396076] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea—a paradigm shift. Cancer Res. 2006;66(4):1883–1890. doi: 10.1158/0008-5472.CAN-05-3153. [DOI] [PubMed] [Google Scholar]
- 44.Meng E, Mitra A, Tripathi K, Finan MA, Scalici J, McClellan S, Madeira da Silva L, Reed E, Shevde LA, Palle K. ALDH1A1 maintains ovarian cancer stem cell-like properties by altered regulation of cell cycle checkpoint and DNA repair network signaling. PLoS One. 2014;9(9) doi: 10.1371/journal.pone.0107142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguewa P, Manrique I, Díaz R, Redrado M, Parrondo R, Perez-Stable C, Calvo A. Id-1B, an alternatively spliced isoform of the inhibitor of differentiation-1, impairs cancer cell malignancy through inhibition of proliferation and angiogenesis. Curr Mol Med. 2014;14(1):151–162. doi: 10.2174/1566524013666131203100643. [DOI] [PubMed] [Google Scholar]
- 46.Manrique I, Nguewa P, Bleau AM, Nistal-Villan E, Lopez I, Villalba M, Gil-Bazo I, Calvo A. The inhibitor of differentiation isoform Id1b, an alternatively spliced isoform of the inhibitor of differentiation-1generated by alternative splicing, maintains cell quiescence and confers self-renewal and cancer stem cell-like properties. Cancer Lett. 2015;356(2 Pt B):899–909. doi: 10.1016/j.canlet.2014.10.035. [DOI] [PubMed] [Google Scholar]
- 47.Karahoca M, Momparler RL. Pharmacokinetic and pharmacodynamic analysis of 5-aza-2'-deoxycytidine (decitabine) in the design of its dose-schedule for cancer therapy. Clin Epigenetics. 2013;5:3. doi: 10.1186/1868-7083-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Momparler RL, Bouffard DY, Momparler LF, Dionne J, Belanger K, Ayoub J. Pilot phase I-II study on 5-aza-2'-deoxycytidine (decitabine) in patients with metastatic lung cancer. Anticancer Drugs. 1997;8:358–368. doi: 10.1097/00001813-199704000-00008. [DOI] [PubMed] [Google Scholar]
- 49.Thibault A, Figg WD, Bergan RC, Lush RM, Myers CE, Tompkins A, Reed E, Samid D. A phase II study of 5-aza-2'deoxycytidine (decitabine) in hormone independent metastatic (D2) prostate cancer. Tumori. 1998;84:87–89. doi: 10.1177/030089169808400120. [DOI] [PubMed] [Google Scholar]