Summary
Controlled differentiation of human embryonic stem cells (hESCs) can be utilized for precise analysis of cell type identities during early development. We established a highly efficient neural induction strategy and an improved analytical platform, and determined proteomic and phosphoproteomic profiles of hESCs and their specified multipotent neural stem cell derivatives (hNSCs). This quantitative dataset (nearly 13,000 proteins and 60,000 phosphorylation sites) provides unique molecular insights into pluripotency and neural lineage entry. Systems-level comparative analysis of proteins (e.g., transcription factors, epigenetic regulators, kinase families), phosphorylation sites, and numerous biological pathways allowed the identification of distinct signatures in pluripotent and multipotent cells. Furthermore, as predicted by the dataset, we functionally validated an autocrine/paracrine mechanism by demonstrating that the secreted protein midkine is a regulator of neural specification. This resource is freely available to the scientific community, including a searchable website, PluriProt.
Highlights
-
•
Controlled neural induction produces pure cultures of PAX6+ neural stem cells
-
•
Most comprehensive (phospho)proteome mapping in pluripotent and multipotent cells
-
•
Prediction and validation of midkine as regulator of neural lineage commitment
-
•
Searchable and publicly available website presenting (phospho)proteomic dataset
Snyder, Brill, Singec, and colleagues demonstrate detailed analysis of human pluripotency and controlled neural lineage entry by using quantitative label-free (phospho)proteomics. The accuracy of the large dataset (13,000 proteins; 60,000 non-redundant phosphorylation sites) allows precise characterization and comparison of pluripotent and multipotent “stemness.” Functional follow-up experiments validate that the understudied protein midkine controls neuralization of hESCs.
Introduction
Human pluripotent stem cells (hPSCs) enable modeling aspects of development and disease, and hold great promise for regenerative medicine and drug discovery (van Hoof et al., 2012, Young, 2011). Previous large-scale analyses of hPSCs shed light on pluripotency, differentiation, and de-differentiation by focusing on transcriptional regulation, epigenetic changes, and non-coding RNAs (Boyer et al., 2005, Brandenberger et al., 2004, Elkabetz et al., 2008, Martinez and Gregory, 2010). However, proteomes contain vast amounts of biological information unobtainable via genomics, transcriptomics, or similar analyses (Wilhelm et al., 2014). Thus, a detailed characterization of pluripotency, lineage specification, and reprogramming by protein profiling is important for complementing other analytical methods and should help to elucidate novel mechanisms.
Regulation of proteins includes quantitative changes and post-translational modifications (PTMs) (Huttlin et al., 2010). A key PTM is reversible phosphorylation of serine (pS), threonine (pT), and tyrosine (pY), which modulates enzyme activities, protein-protein interactions, conformational changes, protein half-life, and signal transduction, among others (Choudhary and Mann, 2010). Multidimensional liquid chromatography (MDLC) coupled with tandem mass spectrometry (MS/MS) enables large-scale analysis of proteomes and phosphoproteomes (Huttlin et al., 2010, Sharma et al., 2014). Although previous reports have provided important insights into the proteomes of hPSCs (Brill et al., 2009, Munoz et al., 2011, Phanstiel et al., 2011, Rigbolt et al., 2011, Swaney et al., 2009, Van Hoof et al., 2009, Van Hoof et al., 2006), none of these studies have applied robustly controlled differentiation strategies in feeder-free monolayer cultures. Hence, proteomic analysis of pluripotent cells compared with their lineage-specific multipotent derivatives has not been reported. Moreover, previous datasets did not reach the depth enabled by recent technical advances (Huttlin et al., 2010, Sharma et al., 2014). Notably, label-free quantification (LFQ) can yield deeper proteome coverage than stable-isotope labeling by amino acids in cell culture while maintaining quantitative accuracy (Collier et al., 2010, Gokce et al., 2011, Sharma et al., 2014).
Here, we employed a controlled and reproducible neural induction strategy to investigate the combined proteomic and phosphoproteomic [termed (phospho)proteomic] changes that occur when hESCs differentiate to a highly pure population of hNSCs. These experiments also include molecular and electrophysiological characterizations of more differentiated cellular progeny, thereby confirming the multipotency of the hNSCs studied. LFQ proteomic methods allowed elucidation of cell type-specific (phospho)proteomes at an unprecedented depth. To demonstrate the utility of the dataset, we performed systems-level analyses of cell-signaling pathways and protein families, and created a map of epigenetic proteins, many of which are regulated during differentiation. Our dataset includes a large (phospho)proteomics resource of transcription factors (n = 487) including previously unidentified phosphorylation sites on OCT4, NANOG, SOX2, and others. Moreover, to demonstrate the utility of the dataset we performed functional experiments showing that the secreted protein midkine (MDK), which our (phospho)proteomic analyses found to be upregulated during neural commitment, instigates neural specification.
Results
Directed Differentiation of hPSCs to Enable (Phospho)Proteomic Profiling of Neural Lineage Commitment
Pluripotent cells were maintained under feeder-free monolayer conditions. For neural induction, exogenous fibroblast growth factor (FGF2) was omitted from the culture medium and a small-molecule cocktail (termed DAP; Figure 1A) was added to suppress pathways that otherwise contribute to pluripotency and/or non-neural differentiation of hESCs (Boles et al., 2014, Chambers et al., 2009, Hasegawa et al., 2012, Pera et al., 2004, Sturgeon et al., 2014). The 6-day DAP treatment that we developed in our laboratory produced highly pure cultures of hNSCs (>97% PAX6+/NESTIN+ cells; Figures 1A and 1B). This neural induction strategy was characterized by demonstrating: inhibition of SMAD phosphorylation sites (Figure S1A); induction of neural markers (Figures S1B, S1D, and S1F); downregulation of pluripotency markers OCT4 and NANOG (Figures S1C and S1F); absence of mesoderm (BRACHYURY), endoderm (SOX17), neural crest (SOX10, TFAP2A, SNAI2), and non-neural ectoderm (MSX1/2) (Figures S1F and S1G); comparison with embryoid body (EB) differentiation (Figure S1F); immunostaining for PAX6/OTX2/NESTIN, normal karyotype; and efficient neuralization using human induced PSCs (hiPSCs) (Figures 1A, S1H, and S1I).
Figure 1.
Controlled Neural Induction and Schematic Diagram of the (Phospho)Proteomic Workflow to Compare Human Pluripotency and Neural Multipotency
(A) Experimental approach for efficient 6-day neural conversion of hESCs into hNSCs using dorsomorphin, A83-01, and PNU-74654 (termed DAP cocktail). Scale bars represent 100 μm.
(B) Fluorescence-activated cell sorting suggests virtually uniform conversion of hPSCs to PAX6+/NESTIN+ (NES+) hNSCs.
(C) Schematic diagram of the workflow for total (phospho)proteome analysis. Fr, fraction; RP, reversed-phase; LC, liquid chromatography; ESI, electrospray ionization; LFQ, label-free quantification. “Precision proteomics” yields accurate results (Choudhary and Mann, 2010).
(D) Reproducibility of protein identification and quantification among experimental replicates was high.
See also Figures S1–S6.
hNSCs expanded in FGF2 for 30 days underwent anterior-to-posterior specification, suggested by induction of GBX2, DACH1, HOXA2, and HOXB4, and downregulation of PAX6, PAX7, PLZF, and OTX1 (Figure S2A), consistent with a previous report (Pankratz et al., 2007). To demonstrate multipotency, we differentiated hNSCs into more mature cell types representing the three main neural lineages (neurons, astrocytes, and oligodendrocytes; Figures S2B–S2E). Moreover, neuronal activity was measured by electrophysiological analyses. Spontaneous synaptic currents, action potential generation, and ion channel response was observed when inhibitory (GABA) and excitatory (NMDA + glycine) stimuli were applied (Figure S3). Together, these results show that highly pure hNSCs and their more differentiated progeny were obtained from 6-day DAP treatment of hPSCs. Because the (phospho)proteomes of hESCs grown under feeder-free conditions and their earliest definitive neural derivatives expressing PAX6 (Pankratz et al., 2007, Zhang et al., 2010a) have not been compared previously, we decided to perform this comparative (phospho)proteome analysis.
Total (Phospho)Proteome Analysis Platform
(Phospho)proteins were digested to peptides and separated by strong cation exchange chromatography (SCX) (Figures 1C and S4; Supplemental Experimental Procedures). Phosphopeptides were enriched from SCX fractions (Gandin et al., 2013, Hou et al., 2013) (Figure 1C). Non-binding and wash fractions (containing primarily non-phosphorylated peptides) and enriched phosphopeptides were subjected to liquid chromatography (LC)-electrospray ionization (ESI)-MS/MS, including optimized LC gradients (Hou et al., 2013). MS/MS data was searched against a human protein database and identifications were filtered at a protein-level false discovery rate (FDR) of <0.01 and peptide-level FDR of <0.006 (Supplemental Experimental Procedures). LFQ was by normalized spectral abundance factor (NSAF) spectral counting (SpC) (Liu et al., 2004, Zhang et al., 2010b), using non-phosphopeptides only, to improve quantitative reliability (Supplemental Experimental Procedures). Analysis of protein phosphorylation included identification of phosphopeptides and phosphorylation sites, and calculation of phosphopeptide spectral counts (PSpCs). The PSpC is the number of MS/MS spectra in a replicate that resulted in identification of a phosphopeptide. Identifications employed “precision proteomics” (Choudhary and Mann, 2010) and additional high-confidence analyses (Supplemental Experimental Procedures).
Variability of protein identifications was only 8.6%–15.0% between replicates (Figure 1D). The number of non-redundant phosphorylation sites is presented in Table S1, whereas Tables S2–S16 are available at http://dx.doi.org/10.17632/v7nt3wm8rk.1. Protein families are presented in Tables S2–S7. Detailed information on all peptide and phosphopeptide identifications is given in Tables S8–S13. Biological data annotation is available at a searchable website, http://pluriprot.org. Figure S5A illustrates how to access information on (phospho)proteins from our dataset. Protein network analyses on PluriProt highlight pathways and biological processes enriched in hESCs or hNSCs (Figure S5B).
(Phospho)Proteins Detected in Pluripotent or Multipotent Cells Only
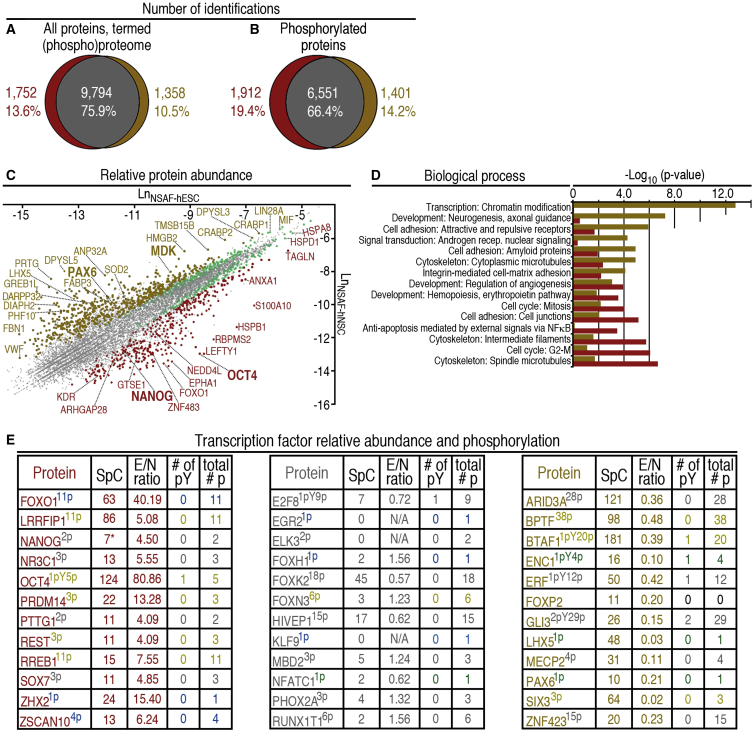
Among the 13.6% of the proteins exclusively detected in hESCs were proteins associated with pluripotency, including ESRRB, UTF1, FOXO1, ALPI (alkaline phosphatase), TERT, and TDGF1 (Figures 2A, 2B, and 2E; Tables S14A and S15A). In contrast, neural markers such as PAX6, PLZF, SOX5, SOX10, LHX1, SEZ6, and ARX were detected in hNSCs only (Tables S14A and S15B). Notably, 76.4% of all detected proteins were phosphorylated (Figures 2A and 2B).
Figure 2.
Comparative Analysis of (Phospho)Proteomes of Pluripotent hESCs and Multipotent hNSCs
(A and B) Venn diagrams illustrating identification of all proteins, termed the (phospho)proteome and phosphorylated proteins in hESCs (red; left portion of each diagram), hNSCs (brown; right portion of each diagram), and both cell populations (gray; overlap).
(C) Proteins with significantly differing relative abundance (t test, three replicates; p ≤ 0.05) are represented as red, green, or brown dots, whereas gray dots represent proteins with p > 0.05. LnNSAF is the mean natural log of the normalized spectral abundance factor (NSAF) for the protein in either hESCs (Table S14, column J) or hNSCs (Table S14, column M).
(D) Process network analysis using differentially abundant proteins suggests distinctions in biological mechanisms between hESCs and hNSCs. Network significance is shown by red bars (hESCs) or brown bars (hNSCs).
(E) Transcription factors more abundant in hESCs (red font; left panel), hNSCs (brown font; right panel), or without detected differences in abundance (gray font; middle panel). SpC designates total spectral counts for the protein in both cell populations; 7∗ designates that NANOG p = 0.00029 (three replicates, t test; Table S14A). N/A, not applicable. The number of localized pY sites (# of pY) and total number of phosphorylation sites (total #p) are shown. Blue superscripts represent phosphorylation detection in hESCs only. Superscripts for proteins with phosphorylation in hESCs and hNSCs are in gold when ≥1 phosphorylation site detection on the protein differed between hESCs and hNSCs; otherwise superscripts are in gray. Green superscripts represent phosphorylation detection in hNSCs only.
See also Figures S6 and S7.
Analysis of Protein Abundance
First, a statistical test (t test) for proteomic data (Zybailov et al., 2006) was used to analyze hESC/hNSC NSAF-SpC ratios (termed E/N ratios). The mean E/N ratio for the dataset was 1.27 and the median was 0.97, providing general support for this approach. Second, the mean E/N ratio of all non-regulated ribosomal proteins in the dataset, 1.13, and the SD (= 0.24; Table S14B) enabled significant difference thresholds of an E/N ratio ≥2.00 or ≤0.50. These thresholds are >3 SD from the mean, and should translate to ca. p < 0.01 when the SpC threshold (≥10; clarified below) is met. Third, the natural log (Ln) of the NSAFs (Zybailov et al., 2006; Table S14A) of the proteins was plotted (Figure 2C). Proteins with an E/N ratio differing significantly between hESCs and hNSCs (t test; p < 0.05) are shown as red, brown, and green dots in Figure 2C. Proteins expected to be more abundant in hESCs with an E/N ratio ≥2.00 are represented by red dots, whereas brown dots indicate those expected to be more abundant in hNSCs, with an E/N ratio ≤0.50 (Figure 2C). Green dots represent proteins with 0.50 < E/N ratio < 2.00. For example, HSPD1, with a significantly differing E/N ratio (1.56; p = 0.00063) was more abundant in hESCs, whereas LIN28A (E/N ratio = 0.63; p = 0.0020) was more abundant in hNSCs (Figure 2C). Independent western blot experiments agreed with E/N ratios (Figure S6A). However, in some cases the t test can miss differentially abundant proteins (e.g., CTCF, Figure S6B), consistent with a recent report (Gokce et al., 2011). Thus, we propose that proteins with p > 0.05 but an E/N ratio ≥2.00 or ≤0.50 (above) and SpC ≥10 in hESCs and/or hNSCs (SpC threshold) should provide reliable information on significant differences. False change rates for proteins with SpC ≥10 were <1% in a previous report (Zhou et al., 2010). Accordingly, protein quantification in the dataset demonstrated high reproducibility (Figure S6C). Furthermore, “housekeeping” proteins GAPDH, HPRT1, and α-tubulin (TUBA1C) (E/N ratios of 0.81, 1.10, and 0.94, respectively) displayed similar relative abundance (pluriprot.org), suggesting quantitative reliability.
Using these parameters, we identified protein networks enriched in hESCs, including structural proteins (e.g., cytoskeleton, cell adhesion) and regulatory proteins (e.g., cell cycle, anti-apoptotic; Figure 2D). Proteins enriched in hNSCs were indicative of chromatin modification, neurogenesis, axonal guidance, cell adhesion, androgen receptor signaling, and microtubules (Figure 2D).
Transcription Factors: Distinct Protein Abundance and Phosphorylation Profiles in Pluripotent and Lineage-Committed Cells
The dataset contains 487 transcription factors, with 416 identified in hESCs and 419 in hNSCs (Figure S6D), including OCT4, NANOG, and PAX6, which serve as positive controls (Figures 2C, 2E, S1C–S1F). Some proteins were detected only via phosphopeptides (Figure 2E, SpC = 0), suggesting enhanced sensitivity for detecting proteins due to phosphopeptide enrichment, consistent with another report (Huttlin et al., 2010).
Most transcription factors were phosphorylated (Figures S6D and S6E; Tables S2–S7). Differential phosphorylation of a protein includes at least one site detected in hESCs or hNSCs only (e.g., LRRFIP1), detection of phosphorylation in hESCs only (e.g., FOXO1), or detection of phosphorylation in hNSCs only (e.g., ENC1) (Figure 2E and Table S15). In contrast, some proteins lacked differing phosphorylation sites (e.g., PTTG1). Repeated identification versus lack of identification implies that the phosphopeptide is likely to be more abundant in the cell population in which it was identified (Brill et al., 2009). The PSpC of individual phosphopeptides was between 1 and >100 (Figure S6F). In addition, pY sites were identified on some transcription factors, e.g., OCT4 (Figure 2E).
Analysis of Kinases and Phosphorylation Motifs
The human genome encodes an estimated 518 protein kinases (Manning et al., 2002), 479 of which (92.5%) were detected in hESCs and hNSCs (Tables S2–S7). Among ∼200 phosphatases encoded by the human genome (Sacco et al., 2012), the dataset contains 154 (∼77%); 144 phosphatases were detected in hESCs and 139 in hNSCs.
Receptor tyrosine kinases (RTKs) ERBB2, PDGFR, KDR, and IGF1R promote pluripotency (Brill et al., 2009, Wang et al., 2007). Differentially phosphorylated ERBB2 contained 23 detected sites, two of which are pY (Table S15C). IGF1R, KDR, and KIT were more abundant in hESCs (Table S14A) and PDGFR, IGF1R, and KDR contained pY sites (Tables S15A and S15C).
We detected 14 Ephrin RTKs, 13 of which were phosphorylated, four with pY sites (Figure S6G). EPHA4 and EPHB2 contained detected phosphorylation in hNSCs only and were upregulated in hNSCs. Among the five Ephrin ligands that were detected, EFNB1 and EFNB2 were more abundant in hNSCs (Figure S6G). These findings suggest regulation of Ephrin signaling when hESCs transition to hNSCs.
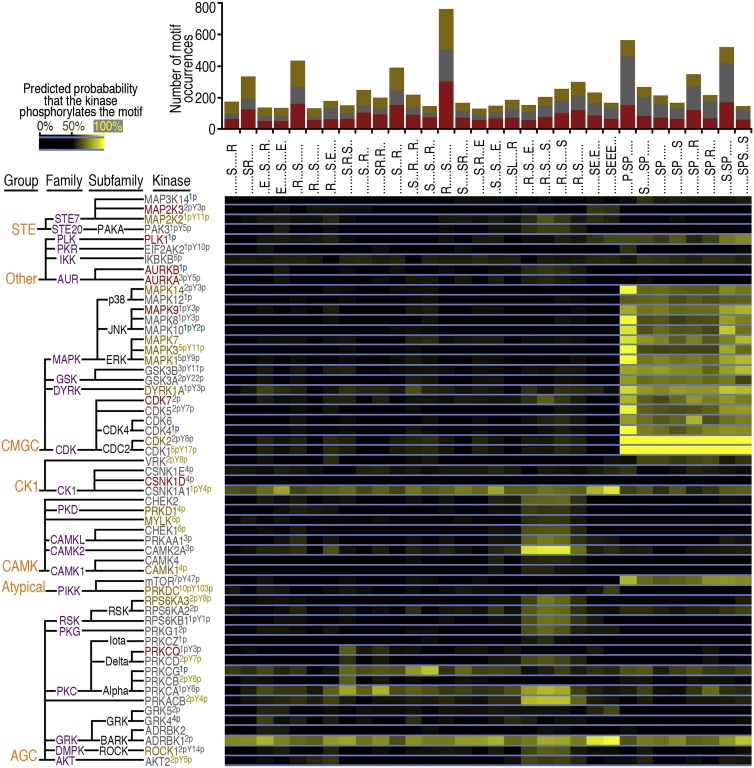
We determined 32 pS sequence motifs in the dataset and predicted S/T kinases in the dataset phosphorylating these motifs (Figure 3). Heatmap analysis suggests functions for numerous S/T kinases in hESCs and hNSCs, including GSK3α/β, CDKs, MAPK (ERK p38, JNK), AKT, mTOR, and AURKA/B. Phosphorylated AURKA was more abundant in hESCs (Figures 3 and S6B), in agreement with a role of Aurora A in sustaining pluripotency in mouse cells (Lee et al., 2012). Hence, comparative kinome analysis in pluripotent and differentiated cells should enable testing of targeted hypotheses.
Figure 3.
S/T Kinases and Their Predicted Substrate Motifs
Motif-X (Schwartz and Gygi, 2005) was used to compute phosphopeptide sequence motifs from localized pS sites. The occurrence of motifs is represented by bar graphs at the top (red, hESCs only; brown, hNSCs only; gray, both cell types). GPS 2.0 (Xue et al., 2008) was used to predict and classify kinases (left) phosphorylating the pS motifs. The colors of kinase symbols (representative of relative abundance) and superscripts (representative of phosphorylation) are defined in Figure 2E. See also Figure S6.
Mapping Cell-Signaling Pathways
The dataset allowed identification of 415 biological pathways (Table S16). Transforming growth factor β (TGF-β) and WNT pathways were examined because they are incompletely characterized in pluripotency and early differentiation, and were modulated by the DAP protocol. Coverage of (phospho)proteins in the canonical TGF-β and WNT pathways was 81% and 89%, respectively (Figures 4A and 4B). Many of these proteins displayed differential abundance and/or phosphorylation. Comparative analysis highlighted (phospho)proteins participating in the regulation of pluripotency and neural conversion (e.g., ID1, LEFTY1, LEFTY2). Additional pathway analyses were performed for Nodal and Notch signaling (Figures S7A and S7B). These examples illustrate that the dataset enables global pathway analysis (Table S16).
Figure 4.
Mapping TGF-β and WNT pathways in hESCs and hNSCs
(Phospho)proteomic data revealed most components of the canonical TGF-β (A) and WNT (B) signaling pathways. Proteins more abundant in hESCs are in red and those more abundant in hNSCs in brown. Proteins without detected differences in abundance are in gray. Proteins not detected are in white. Numbers adjacent to the proteins indicate the E/N ratio. Circles represent protein phosphorylation, with the same colors as the superscript font in Figure 2E. Magenta bands represent ≥1 pY site. Larger circles represent proteins with larger numbers of non-redundant phosphorylation sites. Seven TCF and 14 PRKA proteins were detected (B), respectively (Tables S15A–S15C; for clarity, TCF and PRKA phosphorylation is not shown). See also Figure S7.
Abundance and Phosphorylation of Epigenetic Proteins
Pluripotency is characterized by open chromatin, which is maintained by structural proteins, histone, and DNA modifications that affect gene silencing and activation, whereas lineage commitment of pluripotent cells results in heterochromatin formation (Orkin and Hochedlinger, 2011, Zhu et al., 2013). We reasoned that uniform neural induction is ideal for comparison of the (phospho)proteomic state of epigenetic regulators in pluripotent and multipotent cells. The dataset contains at least 273 proteins involved in epigenetic changes and chromatin modifications (Figure 5). Eighty-seven percent contained identified phosphorylation, including 33% with pY sites (Figure 5, magenta circles).
Figure 5.
(Phospho)Proteome of Epigenetic Regulators in hESCs and hNSCs
Proteins represented as circles were phosphorylated, whereas triangles represent proteins lacking identified phosphorylation (P and NP, respectively). Red or brown represents higher relative protein abundance in hESCs or hNSCs, respectively. Gray indicates that no significant difference in the abundance of the protein was detected. Numbers indicate the E/N ratio. Colors of circular bands indicate comparative phosphorylation between cell types as described for Figures 2E and 4. Symbols of proteins in bold font are mentioned in the text. See also Figure S6.
Previous epigenetic protein characterizations were performed mostly in rodent models. The Polycomb repressive complex 2 (PRC2), containing SUZ12, EZH2, and EED, plays an important role in establishing “bivalent domains” in pluripotent cells that keep developmental genes silent but poised for expression upon differentiation (Young, 2011). All PRC2 proteins were phosphorylated in hESCs, and EZH2 and EED were phosphorylated in hNSCs (Figure 5 and Table S15B). The relative abundance of these proteins was similar to changes described during mouse development (Meissner, 2010). SUZ12 and EZH2 were more abundant in hESCs, whereas EED was more abundant in hNSCs. Other examples of epigenetic regulators are DNMT3B, JARID2, and CHD1, which were more abundant and phosphorylated in hESCs (e.g., pY404 on DNMT3B; pY225 on CHD1).
Non-histone, chromatin-associated high-mobility group (HMG) proteins participate in transcriptional regulation and changes in DNA conformation (Bianchi and Agresti, 2005). The dataset contains 11 HMG proteins, with HMGB2 upregulated in hNSCs (Figure S6B). Neuronal differentiation is initiated by overcoming the repression by REST, thereby inducing neural-specific genes in mouse cells (Bianchi and Agresti, 2005). Consistently, REST was downregulated in hNSCs (Figure 2E). The chromatin insulator CTCF is a key genome organizer that can block the interaction of enhancer elements with unrelated promoters in mouse cells (Handoko et al., 2011). CTCF was more abundant in hNSCs, as measured by MDLC-MS/MS and western blots (Figures 5 and S6B). The heterochromatin protein HP1BP3 was upregulated in hNSCs (www.pluriprot.org).
Validation of (Phospho)Proteomic Data Suggest a Functional Role for MDK During Neural Differentiation
Although progress has been made (Boles et al., 2014, Chambers et al., 2009), our understanding of the mechanisms of neural fate choice from hESCs remains incomplete. Since secreted proteins may contribute to cell differentiation, we focused on protein ligands present in our dataset (Tables S2–S7). Among these, MDK was strongly upregulated in hNSCs (Figure 2C). MDK was identified in differentiating teratocarcinoma cells and is expressed in the mouse neural tube during mid-gestation (Muramatsu, 2011, Uehara et al., 1992). Although MDK was detected in the secretome of hESCs (Bendall et al., 2009), its functional role has not been studied in hPSCs. MDK is expressed by undifferentiated hPSCs and upregulated when hESCs and hiPSCs are differentiated using the EB method or controlled 6-day DAP treatment (Figures 6A and 6B). Upregulation of MDK in hNSCs (Figure 2C) was confirmed by western blots (Figure 6B). MDK was absent from skin fibroblasts (FB) but induced during cellular reprogramming (FB-hiPSC).
Figure 6.
MDK Expression in Pluripotent and Differentiated Cells and Disruption of Neural Conversion by a Monoclonal Antibody against MDK
(A) Representative immunocytochemical analysis of MDK expression by hESCs (WA09), skin fibroblasts (FB, line HS27), and fibroblasts (HS27) after reprogramming to hiPSCs. Scale bars represent 100 μm.
(B) Western blot showing that MDK was not detected in HS27 fibroblasts but was expressed in WA09 hESCs and hiPSCs from HS27 fibroblasts, and was upregulated in differentiating cells (EBs and hNSCs from hESCs). Probing for GAPDH demonstrated similar loading of the lanes.
(C) A monoclonal antibody against MDK did not impair pluripotent self-renewal of hESCs. Antibody concentrations are shown.
(D) The percentage of pluripotent cells expressing OCT4 and NANOG did not differ significantly between anti-MDK and isotype control antibody treatment (3.0 μg/mL). Error bars represent SEM; n = 5.
(E) qRT-PCR showed that a monoclonal antibody against MDK (1.5 μg/mL) inhibits neural gene expression during 6-day DAP treatment. Error bars represent mean + SEM, n = 3. ∗p < 0.05.
(F) Western blot analysis confirmed the inhibitory effect of anti-MDK on early neural marker expression (OTX1, PAX6), despite DAP treatment, in contrast to the isotype control.
(G) Immunocytochemical evidence that neural conversion is inhibited by the anti-MDK antibody (1.5 μg/mL) during 6-day DAP treatment. Few cells induced PAX6 expression in the presence of anti-MDK antibody (right panel). Scale bar represents 200 μm.
(H) Western blot analysis and time course of hESCs differentiated for 6 days with recombinant MDK alone (100 ng/mL) or a combination of the DAP cocktail and MDK (25 or 100 ng/mL) suggest that MDK specifically promotes neural commitment of hPSCs. SOX17, MIXL1, and Brachyury were not induced by MDK. In contrast, controls treated with fetal bovine serum (FBS) for 6 days, which stimulates multi-lineage differentiation of hPSCs, induced these non-neural markers.
We asked whether inhibition of MDK signaling by an anti-MDK monoclonal antibody has functional consequences for pluripotency and/or neural differentiation. hESCs were cultured in the presence of anti-MDK or isotype control antibody. Anti-MDK did not detectably affect pluripotent growth of hESCs, as measured by alkaline phosphatase staining and expression of OCT4 and NANOG (Figures 6C and 6D). However, when anti-MDK was applied during 6-day neural induction, upregulation of OTX1, PAX6, and PLZF mRNAs was impaired (Figure 6E) and neural protein expression was strongly suppressed (Figures 6F and 6G). We also tested whether MDK promotes mesodermal and endodermal lineage commitment. Experiments using recombinant MDK alone or in combination with the DAP cocktail demonstrated that non-neural markers SOX17, BRACHYURY, and MIXL1 were not induced by MDK but detected in controls treated with fetal bovine serum (Figure 6H). Furthermore, MDK alone partially induced expression of PAX6 in hESCs but, remarkably, MDK in combination with DAP resulted in earlier and stronger expression of PAX6 compared with DAP treatment alone (Figure 6H).
Genetic Knockdown of MDK Efficiently Blocks Neural Conversion of Pluripotent Human Cells
To validate that increased MDK expression is important for neural induction, we performed knockdown of MDK in hESCs using retroviral short hairpin RNAs (shRNAs). Four stable cell lines with scrambled control shRNA and two cell lines with efficient MDK knockdown were analyzed by western blotting and immunocytochemistry (Figures 7A–7C). We did not detect differences in self-renewal when hESC lines with scrambled control shRNA or MDK knockdown were expanded in parallel with parental hESCs, similar to the use of anti-MDK antibodies (Figures 6C and 6D). However, when hESCs were treated with DAP for 6 days, PAX6 expression was blocked in both cell lines with MDK knockdown but not in scrambled shRNA controls (Figures 7A–7D). These findings, together with experiments using the anti-MDK antibody (Figures 6E–6G), support the notion that upregulation of MDK is necessary for neural lineage entry of hESCs. Collectively, these results suggest that suppression of TGF-β, bone morphogenetic protein (BMP), and WNT signaling (the rationale for using the DAP cocktail) and the action of autocrine/paracrine MDK are important mechanisms of early neural specification (Figures 4, 6, and 7).
Figure 7.
MDK Knockdown Validates the Importance of MDK Upregulation for Neural Fate Specification of hESCs
(A) Western blot analysis of MDK in control H9-hESC lines (scrambled shRNA) and H9-hESC lines with MDK knockdown (left panel). Quantification (right panel) confirmed efficient MDK knockdown.
(B and C) Immunocytochemical analysis of MDK in control hESCs (scrambled shRNA) (B) and hESCs with MDK shRNA (C) confirmed efficient MDK suppression in the cells containing MDK shRNA. Scale bars represent 100 μm.
(D) Controls and cell lines with MDK knockdown were treated with DAP for 6 days. PAX6 was inhibited in cell lines with MDK knockdown, whereas PAX6 was expressed in scrambled shRNA controls.
(E) Model proposed for controlled neural induction of hESCs, depicting the simultaneous modulation of cell-signaling pathways and a role for MDK. The (phospho)proteomic dataset also suggests changes in relative abundance and phosphorylation of putative MDK receptors (ALK, PTPRZ1, SDC1, and LRP1) in hESCs and hNSCs. Colors are as described in Figure 2E and the sizes of letters reflect E/N and PSpC ratios.
Complementary Analysis of Neural Induction by Transcriptomic and Proteomic Datasets
Transcriptomic data from the same well-defined cell populations was obtained to gain additional insights into gene regulation in pluripotent and multipotent cells. Among the 9,635 genes whose products were identified as mRNAs and proteins, changes in the relative abundance of cognate mRNAs and proteins displayed concordance and discordance (Figure S7C). Overall correlation in changes between both datasets was only r = 0.40. This may reflect the importance of protein translation, stability, and PTMs as part of the regulatory mechanisms that are integrated with epigenetic and transcriptional changes during the switch of cellular identities. The complexity of these coordinated events has been reported in murine ESCs (Lu et al., 2009, Sampath et al., 2008). Hence, combined analysis of proteomic and transcriptomic data is desirable for precise characterization of cell type identities during differentiation and reprogramming (Benevento et al., 2014). The use of both methods allowed the identification of a larger number of significant biological processes than either alone. Cell type-specific biological processes segregated more clearly when using the proteomic dataset (Figures 2D and S7D).
Discussion
To our knowledge, uniformly differentiated, lineage-committed cells derived from hESCs have not been analyzed previously by proteomics. Our data provided insights into the molecular complexity of pluripotency and cell differentiation, and captured specific (phospho)proteomic signatures in direct comparisons of hESCs and hNSCs. The data shed light on cell-signaling pathways and the human kinome, and established an expanded resource of epigenetic proteins.
The dataset allows a well-defined comparison of protein abundance and phosphorylation in pluripotent cells and the earliest definitive hNSCs. This contrasts with previous studies that used hESCs and heterogeneous mixtures of differentiated progeny (Brill et al., 2009, Rigbolt et al., 2011, Van Hoof et al., 2009), or hESCs only (Swaney et al., 2009). Another study compared hESCs and hiPSCs, reporting 6,761 proteins and 19,122 phosphorylation sites (Phanstiel et al., 2011). A proteomic analysis of the membrane fraction of hESCs and heterogeneous cells enriched for NSCs, derived by EB differentiation, yielded 5,105 proteins and 8,560 phosphorylation sites (Melo-Braga et al., 2014). In contrast, we used whole-cell lysates from hESCs and their hNSC derivatives at high purity under feeder-free conditions. Importantly, we did not passage the cells during hNSC induction, so as to capture unperturbed (phospho)proteomic profiles when pluripotent cells transition to the neural lineage. We report nearly 13,000 proteins (Figure 2A), most of which were phosphorylated at nearly 60,000 non-redundant sites (Table S1), revealing a striking depth of (phospho)proteomes and biological pathways. The dataset contains most previously published (phospho)proteins in hPSCs, plus more not reported in those studies (Figures S7E–S7H). In addition, our results suggest previously unknown reproducibility of the hPSC phosphoproteome via comparison with these datasets.
Phosphorylated transcription factors are prominent in hESCs (Brill et al., 2009, Rigbolt et al., 2011, Van Hoof et al., 2009). Our current dataset represents progress toward comprehensive (phospho)proteomic mapping of transcription factors. Rigbolt et al. (2011) found that 83.8% of 216 detected transcription factors were phosphorylated. Similarly, 75.1% of the 487 transcription factors in our dataset were phosphorylated. OCT4 phosphorylation on pT234 and pS235 disrupts its binding to DNA (Brumbaugh et al., 2012). Phosphorylation sites pT225 and pY291 (Table S15A), to the best of our knowledge, have not been identified previously on OCT4. Our dataset also contains two phosphorylation sites on NANOG (pS10 and pS52). In addition, 12 phosphorylation sites were detected on SOX2, five of which were not reported previously (pY2, pT7, pT17 or pS18, pS257, and pS290). These and other examples suggest the importance of post-translational regulation of transcription factors, which is understudied.
Dual SMAD inhibition and modulation of WNT signaling is important for neural conversion of hPSCs, as indicated previously (Chambers et al., 2009, Hasegawa et al., 2012) and by the present study (Figure 1A). NPTX1 plays a role in neural lineage specification from hESCs by reducing NODAL and BMP signaling (Boles et al., 2014), suggesting that NPTX1 further contributes to dual SMAD inhibition (Chambers et al., 2009). However, it remained unclear whether neural induction is regulated by other signals. The secreted protein MDK is an instructive component of neural lineage entry (Figure 7E). Blocking MDK by genetic knockdown and a monoclonal antibody provided independent evidence for a mechanism by which feedforward upregulation of MDK promotes neural induction, consistent with MDK resulting in specific upregulation of PAX6 (Figures 6 and 7). MDK may act through a receptor complex that can include ALK, PTPRZ1, LRP1, and SDC1 (Kadomatsu et al., 2013, Muramatsu, 2011). PTPRZ1 was more abundant in hESCs, whereas LRP1 was more abundant in hNSCs (Figure 7E). Although MDK was reported 26 years ago (Tomomura et al., 1990), MDK and its receptors remain understudied. Hence, the finding that MDK plays a role in PSC differentiation requires detailed follow-up studies.
Collectively, our methods and dataset are resources to leverage diverse studies including further elucidation of pluripotency, differentiation, and reprogramming. The dataset may also prove useful for comparison with stem cells from non-mammalian models. For example, orthologs of all proteins in the COP9 complex in Drosophila germline stem cells (Pan et al., 2014) are in our dataset. We highlighted several analyses that the dataset enables and provided proof of principle for its predictive value. We make this resource freely available to the scientific community as curated tables, an interactive website (www.pluriprot.org), and raw data in the Peptide Atlas database (Institute for Systems Biology).
Experimental Procedures
Cell Culture
Pluripotent cells were cultured under feeder-free conditions on Matrigel with MEF-CM and FGF2. For neural induction, unconditioned medium and 2 μM dorsomorphin, 2 μM A83-01, and 2 μM PNU-74654 (DAP cocktail) were added. A retrovirus-based protocol (Takahashi et al., 2007) was modified to generate hiPSCs. Details are provided in Supplemental Experimental Procedures.
Immunocytochemistry, Fluorescence-Activated Cell Sorting, and Western Blots
For immunocytochemistry, cells were fixed in 4% paraformaldehyde and stained with the antibodies listed in Supplemental Experimental Procedures. Fluorescence-activated cell sorting analysis (Pankratz et al., 2007) and western blots (Brill et al., 2009) were as described previously. Details, including procedures for immunoprecipitations, are provided in Supplemental Experimental Procedures.
Knockdown of MDK by shRNA
hESCs were transduced with either MDK or scrambled control shRNA, and drug selection with puromycin and kanamycin was used to establish stable cell lines.
MDK Inhibition by a Monoclonal Antibody
Medium with either anti-MDK or isotype control antibodies was added daily with the DAP cocktail for 6 days.
Transcriptome Analysis
Illumina HumanHT-12 Expression Arrays were analyzed using a BeadArray Reader. Data were collected using the scanner software. Sample probe files were obtained using GenomeStudio after expression intensities were calculated and quality controlled with detection p values of ≤0.05 for each gene probe. Differentially expressed genes were detected in GeneSpring GX 11.5 by quantile normalization, log transformation, t tests with Benjamini-Hochberg corrected p value of ≤0.05, and fold-change difference of ≥2.0.
Total (Phospho)Proteome Work Flow
Approximately 4 × 107 hESCs or hNSCs were rinsed with PBS and lysed. Lysates were centrifuged, (NH4)2SO4 added to supernatants, mixtures agitated at 4°C, and centrifuged. Protein pellets were stored at −80°C. Pellets were resuspended in 100 mM (NH4)2HCO3/8 M urea containing protease and phosphatase inhibitors and gel filtered, and protein concentrations measured. Protein (1 mg) was reduced, alkylated, digested with trypsin, peptides desalted, dried, and stored at −80°C.
Peptides were separated by SCX, desalted, and dried. Phosphopeptides were enriched from SCX fractions. LC-ESI-MS/MS was performed on each fraction in duplicate with a C18 column, captive spray source, and LTQ Orbitrap Velos mass spectrometer with electron-transfer dissociation.
MS/MS data were searched against a concatenated forward and reverse ipi.HUMAN.v.3.73 database with Sorcerer-SEQUEST. Static carbamidomethylation of C, differential oxidation of M, and differential phosphorylation of S, T, and Y were specified. Protein identifications were filtered at an FDR of <0.01 (ProteinProphet), including stringent phosphopeptide-level verification (Supplemental Experimental Procedures). NSAF-SpCs (Zhang et al., 2010b), Ln-NSAFs (Zybailov et al., 2006), and PSpCs were calculated.
(Phospho)Proteomic Data Mining
Cell-signaling pathways, protein networks, and interaction maps were analyzed and processed using KEGG, Ingenuity, GeneGO, Cytoscape, and Adobe Photoshop. Phosphorylation motifs and kinase predictions were as described previously (Schwartz and Gygi, 2005, Xue et al., 2008) and displayed with Treeview (Saldanha, 2004), and predicted kinases not in the dataset were removed (Figure 3).
Supplemental Material
Table S1 is included in the Supplemental Information, and Tables S2–S16 can be downloaded at http://dx.doi.org/10.17632/v7nt3wm8rk.1. Interactive analysis of the proteomic dataset can be performed by searching for specific proteins of interest (enter protein name on Homepage) and accessing Network diagrams (Networks): http://pluriprot.org.
Source Data
Author Contributions
Concept and experimental design: I.S., J.H., and L.M.B; Experiments performed: I.S., A.M.C., J.H., B.T.D.T., M.T., A.A.W., J.C., E.L., G.J.G., and L.M.B.; Data analysis and discussions: I.S., J.H., K.S.D., X.H., D.M.H., D.A.W., S.A.L., G.J.G., E.Y.S., and L.M.B.; Manuscript writing: I.S., G.J.G., and L.M.B.
Acknowledgments
We thank Toshio Kitamura for PLAT-A cells, and Gregg Duester and Daniel J. Donoghue for discussions. Yoav Altman, Joseph Russo, Edward Monosov, Frederick Lo, Maria Jose Martinez Lopez, Justin D. Blethrow, and Alexey Eroshkin kindly provided technical assistance. Several antibodies used in this study were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology. I.S. received support from the International Bipolar Foundation, CIRM, and Merck and the German Research Council (DFG). A.C. was supported by the Biomedical Sciences Graduate Program at UCSD. B.T. was funded by NIMH T32, UCSD Department of Psychiatry. G.J.G. is funded by BELSPO (IAP-VII/07), Innoviris-Brussels (BB2B), and the FWO. Additional support was provided by NIH P20 GM075059, P01 ES016738-01, CIRM RC1-00125-1, SBMRI NCI CCSG 5 P30 CA30199-28, NINDS 5 P30 NS057096, NIMH RC2 MH090011, P50 GM 085764-03, P01 CA138390, and the Sanford Children's Health Research Center.
Published: August 25, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and 16 tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.07.019.
Contributor Information
Ilyas Singec, Email: ilyas.singec@nih.gov.
Laurence M. Brill, Email: lbrill@san.rr.com.
Evan Y. Snyder, Email: esnyder@sbpdiscovery.org.
Accession Numbers
The (phospho)proteome data can be accessed on the websites specified above, and all raw data are deposited in the Peptide Atlas (accession number PASS00233; https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/PASS_View?identifier=PASS00233; Institute for Systems Biology).
Transcriptomic data are in the Gene Expression Omnibus, accession number GEO: GSE39876.
Supplemental Information
References
- Bendall S.C., Hughes C., Campbell J.L., Stewart M.H., Pittock P., Liu S., Bonneil E., Thibault P., Bhatia M., Lajoie G.A. An enhanced mass spectrometry approach reveals human embryonic stem cell growth factors in culture. Mol. Cell. Proteomics. 2009;8:421–432. doi: 10.1074/mcp.M800190-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevento M., Tonge P.D., Puri M.C., Hussein S.M., Cloonan N., Wood D.L., Grimmond S.M., Nagy A., Munoz J., Heck A.J. Proteome adaptation in cell reprogramming proceeds via distinct transcriptional networks. Nat. Commun. 2014;5:5613. doi: 10.1038/ncomms6613. [DOI] [PubMed] [Google Scholar]
- Bianchi M.E., Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Boles N.C., Hirsch S.E., Le S., Corneo B., Najm F., Minotti A.P., Wang Q., Lotz S., Tesar P.J., Fasano C.A. NPTX1 regulates neural lineage specification from human pluripotent stem cells. Cell Rep. 2014;6:724–736. doi: 10.1016/j.celrep.2014.01.026. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger R., Wei H., Zhang S., Lei S., Murage J., Fisk G.J., Li Y., Xu C., Fang R., Guegler K. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 2004;22:707–716. doi: 10.1038/nbt971. [DOI] [PubMed] [Google Scholar]
- Brill L.M., Xiong W., Lee K.B., Ficarro S.B., Crain A., Xu Y., Terskikh A., Snyder E.Y., Ding S. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J., Hou Z., Russell J.D., Howden S.E., Yu P., Ledvina A.R., Coon J.J., Thomson J.A. Phosphorylation regulates human OCT4. Proc. Nat. Acad. Sci. USA. 2012;109:7162–7168. doi: 10.1073/pnas.1203874109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. Mol. Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- Collier T.S., Sarkar P., Franck W.L., Rao B.M., Dean R.A., Muddiman D.C. Direct comparison of stable isotope labeling by amino acids in cell culture and spectral counting for quantitative proteomics. Anal. Chem. 2010;82:8696–8702. doi: 10.1021/ac101978b. [DOI] [PubMed] [Google Scholar]
- Elkabetz Y., Panagiotakos G., Al Shamy G., Socci N.D., Tabar V., Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–165. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V., Gutierrez G.J., Brill L.M., Varsano T., Feng Y., Aza-Blanc P., Au Q., McLaughlan S., Ferreira T.A., Alain T. Degradation of newly synthesized polypeptides by ribosome-associated RACK1/c-Jun N-Terminal kinase/Eukaryotic Elongation factor 1A2 complex. Mol. Cell. Biol. 2013;33:2510–2526. doi: 10.1128/MCB.01362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokce E., Shuford C.M., Franck W.L., Dean R.A., Muddiman D.C. Evaluation of normalization methods on GeLC-MS/MS label-free spectral counting data to correct for variation during proteomic workflows. J. Am. Soc. Mass. Spectrom. 2011;22:2199–2208. doi: 10.1007/s13361-011-0237-2. [DOI] [PubMed] [Google Scholar]
- Handoko L., Xu H., Li G., Ngan C.Y., Chew E., Schnapp M., Lee C.W., Ye C., Ping J.L., Mulawadi F. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Yasuda S.Y., Teo J.L., Nguyen C., McMillan M., Hsieh C.L., Suemori H., Nakatsuji N., Yamamoto M., Miyabayashi T. Wnt signaling orchestration with a small molecule DYRK inhibitor provides long-term xeno-free human pluripotent cell expansion. Stem Cells Transl. Med. 2012;1:18–28. doi: 10.5966/sctm.2011-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J., Tobe B.T., Lo F., Blethrow J.D., Crain A.M., Wolf D.A., Snyder E.Y., Singec I., Brill L.M. Combined total proteomic and phosphoproteomic analysis of human pluripotent stem cells. Methods Mol. Biol. 2013;1029:163–189. doi: 10.1007/978-1-62703-478-4_12. [DOI] [PubMed] [Google Scholar]
- Huttlin E.L., Jedrychowski M.P., Elias J.E., Goswami T., Rad R., Beausoleil S.A., Villen J., Haas W., Sowa M.E., Gygi S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadomatsu K., Kishida S., Tsubota S. The heparin-binding growth factor midkine: the biological activities and candidate receptors. J. Biochem. 2013;153:511–521. doi: 10.1093/jb/mvt035. [DOI] [PubMed] [Google Scholar]
- Lee D.F., Su J., Ang Y.S., Carvajal-Vergara X., Mulero-Navarro S., Pereira C.F., Gingold J., Wang H.L., Zhao R., Sevilla A. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Sadygov R.G., Yates J.R., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Lu R., Markowetz F., Unwin R.D., Leek J.T., Airoldi E.M., MacArthur B.D., Lachmann A., Rozov R., Ma'ayan A., Boyer L.A. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Martinez N.J., Gregory R.I. MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell. 2010;7:31–35. doi: 10.1016/j.stem.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 2010;28:1079–1088. doi: 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- Melo-Braga M.N., Schulz M., Liu Q., Swistowski A., Palmisano G., Engholm-Keller K., Jakobsen L., Zeng X., Larsen M.R. Comprehensive quantitative comparison of the membrane proteome, phosphoproteome, and sialiome of human embryonic and neural stem cells. Mol. Cell. Proteomics. 2014;13:311–328. doi: 10.1074/mcp.M112.026898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J., Low T.Y., Kok Y.J., Chin A., Frese C.K., Ding V., Choo A., Heck A.J. The quantitative proteomes of human-induced pluripotent stem cells and embryonic stem cells. Mol. Syst. Biol. 2011;7:550. doi: 10.1038/msb.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T. Midkine: a promising molecule for drug development to treat diseases of the central nervous system. Curr. Pharm. Des. 2011;17:410–423. doi: 10.2174/138161211795164167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S.H., Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L., Wang S., Lu T., Weng C., Song X., Park J.K., Sun J., Yang Z.H., Yu J., Tang H. Protein competition switches the function of COP9 from self-renewal to differentiation. Nature. 2014;514:233–236. doi: 10.1038/nature13562. [DOI] [PubMed] [Google Scholar]
- Pankratz M.T., Li X.J., Lavaute T.M., Lyons E.A., Chen X., Zhang S.C. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25:1511–1520. doi: 10.1634/stemcells.2006-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pera M.F., Andrade J., Houssami S., Reubinoff B., Trounson A., Stanley E.G., Ward-van Oostwaard D., Mummery C. Regulation of human embryonic stem cell differentiation by BMP-2 and its antagonist noggin. J. Cell Sci. 2004;117:1269–1280. doi: 10.1242/jcs.00970. [DOI] [PubMed] [Google Scholar]
- Phanstiel D.H., Brumbaugh J., Wenger C.D., Tian S., Probasco M.D., Bailey D.J., Swaney D.L., Tervo M.A., Bolin J.M., Ruotti V. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods. 2011;8:821–827. doi: 10.1038/nmeth.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt K.T., Prokhorova T.A., Akimov V., Henningsen J., Johansen P.T., Kratchmarova I., Kassem M., Mann M., Olsen J.V., Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- Sacco F., Perfetto L., Castagnoli L., Cesareni G. The human phosphatase interactome: an intricate family portrait. FEBS Lett. 2012;586:2732–2739. doi: 10.1016/j.febslet.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A.J. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sampath P., Pritchard D.K., Pabon L., Reinecke H., Schwartz S.M., Morris D.R., Murry C.E. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Schwartz D., Gygi S.P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- Sharma K., D'Souza R.C., Tyanova S., Schaab C., Wisniewski J.R., Cox J., Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of tyr and ser/thr-based signaling. Cell Rep. 2014;8:1583–1594. doi: 10.1016/j.celrep.2014.07.036. [DOI] [PubMed] [Google Scholar]
- Sturgeon C.M., Ditadi A., Awong G., Kennedy M., Keller G. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat. Biotechnol. 2014;32:554–561. doi: 10.1038/nbt.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaney D.L., Wenger C.D., Thomson J.A., Coon J.J. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Nat. Acad. Sci. USA. 2009;106:995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Tomomura M., Kadomatsu K., Matsubara S., Muramatsu T. A retinoic acid-responsive gene, MK, found in the teratocarcinoma system. Heterogeneity of the transcript and the nature of the translation product. J. Biol. Chem. 1990;265:10765–10770. [PubMed] [Google Scholar]
- Uehara K., Matsubara S., Kadomatsu K., Tsutsui J., Muramatsu T. Genomic structure of human midkine (MK), a retinoic acid-responsive growth/differentiation factor. J. Biochem. 1992;111:563–567. doi: 10.1093/oxfordjournals.jbchem.a123797. [DOI] [PubMed] [Google Scholar]
- Van Hoof D., Passier R., Ward-Van Oostwaard D., Pinkse M.W., Heck A.J., Mummery C.L., Krijgsveld J. A quest for human and mouse embryonic stem cell-specific proteins. Mol. Cell. Proteomics. 2006;5:1261–1273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- Van Hoof D., Munoz J., Braam S.R., Pinkse M.W., Linding R., Heck A.J., Mummery C.L., Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- van Hoof D., Krijgsveld J., Mummery C. Proteomic analysis of stem cell differentiation and early development. Cold Spring Harb. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a008177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Schulz T.C., Sherrer E.S., Dauphin D.S., Shin S., Nelson A.M., Ware C.B., Zhan M., Song C.Z., Chen X. Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood. 2007;110:4111–4119. doi: 10.1182/blood-2007-03-082586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M., Schlegl J., Hahne H., Moghaddas Gholami A., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Huang C.T., Chen J., Pankratz M.T., Xi J., Li J., Yang Y., Lavaute T.M., Li X.J., Ayala M. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell. 2010;7:90–100. doi: 10.1016/j.stem.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wen Z., Washburn M.P., Florens L. Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 2010;82:2272–2281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- Zhou J.Y., Schepmoes A.A., Zhang X., Moore R.J., Monroe M.E., Lee J.H., Camp D.G., Smith R.D., Qian W.J. Improved LC-MS/MS spectral counting statistics by recovering low-scoring spectra matched to confidently identified peptide sequences. J. Proteome Res. 2010;9:5698–5704. doi: 10.1021/pr100508p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Adli M., Zou J.Y., Verstappen G., Coyne M., Zhang X., Durham T., Miri M., Deshpande V., De Jager P.L. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B., Mosley A.L., Sardiu M.E., Coleman M.K., Florens L., Washburn M.P. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J. Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.