Summary
Limited availability of human neurons poses a significant barrier to progress in biological and preclinical studies of the human nervous system. Current stem cell-based approaches of neuron generation are still hindered by prolonged culture requirements, protocol complexity, and variability in neuronal differentiation. Here we establish stable human induced neural stem cell (hiNSC) lines through the direct reprogramming of neonatal fibroblasts and adult adipose-derived stem cells. These hiNSCs can be passaged indefinitely and cryopreserved as colonies. Independently of media composition, hiNSCs robustly differentiate into TUJ1-positive neurons within 4 days, making them ideal for innervated co-cultures. In vivo, hiNSCs migrate, engraft, and contribute to both central and peripheral nervous systems. Lastly, we demonstrate utility of hiNSCs in a 3D human brain model. This method provides a valuable interdisciplinary tool that could be used to develop drug screening applications as well as patient-specific disease models related to disorders of innervation and the brain.
Highlights
-
•
Human induced neural stem cell (hiNSC) lines can be passaged and cryopreserved
-
•
Rapid and robust media-independent differentiation in as few as 4 days
-
•
hiNSCs contribute to both central and peripheral nervous systems in vivo
-
•
Demonstration of utility in innervated muscle co-culture and 3D human brain model
In this article, Kaplan and colleagues describe the generation of human induced neural stem cell lines, which undergo rapid neurogenesis independently of media composition. They demonstrate utility in innervated co-culture and a 3D human brain model. This method provides an interdisciplinary tool for developing patient-specific disease models related to disorders of innervation and the brain.
Introduction
Much of our understanding of the human nervous system is derived from animal studies as well as in vitro monoculture of various neural cell types. However, these studies do not truly recapitulate the complexity of human central and peripheral nervous systems, especially at the interface with different cell types. As such, there is a critical need to develop more physiologically relevant in vitro human models of the brain and innervated tissues.
One current cell source for neural tissue engineering applications is digested human brain tissue. Primary human neural cells harvested from fetal and cadaver samples often pose challenging ethical concerns and are unstable in culture. Commercially available cell lines have been produced to overcome this problem through the immortalization of some of these cell types such as ReNcell VM Human Neural Progenitor Cell Line, which is derived from the ventral mesencephalon region of a fetal brain (Donato et al., 2007). However, immortalized lines often function differently from their in vivo counterparts and are not suitable for certain applications such as the generation of specific disease models due to the lack of available brain samples from affected individuals. The discovery of induced pluripotent stem cell (iPSC) technology revolutionized the field of stem cell biology (Takahashi and Yamanaka, 2006). Introduction of the four reprogramming factors, octamer-binding transcription factor 4 (OCT4), SRY (sex-determining region Y) box 2 (SOX2), Kruppel-like factor 4 (KLF4), and c-MYC, into somatic cells under defined conditions results in iPSCs which, similarly to human embryonic stem cells (hESCs), have the capacity to subsequently differentiate into cell types of all three germ layers. A number of protocols have been developed to differentiate iPSCs into various neuronal phenotypes such as motor (Dimos et al., 2008) or dopaminergic (Cai et al., 2010) neurons. However, these protocols are often very time consuming, require multiple complicated intermediate steps such as the formation of non-adherent embryoid bodies and neurospheres (Jiang et al., 2012), and result in a large variability in neuronal differentiation (Hu et al., 2010). These issues associated with iPSC-derived neurons make them unsuitable for many biomedical applications.
Various groups have established methods to directly reprogram induced neurons (iNs) while omitting the PSC stage. Pang et al. (2011) demonstrated that forced expression of BRN2, ASCl1, MYT1l, and NEUROD1 converted human fibroblasts to induced neurons capable of generating action potentials. Pereira et al. (2014) used a similar approach to yield human iNs (hiNs) capable of surviving transplantation into the adult rat brain. This method of direct reprogramming of somatic cells into induced neurons circumvents some of the issues associated with iPSC-derived neurons, including teratoma formation resulting from a pluripotent intermediate and the extended time frame required for differentiation. Nevertheless, hiNs retain some significant challenges. Direct reprogramming into neurons is often inefficient, resulting in a relatively low yield of differentiated neurons (Ambasudhan et al., 2011). Furthermore, because neurons are terminally differentiated, hiNs are also unable to proliferate, thereby posing issues with respect to generating a sufficient supply of cells for subsequent applications.
Further advances in cellular reprogramming have resulted in the generation of induced neural stem cells (iNSCs), which do not pose the risks associated with iPSCs but, unlike terminally differentiated iNs, are capable of self-renewal. Various methods of generating human iNSCs (hiNSCs) have been described that utilize one or more of the standard pluripotent transcription factors (Lee et al., 2015, Wang et al., 2013a, Zhu et al., 2014). While significant progress has been made, there are issues among these techniques, such as the ability to expand and cryopreserve hiNSCs without affecting proliferative and differentiation capacities, relative ease of reprogramming and subsequent differentiation protocols, and discrepancies in overall efficiency.
This study describes a simple and efficient protocol for generating stable hiNSC lines by the direct reprogramming of primary human cells. This method results in the formation of hiNSC colonies that can be expanded indefinitely and cryopreserved without any discernible loss in proliferation or capacity for differentiation. While previous techniques of generating hiNSCs require multiple subsequent differentiation steps with strict media requirements, the ease of the protocol described herein makes it an ideal solution to some of the issues associated with other methods. Once removed from mouse embryonic fibroblasts (MEFs) and dissociated into single-cell suspension, hiNSCs robustly differentiate into ∼80%–90% βIII-tubulin (TUJ1)-, MAP2-, and NEUN-positive neurons in as few as 4 days, independently of media composition. Lastly, hiNSCs injected into an in vivo chick embryo model demonstrated the ability to migrate, engraft, and contribute to the formation of both the central and peripheral nervous systems, suggesting that these hiNSCs retain their neuronal phenotype in vivo even in non-neurogenic microenvironments. As a proof of concept, we demonstrate the utility of these cells in co-culture with skeletal muscle as well as a 3D tissue model of the human brain. Taken together, the positive attributes of reprogrammed hiNSC lines make them ideal for future biomedical applications such as high-throughput drug assays, complex innervated co-cultures, and 3D models using normal and diseased cells.
Results
hiNSC Colonies Exhibit Characteristics of Both hESCs and hNSCs
Reprogramming human neonatal dermis-derived fibroblasts or adult adipose-derived stem cells using a polycistronic virus expressing OCT4, KLF4, SOX2, and c-MYC in defined media on an MEF feeder layer (Figure 1) results in the formation of colonies that exhibit qualities of hESCs as well as hNSCs (for reprogramming efficiency, see Figure S1). Like hESCs, these colonies immunostain positive for OCT4, SOX2, and NANOG; however, they do not express cell-surface markers of pluripotency SSEA4 or TRA-1-81 (Figure 2A), suggesting that these reprogrammed colonies are not truly pluripotent. PCR analysis revealed that while reprogrammed hiNSC colonies express the pluripotent transcription factor NANOG, they also express relatively high levels of endogenous SOX2 expression, but not that of endogenous OCT4 (Figure 2B), providing further evidence of the lack of a pluripotent state. SOX2 has been shown to be a marker of self-renewing, multipotent NSCs (Ellis et al., 2004, Graham et al., 2003), and its endogenous upregulation is suggestive of an NSC fate. It is also important to note that exogenous transgene expression of the polycistronic virus is lost upon increased passage of the colonies and that resulting hiNSCs retain a normal karyotype (Figure S1). While hESCs have the capacity to differentiate into cells of all three germ layers, hiNSCs do not express markers of endoderm or mesoderm (Figure S1), providing further evidence of their specific ectodermal fate. Furthermore, while both undifferentiated hESC and hiNSC grow in tightly packed colonies with clear margins, their respective morphologies differ (Figure 2C). Growing on MEF feeders, hESC colonies appear relatively flat, while hiNSC colonies more closely resemble neurospheres and are dome-like in shape.
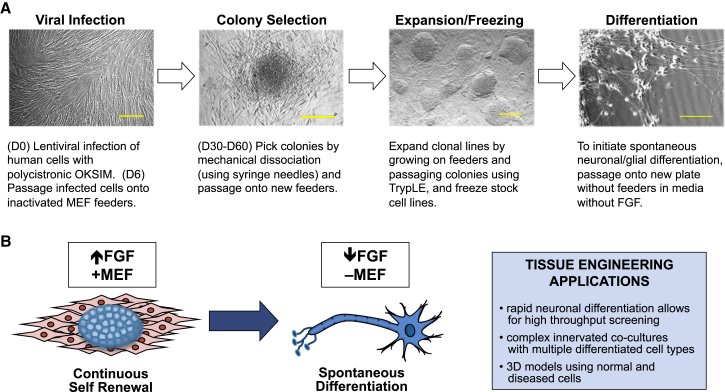
Figure 1.
Method of Reprogramming Human Cells into Induced Neural Stem Cells
(A) Protocol for generating hiNSCs. Scale bars, 100 μm.
(B) hiNSCs cultured as colonies on MEF feeders in the presence of high levels of FGF will proliferate indefinitely. Once removed from feeders, dissociated from colonies into single cells, and subsequently cultured in medium with reduced levels of FGF, hiNSCs exclusively and spontaneously differentiate into neurons and glia. This rapid and robust differentiation makes them uniquely suitable for high-throughput assays and tissue engineering strategies.
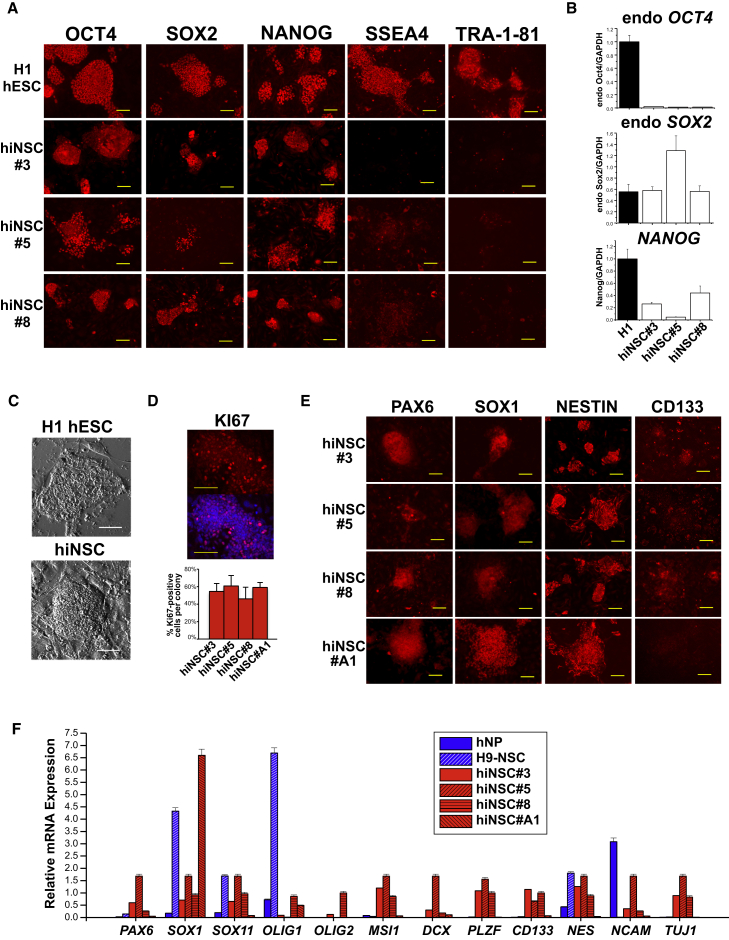
Figure 2.
Clonal hiNSC Colonies Display Characteristics of Both ESCs and NSCs
(A) H1 hESC colonies express all pluripotent markers while reprogrammed hiNSC clonal lines express markers OCT4, SOX2, and NANOG, but not SSEA4 or TRA-1-81.
(B) Gene-expression profile comparing H1 hESC with hiNSC clonal lines. Data represent mean ± SD of three independent experiments.
(C) Morphology of hESC colonies compared with hiNSC. hiNSCs exhibit a domed morphology.
(D) KI67 immunostaining reveals a large percentage of proliferating cells within hiNSC colonies.
(E) hiNSC colonies express NSC markers, PAX6, SOX1, NESTIN, and CD133.
(F) Gene-expression profile comparing expression of NSC markers in commercially available human NSC lines with that of hiNSC clonal lines. Data represent mean ± SD of three independent experiments.
All scale bars, 100 μm. See also Figures S1 and S6.
While hiNSC colonies share certain similarities with hESC, they also share multiple characteristics with neural stem cells. One criterion of NSCs is the ability to self-renew. Reprogrammed hiNSCs grown as colonies on MEFs show an average of between 46% and 60% KI67-positive cells for all clonal lines tested (Figure 2D). Furthermore, immunostaining results demonstrate that clonal lines express various markers typical of NSCs including SOX1, PAX6, NESTIN, and CD133 (Figure 2E). Compared with various established human neural cell lines including H9 hESC-derived NSC (H9-NSC) and immortalized neural progenitors from fetal brain tissue (hNP), clonal hiNSC lines also express a range of standard NSC markers to varying degrees (Figure 2F). While there is inherent variability between reprogrammed clonal hiNSC lines, it is also important to recognize the existing variability in expression between established and commercially available neural cell lines, which suggests that there is not a specific gold standard for distinguishing suitable neural precursors.
hiNSCs Rapidly and Spontaneously Differentiate into Multiple Neuronal and Glial Subtypes
Upon removal of hiNSC colonies from feeders, dissociation into single-cell suspension and subsequent culture in media without basic fibroblast growth factor (bFGF), hiNSCs become mostly TUJ1-positive in as few as 4 days (Figure 3A). They also begin to express glial fibrillary acidic protein (GFAP). By day 4, hiNSCs derived from neonatal dermal fibroblasts exhibited approximately 89% TUJ1-positive and 9% GFAP-positive cells, which increased to 90% and 33%, respectively, by day 14. In hiNSCs generated from adult adipose-derived stem cells, day-4 cells were 78% TUJ1- and 1% GFAP-positive, which increased to 92% and 12%, respectively, by day 14. Spontaneously differentiated hiNSCs also expressed other standard markers of neuronal and glial differentiation including MAP2, NEUN, and S100 (Figure S2).
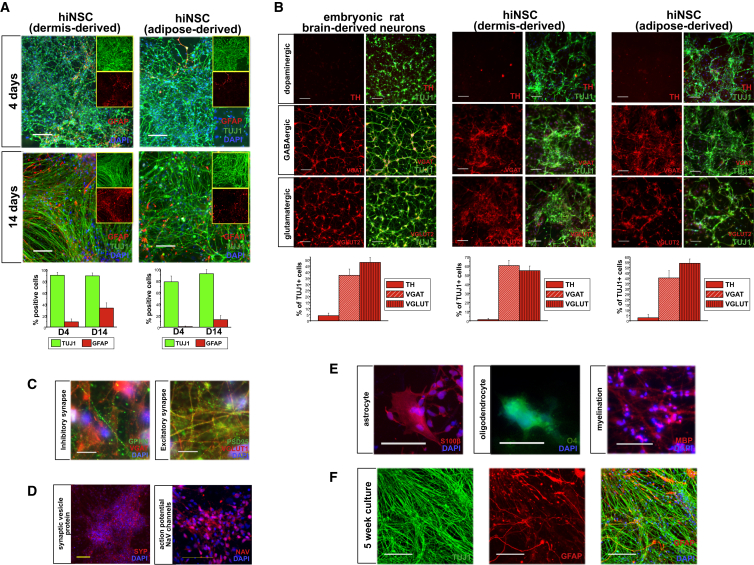
Figure 3.
hiNSCs Rapidly Differentiate into Various Neuronal and Glial Phenotypes
(A) Expression of neuron-specific βIII-tubulin (TUJ1) and glial fibrillary acidic protein (GFAP) in hiNSC clonal lines derived from HFF and hASC at 4 and 14 days. The large panel displays a merged image of TUJ1, GFAP, and DAPI; inlaid insets reveal separate images of TUJ1 (top) and GFAP (bottom). By 4 days, more than 80% of cells stain positive for neuron markers. Scale bars, 100 μm.
(B) hiNSCs spontaneously differentiate into subtypes of neurons. Embryonic rat brain-derived neurons are shown as a positive control. Scale bars, 50 μm.
(C) hiNSCs express post-synaptic proteins at both inhibitory (GEPHYRIN and VGAT) and excitatory (PSD95 and VGLUT1) synapses. Scale bars, 10 μm.
(D) hiNSCs spontaneously express synaptic vesicle protein, SYNAPTOPHYSIN, and voltage-gated sodium channel marker PAN-NAV. Scale bars, 100 μm.
(E) hiNSCs differentiate into multiple types of glia including astrocytes (S100β), oligodendrocytes (O4), and myelin marker positive glia (MBP). Scale bars, 100 μm.
(F) hiNSCs maintain neuronal and glial phenotypes in long-term cultures. Scale bars, 100 μm. Data in (A) and (B) represent mean ± SD of three independent experiments. See also Figures S2–S4 and S6.
While rapid TUJ1 expression is favorable, it is important to further characterize the resulting neuronal phenotypes (Figure 3B). Rat embryonic neurons (RENs) derived from E18 rat brain is a common cell type used in tissue engineering (Tang-Schomer et al., 2014). When cultured under conditions similar to those for hiNSCs, RENs mostly stain positive for vesicular glutamate transporter 2 (VGLUT2) (48%), followed by vesicular GABA transporter (VGAT) (37%), while very few stain positive for tyrosine hydroxylase (TH) (4%). Similarly, hiNSCs generated from neonatal dermal fibroblasts and adult adipose-derived stem cells exhibit a relatively high percentage of VGLUT+ (55% and 54%, respectively) as well as VGAT+ (60% and 40%, respectively).
The ability to form synapses is an important indicator of neuronal maturity. Spontaneously differentiated hiNSCs express inhibitory post-synaptic marker GEPHYRIN, as well as excitatory post-synaptic marker PSD95 (Figure 3C). They also express synaptic vesicle protein SYNAPTOPHYSIN and voltage-gated sodium channels (Figure 3D), which is indicative of their likely ability to fire action potentials.
Characterization of spontaneously differentiated subtypes of glial cells revealed the presence of myelinated neurons (myelin basic protein [MBP] positive). Furthermore, culturing hiNSCs for 8 days in fetal bovine serum (FBS)-based media supplemented with 100 μM retinoic acid (RA) resulted in high levels of S100β astrocyte marker expression and oligodendrocyte marker, O4 (Figure 3E), thereby demonstrating that reprogramming by this method results in tripotent hiNSCs.
For a number of biomedical applications, it is crucial that cells are sustainable in long-term culture. hiNSCs cultured for 5 weeks still express high levels of both TUJ1 and GFAP, and maintain a healthy neuronal morphology with elongated neurite extensions (Figure 3F).
Differentiated hiNSCs Are Functional In Vitro
While differentiated hiNSCs express various markers of mature neurons and glia within a relatively short period of time, it is important to also test functionality. One week after removal from MEFs, hiNSCs cultured on gelatin demonstrated the morphology of immature neurons (Figure 4A), characterized by cell bodies in the early stages of axon development. Fluo-4 calcium imaging of hiNSCs reveals that they have detectable levels of functional calcium signaling at this early time point (Figure 4B). Furthermore, electrophysiology results reveal that 1-week cultured cells have a resting membrane potential of approximately −50 mV (which corresponds to reported Vmem values of immature neurons (Ben-Ari et al., 2007) and that they demonstrate a depolarizing response to the GABA agonist, muscimol (Figure 4C), suggesting that they have functional GABA receptors and that they respond in a manner similar to that of typical immature neurons.
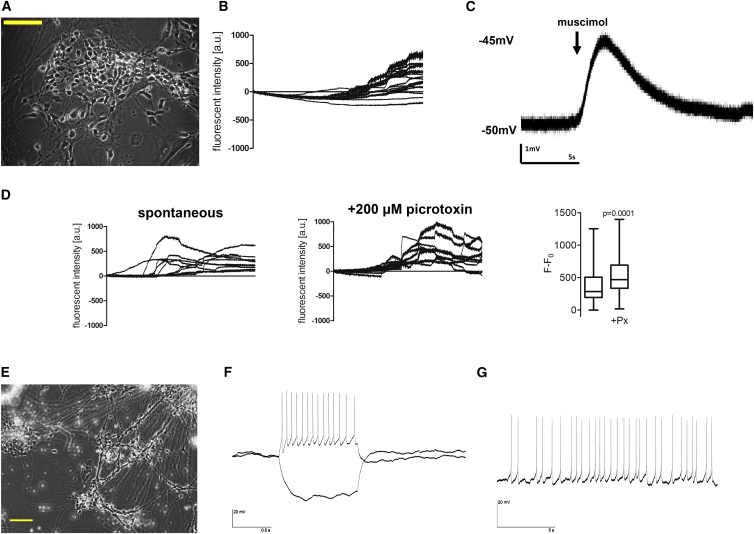
Figure 4.
hiNSCs Are Functional In Vitro
(A) Bright-field image of hiNSCs cultured for 1 week on gelatin. Scale bar, 100 μm.
(B) Calcium imaging of hiNSCs demonstrates active calcium signaling after 1 week.
(C) Electrophysiology recordings show that hiNSCs cultured for 1 week have functional GABA receptors that demonstrate depolarization in response to the GABA agonist muscimol.
(D) At 2 weeks of culture, hiNSCs display a significant increase in calcium signaling in response to picrotoxin. Data represents mean ± SD from eight cell traces.
(E–G) Bright-field image of hiNSCs (E) cultured for 8 weeks on poly-L-lysine-coated coverslips. Scale bar, 100 μm. Patch-clamp electrophysiology results demonstrate that hiNSCs elicit both current-induced (F) and spontaneous (G) action potentials.
Fluo-4 calcium imaging of 2-week-old cultures reveals that spontaneous calcium signaling is increased and that it is significantly activated in response to picrotoxin, an established blocker of inhibitory GABA receptors (Figure 4D). Because activation of GABA receptors inhibits neuronal firing, blockage of these receptors causes an increase in firing. Therefore, hiNSCs cultured for 1–2 weeks respond to well-characterized pharmaceuticals, as expected based on studies in animals and other in vitro neuron systems. Given the short amount of time required to elicit these functional responses, hiNSCs should have future applicability in high-throughput drug studies.
The gold standard for in vitro functionality of neurons is the ability to fire action potentials. hiNSCs were cultured on poly-L-lysine-coated coverslips for 8 weeks (Figure 4E), then subjected to electrophysiological analysis. Current-clamp recordings showed that the average resting membrane potential of recorded cells was between −50 and −70 mV. Differentiated hiNSCs displayed the ability to generate action potentials in response to depolarizing current steps (Figure 4F) as well as spontaneous action potentials (Figure 4G), thereby fully demonstrating maturity and functionality in vitro.
hiNSCs Migrate, Engraft, and Maintain Neuronal Phenotype In Vivo
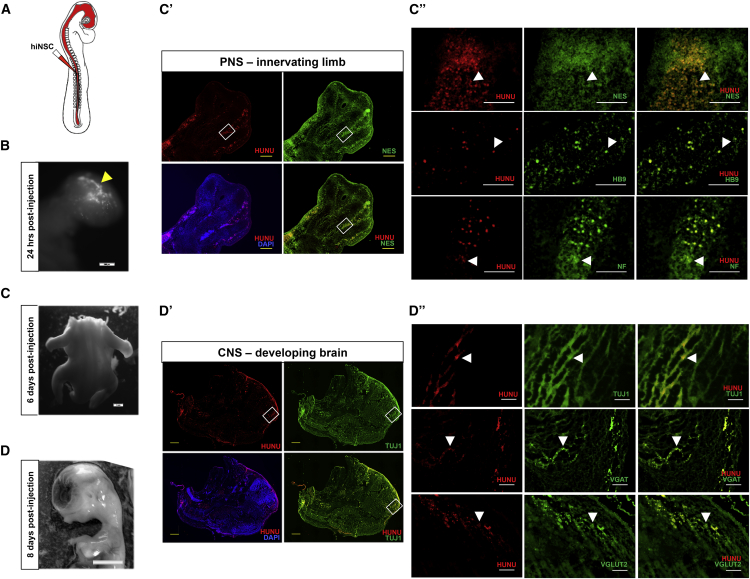
To assay the ability of hiNSCs to survive and differentiate in vivo, we utilized the chick embryo (Figure 5A). In brief, hiNSCs were dissociated, labeled, and injected into the neural tube of a 2.5-day-old chicken embryo. Embryos were allowed to grow for 1–8 days post transplantation and then harvested for analysis. After 24 hr, fluorescently labeled hiNSCs can be visualized within the primitive head region (Figure 5B). Six days after injection of hiNSCs, embryos were harvested and analyzed for incorporation of hiNSCs into the peripheral nervous system (Figures 5C–5C″). Immunostaining results of sagittal cryosections of the developing limb reveal the presence of human cells as indicated by positive staining of human nuclear antigen (HUNU). High-magnification images show that HUNU-positive cells co-localized with the neural stem cell marker, NESTIN. In addition, certain HUNU-positive cells also co-localized with homeobox transcription factor HB9, which plays a critical role in motor neuron differentiation during development, and with E1.9 (NF) antibody staining, which recognizes neurofilament protein expressed in sensory and motor axons (Stainier and Gilbert, 1990). This suggests that hiNSCs can contribute to the formation of the developing peripheral nervous system, and that they maintain their neuronal phenotype even in the presence of a mixed population of non-neuronal cells. Injected hiNSCs also contributed to the developing spine (Figure S5). Eight days after injection of hiNSCs, embryos were harvested and analyzed for incorporation of hiNSCs into the CNS (Figures 5D–5D″). Cryosections of the cranial region show HUNU-positive cells at multiple locations within the developing brain. Higher-magnification images demonstrate that HUNU-positive cells exclusively co-localized with TUJ1. In addition, some HUNU-positive cells present in the brain also co-expressed markers of GABAergic (VGAT) as well as glutamatergic (VGLUT2) neurons. Importantly, this demonstrates that hiNSCs can contribute to the CNS, and can differentiate into various subtypes of neurons in vivo. To date, few studies have demonstrated in vivo incorporation of hiNSCs into the developing CNS as well as the peripheral nervous system (PNS) extending into non-neuronal tissues.
Figure 5.
hiNSCs Migrate, Engraft, and Differentiate into Both the PNS and CNS In Vivo
(A) Schematic of injection method. hiNSC colonies were dissociated into single-cell suspension, fluorescently labeled using DiD, and injected into the entire lumen of the developing neural tube of ∼55-hr-old chicken embryos (in ovo). Embryos were harvested between 1 and 8 days for subsequent analysis.
(B) Arrowhead shows fluorescently labeled hiNSCs in the cranial region 24 hr post injection. Scale bar, 500 μm.
(C) Embryos were harvested at 6 days post injection and the developing limbs were cryosectioned. Scale bar, 1 mm.
(C′ and C″) Rectangular outlines in (C′) reflect magnified areas shown in (C″). Arrowheads indicate the presence of human cells as indicated by human nuclear antigen (HUNU) immunostaining, which co-localizes with NESTIN, HB9 (a marker of developing motor neurons), and NF (neurofilament of sensory and motor axons) in the developing limb. Scale bars, 500 μm in (C′) and 100 μm in (C″).
(D) Embryos were harvested at 8 days post injection and the head region cryosectioned. Scale bar, 5 mm.
(D′ and D″) Rectangular outlines in (D′) reflect magnified areas shown in (D″). Arrowheads reveal the presence of HUNU-positive cells, which broadly co-localized with TUJ1 as well as neuronal subtype-specific markers VGAT (GABAergic) and VGLUT2 (glutamatergic) in the developing brain. Scale bars, 1 mm in (D′) and 100 μm in (D″).
See also Figure S5.
hiNSCs Have Multiple Applications in Tissue Engineering
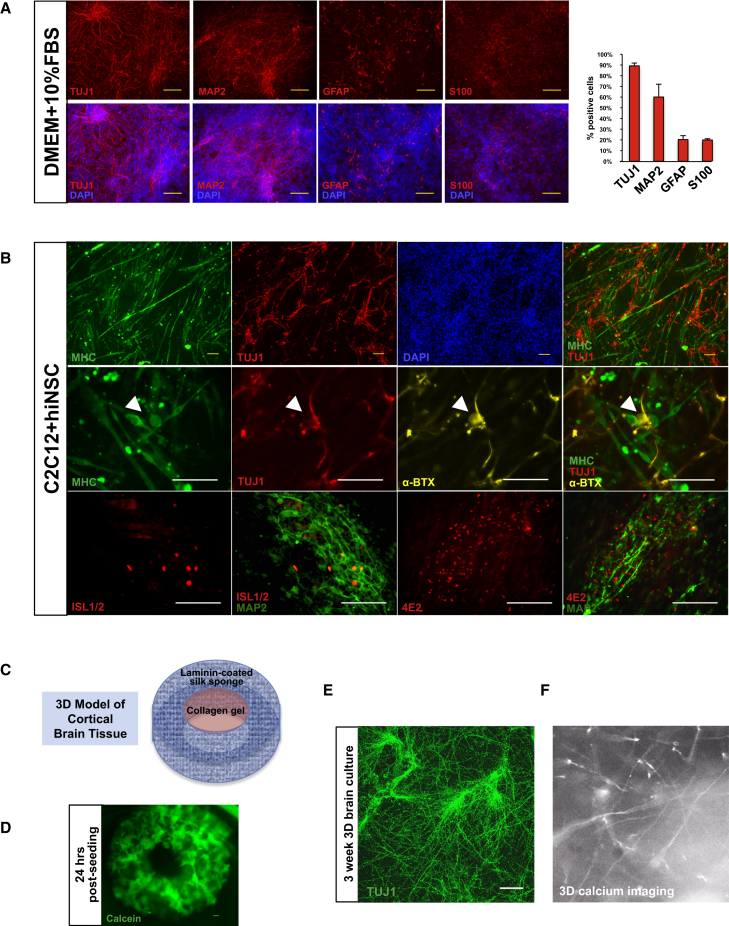
For cultured human neurons to be successfully incorporated into various tissue engineering models, it is crucial that they maintain their phenotype in multiple cell-culture media types and in differentiated co-cultures. hiNSCs grown in generic culture medium DMEM + 10% FBS are also mostly neuronal in phenotype (Figure 6A), with 89% TUJ1+, 60% MAP2+, 20% GFAP+, and 20% S100+. This robust differentiation even in undefined media suggests that hiNSCs can be incorporated into a variety of co-culture models for which FBS is the main supplement.
Figure 6.
Tissue Engineering Applications Using hiNSCs
(A) hiNSCs differentiate into mostly neuronal and glial phenotypes in various media types. Dissociated hiNSCs grown in undefined medium (DMEM + 10% FBS) for 8 days are ∼90% positive for neuronal marker TUJ1. Scale bars, 100 μm. Data represent means ± SD of three independent experiments.
(B) hiNSCs can be co-cultured with other differentiated cell types while still maintaining neuron-specific expression. C2C12, a murine myoblast cell line, was differentiated and co-cultured with hiNSCs for 4–5 days. hiNSCs remained TUJ1-positive in co-culture with differentiating skeletal muscle cells. Co-cultures also exhibited positive α-bungarotoxin (α-BTX) immunostaining (arrowheads), indicative of the presence of nicotinic acetylcholine receptors (AChRs) found in neuromuscular junctions. Differentiating hiNSCs in muscle co-cultures began to express markers of motor neurons (HB9) as well as Schwann cell-associated antigen (4E2/3G2). Scale bars, 100 μm.
(C) Incorporation of hiNSCs into a 3D brain model. This model consists of a silk sponge cut into the shape of a donut, which is coated with laminin. Cells are seeded into this outer ring and allowed to attach. Once attached, a collagen gel is added to the center of the donut, which allows for neurite growth and extension.
(D) Calcein staining of donuts 24 hr post seeding. Scale bar, 300 μm.
(E) TUJ1 immunostaining showing neurite extensions into the collagen gel in 3-week 3D brain cultures. Scale bar, 100 μm.
(F) Snapshot of live calcium signaling in 3D brain cultures.
See also Movie S1.
One example of a common cell type grown in FBS-based media is C2C12, an established murine myoblast cell line. C2C12 cells proliferate in DMEM supplemented with 10% FBS. When grown at high confluence and switched to low serum conditions (1% FBS), cells begin to fuse and form mature skeletal myotubes. C2C12 cells were grown in co-culture with hiNSCs in DMEM + 1% FBS for 4–5 days. Immunostaining analysis reveals the presence of differentiated myosin heavy chain (MHC)-positive myotubes, non-overlapping expression of TUJ1-positive hiNSCs with neurite extensions (Figure 6B), and the formation of α-bungarotoxin-positive neuromuscular junctions. Co-cultured hiNSCs also began to express ISLET1/2, transcription factors that promote motor neuron differentiation, as well as 4E2, a marker of Schwann cell protein found in regenerating nerves at the site of neuromuscular junctions (Astrow et al., 1994).
To test the ability of hiNSCs to integrate into a 3D environment, we incorporated them into the 3D cortical brain tissue model established by our laboratory, which had previously used embryonic rat-derived cortical neurons. Our 3D brain model consists of an aqueous silk scaffold in which the center is removed to create a donut-shaped construct (Figure 6C). This scaffold, which serves as the gray matter of the brain, is then coated with laminin. hiNSCs are seeded and allowed to adhere overnight. Calcein imaging of live cells 24 hr after seeding cells within the scaffold show localization of hiNSCs inside the silk sponge (Figure 6D). The following day, a collagen gel is added to the center to allow for neurite growth, thereby simulating the white matter of the brain. 3D cultures were fixed after 3 weeks in culture and immunostained for TUJ1, revealing the presence of elongated neurite extensions within the collagen gel (Figure 6E). Furthermore, hiNSCs seeded into the 3D brain model exhibited active firing as visualized by Fluo-4 calcium signaling (see also Movie S1), thereby demonstrating functionality of these neurons in 3D.
Discussion
This study describes a simplified protocol for the efficient generation of hiNSCs via direct reprogramming. hiNSC lines grow as colonies on MEF feeder layers. Like hESCs, hiNSCs stain positive for pluripotent transcription factors OCT4, SOX2, and NANOG, but do not express cell-surface markers SSEA4 and TRA-1-81, suggesting that these hiNSC clonal lines are not truly pluripotent. In a previous study using fluorescence-activated cell sorting to derive NSCs from H9 hESCs, only TRA-1-60−/SSEA4− cells could form neurospheres, suggesting that hESCs lacking these markers specifically adopt a neuroectodermal cell fate (Chaddah et al., 2012). All clonal lines tested showed an increase in endogenous SOX2 expression, but not endogenous OCT4 expression. Previous studies of various types of NSCs have demonstrated that the maintained expression of SOX2 and the absence of OCT4 is one of the hallmarks of NSCs (Graham et al., 2003, Mistri et al., 2015). Clonal lines generated by our method result in a highly pure population of hiNSCs lacking these pluripotent markers, providing further evidence of their NSC identity.
Studies have shown that the introduction of pluripotent factors under certain conditions yields iPSCs and not iNSCs. This protocol of directly reprogramming somatic cells into hiNSCs differs from the protocol used to generate iPSCs in several ways. First is the use of xeno-free serum replacement (SR). Rajala et al. (2007) tested nine different types of xeno-free culture media and found that none were able to maintain undifferentiated growth of hESCs, demonstrating a lack of pluripotency. While the xeno-free knockout (KO)-SR may contribute to the adoption of a stable non-pluripotent cell fate, another likely reason is the relatively high levels of bFGF. Neural induction from iPSCs has been shown to require very high levels of FGF (up to 100 ng/mL) (Nemati et al., 2011). Our method of reprogramming hiNSCs results in the formation of colonies that resemble the neurosphere stage, which relies mostly on high levels of FGF. The third factor is the use of MEFs in conjunction with xeno-free KO-SR and high FGF. MEF feeder layers have been shown to be crucial in maintaining the proliferation of undifferentiated stem cells (Thomson et al., 1998). It is plausible that the MEF feeder layers are helping to promote continuous self-renewal of hiNSCs while the xeno-free SR, which lacks the ability to maintain pluripotency of cells, and the high levels of FGF work in combination to promote the neural stem cell fate.
We are not the first investigators to describe a protocol for generating hiNSCs. Most protocols to generate hiNSCs that utilize similar reprogramming genes use them as separate factors (Wang et al., 2013a, Wang et al., 2013b). As such, there is an increased chance of variability with respect to controlling the relative expression of exogenously introduced genes that can accompany the use of multiple expression vectors. By using a singular polycistronic lentivirus, the relative expression and stoichiometry of the introduced factors is held constant in every round of reprogramming. Lastly, because our method could be used to successfully reprogram somatic cells from two different tissues, it is possible that it may also be utilized for other starting cell types. Previous studies have described the generation of hiNSCs; however, few have demonstrated the applicability of their respective protocols in reprogramming multiple cell types (Lee et al., 2015, Wang et al., 2013a, Zhu et al., 2015). For example, a method was described to reprogram urine-derived epithelial cells, but the same protocol could not be used to reprogram fibroblasts (Wang et al., 2013a). We fully acknowledge that herein we have only described the generation of hiNSC lines from two different starting cell types. It remains to be seen whether this method can effectively reprogram other starting cell types or whether it is restricted to human fibroblasts and adipose-derived stem cells.
Aside from the benefits of the actual protocol, our method also results in hiNSCs with multiple favorable properties including a seemingly endless capacity for self-renewal. hiNSC colonies grown on MEF feeders are approximately 50% positive for proliferation marker Ki67, a characteristic that does not change upon increased passage (Figure S6). Many previously described hiNSCs cannot be fully expanded in vitro. Serial passaging of NSCs generated from hematopoietic progenitors showed a decrease in the level of NESTIN staining in cells by passage 7 (Wang et al., 2013b, Wang et al., 2015). Clonal hiNSC lines generated by this method can be expanded indefinitely as colonies (>30 passages to date) and frozen and thawed without any discernible loss in capacity for differentiation (Figure S6). hiNSC lines have been validated by the expression of the NSC markers PAX6, SOX1, NESTIN, and CD133 by immunostaining, as well as other conventional NSC markers quantified by qRT-PCR (Figure 2). Gene-expression analysis demonstrates that these clonal lines all express detectable levels of these markers to varying degrees. It is possible that this variability is functionally relevant. For example, SOX1 expression was considerably higher in hiNSCs derived from adult adipose compared with those derived from neonatal dermis. SOX1 was found to play a critical role in the derivation of NSCs from human adipose-derived mesenchymal stem cells (Feng et al., 2014). Inhibition of SOX1 expression greatly reduced the resulting NSC yield, suggesting that SOX1 may also play a role in the induction of hiNSCs from adipose cells using our method. While there may be some level of variability between clonal hiNSC lines, there is also inherent variability between well-established neural cell lines (Figure 2F), suggesting that there is no benchmark gene-expression profile for human NSCs. All clonal lines tested express NSC-specific markers and do not express markers of other germ layer lineages (Figure S1), indicating that reprogrammed hiNSC colonies are truly neuroectodermal in fate.
While hiNSC colonies have a high proliferative capacity, their subsequent capacity for neuronal and glial differentiation is equally as robust. hiNSCs generated by this method do not require strict post-reprogramming media components or procedures, unlike many recently described hiNSC strategies. While some protocols of hiNSC differentiation require the formation of an intermediate non-adherent neurosphere step (Lee et al., 2015, Wang et al., 2013a), ours does not. For the purposes of tissue engineering this is ideal, as the transition from suspension to adherent substrates can be difficult (Marshall et al., 2008), and the core of neurospheres can often become necrotic (Cahan and Daley, 2013). Furthermore, most methods require specific media components to both differentiate and maintain a neuronal phenotype in culture. While previous hiNSC studies have reported TUJ1 immunostaining after 4 weeks of differentiation in specific media types supplemented with factors such as BDNF and GDNF (Zhu et al., 2014, Zhu et al., 2015), our hiNSC colonies, once trypsinized off of feeders, dissociated, and subcultured on different substrates, begin to differentiate into a high percentage of TUJ1-positive immature neurons and begin to express glial markers in as few as 4 days, independently of media composition. At this early time point they also highly expressed differentiated neuron markers MAP2 and NEUN (Figure S2). Upon differentiation, there was variability between hiNSCs derived from neonatal dermis compared with adult adipose in that dermally derived hiNSCs generated a higher proportion of glia. This discrepancy could be due to random variability between clonal lines, the donor cells being neonatal versus adult, or the different tissues of origin from which starting cells were harvested. Danielyan et al. (2007) reported that GFAP expression is detectable in human skin sections and in cultured fibroblasts. It is possible that dermally derived hiNSCs have a higher capacity for GFAP expression as they may retain cellular memory from their original tissue. Despite this slight variability in glia formation, hiNSCs still have wide utility for multiple applications.
Further analysis of subtype specification of hiNSCs indicated that after 2 weeks in culture, hiNSCs spontaneously differentiate into mostly glutamatergic and GABAergic neurons, and do not express high levels of the dopaminergic neuron marker TH. While these subtypes were obtained from spontaneous differentiation in generic neurobasal media supplemented with B27, we have shown that differentiation can also be directed. Culturing hiNSCs in the presence of certain combinations of Sonic hedgehog (Shh), FGF8, and/or RA resulted in the upregulation of motor neuron marker HB9, dopaminergic neuron marker TH, or glial marker GFAP (Figures S3 and S4), suggesting that while hiNSCs spontaneously differentiate into neurons and glia, this differentiation can be further guided by the addition of growth factors to generate specific neural and glial subtypes. Future studies using hiNSCs could be conducted to identify other potential factors that could be added to basal media to differentiate specific types of neural and/or glial cells, or to develop co-culture systems with other relevant cell types to induce various types of tissue-specific neurons.
At later time points, spontaneously differentiated hiNSCs expressed post-synaptic markers of both inhibitory and excitatory synapses as well as synaptic vesicle protein SYNAPTOPHYSIN, suggesting that hiNSCs can form functional synapses (Tarsa and Goda, 2002). hiNSCs also expressed voltage-gated sodium channels, which is suggestive of their ability to fire action potentials. Not only do hiNSCs spontaneously differentiate into various subtypes of neurons, they also differentiate into multiple types of glia cells including astrocytes and oligodendrocytes.
While hiNSCs express various markers of mature neurons and synapses as detected via immunostaining, it is crucial to validate their functionality. One week after removal from MEFs, hiNSCs exhibit detectable levels of calcium signaling as well as functional GABA receptors, as indicated by a depolarizing response to the GABA agonist muscimol (Figure 4), suggesting that they behave as physiologically immature neurons even at this early time point. hiNSCs differentiated for longer periods displayed the ability to generate both current-induced and spontaneous action potentials, thereby demonstrating their functionality in vitro.
While most in vivo validation studies involve the transplantation of neural cells into the brain of neonatal and/or adult rodent models followed by assay of neural marker expression after several weeks, we wanted to utilize an in vivo model in which we could specifically examine the ability of hiNSCs to rapidly incorporate into both the CNS and PNS. The chick embryo allows for the migration of cells throughout the entire developing neural tube, supporting subsequent integration into both the brain and non-neural tissues, which is not amenable to rodent models. Our study is not the first to utilize a chick embryo model to assay neural cell incorporation in vivo. Kharazi et al. (2013) demonstrated the ability of human fetal brain-derived NSCs to engraft into the lateral ventricle of chick embryonic brain. Another study transplanted mouse ESC-derived motor neurons into chick embryos to visualize extensions from the spinal cord into the periphery (Wichterle et al., 2009). Our model targeted the CNS and PNS by directly injecting hiNSCs into the entire lumen of the neural tube, allowing for growth into both the head and periphery. This in vivo model is more rigorous, as it challenges the cells to maintain their neuronal phenotype even in a microenvironment that may not be conducive to neurogenesis. All human cells assayed that stained positive for human nuclear antigen (HUNU) exclusively co-localized with NESTIN or TUJ1 (n = 227). It is important to note that neural crest cells, which migrate from the neural tube into peripheral tissues, have the capacity to become multiple cell types including melanocytes, cartilage, bone, and smooth muscle, as well as peripheral and enteric neurons and glia (Le Douarin et al., 2007). Upon injection into the chick neural tube, hiNSCs maintained neuronal lineage even when growing outside the brain; however, it is essential to address the limitations of this in vivo system compared with traditional rodent models. While we demonstrate in vitro that hiNSCs cannot be induced to differentiate into endodermal or mesodermal germ layers (Figure S1) and that upon culturing hiNSCs in permissive cell-culture media for the amount of time typically required for teratoma formation there is no spontaneous induction of pluripotent factor OCT4 or germ layer markers (Figure S6), this does not exclude the possibility that hiNSCs could ultimately result in teratoma formation when transplanted in vivo for multiple weeks. It should also be acknowledged that neural stem cells engrafted in rodent brains typically require several months to differentiate. Because hiNSCs generated by our method differentiate so quickly, it will be important to ensure that they also do not result in tumorigenesis and/or neural overgrowth upon transplantation into an adult rodent brain.
Given the robust capacity of these hiNSCs to maintain neuronal phenotype even in non-neuronal conditions, hiNSCs are highly desirable for multiple tissue engineering applications. When removed from MEF feeders and subsequently grown in generic medium DMEM + 10% FBS, hiNSCs express more than 90% TUJ1+ neurons, suggesting that they do not require neuron-specific media to become neurogenic, a feature that has not been explored with other recently described methods of generating iNSCs (Lee et al., 2015, Zhu et al., 2015). This maintenance of neuronal phenotype is very favorable for complex co-cultures where other cell types may have strict media requirements. As proof of principle, we co-cultured differentiating C2C12 skeletal muscle cells with hiNSCs under conditions favorable for the differentiation of C2C12. Both cell types differentiated, expressing their respective markers MHC and TUJ1 in a non-overlapping pattern, suggesting that both cell types can successfully differentiate in co-culture and maintain their individual phenotypes. Muscle neuron co-cultures also expressed α-bungarotoxin, which is suggestive of the presence of nicotinic acetylcholine receptors commonly found at the neuromuscular junction. Furthermore, co-cultured hiNSCs expressed motor neuron-specific ISLET1/2 as well as a marker of Schwann cells, suggesting that perhaps the presence of muscle cells guides the specification of adjacent hiNSCs. hiNSCs can be utilized to develop other physiologically relevant human innervated co-culture systems.
The ability of hiNSCs to grow and demonstrate functionality in a 3D model is also quite remarkable. We have previously described our 3D brain donut model using embryonic rat brain neurons (Chwalek et al., 2015, Tang-Schomer et al., 2014). Recent studies have developed 3D models for the culture of human neural cells, but have their limitations. A 3D human neural cell-culture model of Alzheimer's disease was developed by overexpressing human β-amyloid precursor protein and/or presenilin1 (PSEN1) in immortalized human fetal NSCs (ReNcell) (Choi et al., 2014). However, in vitro differentiation of this model required 6–12 weeks, and this technique does not allow for the incorporation of patient-specific cells. Pasca et al. (2015) described a 3D model of functional cortical neurons and astrocytes derived from human iPSCs; however, the time required to generate these models (between 52 and 137 days) makes this type of 3D culture quite prohibitive for higher-throughput applications. In our 3D brain model seeded with hiNSCs, we see long neurite extensions as early as 2 weeks, which demonstrate functional calcium signaling (see Movie S1).
While this technique of generating stable and rapidly differentiating hiNSC lines has many advantages, in its current form this method of reprogramming may not be suitable for clinical applications due to the presence of MYC and animal products. However, because this lentivirus contains an excisable cassette that can be removed upon addition of Cre-recombinase, it may not pose the risk of incorporating into the host genome. Furthermore, future efforts can be made to further adapt the protocol to be completely xeno-free. It is also important to acknowledge that the requirement of feeder layers for the derivation of these lines adds a layer of complexity that should be addressed upon further optimization of this method. Because the neurons derived from this method are so robust, they are currently amenable to a variety of high-throughput drug screening assays. In addition, future experiments can be performed to incorporate these hiNSCs into other 3D tissue models in which a neuronal component would be relevant, such as the skin and cornea, as well as disease models of the human brain.
Experimental Procedures
Generation of hiNSCs
Human neonatal foreskin fibroblasts (HFFs) (a gift from Dr. Jonathan Garlick, Tufts University) or adult human adipose-derived stem cells (hASCs) isolated as previously described (Abbott et al., 2015) were used to generate hiNSCs. In brief, HFFs or hASCs were plated at a concentration of 105 cells in one gelatin-coated well of a 6-well plate, and cultured in fibroblast medium (DMEM, 10% FBS, and 1% antibiotic-antimycotic). Concentrated virus was used to infect the cells in fibroblast medium with polybrene (Millipore) at an MOI of 1–2. The next day, medium was changed to fresh fibroblast medium. The following day, medium was changed to hiNSC medium: knockout (KO) DMEM supplemented with 20% KO xeno-free SR, 20 ng/mL recombinant bFGF, 1% Glutamax, 1% antibiotic-antimycotic, and 0.1 mM β-mercaptoethanol, which also contained 1% KO growth factor cocktail (GFC) (Invitrogen). Four days later, cells were trypsinized and replated on MEF feeder layers previously inactivated by mitomycin C. hiNSC medium (without KO-GFC) was subsequently changed every 1–3 days. At day 30 or later, colonies with a domed morphology were mechanically picked and passaged onto freshly mitotically inactivated MEFs. Each colony was used to generate one hiNSC line, which was expanded as colonies on MEF feeders. For further expansion, hiNSCs were enzymatically passaged as colonies using trypsin-like enzyme, TrypLE (Invitrogen), onto new MEF feeders for multiple passages, and subsequently frozen to make stocks.
Author Contributions
D.M.C. conceived the idea, performed experiments and data analysis, and wrote the paper. K.C., Y.E.M., M.R.K., and R.D.A. performed experiments and data analysis. D.L.K. and S.M. supervised the project. All authors commented on the manuscript.
Acknowledgments
We thank Erica Palma Kimmerling for helpful discussion. This study was funded by NIH P41 Tissue Engineering Resource Center Grant (EB002520) and R01 NS 092847, and by the German Research Foundation (DFG; CH 1400/2-1, Postdoctoral Fellowship for K.C.).
Published: August 25, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, two tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.07.017.
Supplemental Information
References
- Abbott R.D., Raja W.K., Wang R.Y., Stinson J.A., Glettig D.L., Burke K.A., Kaplan D.L. Long term perfusion system supporting adipogenesis. Methods. 2015;84:84–89. doi: 10.1016/j.ymeth.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambasudhan R., Talantova M., Coleman R., Yuan X., Zhu S., Lipton S.A., Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrow S.H., Son Y.J., Thompson W.J. Differential neural regulation of a neuromuscular junction-associated antigen in muscle fibers and Schwann cells. J. Neurobiol. 1994;25:937–952. doi: 10.1002/neu.480250804. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y., Gaiarsa J.L., Tyzio R., Khazipov R. GABA: a pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol. Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- Cahan P., Daley G.Q. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat. Rev. Mol. Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J.L., Yang M., Poremsky E., Kidd S., Schneider J.S., Iacovitti L. Dopaminergic neurons derived from human induced pluripotent stem cells survive and integrate into 6-OHDA-lesioned rats. Stem Cells Dev. 2010;19:1017–1023. doi: 10.1089/scd.2009.0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaddah R., Arntfield M., Runciman S., Clarke L., van der Kooy D. Clonal neural stem cells from human embryonic stem cell colonies. J. Neurosci. 2012;32:7771–7781. doi: 10.1523/JNEUROSCI.3286-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.H., Kim Y.H., Hebisch M., Sliwinski C., Lee S., D'Avanzo C., Chen H., Hooli B., Asselin C., Muffat J. A three-dimensional human neural cell culture model of Alzheimer's disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwalek K., Tang-Schomer M.D., Omenetto F.G., Kaplan D.L. In vitro bioengineered model of cortical brain tissue. Nat. Protoc. 2015;10:1362–1373. doi: 10.1038/nprot.2015.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielyan L., Tolstonog G., Traub P., Salvetter J., Gleiter C.H., Reisig D., Gebhardt R., Buniatian G.H. Colocalization of glial fibrillary acidic protein, metallothionein, and MHC II in human, rat, NOD/SCID, and nude mouse skin keratinocytes and fibroblasts. J. Invest Dermatol. 2007;127:555–563. doi: 10.1038/sj.jid.5700575. [DOI] [PubMed] [Google Scholar]
- Dimos J.T., Rodolfa K.T., Niakan K.K., Weisenthal L.M., Mitsumoto H., Chung W., Croft G.F., Saphier G., Leibel R., Goland R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Donato R., Miljan E.A., Hines S.J., Aouabdi S., Pollock K., Patel S., Edwards F.A., Sinden J.D. Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neurosci. 2007;8:36. doi: 10.1186/1471-2202-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P., Fagan B.M., Magness S.T., Hutton S., Taranova O., Hayashi S., McMahon A., Rao M., Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev. Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- Feng N., Han Q., Li J., Wang S., Li H., Yao X., Zhao R.C. Generation of highly purified neural stem cells from human adipose-derived mesenchymal stem cells by Sox1 activation. Stem Cells Dev. 2014;23:515–529. doi: 10.1089/scd.2013.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P., Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Hu B.Y., Weick J.P., Yu J., Ma L.X., Zhang X.Q., Thomson J.A., Zhang S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Ren Y., Yuen E.Y., Zhong P., Ghaedi M., Hu Z., Azabdaftari G., Nakaso K., Yan Z., Feng J. Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat. Commun. 2012;3:668. doi: 10.1038/ncomms1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharazi A., Levy M.L., Visperas M.C., Lin C.M. Chicken embryonic brain: an in vivo model for verifying neural stem cell potency. J. Neurosurg. 2013;119:512–519. doi: 10.3171/2013.1.JNS12698. [DOI] [PubMed] [Google Scholar]
- Le Douarin N.M., Brito J.M., Creuzet S. Role of the neural crest in face and brain development. Brain Res. Rev. 2007;55:237–247. doi: 10.1016/j.brainresrev.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Mitchell R.R., McNicol J.D., Shapovalova Z., Laronde S., Tanasijevic B., Milsom C., Casado F., Fiebig-Comyn A., Collins T.J. Single transcription factor conversion of human blood fate to NPCs with CNS and PNS developmental capacity. Cell Rep. 2015;11:1367–1376. doi: 10.1016/j.celrep.2015.04.056. [DOI] [PubMed] [Google Scholar]
- Marshall G.P., 2nd, Ross H.H., Suslov O., Zheng T., Steindler D.A., Laywell E.D. Production of neurospheres from CNS tissue. Methods Mol. Biol. 2008;438:135–150. doi: 10.1007/978-1-59745-133-8_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistri T.K., Devasia A.G., Chu L.T., Ng W.P., Halbritter F., Colby D., Martynoga B., Tomlinson S.R., Chambers I., Robson P., Wohland T. Selective influence of Sox2 on POU transcription factor binding in embryonic and neural stem cells. EMBO Rep. 2015;16:1177–1191. doi: 10.15252/embr.201540467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemati S., Hatami M., Kiani S., Hemmesi K., Gourabi H., Masoudi N., Alaei S., Baharvand H. Long-term self-renewable feeder-free human induced pluripotent stem cell-derived neural progenitors. Stem Cells Dev. 2011;20:503–514. doi: 10.1089/scd.2010.0143. [DOI] [PubMed] [Google Scholar]
- Pang Z.P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D.R., Yang T.Q., Citri A., Sebastiano V., Marro S., Sudhof T.C., Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O'Rourke N.A., Nguyen K.D. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira M., Pfisterer U., Rylander D., Torper O., Lau S., Lundblad M., Grealish S., Parmar M. Highly efficient generation of induced neurons from human fibroblasts that survive transplantation into the adult rat brain. Sci. Rep. 2014;4:6330. doi: 10.1038/srep06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala K., Hakala H., Panula S., Aivio S., Pihlajamaki H., Suuronen R., Hovatta O., Skottman H. Testing of nine different xeno-free culture media for human embryonic stem cell cultures. Hum. Reprod. 2007;22:1231–1238. doi: 10.1093/humrep/del523. [DOI] [PubMed] [Google Scholar]
- Stainier D.Y., Gilbert W. Pioneer neurons in the mouse trigeminal sensory system. Proc. Natl. Acad. Sci. USA. 1990;87:923–927. doi: 10.1073/pnas.87.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tang-Schomer M.D., White J.D., Tien L.W., Schmitt L.I., Valentin T.M., Graziano D.J., Hopkins A.M., Omenetto F.G., Haydon P.G., Kaplan D.L. Bioengineered functional brain-like cortical tissue. Proc. Natl. Acad. Sci. USA. 2014;111:13811–13816. doi: 10.1073/pnas.1324214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarsa L., Goda Y. Synaptophysin regulates activity-dependent synapse formation in cultured hippocampal neurons. Proc. Natl. Acad. Sci. USA. 2002;99:1012–1016. doi: 10.1073/pnas.022575999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wang L., Huang W., Su H., Xue Y., Su Z., Liao B., Wang H., Bao X., Qin D., He J. Generation of integration-free neural progenitor cells from cells in human urine. Nat. Methods. 2013;10:84–89. doi: 10.1038/nmeth.2283. [DOI] [PubMed] [Google Scholar]
- Wang T., Choi E., Monaco M.C., Campanac E., Medynets M., Do T., Rao P., Johnson K.R., Elkahloun A.G., Von Geldern G. Derivation of neural stem cells from human adult peripheral CD34+ cells for an autologous model of neuroinflammation. PLoS One. 2013;8:e81720. doi: 10.1371/journal.pone.0081720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Choi E., Monaco M.C., Major E.O., Medynets M., Nath A. Direct induction of human neural stem cells from peripheral blood hematopoietic progenitor cells. J. Vis. Exp. 2015:52298. doi: 10.3791/52298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H., Peljto M., Nedelec S. Xenotransplantation of embryonic stem cell-derived motor neurons into the developing chick spinal cord. Methods Mol. Biol. 2009;482:171–183. doi: 10.1007/978-1-59745-060-7_11. [DOI] [PubMed] [Google Scholar]
- Zhu S., Ambasudhan R., Sun W., Kim H.J., Talantova M., Wang X., Zhang M., Zhang Y., Laurent T., Parker J. Small molecules enable OCT4-mediated direct reprogramming into expandable human neural stem cells. Cell Res. 2014;24:126–129. doi: 10.1038/cr.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Wang H., Ding S. Reprogramming fibroblasts toward cardiomyocytes, neural stem cells and hepatocytes by cell activation and signaling-directed lineage conversion. Nat. Protoc. 2015;10:959–973. doi: 10.1038/nprot.2015.059. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.