Abstract
The DNA-binding with one zinc finger proteins (Dofs) are a plant-specific family of transcription factors. The Dofs are involved in a variety of biological processes such as phytohormone production, seed development, and environmental adaptation. Dofs have been previously identified in several plants, but not in pepper. We identified 33 putative Dof genes in pepper (CaDofs). To gain an overview of the CaDofs, we analyzed phylogenetic relationships, protein motifs, and evolutionary history. We divided the 33 CaDofs, containing 25 motifs, into four major groups distributed on eight chromosomes. We discovered an expansion of the CaDofs dated to a recent duplication event. Segmental duplication that occurred before the speciation of the Solanaceae lineages was predominant among the CaDofs. The global gene-expression profiling of the CaDofs by RNA-seq analysis showed distinct temporal and pathogen-specific variation during development and response to biotic stresses (two TMV strains, PepMoV, and Phytophthora capsici), suggesting functional diversity among the CaDofs. These results will provide the useful clues into the responses of Dofs in biotic stresses and promote a better understanding of their multiple function in pepper and other species.
Plants constantly encounter biotic and abiotic stresses. Biotic stresses cause serious reduction in crop production and quality. To overcome biotic stresses, plants have various strategies involving the perception of pathogen attacks and the regulation of signaling networks1,2,3. Transcription factors (TFs) are vital proteins that control cellular processes by regulating the expression of downstream target genes through the recognition of specific DNA sequence elements in promoter regions4. TFs also play roles in plant innate immunity5. Multitudinous TF families exist in plants, and more than 60 TF families have been identified by bioinformatics analysis and manual study of plant genomes6. Several plant-specific TF families have been identified, including the DNA-binding with one zinc finger proteins (Dofs).
The Dofs play key roles in many biological process such as seed germination, dormancy, photosynthesis, flowering, light-regulated gene expression, guard cell-specific gene expression, and stress response4,7,8. The Dofs generally have 200–400 amino acids with a highly conserved 52 amino-acid Dof domain at the N-terminal end and a variable domain for transcriptional regulation at the C-terminal end. The Dof domain, is a DNA-binding domain that contains a C2C2-type zinc finger motif that recognizes a cis-regulatory element with a common core sequence of 5′-(A/T)AAAG -3′, except a pumpkin Dof protein designated as AOBP that recognizes an 5′-AGTA-3′ sequence4,7,8. The Dof domain is bifunctional, showing both DNA-binding and protein-protein interaction activities8,9,10,11,12. The variable domain at the Dof C-terminus might be involved in the diverse functions of distinct Dofs via interactions with different regulatory proteins and gene promoters13,14.
Since the first Dof was identified in maize15, many putative Dof genes (Dofs) have been reported in a variety of plant species, such as the 36 Dofs in Arabidopsis16, 30 Dofs in rice16, 41 Dofs in poplar17, 31 Dofs in wheat18, 28 Dofs in sorghum19, 78 Dofs in soybean20, 34 Dofs in tomato21, and 76 Dofs in Chinese cabbage22. Along with the rapid expansion and completion of genomic sequencing, more Dofs in various plants will be identified and could promote a better understanding of Dofs function.
Much research has suggested that Dofs have multiple roles such as plant-specific physiological process, growth, development, and photosynthesis. In maize, Dof1 and 2 are involved in gene expression for carbon metabolism23. ZmDof1/MNB1a regulates the PEPC gene in association with light response and nitrogen assimilation24. In Arabidopsis, DAG1 and DAG2 are involved in seed germination25,26, and CDF1, 2, 3, and 5 are associated with the photoperiodic control of flowering27,28. COG1 and OBP3 regulate phytochrome signaling29,30. In rice, OsDof3, OsDof12, and RPBF are involved in gibberellin-regulated gene expression, the photoperiodic control of flowering, and the regulation of seed-storage protein, respectively9,31,32,33. In potatoes, StDof1 regulates the KST1 gene in association with guard-cell specificity34. In soybean, GmDof4 and GmDof11 are associated with lipid biosynthesis35. Some Dofs are involved in responses to abiotic and biotic stresses. SlCDF1 and SlCDF3 in tomato regulate the COR15, RD29A, and RD10 genes in association with drought and salt tolerances36. TaDof14 and TaDof15 in wheat were significantly up-regulated under drought conditions18. But, reports of Dof family involvement in responses biotic stress have been very limited.
Pepper, a member of Solanaceae plants such as tomato, potato and tobacco, comprise large portion of crops in the world as major vegetable for most global cuisines with pungency, and are exposed to various devastating diseases by Tobacco mosaic virus (TMV), Pepper mottle virus (PepMov), Tomato spotted wilt virus (TSWV), Phytophthora capsici, Collectricum spp. and so on37,38,39. These biotic stresses cause serious reduction in yield and quality. These days, several efforts to clone resistance genes or candidates were reported to overcome pathogen attacks in pepper, but the defense-response mechanism and network still remain unknown. Understanding diverse roles of Dof family in plant biological process could be feasible approach to dissect defense response in biotic stresses and improve crop disease resistance or tolerance. However, in pepper, the function of the Dof family is still largely unknown. To get a comprehensive understanding of Dofs in pepper, a genome-wide identification and expression analysis of the Dof family was performed in the present study. Subsequently, we performed a detailed analysis of gene structures, conserved motifs, duplication status, chromosomal distribution, and phylogenic relationships to explore the evolutionary history of Dof expansion in pepper. To determine the probable functions of the pepper Dofs (CaDofs), we evaluated expression profiles under three pathogen challenges [Tobacco mosaic virus (TMV), Pepper mottle virus (PepMoV), and Phytophthora capsici] to understand the response of CaDofs to biotic stresses using RNA-seq data. The results will provide novel insights into the responses of CaDofs against pathogens and could promote a better understanding Dof function in other species.
Results
Identification of the Dof gene family in pepper
We identified 33 putative CaDofs from the Capsicum annuum ‘CM334’ (hereafter ‘CM334’) genome. Additionally, we obtained 21 putative CaDofs from the Plant Transcription Factor Database (Plant TFDB). We aligned the 33 putative CaDofs from the ‘CM334’ genome with 21 corresponding genes from the Plant TFDB. All of the candidate CaDofs from the Plant TFDB were matched to corresponding genes among the 33 putative CaDofs. Therefore, we used only the 33 CaDofs from C. annuum ‘CM334’ for further investigation. To verify the sequences of the CaDofs, we aligned all of the CaDofs with corresponding transcripts or genes from the ‘CM334’ RNA-seq40, pepper EST41, and C. annuum ‘Zunla-1’ CDS42 databases and manually corrected. We designated the generated CaDofs as CaDof01–33 according to their locations on the chromosomes. The number of Dofs in the pepper genome was similar to those in Arabidopsis (36 Dofs), rice (30 Dofs) and tomato (34 Dofs)16,21. All of the CaDofs contained the highly conserved Dof domain including the zinc finger motif (Supplementary Fig. S1). The conserved Dof domain consists of 52 basic residues located in the N-terminal region. Twenty-six of the 52 residues were perfectly conserved (100% conserved) in all 33 CaDofs. The Dof domains were also highly conserved among the Dofs of other plants, such as Arabidopsis, rice, and tomato (Supplementary Fig. S2). Information about the CaDofs; such as the gene ID, position on the chromosomes, number of nucleotides and amino acids, isoelectric point (pI), and molecular weight (MW); is presented in Table 1. The size of the coding sequence (CDS) of the CaDofs ranged from 525 bp to 1,512 bp, with deduced polypeptide sizes ranging from 173 amino acids to 503 amino acids. The pIs of the deduced CaDofs ranged from 3.61 to 9.79, and the MWs ranged from 15.8 kD to 54.27 kD.
Table 1. The Dof families and related information of each Dof gene in pepper.
| Gene name | Gene ID | chromosome or scaffold | ORF length (bp) | No. of aa | pI | MW (kD) |
|---|---|---|---|---|---|---|
| CaDof01 | CA01g00490 | 1 | 864 | 287 | 8.68 | 32.08 |
| CaDof02 | CA02g01120 | 2 | 1413 | 470 | 5.01 | 51.57 |
| CaDof03 | CA02g01550 | 2 | 579 | 192 | 9.17 | 20.82 |
| CaDof04 | aCapana02g001770 | 2 | 525 | 174 | 8.81 | 19.71 |
| CaDof05 | aCapana02g001919 | 2 | 924 | 307 | 9.71 | 32.95 |
| CaDof06 | CA02g15190 | 2 | 1065 | 354 | 8.75 | 38.45 |
| CaDof07 | CA02g15750 | 2 | 834 | 277 | 5.29 | 31.35 |
| CaDof08 | CA02g25910 | 2 | 864 | 287 | 9.01 | 31.04 |
| CaDof09 | CA02g25980 | 2 | 714 | 237 | 8.98 | 25.03 |
| CaDof10 | aCapana02g003361 | 2 | 1338 | 445 | 7.05 | 48.9 |
| CaDof11 | aCapana03g001966 | 3 | 801 | 266 | 8.55 | 28.84 |
| CaDof12 | CA03g26920 | 3 | 969 | 322 | 3.61 | 35.67 |
| CaDof13 | CA03g29970 | 3 | 1512 | 503 | 5.77 | 54.27 |
| CaDof14 | bKS14005F06 | 4 | 441 | 146 | 8.94 | 15.8 |
| CaDof15 | CA05g08190 | 5 | 1410 | 469 | 6.28 | 51.46 |
| CaDof16 | CA05g18640 | 5 | 702 | 233 | 9.79 | 26.77 |
| CaDof17 | CA06g16390 | 6 | 618 | 205 | 6.57 | 22.58 |
| CaDof18 | CA06g18250 | 6 | 933 | 310 | 5.47 | 33.59 |
| CaDof19 | CA06g18840 | 6 | 1449 | 482 | 5.81 | 52.54 |
| CaDof20 | CA06g19670 | 6 | 924 | 307 | 6.58 | 33.9 |
| CaDof21 | CA06g23590 | 6 | 915 | 304 | 6.78 | 33.78 |
| CaDof22 | CA06g24550 | 6 | 930 | 309 | 9.02 | 33.43 |
| CaDof23 | CA10g08680 | 10 | 972 | 323 | 5.93 | 36.09 |
| CaDof24 | CA10g08690 | 10 | 981 | 326 | 5.89 | 36.44 |
| CaDof25 | CA10g11420 | 10 | 1098 | 365 | 9.11 | 39.69 |
| CaDof26 | CA11g04250 | 11 | 1308 | 435 | 6.54 | 48.33 |
| CaDof27 | CA00g45170 | scaffold1106 | 948 | 315 | 6.63 | 35.38 |
| CaDof28 | CA00g57880 | scaffold1313 | 1008 | 335 | 9.44 | 35.76 |
| CaDof29 | CA00g64610 | scaffold1415 | 906 | 301 | 9.25 | 32.77 |
| CaDof30 | CA00g78480 | scaffold1668 | 642 | 213 | 9.12 | 22.87 |
| CaDof31 | CA00g84150 | scaffold1807 | 828 | 275 | 4.42 | 31.06 |
| CaDof32 | aCapana12g002748 | scaffold1813 | 1113 | 370 | 6.49 | 40.53 |
| CaDof33 | CA00g96300 | scaffold4404 | 648 | 215 | 6.62 | 24 |
ORF, open reading frame; aa, amino acids; pI, isoelectric point; MW, molecular weight.
aGenes are from ‘Zunla-1’ genome.
bGenes are from pepper EST.
Others without marks (a or b) in Gene ID panel are from ‘CM334’ genome.
Phylogenetic relationships of the Dof family in pepper and tomato plants
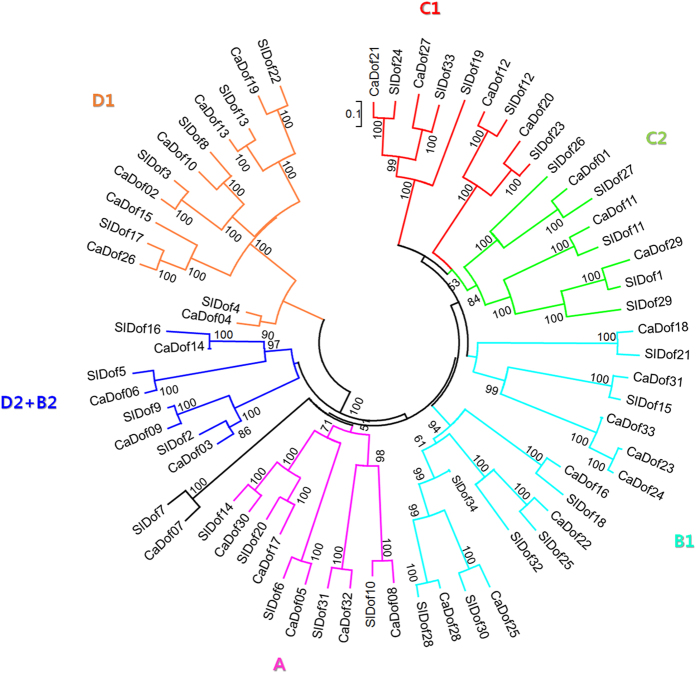
To examine the phylogenetic relationships between the Dof families in pepper and tomato, we performed multiple sequence alignment of the 33 CaDofs and 34 tomato Dofs (SlDofs) using ClustalW. We constructed an unrooted phylogenetic tree using the neighbor-joining method with alignment of the full-length Dofs from tomato and pepper. Based on the resulting phylogenetic tree, we categorized the CaDofs and SlDofs into four major groups (A, B, C, and D) and seven subgroups (Fig. 1 and Supplementary Fig. S3). We named the pepper groups based on the corresponding groups in tomato21. Groups B and C were the largest major groups, each comprising 17 genes (accounting for 25.3%) in pepper and tomato. Groups B and C were divided into two subgroups: B1 and B2 and C1 and C2, respectively. Subgroup B1, which consisted of nine CaDofs and eight SlDofs, was the largest subgroup in both pepper and tomato. Group A and subgroups C1, C2, and D1 contained 10, 9, 8, and 13 members, respectively, from the two plants. Subgroups B2 and D2 were not clearly separated and clustered into the same subgroup with four CaDofs and four SlDofs. Most of the CaDofs were clustered with SlDofs, and each subgroup had a high bootstrap value, suggesting that the CaDofs have a close phylogenetic relationship with that of the SlDofs.
Figure 1. Phylogenetic tree of pepper and tomato Dof genes.
The full length amino acid sequences of CaDofs and SlDofs were aligned by ClustalW. The Neighbor-Joining phylogenetic tree with 1,000 bootstrap replicates was constructed using 33 pepper Dof proteins and 34 tomato Dof proteins. Each specific color represents each Dof subclass.
Identification of conserved motifs and analysis of gene structures among the CaDofs
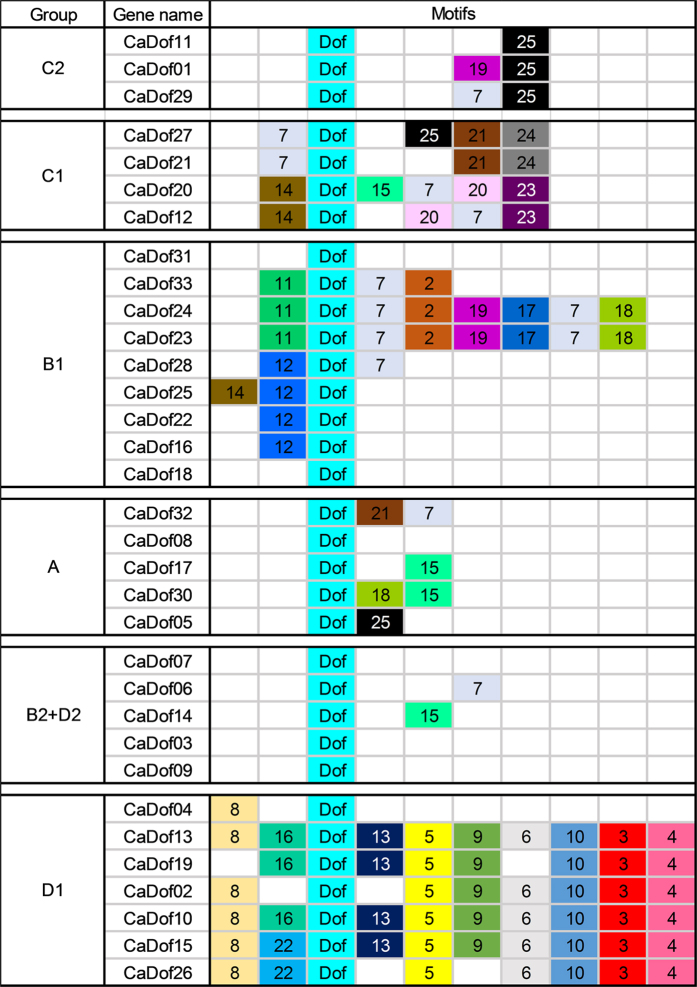
To identify the diversity and conservation among the CaDofs, we investigated the conserved motifs using the MEME suite (http://meme-suite.org/tools/meme). A total of 25 distinct motifs were predicted among all 33 CaDofs (Fig. 2). The size of the identified motifs ranged from 8 amino acids to 113 amino acids. The consensus sequences of the motifs are presented in Supplementary Table S1. The motifs of closely related genes within each group in the phylogenetic tree shared common sequences and positions (Fig. 2). Most of the subgroups included several specific motifs in the C-terminal regions. Motif 1 was uniformly found in all 33 CaDofs and represented the pepper Dof domain (Fig. 2 and Supplementary Table S1). Group A and subgroups B2 and D2 represent the simplest composition of motifs, with no specific motifs except for the Dof domain. Although subgroup B1 had several motifs (motifs 2, 7, 17, 18, and 19) at the C-terminal regions, it appeared to be characterized by motifs 11 and 12 at the N-terminal region. One CaDof (CaDof31) in subgroup B1 did not contain motifs 11 and 12. Subgroup C1 contained four specific motifs on both sides of the Dof domain: motifs 7 and 14 in the N-terminal region and motifs 24 and 23 in the C-terminal region. Subgroup C2 contained one specific motif (motif 25). Subgroup D1, which showed the most diverse composition of motifs among the subgroups, was characterized by five motifs (motifs 3, 4, 5, 8, and 10). Motif 8 was located in the N-terminal region in all of the D1 members except for CaDof19. Conserved motifs 3, 4, 5, and 10 at the C-terminus were detected in seven of the eight Dofs in subgroup D1. CaDof04 in subgroup D1 contained only two motifs (motif 8 and the Dof domain), possibly because it had a shorter gene sequence than the other D1 members.
Figure 2. Schematic diagram of the conserved motifs in pepper 33 Dof genes.
Motifs were identified by MEME program. Each number with colored box means each motif. Light blue boxes labeled Dof represent motif 1. The relative position of each motif in all Dof proteins is shown. Consensus sequence of the defined motifs by MEME are listed in Table S1.
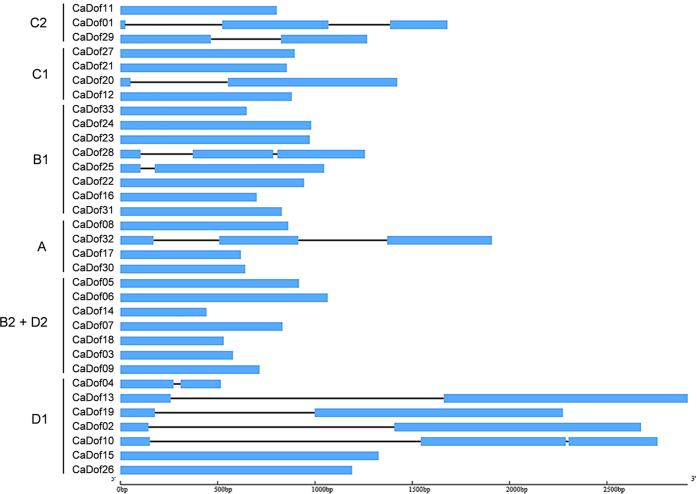
In order to obtain further insight into the structural diversity of the CaDofs, we analyzed the exon-intron organization. As shown in Fig. 3, 22 of the CaDofs had no introns, whereas seven and four CaDofs had one and two introns, respectively. Those exon-intron structures were similar to those of the Dofs in Arabidopsis, rice, and tomato16,21. Most of the introns were generally located upstream of the Dof domain; only three genes (CaDof04, 29, and32) contained introns downstream of the Dof domain. Most of the genes within a given group showed similar organizations of exons and introns. Most of the genes from subgroup D1 contained one intron, and the intron sizes and positions were comparable among the genes in D1. Subgroups B2 and D2 contained no introns. The structures of the CaDofs in group A and subgroups B1, C1, and C2 were more variable than those in subgroups D1, B2, and D2. The distribution of motifs together with the gene structure within each group indicated the phylogenetic relationships among the CaDofs. Those results indicate that the conservation and specificity of gene characters among the subgroups could be related to functional diversity among the Dofs.
Figure 3. Exon and intron structure of Dof genes from pepper.
Blue boxes and black lines represent exons and introns, respectively. The scale bar on the bottom line indicates size of genes.
Chromosomal location and gene duplication of CaDofs
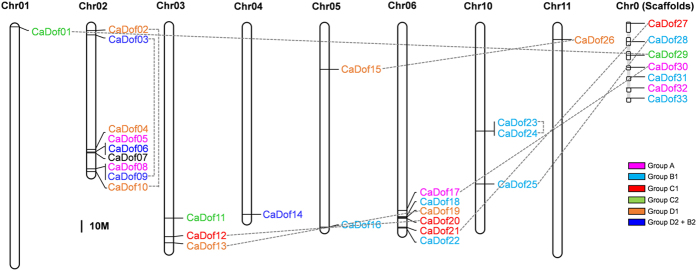
To determine the genomic localization of the CaDofs, we aligned each CaDof as query sequence against the pepper genome database using BLAST. The positions of the CaDofs on the chromosomes had an uneven distribution. Twenty-six genes were located on 8 of the 12 chromosomes, with no genes mapping to chromosomes 7, 8, 9, and 12 (Fig. 4). Nine CaDofs were mapped to chromosome 2. Among them, CaDof05, 06, and 07 and CaDof08 and 09 were clustered together, respectively. Chromosome 6 included six CaDofs, which were clustered in two groups (CaDof18, 19, and 20 and CaDof21 and 22, respectively) on the end of the long arm. Chromosomes 3 and 10 each contained three genes, and CaDof23 and 24 were clustered together. Chromosome 5 contained two genes. Chromosomes 1, 4 and 11 each contained only one gene. Seven of the 33 CaDofs (CaDof27–33) were assigned to chromosome 0 that means unassigned pepper sequences to any of the 12 pepper chromosomes.
Figure 4. Chromosomal locations and duplication events of CaDof genes in pepper.
The locations of CaDofs were based on physical locations. The four chromosomes without CaDofs (chromosome 7, 8, 9, and 12) were excluded in this figure. The numbers on the top indicate each chromosome number. The chromosome 0 (Chr0) means seven different scaffolds containing unassigned CaDofs to any of the 12 pepper chromosomes. Dotted lines indicate 10 pairs of paralogous gene of duplication events. CaDofs have been colored according to their groups of phylogenetic tree. Scale bar represents a 10 Mb distance of chromosome.
To detect potential duplications of the CaDofs, we identified 10 putative pairs of CaDof paralogs based on the chromosomal localization and the neighbor-joining tree (Figs 1 and 4). Each pair of paralogs shared high sequence similarity. The identified paralogs are connected by the lines shown in Fig. 4. Seven of the paralog pairs were putative segmental duplication events, and three pairs were distributed at the same chromosome level. Only one putative pair (CaDof23 and 24) was possibly the result of tandem duplication. Overall, those results suggest that segmental duplication predominated in the expansion of the CaDof family in peppers, although tandem duplication was also involved.
In order to estimate the dates of the duplication events, we estimated the nonsynonymous substitution rate (Ka), the synonymous substitution rate (Ks), and the Ka/Ks ratio. The approximate dates of the duplication events are presented in Table 2. The nine segmental duplications (Fig. 4) of the CaDofs took place from 194.48 million years ago (Mya) to 37.06 Mya, and one tandem duplication (Fig. 4) occurred approximately 1.45 Mya. With the speciation data between pepper and tomato around 19.2 Mya30, all the segmental duplications occurred before the pepper/tomato split, which is also supported by the phylogenic tree shown in Fig. 1. The Ka/Ks ratios of five segmental duplication pairs and the tandem duplication pair were >0.3, suggesting that significant functional divergence among those CaDofs might have occurred after the duplication events. The Ka/Ks ratios of all the duplicate pairs were <1.0, indicating that the CaDofs evolved under negative selection acting against protein-coding changes.
Table 2. Duplicated Dof members and the dates of the duplication times in pepper.
| Gene1 | Gene2 | aKs | Ka | Ka/Ks | bDate(Mya) |
|---|---|---|---|---|---|
| CaDof01 | CaDof29 | 1.17 | 0.50 | 0.43 | 96.30 |
| CaDof02 | CaDof10 | 0.56 | 0.17 | 0.29 | 46.29 |
| CaDof03 | CaDof09 | 1.34 | 0.40 | 0.30 | 109.67 |
| CaDof12 | CaDof20 | 2.37 | 0.25 | 0.10 | 194.48 |
| CaDof13 | CaDof19 | 0.45 | 0.15 | 0.34 | 37.06 |
| CaDof15 | CaDof26 | 0.52 | 0.10 | 0.20 | 42.73 |
| CaDof17 | CaDof30 | 0.95 | 0.36 | 0.38 | 78.24 |
| CaDof21 | CaDof27 | 1.07 | 0.37 | 0.35 | 87.36 |
| CaDof23 | CaDof24 | 0.02 | 0.01 | 0.41 | 1.45 |
| CaDof25 | CaDof28 | 0.96 | 0.37 | 0.38 | 78.89 |
aKa and Ks were calculated using the program K-Estimator 6.1 v.
bThe duplication time was estimated according to formula: T = Ks/2λ. The clock like rate (λ) was 6.1 × 10−9 subsitutions per site per year52. Mya, million years ago.
Expression patterns of CaDofs in various developmental stages
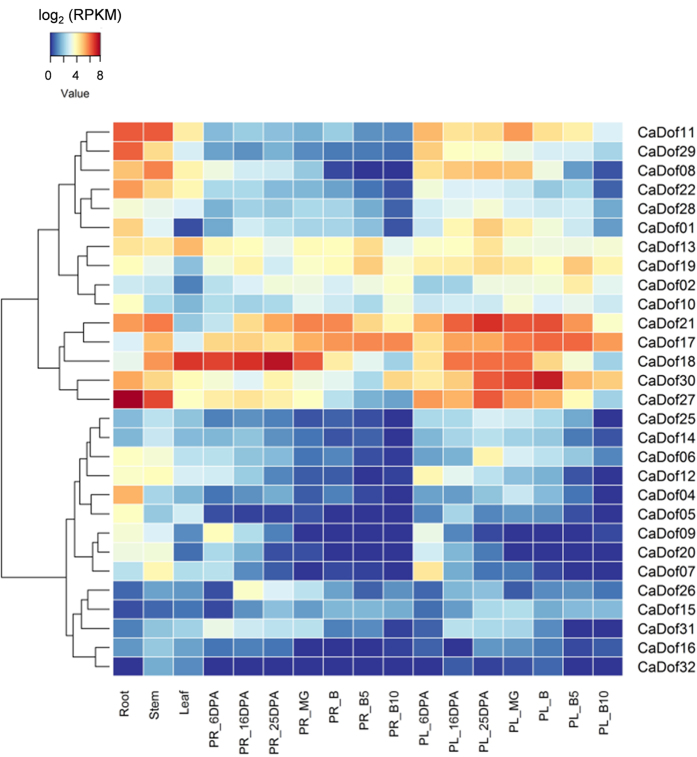
To investigate the involvement of the CaDofs in pepper growth and development, we generated a heat map of the global transcription patterns of the CaDofs based on publicly available RNA-seq data for five tissues (root, stem, leaf, pericarp, and placenta), including seven developmental stages of the pericarp and placenta (Fig. 5). The expression profiles of each CaDof revealed various patterns in different organs and stages. CaDof13, 17, 18, 19, 21, 27, and 30 were expressed at relatively high levels, while CaDof14, 15, 16, 25, 31, and 32 were expressed at relatively low levels in all the tested tissues. The expression levels of CaDof11, 29, and 08 were high in the root and stem and higher in the placenta from 6 days post anthesis (PL_6DPA) to the mature green (MG) stage compared with those in the placenta at other stages, but they were low in the pericarp at all stages. CaDof22 was expressed at higher levels in the root and stem than in the other tissues. Although CaDof21 was expressed at relatively high levels in all the tested tissues, the expression levels in the placenta at several stages [PL_16 and 25DPA, MG, and breaker (B)] were higher than those in the other tissues. In comparison, CaDof30 was also highly expressed in the placenta, especially at the B stage. CaDof18 was constitutively expressed in most of the tested tissues, with especially high expression in some PR stages (PR 6, 16, and 25DPA and MG). CaDof27 showed the highest expression level in the roots among all the CaDofs. CaDof04 was expressed at a high level only in roots and was undetectable in the other organs. Taken together, those results indicate that the CaDofs are possibly involved in biological functions during plant development.
Figure 5. Tissue specific expression analysis of CaDofs.
The heat map was constructed by RNA-seq from C. annuum ‘CM334’. The 29 CaDof genes were used to construct heat map. The 4 CaDof genes (CaDof03, 23, 24, and 33), which are absent data, were excluded. Blue and red colors represent relatively low and high expression (log2 RPKM value), respectively. PR, pericarp; PL, placenta; MG, mature green stage; B, breaker stage; DPA, days post-anthesis; B5 and 10, 5 and 10 days post-breaker.
Responses of CaDofs to TMV, PepMoV, and P. capsici pathogen challenges
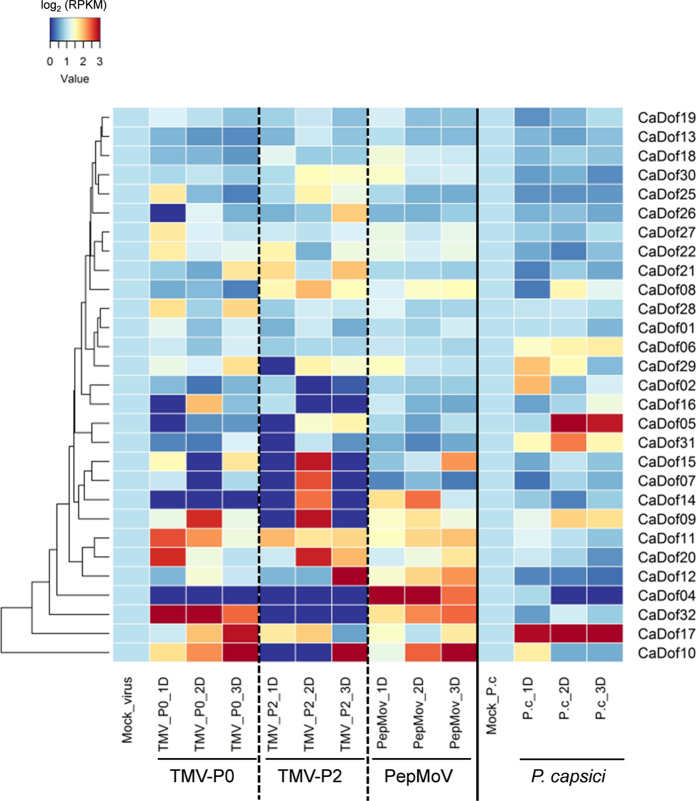
‘CM334’ is well known as a powerful resource for the development of resistant lines against several viruses and P. capsici37,38,39. To examine whether CaDofs are involved in responses to biotic stresses in pepper, we analyzed the expression profiles of the CaDofs using RNA-seq data from ‘CM334’ challenged with TMV Pathotype 0 (TMV-P0), TMV-P2, PepMoV, and P. capsici (Supplementary Table S2). ‘CM334’ is resistant to TMV-P0, PepMoV, and P. capsici but susceptible to TMV-P2. We inoculated ‘CM334’ leaves with TMV-P0, TMV-P2, PepMoV, and P. capsici and then sampled at 0, 1, 2, and 3 days post inoculation. We used the leaf samples to construct RNA-seq libraries, which we subsequently sequenced. We then generated heat maps of the transcript profiles of the CaDofs with each gene expression value normalized by each control sample.
The expression patterns of the CaDofs were differentially regulated in the resistant and susceptible responses against TMV-P0 and TMV-P2, respectively (Fig. 6). CaDof32 was highly up-regulated only in the resistant response against TMV-P0, while the expression levels of CaDof07, 12, 14, and 15 were increased in the susceptible response against TMV-P2. Some of the CaDofs, such as CaDof9, 10, 11, 17, and 20, were up-regulated in both responses but showed temporally different expression patterns. Among those, CaDof10 showed earlier and higher levels of expression in the resistant response. After PepMoV inoculation, some CaDofs were up-regulated, as in the responses to TMV-P0 and TMV-P2, but showed different patterns, such as those of CaDof09, 10, 11, 20, and 32 (Fig. 6). CaDof04 showed earlier and higher levels of expression after PepMoV inoculation but did not show significant changes after other pathogen inoculations. Other CaDofs showed no detectable changes compared with controls and among treatments. In response to P. capsici inoculation, nine CaDofs were expressed at higher levels compared with the controls (Fig. 6). Among those, CaDof17 showed higher expression levels compared with those in control and the other viruses treatments. CaDof04 was significantly down-regulated. In addition, two genes (CaDof05 and 31) showed particularly strong expression levels after P. capsici inoculation compared with those after the other treatments. Overall, the expression profiles in response to the biotic stresses suggest diversity and conservation of the biological functions of CaDofs during plant-microbe interactions.
Figure 6. Biotic stress specific expression analysis of CaDofs.
The heat map was constructed by RNA-seq from three viruses (TMV-P0, TMV-P2 and PepMoV) and Phytophthora capsici inoculated C. annuum ‘CM334’. ‘CM334’ is resistant to TMV-P0, PepMoV, and P. capsici but susceptible to TMV-P2.The 29 CaDof genes were used to construct heat map. The 4 CaDof genes (CaDof03, 23, 24, and 33), which are absent data, were excluded. For expression pattern after virus inoculation, mean of RPKM value in triplicate data sets were used. This data was normalized by control samples (Mock_virus and Mock_P.c). Blue and red colors represent relatively low and high expression (log2 RPKM value), respectively. P.c, Phytophthora capsici; D, days post inoculation.
Discussion
The Dofs are a major group of plant-specific TFs involved in diverse functions4,8. Several studies have addressed the genome-wide identification, function, and evolution of Dofs in Arabidopsis, rice16, poplar17, algae, moss43, wheat18, sorghum19, soybean20, Chinese cabbage22, and tomato21. No previous study has reported the genome-wide identification and transcriptional profiles in response to biotic stresses of Dofs in pepper, an agriculturally important member of the Solanaceae. We identified a total of 33 CaDofs based on two pepper genomes, C. annuum ‘CM334’ and C. annuum ‘Zunla-1’. The number of CaDofs identified was similar to those in other plants, such as the 36 AtDofs in Arabidopsis, the 30 OsDofs in rice16, and the 34 SlDofs in tomato21. In addition, the number of reported Dofs in many plant species ranges from 20 to 50 genes according to information from the Plant TFDB. Taken together, the numbers of Dofs among plant species appear to be regular, regardless of genome size. Consistent with previous results from other plants16,21, the CaDofs were divided into four major groups and seven subgroups based on a neighbor-joining phylogenetic tree. Phylogenetic analysis of the Dofs in pepper and tomato revealed particular clusters of paralogous and orthologous gene pairs within each subgroup supported by high bootstrap values (Fig. 1 and Supplementary Fig. S3). Although the clade patterns are consistent with previous results from Arabidopsis, rice, and tomato, the number of duplicated genes within some groups of Dofs in pepper were different from those in the other three species. Group B contained the most genes in pepper, whereas group C contains the most genes in Arabidopsis and tomato. The numbers of genes in group A and subgroup D1 in pepper were the same as those in tomato. Previous studies indicated that orthologous Dofs in the same clades between monocots and dicots might be derived from common ancestors that evolved before the monocot/dicot split16,21. Taken together, the results suggest that there were unequal gene duplication events in each clade of the Dof family among species, leading to expansion and diversity of the Dof repertoire during the course of evolution.
Structural analysis showed that the CaDofs contained few introns, with intron numbers ranging from zero to two per gene (Fig. 3). Most of the genes had no introns, and those genes all clustered into the same groups. The lengths of the introns within the genes showed similar patterns within the same groups. Similar characteristics of intron/exon structure were observed in Arabidopsis, rice, and tomato16,21, suggesting evolutionary conservation of the Dofs. The chromosomal locations of the CaDofs showed that the CaDofs were located on 8 of the 12 pepper chromosomes, with chromosomes 7, 8, 9, and 12 containing no CaDofs. Chromosome 2 contained nine CaDofs, which was the highest number among the chromosomes. Likewise, chromosomes 7 and 12 in tomato lack Dofs, and the highest number of Dofs (nine SlDofs) in tomato is found on chromosome 2. The chromosomal locations of the orthologous gene pairs between pepper and tomato were compared based on a phylogenetic tree. That comparative analysis revealed that most of the orthologous gene pairs were syntenic between the two plant species. Those results are consistent with previous reports of conserved locations of homologous genes on chromosomes between species40,44. Hence, we assumed that the locations of the seven unassigned CaDofs (CaDof27, 28, 29, 30, 31, 32, and 33) might correspond to syntenic positions of orthologous SlDofs. For instance, CaDof31, which is orthologous to SlDof14, might be located on chromosome 3 in pepper.
The motif analysis of CaDofs revealed that the CaDofs shared conserved motifs in the same subgroup. For example, C1 has specific motifs 7, and 14, whereas C2 contains motif 25. We also found specific motifs in other subgroups: motif 11 and 12 in B1, and motif 3, 4, 5, 8, and 10 in D1 (Fig. 2). The distribution of motifs also showed conserved position of each gene in the same subgroup. These conserved motifs of Dof genes indicated that such structures have been preserved by evolution suggesting that these motifs might play important roles in subgroup functions.
Tandem duplication, segmental duplication, and transposition are major evolutionary mechanisms of gene-family expansion45. In plants, segmental duplication occurs more frequently than tandem duplication and transposition, because most plants are diploidized polyploids and retain numerous duplicated chromosomal blocks within their genomes45. We obtained 10 pairs of paralogs in the pepper Dof family based on chromosomal distribution, phylogenetic analysis, and sequence similarity. One pair of paralogs (CaDof23 and 24) was the result of a putative tandem duplication event. Among the remaining nine pairs, two pairs (CaDof02 and 10 and CaDof03 and 09) were preferentially retained duplicates located within a segmental duplication of a single chromosome, whereas seven pairs (CaDof01 and 29, CaDof12 and 20, CaDof13 and 19, CaDof15 and 26, CaDof17 and 30, CaDof21 and 27, and CaDof25 and 28) were involved in segmental duplications on different chromosomes. Those results indicate that the Dof family in pepper might have evolved mainly via segmental duplications.
We calculated the duplication times of the paralog pairs in the pepper Dof family using the Ks value. The monocots/dicot divergence was estimated to occur around 170–235 Mya46. The Arabidopsis/tomato and pepper/tomato divergences were estimated to occur around 110–130 Mya and 19.2–20 Mya, respectively40,44. We estimated that the duplications of nine of the paralogous Dof sets in pepper occurred from 194.48 (Ks = 2.37) to 37.06 (Ks = 0.45) Mya and that of one the paralogous gene sets (CaDof23 and 24) occurred around 1.45 Mya (Table 2). Therefore, the duplication of most of the CaDofs occurred before the pepper/tomato divergence, and only one paralogous pair was duplicated recently, after the pepper/tomato split. Similar cases have been reported in tomato. The duplication of one SlDof was estimated to have occurred before the pepper/tomato split (19.1–61.1 Mya)21. The Ka/Ks ratio provides a measure of selective pressure. If the gene pair were under purifying selection, it would have been purged by natural selection, presumably because of deleterious effects. Conversely, if the gene pair was under positive selection, it would have been advantageous during the evolution of the two duplicates47. We calculated the Ka/Ks ratio of the duplicated pairs in the CaDof family. The Ka/Ks ratios of all 10 duplicated pairs in the CaDof family were <1.0, suggesting that the duplicated pairs evolved under purifying selection. Duplicate pairs of Dofs in tomato and soybeans were also estimated to have evolved under purifying selection20,21. Overall, the duplication analysis suggests that the segmental expansion of the CaDofs might have taken place before the speciation event separating pepper and tomato and that the functional divergence of the CaDofs was retained after the duplications.
Transcriptional control of gene expression is an important part of plant growth, development, and response to biotic and abiotic stresses48. RNA-seq has been a useful and powerful tool for the analysis of gene expression profiles since next-generation sequencing has become very straightforward. The RNA-seq data showed that most of the CaDofs had distinct tissue-specific expression patterns among the five different tissues tested. The CaDof expression profiles revealed various patterns among different organs and stages (Fig. 5). CaDof17, 18, 21, 27, and 30 were expressed at relatively high levels in various tissues and at several developmental stages. Seventeen of 33 genes showed relatively high expression levels in the root. Similarly, 22 SlDofs in tomato and 22 GmDofs in soybeans were expressed at relatively high levels in the root20,21. Ten (CaDof08, 11, 13, 17, 18, 21, 22, 27, 29, and 30) and seven (CaDof08, 11, 13, 18, 22, 27, and 30) genes showed relatively high expression levels in the stem and leaf, respectively. The expression levels of CaDof11, 21, and 30 in the placenta and that of CaDof18 in the pericarp were higher than those in other organs, suggesting that those genes could play key roles in fruit development, such as capsaicinoid biosynthesis. Taken together, the results indicate that the CaDofs are possibly involved in biological functions during plant development.
Recently, Dof family has also been reported to be possibly involved in biotic stresses49,50. However, still their role associated with biotic stress were largely unknown. Additional clues and works should be needed to understand and confirm their newer roles in biotic stress. In our study, the CaDof family showed distinct expression patterns in response to different biotic stresses (Fig. 6). The transcript levels of the CaDofs showed temporal and pathogen-specific variation. CaDof04, 05, and 32 showed changes in expression levels in the resistant response to TMV-P0 and/or PepMoV and P. capsici, while CaDof07 was up-regulated only in the susceptible response to TMV-P2. In particular, CaDof10 and 11 showed relatively high expression levels in response to each of the viruses, suggesting that they could play a role in defense response against viruses. In addition, some CaDofs such as CaDof09 and 17, showed similar expression patterns and temporal variation following various pathogen inoculations, indicating a shared function in response to biotic stresses. Thus, the differentially regulated CaDofs might play a crucial role in defense against biotic stresses. Those results will provide useful information for the functional characterization of Dofs to understand biological processes in pepper and other plants.
Methods
Identification and annotation of CaDofs
We obtained the consensus amino-acid sequence of the Dof protein as a seed from the Pfam (PF02701) database (http://pfam.sanger.ac.uk/). We then used that sequence to search all of the Dof proteins against the ‘CM334’ genome40 using the HMMER software package (Version 3.0, http://hmmer.org/). We confirmed the collected CaDofs by a BLAST search against NCBI. We then collected the CaDofs manipulated by alignment of the ‘CM334’ genome sequence, the ‘Zunla-1’ genome sequence, the pepper ESTs, and the Dof sequences from the Plant TFDB. If the alignment result showed the same sequence among the Dofs from several databases, we chose the longest sequence as a representative. We computed the pIs and MWs of the predicted protein sequences using the ExPASy Compute pI/MW tool (http://web.expasy.org/compute_pi/). We obtained the full amino-acid sequences of the tomato Dofs, designated SlDofs, from the Plant TFDB.
Motif analysis of the CaDofs and exon/intron structural analysis
We used the MEME suite program (http://meme-suite.org/tools/meme) to identify conserved motifs among the CaDofs. The analysis settings of the MEME suite were a motif length of 6–200 amino acids and 2–120 motif sites. We set the maximum number of motifs to 25. We set the other conditions to the default values. We performed the alignment of the identified CaDof domains using ClustalW (http://www.ebi.ac.uk/Tools/clustalw/). We predicted the zinc finger structure of the Dof domain based on information from the tomato and Arabidopsis Dof domains16,21. We searched and confirmed the exon/intron structures of the CaDofs by aligning the CDSs with the corresponding genomic regions. We presented the exon/intron structures using the online display tool Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/).
Phylogenetic tree analysis
We performed a multiple alignment of the 33 full-length CaDofs and 34 full-length SlDofs using ClustalW. We obtained the Dof sequences for tomato from previous research21 and the Plant TFDB. We used the alignment result to construct a phylogenetic tree using the neighbor-joining method of MEGA6 software (http://www.megasoftware.net/). We created the phylogenetic tree using the following parameters: Poisson correction, pairwise deletion, and a bootstrap value of 1,000.
Chromosomal location and calculation of duplication events
We mapped the CDSs of the Dofs to the ‘CM334’ chromosomes in the pepper genome database40 using BLASTn. Information about the chromosomal locations of the CaDofs was based on the identified physical positions on the chromosomes. We used MEGA6 to make the pairwise alignments of paralogs with the removal of gaps. We estimated the Ka, Ks, and Ka/Ks using K-Estimator 6.1v51. We then used the Ks value of each pair to estimate the duplication time by following formula: duplication time = Ks/2λ, with the clock-like rate (λ) = 6.1 × 10−9 based on previous research52.
Plant materials and pathogen inoculation
We used ‘CM334’ to analyze the responses to TMV-P0, TMV-P2, PepMoV, and P. capsici. We conducted the virus inoculations as described by Kang et al.53. We inoculated Nicotiana benthamiana leaves infected with TMV-P0, TMV-P2, and PepMoV mechanically with 0.1 M phosphate buffer (pH 7.0) into true leaves of ‘CM334’ seedlings. For virus inoculation, three independent experiments of each virus were each performed. We performed the preparation and inoculation of P. capsici as described by Yeom et al.37. We inoculated ‘CM334’ seedlings with 1.0 × 106 zoospores/ml P. capsici. The pepper plants inoculated with viruses and P. capsici were kept in a growth chamber at 23–25 °C under a 16 h/8 h light/dark photoperiod.
Transcriptome analysis
To obtain RNA-seq data from pathogen-infected pepper tissues, we harvested the leaves from various time points after pathogen inoculation to prepare the total RNA. We used RNA samples extracted from three biological replicates of TMV-P0, TMV-P2, and PepMoV inoculated tissues at various time points for library construction using a modified protocol for strand-specific library construction54 (Illumina Inc., San Diego, USA). We sequenced the constructed libraries (insert size: 150–200 bp) using Illumina HiSeq 2000 (Illumina Inc., San Diego, USA). We first aligned the RNA-seq reads to rRNA and tRNA sequences to remove possible contamination. Low-quality reads were filtered by in-house trimming scripts. We then aligned the resulting reads to the reference pepper genome using CLC Assembly Cell (CLC bio, Aarhus, Denmark). Following alignment to each gene model, we normalized the number of mapped reads to the reads per kilobase per million mapped reads (RPKM).
To analyze the expression patterns of the CaDofs in specific tissues and at different developmental stages, we used the RNA-seq data obtained from ‘CM334’40. We generated transcript data for five tissues: root, stem, leaf, pericarp, and placenta. The pericarp and placenta data included seven developmental stages: 6, 16, and 25 days post inoculation; mature green (MG); breaker (B); and 5 and 10 days post B. We illustrated the log2 RPKM values of the expression data for the tissues and developmental stages using the heat-map R package (http://bioconductor.org/). To evaluate the expression patterns in response to the virus and P. capsici inoculations, we compared the normalized transcripts obtained under each biotic stress with each negative control (Supplementary Table S2). For data analysis of virus inoculation, three RNA-seq data sets corresponding three biological replicates of each virus were analyzed. Then, mean of values in triplicate datasets were used for analysis of expression pattern and construction of heat maps. We created the heat map using the heat-map R package (http://bioconductor.org/).
Additional Information
How to cite this article: Kang, W.-H. et al. Genome-wide analysis of Dof transcription factors reveals functional characteristics during development and response to biotic stresses in pepper. Sci. Rep. 6, 33332; doi: 10.1038/srep33332 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ010939062016) from the Rural Development Administration of the Korean government, and the National Research Foundation of the Korea Ministry of Education, Science and Technology (Project number NRF-2015R1A6A1A03031413 and 2015R1A2A1A01002327).
Footnotes
Author Contributions W.-H.K. and S.-I.Y. conceived and designed the research. W.-H.K., H.-A.L, S.K. and S.-I.Y. conducted the experiments, W.-H.K., D.C. and S.-I.Y. analyzed the data. W.-H.K. and S.-I.Y. wrote the manuscript. All authors reviewed the manuscript and approved the final manuscript.
References
- Lee H. A. & Yeom S. I. Plant NB-LRR proteins: tightly regulated sensors in a complex manner. Brief Funct Genomics 14, 233–242, 10.1093/bfgp/elv012 (2015). [DOI] [PubMed] [Google Scholar]
- Yeom S. I., Seo E., Oh S. K., Kim K. W. & Choi D. A common plant cell-wall protein HyPRP1 has dual roles as a positive regulator of cell death and a negative regulator of basal defense against pathogens. Plant J 69, 755–768, 10.1111/j.1365-313X.2011.04828.x (2012). [DOI] [PubMed] [Google Scholar]
- Kunkel B. N. & Brooks D. M. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5, 325–331 (2002). [DOI] [PubMed] [Google Scholar]
- Gupta S. et al. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 241, 549–562, 10.1007/s00425-014-2239-3 (2015). [DOI] [PubMed] [Google Scholar]
- Seo E. & Choi D. & Choi. Functional studies of transcription factors involved in plant defenses in the genomics era. Brief Funct Genomics 14, 260–267, 10.1093/bfgp/elv011 (2015). [DOI] [PubMed] [Google Scholar]
- Jin J., Zhang H., Kong L., Gao G. & Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res 42, D1182–D1187, 10.1093/nar/gkt1016 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguero M., Atif R. M., Ochatt S. & Thompson R. D. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci 209, 32–45, 10.1016/j.plantsci.2013.03.016 (2013). [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci 7, 555–560 (2002). [DOI] [PubMed] [Google Scholar]
- Washio K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiol 133, 850–863, 10.1104/pp.103.027334 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Chen W., Foley R. C., Buttner M. & Singh K. B. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell 7, 2241–2252, 10.1105/tpc.7.12.2241 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. G. & Singh K. B. Characterization of salicylic acid-responsive, arabidopsis Dof domain proteins: overexpression of OBP3 leads to growth defects. Plant J 21, 329–339 (2000). [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J., Moose S. P., Parsons R. L. & Schmidt R. J. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94, 7685–7690 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimofurutani N., Kisu Y., Suzuki M. & Esaka M. Functional analyses of the Dof domain, a zinc finger DNA-binding domain, in a pumpkin DNA-binding protein AOBP. FEBS Lett 430, 251–256 (1998). [DOI] [PubMed] [Google Scholar]
- Kisu Y., Ono T., Shimofurutani N., Suzuki M. & Esaka M. Characterization and expression of a new class of zinc finger protein that binds to silencer region of ascorbate oxidase gene. Plant Cell Physiol 39, 1054–1064 (1998). [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. & Izui K. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. J Biol Chem 268, 16028–16036 (1993). [PubMed] [Google Scholar]
- Lijavetzky D., Carbonero P. & Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evol Biol 3, 17, 10.1186/1471-2148-3-17 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Tuskan G. A. & Cheng M. Z. Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol 142, 820–830, 10.1104/pp.106.083642 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L. M., McIntyre C. L., Gresshoff P. M. & Xue G. P. Members of the Dof transcription factor family in Triticum aestivum are associated with light-mediated gene regulation. Funct Integr Genomics 9, 485–498, 10.1007/s10142-009-0130-2 (2009). [DOI] [PubMed] [Google Scholar]
- Kushwaha H., Gupta S., Singh V. K., Rastogi S. & Yadav D. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol Biol Rep 38, 5037–5053, 10.1007/s11033-010-0650-9 (2011). [DOI] [PubMed] [Google Scholar]
- Guo Y. & Qiu L. J. Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS One 8, e76809, 10.1371/journal.pone.0076809 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cai X. et al. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J Integr Plant Biol 55, 552–566, 10.1111/jipb.12043 (2013). [DOI] [PubMed] [Google Scholar]
- Ma J., Li M. Y., Wang F., Tang J. & Xiong A. S. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 16, 33, 10.1186/s12864-015-1242-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. Plant J 21, 281–288 (2000). [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. & Sheen J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. Plant Cell 10, 75–89 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualberti G. et al. Mutations in the Dof zinc finger genes DAG2 and DAG1 influence with opposite effects the germination of Arabidopsis seeds. Plant Cell 14, 1253–1263 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papi M. et al. Identification and disruption of an Arabidopsis zinc finger gene controlling seed germination. Genes Dev 14, 28–33 (2000). [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T. F., Harmon F. G., Ho L. A. & Kay S. A. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297, 10.1126/science.1110586 (2005). [DOI] [PubMed] [Google Scholar]
- Fornara F. et al. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17, 75–86, 10.1016/j.devcel.2009.06.015 (2009). [DOI] [PubMed] [Google Scholar]
- Park D. H. et al. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. Plant J 34, 161–171 (2003). [DOI] [PubMed] [Google Scholar]
- Ward J. M., Cufr C. A., Denzel M. A. & Neff M. M. The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17, 475–485, 10.1105/tpc.104.027722 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washio K. Identification of Dof proteins with implication in the gibberellin-regulated expression of a peptidase gene following the germination of rice grains. Biochim Biophys Acta 1520, 54–62 (2001). [DOI] [PubMed] [Google Scholar]
- Li D. et al. Functional characterization of rice OsDof12. Planta 229, 1159–1169, 10.1007/s00425-009-0893-7 (2009). [DOI] [PubMed] [Google Scholar]
- Yamamoto M. P., Onodera Y., Touno S. M. & Takaiwa F. Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiol 141, 1694–1707, 10.1104/pp.106.082826 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesch G., Ehrhardt T. & Mueller-Roeber B. Involvement of TAAAG elements suggests a role for Dof transcription factors in guard cell-specific gene expression. Plant J 28, 455–464 (2001). [DOI] [PubMed] [Google Scholar]
- Wang H. W. et al. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J 52, 716–729, 10.1111/j.1365-313X.2007.03268.x (2007). [DOI] [PubMed] [Google Scholar]
- Corrales A. R. et al. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J Exp Bot 65, 995–1012, 10.1093/jxb/ert451 (2014). [DOI] [PubMed] [Google Scholar]
- Yeom S. I. et al. Use of a secretion trap screen in pepper following Phytophthora capsici infection reveals novel functions of secreted plant proteins in modulating cell death. Mol Plant Microbe Interact 24, 671–684, 10.1094/MPMI-08-10-0183 (2011). [DOI] [PubMed] [Google Scholar]
- Caranta C., Thabuis A. & Palloix A. Development of a CAPS marker for the Pvr4 locus: a tool for pyramiding potyvirus resistance genes in pepper. Genome 42, 1111–1116, 10.1139/g99-069 (1999). [DOI] [PubMed] [Google Scholar]
- Wang D. & Bosland P. W. The Genes of Capsicum. HortScience 41, 1169–1187 (2006). [Google Scholar]
- Kim S. et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet 46, 270–278, 10.1038/ng.2877 (2014). [DOI] [PubMed] [Google Scholar]
- Kim H. J. et al. Pepper EST database: comprehensive in silico tool for analyzing the chili pepper (Capsicum annuum) transcriptome. BMC Plant Biol 8, 101, 10.1186/1471-2229-8-101 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C. et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci USA 111, 5135–5140, 10.1073/pnas.1400975111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno M. A., Martinez M., Vicente-Carbajosa J. & Carbonero P. The family of DOF transcription factors: from green unicellular algae to vascular plants. Mol Genet Genomics 277, 379–390, 10.1007/s00438-006-0186-9 (2007). [DOI] [PubMed] [Google Scholar]
- Wang Y. et al. Sequencing and comparative analysis of a conserved syntenic segment in the Solanaceae. Genetics 180, 391–408, 10.1534/genetics.108.087981 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon S. B., Mitra A., Baumgarten A., Young N. D. & May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4, 10, 10.1186/1471-2229-4-10 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G. & Wolfe K. H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16, 1667–1678, 10.1105/tpc.021345 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Selection and gene duplication: a view from the genome. Genome Biol 3, reviews1012.1–reviews1012.3, 10.1186/gb-2002-3-5-reviews1012 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaill P. & Rivas S. Transcriptional control of plant defence responses. Curr Opin Plant Biol 20, 35–46, 10.1016/j.pbi.2014.04.004 (2014). [DOI] [PubMed] [Google Scholar]
- Zheng S. X., Xiao S. & Chye M. L. The gene encoding Arabidopsis acyl-CoA-binding protein 3 is pathogen inducible and subject to circadian regulation. J Exp Bot 63, 2985–3000, 10.1093/jxb/ers009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M. et al. The barley cystatin gene (Icy) is regulated by DOF transcription factors in aleurone cells upon germination. J Exp Bot 56, 547–556, 10.1093/jxb/eri033 (2005). [DOI] [PubMed] [Google Scholar]
- Comeron J. M. K-Estimator: calculation of the number of nucleotide substitutions per site and the confidence intervals. Bioinformatics 15, 763–764, 10.1093/bioinformatics/15.9.763 (1999). [DOI] [PubMed] [Google Scholar]
- Guo M. et al. Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.). BMC Plant Biol 15, 151, 10.1186/s12870-015-0512-7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W. H. et al. Molecular mapping and characterization of a single dominant gene controlling CMV resistance in peppers (Capsicum annuum L.). Theor Appl Genet 120, 1587–1596, 10.1007/s00122-010-1278-9 (2010). [DOI] [PubMed] [Google Scholar]
- Zhong S. et al. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011, 940–949, 10.1101/pdb.prot5652 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.