Abstract
Recently, activating mutations of the hypoxia-inducible factor 2α gene (HIF2A/EPAS1) have been recognized to predispose to multiple paragangliomas (PGLs) and duodenal somatostatinomas associated with polycythemia, and ocular abnormalities. Previously, mutations in the SDHA/B/C/D, SDHAF2, VHL, FH, PHD1, and PHD2 genes have been associated with HIF activation and the development of pseudohypoxic (cluster-1) PGLs. These tumors overlap in terms of tumor location, syndromic presentation, and noradrenergic phenotype to a certain extent. However, they also differ especially by clinical outcome and by presence of other tumors or abnormalities. In the present study, we aimed to establish additional molecular differences between HIF2A and non-HIF2A pseudohypoxic PGLs. RNA expression patterns of HIF2A PGLs (n = 6) from 2 patients were compared with normal adrenal medullas (n = 8) and other hereditary pseudohypoxic PGLs (VHL: n = 13, SDHB: n = 15, and SDHD: n = 14). Unsupervised hierarchical clustering showed that HIF2A PGLs made up a separate cluster from other pseudohypoxic PGLs. Significance analysis of microarray yielded 875 differentially expressed genes between HIF2A and other pseudohypoxic PGLs after normalization to adrenal medulla (false discovery rate 0.01). Prediction analysis of microarray allowed correct classification of all HIF2A samples based on as little as three genes (TRHDE, LRRC63, IGSF10; error rate: 0.02). Genes with the highest expression difference between normal medulla and HIF2A PGLs were selected for confirmatory quantitative reverse transcriptase polymerase chain reaction. In conclusion, HIF2A PGLs show a characteristic expression signature that separates them from non-HIF2A pseudohypoxic PGLs. Unexpectedly, the most significantly differentially expressed genes have not been previously described as HIF target genes.
Introduction
Within the last 5 years, the number of gene mutations associated with paragangliomas (PGLs) and pheochromocytomas (i.e., adrenal PGLs) has more than doubled [1]. Strong genotype-phenotype associations including syndromic presentation; tumor location; malignant potential; and biochemical, metabolomic, and specific imaging phenotypes have been recognized, indicating the need for identification of individualized treatment approaches to hereditary PGLs [2], [3], [4].
At the gene expression level, two main groups of PGLs have been identified: those showing increased expression of hypoxia-related genes (pseudohypoxic PGLs, also referred to as cluster-1) and kinase signaling genes (cluster-2) [5], [6], [7]. Cluster identity of paragangliomas can be well determined based on metanephrine production because almost exclusively cluster-2 paragangliomas produce metanephrine. Regardless of cluster classification, a current concept suggests that inappropriately elevated HIF signaling may be involved in tumorigenesis of most mutation-derived PGLs [8]. Qin et al. showed that joint HIF-1α and HIF-2α stabilization is predominant in all pseudohypoxic PGLs [9].
A recent addition to the list of gene mutations predisposing to pseudohypoxic PGLs are gain-of-function mutations of hypoxia-inducible factor 2 alpha (HIF2A or EPAS1) [10]. HIF2A PGLs are most often multifocal and recurrent, produce norepinephrine, and occur more frequently in females than males (summarized in [11]). At least half of the afflicted patients reported to date show syndromic presentation including polycythemia from early childhood, PGLs at young age, duodenal somatostatinomas [10], [12], and ocular abnormalities [13]. Recently, somatic HIF2A mutations have also been detected in central nervous system hemangioblastomas [14] and duodenal gangliocytic PGLs [15], a rare type of tumor composed of neurons, Schwann cells, and enteric-type neuroendocrine cells that differs from true PGLs by expression of keratins, pancreatic polypeptide, and other intestinal regulatory peptides. In the majority of cases, the mutations were found to be somatic and postzygotic; however, rarely, germline mutations as well as germline mosaicism have also been reported [16], [17].
PGLs have not been previously associated with a comparable syndromic presentation except for cases of von Hippel–Lindau syndrome, in which almost always adrenal PGLs rarely co-occur with polycythemia and/or somatostatinomas [18], [19]. Somatostatinomas have previously been associated with other neuroendocrine syndromes caused by mutations which predispose to cluster-2 PGLs, i.e., multiple endocrine neoplasia 2B (i.e., RET mutations) [20] and neurofibromatosis 1 (NF1 mutations) [21], [22]. Nevertheless, in previous studies, the mRNA expression profiles of nine cases of HIF2A-mutated PGLs from patients with and without syndromic presentation clustered with other pseudohypoxic PGLs [23], [24], [25], [26], whereas, surprisingly, three cases showed more common expression patterns with cluster-2 PGLs [24]. The authors mentioned that several of the reported HIF2A tumors were suspected to carry somatic NF1 mutations; thus, possibly, these three samples were afflicted with both mutations. HIF2A expression was increased even in the latter cases compared with cluster-2 PGLs.
Based on distinct clinical presentations of patients with HIF2A syndrome from patients with other HIF-stabilizing mutations, differences in the tumor biology and clinical outcome are evident. Despite the fact that stabilization of HIF-1α and/or HIF-2α occurs due to mutations in any cluster-1 tumor susceptibility genes, clinical manifestations and outcomes vastly differ. Particularly for patients with HIF2A mutations, who often present early with polycythemia and have a high risk to develop metastatic somatostatinomas and less frequently metastatic PGL and ocular abnormalities, the development of new, targeted approaches to therapy is of the essence. To further elaborate if and how HIF2A-related PGLs differ from non-HIF2A pseudohypoxic PGLs (e.g., SDHx and VHL) on the molecular level and to identify potentially druggable targets, we performed a differential gene expression analysis of cluster-1 PGLs.
Results
Identification of a Differentiating Expression Signature in HIF2A PGLs
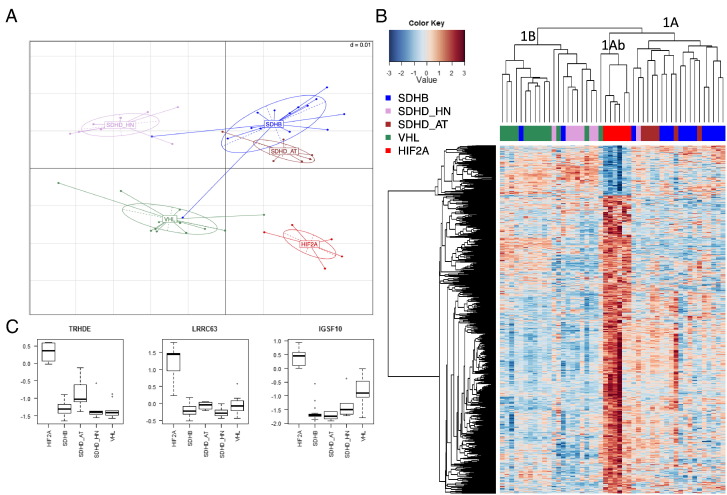
Principal component analysis showed that HIF2A tumor samples have distinct expression characteristics from non-HIF2A pseudohypoxic PGLs (Figure 1A). In agreement with that, unsupervised hierarchical clustering showed that HIF2A PGLs make up a separate subcluster (cluster-1Ab) within the previously described cluster-1A (i.e., a joint cluster of SDHB and SDHD-abdominal/thoracic [AT] PGLs, which is clearly distinguishable from cluster-1B, containing VHL and SDHD head and neck PGLs [HNPs] in two distinct subclusters [27]) (Figure 1B). Significance analysis of microarray with two-class option at a false discovery rate ≤ 0.01 revealed 875 differentially expressed genes in HIF2A PGLs compared with non-HIF2A pseudohypoxic PGLs after normalization to normal adrenal medulla (Figure 1B). Of these 875 genes, 96 were 1.5-fold more highly expressed in HIF2A than non-HIF2A pseudohypoxic PGLs, whereas 27 were 1.5-fold more highly expressed in the latter (Table S1).
Figure 1.
Distinct expression pattern of HIF2A PGLs. (A) Principal component analysis showed that HIF2A PGLs are clearly distinguishable from other pseudohypoxic PGLs based on their expression pattern. (B) Hierarchical clustering of all pseudohypoxic PGLs based on differentially expressed genes observed by significance analysis of microarray showed separate subclustering of HIF2A PGLs in the previously described cluster-1A (a mixed cluster of SDHB and SDHD-AT PGLs). (C) Top three genes, which allow correct classification of HIF2A PGLs with an excellent error rate of merely 2%. The y-axis indicates relative gene expression to normal adrenal medulla. Median, first, and third quartiles of relative expression z-scores of the gene in question are indicated by midline, bottom, and top of the boxes. Whiskers indicate lowest and highest expression values within 1.5 interquartile ranges of the lower and upper quartile, respectively. Extreme values are depicted as dots and may be considered outliers.
Prediction analysis of microarray at a threshold of 2.3 allowed correct classification of all HIF2A samples based on 354 genes with only one misclassification of an SDHD adrenal PGL (D31.1) with an error rate of 0.02 (Table 1). Correct classification among cluster-1 PGLs with this low error rate was even achieved based on the expression of just three genes: TRHDE, LRRC63, and IGSF10 (Figure 1C).
Table 1.
Confusion matrix for classification of HIF-2α samples
| HIF2A | Other Pseudohypoxic | Error Rate | |
|---|---|---|---|
| HIF2A | 6 | 0 | 0.00 |
| Other pseudohypoxic | 1 | 41 | 0.02 |
| Overall error rate= | 0.02 | ||
HIF-α Target Gene Signature
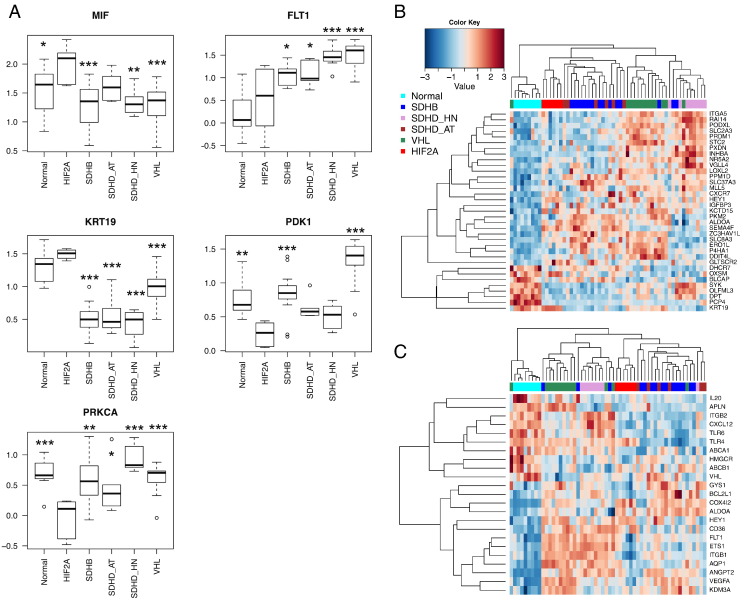
Comparison of previously reported HIF-1α and HIF-2α target gene lists with the 875 genes, which were identified to be differentially expressed between HIF2A and other pseudohypoxic PGLs by significance analysis of microarray, led to few or no matches (0%-8.3%; 0/72 [28], 20/443 [29], 2/24 [30], 7/117 [31], [32], [33], 21/500 [34]). There was no preference for either HIF-1α or HIF-2α target genes. When a fold change (FC) threshold equal or greater than 1.5 or equal or less than −1.5 was chosen, only two reported HIF-1α target genes (MIF: FC = 1.6, FLT1: FC = −1.7), two HIF-1α and HIF-2α target genes (KRT19: FC = 1.8, PDK1: FC = −1.56), and one HIF-2α target gene remained (PRKCA: FC = −1.59). Surprisingly, the expression of the HIF-2α target gene PRKCA was significantly decreased in HIF2A PGLs compared with all other groups.
Moreover, MIF and FLT1 expression changes were in opposing directions, with MIF being more highly expressed in HIF2A than all other groups except for SDHD-AT and FLT1 being expressed at similar levels in HIF2A and normal medulla while being elevated in all other groups (Figure 2A). Similarly, KRT19 and PDK1 were changed in opposing directions. KRT19 was downregulated in all pseudohypoxic PGLs compared with normal medulla and HIF2A samples, whereas PDK1 was downregulated in HIF2A compared with normal medulla, SDHB and VHL PGLs.
Figure 2.
HIF target gene expression in pseudohypoxic PGLs. (A) Five HIF target genes, which were identified to be differentially expressed between HIF2A and other pseudohypoxic PGLs. The y-axis indicates normalized gene expression level. Median, first, and third quartiles of normalized expression z-scores of the gene in question are indicated by midline, bottom, and top of the boxes. Whiskers indicate lowest and highest expression values within 1.5 interquartile ranges of the lower and upper quartile, respectively. Extreme values are depicted as dots and may be considered outliers. Overall ANOVA was performed. Asterisks indicate significantly different expression from HIF2A as determined by post hoc analysis using Dunnett's test (***P ≤ .001, **P ≤ .01, *P ≤ .05). (B and C) Upon restriction of the 1721 differentially expressed genes between normal medulla and pseudohypoxic PGLs to those that have been previously reported as HIF target genes, separate clustering of HIF2A PGLs was maintained, indicating differences in HIF target gene activation between the HIF2A PGLs and other pseudohypoxic PGLs. HIF target genes presented in B are from the transcription factor encyclopedia, and those shown in C are from [29].
Overall, these findings may indicate that the hypoxic expression signatures of HIF2A and other pseudohypoxic PGLs are mainly in agreement as previously suggested and that general upregulation of HIF signaling is a common denominator [5], [7], [35].
To evaluate the overlap in expression between HIF2A and non-HIF2A pseudohypoxic PGLs, we compared the expression patterns of all pseudohypoxic PGLs combined (including HIF2A PGLs) with normal adrenal medulla. Significance analysis of microarray revealed 1721 differentially expressed genes at a q-value of 1%. Ingenuity Pathway Analysis (IPA) of the differentially expressed genes between all pseudohypoxic PGLs and normal medulla with an FC greater than 1.5 predicted transcription factor activation, among others, for HIF-1α, HIF-2α, and ARNT2, an HIFβ subunit (activation z-scores: 2.409, 2.379, and 2.236 and overlap factors: 1.63*10−5, 1.37*10−5, and 2.236*10−3, respectively). Genes and changes in expression which led to the prediction of activation are shown in Table S3. These data suggest that a common activation of both HIF-1α and HIF-2α is likely to be present in all pseudohypoxic PGLs.
Matching the significance analysis of microarray list of differentially expressed genes between normal medulla and all pseudohypoxic PGLs revealed more matches to reported and predicted HIF target gene lists (7.9%-20.1%; 9/72 [28], 35/443 [29], 5/24 [30], 14/117 [31], [32], [33], 50/500 [34]). Interestingly, despite similar expression changes in these HIF targets among all pseudohypoxic PGLs compared with normal medulla, hierarchical clustering of all groups with either of those HIF target gene lists led to separate clustering of HIF2A PGLs from non-HIF2A PGLs in a similar manner as when using all 875 genes (Figure 2, B and C), indicating that HIF2A PGLs are more similar to each other than non-HIF2A PGLs overall but also with respect to HIF target genes. Thus, although all pseudohypoxic PGLs show some agreement in their HIF signature, even when limited to those genes, separation of HIF2A PGLs remains possible. This indicates characteristically different nuances of HIF target gene activation in HIF2A PGLs.
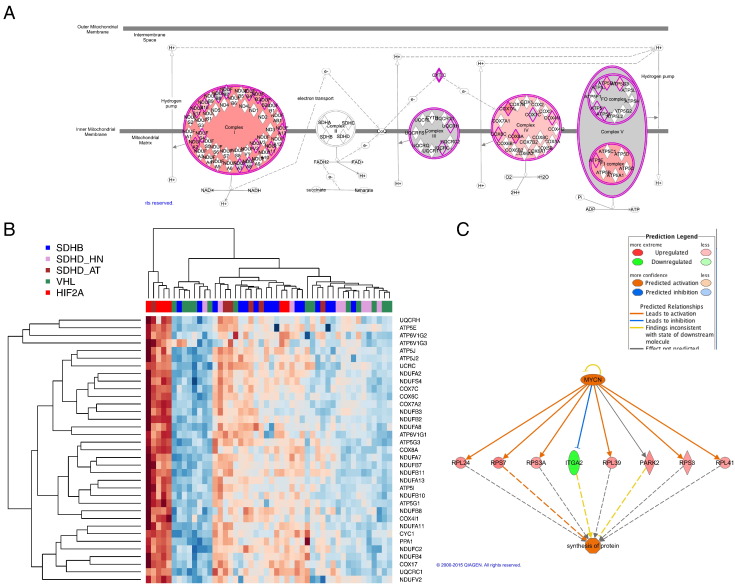
To focus on unique features of HIF2A PGLs on the pathway level, core analysis was performed using IPA. Oxidative phosphorylation was reported as top canonical pathway, with 27 of 93 related genes being more highly expressed in the HIF2A group than the group of non-HIF2A pseudohypoxic PGLs (Figure 3A). This was confirmed by matching the significance analysis of microarray list to the genes listed for oxidative phosphorylation in the Kyoto Encyclopedia of Genes. Upregulation was evident for 35 of 128 listed genes (Figure 3B). However, as is visible in Figure 3B, very strong upregulation of oxydative phosphorylation genes was predominant in only four of six HIF2A samples, which skewed the group median z-score. These four samples were all from patient H48. The two other HIF2A samples from patient H49 subclustered with five SDHB PGLs. Because oxidative phosphorylation was still the top-rated pathway based on group medians of relative expression and we previously showed that SDHB and SDHD-AT PGLs have stronger expression of oxidative phosphorylation genes than VHL and SDHD- HNP, we still consider this finding interesting because it reflects that HIF2A PGLs may depend less on glycolytic energy metabolism. Another pathway reported to be highly activated was EIF2 signaling, with 32 genes from the pathway being upregulated, including 30 ribosomal proteins. In agreement with that, EIF4E was predicted to be activated based on upregulation of nine genes (Table S3, activation z-score = 2.530, overlap P value = .046). Furthermore, translation, expression, and synthesis of proteins were suggested to be elevated in HIF2A PGLs based on IPA downstream analysis. Interestingly, MYCN was predicted to be activated (activation z-score = 3.599, overlap P value = 1.54*10−5), which may explain elevated expression of several genes involved in protein synthesis (Figure 3C).
Figure 3.
Indication of activated pathways in HIF2A PGLs. (A) IPA revealed increased expression of a vast number of genes essential in oxidative phosphorylation in HIF2A PGLs (red indicates upregulation in HIF2A PGLs compared with pseudohypoxic PGLs). (B) Heatmap depicting upregulation of oxidative phosphorylation genes in HIF2A PGLs after comparison of differentially expressed genes in HIF2A compared with other pseudohypoxic PGLs with the oxydative phosphorylation genes listed in the Kyoto Encyclopedia of Genes and Genomes (red indicates upregulation). (C) Upregulation of several genes for ribosomal proteins led to prediction of increased protein synthesis and MYCN activation in HIF2A PGLs by IPA.
Unique Features of HIF2A PGLs
To further specify the characteristic expression signature of HIF2A PGLs, we chose a two-step approach using significance analysis of microarray, first identifying the differentially expressed genes between normal adrenal medulla and HIF2A PGLs (1240 genes, q-value 1%) and then matching it to a list of genes differentially expressed between non-HIF2A and HIF2A PGLs (254 genes, q-value 1%). A heatmap of this characteristic HIF2A-PGL expression signature relative to other pseudohypoxic PGLs is given in Figure S2, and the genes are listed in Table S3.
Genes of Interest
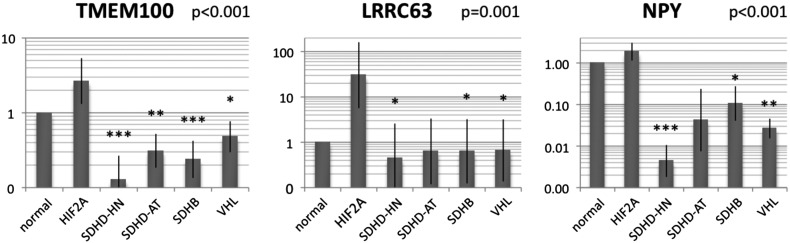
Ten genes of interest were chosen for further exploration by rating the 875 genes we found to be differentially expressed between HIF2A and other pseudohypoxic PGLs based on the magnitude of difference in expression between normal adrenal medulla and HIF2A PGLs and a false discovery rate below 0.01 in significance analysis of microarray. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) for a largely separate set of PGL samples was performed for 9 of the 10 genes of interest and essentially confirmed the microarray data for 6 genes (P ≤ .001: TMEM100, NPY, and LRRC63; P ≤ .05: PTN, GNG11, IL20RA). The three genes most likely to qualify to distinguish HIF2A PGLs from other pseudohypoxic PGLs were thus TMEM100, NPY, and LRRC63 (Figure 4), and they potentially play distinguishing roles in HIF2A PGL tumor biology.
Figure 4.
Relative mRNA expression of genes of interest in HIF2A compared with other pseudohypoxic PGLs as assessed by validatory qRT-PCR. Expression of the candidate markers for HIF2A PGLs, TMEM100, NPY, and LRRC63, was assessed relative to RPLP0 in a vastly separate sample set than the one assessed for the microarray. Bars indicate group means ± SEM. Overall ANOVA was performed. Asterisks indicate significantly different expression from HIF2A as determined by post hoc analysis using Dunnett's test (***P ≤ .001, **P ≤ .01, *P ≤ .05).
Previously, pseudohypoxic PGLs have been reported to be highly vascular and exhibit increased VEGF signaling [36]. Toledo et al. reported similar expression of VEGFA in two cases of HIF2A PGLs and cluster-1 PGLs, which was significantly higher than in cluster-2 PGLs [26]. Within our cohort, VEGFA expression was increased compared with normal medulla in all pseudohypoxic PGL groups but to a lesser extent in HIF2A than the other groups. Expression of the VEGF receptors, NRP1 (VEGF165R), KDR (VEGFR-2), and FLT1 (VEGFR-1), was at similarly low or lower levels in HIF2A PGLs compared with normal medulla. Gene expression was significantly elevated in SDHD-HN and VHL PGLs (KDR); SDHB, VHL, and SDHD-HN PGLs (NRP1); or SDHB, SDHD-AT, SDHD-HN, and VHL PGLs (FLT1) compared with HIF2A samples (Figure S1).
Discussion
Our gene expression data based on 48 pseudohypoxic PGLs and 8 normal adrenal medullas indicate that HIF2A PGLs show expression features that on the one hand unite and on the other hand clearly distinguish them from non-HIF2A pseudohypoxic PGLs.
In support of significant differences in gene expression profiles of HIF2A and non-HIF2A pseudohypoxic PGLs, correct classification of HIF2A PGLs was excellent.
Characteristic expression of TRHDE, LRRC63, and IGSF10 was sufficient to correctly classify all HIF2A PGL samples. In our cohort, LRRC63 was more highly expressed in HIF2A PGL samples than all other tissues, including normal medulla. Thus, LRRC63 may play an essential role in HIF2A PGL development. Currently, the function of LRRC63 is unknown.
Our data further show decreased expression of IGSF10 and TRHDE in all non-HIF2A pseudohypoxic tumors relative to normal medulla and HIF2A PGLs. Interestingly, TRHDE has been shown to be hypermethylated in oral squamous cell cancers and dysplastic tissue compared with adjacent normal tissue [37], which generally leads to decreased transcription. In addition, IGSF10 has been described to be downregulated in radiation-induced rat osteosarcomas relative to normal osteoblasts [38]. Thus, decreased TRHDE and IGSF10 expression, possibly caused by hypermethylation, may contribute to tumorigenesis in non-HIF2A pseudohypoxic PGLs.
Nevertheless, HIF2A-initiated transcription does not seem to play a major role in the observed differential expression patterns. Our data showed minimal overlap of differentially expressed genes between HIF2A and non-HIF2A pseudohypoxic PGLs with previously published HIF target gene lists. Comparison of the combined expression patterns of all pseudohypoxic PGLs (including HIF2A) relative to normal adrenal medulla indicated differential expression of almost always twice as many HIF target genes upon comparison with published and predicted HIF target gene lists [28], [29], [30], [31], [32], [33], [34]. This confirms previous reports suggesting a common HIF pathway-related expression signature for all pseudohypoxic PGLs [5], [7], [35].
Selected HIF2A target genes, however, did show differences in expression level between the analyzed pseudohypoxic PGLs, indicating that slight differences in HIF target gene expression exist and may contribute to the variation in manifestation between the analyzed tumor groups. The HIF1A and HIF2A target gene KRT19 is a key player in epithelial to mesenchymal transition and has been reported to be expressed in neuroendocrine tumors [39]. In addition, it is used as a marker to detect circulating breast cancer cells and correlates with highly proliferating tumors and the risk for metastases [40]. On the contrary, epigenetic downregulation of KRT19 in SDHB-PGLs has been shown [41], [42], and its downregulation contributed to increased cell motility and invasiveness. In our cohort, HIF2A PGLs showed similar KRT19 expression compared with normal medulla and decreased expression in the other pseudohypoxic PGLs. In agreement with that, Toledo et al. showed increased mRNA expression of KRT19 in a HIF2A PGL compared with cluster-1 and cluster-2 PGLs [26]. KRT19 mRNA expression at similar level as seen in the normal medulla may reflect reduced aggressiveness with slower tumor progression observed in our HIF2A patient cohort compared with, e.g., SDHB patients.
Exploration of concerted changes in gene expression in HIF2A-related PGLs revealed oxidative phosphorylation and protein translation/synthesis to be upregulated compared with non-HIF2A pseudohypoxic PGLs. We previously showed increased expression of several oxidative phosphorylation genes, but in a less concerted manner, in SDHB and SDHD-AT PGLs compared with VHL and SDHD-HN PGLs [27], [43]. HIF2A PGLs from patient H48 showed even higher levels of oxidative phosphorylation gene expression, whereas those of H49 fell into a subcluster shared with five SDHB PGLs, which we previously showed to have a tendency for higher expression of oxidative phosphorylation genes, and surprisingly one SDHD-HN PGL. Thus, expression of oxydative phosphorylation genes may be very strong in certain HIF2A PGLs while being comparable to other pseudohypoxic PGLs in others.
Dysfunction of VHL has been reported to cause decreased oxidative phosphorylation complex subunit expression [44] by way of HIF signaling and reactive oxygen species generation [45]. Dysfunction of the SDH complex has been previously associated with reactive oxygen species generation [46], [47], [48] and may thus share a similar mechanism of oxidative phosphorylation gene downregulation for certain mutations. Hervouet et al. [44] used cells which do not express HIF-1α and showed that presence of HIF-2α is essential in downregulation of oxidative phosphorylation genes. In contrast, using the same cell model, Biswas et al. showed that HIF-1α overexpression (with a background of endogeneous HIF-2α) led to a decreased level of mitochondrial activity, whereas HIF-2α overexpression (in absence of HIF-1α) induced mitochondrial activity [49]. In agreement with that, Chiavarina et al. showed that HIF-2α activity increased oxidative phosphorylation gene expression, whereas it was decreased by HIF-1α activity [50]. In HIF2A PGLs, activation of HIF-1α is absent or minimal [12], [51], and thus, a similar picture as seen in the model systems of Biswas and Chiacarina may be present in those tumors, whereas in the other pseudohypoxic tumors, likely a simultaneous activation of HIF-2α and HIF-1α is present [9].
Determination of oxidative phosphorylation function in HIF2A compared with non-HIF2A PGLs or in appropriate PGL cell models is needed to confirm differential regulation of oxidative phosphorylation by HIF-1α and HIF-2α stabilization. Increased expression of oxidative phosphorylation genes in some HIF2A PGLs may indicate that these tumors are not as dependent on the Warburg effect as other pseudohypoxic PGLs. In agreement with that, our own unpublished observations indicate that imaging of elevated glucose turnover via 18-fluoro-deoxyglucose is much less specific for HIF2A PGLs than for other pseudohypoxic PGLs, especially those with SDHB mutations.
Protein translation has previously been reported to be inhibited by hypoxia [52], abnormal pVHL [53], and PHD2 [54]. Thus, decreased protein translation in pseudohypoxic PGLs with PHD2 or VHL dysfunction as well as inhibited PHDs due to SDHx mutations would be expected. Our results show higher expression of 43 ribosomal proteins in HIF2A PGLs compared with the other pseudohypoxic PGLs, indicating that, as discussed above, exclusive HIF-2α stabilization may have effects that differ from general hypoxia or stabilization of all HIF-α subunits.
We identified potential HIF2A PGL markers with distinct expression in HIF2A PGLs compared with the other pseudohypoxic PGLs. Of those, TMEM100, LRRC63, and NPY best qualified as characteristically expressed in HIF2A PGLs. TMEM100 is essential for epithelial to mesenchymal transition [55], which is required for maturation and migration of neural crest precursors. In hepatocellular carcinomas, a tumor suppressor role of TMEM100 by inhibition of proliferation and metastatic spread has been described [56]. In agreement, lack of TMEM100 induces VEGFA expression in myocardial cells [55]. In the HIF2A PGLs, we noticed higher TMEM100 levels and lower VEGFA expression compared with other pseudohypoxic PGLs, in which the opposite pattern was observed. Moreover, the VEGF receptors NRP1, KDR, and FLT1 were expressed at lower levels in adrenal medulla and HIF2A PGLs compared with most non-HIF2A PGLs. Thus, HIF2A PGLs may be less susceptible to antiangiogenic treatment than other pseudohypoxic PGLs.
NPY has been previously shown to be more highly expressed in adrenergic than noradrenergic or RET- than VHL-mutated pheochromocytomas [57]. Here we observed NPY expression that was increased in HIF2A PGLs compared with all other pseudohypoxic PGLs. Toledo et al. showed decreased expression of NPY in an HIF2A PGL compared with NF1 and possibly VHL PGLs, whereas expression level appears similarly low in SDHB as in the HIF2A sample [26].
In conclusion, HIF2A PGLs share certain features of pseudohypoxic PGLs; however, they are also truly distinct. This may be related to a somewhat different activation pattern of HIF target genes, oxidative phosphorylation genes, as well as angiogenesis-related genes. In addition to the indication that HIF2A PGLs are less vascular and less affected by oxidative phosphorylation dysfunction than other pseudohypoxic PGLs, they also show elevated expression of NPY, protein transcription, and MYCN activation genes, as has been shown for cluster-2 PGLs. Thus, unique or even cluster-2–like expression aspects of HIF2A PGLs will have to be factored in when developing new treatment strategies for HIF2A PGLs.
Material and Methods
All tumor and normal tissue samples were collected and processed with informed patient consent as previously reported [27]. Normal adrenal medulla was microdissected from cortex under microscopic guidance as previously described [58]. In addition to the previously reported samples, material of six HIF2A tumors from two different female patients (H48 and H49) were used. Patient H48 underwent surgery at the age of 29 and had 4 paragangliomas and 2 somatostatinomas removed. The patient later developed asynchronous bilateral pheochromocytomas and additional paragangliomas as well as somatostatinomas with corresponding metastases. Patient H49 had a pheochromocytoma and a paraganglioma removed at the age of 18. Within the same year, somatostatinomas of the pancreas and duodenum were resected. Patient information is given in Table 2.
Table 2.
Tissue sample information
| Mutation | Sex | Age at Surgery | Tissue Type | Status | ||
|---|---|---|---|---|---|---|
| N01 | dna | m | 61 | normal | dna | |
| N02 | dna | nk | nk | normal | dna | |
| N03 | dna | f | 53 | normal | dna | |
| N04–1 | dna | f | 72 | normal | dna | |
| N0–2 | dna | f | 72 | normal | dna | |
| N05 | dna | m | 65 | normal | dna | |
| N06 | dna | m | 56 | normal | dna | |
| N07 | dna | m | 62 | normal | dna | |
| B08 | SDHB | f | 30 | PHEO | Pr-NM | |
| B10 | SDHB | m | 24 | PGL | Pr-M | |
| B12 | SDHB | f | 9 | PGL | Mlt-NM | |
| B14 | SDHB | m | 27 | PGL | Met | |
| B15 | SDHB | m | 38 | PGL | Pr-M | |
| B16 | SDHB | f | 47 | PGL | Mlt-NM | |
| B17–1 | SDHB | f | 36 | PGL | Met | |
| B17–2 | SDHB | f | 36 | PGL | Met | |
| B18 | SDHB | m | 52 | PGL | Met | |
| B19 | SDHB | m | 12 | PGL | Pr-NM | |
| B20 | SDHB | m | 31 | PGL | Pr-NM | |
| B21 | SDHB | m | 55 | PGL | Mlt-NM | |
| B22 | SDHB | m | 35 | PGL | Mlt-NM | |
| B23 | SDHB | f | 35 | PGL | Met | |
| B24 | SDHB | m | 17 | PHEO | Mlt-NM | |
| D25 | SDHD | m | 24 | HNP | Pr-NM | |
| D26 | SDHD | f | 34 | HNP | Bi-M | |
| D27 | SDHD | f | 49 | HNP | Pr-NM | |
| D28 | SDHD | f | 61 | HNP | Pr-NM | |
| D29 | SDHD | m | 16 | PHEO | Pr-NM | |
| D30 | SDHD | f | 31 | PHEO | Pr-NM | |
| D31–1 | SDHD | f | 27 | PHEO | Mlt-NM | |
| D31–2 | SDHD | f | 27 | HNP | Mlt-NM | |
| D31–3 | SDHD | f | 27 | HNP | Mlt-NM | |
| D32 | SDHD | m | 48 | HNP | Bi-NM | |
| D33 | SDHD | m | 61 | PHEO | Pr-NM | |
| D44 | SDHD | f | 29 | HNP | Mlt-NM | |
| D35–1 | SDHD | m | 32 | PHEO | Mlt-NM | |
| D35–2 | SDHD | m | 33 | PGL | Mlt-NM | |
| V36 | VHL | f | 25 | PHEO | Bi-NM | |
| V37–1 | VHL | m | 23 | PHEO | Bi/Mlt-NM | |
| V37–2 | VHL | m | 23 | PHEO | Bi/Mlt-NM | |
| V38 | VHL | M | 16 | PHEO | Bi/Rec-NM | |
| V39 | VHL | m | 29 | PHEO | Pr-NM | |
| V40 | VHL | m | 13 | PHEO | Bi-NM | |
| V41 | VHL | f | 43 | PHEO | Pr-NM | |
| V42 | VHL | m | 29 | PHEO | Bi/Mlt-NM | |
| V43 | VHL | f | 43 | PHEO | Bi-NM | |
| V44 | VHL | m | 39 | PHEO | Pr-NM | |
| V45 | VHL | m | 31 | PHEO | Bi-NM | |
| V46 | VHL | m | 33 | PHEO | Bi/Mlt/Rec-NM | |
| V47 | VHL | m | 19 | PHEO | Bi/Mlt-NM | |
| H48–1 | HIF2A | f | 29 | PGL | Mlt-NM | |
| H48–4 | HIF2A | f | 29 | PGL | Mlt-NM | |
| H48–5 | HIF2A | f | 29 | PGL | Mlt-NM | |
| H48–6 | HIF2A | f | 29 | PGL | Mlt-NM | |
| H49–1 | HIF2A | f | 18 | PHEO | Mlt-NM | |
| H49–2 | HIF2A | f | 18 | PGL | Mlt-NM |
Abbreviations: dna, does not apply; f, female; m, male; nk, not known; PGL, paraganglioma; PHEO, pheochromocytoma (i.e. adrenal PGL); HNP, head and neck paraganglioma; pr, solitary PHEO/PGL; mlt, multiple PGLs; bi, bilateral PHEO; m, metastatic disease; met, metastases; nm, nonmetastatic disease.
GeneChip Human Gene 1.0 ST Array (Affymetrix)
“Core” probe sets were used to perform “gene-level” probe set summarization, background subtraction, and quantile normalization using the RMA option in Expression Console 1.0 (Affymetrix). Data analysis was performed using R packages from the Bioconductor project (http://www.bioconductor.org), as previously described [27].
Differential expression analysis was done by significance analysis of microarray. Class prediction analysis using prediction analysis for microarray was done to predict the genotypes.
IPA
Data were analyzed through the use of QIAGEN's IPA (QIAGEN Redwood City, www.qiagen.com/ingenuity).
qRT-PCR for Genes of Interest
qRT-PCR was performed for nine genes of interest using Taqman primer/probes (Life Technologies, Table S1) on a widely independent sample set of HIF2A (n = 6), SDHB (n = 9), SDHD (n = 12), VHL (n = 10), and normal adrenal medulla (n = 5). Patient information is given in Table S2.
Acknowledgements
We would like to thank Dr. Dimitrios Koutsimpelas for his help with retrieving patient information.
Footnotes
Support: This study was funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, USA.
Disclosure statement: The authors have nothing to disclose.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2016.07.008.
Contributor Information
Stephanie M.J. Fliedner, Email: stephanie.fliedner@uksh.de.
Karel Pacak, Email: karel.pacak@nih.gov.
Appendix A. Supplementary data
Expression of VEGFA and VEGF receptors in pseudohypoxic PGLs and normal medulla. VEGFA expression is mildly upregulated in HIF2A PGLs, whereas FLT1/VEGFR-1 and KDR/VEGFR-2 are expressed at similar levels as in normal adrenal. Moreover, NRP1, a co-receptor for KDR which enhances receptor activation upon ligand binding, is expressed at similar levels in HIF2A PGLs and normal medulla. The y-axis indicates normalized gene expression level. Median, first, and third quartiles of normalized expression z-scores of the gene in question are indicated by midline, bottom, and top of the boxes. Whiskers indicate lowest and highest expression values within 1.5 interquartile ranges of the lower and upper quartile, respectively. Extreme values are depicted as dots and may be considered outliers. Overall analysis of variance (ANOVA) was performed. Asterisks indicate significantly different expression from HIF2A as determined by post hoc analysis using Dunnett's test (***P ≤ .001, **P ≤ .01, *P ≤ .05).
Heatmap of a characteristically expressed gene set of 254 genes.
Table S1. Taqman Assays Used for qRT-PCR Validation
Table S2. Patient Information for Samples Used in qRT-PCR Validation. Samples Which Were Also Used for the Microarray Are Marked by an Asterisk.
Table S3. Genes and Changes in Expression, Which Led to the Prediction of Activation of HIF1A, HIF2A, and ARNT2.
References
- 1.Jochmanova I, Zhuang Z, Pacak K. Pheochromocytoma: gasping for air. Horm Cancer. 2015;6:191–205. doi: 10.1007/s12672-015-0231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao JU, Engelke UF, Sweep FC, Pacak K, Kusters B, Goudswaard AG, Hermus AR, Mensenkamp AR, Eisenhofer G, Qin N. Genotype-specific differences in the tumor metabolite profile of pheochromocytoma and paraganglioma using untargeted and targeted metabolomics. J Clin Endocrinol Metab. 2015;100:E214–E222. doi: 10.1210/jc.2014-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richter S, Peitzsch M, Rapizzi E, Lenders JW, Qin N, de Cubas AA, Schiavi F, Rao JU, Beuschlein F, Quinkler M. Krebs cycle metabolite profiling for identification and stratification of pheochromocytomas/paragangliomas due to succinate dehydrogenase deficiency. J Clin Endocrinol Metab. 2014;99:3903–3911. doi: 10.1210/jc.2014-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eisenhofer G, Lenders JW, Timmers H, Mannelli M, Grebe SK, Hofbauer LC, Bornstein SR, Tiebel O, Adams K, Bratslavsky G. Measurements of plasma methoxytyramine, normetanephrine, and metanephrine as discriminators of different hereditary forms of pheochromocytoma. Clin Chem. 2011;57:411–420. doi: 10.1373/clinchem.2010.153320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenhofer G, Huynh TT, Pacak K, Brouwers FM, Walther MM, Linehan WM, Munson PJ, Mannelli M, Goldstein DS, Elkahloun AG. Distinct gene expression profiles in norepinephrine- and epinephrine-producing hereditary and sporadic pheochromocytomas: activation of hypoxia-driven angiogenic pathways in von Hippel–Lindau syndrome. Endocr Relat Cancer. 2004;11:897–911. doi: 10.1677/erc.1.00838. [DOI] [PubMed] [Google Scholar]
- 6.Dahia PL, Ross KN, Wright ME, Hayashida CY, Santagata S, Barontini M, Kung AL, Sanso G, Powers JF, Tischler AS. A HIF1alpha regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genet. 2005;1:72–80. doi: 10.1371/journal.pgen.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Favier J, Briere JJ, Burnichon N, Riviere J, Vescovo L, Benit P, Giscos-Douriez I, De Reynies A, Bertherat J, Badoual C. The Warburg effect is genetically determined in inherited pheochromocytomas. PLoS One. 2009;4:e7094. doi: 10.1371/journal.pone.0007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jochmanova I, Yang C, Zhuang Z, Pacak K. Hypoxia-inducible factor signaling in pheochromocytoma: turning the rudder in the right direction. J Natl Cancer Inst. 2013;105:1270–1283. doi: 10.1093/jnci/djt201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin N, de Cubas AA, Garcia-Martin R, Richter S, Peitzsch M, Menschikowski M, Lenders JW, Timmers HJ, Mannelli M, Opocher G. Opposing effects of HIF1alpha and HIF2alpha on chromaffin cell phenotypic features and tumor cell proliferation: Insights from MYC-associated factor X. Int J Cancer. 2014;135:2054–2064. doi: 10.1002/ijc.28868. [DOI] [PubMed] [Google Scholar]
- 10.Zhuang Z, Yang C, Lorenzo F, Merino M, Fojo T, Kebebew E, Popovic V, Stratakis CA, Prchal JT, Pacak K. Somatic HIF2A gain-of-function mutations in paraganglioma with polycythemia. N Engl J Med. 2012;367:922–930. doi: 10.1056/NEJMoa1205119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jochmanova I, Zelinka T, Widimsky J, Jr., Pacak K. HIF signaling pathway in pheochromocytoma and other neuroendocrine tumors. Physiol Res. 2014;63(Suppl 2):S251–S262. doi: 10.33549/physiolres.932789. [DOI] [PubMed] [Google Scholar]
- 12.Pacak K, Jochmanova I, Prodanov T, Yang C, Merino MJ, Fojo T, Prchal JT, Tischler AS, Lechan RM, Zhuang Z. New syndrome of paraganglioma and somatostatinoma associated with polycythemia. J Clin Oncol. 2013;31:1690–1698. doi: 10.1200/JCO.2012.47.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pacak K, Chew EY, Pappo AS, Yang C, Lorenzo FR, Wilson MW, Aronow MB, Young JA, Popovic V, Zhuang Z. Ocular manifestations of hypoxia-inducible factor-2alpha paraganglioma-somatostatinoma-polycythemia syndrome. Ophthalmology. 2014;121:2291–2293. doi: 10.1016/j.ophtha.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taieb D, Barlier A, Yang C, Pertuit M, Tchoghandjian A, Rochette C, Zattara-Canoni H, Figarella-Branger D, Zhuang Z, Pacak K. Somatic gain-of-function HIF2A mutations in sporadic central nervous system hemangioblastomas. J Neurooncol. 2016;126:473–481. doi: 10.1007/s11060-015-1983-y. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang Z, Yang C, Ryska A, Ji Y, Hou Y, Graybill SD, Bullova P, Lubensky IA, Kloppel G, Pacak K. HIF2A gain-of-function mutations detected in duodenal gangliocytic paraganglioma. Endocr Relat Cancer. 2016;23:L13–16. doi: 10.1530/ERC-16-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenzo FR, Yang C, Ng Tang Fui M, Vankayalapati H, Zhuang Z, Huynh T, Grossmann M, Pacak K, Prchal JT. A novel EPAS1/HIF2A germline mutation in a congenital polycythemia with paraganglioma. J Mol Med. 2013;91:507–512. doi: 10.1007/s00109-012-0967-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buffet A, Smati S, Mansuy L, Menara M, Lebras M, Heymann MF, Simian C, Favier J, Murat A, Cariou B. Mosaicism in HIF2A-related polycythemia-paraganglioma syndrome. J Clin Endocrinol Metab. 2014;99:E369–E373. doi: 10.1210/jc.2013-2600. [DOI] [PubMed] [Google Scholar]
- 18.Karasawa Y, Sakaguchi M, Minami S, Kitano K, Kawa S, Aoki Y, Itoh N, Sakurai A, Miyazaki M, Watanabe T. Duodenal somatostatinoma and erythrocytosis in a patient with von Hippel–Lindau disease type 2 A. Intern Med. 2001;40:38–43. doi: 10.2169/internalmedicine.40.38. [DOI] [PubMed] [Google Scholar]
- 19.Capodimonti S, Teofili L, Martini M, Cenci T, Iachininoto MG, Nuzzolo ER, Bianchi M, Murdolo M, Leone G, Larocca LM. Von hippel–lindau disease and erythrocytosis. J Clin Oncol. 2012;30:e137–e139. doi: 10.1200/JCO.2011.38.6797. [DOI] [PubMed] [Google Scholar]
- 20.Moline J, Eng C. Multiple endocrine neoplasia type 2: an overview. Genet Med. 2011;13:755–764. doi: 10.1097/GIM.0b013e318216cc6d. [DOI] [PubMed] [Google Scholar]
- 21.Garbrecht N, Anlauf M, Schmitt A, Henopp T, Sipos B, Raffel A, Eisenberger CF, Knoefel WT, Pavel M, Fottner C. Somatostatin-producing neuroendocrine tumors of the duodenum and pancreas: incidence, types, biological behavior, association with inherited syndromes, and functional activity. Endocr Relat Cancer. 2008;15:229–241. doi: 10.1677/ERC-07-0157. [DOI] [PubMed] [Google Scholar]
- 22.Relles D, Baek J, Witkiewicz A, Yeo CJ. Periampullary and duodenal neoplasms in neurofibromatosis type 1: two cases and an updated 20-year review of the literature yielding 76 cases. J Gastrointest Surg. 2010;14:1052–1061. doi: 10.1007/s11605-009-1123-0. [DOI] [PubMed] [Google Scholar]
- 23.Welander J, Andreasson A, Brauckhoff M, Backdahl M, Larsson C, Gimm O, Soderkvist P. Frequent EPAS1/HIF2alpha exons 9 and 12 mutations in non-familial pheochromocytoma. Endocr Relat Cancer. 2014;21:495–504. doi: 10.1530/ERC-13-0384. [DOI] [PubMed] [Google Scholar]
- 24.Comino-Mendez I, de Cubas AA, Bernal C, Alvarez-Escola C, Sanchez-Malo C, Ramirez-Tortosa CL, Pedrinaci S, Rapizzi E, Ercolino T, Bernini G. Tumoral EPAS1 (HIF2A) mutations explain sporadic pheochromocytoma and paraganglioma in the absence of erythrocytosis. Hum Mol Genet. 2013;22:2169–2176. doi: 10.1093/hmg/ddt069. [DOI] [PubMed] [Google Scholar]
- 25.Favier J, Buffet A, Gimenez-Roqueplo AP. HIF2A mutations in paraganglioma with polycythemia. N Engl J Med. 2012;367:2161. doi: 10.1056/NEJMc1211953. [author reply 2161-2162] [DOI] [PubMed] [Google Scholar]
- 26.Toledo RA, Qin Y, Srikantan S, Morales NP, Li Q, Deng Y, Kim SW, Pereira MA, Toledo SP, Su X. In vivo and in vitro oncogenic effects of HIF2A mutations in pheochromocytomas and paragangliomas. Endocr Relat Cancer. 2013;20:349–359. doi: 10.1530/ERC-13-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shankavaram U, Fliedner SM, Elkahloun AG, Barb JJ, Munson PJ, Huynh TT, Matro JC, Turkova H, Linehan WM, Timmers HJ. Genotype and tumor locus determine expression profile of pseudohypoxic pheochromocytomas and paragangliomas. Neoplasia. 2013;15:435–447. doi: 10.1593/neo.122132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 29.Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood. 2011;117:e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yusuf D, Butland SL, Swanson MI, Bolotin E, Ticoll A, Cheung WA, Zhang XY, Dickman CT, Fulton DL, Lim JS. The transcription factor encyclopedia. Genome Biol. 2012;13:R24. doi: 10.1186/gb-2012-13-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camenisch G, Wegner RH, Peso L. In: HIF1A. Yusuf D, editor. vol. 13. 2012. p. R24. (Genome Biol). [Google Scholar]
- 33.Olechnowicz SWZ, Peet DJ. In: EPAS1. Yusuf D, editor. vol. 13. 2012. p. R24. (Genome Biol). [Google Scholar]
- 34.Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies hypoxia inducible factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37:4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter S, Qin N, Pacak K, Eisenhofer G. Role of hypoxia and HIF2alpha in development of the sympathoadrenal cell lineage and chromaffin cell tumors with distinct catecholamine phenotypic features. Adv Pharmacol. 2013;68:285–317. doi: 10.1016/B978-0-12-411512-5.00014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Favier J, Igaz P, Burnichon N, Amar L, Libe R, Badoual C, Tissier F, Bertherat J, Plouin PF, Jeunemaitre X. Rationale for anti-angiogenic therapy in pheochromocytoma and paraganglioma. Endocr Pathol. 2012;23:34–42. doi: 10.1007/s12022-011-9189-0. [DOI] [PubMed] [Google Scholar]
- 37.Towle R, Truong D, Hogg K, Robinson WP, Poh CF, Garnis C. Global analysis of DNA methylation changes during progression of oral cancer. Oral Oncol. 2013;49:1033–1042. doi: 10.1016/j.oraloncology.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Daino K, Ugolin N, Altmeyer-Morel S, Guilly MN, Chevillard S. Gene expression profiling of alpha-radiation–induced rat osteosarcomas: identification of dysregulated genes involved in radiation-induced tumorigenesis of bone. Int J Cancer. 2009;125:612–620. doi: 10.1002/ijc.24392. [DOI] [PubMed] [Google Scholar]
- 39.Jain R, Fischer S, Serra S, Chetty R. The use of cytokeratin 19 (CK19) immunohistochemistry in lesions of the pancreas, gastrointestinal tract, and liver. Appl Immunohistochem Mol Morphol. 2010;18:9–15. doi: 10.1097/PAI.0b013e3181ad36ea. [DOI] [PubMed] [Google Scholar]
- 40.Skondra M, Gkioka E, Kostakis ID, Pissimissis N, Lembessis P, Pectasides D, Koutsilieris M. Detection of circulating tumor cells in breast cancer patients using multiplex reverse transcription-polymerase chain reaction and specific primers for MGB, PTHRP and KRT19 correlation with clinicopathological features. Anticancer Res. 2014;34:6691–6699. [PubMed] [Google Scholar]
- 41.Loriot C, Burnichon N, Gadessaud N, Vescovo L, Amar L, Libe R, Bertherat J, Plouin PF, Jeunemaitre X, Gimenez-Roqueplo AP. Epithelial to mesenchymal transition is activated in metastatic pheochromocytomas and paragangliomas caused by SDHB gene mutations. J Clin Endocrinol Metab. 2012;97:E954–E962. doi: 10.1210/jc.2011-3437. [DOI] [PubMed] [Google Scholar]
- 42.Loriot C, Domingues M, Berger A, Menara M, Ruel M, Morin A, Castro-Vega LJ, Letouze E, Martinelli C, Bemelmans AP. Deciphering the molecular basis of invasiveness in Sdhb-deficient cells. Oncotarget. 2015;6:32955–32965. doi: 10.18632/oncotarget.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fliedner SM, Kaludercic N, Jiang XS, Hansikova H, Hajkova Z, Sladkova J, Limpuangthip A, Backlund PS, Wesley R, Martiniova L. Warburg effect's manifestation in aggressive pheochromocytomas and paragangliomas: insights from a mouse cell model applied to human tumor tissue. PLoS One. 2012;7:e40949. doi: 10.1371/journal.pone.0040949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hervouet E, Demont J, Pecina P, Vojtiskova A, Houstek J, Simonnet H, Godinot C. A new role for the von Hippel–Lindau tumor suppressor protein: stimulation of mitochondrial oxidative phosphorylation complex biogenesis. Carcinogenesis. 2005;26:531–539. doi: 10.1093/carcin/bgi001. [DOI] [PubMed] [Google Scholar]
- 45.Hervouet E, Cizkova A, Demont J, Vojtiskova A, Pecina P, Franssen-van Hal NL, Keijer J, Simonnet H, Ivanek R, Kmoch S. HIF and reactive oxygen species regulate oxidative phosphorylation in cancer. Carcinogenesis. 2008;29:1528–1537. doi: 10.1093/carcin/bgn125. [DOI] [PubMed] [Google Scholar]
- 46.Goffrini P, Ercolino T, Panizza E, Giache V, Cavone L, Chiarugi A, Dima V, Ferrero I, Mannelli M. Functional study in a yeast model of a novel succinate dehydrogenase subunit B gene germline missense mutation (C191Y) diagnosed in a patient affected by a glomus tumor. Hum Mol Genet. 2009;18:1860–1868. doi: 10.1093/hmg/ddp102. [DOI] [PubMed] [Google Scholar]
- 47.Chang YL, Hsieh MH, Chang WW, Wang HY, Lin MC, Wang CP, Lou PJ, Teng SC. Instability of succinate dehydrogenase in SDHD polymorphism connects reactive oxygen species production to nuclear and mitochondrial genomic mutations in yeast. Antioxid Redox Signal. 2015;22:587–602. doi: 10.1089/ars.2014.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, Lemire BD. Mutations in the C. elegans succinate dehydrogenase iron-sulfur subunit promote superoxide generation and premature aging. J Mol Biol. 2009;387:559–569. doi: 10.1016/j.jmb.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 49.Biswas S, Troy H, Leek R, Chung YL, Li JL, Raval RR, Turley H, Gatter K, Pezzella F, Griffiths JR. Effects of HIF-1alpha and HIF2alpha on growth and metabolism of clear-cell renal cell carcinoma 786-0 xenografts. J Oncol. 2010;2010:757908. doi: 10.1155/2010/757908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiavarina B, Martinez-Outschoorn UE, Whitaker-Menezes D, Howell A, Tanowitz HB, Pestell RG, Sotgia F, Lisanti MP. Metabolic reprogramming and two-compartment tumor metabolism: opposing role(s) of HIF1alpha and HIF2alpha in tumor-associated fibroblasts and human breast cancer cells. Cell Cycle. 2012;11:3280–3289. doi: 10.4161/cc.21643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang C, Zhuang Z, Fliedner SM, Shankavaram U, Sun MG, Bullova P, Zhu R, Elkahloun AG, Kourlas PJ, Merino M. Germ-line PHD1 and PHD2 mutations detected in patients with pheochromocytoma/paraganglioma-polycythemia. J Mol Med. 2015;93:93–104. doi: 10.1007/s00109-014-1205-7. [DOI] [PubMed] [Google Scholar]
- 52.Liu L, Cash TP, Jones RG, Keith B, Thompson CB, Simon MC. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao WT, Zhou CF, Li XB, Zhang YF, Fan L, Pelletier J, Fang J. The von Hippel–Lindau protein pVHL inhibits ribosome biogenesis and protein synthesis. J Biol Chem. 2013;288:16588–16597. doi: 10.1074/jbc.M113.455121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Romero-Ruiz A, Bautista L, Navarro V, Heras-Garvin A, March-Diaz R, Castellano A, Gomez-Diaz R, Castro MJ, Berra E, Lopez-Barneo J. Prolyl hydroxylase–dependent modulation of eukaryotic elongation factor 2 activity and protein translation under acute hypoxia. J Biol Chem. 2012;287:9651–9658. doi: 10.1074/jbc.M111.299180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizuta K, Sakabe M, Hashimoto A, Ioka T, Sakai C, Okumura K, Hattammaru M, Fujita M, Araki M, Somekawa S. Impairment of endothelial-mesenchymal transformation during atrioventricular cushion formation in Tmem100 null embryos. Dev Dyn. 2015;244:31–42. doi: 10.1002/dvdy.24216. [DOI] [PubMed] [Google Scholar]
- 56.Ou D, Yang H, Hua D, Xiao S, Yang L. Novel roles of TMEM100: inhibition metastasis and proliferation of hepatocellular carcinoma. Oncotarget. 2015;6:17379–17390. doi: 10.18632/oncotarget.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cleary S, Phillips JK, Huynh TT, Pacak K, Elkahloun AG, Barb J, Worrell RA, Goldstein DS, Eisenhofer G. Neuropeptide Y expression in phaeochromocytomas: relative absence in tumours from patients with von Hippel–Lindau syndrome. J Endocrinol. 2007;193:225–233. doi: 10.1677/JOE-06-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fliedner SM, Breza J, Kvetnansky R, Powers JF, Tischler AS, Wesley R, Merino M, Lehnert H, Pacak K. Tyrosine hydroxylase, chromogranin A, and steroidogenic acute regulator as markers for successful separation of human adrenal medulla. Cell Tissue Res. 2010;340:607–612. doi: 10.1007/s00441-010-0965-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of VEGFA and VEGF receptors in pseudohypoxic PGLs and normal medulla. VEGFA expression is mildly upregulated in HIF2A PGLs, whereas FLT1/VEGFR-1 and KDR/VEGFR-2 are expressed at similar levels as in normal adrenal. Moreover, NRP1, a co-receptor for KDR which enhances receptor activation upon ligand binding, is expressed at similar levels in HIF2A PGLs and normal medulla. The y-axis indicates normalized gene expression level. Median, first, and third quartiles of normalized expression z-scores of the gene in question are indicated by midline, bottom, and top of the boxes. Whiskers indicate lowest and highest expression values within 1.5 interquartile ranges of the lower and upper quartile, respectively. Extreme values are depicted as dots and may be considered outliers. Overall analysis of variance (ANOVA) was performed. Asterisks indicate significantly different expression from HIF2A as determined by post hoc analysis using Dunnett's test (***P ≤ .001, **P ≤ .01, *P ≤ .05).
Heatmap of a characteristically expressed gene set of 254 genes.
Table S1. Taqman Assays Used for qRT-PCR Validation
Table S2. Patient Information for Samples Used in qRT-PCR Validation. Samples Which Were Also Used for the Microarray Are Marked by an Asterisk.
Table S3. Genes and Changes in Expression, Which Led to the Prediction of Activation of HIF1A, HIF2A, and ARNT2.