Abstract
The THO/TREX complex consists of several conserved subunits and is required for mRNA export. In metazoans, THO/TREX binds a subset of mRNAs during RNA splicing, and facilitates their nuclear export. How THO/TREX selects RNA targets is, however, incompletely understood. In our recent study, we reported that THO is loaded onto Piwi-interacting RNA (piRNA) precursor transcripts independent of splicing, and facilitates convergent transcription in Drosophila ovary. The precursors are later processed into mature piRNAs, small noncoding RNAs that silence transposable elements (TEs). We observed that piRNAs originating from dual-strand clusters, where precursors are transcribed from both strands, were specifically affected by THO mutation. Analysis of THO-bound RNAs showed enrichment of dual-strand cluster transcripts. Interestingly, THO loading onto piRNA precursors was dependent on Cutoff (Cuff), which comprises the Rhino-Deadlock-Cutoff (RDC) complex that is recruited to dual-strand clusters by recognizing H3K9me3 and licenses convergent transcription from he cluster. We also found that THO mutation affected transcription from dual-strand clusters. Therefore, we concluded that THO/TREX is recruited to dual-strand piRNA clusters, independent of splicing events, via multi-protein interactions with chromatin structure. Then, it facilitates transcription likely by suppressing premature termination to ensure adequate expression of piRNA precursors. [BMB Reports 2016; 49(7): 355-356]
Keywords: Drosophila, Dual-strand piRNA cluster, piRNA, RDC (Rhino-Deadlock-Cutoff) complex, THO/TREX
TREX (transcription/export) is an evolutionarily conserved multi-protein complex. TREX can be sub-divided into THO and additional factors. THO consists of six subunits; three have homologs in yeast (Hpr1/Thoc1, Thoc2, and Thoc3/Tex1) and the other three are metazoan-specific (Thoc5/FMIP, Thoc6, and Thoc7). Two additional conserved proteins, Yra1/REF/Aly and Sub2/UAP56, comprise TREX with THO. In yeast, THO/TREX is recruited by interaction with RNA pol II and facilitates transcriptional elongation. THO/TREX is loaded on nascent transcripts during transcription, and then Yra1 brings Mex67p, which in turn transports the mRNA through nuclear pores into the cytoplasm. Interestingly, the function of metazoan THO/TREX is somewhat different from that of yeast. Although metazoan THO/TREX is also loaded onto nascent transcripts, it does not interact with RNA pol II; its loading is dependent on splicing. Cap-binding proteins (CBC20 and CBC80) and exon junction complexes facilitate THO/TREX recruitment onto mRNA. However, TREX can be loaded onto intron-less genes, such as Hsp70, in fly and human. It is, however, not fully understood how THO/TREX is loaded onto nascent transcripts in a splicing-independent manner. In our recent paper, we reported that THO/TREX could be loaded onto transcripts from distinct genomic loci called dual-strand piRNA clusters in Drosophila ovary, suggesting a novel splicing-independent mechanism of THO/TREX loading onto target RNAs.
piRNA is a class of small noncoding RNA that silences TEs in animal germlines. Unlike other small noncoding RNAs, most piRNAs originate from discrete genomic source loci called piRNA clusters. piRNA clusters can be divided into two types based on the orientation of their transcription. In uni-strand clusters, piRNA precursors are transcribed in only one direction whereas dual-strand clusters are transcribed convergently from both ends.
In the paper, we found that flies with defective THO shared several phenotypes with piRNA mutants, such as dorso-ventral patterning defect and genomic instability. To determine whether THO is involved in piRNA biogenesis, we conducted total RNA-seq of the ovaries from wild type and thoc5 mutant flies. We found no significant differences in the expression profiles of protein-coding genes. However, most TEs were upregulated in thoc5 mutant ovary; the de-repression was especially prominent in TEs that are expressed in germ cells.
Genome-wide analysis of small RNAs showed that loss of THO did not affect miRNA or endo-siRNA levels, while significantly decreasing the levels of piRNAs that are complementary to TEs expressed in germ cells. We further analyzed the genomic loci from which the piRNAs originated. piRNAs from dual-strand clusters but not uni-strands were significantly decreased in thoc5. This is reminiscent of some piRNA-related proteins (Cuff, a heterochromatin protein 1 family protein Rhino, and UAP56), loss of which decreases piRNA levels in a dual-strand-specific manner.
Next, we examined the function of THO in piRNA biogenesis by visualizing its subunits in ovary germ cells. We noted that Thoc5 co-localized in discrete nuclear foci with UAP56 and Rhino. Moreover, the localization of THO subunits (Thoc5, Thoc2, and Thoc7) and UAP56 was co-dependent. The localization of THO/TREX as nuclear foci suggested that the complexes were loaded onto dual-strand piRNA precursors. Therefore, we purified Thoc5 from fly ovary and analyzed co-purified RNA by deep sequencing. Consistent with piRNA expression changes, we observed specific enrichment of dual-strand precursor RNAs in Thoc5 pull-down. Uni-strand precursors were not enriched, and only a small subset of mRNAs, such as Hsp70, was enriched.
The THO/TREX localization to dual-strand piRNA precursors could not be explained by splicing-dependent loading because the precursor contains no intron. So, we determined whether THO/TREX localization was mediated by proteins that associate with chromatin markers present at the loci. Co-immunoprecipitation showed that Thoc5 interacts with Cuff, and immunofluorescence results suggested that Cuff and Rhino are required for Thoc5 localization. Notably, tethering of Cuff to a reporter RNA was sufficient to recruit THO to the reporter RNA.
The total RNA-seq results showed that thoc5 mutation reduced dual-strand piRNA precursor levels, suggesting that THO/TREX is necessary for precursor transcription. To determine whether THO/TREX is required for transcription, we assayed nascent transcript levels by chromatin-associated RNA-seq, and found that dual-strand cluster transcripts were indeed decreased in thoc5. RNA-FISH and RNA Pol II chromatin immunoprecipitation results further supported the decreased levels of dual-strand cluster transcripts in thoc5.
Together, the results suggest that THO/TREX is recruited to dual-strand piRNA clusters by chromatin-associated factors, and facilitates transcription. This could be a requirement for long transcripts from piRNA clusters, for which splicing should be suppressed. On the basis of these findings, we proposed a new splicing-independent loading model of THO/TREX in which the chromatin state is the key regulator (Fig. 1). The loading process is distinct from both previously reported processes: splicing-dependent loading in metazoans and RNA pol II-dependent loading in yeasts.
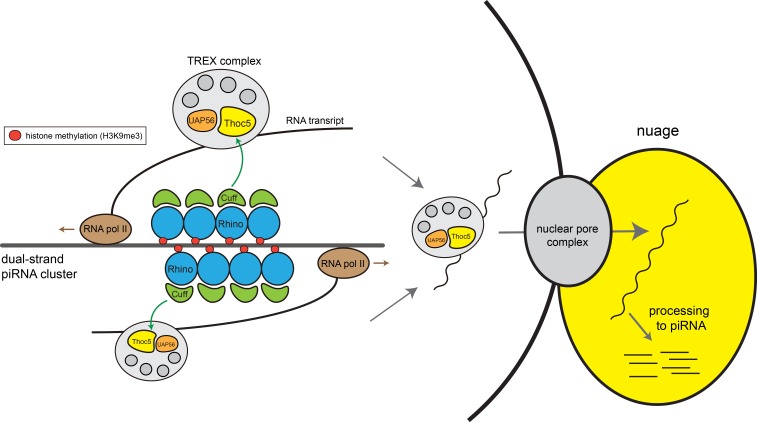
Fig. 1. A model of splicing-independent loading of THO/TREX onto nascent RNAs from a dual-strand piRNA cluster. The RDC complex, which is composed of Rhino, Deadlock (not shown in the figure) and Cuff, is recruited to dual-strand piRNA cluster by recognizing H3K9me3 marks, and facilitates convergent transcription. THO/TREX is loaded onto dual-strand precursor RNAs via interaction of Thoc5 with Cuff. The loading of THO/TREX not only facilitates efficient transcription of precursor RNAs but also mediates their export through nuclear pores into the nuage, a germ-cell specific cytoplasmic structure where piRNA precursors are processed into mature piRNAs.
The model immediately raises a number of questions. First, THO/TREX is recruited by Cuff, which comprises the RDC complex with Rhi to read heterochromatin histone modification, H3K9me3. Usually H3K9me3 marks silenced genes and TEs, yet active transcription of the dual-strand piRNA cluster requires the repressive histone modification. Further studies may elucidate how THO/TREX discriminates dual-strand piRNA cluster from other epigentically repressed genetic loci. Second, THO/TREX is ubiquitously expressed, while the RDC complex is ovarian germline-specific. It would be interesting to investigate whether splicing-independent loading of THO/TREX also occurs in other tissues in which RDC is not expressed. Specifically, testis has been shown to be affected by loss of THO/TREX function. Further investigation is required to understand how THO/TREX modulates gene and TE expression in testes. Third, as TREX may be a more dynamic complex than THO, THO and TREX may play distinct roles by regulating different subsets of genes. Fourth, recent studies showed that piRNAs are also expressed in somatic cells and are proposed modulators of global gene expression profile. Computational analysis of piRNAs matched to genes would provide more information on somatic non-coding small RNAs.
Acknowledgments
This work was supported by grants from the Basic Science Research Program through the National Research Foundation of Korea (NRF) (no. 2015R1A2A2A01003598) to YDC.