Abstract
Objectives
MDR MRSA isolates cultured from primates, their facility and primate personnel from the Washington National Primate Research Center were characterized to determine whether they were epidemiologically related to each other and if they represented common local human-associated MRSA strains.
Methods
Human and primate nasal and composite environmental samples were collected, enriched and selected on medium supplemented with oxacillin and polymyxin B. Isolates were biochemically verified as Staphylococcus aureus and screened for the mecA gene. Selected isolates were characterized using SCCmec typing, MLST and WGS.
Results
Nasal cultures were performed on 596 primates and 105 (17.6%) were MRSA positive. Two of 79 (2.5%) personnel and two of 56 (3.6%) composite primate environmental facility samples were MRSA positive. Three MRSA isolates from primates, one MRSA from personnel, two environmental MRSA and one primate MSSA were ST188 and were the same strain type by conventional typing methods. ST188 isolates were related to a 2007 ST188 human isolate from Hong Kong. Both MRSA isolates from out-of-state primates had a novel MLST type, ST3268, and an unrelated group. All isolates carried ≥1 other antibiotic resistance gene(s), including tet(38), the only tet gene identified.
Conclusions
ST188 is very rare in North America and has almost exclusively been identified in people from Pan-Asia, while ST3268 is a newly reported MRSA type. The data suggest that the primate MDR MRSA was unlikely to come from primate centre employees. Captive primates are likely to be an unappreciated source of MRSA.
Introduction
MRSA is increasingly prevalent in community settings and is a major nosocomial pathogen.1 MRSA is known to colonize and cause infections in humans, livestock, pets, zoo animals and marine mammals.1,2 However, relatively little is known about the dynamics of Staphylococcus aureus or MRSA colonization or/and infection in non-human primates, which are important research models for human disease.3–6
The Washington National Primate Research Center (WaNPRC) is one of seven National Primate Research Centers in the USA. Historically, MRSA infections in WaNPRC primates have not been considered a major concern, because they have not been frequently identified, or the cause of significant clinical disease. However, between February and September 2014, there were nine cases of MRSA infection in primates with cranial implants, including two animals with cranial implant chamber infections with MRSA and neurological symptoms. Moreover, in December 2014, an animal technician sustained a bite wound from a MRSA colonized primate, and developed a severe MRSA skin infection, which required intravenous daptomycin treatment. This suggests MRSA transmission from a primate to staff personnel.6 Initial antimicrobial susceptibility testing of MRSA isolated from primates and the human skin infection showed that they had similar MDR profiles with susceptibility to daptomycin and vancomycin, and varied susceptibility to trimethoprim/sulfamethoxazole and tetracycline. These phenotypic similarities, the closed environment and spread of MRSA from one area to another within the three areas of the facilities suggested that the isolates obtained from primates and personnel could represent a single MRSA strain or a single clone, underscoring the need for a better understanding of primate MRSA isolates and their transmission.
In 2015, the WaNPRC implemented screening of all primates, including Macaca fascicularis, Macaca mulatta and Macaca nemestrina, for nasal carriage of MRSA, providing an opportunity to explore the strain characteristics and molecular epidemiology of primate-associated MRSA. The aims of this study were to: (i) determine the prevalence of MRSA carriage among primates, staff and the primate centre environment; and (ii) characterize a selected number of MRSA isolates to determine their relatedness to each other and the potential source of the strain or strains recovered. In addition, the study would determine whether transmission of MRSA was occurring within the WaNPRC.
Materials and methods
WaNPRC Facility
The WaNPRC facility is composed of three sections: section 1 includes five floors, and was the first area where MRSA was identified; section 2 is within the same building, but physically separated ∼1 km from the first section; and section 3 is on a separate campus within the city. The main surgical suites for the centre are housed in section 1, and primates from all three sections may undergo procedures there.
Primate, human and environmental samples
Nasal samples (n = 596) were taken from ketamine-sedated primates (M. fascicularis, M. mulatta and M. nemestrina) using standard microbiological swabs; BD BBL CultureSwab Plus Amies Medium (Becton Dickinson, Franklin Lakes, NJ, USA) and Starplex Starswab II (Starplex Scientific, Etobicoke, ON, Canada). Skin swabs from 53 animals, 4 wound samples, 2 rectal samples from animals with diarrhoea and 2 oral samples were additionally processed for the presence of MRSA. All samples were processed the same day that they were collected.
Nasal samples were taken from volunteer primate personnel (n = 79) using SANICULT™ Transport Swabs (Starplex Scientific) gently placed into the anterior nares and rotated twice in each nostril as previously described.7,8
Composite environmental surface samples (n = 56) were collected from the five floors of section 1, where the MRSA was first noted in the primates, as previously described.7 No environmental samples from sections 2 and 3 were collected.
Ethics
Human samples were collected under University of Washington IRB approval no. 35382. Primate samples were taken as part of the general care of the animals.
Isolation and verification of S. aureus and MRSA
Samples were enriched and positive cultures were plated on oxacillin-resistant S. aureus Base® medium (ORSAB; Oxoid Limited, Basingstoke, UK) and incubated at 36.5°C for 48 h as previously described.7 Dark blue colonies were isolated and plated on Brucella agar (Difco Laboratories, Division BD Sparks, MD, USA) supplemented with 5% sterile sheep blood and incubated at 36.5°C in 5% CO2 for 24 h. Colonies that produced β-haemolysis were verified as S. aureus using the Staphaurex® test (Remel, Lenexa, KS, USA) according to the manufacturer's instructions. Verification of MRSA was determined using the Thermo Scientific PBP2' latex agglutination test kit® according to the manufacturer's instructions (Thermo Fisher Scientific, Remel Products, Lenexa, KS, USA). In addition, a subset of isolates representing each of the three areas within the facility, had the mecA gene verified by PCR assay as previously described.9 This method identified MRSA and mecA-positive coagulase-negative Staphylococcus spp., the latter of which were not further characterized. However, on occasion MSSA isolates were identified. Both MRSA and MSSA were stored frozen at −80°C.
MLST and SCCmec typing
Twelve isolates (five MRSA and two MSSA from primates, two MRSA and one MSSA from personnel, and two environmental MRSA from the primate facility) were selected for characterization using molecular methods. In addition, two environmental MRSA isolates (one from a local wastewater treatment plant and one from a wetland) were included as epidemiologically unrelated controls (Table 1). The isolates from the animals were randomly selected to represent all three areas of the facility at the beginning of the study and include isolates from animals sent from other locations. All isolates were screened for SCCmec types I–V by multiplex PCR as previously described, using controls for each type (I–V).10 The SCCmec type was confirmed by amplification with appropriate single primer sets. MLST was performed using previously published PCR assays, primers and cycling parameters11 with PCR products sequenced bi-directionally at the University of Washington's Genome Sciences High-Throughput Sequencing facility. MLST alleles were assigned in accordance with the S. aureus MLST database (http://saureus.mlst.net/). Novel MLST types were submitted to the MLST web site for new ST assignments.
Table 1.
Characterization of MRSA and MSSA isolates
| Strain ID | Primate centre location | Source | S. aureus type | Date isolated | MLST type | SCCmec type | Antibiotic resistance genes |
|---|---|---|---|---|---|---|---|
| PC2A | 1 | environment | MRSA | 16/1/2015 | ST188 | IVa | aacB-le-aph2-la, blaZ, erm(B), tet(38), gyrA Ser84Leu |
| PC13A | 1 | environment | MRSA | 16/1/2015 | ST188 | IVa | aacB-le-aph2-la, blaZ, erm(B), tet(38), gyrA Ser84Leu |
| Z224 | 1 | primate nasal | MRSA | 24/2/2015 | ST188 | IVa | aacB-le-aph2-la, ant4-IA, blaZ, erm(B), tet(38), gyrA Ser84Leu |
| A212 | 2 | primate nasal | MRSA | 12/2/2015 | ST188 | IVa | aacB-le-aph2-la, blaZ, erm(B), tet(38), gyrA Ser84Leu |
| A213 | 3 | primate nasal | MRSA | 13/2/2015 | ST188 | IVa | aacB-le-aph2-la, blaZ, erm(B), tet(38), gyrA Ser84Leu |
| NPC104A | personnel nasal | MRSA | 27/1/2015 | ST188 | IVa | aacB-le-aph2-la, blaZ, erm(B), tet(38), gyrA Ser84Leu | |
| Z1224 | 1 | primate nasal | MSSA | 24/2/2015 | ST188 | negative | aacB-le-aph2-la, blaZ, erm(B), tet(38), gyrA Ser84Leu |
| TXA | 3 | primate nasal | MRSA | 19/5/2015 | ST3268 | V | aacB-le-aph2-la, blaZ, erm(B), gyrA Ser84Leu |
| TXB | 3 | primate nasal | MRSA | 19/5/2015 | ST3268 | V | aacB-le-aph2-la, blaZ, erm(B), gyrA Ser84Leu |
| A217 | 1 | primate nasal | MSSA | 17/2/2015 | ST226 | negative | tet(38) |
| NPC132A | personnel nasal | MRSA | 3/2/2015 | ST8 | IV | aacB-le-aph2-la, blaZ, erm(B) | |
| NPC144 | personnel nasal | MSSA | 3/2/2015 | ST15 | negative | aacB-le-aph2-la, ant4-IA, blaZ, erm(B), tet(38) | |
| WW2014 | non-primate control: wastewater effluent | MRSA | 2014 | ST8 | IVa | aph(3)-IIIa, blaZ, tet(38), gyrA Ser84Leu | |
| UWB-405 | non-primate control: wetland | MRSA | 2014 | ST133 | IVc | tet(38) |
tet(38) PCR confirmatory assay
Selected isolates that were tet(38) positive by WGS were verified using the PCR assay previously described.12 PCR amplicons were sequenced as described above. Sequences were compared using the NCBI non-redundant database (accessed 29 January 2016) using a BLASTN search.13
WGS and analysis
Libraries were prepared and sequenced on an Illumina MiSeq as previously described14 with the addition of size selection for library fragments in the 400–800 bp range. De novo genome assemblies were generated as previously described.14 The average read depth obtained was 42× (range, 33.7–51.4× coverage). To select the most appropriate genome for epidemiological analysis, contigs in each isolate's assembly were compared with the NCBI non-redundant database (accessed 20 March 2015) using a BLASTN search.13 The organism(s) matched with the highest bitscore for each contig was recorded, and weighted proportionally to the length of the contig. The cumulative weights of top matches were then integrated across isolates. Based on this analysis, S. aureus CN1 (GenBank accession no. 537389513) was selected as the reference complete genome most closely related to all isolates. Alternatively, a draft genome of ST188 SCCmec V CUHK_HK188 (GenBank accession no. JFFV00000000), isolated from a human in Hong Kong, was also used for comparison. Sequence reads generated in this study are available from the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession no. SRP067697. Sequence reads were aligned to the CN1 genome, and variant calling (SNPs and INDELs) was performed using bwa-mem (v0.7.12)15 and samtools (v1.1),16 except that minimum gapped reads of 3 and minimum fraction of gapped reads of 0.2 were used. All-by-all pairwise distance matrices were constructed as previously described.14 Approximately maximum-likelihood phylogenetic trees were constructed using FastTree 2.18.17
Antibiotic resistance gene content
We examined isolates with respect to a set of 48 antibiotic resistance genes (Table S1, available as Supplementary data at JAC Online). We performed a BLASTX search of the de novo assemblies of all isolates against these factors and scored factors as present if a match with ≥80% identity and ≥80% coverage was identified, as described previously.18 Mutations in gyrA and gyrB were identified through examination of the primary variant calls.
Results
MRSA isolates from primates, personnel and the primate facility
A total of 596 primates were cultured for nasal carriage of MRSA, including three groups of animals that were imported from outside the state. A total of 105 (17.6%) animals had MRSA-positive nasal cultures. MRSA-positive primates included all three species from all three sections of the facility, and one of the three groups of primates from outside the state. In addition, MRSA was isolated from 20 (37.7%) of 53 skin, 2 of 4 (50%) wound and 1 of 2 rectal samples. One of two (50%) primate oral samples was positive for MSSA, but not MRSA. MRSA was detected from two of 79 (2.5%) primate centre personnel: both individuals were researchers; no husbandry, office or veterinarian staff was MRSA positive of those tested. Two (3.6%) of the composite environmental samples were MRSA positive: a primate chair that animals were placed in during research sessions, door knobs, and outside of the primate centre elevator buttons.
MLST and SCCmec typing of MRSA strains
Twelve representative isolates were chosen for further molecular characterization (Table 1). Three MRSA primate isolates from the WaNPRC, including one from each part of the WaNPRC (sections 1–3), two MRSA isolates from animals originally from outside of Washington state, two MRSA isolates from primate research personnel and two environmental MRSA isolates were selected, plus two MSSA isolated from primate nasal cultures and one MSSA isolated from a third employee. Two unrelated MRSA isolates taken from the local outside environment were also included as controls (Table 1). MLST identified two environmental MRSA isolates, three primate MRSA isolates, one personnel MRSA isolate and one primate MSSA isolate as ST188. All six MRSA isolates carried SCCmec type IVa. The two MRSA isolates from animals originating outside of Washington state and shipped to WaNPRC and sampled while in quarantine (TXA, TXB), both had the same novel MLST type, which has been assigned ST3268 and carried SCCmec type V. One MSSA from primate nasal culture (A217) was a different, previously unreported MLST type, which has now been assigned ST226 (http://saureus.mlst.net/). The ST226 strain was negative for SCCmec types I–V. The remaining MRSA isolate (NPC132A) from one researcher, was ST8 and SCCmec IV, while the MSSA isolate (NPC144), from a separate primate researcher, was ST15. The control MRSA isolate (WW2014), from a Seattle wastewater treatment plant, was ST8 SCCmec IVa and the control (UWB-405), from a restored wetland, was ST133 SCCmec IVc.
Whole-genome molecular epidemiology
The 14 isolates were shotgun sequenced and compared against the draft genome of a previously sequenced S. aureus ST188 isolate (CUHK_HK188, Figure 1a), as well as the S. aureus CN1 reference genome (Table S2), which was more phylogenomically central to the strains studied.
Figure 1.
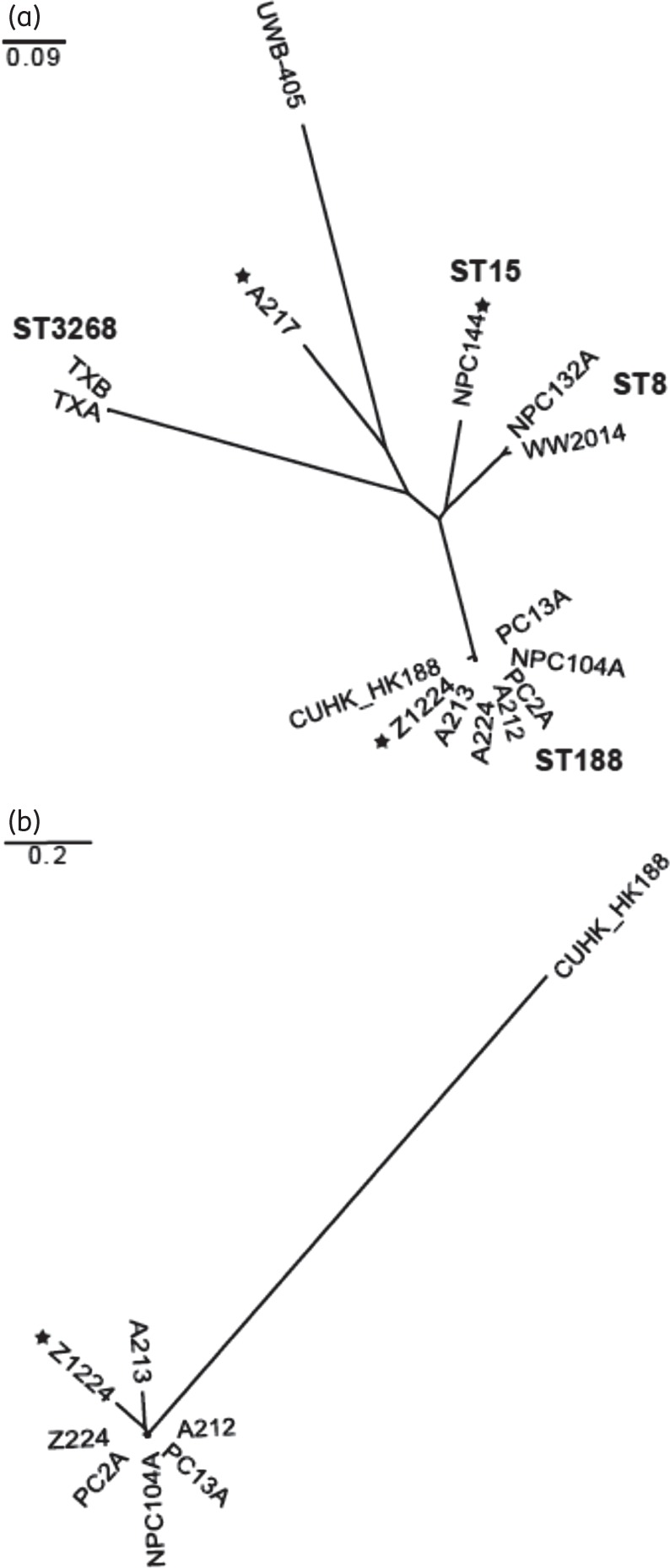
Phylogenomic relationships of S. aureus isolates. Molecular epidemiology of (a) all S. aureus isolates and (b) ST188 S. aureus isolates. Relationships are shown with respect to the draft genome of ST188 strain CUHK_HK188. The scale bars are expressed in changes per site in both panels. STs of select clades are indicated in (a), in bold. MSSA strains are indicated by a black star; the remaining strains are MRSA. MSSA strain A217 has recently been assigned MLST type ST226.
The ST188 strains comprised a distinct clade, which was substantially removed from the other isolates (Figure 1a). Although distinguishable at the genomic level, these isolates were both highly similar to each other with respect to the number of distinguishing polymorphisms they carried (Table S3) and were easily discernible from the previously reported ST188 draft genome (Figure 1b), suggesting a recent shared ancestry.14 Notably, both the ST188 primate MSSA nasal isolate (Z1224) and the ST188 MRSA obtained from a person's nasal culture (NPC104A) were included in this group, suggesting that despite their different origin and phenotypes, they both belong to the same population as the other primate MRSA isolates (Figure 1a and b).
The two ST3268 isolates, which originated from an outside facility, similarly formed a distinct and isolated clade. Like the ST188 isolates, the ST3268 isolates were genomically distinguishable from one another, but demonstrated only a small absolute number of distinguishing variants. This limited number of genomic differences suggests that the ST3268 isolates have a common ancestry.
Control strains and other isolates obtained from primate facility personnel were distributed more broadly throughout the phylogeny, indicating greater degrees of phylogenomic divergence (Figure 1a).
Antibiotic resistance genes
The genomes were examined for the presence of 48 known S. aureus antibiotic resistance genes (Table 1 and Table S1). Consistent with their MDR phenotype, the WaNPRC MRSA ST188 isolates and MSSA Z1224 carried aminoglycoside resistance gene aacB-le-aph2-la, β-lactamase gene blaZ, rRNA methylase gene erm(B), conferring resistance to macrolides, lincosamide and streptogramin B, and tetracycline resistance gene tet(38). The ST3268 MRSA isolates TXA and TXB similarly carried aacB-le-aph2-la, blaZ and erm(B). The human ST8 MRSA isolate NPC132A carried aacB-le-aph2-la, blaZ and erm(B). The ST15 MSSA carried aacB-le-aph2-la, blaZ, erm(B) and tet(38), while MSSA ST226 carried tet(38) (Table 1). MRSA control strain WW2014 carried blaZ, tet(38) and aminoglycoside resistance gene aph(3)-llla, while the UWB-405 isolate ST133 carried only tet(38). Because the tet(38) gene has not been commonly identified in MRSA, we verified its presence in these isolates using targeted PCR and conventional sequencing to verify its presence.
We separately examined WGS data for gyrA and gyrB mutations, which could confer resistance to fluoroquinolones. Ten of the strains, including all of the primate MRSA isolates, carried the Ser84Leu gyrA mutation previously reported in S. aureus (Table 1).19
Discussion
SCCmec typing, MLST and WGS indicate that the primate MRSA isolates examined in this study comprised two different groups of genomically related strains (ST188 and ST3268), each of which were the same strain type by conventional typing methods.20 We have continued to isolate MRSA over the last year from the primates and have only identified ST188 and ST3268 in the colony. All the primate MRSA isolates were MDR carrying both acquired genes and resistance-associated mutations (Table 1). The majority of primate MRSA isolates were ST188 SCCmec type IVa, which is very uncommon in North America. MRSA ST188 has previously been isolated almost exclusively from Pan-Asian humans and often carries other SCCmec types.21,22 There have been few reports of MRSA ST188 strains isolated from people in the USA, or from animals and/or their environment.23–25 A recent study reported three MSSA ST188 from sanctuary chimpanzees isolated in Uganda and 10 MSSA ST188 isolated from wild Madagascar lemurs.5 None of the African MSSA ST188 isolates was phenotypically resistant to the three antibiotics tested.5
The WaNPRC periodically hosts guest researchers from Pan-Asia, and we cannot rule out the possibility that a foreign researcher or another person inadvertently introduced MRSA ST188 into the primate colony. However, several observations make it more likely that ST188 strains were brought into the USA by a primate. The predominance of ST188 in the primates studied and the paucity of that ST within the local human community suggest that MSSA and/or MRSA ST188 could be part of the normal primate microbiota, particularly since MSSA ST188 has been identified in 13 (15%) of 84 MSSA isolated in African primates.5 We note that MRSA ST188 has also been isolated from non-human primates from another primate facility on the East Coast (J. Lane, WaNPRC, personal communication): the probability of multiple, independent human-to-primate transmission events in independent facilities is expected to be low. However, it is possible that the primates became colonized with MRSA ST188 through contact with humans in Asia before being imported to domestic animal facilities. The relatively low prevalence of human ST188 even in endemic Pan-Asian regions makes this possibility less likely.21,22 We must emphasize that the timing of ST188 introduction cannot be determined from the present study. ST188 strains may have initially been carried for an indeterminate period as commensal bacteria in one or a few animals, either nationally or within the WaNPRC colony, and have more recently acquired virulence factors, which enabled them to spread and begin to cause disease.
The second MRSA group belonged to a new MLST type, ST3268, and was associated with animals from one out-of-state facility. This strain appears to be unrelated to other S. aureus STs by eBURST analysis (data not shown). Although not sequenced in this study, isolates of ST3268 were additionally identified from nasal culture of three WaNPRC animals experimentally infected with Simian immunodeficiency virus. A fourth animal had the same ST3268 from a shoulder abscess caused by a bite wound. Because the resident WaNPRC animals did not have direct contact with the out-of-state primates, the most direct transmission route connecting all four of these strains would be through environmental contamination from a common procedure room. ST3268 was not recovered from environmental samples, but given that only one of the three sites within the facility was sampled, it could easily have been missed by this screen. Consequently, it is unlikely that employees of WaNPRC were the original source of the ST3268 isolates.
It is noteworthy that an MRSA isolate from one researcher was ST8, which is common in the community, but was not cultured from any primates or the environment.26 Our failure to identify this or other common human MRSA STs from primates also speaks against the likelihood of human to primate transmission within the facility. This is in contrast to the recent Hanley et al.6 publication, which identified USA300/ST8, a community-associated strain, as the predominant MRSA strain at the Texas facility. A report from a Greek zoological park also identified both human-associated MRSA ST8 and ST15 within their monkey populations.27 The MRSA strains from infected M. mulatta, originally from China, were not characterized, thus their MLST type is unknown at this time.3
To guard against the potential spread of MRSA among primates and to protect workers at the centre further, extensive decontamination of the WaNPRC facility has been implemented using hydrogen peroxide cleaner and UV light systems (Chlordisys Torch Tower). In addition, MRSA appropriate personnel protection equipment is required. Further, MRSA-positive animals are now segregated from other primates when possible, and extensive decontamination of the primates themselves has been carried out with >90% success rate at 4 weeks post-decolonization (data not shown).
MRSA infections have the potential to disrupt primate experiments and, in rare cases cause occupational infections in personnel, at both this and other facilities. The current study suggests that primates in US primate centres and from commercial suppliers are likely an unappreciated source and reservoir of MRSA, and should consequently be screened when animals are moved between and within facilities.5 We also recommend that a baseline level of MRSA carriage be determined at all primate facilities. Studies have isolated MSSA from wild primate populations, but no studies have yet been published showing MRSA in wild primate populations, or what the risk to humans the MRSA-positive primates are. This is particularly important in locations where primates and humans interact closely.5 Future work on these primate populations would be helpful in determining the ultimate origins and population dynamics of primate-associated MRSA strains.
Funding
This project was supported in part by the Office of Research Infrastructure Programs (ORIP) of the National Institutes of Health through Grant Number P51OD010425 through the Washington National Primate Research Center.
Transparency declarations
None to declare.
Supplementary data
Tables S1 to S3 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
We thank Dr Stephen Salipante, Adam Waalkes and Kelsi Penewit for performing WGS and genome sequence analysis. We also thank all WaNPRC personnel who worked with us to obtain the primate samples for this study.
References
- 1.Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis 2008; 46 Suppl 5: S344–9. [DOI] [PubMed] [Google Scholar]
- 2.Weese JS. Methicillin-resistant Staphylococcus aureus in animals. ILAR J 2010; 51: 233–44. [DOI] [PubMed] [Google Scholar]
- 3.Lee JI, Kim KS, Oh B-C et al. Acute necrotic stomatitis (noma) associated with methicillin-resistant Staphylococcus aureus infection in a newly acquired rhesus macaque (Macaca mulatta). J Med Primatol 2011; 40: 188–93. [DOI] [PubMed] [Google Scholar]
- 4.Taylor WM, Grady AW. Catheter-tract infections in rhesus macaques (Macaca mulatta) with indwelling intravenous catheters. Lab Anim Sci 1998; 48: 448–54. [PubMed] [Google Scholar]
- 5.Schaumburg F, Mugisha L, Kappeller P et al. Evaluation of non-invasive biological samples to monitor Staphylococcus aureus colonization in great apes and lemurs. PLoS One 2013; 10: e78048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanley PW, Barnhart KF, Abee CR et al. Methicillin-resistant Staphylococcus aureus prevalence among captive chimpanzees, Texas, USA, 2012. Emerg Infect Dis 2015; 212: 158–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts MC, Soge OO, No D et al. Isolation and characterization of methicillin-resistant Staphylococcus aureus from fire stations in two northwest fire districts. Am J Infect Control 2011; 39: 382–9. [DOI] [PubMed] [Google Scholar]
- 8.Roberts MC, Soge OO, Horst JA et al. Methicillin-resistant Staphylococcus aureus from dental school clinic surfaces and students. Am J Infect Control 2011; 39: 628–32. [DOI] [PubMed] [Google Scholar]
- 9.Soge OO, Meschke JS, No DB et al. Characterization of methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative Staphylococcus spp. isolated from US West Coast public marine beaches. J Antimicrob Chemother 2009; 64: 1148–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang K, McClure JA, Elsayed S et al. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 2005; 43: 5026–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enright MC, Day NP, Davies CE et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 2000; 38: 1008–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troung-Bolduc QC, Dunman PM, Strahilevitz J et al. MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J Bacteriol 2005; 187: 2395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W et al. Basic local alignment search tool. J Mol Biol 1990; 215: 403–10. [DOI] [PubMed] [Google Scholar]
- 14.Salipante SJ, SenGupta DJ, Cummings LA et al. Application of whole genome sequencing for bacterial strain typing in molecular epidemiology. J Clin Microbiol 2015; 31: 1072–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25: 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Handsaker B, Wysoker A et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 2010; 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salipante SJ, Roach DJ, Kitzman JO et al. Large-scale genomic sequencing of extra-intestinal pathogenic Escherichia coli strains. Genome Res 2015; 25: 119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka M, Wang T, Onodera Y et al. Mechanism of quinolone resistance in Staphylococcus aureus. J Infect Chemother 2000; 6: 131–9. [DOI] [PubMed] [Google Scholar]
- 20.SenGupta DJ, Cummings LA, Hoogestraat DR et al. Whole-genome sequencing for high-resolution investigation of methicillin-resistant Staphylococcus aureus epidemiology and genome plasticity. J Clin Microbiol 2014; 52: 2787–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu F, Li T, Huang X et al. Virulence gene profiling and molecular characterization of hospital-acquired Staphylococcus aureus isolates associated with bloodstream infection. Diagn Microbiol Infect Dis 2012; 74: 363–8. [DOI] [PubMed] [Google Scholar]
- 22.Ghaznavi-Rad E, Nor Shamsudin M, Sekawi Z et al. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J Clin Microbiol 2010; 48: 867–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David MZ, Siegel JD, Henderson J et al. Hand and nasal carriage of discordant Staphylococcus aureus isolates among urban jail detainees. J Clin Microbiol 2014; 52: 3422–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson LO, Reynolds C, Spratt BG et al. Diversity of methicillin-resistant Staphylococcus aureus strains isolated from residents of 26 nursing homes in Orange County, California. J Clin Microbiol 2013; 51: 3788–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larson LKR, Harper AL, Hanson BM et al. Methicillin-resistant Staphylococcus aureus in pork production shower facilities. Appl Environ Microbiol 2011; 77: 696–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witte W. Community-acquired methicillin-resistant Staphylococcus aureus: what do we need to know? Clin Microbiol Infect 2009; 15 Suppl 7: 17–25. [DOI] [PubMed] [Google Scholar]
- 27.Drougka E, Posantzis D, Giormezis N et al. Human Staphylococcus aureus lineages among zoological park residents in Greece. Open Vet J 2015; 5: 148–53. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


