Abstract
Objectives
Recent surveillance of MRSA colonizing patients and healthcare workers in two African countries (Angola and São Tomé and Príncipe) reported the frequent recovery of oxacillin-susceptible MRSA (OS-MRSA): Staphylococcus aureus strains that gave positive results with the mecA DNA probe, but had low oxacillin MIC values characteristic of susceptible S. aureus. This apparent dissociation of the drug-resistant phenotype from mecA—the primary genetic determinant of resistance—prompted us to perform a more detailed analysis on nine of the African OS-MRSA strains.
Methods
Oxacillin MIC values were determined by Etest and population analysis profiles with and without induction of the stringent stress response by mupirocin. Biochemical profiling using SDS–PAGE followed by western blotting was used for the detection of PBP2A protein produced.
Results
Cultures of the African MRSA strains (ST88-IVa and ST8-V) showed heterogeneous oxacillin resistance in which the majority of cells exhibited low oxacillin MICs (≤0.75 mg/L), but highly resistant subpopulations were also present with oxacillin MIC values up to several hundred mg/L and with frequencies of 10−4 to 10−6. The same strains after induction of the stringent stress response by mupirocin ‘converted’ the heterogeneous phenotypes into a more homogeneous and higher level resistance. After induction by oxacillin and mupirocin, each of the nine African OS-MRSA strains produced PBP2A—the protein product of mecA.
Conclusions
The resistant phenotype of OS-MRSA resembles the phenotypes of historically early MRSA clones. The nature of genetic determinants responsible for the heterogeneous phenotypes of OS-MRSA remains to be determined.
Introduction
Oxacillin-susceptible MRSA (OS-MRSA) are defined as Staphylococcus aureus possessing the antibiotic resistance gene mecA and, yet, exhibiting a low MIC value of oxacillin (<2 mg/L), characteristic of fully susceptible strains.1
It seems that the first use of the term OS-MRSA was in 2007 by Hososaka et al.,1 who described a surveillance study in 11 Japanese hospitals and found that 1.25% of the MRSA were OS-MRSA. However, strains with the OS-MRSA phenotype had been described previously in the USA by Sakoulas et al.2 in 2001 and in Germany by Kampf et al.3 in 2003. More recently, OS-MRSA strains have been reported in Europe (in the UK, Germany and Greece),4–6 in the USA7,8 and in Asia.9–11
In a recent surveillance study,12 164 S. aureus strains were recovered from nasal carriage of patients and healthcare workers in African hospitals and 29 (17.7%) of the strains showed properties typical of OS-MRSA: they produced a positive reaction with a DNA probe indicating the presence of the mecA determinant, but showed oxacillin MIC values of ≤3 mg/L, suggesting that they expressed oxacillin-susceptible phenotypes.
In the study described here, nine African OS-MRSA strains12 and a control strain, SA110,2 were characterized by microbiological and biochemical techniques. Antibiotic-resistant phenotypes were determined by population analysis of the cultures without and with the presence of mupirocin (an agent capable of inducing a stringent stress response and high-level resistance in the bacteria).13 The heterogeneous phenotype of the cultures could be converted into high and homogeneous resistance by the inclusion of mupirocin—an isoleucyl-tRNA synthetase inhibitor—in the medium.13–15 Protein expression profiles of the strains were compared using SDS–PAGE without and with treatment with sub-MIC concentrations of oxacillin and mupirocin. The amounts of PBP2A were compared using a monoclonal antibody against PBP2A.
The aim of our study was to better characterize and understand the mechanism of this unique antibiotic-resistant phenotype of the OS-MRSA strains.
Materials and methods
Bacterial strains and growth conditions
Bacterial strains, which include representatives of the two major OS-MRSA clones circulating in Angola and São Tomé and Príncipe, are described in Table 1. S. aureus strains were grown in tryptic soy broth (TSB; Difco Laboratories, Detroit, MI, USA) at 37°C with aeration or on tryptic soy agar (TSA; Difco Laboratories, Detroit, MI, USA). Bacterial growth was monitored by measurement of the OD at 620 nm (OD620) with a spectrophotometer (model 2800; UNICO, Dayton, NJ, USA).
Table 1.
Strains, isolation origins and dates, clonal types and oxacillin susceptibility (without and with mupirocin)
| Strain | Year | Country | PFGE | spa type | MLST (ST) | SCCmec | Oxacillin MIC (mg/L) |
|
|---|---|---|---|---|---|---|---|---|
| without MUP | with MUP | |||||||
| ANG44 | 2012 | Angola | B2 | t186 | 88 | IVa | 1 | 24 |
| ANG49 | 2012 | Angola | B4 | t1951 | 88 | IVa | 0.5 | 96 |
| ANG278B | 2012 | Angola | B1 | t325 | 88 | IVa | 1 | >256 |
| ANG293A | 2012 | Angola | B4 | t786 | 88 | IVa | 0.75 | 96 |
| STP33 | 2010 | São Tomé and Príncipe | B2 | t786 | 88 | IVa | 0.28 | >256 |
| STP74A | 2010 | São Tomé and Príncipe | B3 | t186 | 88 | IVa | 0.5 | 192 |
| STP254 | 2012 | São Tomé and Príncipe | B1 | t786 | 88 | IVa | 0.38 | >256 |
| STP292 | 2012 | São Tomé and Príncipe | C4 | t451 | 8 | V | 0.5 | 8 |
| STP59 | 2010 | São Tomé and Príncipe | C12 | t451 | 8 | V | 0.75 | 1.5 |
| SA110 | 2000 | USA | t064 | 8 | IVd | 1.5 | >256 | |
MUP, mupirocin.
SA110 was an OS-MRSA isolate from a patient at the Beth Israel Deaconess Medical Center in 2000.2 Typing of the isolate was carried out in the current study.
Antibiotic susceptibility
Antibiotic susceptibility was determined by Etest (bioMérieux) and population analysis profiles (PAPs). The Etest was done by spreading a small aliquot of overnight culture diluted to an OD620 of 0.08 onto TSA plates with and without a sub-MIC concentration of mupirocin (0.03 mg/L) and adding an oxacillin Etest strip onto the surface of the plates.13 MIC values of oxacillin were determined after 48 h of incubation at 37°C.
PAPs were done on overnight cultures diluted with TSB and plated onto TSA plates with and without a sub-MIC concentration of mupirocin (0.03 mg/L)13 and serial (2-fold) dilutions of oxacillin according to the population analysis method described previously.16
Typing methods
Chromosomal DNA for PFGE was prepared as described previously.17 Chromosomal DNA for PCR was prepared as previously described.18,19 MLST, based on the sequences of seven house-keeping genes,20,21 was performed in accordance with published procedures. SCCmec types were determined using described protocols.16,17
Protein analysis
Staphylococcal membrane proteins were prepared following the method described previously.13 SDS–PAGE and western blotting were performed for the detection of PBP2A in membrane preparations of the nine African OS-MRSA strains and the reference strain SA110, using methods described previously.13 In order to induce the mecA gene and optimize expression of the resistant phenotype, 0.5 mg/L oxacillin and 0.03 mg/L mupirocin were added to the growth medium. For detection of β-lactamase, a nitrocefin assay was performed in a 96-well plate. Colour change of nitrocefin from yellow to red was examined with 50 µL of each bacterial culture at an OD620 of 0.5 by adding 2 µL of nitrocefin solution (stock: 1 mg/mL in DMSO).
Results and discussion
The properties of the nine OS-MRSA strains originating in Africa and the OS-MRSA strain SA110 (often used as a reference for MRSA with this phenotype) are shown in Table 1. The four isolates from Angola (ANG44, ANG49, ANG278B and ANG293A) and three of the five isolates from São Tomé and Príncipe (STP33, STP74A and STP254) showed the characteristics of the so-called ‘African clone’ (ST88-SCCmec type IVa) widely spread in this continent, while the two remaining isolates from São Tomé and Príncipe (STP59 and STP292) belonged to ST8-V, the major clone circulating in the country.
The oxacillin-resistant phenotype of the nine African strains was evaluated by Etest (Figure S1, available as Supplementary data at JAC Online) on agar medium with or without supplementation by mupirocin, an agent that was shown to boost levels of oxacillin resistance by triggering the stringent stress response in bacteria.13,20 All African strains exhibited substantially increased levels of oxacillin resistance in the presence of mupirocin, except for two strains—strains STP292 and STP59, which showed moderate or little effect by mupirocin (Figure S1H and I). These two strains share a common MLST type ST8, SCCmec type V, spa type t451 and show PFGE subtypes that differ in a single band. The other strains belonged to ST88-SCCmec IVa.
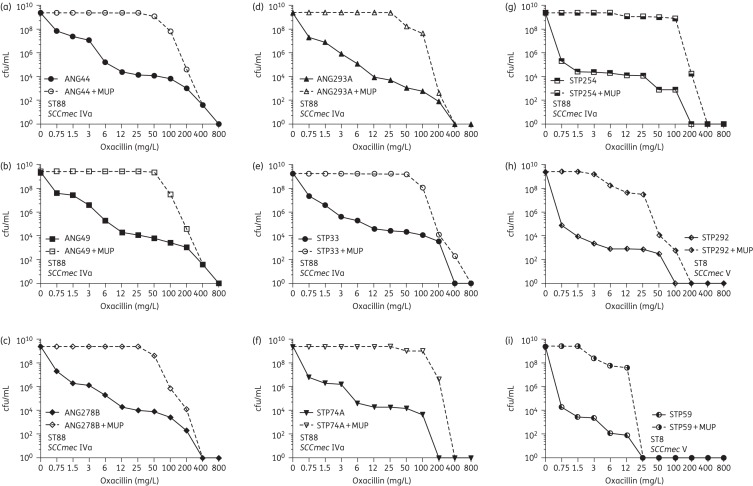
The antibiotic-resistant phenotype of the nine African strains was also evaluated by PAPs (Figure 1). In cultures of each of the strains, the majority of the bacteria had low oxacillin MICs: cells began to lose viability when exposed to even low concentrations of oxacillin (<1.5 mg/L). However, highly resistant subpopulations of bacteria were also present in cultures of each of the African strains with frequencies varying from 10−4 to 10−6 and the resistance level of these subpopulations was quite high with MIC values often exceeding 100 mg/L oxacillin.
Figure 1.
PAPs of the nine African strains on oxacillin plates with and without supplementation by a sub-MIC concentration of mupirocin. MUP, mupirocin.
Figure 1 shows that in the presence of mupirocin, the phenotype of seven of the nine African strains was converted from a heterogeneous into a highly homogeneous phenotype in which the great majority of bacteria expressed very high levels of oxacillin resistance. In two strains, STP292 and STP59 (both ST8-V), the resistance level was increased, but the phenotype remained heterogeneous. The results of the PAPs were consistent with the findings obtained by Etest (Figure S1).
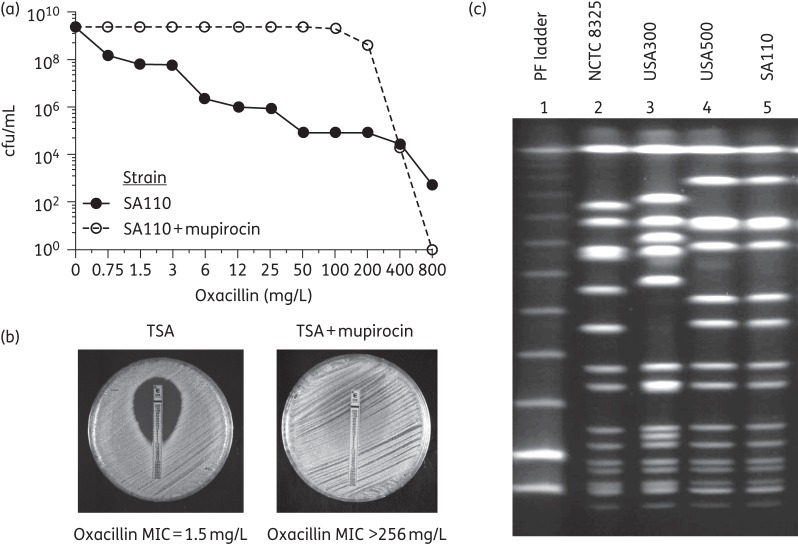
A similar, profound, effect of induction of the stringent stress response by mupirocin was also demonstrated in MRSA strain SA110 (ST8-IVd), frequently used as a prototype of OS-MRSA.2 Similarly to the case of the African strains, the heterogeneous resistant phenotype of strain SA110 was converted into a high and homogeneous resistance in the presence of mupirocin (Figure 2a and b). PFGE analysis revealed that SA110 had the same PFGE pattern as a representative of the lineage USA500 (ST8-IV) (Figure 2c).22 The results of Etest and PAPs suggest that MRSA strains with ST8-SCCmec V such as STP292 and STP59 may be either more relaxed to the stringent stress response or less susceptible to mupirocin as compared with the other African strains.
Figure 2.
Oxacillin resistance phenotype of ‘OS-MRSA’ strain SA110—determined by population analysis (a) or Etest (b)—in the absence and presence of mupirocin added to the growth medium. (c) PFGE profiles of strain SA110 and two strains representing the MRSA clones USA500 and USA300. The PFGE profile of strain NCTC 8325 was also included as a control. Noteworthy, according to Li et al.,22 USA300 is derived from USA500.
In the next series of experiments, the nine African MRSA strains were analysed by SDS–PAGE with and without induction of resistance by oxacillin and/or mupirocin (Figure S2A–C). Figure S2(A–C) and Figure 3 document the production of PBP2A (detected by the monoclonal antibody specific to this protein) by each of the African OS-MRSA strains and by the control OS-MRSA strain SA110 in the presence of oxacillin and mupirocin.
Figure 3.
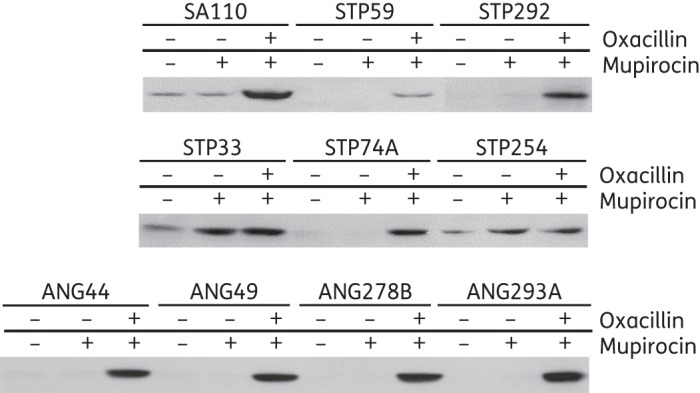
Titration of the amounts of PBP2A produced by the OS-MRSA isolates (SA110 and the nine MRSA isolates from Africa).
In seven of the nine African OS-MRSA [STP59/ST8-V, STP74A/ST88-IVa, STP292/ST8-V and the four ST88-IVa strains from Angola (ANG44, ANG49, ANG278B and ANG293A)], production of PBP2A was only detectable after induction with both oxacillin and mupirocin (Figure S2A–C and Figure 3). In the two remaining strains (STP254 and STP33), the amount of PBP2A was increased by the addition of mupirocin; the addition of oxacillin had no impact (Figure S2B and Figure 3), which correlates with the absence of β-lactamase in these strains (Figure S2B; the fifth arrow from the top). The absence of expression of β-lactamase in STP33 and STP254 was also confirmed with a nitrocefin assay (data not shown). The production of PBP2A in control strain SA110 was greatly increased by mupirocin and oxacillin (Figure S2A and Figure 3).
Although each of the nine African OS-MRSA strains were able to change their phenotypes from susceptible to resistant under stress conditions, these strains appear to be intrinsically more susceptible to oxacillin—without induction of the mecA gene—as compared with most MRSA strains.
The properties of the African MRSA strains belonging to the two main lineages widely spread in Angola and São Tomé and Príncipe (ST88-IVa and ST8-V) suggest that they represent resurgence of clonal types of MRSA, which express antibiotic resistance in a highly heterogeneous fashion. Strains with these particular resistance phenotypes have already been seen among MRSA clinical strains in earlier surveillance studies in Europe and the USA.20,23,24 While the antibiotic resistance level (oxacillin MIC) of the African strains was low, bordering on susceptibility, data presented in this communication show that the borderline resistance of these strains is due to their unusual and extremely heterogeneous phenotypes.
While MRSA with comparable highly heterogeneous resistance have been described before in the literature,2–11 the appearance of MRSA lineages with these properties among clinical strains recovered with high frequency in a specific geographical area seems to represent the first case in which MRSA strains with such highly heterogeneous phenotypes have a significant representation among clinical specimens.
Funding
This work was supported by a grant from the National Institutes of Health (NIH), 2RO1 AI457838-15, awarded to A. T.
Transparency declarations
None to declare.
Supplementary data
Figures S1 and S2 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
We thank Dr Sakoulas for sending us strain SA110 and Dr Maria Pardos de la Gandara for typing of this strain.
References
- 1.Hososaka Y, Hanaki H, Endo H et al. Characterization of oxacillin-susceptible mecA-positive Staphylococcus aureus: a new type of MRSA. J Infect Chemother 2007; 13: 79–86. [DOI] [PubMed] [Google Scholar]
- 2.Sakoulas G, Gold HS, Venkataraman L et al. Methicillin-resistant Staphylococcus aureus: comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J Clin Microbiol 2001; 39: 3946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kampf G, Adena S, Ruden H et al. Inducibility and potential role of mecA-gene-positive oxacillin-susceptible Staphylococcus aureus from colonized healthcare workers as a source for nosocomial infections. J Hosp Infect 2003; 54: 124–9. [DOI] [PubMed] [Google Scholar]
- 4.Ikonomidis A, Michail G, Vasdeki A et al. In vitro and in vivo evaluations of oxacillin efficiency against mecA-positive oxacillin-susceptible Staphylococcus aureus. Antimicrob Agents Chemother 2008; 52: 3905–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saeed K, Ahmad N, Dryden M et al. Oxacillin-susceptible methicillin-resistant Staphylococcus aureus (OS-MRSA), a hidden resistant mechanism among clinically significant isolates in the Wessex region/UK. Infection 2014; 42: 843–7. [DOI] [PubMed] [Google Scholar]
- 6.Witte W, Pasemann B, Cuny C. Detection of low-level oxacillin resistance in mecA-positive Staphylococcus aureus. Clin Microbiol Infect 2007; 13: 408–12. [DOI] [PubMed] [Google Scholar]
- 7.Bearman GM, Rosato AE, Assanasen S et al. Nasal carriage of inducible dormant and community-associated methicillin-resistant Staphylococcus aureus in an ambulatory population of predominantly university students. Int J Infect Dis 2010; 14 Suppl 3: e18–24. [DOI] [PubMed] [Google Scholar]
- 8.Forbes BA, Bombicino K, Plata K et al. Unusual form of oxacillin resistance in methicillin-resistant Staphylococcus aureus clinical strains. Diagn Microbiol Infect Dis 2008; 61: 387–95. [DOI] [PubMed] [Google Scholar]
- 9.Chen FJ, Huang IW, Wang CH et al. mecA-positive Staphylococcus aureus with low-level oxacillin MIC in Taiwan. J Clin Microbiol 2012; 50: 1679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He W, Chen H, Zhao C et al. Prevalence and molecular typing of oxacillin-susceptible mecA-positive Staphylococcus aureus from multiple hospitals in China. Diagn Microbiol Infect Dis 2013; 77: 267–9. [DOI] [PubMed] [Google Scholar]
- 11.Jannati E, Arzanlou M, Habibzadeh S et al. Nasal colonization of mecA-positive, oxacillin-susceptible, methicillin-resistant Staphylococcus aureus isolates among nursing staff in an Iranian teaching hospital. Am J Infect Control 2013; 41: 1122–4. [DOI] [PubMed] [Google Scholar]
- 12.Conceição T, Coelho C, de Lencastre H et al. Frequent occurrence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) strains in two African countries. J Antimicrob Chemother 2015; 70: 3200–4. [DOI] [PubMed] [Google Scholar]
- 13.Kim C, Mwangi M, Chung M et al. The mechanism of heterogeneous β-lactam resistance in MRSA: key role of the stringent stress response. PLoS One 2013; 8: e82814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mwangi MM, Kim C, Chung M et al. Whole-genome sequencing reveals a link between β-lactam resistance and synthetases of the alarmone (p)ppGpp in Staphylococcus aureus. Microb Drug Resist 2013; 19: 153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aedo S, Tomasz A.. Role of the stringent stress response in the antibiotic resistance phenotype of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2016; 60: 2311–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother 1991; 35: 124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung M, de Lencastre H, Matthews P et al. Molecular typing of methicillin-resistant Staphylococcus aureus by pulsed-field gel electrophoresis: comparison of results obtained in a multilaboratory effort using identical protocols and MRSA strains. Microb Drug Resist 2000; 6: 189–98. [DOI] [PubMed] [Google Scholar]
- 18.Milheiriço C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother 2007; 51: 3374–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milheiriço C, Oliveira DC, de Lencastre H. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: ‘SCCmec IV multiplex’. J Antimicrob Chemother 2007; 60: 42–8. [DOI] [PubMed] [Google Scholar]
- 20.Crisóstomo MI, Westh H, Tomasz A et al. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc Natl Acad Sci USA 2001; 98: 9865–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright MC, Day NP, Davies CE et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 2000; 38: 1008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li M, Diep BA, Villaruz AE et al. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci USA 2009; 106: 5883–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lencastre H, Sa Figueiredo AM, Urban C et al. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother 1991; 35: 632–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dordel J, Kim C, Chung M et al. Novel determinants of antibiotic resistance: identification of mutated loci in highly methicillin-resistant subpopulations of methicillin-resistant Staphylococcus aureus. MBio 2014; 5: e01000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.