Summary
While Cas9 nucleases permit rapid and efficient generation of gene-edited cell lines, the CRISPR-Cas9 system can introduce undesirable “on-target” mutations within the second allele of successfully modified cells via non-homologous end joining (NHEJ). To address this, we fused the Streptococcus pyogenes Cas9 (SpCas9) nuclease to a peptide derived from the human Geminin protein (SpCas9-Gem) to facilitate its degradation during the G1 phase of the cell cycle, when DNA repair by NHEJ predominates. We also use mRNA transfection to facilitate low and transient expression of modified and unmodified versions of Cas9. Although the frequency of homologous recombination was similar for SpCas9-Gem and SpCas9, we observed a marked reduction in the capacity for SpCas9-Gem to induce NHEJ-mediated indels at the target locus. Moreover, in contrast to native SpCas9, we demonstrate that transient SpCas9-Gem expression enables reliable generation of both knockin reporter cell lines and genetically repaired patient-specific induced pluripotent stem cell lines free of unwanted mutations at the targeted locus.
Highlights
-
•
“On-target” deleterious mutations are often encountered with CRISPR-Cas9 gene editing
-
•
SpCas9-Gem retains gene-targeting efficiency with reduced NHEJ activity
-
•
SpCas9-Gem facilitates derivation of “indel-free” gene-edited iPSCs
The use of the Cas9/CRISPR system for efficient generation of precisely modified human pluripotent stem cells without secondary deleterious mutations in the untargeted allele can be challenging. In this article, Howden and colleagues describe a variant of Cas9 that has been fused to a peptide derived from human Geminin to facilitate its degradation during G1 phase of the cell cycle when DNA repair by NHEJ predominates. Using this variant (SpCas9-Gem) they demonstrate reliable and efficient derivation of both knockin reporter iPSCs and genetically repaired patient-specific iPSC lines free of NHEJ-mediated indels at the target locus.
Introduction
The utility of human pluripotent stem cells (hPSCs) is significantly enhanced by an ability to precisely modify a chosen locus with minimal impact on the remaining genome. Ideally this is achieved by gene targeting, which historically has proved challenging in hPSCs due to the low frequency of homologous recombination (HR) (Zwaka and Thomson, 2003). More recently, genetic modification of hPSCs has been facilitated through the use of nucleases to induce DNA double-strand breaks (DSBs) at a target locus, which are subsequently repaired by HR or the error-prone non-homologous end-joining (NHEJ) pathway. Zinc finger nucleases and transcription activator-like endonucleases (TALENs) were the first sequence-specific nucleases employed for gene targeting in hPSCs (Hockemeyer et al., 2009, Hockemeyer et al., 2011, Zou et al., 2009). However, both approaches require the design, expression, and validation of a new pair of proteins for every targeted locus, rendering both of these platforms time consuming and labor intensive. Due to its simplicity, ease of manipulation, and high efficiency, the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system is fast becoming the preferred method for the precise modification of eukaryotic genomes (Cho et al., 2013, Cong et al., 2013, Jinek et al., 2012, Jinek et al., 2013, Mali et al., 2013). However, a potential limitation to this system is the introduction of “on-target” disruptive mutations caused by Cas9-induced NHEJ occurring within the untargeted allele of knockin or gene-corrected hPSC clones (Howden et al., 2015, Merkle et al., 2015). Although reducing the Cas9 dosage may limit both off-target and on-target NHEJ activity (Cradick et al., 2013), a corresponding decrease in gene-targeting efficiency is also expected.
Previous studies, including one in normal human fibroblasts, have demonstrated that HR activity is virtually absent during G1 phase, increases sharply in S phase, and begins to decrease by G2/M, whereas NHEJ activity occurs throughout the cell cycle (Mao et al., 2008, Saleh-Gohari and Helleday, 2004). Hence, controlling the presence of Cas9 protein during specific stages of the cell cycle could minimize the formation of undesirable insertion or deletion mutations (indels) induced by NHEJ. We hypothesized that the presence of Cas9 in G1 would induce DNA DSBs that would invariably be repaired by error-prone NHEJ, whereas elimination of the nuclease within this phase could potentially limit the introduction of disruptive indels at the target locus.
Previous studies have shown that fusion of fluorescent reporters to an N-terminal peptide derived from the human Geminin protein resulted in their proteolytic destruction in the late M and G1 phases of the cell cycle (Pauklin and Vallier, 2013, Sakaue-Sawano et al., 2008), a process mediated by the APCCdh1 E3 ligase complex (Nishitani et al., 2004). In this study, an identical Geminin-derived peptide was fused to SpCas9 (designated SpCas9-Gem) to facilitate nuclease degradation during G1. We show that the frequency of HR is not impaired by fusion of the Geminin peptide to SpCas9, but observe a significant decrease in NHEJ frequency, as evidenced by the absence of indels within the target locus. Together, these features of SpCas9-Gem permit efficient and reliable generation of both knockin cell lines and gene-corrected patient-specific induced pluripotent stem cell (iPSC) lines that are free of disruptive mutations within the untargeted allele.
Results
SpCas9-Gem mRNA Transfection Results in Transient and Cell-Cycle-Dependent Protein Expression
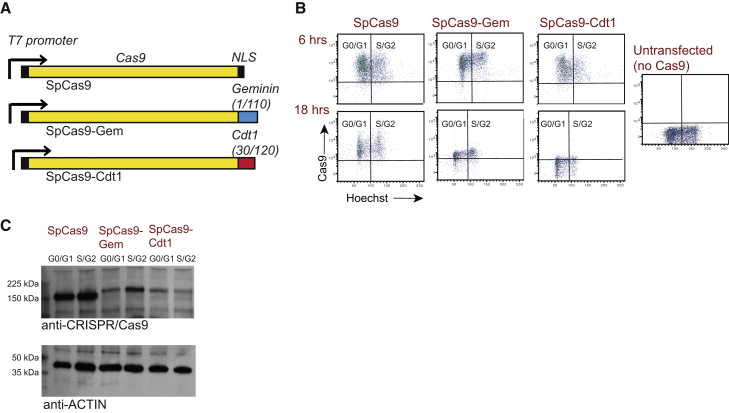
SpCas9-Gem was generated by fusing the first 110 amino acids of Geminin to the C-terminal region of SpCas9, following removal of the C-terminal nuclear localization signal (Figure 1A). Because levels of Geminin and Cdt1 oscillate inversely, whereby Cdt1 levels are highest during G1 and Geminin levels are highest during the S and G2 phases (Nishitani et al., 2004, Sakaue-Sawano et al., 2008), we also generated a second variant of SpCas9 (termed SpCas9-Cdt1) by fusing SpCas9 to amino acids 30–120 of Cdt1. Fusion of this peptide to fluorescent reporters has previously been shown to be sufficient for marking cells in G1 (Pauklin and Vallier, 2013, Sakaue-Sawano et al., 2008). An upstream T7 promoter was used for in vitro transcription of mRNA encoding SpCas9, SpCas9-Gem and SpCas9-Cdt1 (Figure 1A). We performed intracellular staining and flow cytometric analysis following transfection of mRNA encoding each of the Cas9 variants in the human embryonic stem cell line, H9, to examine their abundance and stability over time. Prior to analysis, cells were incubated in the presence of Hoechst 33342 to distinguish cells in G0/G1 and S/G2 phases of the cell cycle. Although at 6 hr post transfection all three Cas9 variants were present at similar levels, by 18 hr post transfection a marked reduction in both SpCas9-Gem and SpCas9-Cdt1 abundance was observed relative to native SpCas9 (Figure 1B). Furthermore, at both time points SpCas9-Gem appeared to be expressed at higher levels in the S/G2 subpopulation, which was not observed for the native SpCas9 or SpCas9-Cdt1 variant. Although significant expression of SpCas9-Gem in the G0/G1 subpopulation was also noted, we hypothesized that this could be due to the binding of CRISPR-Cas9 antibody to not just the intact Cas9 protein but also fragmented protein that was undergoing or had recently undergone proteolytic destruction. To address this concern we performed western blot analysis on the G0/G1 and S/G2 subpopulations 12 hr after the introduction of mRNA encoding each of the Cas9 variants into H9 cells, which were subsequently sorted by fluorescence-activated cell sorting based on DNA content. Using this analysis we clearly detected higher SpCas9-Gem protein levels in the S/G2 population relative to the G0/G1 (Figure 1C), confirming cell-cycle-dependent expression of the SpCas9-Gem variant.
Figure 1.
Analysis of SpCas9-Gem Protein
(A) Schematic diagram of the Cas9 variants generated in this study. Unmodified SpCas9, and the SpCas-Gem and SpCas9-Cdt1 variants were cloned downstream of a T7 promoter to permit their in vitro transcription. NLS, nuclear localization signal.
(B) Flow cytometric analysis following intracellular staining of Cas9 protein in H9 cells transfected with mRNA encoding SpCas9, SpCas9-Gem, or SpCas9-Cdt1. Cells were analyzed at 6 hr and 18 hr post transfection and incubated in the presence of Hoechst 33342 prior to harvesting. Untransfected H9 cells are shown as a negative control.
(C) Western blot analysis of Cas9 protein expression 12 hr post transfection of SpCas9, SpCas9-Gem, or SpCas9-Cdt1 in H9 cells. Prior to harvesting, cells were incubated in the presence of Hoechst 33342 and the G0/G1 and S/G2 subpopulations were sorted based on DNA content. Anti-actin was included as a loading control.
Generation of Indel-Free Knockin Reporter iPSCs from Human Fibroblasts
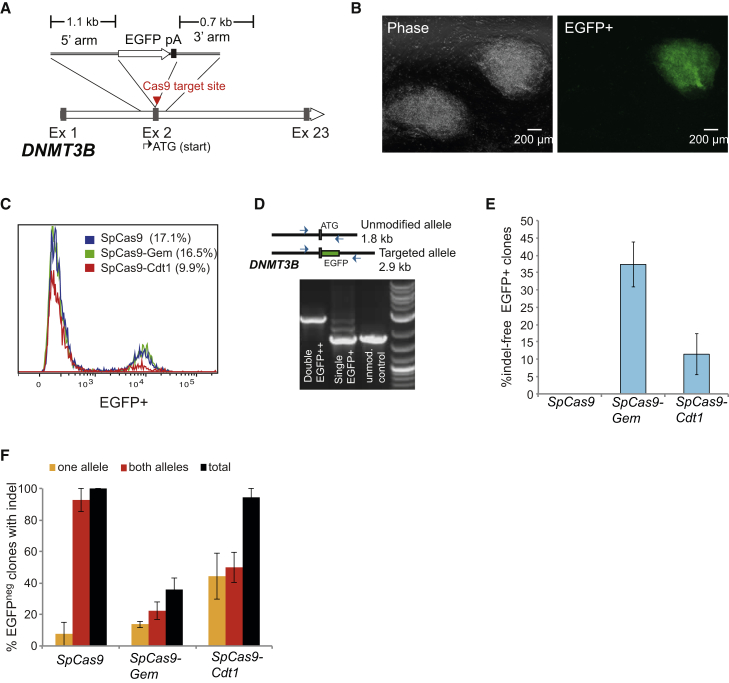
To evaluate the HR and NHEJ activities associated with SpCas9-Gem and SpCas9-Cdt1, we used a recently described method that permits both rapid and efficient generation of gene-edited induced pluripotent stem cell (iPSC) lines following a single electroporation of human fibroblasts (Howden et al., 2015). As in the previous study, we chose to target the DNMT3B gene with an EGFP reporter (Figure 2A), since DNMT3B is highly expressed in pluripotent cells and quickly downregulated following differentiation, allowing targeted iPSC colonies to be easily identified by fluorescent microscopy (Figure 2B) and flow cytometry (Figure 2C). Episomal reprogramming vectors, a plasmid encoding a single-guide RNA (sgRNA) specific to sequence spanning the DNMT3B start codon and a plasmid encoding an EGFP reporter flanked by 1.1 kb and 0.7 kb and homology arms specific to sequences upstream and downstream of the DNMT3B start codon (Figure 2A) were co-transfected with mRNA encoding SpCas9, SpCas9-Gem, or SpCas9-Cdt1. No observable difference in cell viability was associated with transfection of mRNA encoding SpCas9-Gem or SpCas9-Cdt1 compared with the native SpCas9 (Figure S1A). To estimate targeting efficiency associated with each of the Cas9 variants, we passaged cells approximately 3 weeks post transfection using EDTA to selectively remove residual fibroblasts from iPSCs (Howden et al., 2015). After three passages the total cell population was analyzed by flow cytometry. As measured by the number of EGFP-expressing cells, targeting efficiency was comparable for both the SpCas9 and SpCas9-Gem variants (17.1% versus 16.5%) but slightly lower for the SpCas9-Cdt1 variant (9.9%) (Figure 2C). We did not observe any EGFP-expressing cells when Cas9 mRNA was omitted from the transfection (data not shown). At 2–3 weeks post transfection, randomly selected EGFP-expressing and EGFP non-expressing (EGFPneg) colonies were picked and expanded for further analysis (Figure 2B and Table 1). EGFP-expressing clones were further categorized as either having a single allele (EGFP+) or both DNMT3B alleles (EGFP++) targeted, based on flow cytometry and PCR analysis (Figures 2D and S1). Of 28 EGFP-expressing clones isolated from transfections performed with SpCas9 or SpCas9-Gem mRNA, we observed 11 (39%) and 6 (21%) EGFP++ clones, respectively (Table 1). The SpCas9-Cdt1 variant resulted in only a single (6%) EGFP++ clone out of 17 clones analyzed. Next we analyzed the untargeted allele of single-targeted EGFP+ clones by Sanger sequencing of the PCR product amplified from genomic DNA using primers flanking the DNMT3B start codon (Figure 2D). With respect to unmodified SpCas9, NHEJ activity could be detected within the untargeted allele in all 17 clones analyzed (Figure 2E), as evidenced by the presence of indels ranging from 1 bp to over 1 kb in size (Table S1). In contrast, of 22 clones analyzed, we identified 8 (36%) “indel-free” clones derived using the SpCas9-Gem variant, which did not contain mutations within the untargeted DNMT3B allele. Interestingly, indel-free clones could also be generated with the SpCas9-Cdt1 variant, albeit with a lower efficiency (2 out of 16 EGFP+ clones [12%] analyzed). We next investigated the NHEJ activity associated with each of the Cas9 variants in EGFPneg clones, again by analysis of PCR amplicons encompassing the DNMT3B start codon and surrounding sequence. Sequencing analysis revealed indels in all 25 EGFPneg clones expanded and analyzed from experiments performed with SpCas9, with all but one clone carrying disruptions in both DNMT3B alleles (Figure 2F and Table S2). Mutations in DNMT3B could also be found in the vast majority (95%) of EGFPneg clones isolated and expanded from experiments performed using SpCas9-Cdt1 mRNA; however, approximately half of these had only one allele disrupted by NHEJ with the other allele remaining intact. Conversely, only 35% of EGFPneg clones generated with the SpCas9-Gem variant contained mutations in DNMT3B, with single allele and biallelic disruption in 13% and 22% of EGFPneg clones, respectively.
Figure 2.
Generation of Knockin Reporter iPSC Lines Using Cas9 Variants
(A) Schematic diagram of the DNMT3B locus and the homologous template used for gene targeting. pA, polyA signal; Ex, exon.
(B) Representative phase contrast and fluorescent images depicting an EGFP-expressing and non-expressing iPSC colony emerging approximately 2 weeks after simultaneous reprogramming and gene targeting of DNMT3B in human fibroblasts.
(C) Targeting efficiency associated with SpCas9, SpCas9-Gem and SpCas9-Cdt1 assessed by flow cytometric analysis of the total iPSC population after EDTA passaging, initiated approximately 3 weeks post electroporation.
(D) Schematic of the DNMT3B locus showing location of PCR primers used to screen EGFP-expressing clones, and representative gel analysis of the PCR products amplified from EGFP-expressing clones. These primers preferentially amplify the untargeted allele in monoallelic (EGFP+) clones, whereas only the larger 2.9-kb band is visible following PCR analysis of biallelic (EGFP++) clones.
(E) Proportion of single-targeted EGFP+ clones identified without disruption of the other untargeted DNMT3B allele.
(F) Proportion of EGFPneg (untargeted) iPSC clones with one or both DNMT3B alleles disrupted by NHEJ. EGFP+ and EGFPneg colonies were isolated and expanded from three independent experiments, performed with each version of Cas9. Data represent mean ± SEM from three independent experiments.
Table 1.
Number of iPSC Clones Isolated, Expanded, and Characterized Following Targeting of the DNMT3B Locus
| Targeted (EGFP+) Clones |
Non-targeted (EGFPneg) Clones | Total Clones Analyzed | ||
|---|---|---|---|---|
| One Allele | Both Alleles | |||
| SpCas9 | 17 (0) | 11 | 25 | 53 |
| SpCas9-Gem | 22 (8) | 6 | 23 | 51 |
| SpCas9-Cdt1 | 16 (2) | 1 | 22 | 39 |
The number of clones with EGFP targeted to one allele without an additional mutation in the untargeted allele is shown in parentheses.
Generation of Indel-Free Gene-Corrected iPSCs from Patient Fibroblasts
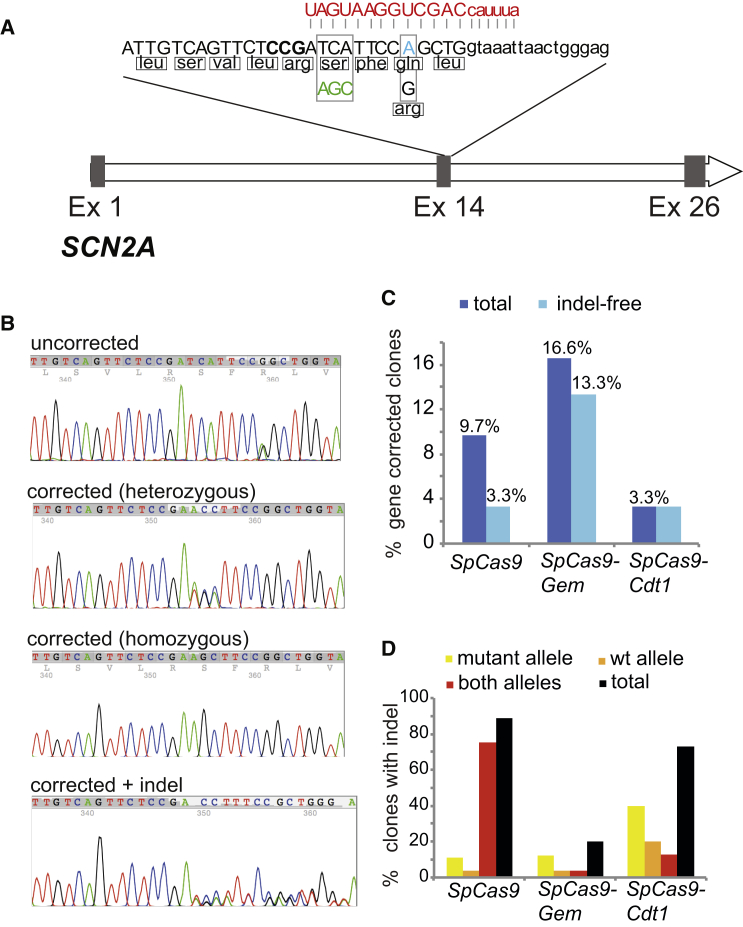
Next we assessed the capacity of SpCas9-Gem to facilitate the derivation of gene-corrected iPSCs lacking deleterious indels within the second allele. To demonstrate this, we chose to correct an autosomal dominant G > A transition (R853Q) in exon 14 of the SCN2A gene identified in a pediatric patient with severe epilepsy. We attempted to simultaneously reprogram and correct the disease-causing mutation in the patient fibroblasts using our one-step protocol and mRNA encoding SpCas9, SpCas9-Gem, or SpCas9-Cdt1. To facilitate gene repair we used an sgRNA encompassing the patient-specific mutation and a plasmid encoding wild-type SCN2A sequence, comprising ≈0.9-kb arms flanking the intended DNA DSB. A silent 3-bp change was also included to aid in identification of correctly modified clones and to minimize Cas9 recutting following HR with the homologous template (Figure 3A). To assess HR and NHEJ activities, we extracted genomic DNA from randomly selected iPSC colonies, followed by sequencing analysis of the PCR product amplified using primers flanking exon 14 of SCN2A. With respect to the SpCas9-Gem variant, gene repair was observed in 5 of 30 clones (17%) screened, as evidenced by the loss of the disease-causing mutation and gain of the 3-bp synonymous change carried by the corrective plasmid (Figures 3B and 3C). Indel formation at the second SCN2A allele could not be detected in four of the gene-corrected clones, although biallelic HR with the repair template was observed, as evidenced by homozygosity for the plasmid-specific 3-bp change (TCA > AGC). Targeted correction of SCN2A was also observed with the SpCas9 and SpCas9-Cdt1 variants, although at a lower frequency (9.7% and 3.3%, respectively). Furthermore, only one gene-corrected clone out of 31 (3.3%) iPSC colonies screened for both the SpCas9 and SpCas9-Cdt1 variants did not contain a secondary mutation at the other SCN2A allele. As seen in the previous DNMT3B targeting experiment, sequencing analysis of uncorrected iPSC clones revealed a much lower incidence of NHEJ associated with the SpCas9-Gem variant compared with unmodified SpCas9 and the SpCas9-Cdt1 variant (Figure 3D). Although our sgRNA was designed to recognize the mutant over wild-type SCN2A allele, indels could be detected in both alleles for each of the Cas9 variants tested. This is not unexpected, since single mismatches in sgRNA are often well tolerated, even within the 3′ end of the protospacer sequence (Fu et al., 2013, Hsu et al., 2013). However, analysis of all clones with only a single allele disrupted by NHEJ did reveal a preference for the mutant allele (Figure 3D and Table S3).
Figure 3.
Generation of Gene-Corrected iPSC Lines Using Cas9 Variants
(A) Schematic diagram of the SCN2A gene, with mutation in exon 14. The crRNA sequence used (red), PAM location (bold), and the patient-specific autosomal dominant mutation (blue) along with the 3-bp synonymous change (green) incorporated into the plasmid template used for repair are shown. Exon sequences are in uppercase, introns in lowercase.
(B) Sequencing analysis of exon 14 of the SCN2A gene from a representative uncorrected patient-specific iPSC clone and three gene-corrected iPSC clones with either one allele targeted (heterozygote), both alleles targeted (homozygote), or one allele targeted and the other disrupted by NHEJ.
(C) Proportion of gene-corrected iPSC clones without disruption of the second SCN2A allele.
(D) Proportion of uncorrected iPSC clones with one or both SCN2A alleles disrupted by NHEJ.
SpCas9-Gem Retains HR Capacity with Reduced NHEJ Activity in hPSCs
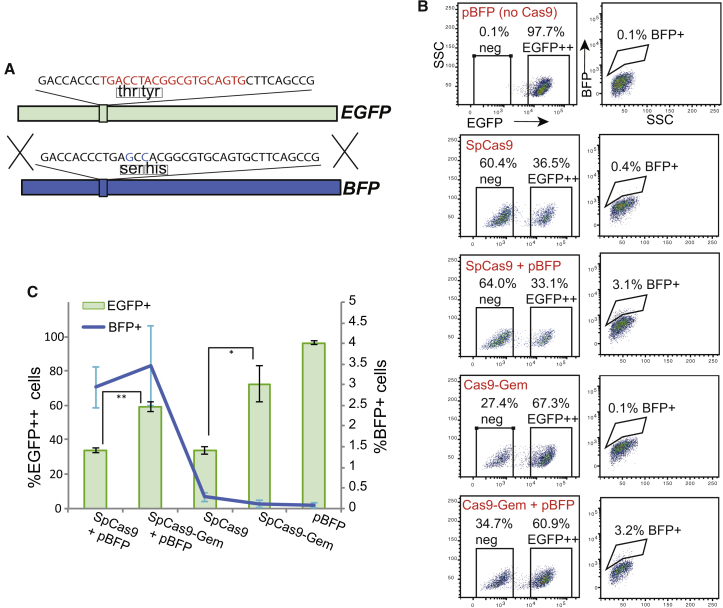
To evaluate the SpCas9-Gem variant in a cell type other than human fibroblasts, we utilized iPSC lines generated in the previous experiments (see Figure S1) that harbor EGFP targeted to both DNMT3B alleles. For a homologous template we used a plasmid encoding the full-length BFP sequence that was identical to EGFP except for C > G and T > C transitions, corresponding to two consecutive amino acid changes (T67S and Y68H) (Figure 4A). This is sufficient for conversion of EGFP to BFP reporter expression (Glaser et al., 2016). We also included a plasmid encoding an sgRNA that specifically recognizes EGFP over BFP. Using this system in an iPSC line constitutively expressing EGFP, we simultaneously estimated both HR (as determined by the number of BFP+ cells) and NHEJ frequencies (as determined by the number of EGFPneg cells) in the total population of transfected cells after 3 days by flow cytometry (Figure 4B). SpCas9 or SpCas9-Gem mRNA resulted in similar levels of HR, as determined by the number of BFP+ cells, when co-transfected with BFP plasmid DNA and EGFP-specific sgRNA into two independent EGFP++ iPSC lines (Figures 4B and 4C). However, a significantly reduced proportion of EGFPneg cells, caused by disruption of the EGFP gene, was consistently associated with the SpCas9-Gem variant. This approximately 2-fold reduction in NHEJ activity was observed in both the presence and absence of the BFP donor template (Figure 4C). To determine whether simply lowering the nuclease dosage could also achieve reduced indel formation while maintaining HR efficiency, we co-transfected 50, 100, 250, or 500 ng of mRNA encoding SpCas9 with the BFP plasmid and EGFP-specific sgRNA plasmid into iPSCs constitutively expressing EGFP. In agreement with previous studies, lower concentrations of Cas9 did indeed reduce NHEJ frequency, although a consequential decrease in HR efficiency was also observed. Specifically, the lowest dose of SpCas9 mRNA (50 ng) reduced NHEJ to rates observed with 500 ng of the SpCas9-Gem mRNA but resulted in more than 5-fold reduction in HR frequency (Figure S2A). A positive correlation between Cas9 mRNA concentration and HR efficiency was also observed when targeting EGFP to the DNMT3B locus in H9 cells (Figure S2B). Furthermore, no significant difference in targeting efficiency was observed following co-transfection of mRNA encoding SpCas9 or SpCas9-Gem with the DNMT3B-EGFP homologous template and DNMT3B-specific sgRNA in H9 cells (Figure S3A). We also evaluated the “on-target” indel frequency of the SpCas9-Gem variant following targeting of the DNMT3B and GAPDH loci in H9 cells. In these experiments, we used targeting constructs that contain a drug-selectable marker to facilitate rapid isolation, expansion, and characterization of individual colonies (Figures S3B and S3C). Consistent with the EGFP-to-BFP conversion assay in the previous experiments, an approximately 2- to 3-fold reduction in NHEJ activity was associated with the SpCas9-Gem variant compared with native SpCas9.
Figure 4.
Evaluation of SpCas9-Gem in Pluripotent Stem Cells
(A) Schematic summary of EGFP-to-BFP conversion targeting strategy. The location of the two amino acid changes and the Cas9 target site (red) are indicated.
(B and C) Representative flow cytometry plots (B) and graphical plot (C) depicting the proportion of BFP+ (targeting efficiency) and EGFPneg cells (NHEJ frequency) assessed 3 days after the introduction of SpCas9 and SpCas9-Gem with and without a BFP homologous template in iPSCs constitutively expressing EGFP. Data represent the average ± SD of four independent experiments in two EGFP++ iPSC lines. ∗p < 0.0001, ∗∗p < 0.000001.
Discussion
The recent development of the CRISPR-Cas9 system will undoubtedly have an enormous impact on the utility of hPSCs for the elucidation of developmental processes, disease modeling, or even personalized regenerative medicine. However, the reliable and efficient generation of knockin or gene-corrected hPSCs without secondary deleterious mutations has been somewhat elusive. Because cell-cycle phase largely determines the frequency of the alternative HR and NHEJ DNA repair pathways, it is reasonable to expect that controlling the timing of a DNA DSB may selectively favor one pathway over the other. Chemical-mediated cell synchronization followed by timed delivery of Cas9-guide RNA ribonucleoprotein complexes has recently been shown to be effective for increasing HR efficiency in the human cell line HEK-293, although whether this led to decreased NHEJ activity in alleles that had not undergone HR was not reported (Lin et al., 2014). Moreover, different cell types, such as hPSCs, have distinct requirements for synchronization, which do not always translate to effective increases in HR. Recent studies have also shown that small molecules such as SCR7 may inhibit NHEJ and consequently promote HR (Maruyama et al., 2015). However, these kinds of approaches are likely to have variable effects across different cell types and may potentially exhibit undesirable cytotoxic effects, rendering them unsuitable in therapeutic contexts. In an attempt to address some of these issues, we generated a variant of the SpCas9 nuclease (SpCas9-Gem) that persists during stages of the cell cycle when the HR pathway is active, but is degraded during G1, when NHEJ activity predominates. An attractive feature of the SpCas9-Gem variant is its potential applicability to different cell types without a requirement for cells to be manipulated or specially prepared prior to introduction of the gene-editing factors. In the current study we have demonstrated the versatility of SpCas9-Gem in primary human fibroblasts and hPSCs, two cell types known to exhibit very different cell-cycle profiles. Although the lower frequency of NHEJ associated with SpCas9-Gem was less pronounced in hPSCs, we hypothesize that this may be attributed to the dramatically shorter G0/G1 phase that is characteristic of hPSCs, compared with somatic cell types such as fibroblasts.
While this manuscript was in preparation, Gutschner et al. (2016) also described a version of SpCas9 fused to the N-terminal region of human Geminin. Although the authors reported a modest increase (28%–87%) in HR compared with unmodified SpCas9 in HEK293T cells, their Cas9-Geminin fusion provoked similar levels of indel formation. This may be explained by their use of plasmid DNA to express Cas9 from a β-actin promoter, resulting in sustained levels of nuclease transcription that likely persists over several days and during the course of multiple cycles of cell division. In the study presented here, transfection of in vitro transcribed mRNA was utilized, enabling uniform and transient expression of SpCas9-Gem protein during S/G2. Following degradation during G1, nuclease levels are drastically reduced, even in subsequent S/G2 phases, which can most likely be attributed to the transient and unstable properties of mRNA.
Of note, the SpCas9-Cdt1 variant also enabled the generation of correctly targeted lines, albeit with a lower efficiency than the unmodified or SpCas9-Gem variants. Although this may be due to the fact that proteins fused to the Cdt1 peptide can persist during the initial stages of S phase, when HR becomes a potential option for DNA repair, this may also be the result of incomplete degradation of SpCas9-Cdt1 during S/G2/M (see Figure 1). The SpCas9-Cdt1 fusion induced indel formation significantly more often than SpCas9-Gem but at a lower frequency than unmodified SpCas9, which was particularly evident when assessing the total number of alleles disrupted by NHEJ. It is difficult to determine whether this is due to lower overall levels of SpCas9-Cdt1 or perhaps a reduced nuclease activity relative to unmodified SpCas9.
Although any off-target effects associated with the Cas9 variants generated in this study were not analyzed, the lower NHEJ activity of SpCas9-Gem would likely translate to a reduction in unwanted mutations across the genome. Nonetheless, the incorporation of the Geminin peptide into the recently described high-fidelity versions of SpCas9 (SpCas9-HF1 and eCas9) (Kleinstiver et al., 2016, Slaymaker et al., 2016) may reduce the likelihood of off-target NHEJ further.
In conclusion, the SpCas9-Gem variant described here facilitates efficient and reliable generation of knockin and gene-corrected hPSC lines free of unwanted mutations at the targeted locus. Our results demonstrate the utility of SpCas9-Gem for achieving effective and precise genome modifications while minimizing the “on-target” effects of NHEJ in both somatic cells types, such as fibroblasts, and pluripotent cell types, such as embryonic stem cells and iPSCs. This is a significant advance in the downstream application of hPSCs for disease modeling and functional genomics, and prepares the field for gene-correction applications in regenerative medicine. The use of additional peptides or sequences that promote greater cell-cycle specificity of Cas9 may permit even further reductions in “on-target” NHEJ while increasing desirable HR activity.
Experimental Procedures
Oligonucleotide and gBlock Sequences
A list of oligonucleotide (ODN) sequences for PCR amplification and generation of sgRNA plasmids (Table S4) and complete sequences of the gBlocks (Integrated DNA Technologies) used in this study can be found in Supplemental Sequencing Data.
Vector Construction
To enable in vitro transcription of mRNA encoding SpCas9, an XbaI-NsiI fragment from plasmid hCas9 (Addgene plasmid #41815) containing the entire SpCas9 sequence was inserted into the NheI and SbfI sites of the pDNR-Dual vector (Clontech), to generate pDNR-SpCas9. The SpCas9-Gem variant was generated by PCR-amplifying sequence from hCas9 using ODNs 1 and 2 and sequence from pTRE-CellCycle (Clontech) using ODNs 3 and 4 followed by insertion into the EcoRI and AgeI restriction sites in pDNR-SpCas9. The pSpCas9-CdtI variant was generated by PCR-amplifying sequence from hCas9 using ODNs 1 and 2 and sequence from pTRE-CellCycle (Clontech) using ODNs 3 and 5 followed by insertion into the EcoRI and AgeI restriction sites in pDNR-SpCas9. For construction of the DNMT3B targeting vector, a gBlock encoding 5′ BglII and NdeI sites, nucleotides 150–724 of the EGFP gene, an SV40 polyA signal, and 700 bp of sequence beginning at the DNMT3B start codon was cloned into the HincII site of the pSMARTHCKan plasmid vector (Lucigen). ODNs 6 and 7 were used to PCR amplify (from a previously generated BAC clone carrying an EGFP reporter inserted at the DNMT3B start codon) a DNA fragment carrying an approximately 1.1-kb sequence upstream of the DNMT3B start codon and the first 150 bp of EGFP, which was subsequently inserted into the BglII and NdeI sites of the plasmid described above. A plasmid designed for insertion and expression of sgRNA sequences was constructed by inserting a gBlock encoding a U6 promoter, two BbsI sites, and SpCas9-specific tracrRNA sequence into the HincII site of pSMARTHCKan. ODNs specific to each target locus were annealed and inserted into the BbsI sites of the pSMART-sgRNA vector. For repair of the patient-specific mutation, two gBlocks encoding a total of 1750 bp of the SCN2A gene was inserted into the AatII and EcoRI sites of the pDNR-Dual vector. A serine residue within the region recognized by our SCN2A-specific sgRNA was changed from TCA to AGT to minimize Cas9-induced cutting of the plasmid following its introduction into cells. A BFP targeting vector was generated by inserting a gBlock encoding the full-length BFP gene into the AatII and EcoRI sites of pDNR-Dual. The DNMT3B gene-targeting construct containing the EGFP and puromycin resistance genes was generated as previously described (Howden et al., 2015). The plasmids used for in vitro transcription of native SpCas9 (plasmid 80425), SpCas9-Gem (plasmid 80424), and SpCas9-Cdt1 (plasmid 80426) along with the plasmid used for sgRNA expression (plasmid 80427) are all available from Addgene upon request.
In Vitro Transcription
Capped and polyadenylated in vitro transcribed mRNA encoding SpCas9, SpCas9-Gem, and SpCas9-Cdt1 was generated using the mMESSAGE mMACHINE T7 ULTRA transcription kit (Thermo Fisher) according to the manufacturer's recommendations. Plasmid template was linearized with PmeI prior to transcription. LiCl was used to precipitate mRNA before resuspension. A truncated version of the EBNA1 protein was transcribed using the mMESSAGE mMACHINE SP6 transcription kit, as previously described (Howden et al., 2006).
Transfection
All transfections were performed using the Neon Transfection System (Thermo Fisher). For simultaneous reprogramming and gene editing of human fibroblasts, cells were harvested with TrypLE (Thermo Fisher) 2 days after passaging and resuspended in Buffer R at a final concentration of 1 × 107 cells/mL. One hundred microliters of the cell suspension was added to a tube containing plasmids necessary for both reprogramming and gene targeting as well as mRNA encoding either SpCas9, SpCas9-Gem, or SpCas9-Cdt1. In vitro transcribed mRNA encoding a truncated version of the EBNA1 protein was also included to enhance nuclear uptake of the reprogramming plasmids (Chen et al., 2011, Howden et al., 2006). See Table S5 for DNA/mRNA concentrations used in each transfection. Electroporation was performed in 100- μL tips using the following conditions: 1,400 V, 20 ms, two pulses. Following electroporation, cells were plated on a single 10-cm Matrigel-coated (Corning) plate and maintained in fibroblast medium until 4 days post transfection, then switched to E7 medium (E8 medium without transforming growth factor β) supplemented with 100 μM sodium butyrate and changed every other day as described previously (Chen et al., 2011). Sodium butyrate was removed from the medium after the appearance of the first iPSC colonies at around day 10. iPSC and H9 transfections were performed in 10- μL and 100- μL tips, respectively, according to the manufacturer's recommendation using the following conditions: 1,100 V, 30 ms, one pulse. Following transfection, cells were transferred to Matrigel-coated plates containing E8 medium supplemented with 5 μM Y-27632 (Tocris), which was omitted in subsequent media changes.
Cell Culture
Fibroblast lines were derived from donors with informed parental consent and approval from the local research ethics committee. Fibroblasts were cultured in DMEM (Thermo Fisher) supplemented with 15% fetal bovine serum (HyClone) at 37°C, 5% CO2, and 5% O2. All hPSC lines were maintained and expanded in E8 medium with daily media changes and passaged every 3–4 days with EDTA in 1× PBS as previously described (Chen et al., 2011).
PCR, RT-PCR, and Sanger Sequencing
Total genomic DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen). PCR was performed using GoTaq Green PCR Mastermix (Promega). Sequence encompassing the DNMT3B start codon and surrounding region PCR was amplified using ODNs 8 and 9. Exon 14 of SCN2A and surrounding sequence was amplified using ODNs 15 and 16. Prior to sequencing, PCR amplicons were treated with rAPid Alkaline Phosphatase (Roche Life Science) and Endonuclease I (NEB) for 30 min at 37°C followed by heat inactivation at 80°C for 15 min. Sequencing reactions were performed using BigDye Terminator v3.1 (Thermo Fisher). Reactions were purified and sequenced by the Australian Genome Research Facility.
Flow Cytometry
Cells were analyzed for EGFP and BFP expression by flow cytometry using a LSRFortessa Call Analyzer (BD Biosciences). Data acquisition and analysis were performed using FACSDiva (Becton Dickinson) and FlowLogic software (Inivai). Gating was performed on live cells based on forward and side scatter analysis.
Cell Sorting
Prior to sorting, cells were harvested with TrypLE and resuspended in 2 mL of E8 medium containing Hoechst 33,342 (10 μg/mL) and incubated at 37°C for 30 min. Cells were sorted according to their DNA content using a FACSAria Fusion Cell Sorter (BD Biosciences).
Western Blot
Western blots were performed by standard Tris-glycine SDS-PAGE followed by transfer to polyvinylidene fluoride membranes. Following blocking with 10% skim milk in TBS-T (20 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.1% Tween 20), membranes were probed with anti-CRISPR/Cas9 (Abcam, ab191468) at 1 μg/mL and anti-β-actin (Cell Signaling Technology, #4970) at 1:5,000 and visualized by chemiluminescence.
Intracellular Staining
Prior to harvest, cells were incubated in medium containing Hoechst 33342 (10 μg/mL) at 37°C for 30 min. Cells were harvested, fixed with 2% paraformaldehyde, and permeabilized with 0.1% Triton X-100. Cells were stained with anti-CRISPR/Cas9 (AbCam, ab191468) for 1 hr followed by staining with a goat immunoglobulin G anti-mouse secondary antibody directly conjugated to allophycocyanin. Cells were analyzed by flow cytometry as described above.
Author Contributions
S.E.H. conceived the experiments and wrote the paper; B.M and J.V performed western blot analysis; A.G. conceptualized EGFP-to-BFP conversion experiments; S.P. facilitated SCN2A gene-correction experiments; A.G.E., M.H.L., and E.G.S. provided supervision and assisted in manuscript preparation.
Acknowledgments
S.E.H. is supported by a National Health and Medical Research Council (NHMRC) Overseas Biomedical Fellowship. A.G.E. and E.G.S. are Senior Research Fellows and M.H.L. Senior Principal Research Fellow of the NHMRC. Murdoch Children’s Research Institute is supported by the Victorian Government's Operational Infrastructure Support Program.
Published: August 4, 2016
Footnotes
Supplemental Information includes Supplemental Sequencing Data, three figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.07.001.
Supplemental Information
References
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.W., Kim S., Kim J.M., Kim J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick T.J., Fine E.J., Antico C.J., Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser A., McColl B., Vadolas J. GFP to BFP conversion: a versatile assay for the quantification of CRISP/Cas9-mediated genome editing. Mol. Ther. Nucleic Acids. 2016;5:e334. doi: 10.1038/mtna.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T., Haemmerle M., Genovese G., Draetta G.F., Chin L. Post-translational regulation of Cas9 during G1 enhances homology-directed repair. Cell Rep. 2016;14:1555–1566. doi: 10.1016/j.celrep.2016.01.019. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D., Wang H., Kiani S., Lai C.S., Gao Q., Cassady J.P., Cost G.J., Zhang L., Santiago Y., Miller J.C. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden S.E., Wardan H., Voullaire L., McLenachan S., Williamson R., Ioannou P., Vadolas J. Chromatin-binding regions of EBNA1 protein facilitate the enhanced transfection of Epstein-Barr virus-based vectors. Hum. Gene Ther. 2006;17:833–844. doi: 10.1089/hum.2006.17.833. [DOI] [PubMed] [Google Scholar]
- Howden S.E., Maufort J.P., Duffin B.M., Elefanty A.G., Stanley E.G., Thomson J.A. Simultaneous reprogramming and gene correction of patient fibroblasts. Stem Cell Rep. 2015;5:1109–1118. doi: 10.1016/j.stemcr.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Staahl B.T., Alla R.K., Doudna J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife. 2014;3:e04766. doi: 10.7554/eLife.04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Bozzella M., Seluanov A., Gorbunova V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle. 2008;7:2902–2906. doi: 10.4161/cc.7.18.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle F.T., Neuhausser W.M., Santos D., Valen E., Gagnon J.A., Maas K., Sandoe J., Schier A.F., Eggan K. Efficient CRISPR-Cas9-mediated generation of knockin human pluripotent stem cells lacking undesired mutations at the targeted locus. Cell Rep. 2015;11:875–883. doi: 10.1016/j.celrep.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H., Lygerou Z., Nishimoto T. Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J. Biol. Chem. 2004;279:30807–30816. doi: 10.1074/jbc.M312644200. [DOI] [PubMed] [Google Scholar]
- Pauklin S., Vallier L. The cell-cycle state of stem cells determines cell fate propensity. Cell. 2013;155:135–147. doi: 10.1016/j.cell.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Saleh-Gohari N., Helleday T. Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells. Nucleic Acids Res. 2004;32:3683–3688. doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J., Maeder M.L., Mali P., Pruett-Miller S.M., Thibodeau-Beganny S., Chou B.K., Chen G., Ye Z., Park I.H., Daley G.Q. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaka T.P., Thomson J.A. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.