Abstract
Parathyroid hormone-related peptide (PTHrP) is recognized as the major causative agent of humoral hypercalcemia of malignancy (HHM). The paraneoplastic PTHrP has also been implicated in tumor progression and metastasis of many human cancers. Conventional PTHrP detection methods like immunoradiometric assay (IRMA) lack the sensitivity required to measure target peptide levels prior to the development of hypercalcemia. In general, sensitive, multiplexed peptide measurement by immunoassay represents challenges that we address in this paper. We describe here the first ultrasensitive multiplexed peptide assay to measure intact PTHrP 1-173 as well as circulating N-terminal and C-terminal peptide fragments. This versatile approach should apply to almost any collection of peptides that are long enough to present binding sites for two antibodies. To target PTHrP, we employed a microfluidic immunoarray featuring a chamber for online capture of the peptides from serum onto magnetic beads decorated with massive numbers of peptide-specific antibodies and enzyme labels. Magnetic bead-peptide conjugates were then washed and sent to a detection chamber housing an antibody-modified 8-electrode array fabricated by inkjet printing of gold nanoparticles. Limits of detection (LODs) of 150 aM (∼1000-fold lower than IRMA) in 5 μL of serum were achieved for simultaneous detection of PTHrP isoforms and peptide fragments in 30 min. Good correlation for patient samples was found with IRMA (n = 57); r2 = 0.99 assaying PTHrP 1-86 equiv fragments. Analysis by a receiver operating characteristic (ROC) plot gave an area under the curve of 0.96, 80–83% clinical sensitivity, and 96–100% clinical specificity. Results suggest that PTHrP1-173 isoform and its short C-terminal fragments are the predominant circulating forms of PTHrP. This new ultrasensitive, multiplexed assay for PTHrP and fragments is promising for clinical diagnosis, prognosis, and therapeutic monitoring from early to advanced stage cancer patients and to examine underlying mechanisms of PTHrP overproduction.
Parathyroid hormone (PTH)-like factor was first postulated by Albright over 60 years ago1 as a humoral factor responsible for development of hypercalcemia in cancer patients and later described as humoral hypercalcemia of malignancy (HHM).2 The true nature of this PTH-like factor remained elusive since it escaped detection by immunoassays using antibodies raised against PTH3,4 but could be detected in bioassays using PTH receptor.5,6 This led to cloning and characterization of this PTH-like factor now known as parathyroid hormone-related peptide (PTHrP)7,8 and to development of specific immunoassays.9,10
Human PTHrP has three isoforms of 139, 141, and 173 amino acids and is widely expressed in normal and cancerous human tissues.11,12 All isoforms have identical sequences through residue 139 and undergo post-translational cleavage generating N-terminal, midregion, and C-terminal peptides with distinct physiological functions. PTHrP exerts PTH-like actions in bone and kidney by binding and activating the guanylyl nucleotide-binding (G) protein-linked receptor (PTH1R) causing hypercalcemia.13,14
Distinct from PTH, PTHrP acts as endocrine, autocrine, paracrine, or intracrine factor in a vast range of important physiological roles including skeletal development, placental calcium transport, muscle relaxation, and mammary gland development.15,16 Circulating levels of PTHrP correlate with disease progression in cancers including breast, prostate, melanoma,17−20 and bone metastasis.21−23 Currently PTHrP can only be detected in the blood when hypercalcemia develops,9,10 when there is ∼50% chance of mortality in 30 days.24 Thus, existing PTHrP assays are limited to confirming the humoral origin of hypercalcemia but cannot provide early detection of PTHrP-producing tumors, which requires assays with much higher sensitivity.
Peptides have been particularly difficult targets for ultrasensitive multiplexed immunoassays. Enzyme linked immunosorbent assay (ELISA),25 immunofluorometric assays (IFMA),26 and mass spectrometry27,28 can be used to measure PTHrP but have limits of detection (LOD) in the picomolar range that are not low enough to measure serum PTHrP representative of early stage cancers. In addition, immunoradiometric (IRMA) and radioimmunoassay assays (RIA)9,10,29 commonly used for PTHrP employ high energy isotopes such as 125I that pose health hazards and have short shelf-lives.28,30 None of these assays measure specific PTHrP isoforms and in particular the human specific PTHrP 1-173 isoform. These assays mostly target the 1-86 peptide fragment. IRMA and RIA have LODs ranging from 0.3 to 4 pM,29,31,32 ELISA (Elabscience Biotechnology) 0.5 pM,25 IFMA 0.5 pM,26 and mass spectrometry 10 pM.28 Furthermore, these assays lack multiplexing capability and detect only a single PTHrP isoform.
Multiplexed assays have been developed for other peptides. Zhong et al.33 reported an electrochemical multiplex immunoassay using liposomes which contained electrochemically active molecules as signal enhancers for simultaneous detection of neuron-specific enolase (NSE) and pro-gastrin-releasing peptide (ProGRP). This immunoassay has one of the best non-PTHrP peptide LODs so far, 10 pg/mL (picomolar range) for ProGRP and 0.18 ng/mL (nanomolar range) for NSE. Using peptide immunoaffinity enrichment coupled with stable isotope dilution mass spectrometry (SISCAPA-MRM), Kuhn et al.34 developed a multiplexed assay for peptides troponin I (cTnI) and interleukin-33 (IL-33) trypsin-digested peptide standards. LODs of 2.5 μg/L (83 pM) for IL-33 Pep-1 and 1.0 μg/L (33 pM) for IL-33 Pep-2 were achieved. However, these methods applied to PTHrP would still be expected to have insufficient LODs for early cancer diagnostics.
In this paper, we describe the first ultrasensitive immunoarray to detect PTHrP 1-173 and smaller peptide fragments using a novel semiautomated microfluidic device that we previously developed for full proteins.35 The microfluidic system delivers samples to a capture chamber where massively enzyme-labeled magnetic beads equipped with multiple antibodies capture target peptides. These beads are washed and delivered to an 8-sensor inkjet printed 4 nm gold nanoparticle immunoarray36 decorated with a second set of antibodies that recognize and bind bead-bound target peptides. Peptides are measured simultaneously by activation of enzyme labels and electrochemical detection. Exquisite LODs and sensitivities are achieved because of these design factors: (1) highly efficient capture of target peptides from the samples onto magnetic beads with 120 000 ± 8 000 antibodies, (2) the sensors see only the beads and never contact the full sample to limit nonspecific binding, and (3) 250 000 ± 15 000 HRP detection labels are present on each magnetic bead. The number antibodies and enzyme labels (HRP) on the magnetic beads are comparable to those previously reported.36 Intact PTHrP isoforms as well as N- and C-terminal fragments were detected simultaneously in serum with ultralow LODs of 150 aM, 1000-fold lower than commercial PTHrP assays. Good correlation between microfluidic immunoarray and IRMA results in cancer patient serum were found. Statistical analysis of limited patient sample data predicts good cancer diagnostic potential, with 80–83% sensitivity (true positive rate) and 96–100% specificity (0−4% false positive rate).
Experimental Section
Chemicals and Serum Samples
Chemicals and materials are listed in the Supporting Information. Human serum samples from cancer patients with solid tumors and healthy individuals were obtained from McGill University Health Center Biorepository. Blood was drawn in regular tubes, which were put on ice immediately, separated within 60 min, aliquoted, and stored at −80 °C prior to assay. The samples were stored for no longer than 12 months prior to assay (Table S1). All samples used in this study were acquired under a McGill University Institutional Review Board (IRB)-approved protocol, and informed consent was obtained from all study participants.
PTHrP Peptides and Antibodies
PTHrP 1-173 was produced from cDNA encoding PTHrP 1-173. Human PTHrP fragments 1-33, 151-169, 140-173 were from Sheldon Biotechnology Center (McGill University, CA). Human recombinant PTHrP 1-86 was from Bachem (Torrance, CA, catalog no. H-9815). Monoclonal antibodies M45 (IgM) and PA158 (IgG) were raised against PTHrP1-33; monoclonal antibody PA104 (IgG) was raised against PTHrP 140-173; monoclonal antibody PA6 (IgG) was raised against PTHrP 151-169, PA104, PA158.37 Polyclonal antibodies against human PTHrP 1-173 (IgY lots 3103 and 3104 were raised in chicken and purified commercially (Genway Biotech, San Diego, CA).
Stock concentration of peptide standards (200–500 ng for 1-33, 151-169, 140-173, and 1-173 and PTHrP 1-86 were first diluted in water or PBS buffer pH 7.4 to 50 pM and stored at −80 °C (according to the manufacturer’s specifications). The antibodies were reconstituted in PBS buffer pH 7.4 down to the working concentration and stored at −80 °C. The peptides and antibodies were stable for 12 months. On assay days, one vial of 50 pM peptide standard was then diluted to 1 pM followed by serial dilution in 5× diluted calf serum in PBS buffer pH 7.4. The diluted standards were used the same day they were prepared and any left-over standards were discarded. In all the calibration curves, 5× diluted calf serum was employed as the assay diluent for serial dilutions. Electrode surface area was calculated by cycling the gold arrays in 0.18 M H2SO4 between 1.5 V and −0.2 V at 100 mV s–1 (see Supporting Information). Current density was used for quantitation of the standards. The peak height (I, nA) was divided by the surface area of the electrode to yield the current density that was plotted against the concentration of the peptide fragments. Log fitting was used to plot the data.
Array Fabrication
Immunosensor arrays were inkjet printed from 4 nm dodecanethiol-gold nanoparticles (AuNPs) on Kapton film as previously described36 (see the Supporting Information). After monoclonal antibodies (Ab1) were attached to sensors, they were washed with PBS-T20 and incubated with 2% BSA to minimize nonspecific binding (NSB) (see the Supporting Information). Arrays were then fitted into the detection chamber. Multiple horseradish peroxidase (HRP) and antibodies (Ab2) were attached onto 1 μm magnetic beads (HRP-MB-Ab2) as previously described36b (Table S2).
Detection of PTHrP Isoforms and Fragments
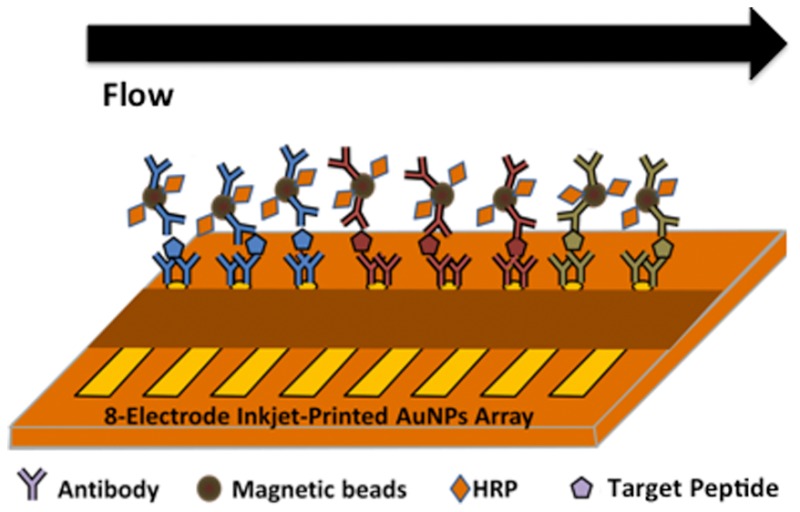
The microfluidic immunoassay system (Figure 1)35 was conditioned by flowing PBS-T20 for 4 min before and after each standard run to minimize NSB and carry-over effect. HRP-MB-Ab2 beads were dispersed in PBS buffer (pH 7.4) and injected into the capture chamber followed by injection of sample or PTHrP standard in 5× diluted calf serum (Figure S1). HRP-MB-Ab2 beads were held in the capture chamber with a magnet while the sample was injected. For simultaneous multiplexed detection of peptides, mixed HRP-MB-Ab2 beads for each peptide in PBS buffer were injected into the capture chamber followed by injection of sample or standard mixtures. Flow was stopped, magnet removed, and capture chamber stirred 30 min for peptide capture by HRP-MB-Ab2.
Figure 1.
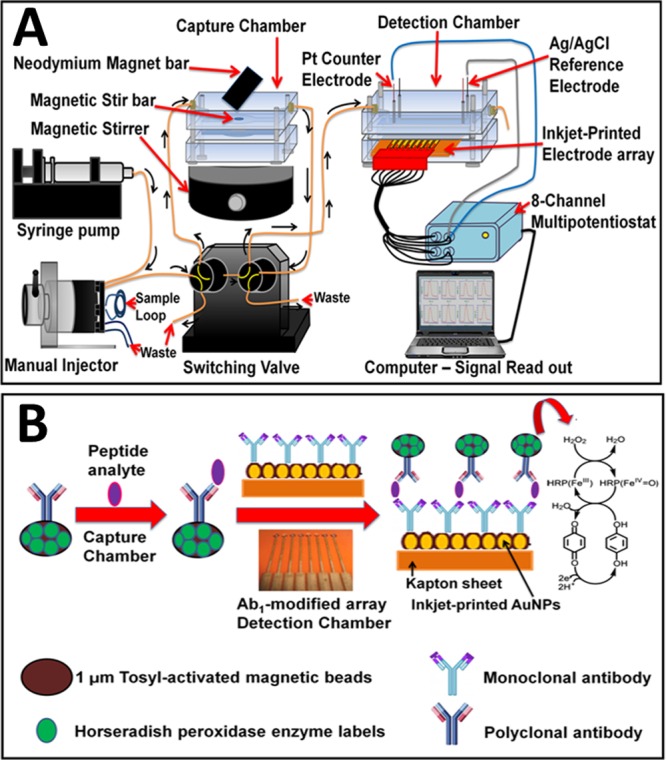
Immunoarray with online peptide capture: (A) microfluidic device and (B) detection pathway.
The resulting beads were washed with PBS-T20, the magnet removed, and flow switched to transport peptide-Ab2-MB-HRP beads into the detection chamber. Flow was stopped for 15 min to allow sensors to capture peptide-bead conjugates. After washing, amperometric detection at −0.3 V vs Ag/AgCl was enabled by flowing 1 mM hydroquinone + 0.1 mM H2O2 through the detection chamber.36 A commercial IRMA immunoassay was used as a reference method (see the Supporting Information). Total assay time is ∼50 min (30 min incubation in the capture chamber; 15 min incubation in the detection chamber; 5 min total wash time). However, since incubation of sample 1 in the detection chamber and incubation of sample 2 in the capture chamber can be done simultaneously after the first standard run, the total cycle time for the analysis is 30 min. Up to 8 different peptide fragments can be assayed simultaneously.
Results
Single Peptide Detection
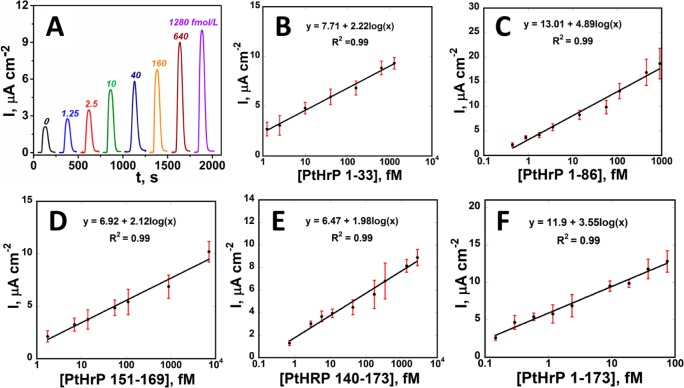
PTHrP undergoes post-translational cleavage at lysine or arginine to yield N-terminal, midregion, and C-terminal peptide fragments.7,10 We first designed single peptide assays for intact PTHrP 1-173, N-terminal (1-33 and 1-86) and C-terminal fragments (151-169 and 140-173). PA158, PA6, and PA104 were employed as capture antibodies on the sensors while M45, IgY3103, and IgY3104 were attached onto magnetic beads as detection antibodies (Table S2 and Figure S2). Calf serum diluted 5× in PBS pH 7.4, a good surrogate for human serum in immunoassays,38 was employed as peptide standards diluent. Calf serum as reported by the manufacturer (Sigma-Aldrich) has a total protein concentration of 5.5–8.0%, which is similar to human serum (4.0–9.0%). Analytical performance of the immunoassay was evaluated including its linearity, precision (intra-assay and interassay), sensitivity, specificity, limit of detection, carry-over effect, and method comparison using IRMA (Figure S3 and Table S3).
Calibrations for peptide fragments are shown in Figure 2. Signals for peptide-free controls result from residual NSB of magnetic beads and direct reduction of H2O2. Peaks increased linearly with log C for peptide fragments from 150 aM to 7 pM. LODs measured as 3-SDs above control were 150–170 aM (3–5 fg/mL) for all peptides (Table S4 and Figure S4). LODs were 1000-fold better than those of commercial IRMA and ELISA kits (0.3–4 pM). Good reproducibility is indicated by small error bars (Figure 2). Sensitivities (as slopes of calibration plots) were 1.98–2.12 μA cm–2 [log C]−1 for 1-33, 140-173, and 151-169 but increased to 3.55–4.98 μA cm–2 [log C]−1) for larger peptides (1-86 and 1-173) (Table S4).
Figure 2.
Array results: (A) for 1-33 at −0.3 V vs Ag/AgCl. Calibrations for PTHrP fragments in 5× diluted calf serum (n = 8): (B) 1-33, (C) 1-86, (D) 151-169, (E) 140-173, and (F) intact PTHrP 1-173.
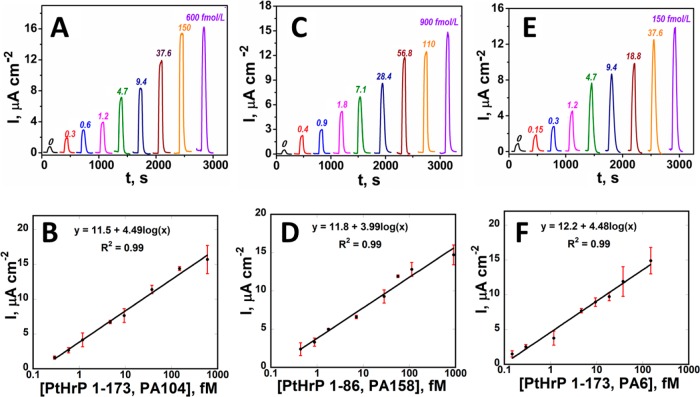
Multiplexed Peptide Detection
Peptide fragments with the best sensitivities (1-86 and 1-173) were selected for multiplexed detection (Figure S5A and Table S4). Minimal cross-reactivity was found between antibodies for PTHrP 1-173 and 1-86 (Figure S5B). Calibration plots for detection of PTHrP 1-173 and 1-86 standard mixtures (Figure 3) show linear dynamic ranges from 150 aM to 600 fM. LODs were 400 aM for 1-86, 300 aM for 1-173 using PA104 antibody, and 150 aM for 1-173 using PA6. Good reproducibility is illustrated by small error bars (Figure 3B,D,F). Combination of PA104 and IgY3104 and PA6 and IgY3103 gave similar sensitivity for detection of intact PTHrP 1-173 isoform (4.49 vs 4.48 μA cm–2 [log C]−1, Figure 3B,F), consistent with separate specificity tests (Figure S5B) and binding studies.37
Figure 3.
Array results for standard peptide mixtures in 5× diluted calf serum at −0.3 V vs Ag/AgCl for (A) intact PTHrP 1-173 using PA104, (C) 1-86 peptide fragment (E) intact PTHrP 1-173 using PA6, and calibration plots for intact PTHrP 1-173 (B and F) and 1-86 fragment (D) (n = 3).
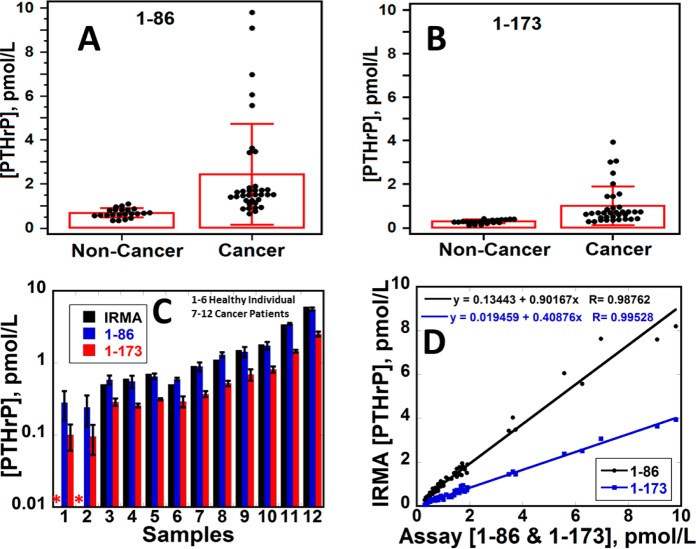
Validation of Accuracy
Serum samples from cancer patients with solid tumors and cancer-free individuals were assayed and compared with IRMA results. The antibody used for 1-86 peptide binds all three PTHrP isoforms and their N-terminal fragments. The antibody used for 1-173 binds PTHrP 1-173 and shorter C-terminal fragments including 140-173 and 151-169.37 Significant differences in PTHrP levels between cancer patients and cancer-free controls were observed. Cancer patient samples had larger amounts of PTHrP up to 9 pM compared to healthy individuals (<1 pM, Figure 4A,B), with statistical difference between means confirmed by t tests (P < 0.001) (Table S5). Assays by the immunoarray (1-86) and IRMA gave similar levels of PTHrP and t tests (P < 0.001) confirmed no significant difference between the two methods (Figure 4C). The immunoarray detected PTHrP in all samples including 4 samples with PTHrP levels that were too low to be measured by IRMA. Immunoarray results for 1-86 also gave good linear correlation with IRMA for 57 samples (22 controls and 35 cancer subjects) with slopes close to 1 (0.90 ± 0.02), intercepts near 0 (1.33 ± 0.51) and r2 = 0.99 (Figure 4D and Table S6). These results confirm the accuracy of our immunoarray protocol. Values obtained with microfluidic assays measuring intact PTHrP 1-173 and its (C-terminal) fragments (red) were only slightly lower than those from the 1-86 assay (blue) recognizing all three isoforms and short N-terminal fragments, suggesting that PTHrP 1-173 and its short C-terminal fragments are the major forms of PTHrP in serum (Figure 4C).
Figure 4.
Distributions of PTHrP levels in cancer patient serum (37) and cancer-free individuals (22) for (A) 1-86; (B) PTHrP 1-173; (C) bar graph comparing IRMA and immunoarray (1-86 and 1-173) results for PTHrP (n = 12) and (D) correlation plot of IRMA and immunoarray data (1-86 and 1-173) (n = 57). Asterisk (*) denotes value below IRMA LOD.
Data were also analyzed using receiver operating characteristic (ROC) plots to predict diagnostic accuracy. Here, sensitivity (true positive rate) is plotted against 100-specificity (false positive rate) for different cutoff points. A test with perfect discrimination has a ROC curve that passes through the upper left corner (100% sensitivity, 100% specificity).39 The area under a ROC curve (AUC) quantifies the overall ability of the test to discriminate between individuals with and without the disease. Data with zero false positives and zero false negatives has an AUC of 1.00.
For PTHrP (n = 57) the ROC plot had AUC 0.96 for the 1-86 fragment assay and 0.94 for PTHrP 1-173. The 1-86 fragments gave 80% sensitivity and 100% specificity while intact PTHrP 1-173 gave 82.9% sensitivity and 95.5% specificity. The cancer vs noncancer cutoff PTHrP was 1.1 pM using the 1-86 assay, in agreement with IRMA results. Curves for individual peptides (Figure 5A) gave relatively similar sensitivity and specificity when using normalized, mean values of the two peptides (Figure 5B).
Figure 5.
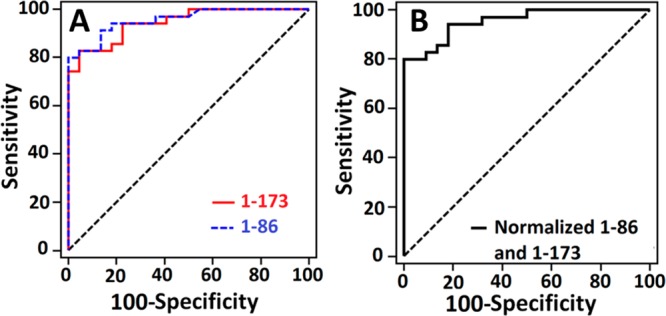
Receiver operating characteristic (ROC) curves for (A) serum assays for 1-173 (red) with AUC 0.94, 95.5% specificity and 82.9% sensitivity and 1-86 (blue) with AUC 0.96, 100% specificity and 80% sensitivity and (B) normalized value for both 1-86 and 1-173 with AUC 0.96, 100% specificity and 80% sensitivity.
Discussion
Results described above demonstrate the first assay for simultaneous detection of PTHrP and its peptide fragments in serum with ultralow LODs of 150 aM. Compared to commercial clinical assays such as IRMA and ELISA kits (0.3–4 pM), our microfluidic immunoarray assay represents more than 1000-fold better detection limits for PTHrP or indeed any peptides (see introduction). In addition, this method offers a degree of automation as well as simplicity to bring assays to a point of care setting. Judged against competing analytical techniques such as mass spectrometry, the excellent sensitivity (2–5 μA cm–2 [log C]−1), low cost, and reasonable speed reflect key advantages of this electrochemical assay. The 1 μm superparamagnetic beads with 250 000 ± 15 000 HRP labels and 120 000 ± 8 000 antibodies (Ab2) per bead enabled high capture efficiency and ultrahigh sensitivity in 30 min assays. To reiterate, the 1-86 peptide assays measure the level of all three PTHrP isoforms (PTHrP 1-139, 1-141, and 1-173) and their fragments containing N-terminal ends. The assay using 1-173 as a standard measures the complete isoform PTHrP 1-173 and its short C-terminal fragments. The results in Figure 4D are consistent with this fact since the slope of the IRMA-microfluidic assay correlation plot for the I-173 standard is less than that of 1-86, since IRMA measured I-86. A novel and interesting observation from Figure 4C is that circulating concentrations of the PTHrP 1-173 isoform and its C-terminal fragments were only slightly smaller that the concentrations of all three isoforms and their fragments measured by the I-86 assay, suggesting that PTHrP 1-173 and its short C-terminal fragments are the predominant circulating forms of PTHrP. This preliminary finding requires further study in larger cohorts.
Sensitivities of single-detection assays ranged from 2 to 5 μA cm–2 [log C]−1. The highest sensitivities were obtained for peptide fragments 1-86 and 1-173. Assay results revealed a significant difference between the levels of PTHrP in healthy individuals (<1 pM) (Figure S6) compared to cancer patients. Our assays could also detect PTHrP levels in all the samples (Figure 4). In agreement, ROC analyses (Figure 5) gave 80–83% clinical sensitivity and 96–100% clinical specificity for detection of cancer using the combined PTHrP 1-86 and 1-173 assay. While clearly more samples need to be analyzed for confirmation, results suggest a high potential of the PTHrP 1-86 and 1-173 immunoassay for early stage cancer diagnostics.
Fragments of PTHrP are under investigation to determine their diagnostic potential in a variety of human cancers. Washam et al. identified N-terminal fragment of PTHrP 12-48 as a plasma biomarker for breast cancer bone metastasis with a sensitivity of 91% and specificity of 93%.40 Using mass spectrometry, PTHrP 12-48 was significantly elevated in plasma of breast cancer patients with bone metastasis compared to controls without metastasis (P < 0.0001). Combination of a clinical serum marker N-telopeptide of type I collagen (NTx) with plasma PTHrP 12-48 greatly increased the diagnostic specificity and accuracy (AUC = 0.99). The LOD of the mass spectrometry method was in ng/mL (nanomolar) range. Our assay, on the other hand, can detect PTHrP levels and both large and small fragment sizes at levels as low as 150 aM and at much lower cost than mass spectrometry, which cannot approach such low detection levels.25,26
The microfluidic immunoarray offers a simple, rapid, low cost way to simultaneously detect PTHrP peptide fragments. Inkjet printing technology offers both a simple and elegant way to fabricate disposable low-cost sensor electronics for the immunoarray. A single 8-electrode array costs ∼$0.2 in materials and up to 56 arrays can be printed in a single run.36 Thus, ease of fabrication and utilization of commercial components makes this approach accessible to virtually any biomedical laboratory at low cost. Capture and detection chamber are made by templating PDMS channels on machined aluminum molds to avoid lithography and mounted on hard plastic PMMA housings with inlet and outlet lines. The microfluidic device requires only a small sample volume (5 μL) and offers a degree of automation and reliability to enhance reproducibility and throughput. These advantages make the microfluidic immunoarrays a promising tool for development of sensitive, integrated, portable, clinical diagnostic devices in a short time with minimal sample and reagent requirements. Additional design improvements are underway with a goal of achieving a pump-free, automated microfluidic assay for point-of-care diagnostics.
In summary, we describe above a novel approach for simultaneous detection of isoforms of PTHrP in an assay suitable for comparing circulating forms. The assay provides the best detection limit for peptide detection to date and enables accurate analysis of normal and pathological clinical samples with numerous potential applications in pathologies and physiological conditions in which PTHrP has been implicated. Results from cancer patient sample analyses support the potential diagnostic utility of such assays, although a much larger sample cohort will be required for full clinical validation. The analytical approach is general and should be applicable to measurement of any peptides for which two appropriate antibodies exist.
Acknowledgments
This work was supported by Grants EB016707 and EB014586 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB) at the National Institutes of Health. We thank Dr. Benoett Ochietti (McGill University, Montreal Canada) for assistance with the peptide synthesis.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.6b02637.
Experimental details and supplementary figures and tables (PDF)
Author Contributions
B.A. Otieno and C.E. Krause contributed equally to this article. All authors confirmed they have contributed to the intellectual content of this paper and have met the following three requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
The authors declare the following competing financial interest(s): R. B. Kremer and D. C. Huang: PTHRP, its isoforms and antagonists thereto in the diagnosis and treatment of disease, U.S. Patent 7,897,139 B2, issued March 1, 2011. The remaining authors have no conflicts of interest related to this work.
Supplementary Material
References
- Albright F. N. Engl. J. Med. 1941, 225, 789–791. 10.1056/NEJM194111132252007. [DOI] [Google Scholar]
- Stewart A. F.; Horst R.; Deftos L. J.; Cadman E. C.; Lang R.; Broadus A. E. N. Engl. J. Med. 1980, 303, 1377–1383. 10.1056/NEJM198012113032401. [DOI] [PubMed] [Google Scholar]
- Riggs B. L.; Arnaud C. D.; Reynolds J. C.; Smith L. H. J. Clin. Invest. 1971, 50, 2079–2083. 10.1172/JCI106701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell D.; Singer F. R.; Murray T. M.; Minkin C.; Potts J. T. Jr. N. Engl. J. Med. 1973, 289, 176–181. 10.1056/NEJM197307262890403. [DOI] [PubMed] [Google Scholar]
- Rodan S. B.; Insogna K. L.; Vignery A. M.; Stewart A. F.; Broadus A. E.; D’Souza S. M.; Bertolini D. R.; Mundy G. R.; Rodan G. A. J. Clin. Invest. 1983, 72, 1511–1515. 10.1172/JCI111108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart A. F.; Insogna K. L.; Goltzman D.; Broadus A. E. Proc. Natl. Acad. Sci. U. S. A. 1983, 80, 1454–1458. 10.1073/pnas.80.5.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva L. J.; Winslow G. A.; Wettenhall R. E.; Hammonds R. G.; Moseley J. M.; Diefenbach-Jagger H.; Rodda C. P.; Kemp B. E.; Rodriguez H.; Chen E. Y.; et al. Science 1987, 237, 893–896. 10.1126/science.3616618. [DOI] [PubMed] [Google Scholar]
- Strewler G. J.; Stern P. H.; Jacobs J. W.; Eveloff J.; Klein R. F.; Leung S. C.; Rosenblatt M.; Nissenson R. A. J. Clin. Invest. 1987, 80, 1803–1807. 10.1172/JCI113275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe W. A.; Norbury S.; Heath D. A.; Ratcliffe J. G. Clin. Chem. 1991, 37, 678–685. [PubMed] [Google Scholar]
- Suehiro M.; Murakami M.; Fukuchi M. Ann. Nucl. Med. 1994, 8, 231–7. 10.1007/BF03165025. [DOI] [PubMed] [Google Scholar]
- Kremer R.; Woodworth C. D.; Goltzman D. Am. J. Physiol. 1996, 271, C164–171. [DOI] [PubMed] [Google Scholar]
- El Abdaimi K.; Papavasiliou V.; Goltzman D.; Kremer R. Am. J. Physiol. Cell Physiol. 2000, 279, C1230–1238. [DOI] [PubMed] [Google Scholar]
- Luparello C. Cancers 2011, 3, 396–407. 10.3390/cancers3010396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds C. S.; Kovacs C. S. Crit. Rev. Eukaryotic Gene Expression 2010, 20, 235–273. 10.1615/CritRevEukarGeneExpr.v20.i3.40. [DOI] [PubMed] [Google Scholar]
- Sebag M.; Henderson J.; Goltzman D.; Kremer R. Am. J. Physiol. 1994, 267, C723–730. [DOI] [PubMed] [Google Scholar]
- McCauley L. K.; Martin T. J. J. Bone Miner. Res. 2012, 27, 1231–1239. 10.1002/jbmr.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecherstorfer M.; Schilling T.; Blind E.; Zimmer-Roth I.; Baumgartner G.; Ziegler R.; Raue F. J. Clin. Endocrinol. Metab. 1994, 78, 1268–1270. 10.1210/jcem.78.5.8175989. [DOI] [PubMed] [Google Scholar]
- Li J.; Karaplis A. C.; Huang D. C.; Siegel P. M.; Camirand A.; Yang X. F.; Muller W. J.; Kremer R. J. Clin. Invest. 2011, 121, 4655–4669. 10.1172/JCI46134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. C.; Yang X. F.; Ochietti B.; Fadhil I.; Camirand A.; Kremer R. Endocrinology 2014, 155, 3739–3749. 10.1210/en.2013-1803. [DOI] [PubMed] [Google Scholar]
- Dougherty K. M.; Blomme E. A.; Koh A. J.; Henderson J. E.; Pienta K. J.; Rosol T. J.; McCauley L. K. Cancer Res. 1999, 59, 6015–6022. [PubMed] [Google Scholar]
- Rabbani S. A.; Gladu J.; Harakidas P.; Jamison B.; Goltzman D. Int. J. Cancer 1999, 80, 257–264. . [DOI] [PubMed] [Google Scholar]
- Guise T. A.; Yin J. J.; Taylor S. D.; Kumagai Y.; Dallas M.; Boyce B. F.; Yoneda T.; Mundy G. R. J. Clin. Invest. 1996, 98, 1544–1549. 10.1172/JCI118947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T.; Yano S.; Hanibuchi M.; Kanematsu T.; Muguruma H.; Sone S. Int. J. Cancer 2004, 108, 511–515. 10.1002/ijc.11586. [DOI] [PubMed] [Google Scholar]
- Stewart A. F. N. Engl. J. Med. 2005, 352, 373–379. 10.1056/NEJMcp042806. [DOI] [PubMed] [Google Scholar]
- Nordholm A.; Rix M.; Olgaard K.; Lewin E. Scand. J. Clin. Lab. Invest. 2014, 74, 206–212. 10.3109/00365513.2013.876656. [DOI] [PubMed] [Google Scholar]
- Canario A. V. M; Rotllant J.; Fuentes J.; Guerreiro P. M.; Rita Teodósio H.; Power D. M.; Clark M. S. FEBS Lett. 2006, 580, 291–299. 10.1016/j.febslet.2005.12.023. [DOI] [PubMed] [Google Scholar]
- Kushnir M. M.; Rockwood A. L.; Strathmann F. G.; Frank E. L.; Straseski J. A.; Meikle A. W. Clin. Chem. 2016, 62, 218–226. 10.1373/clinchem.2015.244012. [DOI] [PubMed] [Google Scholar]
- Lu C. M.; Burton W. D.; Fitzgerald R. L.; Deftos L. J.; Buchholz B. A.; Vogel J. S.; Herold D. A. Anal. Chem. 2002, 74, 5507–5512. 10.1021/ac020182a. [DOI] [PubMed] [Google Scholar]
- Takahashi S.; Hakuta M.; Aiba K.; Ito Y.; Horikoshi N.; Miura M.; Hatake K.; Ogata E. Endocr.-Relat. Cancer 2003, 10, 403–407. 10.1677/erc.0.0100403. [DOI] [PubMed] [Google Scholar]
- Shan G.; Huang W.; Gee S. J.; Buchholz B. A.; Vogel J. S.; Hammock B. D. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 2445–2449. 10.1073/pnas.040575997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong N. U.; deB Edwardes M. D.; Papavasiliou V.; Goltzman D.; Kremer R. Am. J. Med. 2003, 115, 115–121. 10.1016/S0002-9343(03)00310-3. [DOI] [PubMed] [Google Scholar]
- Deftos L. J.; Barken I.; Burton D. W.; Hoffman R. M.; Geller J. Biochem. Biophys. Res. Commun. 2005, 327, 468–472. 10.1016/j.bbrc.2004.11.162. [DOI] [PubMed] [Google Scholar]
- Zhong Z.; Peng N.; Qing Y.; Shan J.; Li M.; Guan W.; Dai N.; Gu X.; Wang D. Electrochim. Acta 2011, 56, 5624–5629. 10.1016/j.electacta.2011.04.012. [DOI] [Google Scholar]
- Kuhn E.; Addona T.; Keshishian H.; Burgess M.; Mani D. R.; Lee R. T.; Sabatine M. S.; Gerszten R. E.; Carr S. A. Clin. Chem. 2009, 55, 1108–1117. 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otieno B. A.; Krause C. E.; Latus A.; Chikkaveeraiah B. V.; Faria R. C.; Rusling J. F. Biosens. Bioelectron. 2014, 53, 268–274. 10.1016/j.bios.2013.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Jensen G. C.; Krause C. E.; Sotzing G. A.; Rusling J. F. Phys. Chem. Chem. Phys. 2011, 13, 4888–4894. 10.1039/c0cp01755h. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Krause C. E.; Otieno B. A.; Latus A.; Faria R. C.; Patel V.; Gutkind J. S.; Rusling J. F. ChemistryOpen 2013, 2, 141–145. 10.1002/open.201300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer R.; Huang D. C.. PTHRP, its isoforms and antagonists thereto in the diagnosis and treatment of disease. U.S. Patent 7,897,139 B2, March 1, 2011.
- Yu X.; Munge B.; Patel V.; Jensen G.; Bhirde A.; Gong J. D.; Kim S. N.; Gillespie J.; Gutkind J. S.; Papadimitrakopoulos F.; Rusling J. F. J. Am. Chem. Soc. 2006, 128, 11199–11205. 10.1021/ja062117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweig M. H.; Campbell G. Clin. Chem. 1993, 39, 561–577. [PubMed] [Google Scholar]
- Washam C. L.; Byrum S. D.; Leitzel K.; Ali S. M.; Tackett A. J.; Gaddy D.; Sundermann S. E.; Lipton A.; Suva L. J. Cancer Epidemiol., Biomarkers Prev. 2013, 22, 972–983. 10.1158/1055-9965.EPI-12-1318-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.