Summary
Pre-clinical studies indicate that neural stem cells (NSCs) can limit or reverse CNS damage through direct cell replacement, promotion of regeneration, or delivery of therapeutic agents. Immortalized NSC lines are in growing demand due to the inherent limitations of adult patient-derived NSCs, including availability, expandability, potential for genetic modifications, and costs. Here, we describe the generation and characterization of a new human fetal NSC line, immortalized by transduction with L-MYC (LM-NSC008) that in vitro displays both self-renewal and multipotent differentiation into neurons, oligodendrocytes, and astrocytes. These LM-NSC008 cells were non-tumorigenic in vivo, and migrated to orthotopic glioma xenografts in immunodeficient mice. When administered intranasally, LM-NSC008 distributed specifically to sites of traumatic brain injury (TBI). These data support the therapeutic development of immortalized LM-NSC008 cells for allogeneic use in TBI and other CNS diseases.
Highlights
-
•
The generation of a new human fetal L-MYC-immortalized NSC line is described
-
•
These NSCs display self-renewal and can differentiate into neurons and glia
-
•
The NSCs can target glioma xenografts and sites of traumatic brain injury in mice
-
•
This NSC line may become applicable in therapy of various CNS diseases
Neural stem cell-based therapies are an emerging and promising approach for treatment of CNS pathologies. In this article, Gutova and colleagues describe the generation and characterization of an L-MYC-immortalized human fetal NSC line (LM-NSC008). These cells displayed in vitro self-renewal and differentiation into neurons, oligodendrocytes, and astrocytes, and migrated in vivo to orthotopic glioma xenografts in immunodeficient mice. After intranasal administration, LM-NSC008 distributed specifically to sites of traumatic brain injury in mice. These data support the development of immortalized LM-NSC008 cells for allogeneic use in targeted therapy of brain tumors, traumatic brain injury, and potentially other CNS diseases.
Introduction
Despite decades of research, treatments for patients with diseased or damaged regions of the CNS remain palliative at best (Pathan et al., 2009). Cell-based therapies are emerging as a novel and potentially powerful approach for the treatment of CNS pathologies, and multipotent neural stem cells (NSCs) in particular are an attractive cell type for use in CNS therapies. Recent pre-clinical proof-of-concept studies have demonstrated the potential of NSC-based treatments for disorders requiring neural cell replacement (Begum et al., 2015), protection from external insult (Umeda et al., 2016), antibody production (Kanojia et al., 2015), and targeted delivery of therapeutic agents (Aboody et al., 2013), including prodrug-activating enzymes (Metz et al., 2013). Despite these early promising results, there are still major practical limitations that must be addressed before widespread clinical use of NSC-based therapeutics is possible (Daniela et al., 2007). One constraint is the limited number of NSCs showing consistent in vivo behaviors and available in numbers sufficient for genetic modification prior to administration to patients.
Practical considerations limit the use of autologous NSCs for cell-based therapy. Allogeneic donor cells remain an attractive possibility if an appropriate source can be identified. Although the self-renewing NSCs present in developing brain tissue could be used as a renewable cell population, culture conditions have yet to be identified that reproducibly permit continuous propagation of primary NSCs. One common approach is to expand NSC pools by repeated subculture of polyclonal neurospheres. However, progressive passages lead to decreased capacity for cellular self-renewal, decreased differentiation potential, and increased accumulation of chromosomal and functional instabilities (Reynolds and Weiss, 1992, Kallos and Behie, 1999, Nakagawa et al., 2008). Thus a new source of primary tissue must be obtained for each production cycle, which makes process scale-up, regulatory approval, and clinical translation substantially more difficult and costly. A more practical approach has been to generate stable, immortal NSC lines by retroviral transduction of an MYC gene into early gestational NSC pools (Kim et al., 2008). These MYC-immortalized NSC lines retain their self-renewal capabilities for substantial numbers of passages (>30 as opposed to five to six passages for non-immortalized NSCs), which exponentially increases the number of NSCs available for use in potential therapies (Kim, 2004).
There have been safety-related concerns, as yet unrealized, related to MYC-based NSC immortalization, including the risk that an MYC transgene could render the NSC line tumorigenic upon transplantation (Nakagawa et al., 2010). However, the clonal v-MYC-immortalized human NSC line, HB1.F3.CD21, has shown chromosomal and functional stability over multiple passages and good manufacturing practice (GMP) scale-up (Aboody et al., 2013), and demonstrated clinical safety and non-tumorigenicity in patients with recurrent glioblastoma (NCT02015819). Another NSC line (CTX0E03) was established in 2006 using the c-MYC gene commonly used in generation of induced pluripotent stem cells (iPSCs) (Pollock et al., 2006, Nakagawa and Yamanaka, 2010, Hicks et al., 2013). In this case, a conditional technology was used to enable suppression of c-MYC via systemic tamoxifen administration, if necessary, to ensure that c-MYC expression could be controlled upon transplantation (Pollock et al., 2006).
These two MYC-immortalized NSC lines exhibit properties that cause them to differ in their utility for treating diseases. Specifically, HB1.F3.CD21 NSCs exhibit superior tumor tropism but more limited differentiation capacity compared with CTX0E03 NSCs. These cells secrete bioactive factors that are useful for promoting host neuronal plasticity or vascular growth into damaged brain areas and have demonstrated clinical efficacy in stroke patients (Stroemer et al., 2009). The reasons for these differences are unknown, but may arise from differences in the initial progenitor pools chosen (gestational age), specifics of culture and transduction conditions, and chromosomal insertion points.
Thus while the potential of MYC immortalization for the production of therapeutic NSC lines has been demonstrated, realizing this potential will require generation and validation of multiple lines optimized for particular clinical applications. To facilitate this effort, we have developed a protocol for producing and characterizing new MYC-immortalized NSC lines that can be propagated up to at least passage 15 (p15). This protocol incorporates two recent technological advances that should further improve safety profiles relative to older MYC-immortalized NSC lines: (1) culture and freezing conditions that exclusively use human-derived supplements, and, most importantly, (2) transduction with L-MYC to reduce the risk of transformation (Nakagawa et al., 2008). L-MYC has significantly lower transformation activity in cultured cells than the other MYC members (Oster et al., 2003), and only a small number of human cancers have been associated with the aberrant expression of L-MYC (Nakagawa et al., 2010).
Here we describe the generation of the first L-MYC-immortalized human NSC line derived from human fetal brain, for brevity referred to as LM-NSC008. We find that the LM-NSC008 line is chromosomally normal, and overexpresses several stem cell-associated genes. LM-NSC008 cells proliferate faster than the parental cells, and can be expanded under both suspension and adherent culture conditions (an advantage for eventual clinical scale-up). The LM-NSC008 stem cell line is non-tumorigenic when injected to non-tumor-bearing mouse brain for up to 3 months (6- and 12-month studies are in progress). Furthermore, these cells exhibit tumor-specific tropism, and, on removal of growth factors, differentiate into neural, astrocytic, and oligodendroglial lineages, as do normal non-immortalized NSCs. Once developed into a “clinical-grade” stem cell line, LM-NSC008 may find widespread use in stem cell-mediated therapies for CNS pathologies.
Results
Generation and Characterization of L-MYC Transduced NSC Clones
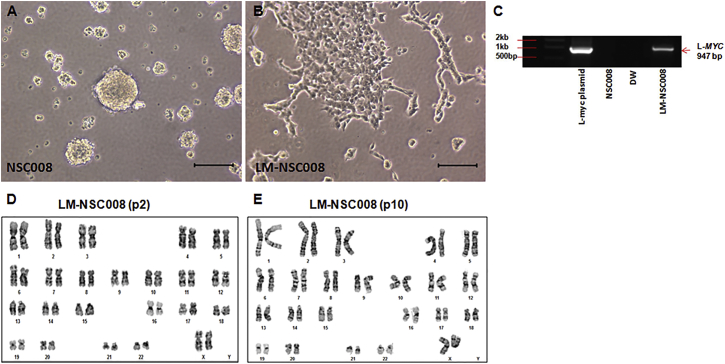
Cultures of dissociated NSCs were generated from human fetal brain tissue of 10–14 weeks gestation. NSCs were cultured under hypoxic conditions (4% O2) in a humidified incubator (Binder). In growth factor-supplemented stem cell medium, the primary hNSCs (NSC008) grew in suspension and formed neurospheres (Figure 1A). At p2, we transduced the primary NSC008 cells with retrovirus carrying L-MYC and puromycin resistance gene (MOI of 2.5). After 24–48 hr, transduced cells were grown in selection culture media containing puromycin (0.5 μg/mL), which eliminated all untransduced cells within 2–3 days. Cells were grown in puromycin for 28 days to select for NSCs stably expressing the L-MYC gene. The LM-NSC008 cells grown under the same culture conditions as primary NSC008 cells grew as a monolayer after two to three passages in vitro (Figure 1B). This makes the LM-NSC008 cells more amenable to microscopic testing, cytology examination, and scale-up of cell production. Expression of the L-MYC gene was confirmed by genomic PCR analysis (Figure 1C). LM-NSC008 cells had normal karyotype at p2 and p10 in vitro (Figures 1D and 1E), and did not show abnormal growth or tumorigenicity in vitro (Figure S1). Soft agar colony formation assays in 1% agarose hydrogels were performed to assess cellular anchorage-independent growth of L-MYC (LM-NSC008)-transduced and v-MYC-transduced (HB1.F3.CD) hNSC cells in vitro compared with MCF7 breast cancer and 5,637 bladder cancer lines (Figure S1). LM-NSC008 and HB1.F3.CD cells, in contrast to the cancer cell lines (Figure S1A), did not form colonies in soft agar, but rather remained viable as single cells (Figure S1B). These observations are quantified in Figure S1C.
Figure 1.
Visualization of Primary NSCs in Culture
(A) Primary NSCs (NSC008) were isolated from the brain of a 10-week-old fetus and cultured in flasks (passage 5). Primary NSCs grew as neurospheres in suspension. Scale bar, 100 μm.
(B) Primary NSCs were transduced with a retroviral virus expressing the L-MYC gene. NSC.Lmyc cells (LM-NSC008) were cultured under the same conditions as untransduced primary NSCs. LM-NSC008 cells preferentially grew as a monolayer. Scale bar, 100 μm.
(C) PCR analysis of genomic DNA derived from LM-NSC008 cells. The experiment was repeated three times.
(D and E) Karyotype analysis of the LM-NSC008 cell line at passage 2 (D) and passage 10 (E).
In Vitro Growth Kinetics, Tumor-Specific Migration, and Multilineage Differentiation of LM-NSC008 Cells
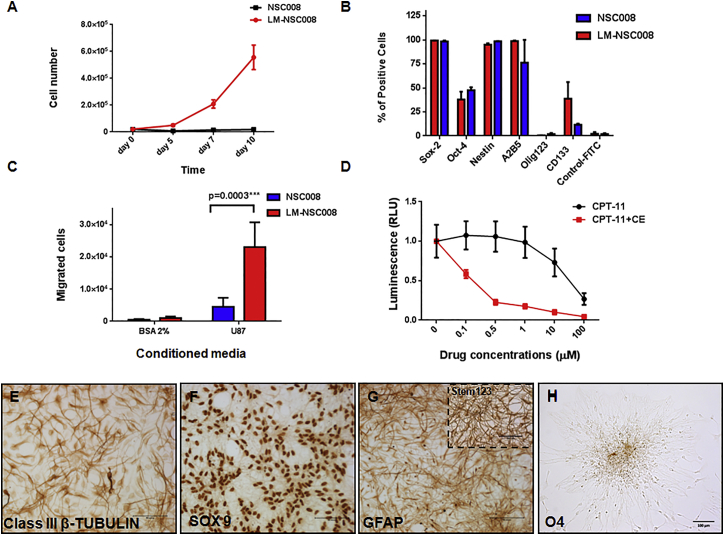
To determine whether the characteristics of these newly derived NSCs changed after expansion and passage in vitro, we compared primary NSC008 and LM-NSC008 grown through p6 with regard to viability, growth kinetics, and NSC marker expression (Figure 2). Both transduced and untransduced cells showed >90% viability grown in neurobasal medium under hypoxic conditions (4% O2). The growth kinetics of primary NSC008 and LM-NSC008 cells differed significantly when tested in culture over 10 days (using p6 NSC008 and LM-NSC008), showing significant differences in proliferation rate of NSC008 and LM-NSC008 cells (p = 0.005 on day 5 and p = 0.002 on days 7 and 10 as determined by multiple t test analysis with Holm-Sidak adjustment for multiple comparisons) (Figure 2A). The untransduced NSC008 did not proliferate (initial cell number at plating on day 1 = 2 × 104/well, final cell number on day 10 = 1.8 × 104) (Figure 2A, blue line), whereas the LM-NSC008 cells showed robust proliferation (initial cell number at plating on day 1 = 2 × 104, final cell number on day 10 = 6.6 × 105; cell population doubling time ∼30 hr for growth between days 5 and 10, after an initial lag time from days 0–5, Figure 2A, red line). The untransduced NSC008 stopped dividing approximately at p6 to p10, while the LM-NSC008 cells continued to display robust growth even at p15 (the highest passage number we tested).
Figure 2.
Growth Kinetics and Migration of Primary NSC008 and LM-NSC008 Cells in Culture
(A) Proliferation of primary NSC008 and LM-NSC008 cells (passage 6) grown for 10 days in culture media (serum-free media containing FGF and EGF). Mean values ± SD of two independent experiments in triplicate measurements are shown.
(B) Flow cytometry analysis of NSC008 and LM-NSC008 expressing stem cell markers. Mean values ± SD of four independent experiments in triplicates are shown.
(C) Tropism of NSC008 and LM-NSC008 cells to conditioned media from U87 human glioma cells as assessed by Boyden chamber migration assay. Mean values ± SD of three independent experiments in triplicate measurements are shown.
(D) Dose response of U251 human glioma cells to CPT-11 and to CPT-11 in the presence of hCE1m6 expressed by LM-NSC008 cells. Mean values ± SD of two independent experiments in quadruplicates are shown. RLU, relative light unit.
(E–H) Immunocytochemical staining of LM-NSC008 cells grown for 10 days in differentiation media. Cells were stained for neuronal class III β-TUBULIN (E), the astrocyte markers SOX9 (F) and GFAP (G) (inset Stem123), and oligodendrocyte marker O4 (H). Scale bars, 100 μm.
We used flow cytometry analysis and immunocytochemistry to detect stem cell and neural differentiation markers expressed by LM-NSC008 cells (Figure 2B). Flow cytometry revealed that virtually all undifferentiated primary NSCs and LM-NSC008 cells expressed the neural stem cell markers SOX2 and NESTIN, fewer cells expressed OCT4, and virtually no cells were Olig-1, -2, or -3 positive, consistent with a neural stem cell profile (Figure 2B). LM-NSC008 and primary NSC008 differed in expression of the stem cell marker CD133, perhaps reflecting arrested development related to L-MYC expression in proliferation media (supplemented with fibroblast growth factor [FGF] and epidermal growth factor [EGF]), LM-NSC008 cells divided approximately every 36 hr, whereas they stopped dividing under pro-differentiation conditions as previously reported (Nunes et al., 2003) (Figure S2G).
To explore the therapeutic potential of LM-NSC008 cells, we characterized their migration to tumor-conditioned media in vitro. In Boyden chamber cell migration assays, untransduced primary NSC008 at p6 exhibited little migration toward tumor cell-conditioned media compared with LM-NSC008 cells at the same passage (Figure 2C). These data suggest that after several passages in vitro, the untransduced parental NSC008 lack the abilities to proliferate and migrate toward tumor-conditioned media, whereas the LM-NSC008 cells possess proliferative and migratory properties through at least p15 (Figure 2C). To demonstrate the potential utility of LM-NSC008 cells in cancer-targeted therapies, we adenovirally transduced LM-NSC008 cells to express a modified human CARBOXYLESTERASE (CE; hCE1m6), which converts irinotecan (CPT-11) to SN-38, a potent topoisomerase I inhibitor (Wierdl et al., 2008, Metz et al., 2013). We performed in vitro cytotoxicity assays using human U251 glioma cells and various concentrations of irinotecan alone or in combination with CE derived from LM-NSC008 cells adenovirally transduced to express hCE1m6. We observed enhanced irinotecan toxicity (500-fold) in the presence of hCE1m6 in the culture media when compared with CPT-11 alone. CE (hCE1m6) activity was measured at 995 nmol/min/mL in the conditioned media derived from LM-NSC008 cells prior to the cytotoxicity experiment (Figure 2D). This suggests a potential use of LM-NSC008 cells for enzyme/prodrug therapy to treat glioma and other tumors.
To probe the differentiation potential of LM-NSC008 cells in vitro (Figures 2E–2H), we immunostained differentiated LM-NSC008 (grown for 10 days in differentiation media) for class III β-tubulin, SOX9, glial fibrillary acidic protein (GFAP), and O4. The immunoreactivity observed indicated that LM-NSC008 cells were able to differentiate into neurons (class III β-tubulin), astrocytes (SOX9, GFAP), and oligodendrocytes (O4) (Figures 2E–2H). We also compared differentiation of LM-NSC008 with untransduced primary NSC008 at p5 (Figure S3). Differentiated LM-NSC008 cells and untransduced NSC008 cells showed similar immunoreactivity for neuronal and glial markers class III β-TUBULIN, SOX9, and GFAP (Figure S3).
Tumor-Directed Migration of hNSCs In Vivo
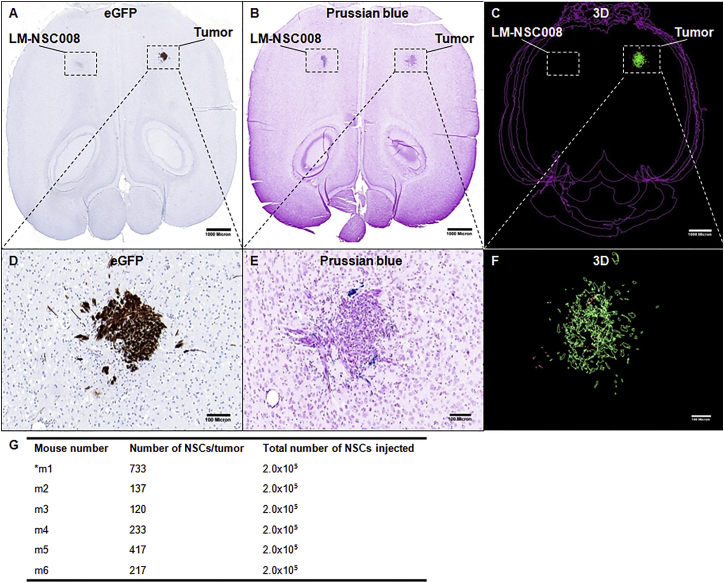
An orthotopic glioma xenograft model was generated by injecting U251.EGFP.ffluc human glioma cells (1 × 105/2 μL PBS) into the right frontal hemisphere of adult NSG mice. Seven days later, iron-labeled LM-NSC008 cells (2 × 105/3 μL PBS) were injected intracranially and contralateral to the tumor injection site (left frontal lobe, n = 6 tumor-bearing mice, n = 2 control non-tumor-bearing mice). Control mice received sham injections of PBS into the right frontal lobe instead of tumor. Mice were euthanized 4 days after receiving LM-NSC008 injections, and their brains were harvested, fixed, sectioned, and stained for histological examination. H&E and Prussian blue staining were used to visualize tumors and LM-NSC008 cells, respectively. Tumor sites were confirmed by immunostaining for EGFP (Figure 3A, with tumor area enlarged in Figure 3D), which revealed the presence of compact tumor nodules located in the deep cortex and caudate-putamen, ranging in diameter between 0.6 and 1 mm. Using Prussian blue staining, we could often identify the injection site of iron-labeled NSCs as a distinct, compact cellular focus located contralateral to the tumor site. Iron-labeled LM-NSC008 cells were found at the tumor site by 4 days post injection (Figure 3B, with tumor area enlarged in Figure 3E) having migrated from the contralateral hemisphere. LM-NSC008 cells were mostly found in peripheral areas of the tumor (Figure 3E), dispersed within tumor nodules (Figure 3F) and in proximity to infiltrating tumor cells. Three-dimensional (3D) reconstruction was used to quantify the number of NSCs at the tumor site and their spatial distribution within the tumor (Figures 3C and 3F show a z-axis projection of 8–10 slices, 200 μm apart). Quantification of LM-NSC008 cell numbers at the tumor site is summarized in Figure 3G. We estimated that the average number of LM-NSC008 cells within the tumor that migrated from the opposite hemisphere was 310 ± 233. Immunostaining for L-MYC confirmed its presence at the LM-NSC008 injection site and the tumor site in the contralateral hemisphere (Figure S4A, enlarged in Figures S4B and S4C). Human kidney tissue was used as a positive control for L-MYC staining (Figure S4D). Negative control was obtained from omitted primary antibody staining in the LM-NSC008 injection site (Figure S4E) and tumor site (Figure S4F).
Figure 3.
Tumor-Specific Migration of LM-NSC008 Cells from Contralateral Hemisphere In Vivo
(A, B, D, E) U251 human glioma xenografts were established in adult NSG mice. Consecutive horizontal brain sections from a tumor-bearing mouse that received an intracranial (contralateral to the tumor) injection of LM-NSC008 cells (2 × 105) labeled with feraheme. (A) Immunohistochemical staining of adjacent brain sections with EGFP antibodies to visualize tumor xenografts (scale bar, 1000 μm). (D) Enlargement of tumor area shown as inset in (A) (scale bar, 100 μm). (B) Prussian blue staining of axial brain sections, feraheme-labeled NSCs (blue) (scale bar, 1000 μm). (E) Higher-magnification of tumor site shown as inset in (B) with blue LM-NSC008 cells (scale bar, 100 μm).
(C and F) Three-dimensional reconstruction of tumors (green) and LM-NSC008 cells (red) (n = 6). (F) 3D enlargement of tumor area shown as inset in (C).
(G) A summary of 3D reconstructions of tumors and LM-NSC008 cells at the tumor site.
Scale bars 1,000 μm (A–C) and 100 μm (D–F). Asterisk indicates a mouse number.
Migration of LM-NSC008 Cells to Sites of Traumatic Brain Injury
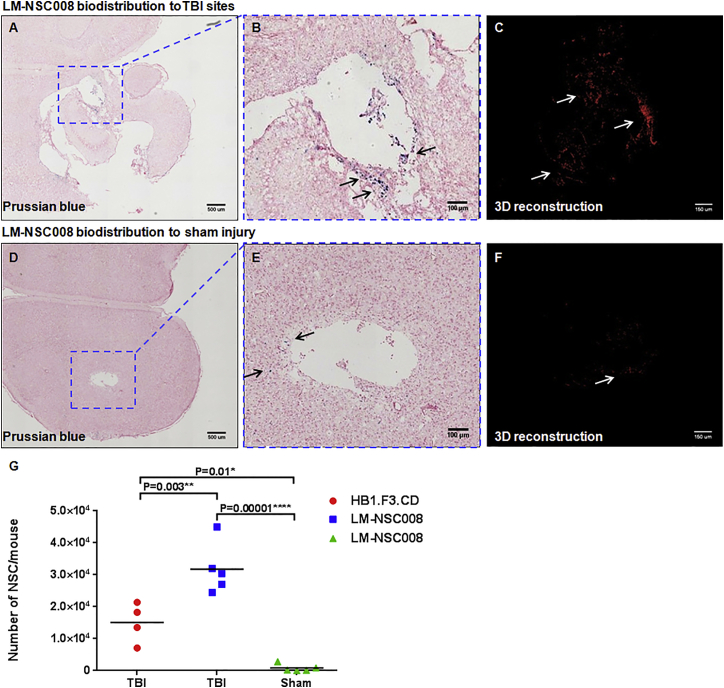
To show the potential of LM-NSC008 cells to target injured brain, we performed in vivo biodistribution experiments using a controlled cortical impact (CCI) traumatic brain injury (TBI) model. TBI was generated in adult C57BL/6 mice using an impactor device as described previously (Brody et al., 2007). Sham-operated mice (without TBI) were used as controls. The LM-NSC008 and HB1.F3.CD NSCs (Kim et al., 2008) were administered intranasally on days 9 and 11 following generation of moderate TBI or a sham operation (n = 5 per group), as described previously (Gutova et al., 2015). Mice received intranasal drops of hNSCs (5 × 105 cells in 12 μL of PBS), equally divided and administered into both nostrils. LM-NSC008 and HB1.F3.CD NSCs were pre-labeled with Molday ION rhodamine B as described previously (Addicott et al., 2011). Brain tissue was harvested 2–7 days after the last NSC administration, and iron-labeled hNSCs were visualized by Prussian blue staining. 3D reconstruction was performed using Prussian blue-stained sections. LM-NSC008 cells showed robust migration to TBI injury sites (Figures 4A and 4B), while sham-operated control mice had fewer LM-NSC008 stem cells at the surgery sites (Figures 4D and 4E). Numbers of LM-NSC008 and HB1.F3.CD hNSCs were quantified at TBI sites from z-axis projections of Prussian blue-stained brain sections (Figures 4C and 4F), and total numbers of hNSCs were calculated for each mouse. A summary is presented in Figure 4G, comparing the results of: (1) LM-NSC008 cell migration to TBI and sham injury sites, ∗∗∗∗p = 0.00001; and (2) HB1.F3.CD migration to TBI and sham sites, ∗p = 0.01. Comparison of TBI-directed migration of both hNSCs (LM-NSC008 and HB1.F3.CD) showed significant difference among TBI-specific migration of these lines, ∗∗p = 0.003. These data suggest that LM-NSC008 administered intranasally can reach sites of CNS injury in higher numbers than HB1.F3.CDs and have the potential to be used for brain tissue repair and/or regeneration.
Figure 4.
Distribution of LM-NSC008 Cells to TBI Sites
(A) Prussian blue staining of brain section with TBI, demonstrating iron-labeled LM-NSC008 cells (blue) distributed throughout injury site. Scale bar, 500 μm.
(B) Magnified area of TBI described in (A). Scale bar, 100 μm.
(C) 3D reconstruction of LM-NSC008 cells distributed in the vicinity of TBI site. Scale bar, 150 μm.
(D) Prussian blue staining of sham-injured brain section demonstrating rare (10–20) LM-NSC008 cells occasionally visualized around sham surgery sites. Scale bar, 500 μm.
(E) Magnified area of injury site described in (D) Scale bar, 100 μm.
(F) 3D reconstruction of LM-NSC008 cell distribution in sham-injured brain. Scale bar, 150 μm.
(G) Quantification of average numbers of HB1.F3.CD and LM-NSC008 cells at TBI and sham injury sites (n = 5).
The arrows in (B, C, E, F) indicate LM-NSC008 cells in the vicinity of TBI or sham injury site. Statistical analysis of the treated and control groups was performed using one-way ANOVA. Tukey's adjusted p values are reported for pairwise comparisons of migration of hNSCs to TBI and sham injury sites.
LM-NSC008 Cell Fate in Normal Brain
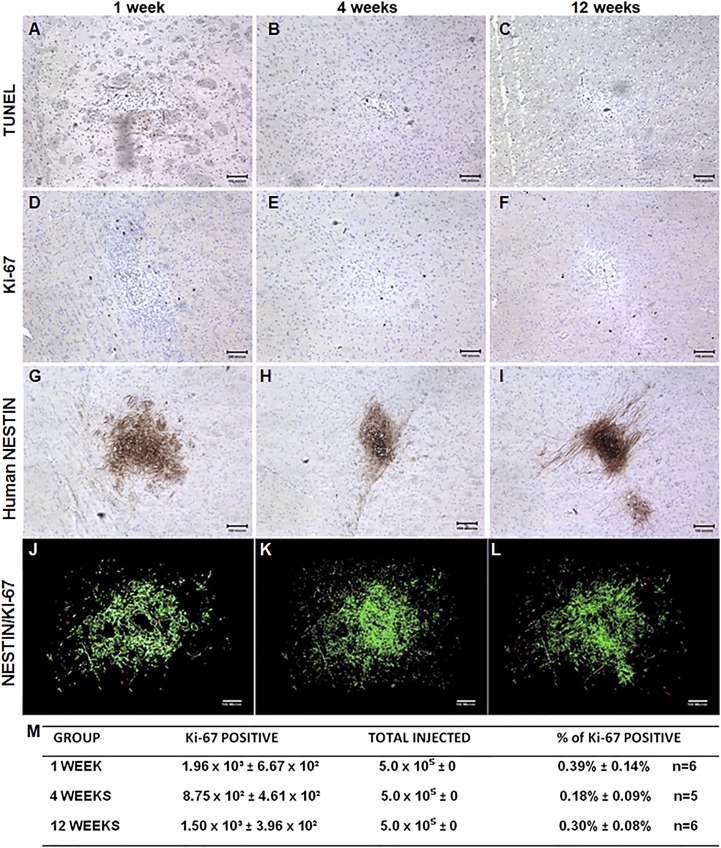
To characterize the behavior of LM-NSC008 cells in normal brain (no tumor or TBI), we injected 5 × 105 cells in 4 μL of PBS into the right frontal lobe of NSG mice (n = 6) (Figure 5). Mice were placed into three groups that were euthanized at 1, 4, or 12 weeks after LM-NSC008 injections. Brain sections were examined for the presence of viable LM-NSC008 cells, for the presence of markers indicating their fates (proliferation, differentiation, or apoptosis). H&E staining of paraffin-embedded sections showed foci of architectural distortion associated with the presence of LM-NSC008 cells at the injection site (not shown). Immunohistochemical staining for human NESTIN confirmed the presence of LM-NSC008 cells at the injection sites and minimal migration away from the site (Figures 5G–5I). TUNEL staining indicated apoptosis of LM-NSC008 cells in normal brain at 1, 4, and 12 weeks (Figures 5A–5C). Ki-67 staining (human-specific) indicated in vivo division of LM-NSC008 cells at 1, 4, and 12 weeks without formation of masses caused by uncontrolled LM-NSC008 proliferation (Figures 5D–5F). We did not observe neurological symptoms in mice that might be related to the injection of LM-NSC008 cells.
Figure 5.
LM-NSC008 Cell Fate in Non-Tumor-Bearing Brain
NSG mice were injected intracerebrally with LM-NSC008 cells (5 × 105, right frontal lobe) and euthanized 1 week (A, D, G), 4 weeks (B, E, H), and 12 weeks (C, F, I) (n = 6) after injection.
(A–C) Immunohistochemical (IHC) detection of apoptotic NSCs by using TUNEL staining.
(D–F) IHC detection of the human-specific cell proliferation marker Ki-67.
(G–I) IHC of mouse brain tissue using human-specific NESTIN antibodies.
(J–L) z-Axis projection of all Ki-67- and hNESTIN-stained sections. NESTIN staining pseudocolored green and Ki-67 stained red.
(M) Quantification of the percentile of Ki-67-positive cells compared with the total injected dose of LM-NSC008 cells.
Scale bars, 100 μm.
To further demonstrate the lack of tumorigenicity, we determined from IHC-stained brain sections the numbers of Ki-67-positive cells per mouse brain and their locations. Human NESTIN-positive cells (pseudocolored green) are highlighted and merged with Ki-67-positive cells (red) in Figures 5J–5L. The number of Ki-67-positive cells for each mouse was compared with the total number of injected cells (5 × 105/mouse), and their percentiles are presented in Figure 5M. Quantification of human NESTIN expression intensity is presented in Figure S5 and demonstrated a time-dependent decrease in NESTIN expression at the LM-NSC008 injection site.
Gene-Expression Profiling and Genome-wide Analysis of Primary NSC008 and LM-NSC008 Cells
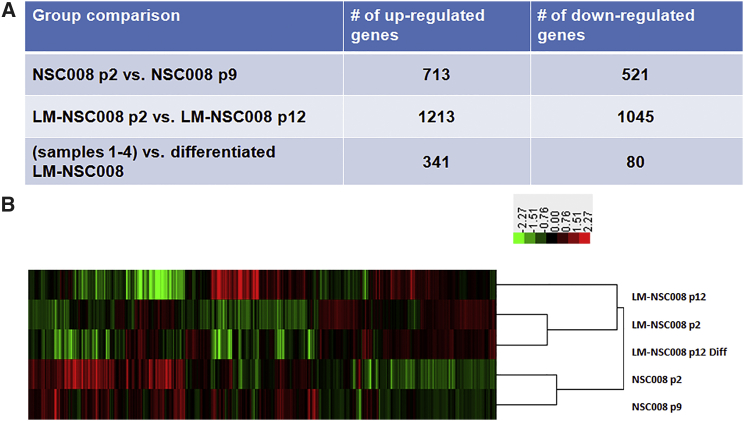
We performed RNA-sequencing (RNA-seq) analysis to determine the transcriptional changes that occurred in the newly derived NSCs in vitro with and without expression of L-MYC. To examine early and late passages, we isolated RNA from primary untransduced NSC008 at p2 and p9 (hNSC p2 and hNSC p9, respectively) and from LM-NSC008 cells at p2 and p12 (LM-NSC008 p2 and LM-NSC008 p12, respectively). We also differentiated the LM-NSC008 cells in vitro, using a published protocol (Nunes et al., 2003), and isolated RNA for analysis (LM-NSC008 p12.Diff). We then performed RNA-seq expression profiling on the samples. To probe for differential gene expression, we selected genes that showed either increased or decreased expression based on results using EdgeR (Figure 6A), a Bioconductor package for differential expression (DE) analyses of RNA-seq reads (Meng et al., 2014). We performed three group comparisons using DE gene-selection criteria: (1) absolute fold change (FC) ≥ 3 (for comparisons of hNSC p2 versus p9 and LM-NSC008 p2 versus p12); and (2) absolute FC ≥ 3 and p ≤ 0.05 (for comparison of all previous samples with the LM-NSC008 p12 differentiated sample) (Figure 6A). Hierarchical clustering analysis was performed using Cluster 3.0 and the average linkage clustering method (de Hoon et al., 2004). The results were visualized in Treeview and a heatmap was generated (Saldanha, 2004) (Figure 6B). All up- and downregulated genes were further analyzed using DAVID functional annotation analysis to compare the same groups (LM-NSC008 p2 versus LM-NSC008 p12) (Table 1). Further analysis compared hNSC008 p2 with hNSC008 p9 (Table S1) and LM-NSC008 at p2 and p12 were also compared with the LM-NSC008 differentiated sample (Table S2).
Figure 6.
Summary of Up- and Downregulated Genes within Samples Analyzed
(A) Untransduced NSC008 at passage 2 were compared with NSC008 at passage 9, L-MYC transduced cells (LM-NSC008) were compared at passages 2 versus 12, and NSC008 and LM-NSC008 cells were compared with differentiated LM-NSC008 cells.
(B) Heatmap showing upregulated (red) and downregulated (green) genes.
Table 1.
DAVID Functional Annotation Analysis of LM-NSC008 at Passage 2 versus LM-NSC008 at Passage 12
| Term | Fold Enrichment | Count | p Value |
|---|---|---|---|
| Top 5 Enriched Pathways Upregulated | |||
| Neuron differentiation | 3.154299 | 72 | 3.73 × 10−18 |
| Transmission of nerve impulse | 3.125009 | 57 | 3.55 × 10−14 |
| Synaptic transmission | 3.219573 | 50 | 5.28 × 10−13 |
| Cell-cell signaling | 2.430563 | 76 | 8.98 × 10−13 |
| Cell adhesion | 2.247814 | 82 | 6.21 × 10−12 |
| Top 5 Enriched Pathways Downregulated | |||
| Cell adhesion | 2.77224 | 104 | 8.81 × 10−22 |
| Biological adhesion | 2.768286 | 104 | 9.83 × 10−22 |
| Regulation of cell motion | 4.447297 | 46 | 1.65 × 10−17 |
| Cell motion | 2.906924 | 74 | 1.13 × 10−16 |
| Actin cytoskeleton organization | 3.963039 | 48 | 4.85 × 10−16 |
Comparison of gene expression in untransduced NSC008 at p2 versus p9 revealed upregulation of genes involved in pathways regulating neurogenesis, transmission of nerve impulses, synaptic transmission, nervous system development, and cell-cell signaling (Tables S1 and S5). Genes involved in immune response and cell-adhesion pathways were downregulated (Table S1). Upregulated genes in the LM-NSC008 cells included those involved in pathways responsible for regulating the development of the neural system, while downregulated genes were involved in cell adhesion and regulation of cell migration (Table 1). When NSC008 and LM-NSC008 cells were compared with the differentiated LM-NSC008 cells, we observed upregulation of regulators of cell growth and wound healing, while metabolic pathways were downregulated, indicating slower proliferation rates and differentiation of LM-NSC008 cells cultured under pro-differentiation conditions (Tables S2 and S5).
Analyses of somatic mutations and copy-number variations with whole-exome sequencing were also performed to check for potential mutation events or changes in the karyotype as a result of L-MYC expression. Only eight somatic mutations were identified when comparing LM-NSC008 p12 with LM-NSC008 p2. Of these eight mutations, six were located in introns and two were synonymous, so none were considered functional and most likely represent germline mutations. We also did not observe any large copy-number variations, with only two regions covering two coding exons of gene CDKL1 and MYLK showing possible loss of one allele in the p12 sample (Table S4). Overall, we concluded that there were no significant differences in the genomic sequences between the p12 and p2 samples, indicating that L-MYC gene expression did not contribute to increased genomic instability in LM-NSC008 cells.
Discussion
We chose L-MYC to generate immortalized hNSC lines for therapeutic purposes based on its low tumorigenicity, as reported by Nakagawa et al. (2010). In this study we have shown that expression of the L-MYC gene in human fetal brain-derived NSCs can promote self-renewal and multipotency in these cells. Untransduced NSCs from the same primary pool proliferate for only a limited number of passages in vitro before undergoing apoptosis. Our findings demonstrate that stable expression of the L-MYC gene in NSC008 cells promotes their survival and proliferation while preserving their migration and differentiation properties in vitro and in vivo. In this way, the LM-NSC008 cells will provide for GMP grade production as well as modification for therapeutic purposes. RNA- and DNA-sequencing analyses suggest that the L-MYC gene regulates distinct sets of target genes (Nakagawa et al., 2008, Nakagawa et al., 2010). Most likely the L-MYC gene acts to suppress expression of genes involved in differentiation, as was demonstrated previously (Nakagawa et al., 2010). At the same time, analysis of specific somatic mutations demonstrated that L-MYC expression in LM-NSC008 cells did not increase their genetic instability.
The LM-NSC008 cells also recapitulated important phenotypes of NSCs: after differentiation, the LM-NSC008 cells expressed neuronal class III β-TUBULIN, astroglial markers SOX9 and GFAP, and oligodendrocyte marker O4, which coincided with neuronal, astrocytic, and oligodendroglial morphologies, respectively (Figures 2E–2H) (Wegner and Stolt, 2005, Sofroniew and Vinters, 2010). These are key characteristics that stem cells must display if they are to be used in cell-replacement therapies for neurodegenerative diseases or brain injury (Lemmens and Steinberg, 2013, Tang et al., 2013).
The MYC genes orchestrate diverse cellular functions in the normal cells, including self-renewal and proliferation, morphological maturation, apoptosis, energy utilization, and multipotent differentiation (Conacci-Sorrell et al., 2014). Nakagawa et al. (2010) previously showed that L-MYC has the lowest transformational activity of the MYC isoforms in iPSCs, as well as a robust ability to reprogram cells for generation of iPSCs. Our gene-expression profiling data are in accord with the reported wide-ranging effects of L-MYC gene expression in hNSCs. The large number of upregulated genes (1,213) and downregulated genes (1,045) in LM-NSC008 p2 versus LM-NSC008 p12 (Figure 6) may reflect the complex protein interactome in which L-MYC participates directly or indirectly via its DNA-binding and transactivation domains (Tu et al., 2015). The robust changes in the gene-expression profile also suggest that the introduction of the L-MYC gene into hNSCs may lead to epigenetic plasticity that could underlie the large-scale changes in gene expression (Arrowsmith et al., 2012, Watanabe et al., 2013).
Research on the “druggable epigenome” and the use of small-molecule drugs to break the epigenetic barrier by modulating the activity of enzymes acting on the epigenome (e.g., DNA methylases/demethylases, histone methyltransferases/demethylases, acetyltransferases/deacetylases, and RNAi) has led to advances in basic as well as translational science, including clinical development (Arrowsmith et al., 2012). We surmise that in future research epigenome-modulating drugs may be applicable for optimization of the tumor-targeting and therapeutic properties of therapeutic NSCs, including the LM-NSC008 cells. Such studies could use LM-NSC008 cells deposited in a master cell bank, from which various subclones obtained by epigenetic reprogramming could be generated. In addition, analysis of the immunological parameters of the LM-NSC008 cells will be a crucial component of future pre-clinical studies using this cell line. Such immunological parameters could include the levels of major histocompatibility complex class I and class II gene expression (that could determine the immunological interactions between the transplanted NSCs and the host immune system), as well as expression profiling of various cytokine receptor genes in the LM-NSC008 cells, which could underlie the signaling pathways responsible for tumor tropism and targeting of the nervous system affected by various pathological conditions.
In summary, we have generated a stable, expandable human NSC line (LM-NSC008) by overexpression of the L-MYC gene. We have characterized the LM-NSC008 cell line in vitro and in vivo. Our findings demonstrate the potential therapeutic utility of LM-NSC008 cells for therapeutic drug delivery to CNS tumors, restorative therapies for TBI, and possibly other diseases of the CNS. Further pre-clinical studies in CNS disease models are warranted for translational development and eventual clinical use of L-MYC-transduced NSC lines.
Experimental Procedures
Generation and Characterization of Primary NSC Cultures
Fetal brain from elective abortions was obtained as discarded tissue from Advanced Bioscience Resources under a City of Hope Institutional Review Board approved protocol (City of Hope IRB #10079). All procedures that involved stem cell isolation and propagation were performed under approved protocols (City of Hope SCRO #11002, and IBC #11016). Human brain tissue (from fetuses of 10–14 weeks of gestation) was dissociated using 0.1% collagenase and hyaluronidase solution (3,000 U/mL collagenase, 1,000 U/mL hyaluronidase in Hank's balanced salt solution), and isolated NSCs were cultured in serum-free NSC medium (RHB-A medium; Stem Cell Science) supplemented with 10 ng/mL basic FGF (bFGF), 10 ng/mL EGF, 2 mM L-glutamine (Invitrogen), Gem21 NeuroPlex Serum-Free Supplement (Gemini Bio-Products, #400-160), and penicillin-streptomycin (Mediatech, 30-002-CI). Growth factors (bFGF and EGF) were added every other day and media were completely changed every 7 days. Cells were passaged by gentle trituration of neurospheres.
RNA and DNA were isolated from LM-NSC008 p2 and p12, NSC008 p2 and p9, and differentiated LM-NSC008 cells to characterize gene expression. LM-NSC008 cells were differentiated in vitro by culturing in DMEM medium supplemented with 1% fetal bovine serum for 10 days as previously described (Nunes et al., 2003). Samples for karyotype determination were prepared for G-banding karyotyping analysis using standard cytogenetic methods at City of Hope Cytogenetics Core Facility.
Generation of NSC Lines that Stably Express the L-MYC Transgene
The L-MYC expression vector (pMXs-hu-L-Myc) was obtained from Addgene (Okita et al., 2013). The L-MYC gene from pMXs-hu-L-Myc was recloned into a retroviral expression vector (pCMV-LL-PGK-puro) containing the puromycin selection marker. Retrovirus carrying the L-MYC gene was used for transduction of human fetal-derived NSCs at MOI 2.5.
Boyden Chamber Cell Migration Assay
In vitro chemotaxis assays were conducted using 24-well cell-culture plates with polycarbonate inserts (8-μm pore size; Millipore) as described previously (Gutova et al., 2010). An independent t test was performed to determine the statistically significant difference between NSC008 and LM-NSC-008 migration to U87 conditioned media (p = 0.0003).
Cytotoxicity Assays
Detailed methodology is presented in Supplemental Experimental Procedures.
Colony Formation Assay
Detailed methodology is presented in Supplemental Experimental Procedures.
Iron Labeling of NSCs
Human NSCs (HB1.F3.CD and LM-NSC008) were labeled with Molday ION rhodamine B prior to intranasal administration. LM-NSC008 cells and HB1.F3.CDs were visualized within brain tissue by Prussian blue staining using an Accustain Iron Stain Kit (Sigma-Aldrich).
Immunohistochemistry and Immunocytochemistry
Detailed methodology is presented in Supplemental Experimental Procedures.
Flow Cytometry Analysis of Primary NSC008 and LM-NSC008 Cells
Detailed methodology is presented in Supplemental Experimental Procedures.
Serial Section Reconstruction and Measurement
Reconstruction and measurement was performed using Reconstruct (version 1.1.0.1) (Fiala, 2005) software as described previously (Gutova et al., 2013). In brief, 7–11 serial brain sections (10 μm each) separated by 200 μm were imported into the Reconstruct program and aligned manually. NSC quantification was performed using bright-field Prussian blue images, and autotracing of the tumor was done on adjacent EGFP-stained images. From the number of NSCs on each section analyzed, the total number of cells per brain was calculated using spacing between stained sections and approximation of NSC numbers on unstained sections.
Animal Studies of LM-NSC008 Cell Distributions
All animal studies were performed under approved City of Hope Institutional Animal Care and Use Committee protocols (IACUC 12025). Animal studies used NOD scid IL2Rgammanull (NSG) mice, 8–12 weeks old regardless of sex. For distribution studies, orthotopic tumor xenografts were generated by injecting human U251.EGFP.ffluc glioma cells (1 × 105 in 2 μL of serum-free DMEM) into the right frontal lobe of NSG mice (study day 1). LM-NSC008 cells (2 × 105 in 3 μL of PBS) were injected contralaterally to the tumor (n = 6 mice) on day 7 of the study. Control mice (n = 2) did not receive tumor injection, but did receive sham injection of PBS (to mimic injection of tumor cells) and NSCs on the opposite side of the brain. Mice were evaluated for clinical signs before and after tumor or NSC injections. Experimental animals were monitored daily for any debilitating signs secondary to tumor growth, including: labored breathing, weight loss (>20% body weight), scruffy coat, hunched posture, hypo- or hyperthermia, impaired ambulatory movement, and inability to remain upright. Tumors were confirmed by Xenogen imaging prior to NSC administration. All mice were euthanized on day 4 after NSC administration. Brains were removed without perfusion and processed for histological examination using H&E and EGFP immunostaining to detect tumors and tissue morphology. Prussian blue staining was done to identify Molday ION-labeled NSCs, as described previously (Addicott et al., 2011).
TBI was generated in adult (C57BL/6) Cdh23ahl mice via impactor device as described previously (Brody et al., 2007). In brief, mice were subjected to a single moderate left lateral (CCI) injury with craniotomy under isoflurane anesthesia. An electromagnetic impact device was used for generation of moderate contusion TBI using the following bregma coordinates: x axis, 1.2 mm left of midline; y axis, 1.5 mm anterior to lambda and 2 mm deep. After the probe was inserted into the brain (2 mm deep), the dwell time was 100 ms. The hole was then covered with a plastic skull cap and the skin was sutured. Sham TBI injury was performed on a control group of mice, which included craniotomy but no impactor stimulation, as described previously (Brody et al., 2007).
Intranasal Administration of NSCs
LM-NSC008 and HB1.F3.CD cells were administered intranasally on days 9 and 11 after mild TBI. Mice received intranasal drops of either LM-NSC008 or HB1.NSC.CD cells (5 × 105 cells per 12 μL/mouse) into both nostrils divided equally (6 μL) with 2-min intervals pre-labeled with Molday ION rhodamine B (Addicott et al., 2011, Gutova et al., 2015). Mice were euthanized 2 weeks after the last NSC administration. Brain tissue was harvested and fixed in 4% paraformaldehyde. Paraffin-embedded sections were prepared and Prussian blue staining was performed to visualize iron-labeled hNSCs. Prussian blue-stained sections were used to quantify the number of hNSC cells in each brain for TBI and sham groups.
In Vivo Tumorigenicity Studies
For determination of the fate of NSCs in non-tumor-bearing mouse brain, NSG mice were injected with 5 × 105 NSCs (n = 6) intracranially and were euthanized at 1, 4, and 12 weeks after injection, after which their brains were harvested. The brains were then isolated, fixed, and stained (every tenth section) with H&E (American Master Tech Modified Mayer's H&E), Ki-67 (Dako), TUNEL (Chemicon), and human NESTIN (Millipore). Quantification of percentile Ki-67-positive cells was performed using previously stained sections relative to the dose on LM-NSC008 cells injected.
mRNA Deep Sequencing with Illumina Hiseq2500
Detailed methodology is presented in Supplemental Experimental Procedures.
RNA-Seq Data Analysis
Detailed methodology is presented in Supplemental Experimental Procedures. The records of RNA-seq data have been approved and assigned the accession number GEO: GSE84166.
Whole-Exome Sequencing for Somatic Mutations and Copy-Number Variation Analysis
Detailed methodology is presented in Supplemental Experimental Procedures.
Author Contributions
Z.L. and D.O. designed and carried out experiments, and analyzed data. M.J.C., D.S., S.A., L.T., J.B., and X.R. carried out experiments and analyzed data. R.M., P.M.P., J.N., Y.L., X.W., and D.L.B. generated and analyzed data, and wrote the manuscript. A.J.A. provided fetal brain tissue for experiments. K.S.A. and M.E.B. provided advice on experimental design and edited the manuscript. M.G. led the project, carried out data analysis and interpretation, and prepared the final manuscript.
Acknowledgments
The authors acknowledge the technical expertise and advice of Sofia Loera of the City of Hope Pathology Core, and the editorial assistance of Dr. Keely L. Walker and Andrea Lynch (City of Hope). We also thank Dr. Shinya Yamanaka for providing the L-MYC plasmids. This work was supported by funding from NIH (R01 NS065069-07), Accelerate Brain Cancer Cure (ABC2), Alex’s Lemonade Stand Foundation, Altschul foundation, and Pediatric Cancer Research Foundation. Research was also supported by work performed in the Bioinformatics, Cytogenetics and Pathology Cores supported by the National Cancer Institute of the NIH under grant number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. K.S.A. and A.J.A. are uncompensated Board Members, Chief Scientific and Chief Financial Officers, and shareholders of TheraBiologics.
Published: August 18, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.07.013.
Accession Numbers
The RNA-seq data discussed in this publication was deposited in NCBI's Gene Expression Omnibus and are accessible through accession numbers GEO: GSE84166, GSM2228431, GSM2228432, GSM2228433, GSM2228434, and GSM2228435.
Supplemental Information
References
- Aboody K.S., Najbauer J., Metz M.Z., D'Apuzzo M., Gutova M., Annala A.J., Synold T.W., Couture L.A., Blanchard S., Moats R.A. Neural stem cell-mediated enzyme/prodrug therapy for glioma: preclinical studies. Sci. Transl Med. 2013;5:184ra159. doi: 10.1126/scitranslmed.3005365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addicott B., Willman M., Rodriguez J., Padgett K., Han D., Berman D., Hare J.M., Kenyon N.S. Mesenchymal stem cell labeling and in vitro MR characterization at 1.5 T of new SPIO contrast agent: Molday ION Rhodamine-B. Contrast Media Mol. Imaging. 2011;6:7–18. doi: 10.1002/cmmi.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrowsmith C.H., Bountra C., Fish P.V., Lee K., Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- Begum A.N., Guoynes C., Cho J., Hao J., Lutfy K., Hong Y. Rapid generation of sub-type, region-specific neurons and neural networks from human pluripotent stem cell-derived neurospheres. Stem Cell Res. 2015;15:731–741. doi: 10.1016/j.scr.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody D.L., Mac Donald C., Kessens C.C., Yuede C., Parsadanian M., Spinner M., Kim E., Schwetye K.E., Holtzman D.M., Bayly P.V. Electromagnetic controlled cortical impact device for precise, graded experimental traumatic brain injury. J. Neurotrauma. 2007;24:657–673. doi: 10.1089/neu.2006.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conacci-Sorrell M., McFerrin L., Eisenman R.N. An overview of MYC and its interactome. Cold Spring Harb Perspect. Med. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniela F., Vescovi A.L., Bottai D. The stem cells as a potential treatment for neurodegeneration. Methods Mol. Biol. 2007;399:199–213. doi: 10.1007/978-1-59745-504-6_14. [DOI] [PubMed] [Google Scholar]
- de Hoon M.J., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Fiala J.C. Reconstruct: a free editor for serial section microscopy. J. Microsc. 2005;218:52–61. doi: 10.1111/j.1365-2818.2005.01466.x. [DOI] [PubMed] [Google Scholar]
- Gutova M., Najbauer J., Chen M.Y., Potter P.M., Kim S.U., Aboody K.S. Therapeutic targeting of melanoma cells using neural stem cells expressing carboxylesterase, a CPT-11 activating enzyme. Curr. Stem Cell Res. Ther. 2010;5:273–276. doi: 10.2174/157488810791824421. [DOI] [PubMed] [Google Scholar]
- Gutova M., Shackleford G.M., Khankaldyyan V., Herrmann K.A., Shi X.H., Mittelholtz K., Abramyants Y., Blanchard M.S., Kim S.U., Annala A.J. Neural stem cell-mediated CE/CPT-11 enzyme/prodrug therapy in transgenic mouse model of intracerebellar medulloblastoma. Gene Ther. 2013;20:143–150. doi: 10.1038/gt.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutova M., Shahmanyan D., Oganesyan D., Abramyants Y., Danielyan L., Frey W.H., II, Khankaldyyan V., Najbauer J., Balyasnikova I.V., Moats R.A. Intranasal delivery of therapeutic neural stem cells to target intracerebral glioma. J. Stem Cells Regen. Med. 2015;1:007. [Google Scholar]
- Hicks C., Stevanato L., Stroemer R.P., Tang E., Richardson S., Sinden J.D. In vivo and in vitro characterization of the angiogenic effect of CTX0E03 human neural stem cells. Cell Transpl. 2013;22:1541–1552. doi: 10.3727/096368912X657936. [DOI] [PubMed] [Google Scholar]
- Kallos M.S., Behie L.A. Inoculation and growth conditions for high-cell-density expansion of mammalian neural stem cells in suspension bioreactors. Biotechnol. Bioeng. 1999;63:473–483. doi: 10.1002/(sici)1097-0290(19990520)63:4<473::aid-bit11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Kanojia D., Balyasnikova I.V., Morshed R.A., Frank R.T., Yu D., Zhang L., Spencer D.A., Kim J.W., Han Y., Yu D. Neural stem cells secreting anti-HER2 antibody improve survival in a preclinical model of HER2 overexpressing breast cancer brain metastases. Stem Cells. 2015;33:2985–2994. doi: 10.1002/stem.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.U. Human neural stem cells genetically modified for brain repair in neurological disorders. Neuropathology. 2004;24:159–171. doi: 10.1111/j.1440-1789.2004.00552.x. [DOI] [PubMed] [Google Scholar]
- Kim S.U., Nagai A., Nakagawa E., Choi H.B., Bang J.H., Lee H.J., Lee M.A., Lee Y.B., Park I.H. Production and characterization of immortal human neural stem cell line with multipotent differentiation property. Methods Mol. Biol. 2008;438:103–121. doi: 10.1007/978-1-59745-133-8_10. [DOI] [PubMed] [Google Scholar]
- Lemmens R., Steinberg G.K. Stem cell therapy for acute cerebral injury: what do we know and what will the future bring? Curr. Opin. Neurol. 2013;26:617–625. doi: 10.1097/WCO.0000000000000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Lu Z., Liu H., Zhang L., Zhang S., Chen Y., Rao M.K., Huang Y. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz M.Z., Gutova M., Lacey S.F., Abramyants Y., Vo T., Gilchrist M., Tirughana R., Ghoda L.Y., Barish M.E., Brown C.E. Neural stem cell-mediated delivery of irinotecan-activating carboxylesterases to glioma: implications for clinical use. Stem Cells Transl Med. 2013;2:983–992. doi: 10.5966/sctm.2012-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Yamanaka S. Reprogramming of somatic cells to pluripotency. Adv. Exp. Med. Biol. 2010;695:215–224. doi: 10.1007/978-1-4419-7037-4_14. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Takizawa N., Narita M., Ichisaka T., Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. USA. 2010;107:14152–14157. doi: 10.1073/pnas.1009374107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes M.C., Roy N.S., Keyoung H.M., Goodman R.R., McKhann G., 2nd, Jiang L., Kang J., Nedergaard M., Goldman S.A. Identification and isolation of multipotential neural progenitor cells from the subcortical white matter of the adult human brain. Nat. Med. 2003;9:439–447. doi: 10.1038/nm837. [DOI] [PubMed] [Google Scholar]
- Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- Oster S.K., Mao D.Y., Kennedy J., Penn L.Z. Functional analysis of the N-terminal domain of the Myc oncoprotein. Oncogene. 2003;22:1998–2010. doi: 10.1038/sj.onc.1206228. [DOI] [PubMed] [Google Scholar]
- Pathan S.A., Iqbal Z., Zaidi S.M., Talegaonkar S., Vohra D., Jain G.K., Azeem A., Jain N., Lalani J.R., Khar R.K. CNS drug delivery systems: novel approaches. Recent Pat. Drug Deliv. Formul. 2009;3:71–89. doi: 10.2174/187221109787158355. [DOI] [PubMed] [Google Scholar]
- Pollock K., Stroemer P., Patel S., Stevanato L., Hope A., Miljan E., Dong Z., Hodges H., Price J., Sinden J.D. A conditionally immortal clonal stem cell line from human cortical neuroepithelium for the treatment of ischemic stroke. Exp. Neurol. 2006;199:143–155. doi: 10.1016/j.expneurol.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Saldanha A.J. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sofroniew M.V., Vinters H.V. Astrocytes: biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroemer P., Patel S., Hope A., Oliveira C., Pollock K., Sinden J. The neural stem cell line CTX0E03 promotes behavioral recovery and endogenous neurogenesis after experimental stroke in a dose-dependent fashion. Neurorehabil. Neural Repair. 2009;23:895–909. doi: 10.1177/1545968309335978. [DOI] [PubMed] [Google Scholar]
- Tang H., Sha H., Sun H., Wu X., Xie L., Wang P., Xu C., Larsen C., Zhang H.L., Gong Y. Tracking induced pluripotent stem cells-derived neural stem cells in the central nervous system of rats and monkeys. Cell Reprogram. 2013;15:435–442. doi: 10.1089/cell.2012.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu W.B., Helander S., Pilstal R., Hickman K.A., Lourenco C., Jurisica I., Raught B., Wallner B., Sunnerhagen M., Penn L.Z. Myc and its interactors take shape. Biochim. Biophys. Acta. 2015;1849:469–483. doi: 10.1016/j.bbagrm.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Umeda K., Saida S., Yamaguchi H., Okamoto S., Okamoto T., Kato I., Hiramatsu H., Imai T., Kodaira T., Heike T. Central nervous system recurrence of desmoplastic small round cell tumor following aggressive multimodal therapy: a case report. Oncol. Lett. 2016;11:856–860. doi: 10.3892/ol.2015.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A., Yamada Y., Yamanaka S. Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120292. doi: 10.1098/rstb.2012.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M., Stolt C.C. From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Wierdl M., Tsurkan L., Hyatt J.L., Edwards C.C., Hatfield M.J., Morton C.L., Houghton P.J., Danks M.K., Redinbo M.R., Potter P.M. An improved human carboxylesterase for enzyme/prodrug therapy with CPT-11. Cancer Gene Ther. 2008;15:183–192. doi: 10.1038/sj.cgt.7701112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.