Summary
A cardiomyocyte differentiation in vitro system from zebrafish embryos remains to be established. Here, we have determined pluripotency window of zebrafish embryos by analyzing their gene-expression patterns of pluripotency factors together with markers of three germ layers, and have found that zebrafish undergoes a very narrow period of pluripotency maintenance from zygotic genome activation to a brief moment after oblong stage. Based on the pluripotency and a combination of appropriate conditions, we established a rapid and efficient method for cardiomyocyte generation in vitro from primary embryonic cells. The induced cardiomyocytes differentiated into functional and specific cardiomyocyte subtypes. Notably, these in vitro generated cardiomyocytes exhibited typical contractile kinetics and electrophysiological features. The system provides a new paradigm of cardiomyocyte differentiation from primary embryonic cells in zebrafish. The technology provides a new platform for the study of heart development and regeneration, in addition to drug discovery, disease modeling, and assessment of cardiotoxic agents.
Keywords: cardiomyocytes, in vitro differentiation, embryogenesis, pluripotency, zebrafish
Graphical Abstract
Highlights
-
•
Zebrafish embryos may start to exit from pluripotency shortly after the oblong stage
-
•
Beating cell clusters are efficiently generated from zebrafish blastomeres
-
•
Beating cell clusters contain specific cardiomyocyte subtypes
-
•
Induced cardiomyocytes possess normal electrophysiological features
In this article, Zhou, Cheng, and colleagues describe a method of cardiomyocyte differentiation from primary embryonic cells in zebrafish after determining a pluripotency window of embryos. The induced cardiomyocytes differentiated into functional and specific cardiomyocyte subtypes, which exhibited typical contractile kinetics and electrophysiological features. The technology provides a new platform for the study of heart development and regeneration.
Introduction
Cardiovascular diseases rank the first in cause of death, which account for ∼30% of all deaths worldwide (Mozaffarian et al., 2015). Unlike robust cardiac regeneration observed in adult zebrafish and embryonic and neonatal mice (Porrello et al., 2011, Poss et al., 2002, Sturzu et al., 2015), adult mammalian hearts are proposed to be postmitotic (Rumiantsev and Carlson, 1991) and possess extremely low renewal potency (no more than 1%–2% per year) (van Berlo and Molkentin, 2014). Thus, the human heart is scarcely able to compensate the lost cardiomyocytes after heart failure (Bergmann et al., 2015, Laflamme and Murry, 2011). Promising therapeutic strategies to cure the injured heart include activation of endogenous cardiac progenitor/stem cell differentiation, stimulation of pre-existing cardiomyocyte proliferation via administration of chemical compounds, modified mRNAs, genes, and recombinant proteins (Eulalio et al., 2012, Sahara et al., 2015, Zangi et al., 2013), and transplantation of cardiac progenitor/stem cells/cardiomyocytes (Sahara et al., 2015). For the application of cardiomyocyte-based therapy to humans in the clinic, it is essential to clearly understand molecular mechanisms of differentiation from various cell sources to functional cardiomyocytes.
Several approaches have been established in mouse and human to obtain functional cardiomyocytes, including directed reprogramming of fibroblasts to cardiomyocytes using developmental transcription factors, such as GATA4, MEF2C, and TBX5, in mice (Ieda et al., 2010), activating resident stem or progenitor cells by WT1/THYMOSIN to induce the proliferation of endogenous cardiomyocytes in mice (Smart et al., 2011), and inducing differentiation of cardiomyocytes from embryonic stem cells in humans (Chong et al., 2014) or induced pluripotent stem cells (PSCs) from human fibroblasts and patients with heart failure (Burridge et al., 2014, Feaster et al., 2015, Zwi-Dantsis et al., 2013). Nevertheless, there are many factors and pathways, including genetic and epigenetic regulations (Burridge et al., 2015), which affect regenerative function of these cells. Establishment of novel differentiation models will undoubtedly help our understanding of molecular mechanisms of cardiomyocyte differentiation.
Zebrafish hearts can rapidly regenerate without scarring after 20% ventricular resection (Poss et al., 2002). Thus, zebrafish cardiac muscle differentiation is an ideal model for the interpretation of molecular mechanisms of cardiac regeneration in the mammalian heart. Recently, several factors and pathways in the regulation of cardiomyocyte regeneration have been identified in zebrafish, such as Caveolin-1, Cdk9, nuclear factor κB, Telomerase, Neuregulin 1 (Nrg1), microRNA-101a, and bone morphogenetic protein signaling (Beauchemin et al., 2015, Bednarek et al., 2015, Cao et al., 2016, Gemberling et al., 2015, Karra et al., 2015, Matrone et al., 2015, Uygur and Lee, 2016, Wu et al., 2016). Contractile cells were first observed under spontaneous differentiation conditions in vitro using late-gastrula cells (Huang et al., 2012). Heart aggregates were generated spontaneously, in vitro, from larval zebrafish 3 days post fertilization with an efficiency of 0.4 heart aggregate per larval fish (Grunow et al., 2015). Furthermore, several questions remain open, such as which conditions are required for efficient in vitro cardiomyocyte differentiation and whether cardiomyocyte-like cells produced in vitro are functional. Therefore, systematic modeling of cardiomyocyte differentiation with high efficiency is especially necessary in zebrafish. In this study, we established a rapid and efficient method for cardiomyocyte differentiation from zebrafish primary embryonic cells after determination of their pluripotency timing at different developmental stages. Crucial factors affecting in vitro generation of cardiomyocytes were then characterized. Contractile kinetics and sarcomere formation were also investigated. Lastly, functional electrophysiological features of beating cell clusters (BCCs) were identified. These findings are valuable for the development of high-throughput strategic screening of agents for drug discovery, disease modeling, and assessment of cardiotoxic agents, in addition to dissecting the molecular mechanisms of heart development and regeneration.
Results
Pluripotency Timing of Early Embryonic Cells in Zebrafish
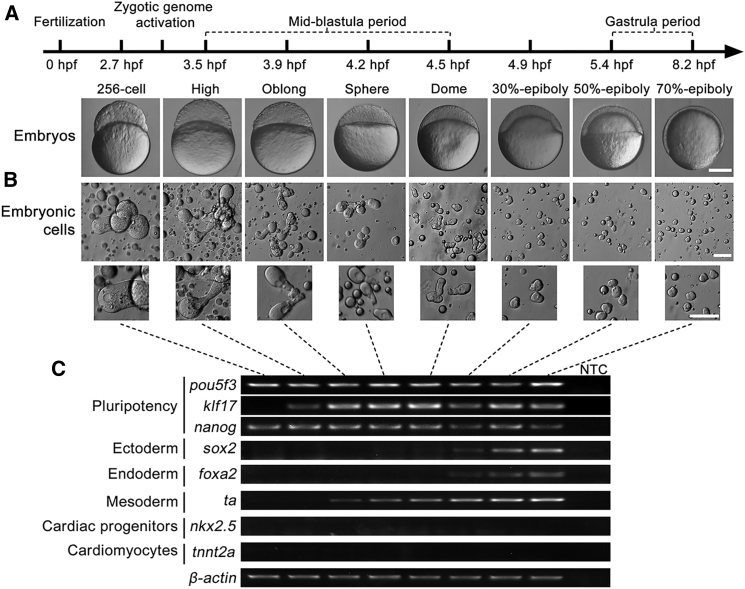
To determine the pluripotency of embryonic cells in zebrafish, we first detected the morphological changes in early embryonic cells at different developmental stages (Figures 1A and 1B). Calabash-like cells with a large cell size were clearly observed before dome stage in early embryonic cells (Figure 1B). The cells became gradually smaller in size until the stage of 50% epiboly. Notably, a cytoplasmic promontory was observed in all stages of development, indicating an asymmetric spread of the cytoplasm. We then performed expression analysis of both pluripotency and germ layer markers in primary embryonic cells in zebrafish at different developmental stages (Figure 1C). RT-PCR showed that pluripotency markers pou5f3 (POU domain class 5 transcription factor 3, also called oct4) and nanog (Nanog homeobox) were expressed at all stages, implying both maternal and zygotic expressions, while klf17 (Kruppel-like factor 17, also called klf4b) started to express at the high stage (3.5 hr post fertilization [hpf]), indicating an expression pattern after zygotic genome activation. To determine the stage of embryonic cell differentiation, we further investigated gene-expression patterns of three germ layers (ectoderm, mesoderm, and endoderm) (Figure 1C). Both ectoderm marker sox2 (SRY box 2) and endoderm marker foxa2 (forkhead box A2, also known as hnf3b) were expressed from the stage of 30% epiboly (4.9 hpf), while mesoderm marker ta (T brachyury homolog a, also known as ntla) was detectable from the oblong stage (3.9 hpf). Gene-expression patterns of germ layer markers together with asymmetric spread of the cytoplasm suggested that differentiation of cell lineages in zebrafish embryos occurs as early as the oblong stage. Thus, zebrafish embryonic cells undergo a very short pluripotent state from zygotic genome activation to a brief moment after the oblong stage.
Figure 1.
Pluripotency Timing during Embryogenesis in Zebrafish
(A) Representative morphology of early embryos at 256-cell, high, oblong, sphere, dome, 30%-epiboly, 50%-epiboly, and 70%-epiboly stages. hpf, hours post fertilization. Scale bar, 250 μm.
(B) Morphology of isolated embryonic cells from different stages. Asymmetric spread of cytoplasm was observed in all stages (lower panel). Scale bar, 50 μm.
(C) RT-PCR analysis of gene expression patterns in different stages of the embryos. Primer sequences and PCR conditions are listed in Table S1. NTC, no template control.
Efficient Cardiomyocyte Differentiation from Primary Embryonic Cells
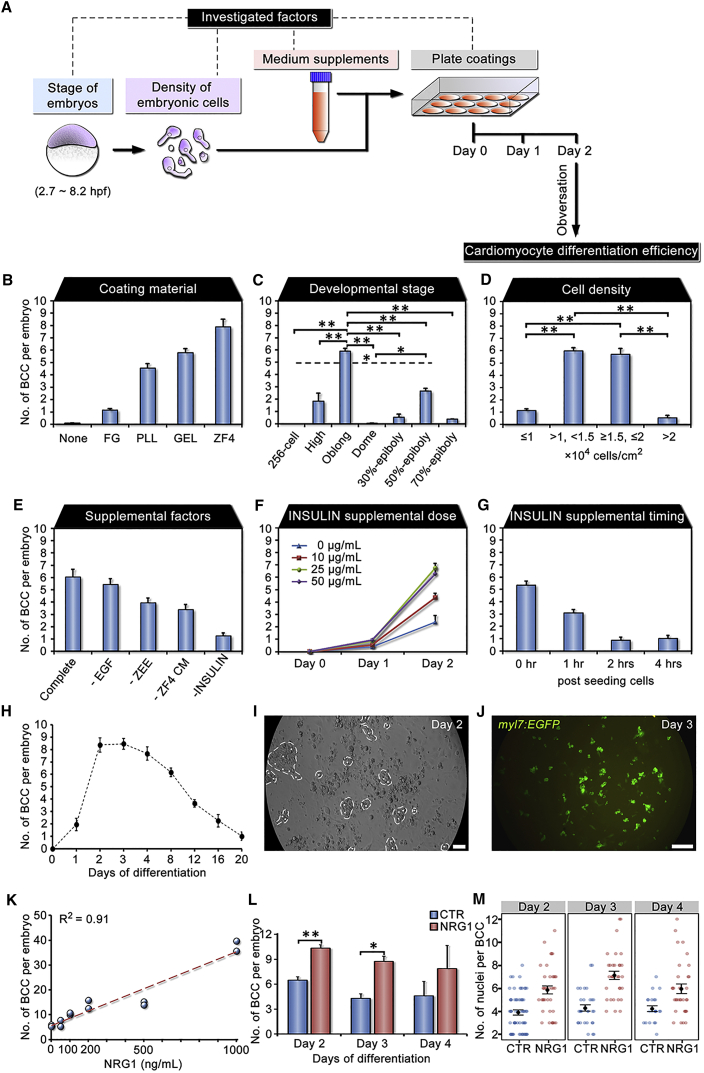
To develop an efficient method for in vitro cardiomyocyte differentiation using primary embryonic cells in zebrafish, we identified four groups of factors that all affected generation efficiency of BCCs, including coating materials, developmental stages, seeding density of the cells, and medium supplements (Figure 2A). A cell cluster exhibiting independent contraction activity was counted as one BCC. Total numbers of BCCs generated in the culture were recorded on day 2 of differentiation regardless of their size.
Figure 2.
Factors and Conditions that Affect Cardiomyocyte Differentiation Efficiency from Primary Embryonic Cells
(A) Selected factors affecting BCC (beating cell clusters) generation efficiency at different experimental steps, including stage of embryos, seeding density of embryonic cells, factors supplemented in the medium, and culture-ware coatings.
(B) Effect of culture-ware coatings on BCC generation efficiency. Oblong-stage embryonic cells were used for cardiomyocyte differentiation. Coating materials, fibrin gel (FG), poly-L-lysine (PLL, 0.1 mg/mL), gelatin (GEL, 2 mg/mL), ZF4 cells (ZF4, 2 × 104 cells/cm2), and control (None). Experiment repeated once, n = 3 wells of cells/group.
(C) Effect of developmental stages of embryonic cells on BCC generation efficiency. The culture plates were coated with gelatin. The dashed line indicates the comparison between 256-cell and 50% epiboly. Five independent experiments, n = 3–20 wells of cells/group.
(D) Effect of seeding density of embryonic cells on BCC generation efficiency. Embryonic cells from oblong stage were seeded in gelatin-coated plates at different cell densities during in vitro cardiomyocyte differentiation. Eight independent experiments, n = 3–10 wells of cells/group. In the following investigations (E–G), oblong-stage cells were seeded at a density of 1–2 × 104 cells/cm2 in gelatin-coated plates during in vitro cardiomyocyte differentiation.
(E) Evaluating effect of supplementation of epidermal growth factor (EGF), zebrafish embryonic extract (ZEE), ZF4 cell-conditioned medium (ZF4 CM), INSULIN (INS), or control (complete medium) on BCC generation efficiency by removing single factor. Experiment repeated once, n = 3 wells of cells/group.
(F) Effect of INSULIN supplemental doses on BCC generation efficiency at days 0, 1, and 2 of culture. INSULIN was supplemented at a concentration of 0, 10, 25, or 50 μg/mL. Experiment repeated once, n = 3 wells of cells/group.
(G) Effect of INSULIN addition (50 μg/mL) at 0, 1, 2, and 4 hr post seeding cells on BCC generation efficiency. Experiment repeated once, n = 3 wells of cells/group.
(H) Numbers of BCCs per embryo at different days of differentiation under optimum conditions from (B–G). Two independent experiments, n = 3–4 wells of cells/group.
(I) Representative image of efficient BCC generation at day 2 of differentiation. Dashed lines, beating areas; dots, contracting sites. The image was extracted from a video (Movie S1). Scale bar, 50 μm.
(J) EGFP expression driven by myl7 promoter in myl7:EGFP transgenic embryonic cells on day 3 of differentiation. Scale bar, 200 μm.
(K–M) Effects of NRG1 on cardiomyocyte proliferation using in vitro cardiac differentiation system in zebrafish. (K) A dose-response evaluation of NRG1 for BCC generation (NRG1 at 0, 50, 100, 200, 500 and 1,000 ng/mL). The linear regression line was y = 0.0297x + 5.6657. Two independent experiments, n = 2 wells of cells/group. (L) Effects of NRG1 treatment (100 ng/mL) on BCC formation on days 2, 3, and 4 of differentiation. Three independent experiments, n = 3–8 wells of cells/group. CTR, 0 ng/mL of NRG1. (M) Proliferative effects of NRG1 on cardiomyocytes. Cell culture was stained with Hoechst 33342 prior to observation under an inverted fluorescent microscope. Numbers of nuclei within each BCC (0 or 100 ng/mL of NRG1 treatment) were recorded on days 2, 3, and 4 of differentiation. Two independent experiments, n = 23–66 BCCs/group.
Data are shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01.
Firstly, we evaluated the effect of coating materials on plates, including fibrin gel (FG), poly-L-lysine (PLL), gelatin (GEL), feeder ZF4 cells (ZF4), or control (none), on cardiomyocyte differentiation efficiency from embryonic cells at the oblong stage by comparing the number of BCCs generated per embryo in each group. Results showed that ZF4 cell co-culture was the most efficient for BCC generation, and both PLL and GEL groups produced greater numbers of BCCs than the control group (Figure 2B).
Secondly, we compared BCC generation efficiency of the embryonic cells seeding at different developmental stages, including 256-cell, high, oblong, dome, 30% epiboly, 50% epiboly, and 70% epiboly, on gelatin-coated plates to determine an optimum stage for cardiomyocyte differentiation. Embryonic cells at the oblong stage showed the greatest efficiency for cardiomyocyte generation in comparison with the other stages (p < 0.01; Figure 2C).
Thirdly, since seeding density of embryonic stem-like cells altered their fates for differentiation in a previous study (Ho et al., 2014), we investigated the effect of seeding density of the cells on their cardiomyocyte induction potential. We observed that cells seeding at a density ranging from 1–2 × 104 cells/cm2 had higher BCC yield than the other densities (p < 0.01; Figure 2D). High density of primary embryonic cells led to the formation of large cell aggregates, which eventually did not differentiate into cardiomyocytes. Thus, the seeding density of embryonic cells is important for efficient BCC generation.
Finally, we evaluated the effect of supplemental factors on the cardiomyocyte induction, including epidermal growth factor (EGF), zebrafish embryonic extract (ZEE), ZF4 cell-conditioned medium (ZF4 CM), and INSULIN. On removal of a single factor from the recipe of the medium in each group, INSULIN affected the BCC generation efficiency, ZEE or ZF4 CM deduction also decreased the efficiency, while EGF did not (Figure 2E). INSULIN addition had a dose-dependent effect on the induction efficiency at concentrations of 0, 10, 25, and 50 μg/mL with a greater efficiency when added at the beginning of the induction (Figures 2F and 2G). Thus, maximum induction efficiency for cardiomyocyte differentiation can be achieved using the combination of oblong-stage embryonic cells at a density from 1–2 × 104 cells/cm2, ZF4 feeder cells, and supplements of ZEE, ZF4 CM, and INSULIN. Using this condition, we observed that the BCCs can present within as early as 28 hr of the induction, and the number of BCCs reached a peak on day 2 (8.4 ± 0.6 BCCs per embryo) (Figures 2H and 2I; Movie S1). The contraction activity was decreased in some BCCs after 8 days of differentiation while the beats were retained in the others for up to 20 days (Figure 2H). In addition, cardiac marker Myl7 can be detected in these induced cardiomyocytes from myl7:EGFP transgenic zebrafish (Figure 2J). These results indicate that this culture process for cardiomyocyte induction is close to in vivo cardiogenesis timing (24 hpf) in zebrafish (Kimmel et al., 1995).
Nrg1 has been recently reported as a cardiac mitogen, which promoted endogenous heart regeneration in zebrafish (Gemberling et al., 2015). To explore the regenerative responses of cardiomyocytes to the mitogen, we investigated the effects of NRG1 treatment on formation and proliferation of zebrafish cardiomyocytes using our culture system in vitro. We observed that efficiencies of generating BCCs from embryonic cells were increased linearly (R2 = 0.91) with an increasing amount of NRG1 (Figure 2K). NRG1 significantly promoted the cardiac differentiation of embryonic cells on day 2 (p < 0.01) and day 3 (p < 0.05; Figure 2L). In addition, NRG1-treated BCCs showed more nuclei than control BCCs, and numbers of nuclei NRG1-treated BCCs increased from day 2 to day 3 (Figure 2M). These data suggested that Nrg1 promoted proliferation and formation of cardiomyocytes in zebrafish.
Morphology and Contractile Kinetics of Beating Cell Clusters
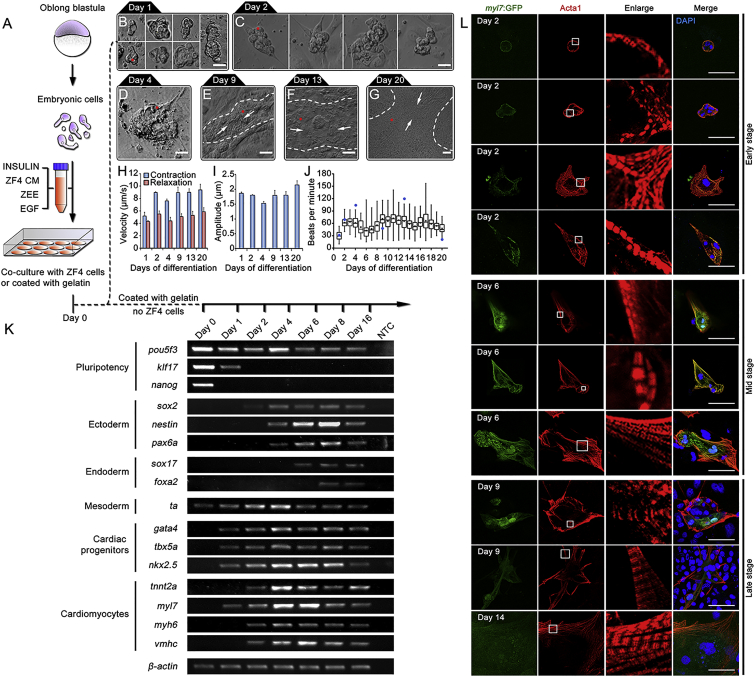
For morphological observations, we performed induction culture using the optimal condition mentioned above, and observed morphological changes in the BCCs at different stages (Figure 3A). Most of BCCs on days 1 and 2 were irregular spheres, fusiform, or long-shuttle in shape (Figures 3B and 3C). Afterward the BCCs gradually spread, their contracting area enlarged (Figures 3D and 3E), and they finally became rhythmically beating cell sheets, which lasted for more than 20 days (Figures 3F and 3G).
Figure 3.
Morphology and Beating Characteristics of BCCs Differentiated from Primary Embryonic Cells
(A) Schematic diagram of in vitro differentiation procedures.
(B–G) Morphology of BCCs at different days of differentiation when co-culturing with ZF4 cells. Dashed lines, contracting area; arrows, contracting directions; Red dots show the sites that were used for track analysis of BCC contractile kinetics in (H) and (I). Scale bar, 20 μm.
(H and I) Contraction and relaxation velocity (H) and contraction amplitudes (I) of BCCs at different days of differentiation. Three contractions of each BCC pointed by red dots in (B–G) were tracked. Data are shown as mean ± SEM.
(J) Beating rates of BCCs at day 1–20. Cells were cultured in gelatin-coated plates. Two independent experiments, n = 52–101 BCCs/group. Blue dots show the beating rates of the BCC in (B–G).
(K) Gene-expression analysis by RT-PCR during cardiomyocyte differentiation of primary embryonic cells. Primer sequences and PCR conditions are listed in Table S1. NTC, no template control.
(L) Cellular and molecular characteristics of Tg (myl7:EGFP) BCCs at different stages of differentiation. Early stage, day 2; middle stage, day 6; late stage, days 9 and 14. Cardiomyocytes marker, Myl7 (green); sarcomeres maker, α-skeletal actin (Acta1, red). Enlarged squares indicate the process of sarcomere formation during cardiomyocyte maturation. Nuclei were stained with DAPI (blue). Scale bars, 50 μm.
We then measured the contraction/relaxation velocity and amplitude of BCCs. Contraction velocity was higher than the relaxation velocity (8.2 ± 0.7 μm/s versus 5.1 ± 0.3 μm/s; Figure 3H), while their contractile amplitude was maintained at 1.8 ± 0.1 μm for 20 days (Figure 3I). These BCCs beat slowly during the first day (29.6 ± 1.3 bpm) and then faster at a rate of 59.0 ± 0.5 bpm (Figure 3J). These kinetics features are comparable with those of the cardiomyocytes derived from human stem cells (Asp et al., 2010, Hayakawa et al., 2014).
Cardiac Marker Expression and Sarcomere Formation
To investigate gene-expression patterns of the embryonic cells during differentiation induction, we analyzed four types of markers for pluripotency, germ layers, cardiac progenitors, and cardiomyocytes. RT-PCR showed that klf17 and nanog were expressed only for the first 2 days, while pou5f3 expression gradually decreased. For the germ layer markers, ectoderm genes (sox2, nestin, and pax6a) started to express on day 4 and then endoderm (sox17, and foxa2) genes followed their expression, whereas expression of mesoderm marker ta was detectable from days 0–16 (Figure 3K). We further investigated marker expressions of cardiac progenitors and cardiomyocytes. The genes for cardiac progenitors (gata4, tbx5a, and nkx2.5) started to express from day 1, while cardiomyocyte markers (tnnt2a, myh6, and vmhc) appeared on the second day except for myl7, which was expressed from day 1 (Figure 3K). Indeed these makers (nkx2.5 and tnnt2a) were not expressed during early stages of embryogenesis (Figure 1C). These results indicated that cardiac progenitor cells and cardiomyocytes had generated and that pluripotency decreased during cardiomyocyte induction.
To characterize sarcomere formation, we used myl7:EGFP transgenic primary embryonic cells for BCC generation, which enabled us to evaluate the expression of cardiac marker myl7 during cardiomyocyte differentiation. Immunostaining of α-skeletal muscle actin (Acta1, a marker for I bands of sarcomeres) showed continuous dot-like signals in BCCs on day 2, indicating an early phase of sarcomeric assembly. Continuous band-like signals could be observed in some BCCs at day 6, and a typical pattern of sarcomeric I bands was detected in some spread BCCs at days 9 and 14 (Figure 3L), indicating mature sarcomere formation. However, mature and well-organized sarcomeres were scarcely detected in ball-like BCCs (upper panel of day 9 in Figure 3L). These data demonstrated that the BCCs derived from primary embryonic cells expressed cardiac markers of both progenitor cells and cardiomyocytes and presented a dynamic sarcomeric maturation process within the induced cardiomyocytes.
BCCs Display Functional Electrophysiological Features
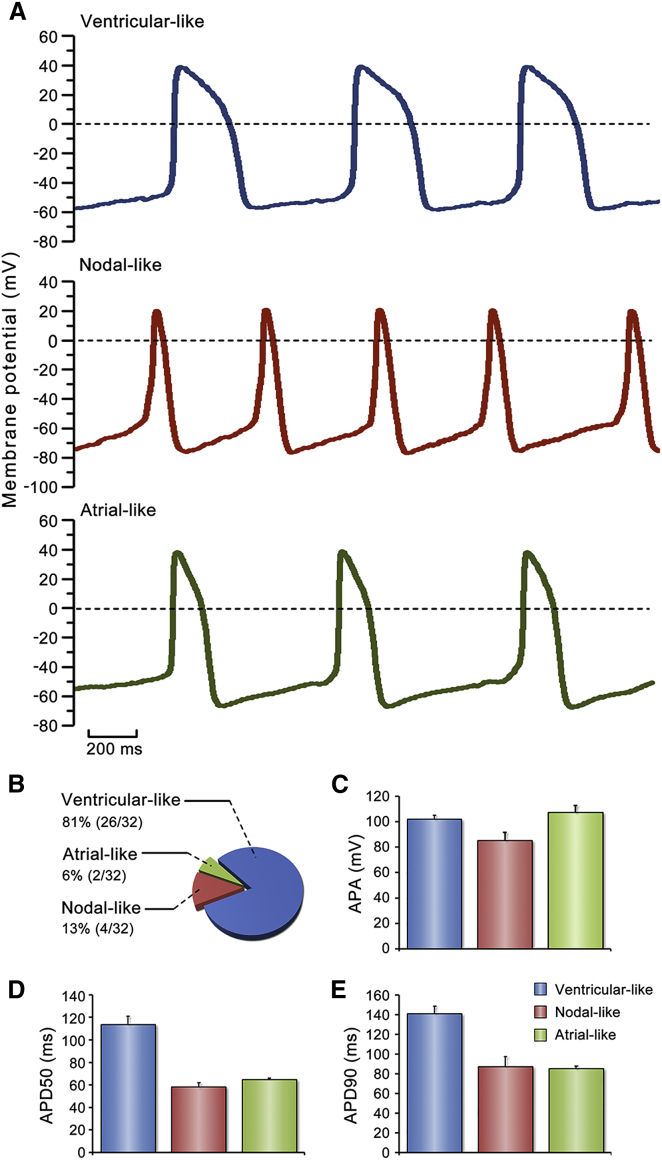
To address the question of whether the generated BCCs have functional features of cardiomyocytes, we first recorded their spontaneous whole-cell action potentials (APs) using a patch-clamp amplifier. Three typical AP patterns, namely ventricular-like, nodal-like, and atrial-like, were observed in the BCCs (Figure 4A). The BCCs with ventricular-like APs exhibited a rapid upstroke and a distinct plateau phase. Nodal-like APs showed a typical phase 4 of pacemaker potential, which depolarized slowly up to the threshold potential followed by a more gradual upstroke and then a rapid repolarization phase. Atrial-like BCCs indicated a fast depolarization pattern similar to that of ventricular-like BCCs, but with no plateau phase during their repolarization. No membrane potential upstrokes were detected in nonbeating embryonic cells. Among tested BCCs, 81% cells were ventricular-like, 13% were nodal-like, and 6% were atrial-like subtype cardiomyocytes (Figure 4B). All three subtypes had similar AP amplitude (APA; Figure 4C). However, the ventricular-like subtype showed longer AP durations as indicated in both APD50 (AP duration at 50% repolarization) and APD90 than both nodal-like and atrial-like subtype cardiomyocytes (Figures 4D and 4E). Thus, the induction can generate three typical subtypes of cardiomyocytes with distinct cardiac spontaneous AP patterns.
Figure 4.
Electrophysiological Characterization of BCCs
(A) Representative action potential (AP) patterns of three subtypes of cardiomyocytes. AP was recorded by whole-cell patch-clamp method on day 5 of differentiation.
(B) Proportions of ventricular-like, nodal-like, and atrial-like BCCs.
(C–E) AP amplitude (APA), AP duration at 50% repolarization (APD50), and AP duration at 90% repolarization (APD90) in three types of BCCs. Two independent experiments: ventricular-like, n = 22 BCCs; nodal-like, n = 4 BCCs; atrial-like, n = 2 BCCs.
Data are shown as mean ± SEM.
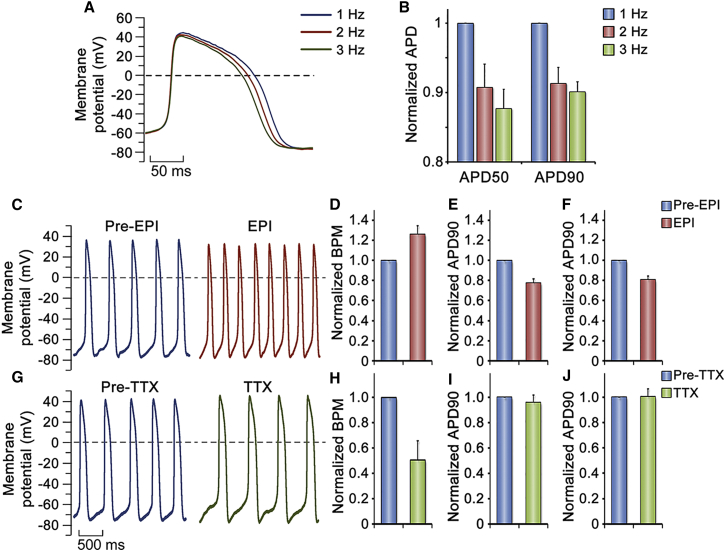
Next, we evaluated the response of the BCCs to current pulse stimulation at an increasing frequency. The tested cells exhibited shortened APD as the frequency of stimulation increased (1, 2, and 3 Hz) (Figure 5A). Both APD50 and APD90 declined when stimulated at 3 Hz, and 2-Hz stimulation reduced APD90 of BCCs compared with 1-Hz stimulation (Figure 5B). Therefore, induced cardiomyocytes were able to efficiently respond to current pulse stimulation.
Figure 5.
Stimulation Rates Adaptation and Drug Responses of BCCs
(A) APs of day-5 ventricular-like BCCs were recorded at 5-ms current pulse stimulation at 1, 2, and 3 Hz. All APs were from the same BCC stimulated at 1, 2, and 3 Hz.
(B) Fractional changes of AP durations at 50% and 90% repolarization. The values were normalized to the values observed at 1-Hz stimulation. Experiment repeated once, n = 5 BCCs.
(C–J) Effect of adrenergic receptor agonist and Na+ channel blocker on spontaneous AP of day-5 BCCs. (C) Representative APs of the BCC before (Pre-EPI) and during (EPI) perfusion of bath solution containing 10 μM adrenergic receptor agonist epinephrine (EPI). (D–F) Fractional changes of beating rates (D), AP durations at 50% repolarization (E), and AP durations at 90% repolarization (F). The values were normalized to the values obtained before EPI applied (Pre-EPI). Experiment repeated once, n = 8 BCCs. (G) Representative APs of the BCC before (Pre-TTX) and during (TTX) perfusion of bath solution containing 10 nM Na+ channel blocker tetrodotoxin (TTX). (H–J) Fractional changes of beating rates (H), AP durations at 50% repolarization (I), and AP durations at 90% repolarization (J). The values were normalized to the values obtained before TTX applied (Pre-TTX). Experiment repeated once, n = 4 BCCs.
Data are shown as mean ± SEM.
Because response to epinephrine (EPI) is an essential feature of the heart, we then tested whether BCCs could respond to epinephrine treatment. An accelerating frequency of membrane potential was detected upon EPI treatment (Figures 5C and 5D). Correspondingly, both APD50 and APD90 were reduced by the treatment (Figures 5E and 5F), indicating functional existence of β-adrenergic receptors in the BCCs.
As tetrodotoxin (TTX) can block cellular Na+ channels, we finally examined whether induced cardiomyocytes possessed TTX-sensitive Na+ channels. AP records showed that TTX had no effect on APA but reduced AP frequencies (Figures 5G and 5H). Neither APD50 nor APD90 of the BCCs was affected by the TTX treatment (Figures 5I and 5J). These data demonstrated that TTX-sensitive Na+ channels were functional in BCCs.
Discussion
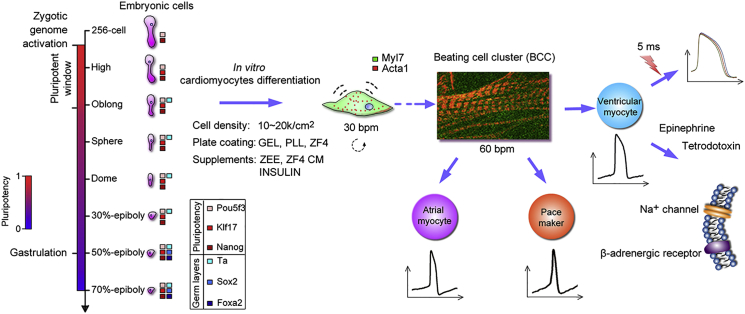
Given the physiological similarities of cardiomyocytes between zebrafish and human (Brette et al., 2008, Nemtsas et al., 2010), the zebrafish has emerged as an outstanding system for studying cardiac development and regeneration, performing cardiac-associated phenotypic screening, testing cardiotoxicity, and modeling heart diseases (Arnaout et al., 2007, Asnani and Peterson, 2014, Bakkers, 2011, Jopling et al., 2010, Kikuchi, 2014, Liu and Stainier, 2012, Ni et al., 2011). In the present study, we have determined the pluripotency window of zebrafish embryos by analyzing gene-expression patterns of pluripotency factors in combination with markers of three germ layers. Based on the pluripotency, we established a rapid and efficient method for cardiomyocyte generation in vitro from primary embryonic cells. The induced cardiomyocytes differentiated into specific cardiomyocyte subtypes including ventricular, atrial, and pacemaker cells. We then characterized the contractile kinetics and electrophysiological functions of these in vitro generated cardiomyocytes. The system depicts a new paradigm of cardiomyocyte differentiation from primary embryonic cells (Figure 6).
Figure 6.
A Working Model for In Vitro Cardiomyocyte Differentiation from Primary Embryonic Cells in Zebrafish
A pluripotent window of embryogenesis in zebrafish, from zygotic genome activation to a short moment after the oblong stage (<2 hr), allows us to efficiently induce the oblong-stage cells to differentiate into functional cardiomyocytes under a combination of optimal conditions. The induced cardiomyocytes can differentiate into specific cardiomyocyte subtypes, including ventricular, atrial, and pacemaker cells.
The pluripotency of zebrafish embryos remains elusive. Several stem-like cell lines have been derived from oblong-stage embryos (Fan et al., 2004, Ho et al., 2014, Hong et al., 2014). Blastomere or cultured embryonic cell transplant studies showed the existence of uncommitted cells in gastrula (Fan et al., 2004, Ho and Kimmel, 1993). It is possible in zebrafish that some embryonic cells remain in a pluripotent state when the mesoderm starts to be specified in midblastula embryos. Indeed, recent single-cell analysis showed that genome methylation levels are quite different from zygotes to blastula in mammals (Guo et al., 2014), indicating a differential pluripotent state among embryonic cells. In addition, few studies have addressed when zebrafish embryos start to exit the pluripotent state. Monitoring dynamic expression of pluripotency-associated markers in different stages of embryos may be a better solution. Nevertheless, pluripotency markers widely used in mammals, such as SOX2, did not serve well for zebrafish in this or previous (Robles et al., 2011) studies. Recent studies showed that pluripotency factors Pou5f3 and Nanog played crucial roles in pluripotency establishment in zebrafish. Both pou5f3 and nanog are expressed maternally and zygotically (Takeda et al., 1994, Xu et al., 2012), playing key roles in activation of zygotic genes in zebrafish (Lee et al., 2013, Leichsenring et al., 2013). The zygotic genome is transcriptionally quiescent after fertilization until the first large wave of zygotic transcript activation and maternal transcript degradation, which is termed maternal-to-zygotic transition (Tadros and Lipshitz, 2009). The zygotic genome activation initiates at the 1,000-cell stage (3 hpf) in zebrafish (Mathavan et al., 2005) and at the 2-cell stage (24 hpf) in mouse (Hamatani et al., 2004). For zygotic genome activation in zebrafish, Nanog, Pou5f3, and Soxb1 bind to and activate global expression of many embryonic genes including miR-430, which is responsible for maternal RNA clearance (Lee et al., 2013, Lee et al., 2014, Leichsenring et al., 2013). Zygotic genome activation in mammals is probably different from that in zebrafish, as establishment of totipotency does not depend on OCT4A in mice (Wu et al., 2013). Three germ layers emerge after gastrulation in mice. However, a mesoderm marker (ta) starts to express as early as the oblong stage (midblastula) in zebrafish, indicating a much earlier differentiation of embryogenesis in zebrafish than in mouse. This was confirmed by the decrease in efficiency of cardiomyocyte generation when late-blastula and gastrula embryonic cells were used for induction in our study. Therefore, pluripotency of embryonic cells in zebrafish cannot be determined only by pluripotency factors such as Pou5f3 and Nanog. The onset of germ layer differentiation is another critical factor. Thus, based on expression patterns of markers of both pluripotency and germ layers, we determine the pluripotent window of embryogenesis in zebrafish as from zygotic genome activation to a brief moment after the oblong stage (less than 2 hr), which is a very narrow period for pluripotency maintenance in the cell priming of mesoderm differentiation.
A better understanding of pluripotent state of zebrafish embryos allows us to accurately induce differentiation to cardiomyocytes from primary embryonic cells. A central feature of our method is the use of a combination of appropriate conditions including cell density, ZF4 feeder cells, and supplements to facilitate the cardiomyocyte induction, in addition to utilizing pluripotency characteristics. This induction process is similar to in vivo cardiogenesis timing (24 hpf) in zebrafish (Kimmel et al., 1995). Among the supplements (ZEE, ZF4 CM, and INSULIN), INSULIN is an essential factor. Indeed, INSULIN and its receptor are both maternally and zygotically expressed, and knockdown of insulin receptor resulted in cardiac abnormalities in zebrafish, suggesting that INSULIN plays important roles in cardiogenesis (Papasani et al., 2006, Toyoshima et al., 2008). Treatment of mouse embryonic stem cells with INSULIN or insulin-like growth factor (IGF) 1 or 2 promoted proliferation of mesodermal cells and cardiomyocyte generation via the PI3K/AKT/TOR signaling pathway (Engels et al., 2014). IGF signaling also promotes a continued expansion of cardiac progenitor cells derived from human PSCs (Birket et al., 2015).
Another feature of the induction system is that the induced cardiomyocytes have typical contractile kinetics and electrophysiological characteristics. Spontaneous beats of cardiomyocytes are driven by depolarization-repolarization cycles of the cell membrane potential. AP patterns of in vitro generated BCCs in this study resemble those of ventricles, atria, or pacemakers in adult zebrafish hearts (Nemtsas et al., 2010, Tessadori et al., 2012), and are also similar to those of human (Nemtsas et al., 2010). Functional properties of cardiomyocytes can further be manifested by rate adaptation tests and drug treatments. In rate adaptation tests, BCCs adapted to the stimulations at increasing frequencies by shortening their APD, which is consistent with zebrafish ventricular myocytes (Brette et al., 2008) and human PSC-derived cardiomyocytes (He et al., 2003, Lian et al., 2012, Zhang et al., 2009). Another property is functional response to β-adrenergic stimulations, which is comparable in zebrafish hearts (Parker et al., 2014, Steele et al., 2011) and human PSC-derived cardiomyocytes (Zhang et al., 2009), suggesting the presence of functional β-adrenergic receptors in the BCCs. A more important feature is the presence of a sodium channel in the induced cardiomyocytes, as revealed by TTX response. A similar effect of TTX on the induced cardiomyocytes was also observed in adult hearts of zebrafish (Chopra et al., 2010, Nemtsas et al., 2010).
Robust regenerative ability in adult cardiomyocytes makes zebrafish an ideal model for studying heart regeneration (Jopling et al., 2010, Poss et al., 2002, Raya et al., 2003). Since cardiomyocyte proliferation is a key step of zebrafish heart regeneration (Jopling et al., 2010), it is important to discover essential components that trigger or promote this process. An efficient in vitro culture of adult zebrafish cardiomyocytes has been established recently (Sander et al., 2013). In this study, we have provided a detailed method of in vitro cardiomyocyte differentiation from embryonic cells. Differentiated cardiomyocytes are functional and capable of recapitulating proliferative responses to mitogens. Generation and proliferation of cardiomyocytes were both substantially enhanced by NRG1 treatment, consistent with previous studies in mammalian and zebrafish heart regeneration (Bersell et al., 2009, Gemberling et al., 2015). Combining utilization of transgenic fish lines that specifically express fluorescent protein in cardiomyocyte nuclei, our system possesses potential in exploring novel mitogens or factors involved in heart regeneration. The advantage of our system is that it is primed to generate cardiomyocytes from lethal mutant lines that cannot survive to adulthood. As long as the mutant embryos remain viable at the oblong stage, generation of differentiated cardiomyocytes can be achieved by using the protocols provided herein, and thus gene functional studies can be carried out based on regenerative responses of the mutant cardiomyocytes. Therefore, we have determined an alternative way to gain insight into heart regeneration mechanisms in zebrafish. The primary embryonic cell-based system can be used to perform high-throughput screening of factors for heart study not merely limited to small molecules (Huang et al., 2012, Xu et al., 2013) to also include nucleic acids, proteins and lipids, and so forth, which is difficult to achieve in the zebrafish in vivo screening model. The system is also promising for the dissection of pathways of cardiac development and regeneration in lethal mutants.
Experimental Procedures
See also Supplemental Experimental Procedures.
Zebrafish, Cells, and Antibodies
All animal experiments and methods were performed in accordance with the relevant approved guidelines and regulations, as well as under the approval of the Ethics Committee of Wuhan University. The wild-type AB zebrafish (Danio rerio), the Tg (myl7:EGFP) transgenic line, and ZF4 cell line CRL-2050 were used in this study. Antibodies used in the immunofluorescence assay were polyclonal antibody anti-Acta1 (1:10 dilution; AP14779, Abgent) and Cy3-conjugated secondary antibody (1:100 dilution; E031620-01, EarthOx).
Isolation of Primary Embryonic Cells
Zebrafish embryos were bleached in 0.003% sodium hypochlorite for 2 min, rinsed with sterilized 0.5× E2 medium, and dechorionated in 0.1 mg/mL pronase E (P5147, Sigma-Aldrich) with occasional swirling, then rinsed with sterilized 0.5× E2 again. Embryonic cells were cultured in 24-well plates. The cells seeded in each well were isolated from 40–50 oblong embryos to ensure a consistent density of cells among wells.
Induction of Cardiomyocyte Differentiation
Embryonic cells at oblong stage or other stages were induced for cardiomyocyte differentiation at 28.5°C. Cells were plated onto plates at different cell densities ranging from 1 to 4.5 × 104 cells/cm2. ZF4 cells (zebrafish embryonic fibroblast cell line) were cultured as feeder cells at a density of 2 × 104 cells/cm2. Complete medium used for cardiomyocyte differentiation from primary embryonic cells was the basal medium supplemented with 1% fish serum, 85 μg protein/mL ZEE, 30% conditioned medium (ZF4 CM), 25 μg/mL INSULIN (I5500, Sigma-Aldrich), and 50 ng/mL EGF (315-09, Pepro Tech). During evaluating proliferative responses of BCCs to mitogens, recombinant NRG1 (11609H01H50, Thermo Fisher Scientific) was supplemented to culture the oblong embryonic cells at a concentration of 0, 50, 100, 200, 500 and 1,000 ng/mL, respectively. BCCs emerging in the culture were recorded under an inverted microscope (Leica DM IRB).
RT-PCR
Total RNAs of embryonic cells at different stages were isolated using TRIzol reagent (15596026, Thermo Fisher). Extracted RNAs were treated with DNase treatments (M610A, Promega). The first strands of cDNAs were synthesized following the protocol (M170A, Promega). Primer sequences and PCR conditions are listed in Table S1.
Contraction Kinetics
Videos of BCCs were recorded with a camera equipped on an inverted microscope (Leica DM IRB). Images were extracted from the videos at a frequency of 10 frames per second. A contracting site was tracked through a series of images. x and y coordinates of the sites were recorded using ImageJ software (http://imagej.nih.gov/ij/). Contraction amplitudes were calculated based on the coordinates of the contracting site before and after the contraction. The contraction or relaxation velocity was calculated by dividing contraction or relaxation distance by the duration, respectively.
Electrophysiology
BCCs were picked up by a glass capillary connected to a mouth pipette at day 2 of differentiation, replated onto coverslips coated with PLL, and maintained in complete cardiomyocyte differentiation medium before recording the AP. AP activities of cardiomyocytes were recorded in a whole-cell patch-clamp configuration using a HEKA EPC10 amplifier driven by PatchMaster software (HEKA).
Author Contributions
R.Z. and H.C. conceived and designed research. Y.X. and R.Z. analyzed and interpreted the data and wrote the paper. Y.X., M.G., Y.Z., L.G., Z.L., and Q.H. carried out the experimental work. J.Y., Y.X., and L.G. analyzed the data. All authors read and approved the final manuscript.
Acknowledgments
We thank Dr. Yun Deng for providing Tg (myl7:EGFP) zebrafish, Dr. Niels C. Bols and Dr. Peng Hu for their technical support with zebrafish. This work was supported by the National Natural Science Foundation of China (31571280, 31272648, and 31471182), the National Key Basic Research project (2010CB126306), National Key Technologies R&D Program (2012BAD26B01), Hubei Province Science and Technology project, and the Chinese 111 project.
Published: August 25, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, one table, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.07.020.
Contributor Information
Hanhua Cheng, Email: hhcheng@whu.edu.cn.
Rongjia Zhou, Email: rjzhou@whu.edu.cn.
Supplemental Information
References
- Arnaout R., Ferrer T., Huisken J., Spitzer K., Stainier D.Y., Tristani-Firouzi M., Chi N.C. Zebrafish model for human long QT syndrome. Proc. Natl. Acad. Sci. USA. 2007;104:11316–11321. doi: 10.1073/pnas.0702724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asnani A., Peterson R.T. The zebrafish as a tool to identify novel therapies for human cardiovascular disease. Dis. Model. Mech. 2014;7:763–767. doi: 10.1242/dmm.016170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp J., Steel D., Jonsson M., Ameen C., Dahlenborg K., Jeppsson A., Lindahl A., Sartipy P. Cardiomyocyte clusters derived from human embryonic stem cells share similarities with human heart tissue. J. Mol. Cell Biol. 2010;2:276–283. doi: 10.1093/jmcb/mjq022. [DOI] [PubMed] [Google Scholar]
- Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc. Res. 2011;91:279–288. doi: 10.1093/cvr/cvr098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchemin M., Smith A., Yin V.P. Dynamic microRNA-101a and Fosab expression controls zebrafish heart regeneration. Development. 2015;142:4026–4037. doi: 10.1242/dev.126649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Bednarek D., Gonzalez-Rosa J.M., Guzman-Martinez G., Gutierrez-Gutierrez O., Aguado T., Sanchez-Ferrer C., Marques I.J., Galardi-Castilla M., de Diego I., Gomez M.J. Telomerase is essential for zebrafish heart regeneration. Cell Rep. 2015;12:1691–1703. doi: 10.1016/j.celrep.2015.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O., Zdunek S., Felker A., Salehpour M., Alkass K., Bernard S., Sjostrom S.L., Szewczykowska M., Jackowska T., Dos Remedios C. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- Bersell K., Arab S., Haring B., Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- Birket M.J., Ribeiro M.C., Verkerk A.O., Ward D., Leitoguinho A.R., den Hartogh S.C., Orlova V.V., Devalla H.D., Schwach V., Bellin M. Expansion and patterning of cardiovascular progenitors derived from human pluripotent stem cells. Nat. Biotechnol. 2015;33:970–979. doi: 10.1038/nbt.3271. [DOI] [PubMed] [Google Scholar]
- Brette F., Luxan G., Cros C., Dixey H., Wilson C., Shiels H.A. Characterization of isolated ventricular myocytes from adult zebrafish (Danio rerio) Biochem. Biophys. Res. Commun. 2008;374:143–146. doi: 10.1016/j.bbrc.2008.06.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11:855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P.W., Sharma A., Wu J.C. Genetic and epigenetic regulation of human cardiac reprogramming and differentiation in regenerative medicine. Annu. Rev. Genet. 2015;49:461–484. doi: 10.1146/annurev-genet-112414-054911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Navis A., Cox B.D., Dickson A.L., Gemberling M., Karra R., Bagnat M., Poss K.D. Single epicardial cell transcriptome sequencing identifies Caveolin-1 as an essential factor in zebrafish heart regeneration. Development. 2016;143:232–243. doi: 10.1242/dev.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J.J., Yang X., Don C.W., Minami E., Liu Y.W., Weyers J.J., Mahoney W.M., Van Biber B., Cook S.M., Palpant N.J. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature. 2014;510:273–277. doi: 10.1038/nature13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra S.S., Stroud D.M., Watanabe H., Bennett J.S., Burns C.G., Wells K.S., Yang T., Zhong T.P., Roden D.M. Voltage-gated sodium channels are required for heart development in zebrafish. Circ. Res. 2010;106:1342–1350. doi: 10.1161/CIRCRESAHA.109.213132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels M.C., Rajarajan K., Feistritzer R., Sharma A., Nielsen U.B., Schalij M.J., de Vries A.A., Pijnappels D.A., Wu S.M. Insulin-like growth factor promotes cardiac lineage induction in vitro by selective expansion of early mesoderm. Stem Cells. 2014;32:1493–1502. doi: 10.1002/stem.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S., Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- Fan L., Crodian J., Liu X., Alestrom A., Alestrom P., Collodi P. Zebrafish embryo cells remain pluripotent and germ-line competent for multiple passages in culture. Zebrafish. 2004;1:21–26. doi: 10.1089/154585404774101644. [DOI] [PubMed] [Google Scholar]
- Feaster T.K., Cadar A.G., Wang L., Williams C.H., Chun Y.W., Hempel J.E., Bloodworth N., Merryman W.D., Lim C.C., Wu J.C. Matrigel mattress: a method for the generation of single contracting human-induced pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2015;117:995–1000. doi: 10.1161/CIRCRESAHA.115.307580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemberling M., Karra R., Dickson A.L., Poss K.D. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife. 2015;4:e05871. doi: 10.7554/eLife.05871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunow B., Mohamet L., Shiels H.A. Generating an in vitro 3D cell culture model from zebrafish larvae for heart research. J. Exp. Biol. 2015;218:1116–1121. doi: 10.1242/jeb.118224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Zhu P., Yan L., Li R., Hu B., Lian Y., Yan J., Ren X., Lin S., Li J. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–610. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- Hamatani T., Carter M.G., Sharov A.A., Ko M.S. Dynamics of global gene expression changes during mouse preimplantation development. Dev. Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Hayakawa T., Kunihiro T., Ando T., Kobayashi S., Matsui E., Yada H., Kanda Y., Kurokawa J., Furukawa T. Image-based evaluation of contraction-relaxation kinetics of human-induced pluripotent stem cell-derived cardiomyocytes: correlation and complementarity with extracellular electrophysiology. J. Mol. Cell Cardiol. 2014;77:178–191. doi: 10.1016/j.yjmcc.2014.09.010. [DOI] [PubMed] [Google Scholar]
- He J.Q., Ma Y., Lee Y., Thomson J.A., Kamp T.J. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ. Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- Ho R.K., Kimmel C.B. Commitment of cell fate in the early zebrafish embryo. Science. 1993;261:109–111. doi: 10.1126/science.8316841. [DOI] [PubMed] [Google Scholar]
- Ho S.Y., Goh C.W., Gan J.Y., Lee Y.S., Lam M.K., Hong N., Hong Y., Chan W.K., Shu-Chien A.C. Derivation and long-term culture of an embryonic stem cell-like line from zebrafish blastomeres under feeder-free condition. Zebrafish. 2014;11:407–420. doi: 10.1089/zeb.2013.0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong N., Schartl M., Hong Y. Derivation of stable zebrafish ES-like cells in feeder-free culture. Cell Tissue Res. 2014;357:623–632. doi: 10.1007/s00441-014-1882-0. [DOI] [PubMed] [Google Scholar]
- Huang H., Lindgren A., Wu X., Liu N.A., Lin S. High-throughput screening for bioactive molecules using primary cell culture of transgenic zebrafish embryos. Cell Rep. 2012;2:695–704. doi: 10.1016/j.celrep.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C., Sleep E., Raya M., Marti M., Raya A., Izpisua Belmonte J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karra R., Knecht A.K., Kikuchi K., Poss K.D. Myocardial NF-kappaB activation is essential for zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA. 2015;112:13255–13260. doi: 10.1073/pnas.1511209112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K. Advances in understanding the mechanism of zebrafish heart regeneration. Stem Cell Res. 2014;13:542–555. doi: 10.1016/j.scr.2014.07.003. [DOI] [PubMed] [Google Scholar]
- Kimmel C.B., Ballard W.W., Kimmel S.R., Ullmann B., Schilling T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Laflamme M.A., Murry C.E. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.T., Bonneau A.R., Takacs C.M., Bazzini A.A., DiVito K.R., Fleming E.S., Giraldez A.J. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.T., Bonneau A.R., Giraldez A.J. Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 2014;30:581–613. doi: 10.1146/annurev-cellbio-100913-013027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M., Maes J., Mossner R., Driever W., Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341:1005–1009. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- Lian X., Hsiao C., Wilson G., Zhu K., Hazeltine L.B., Azarin S.M., Raval K.K., Zhang J., Kamp T.J., Palecek S.P. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA. 2012;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Stainier D.Y. Zebrafish in the study of early cardiac development. Circ. Res. 2012;110:870–874. doi: 10.1161/CIRCRESAHA.111.246504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavan S., Lee S.G., Mak A., Miller L.D., Murthy K.R., Govindarajan K.R., Tong Y., Wu Y.L., Lam S.H., Yang H. Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet. 2005;1:260–276. doi: 10.1371/journal.pgen.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrone G., Wilson K.S., Maqsood S., Mullins J.J., Tucker C.S., Denvir M.A. CDK9 and its repressor LARP7 modulate cardiomyocyte proliferation and response to injury in the zebrafish heart. J. Cell Sci. 2015;128:4560–4571. doi: 10.1242/jcs.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffarian D., Benjamin E.J., Go A.S., Arnett D.K., Blaha M.J., Cushman M., de Ferranti S., Despres J.P., Fullerton H.J., Howard V.J. Heart disease and stroke statistics—2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–e322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- Nemtsas P., Wettwer E., Christ T., Weidinger G., Ravens U. Adult zebrafish heart as a model for human heart? An electrophysiological study. J. Mol. Cell Cardiol. 2010;48:161–171. doi: 10.1016/j.yjmcc.2009.08.034. [DOI] [PubMed] [Google Scholar]
- Ni T.T., Rellinger E.J., Mukherjee A., Xie S., Stephens L., Thorne C.A., Kim K., Hu J., Lee E., Marnett L. Discovering small molecules that promote cardiomyocyte generation by modulating Wnt signaling. Chem. Biol. 2011;18:1658–1668. doi: 10.1016/j.chembiol.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papasani M.R., Robison B.D., Hardy R.W., Hill R.A. Early developmental expression of two insulins in zebrafish (Danio rerio) Physiol. Genomics. 2006;27:79–85. doi: 10.1152/physiolgenomics.00012.2006. [DOI] [PubMed] [Google Scholar]
- Parker T., Libourel P.A., Hetheridge M.J., Cumming R.I., Sutcliffe T.P., Goonesinghe A.C., Ball J.S., Owen S.F., Chomis Y., Winter M.J. A multi-endpoint in vivo larval zebrafish (Danio rerio) model for the assessment of integrated cardiovascular function. J. Pharmacol. Toxicol. Methods. 2014;69:30–38. doi: 10.1016/j.vascn.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., Sadek H.A. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K.D., Wilson L.G., Keating M.T. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Raya A., Koth C.M., Buscher D., Kawakami Y., Itoh T., Raya R.M., Sternik G., Tsai H.J., Rodriguez-Esteban C., Izpisua-Belmonte J.C. Activation of Notch signaling pathway precedes heart regeneration in zebrafish. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl 1):11889–11895. doi: 10.1073/pnas.1834204100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles V., Marti M., Izpisua Belmonte J.C. Study of pluripotency markers in zebrafish embryos and transient embryonic stem cell cultures. Zebrafish. 2011;8:57–63. doi: 10.1089/zeb.2010.0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumiantsev P.P., Carlson B.M. Harwood Academic Publishers; Chur, Switzerland; New York, New York: 1991. Growth and Hyperplasia of Cardiac Muscle Cells. [Google Scholar]
- Sahara M., Santoro F., Chien K.R. Programming and reprogramming a human heart cell. EMBO J. 2015;34:710–738. doi: 10.15252/embj.201490563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander V., Sune G., Jopling C., Morera C., Izpisua Belmonte J.C. Isolation and in vitro culture of primary cardiomyocytes from adult zebrafish hearts. Nat. Protoc. 2013;8:800–809. doi: 10.1038/nprot.2013.041. [DOI] [PubMed] [Google Scholar]
- Smart N., Bollini S., Dube K.N., Vieira J.M., Zhou B., Davidson S., Yellon D., Riegler J., Price A.N., Lythgoe M.F. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele S.L., Yang X., Debiais-Thibaud M., Schwerte T., Pelster B., Ekker M., Tiberi M., Perry S.F. In vivo and in vitro assessment of cardiac beta-adrenergic receptors in larval zebrafish (Danio rerio) J. Exp. Biol. 2011;214:1445–1457. doi: 10.1242/jeb.052803. [DOI] [PubMed] [Google Scholar]
- Sturzu A.C., Rajarajan K., Passer D., Plonowska K., Riley A., Tan T.C., Sharma A., Xu A.F., Engels M.C., Feistritzer R. Fetal mammalian heart generates a robust compensatory response to cell loss. Circulation. 2015;132:109–121. doi: 10.1161/CIRCULATIONAHA.114.011490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W., Lipshitz H.D. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Takeda H., Matsuzaki T., Oki T., Miyagawa T., Amanuma H. A novel POU domain gene, zebrafish pou2: expression and roles of two alternatively spliced twin products in early development. Genes. Dev. 1994;8:45–59. doi: 10.1101/gad.8.1.45. [DOI] [PubMed] [Google Scholar]
- Tessadori F., van Weerd J.H., Burkhard S.B., Verkerk A.O., de Pater E., Boukens B.J., Vink A., Christoffels V.M., Bakkers J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS One. 2012;7:e47644. doi: 10.1371/journal.pone.0047644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima Y., Monson C., Duan C., Wu Y., Gao C., Yakar S., Sadler K.C., LeRoith D. The role of insulin receptor signaling in zebrafish embryogenesis. Endocrinology. 2008;149:5996–6005. doi: 10.1210/en.2008-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uygur A., Lee R.T. Mechanisms of cardiac regeneration. Dev. Cell. 2016;36:362–374. doi: 10.1016/j.devcel.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Berlo J.H., Molkentin J.D. An emerging consensus on cardiac regeneration. Nat. Med. 2014;20:1386–1393. doi: 10.1038/nm.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Han D., Gong Y., Sebastiano V., Gentile L., Singhal N., Adachi K., Fischedick G., Ortmeier C., Sinn M. Establishment of totipotency does not depend on Oct4A. Nat. Cell Biol. 2013;15:1089–1097. doi: 10.1038/ncb2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Kruse F., Vasudevarao M.D., Junker J.P., Zebrowski D.C., Fischer K., Noël E.S., Grün D., Berezikov E., Engel F.B. Spatially resolved genome-wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev. Cell. 2016;36:36–49. doi: 10.1016/j.devcel.2015.12.010. [DOI] [PubMed] [Google Scholar]
- Xu C., Fan Z.P., Muller P., Fogley R., DiBiase A., Trompouki E., Unternaehrer J., Xiong F., Torregroza I., Evans T. Nanog-like regulates endoderm formation through the Mxtx2-Nodal pathway. Dev. Cell. 2012;22:625–638. doi: 10.1016/j.devcel.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Tabebordbar M., Iovino S., Ciarlo C., Liu J., Castiglioni A., Price E., Liu M., Barton E.R., Kahn C.R. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell. 2013;155:909–921. doi: 10.1016/j.cell.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Spater D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wilson G.F., Soerens A.G., Koonce C.H., Yu J., Palecek S.P., Thomson J.A., Kamp T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009;104:e30–e41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwi-Dantsis L., Huber I., Habib M., Winterstern A., Gepstein A., Arbel G., Gepstein L. Derivation and cardiomyocyte differentiation of induced pluripotent stem cells from heart failure patients. Eur. Heart J. 2013;34:1575–1586. doi: 10.1093/eurheartj/ehs096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.