Summary
Frontotemporal dementia (FTD) and other tauopathies characterized by focal brain neurodegeneration and pathological accumulation of proteins are commonly associated with tau mutations. However, the mechanism of neuronal loss is not fully understood. To identify molecular events associated with tauopathy, we studied induced pluripotent stem cell (iPSC)-derived neurons from individuals carrying the tau-A152T variant. We highlight the potential of in-depth phenotyping of human neuronal cell models for pre-clinical studies and identification of modulators of endogenous tau toxicity. Through a panel of biochemical and cellular assays, A152T neurons showed accumulation, redistribution, and decreased solubility of tau. Upregulation of tau was coupled to enhanced stress-inducible markers and cell vulnerability to proteotoxic, excitotoxic, and mitochondrial stressors, which was rescued upon CRISPR/Cas9-mediated targeting of tau or by pharmacological activation of autophagy. Our findings unmask tau-mediated perturbations of specific pathways associated with neuronal vulnerability, revealing potential early disease biomarkers and therapeutic targets for FTD and other tauopathies.
Graphical Abstract

Highlights
-
•
Upregulation of tau and phospho-tau in FTD patient iPSC-derived tau A152T neurons
-
•
Upregulation of insoluble tau in A152T neurons
-
•
Altered proteostasis stress-inducible pathways in tau A152T neurons
-
•
Tau-dependent vulnerability to stress in A152T neurons reverted by tau downregulation
Haggarty and colleagues show in-depth phenotypic characterization of a human iPSC-derived neuronal model of tau-A152T associated with FTD. This study reveals upregulation of phospho-tau and detergent-insoluble oligomeric tau, dysregulation of proteostasis pathways, and consequent increased cell vulnerability to stress, unmasking potential disease biomarkers and therapeutic targets. This study further demonstrates that tau toxicity can be rescued by genetic and pharmacological downregulation of tau.
Introduction
Frontotemporal dementia (FTD) refers to a group of neurodegenerative diseases caused by focal but progressive neuronal loss, astrogliosis, and spongiosis in the frontal and temporal cortices associated with abnormal intracellular accumulation of proteins, most commonly tau or TDP43 (TAR DNA-binding protein 43) (Karageorgiou and Miller, 2014, Neumann et al., 2015). Currently there are no effective disease-modifying therapies for FTD, making the treatment and prevention of FTD an area of significant unmet medical need.
Tau is expressed ubiquitously in the brain and locates predominantly in neuronal axons, where it regulates microtubule polymerization and guides the transport of proteins and organelles (Kosik et al., 1989, Morris et al., 2011). Alternative splicing of MAPT exons 2, 3, and 10 originates six tau isoforms that differ from one another by 29- or 58-amino-acid inserts at the N terminus, and by the presence of either three (3R-tau) or four (4R-tau) tandem-repeat sequences at the C terminus. Tau function and localization are regulated by post-translational modifications (PTMs); for example, phosphorylation, acetylation, and proteolysis (Johnson and Stoothoff, 2004, Min et al., 2010, Wang et al., 2009). In FTD, sporadic or autosomal dominant forms caused by MAPT mutations, inclusions containing hyperphosphorylated tau (P-tau) are detected within neurons and glia of affected brain regions.
Although these inclusions are key pathological features, the events leading to neuronal loss may start even earlier, but the tau species and precise molecular events causing cell death are poorly understood. Therefore it is crucial to investigate the early molecular events of disease, such as alterations in tau biochemistry and affected cellular pathways (Gerson et al., 2014, Johnson and Stoothoff, 2004). In this context, human induced pluripotent stem cell (iPSC)-derived neurons allow exploring the molecular basis of tau pathogenesis in a disease-relevant genetic background (Ehrlich et al., 2015, Haggarty et al., 2016, Iovino et al., 2015).
Here, we investigated the underlying molecular and cellular mechanisms of pathogenicity associated with the rare tau variant A152T in a human neuronal context. Although the role of tau A152T in disease is still debated, it has been shown to affect tau function and PTMs, promote oligomerization and postmortem detection of inclusions, cause neuronal dysfunction independent of aggregation and neuroinflammation in animal models, and increase significantly the risk for FTD and other neurodegenerative diseases (Coppola et al., 2012, Kara et al., 2012, Labbe et al., 2015, Lee et al., 2013, Maeda et al., 2016, Decker et al., 2016, Pir et al., 2016, Sydow et al., 2016). We utilized iPSCs derived from A152T carriers and derived neural progenitor cells (NPCs) and differentiated neuronal cells (Figure 1A). These cells represent ex vivo models of human neurons, with tau expression at endogenous physiologically relevant levels and in the context of the genomic background associated with disease. Overall, our results reveal potential targets for disease-modifying therapeutics to affect FTD and other tauopathies.
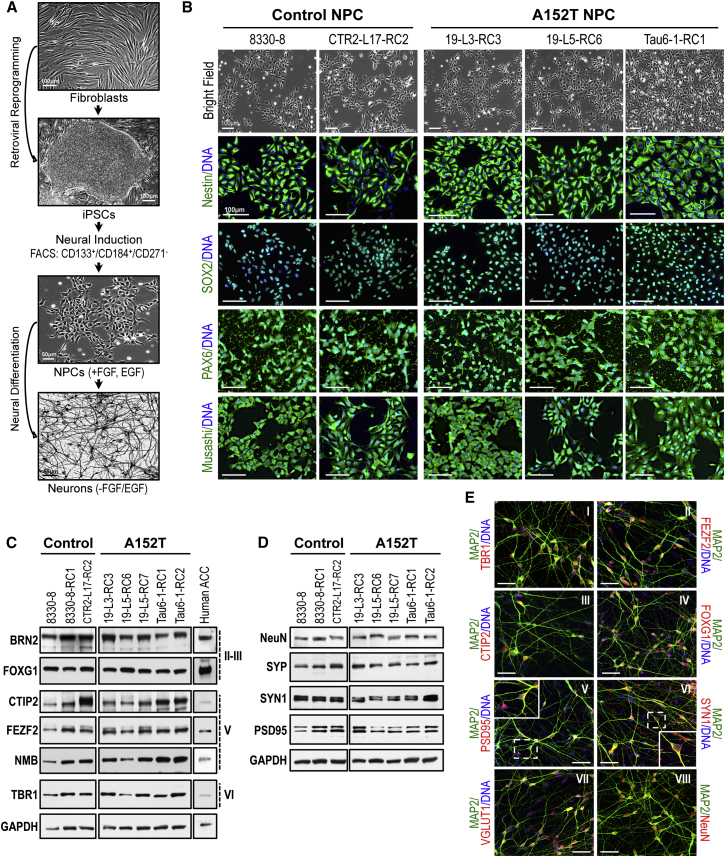
Figure 1.
Human iPSC-Derived NPCs, and Synaptic and Cortical Markers in NPC-Derived Neurons
(A) Summary of the experimental strategy followed. See also Figure S1.
(B) Bright-field and IF analysis of NPC markers (green) in control and A152T lines. Scale bar, 100 μm.
(C and D) Western blot analysis of cortical layers II–III, V, and VI markers (C) and synaptic proteins (D), in 5-week control and A152T neurons. Human ACC is a positive control for cortical markers. GAPDH is the loading control. Samples ran in the same blot and are cropped for clarity (n ≥ 3 independent experiments). See also Figure S2.
(E) IF analysis of cortical (I–IV), synaptic (V–VI), glutamatergic (VII), and neuronal (VIII) markers (red) in 5-week neurons (representative of control and A152T). Dotted insets correspond to zoomed-in images. Scale bars, 20 μm. See also Figure S3.
Results
Human iPSC Lines
Cells from two individuals carrying the MAPT heterozygous variant A152T (c.1407G > A; NCBI RefSeq NM_001123066; rs143624519) were studied. The first individual was diagnosed with a form of FTD, progressive supranuclear palsy, at the time of skin biopsy (FTD19, Table S1), whose iPSCs were generated using standard retroviral vectors and the Yamanaka factors OCT3/4, SOX2, KLF4, and c-MYC (Biswas et al., 2016). The second was an asymptomatic individual (Tau6, Table S1) whose iPSCs and neuronal phenotypes have been in part characterized by Fong et al. (2013). Two age-matched individuals were also included as non-mutant controls: 8330-8 (Sheridan et al., 2011) and CTR2-L17 (Almeida et al., 2012). Two iPSC clones from the first A152T carrier (19-L3, 19-L5), one clone from the second A152T carrier (Tau6-1), and two control iPSC clones (8330-8 and CTR2-L17) were selected for further studies (Table S1).
All iPSCs had typical characteristics of PSCs (Biswas et al., 2016, Fong et al., 2013), including: colony-type morphology; expression of the stem cell surface marker SSEA-4 and pluripotency transcription factors OCT4/POU5F1 and NANOG (Figure S1A); elevated expression of endogenous OCT4/POU5F1, NANOG, and the zinc-finger protein pluripotency marker ZFP42/REX1, relative to the corresponding fibroblasts (Figure S1B); and ability for iPSC in vitro formation of embryoid bodies (EBs) and spontaneous differentiation of EBs into the three germ layers ectoderm (TUJ1 expression), mesoderm (SMA expression), and endoderm (AFP expression) (Figure S1C) (Itskovitz-Eldor et al., 2000). Sanger sequencing of the MAPT locus confirmed the c.1407G > A mutation in A152T iPSCs (Figure S1D). Preservation of normal karyotype was confirmed (Figure S1E). Based on cell proliferation, morphology, and ability to remain undifferentiated in culture over >50 passages, there were no qualitative differences between control and A152T iPSCs.
Derivation and Characterization of Human NPCs
NPC lines were generated from iPSCs through isolation of neural rosettes (Nemati et al., 2011, Ungrin et al., 2008) (Table S1). Newly generated NPCs were immunopositive for the neural stem cell markers Nestin, SOX2, and Musashi-1, and the neuroepithelial marker PAX6 (Figure 1B), and were maintained in proliferative state under defined growth media with EGF (epidermal growth factor) and FGF (fibroblast growth factor ) (Figure 1A) (Nemati et al., 2011, Sheridan et al., 2011, Yuan et al., 2011). Based on proliferation, cell morphology, and viability, there were no qualitative differences between control and A152T NPCs (Figure 1B).
NPC Differentiation into Neurons
Based on an initial time-course analysis of NPC differentiation (Figures S2A–S2F), we examined cortical and synaptic markers in 5-week neurons from two control and two A152T lines. Both control and A152T neurons expressed markers of the cortical layers II–VI, with some variability between cell lines (Figure 1C) that was not correlated with MAPT genotype (Figure S2G), and comparable overall with a human anterior cingulate cortex (ACC) sample. Expression of cortical markers was also confirmed by immunofluorescence (IF) imaging of TBR1-, FEZF2-, CTIP2-, and FOXG1-positive neurons, with highest detection in the nucleus and cell body (Figures 1EI–1EIV). Expression of synaptic proteins PSD95, SYN1, and SYP was also detected at comparable levels in all neuronal cells (Figures 1D and S2H), with appreciable detection in neuronal processes but greater detection in the cell body, as expected at an early stage of neuronal maturity (Figures 1EV and 1EVI). NeuN is a neuronal-specific nuclear/perinuclear protein and MAP2 is a microtubule-associated protein, both utilized as neuronal markers (Figures 1D and 1EVIII). The diversity of neuronal cell types obtained from NPC differentiation was examined based on expression of specific markers. We detected glutamatergic neurons, which are highly affected in FTD (VGLUT1, Figures 1EVII and S3) (Huey et al., 2006), as well as cholinergic, GABAergic, dopaminergic/adrenergic, and serotonergic neurons, in addition to glia, with no detection of neural crest or cardiomyocyte lineage cells (Figure S3).
These results revealed that NPC-differentiated cells display neuron-specific morphology, and expression of protein markers reminiscent of in vivo neurogenesis and human cortical neurons. The variability captured between cell lines (Figures 1C and 1D) was mostly due to inherent differences between iPSC-derived clonal lines and was not correlated with MAPT genotype (Figures S2G and S2H).
Upregulation of Tau in A152T Neurons
FTD-associated MAPT mutations, including the variant A152T, have been shown to promote accumulation of tau and neurodegeneration (Kara et al., 2012, Lee et al., 2013). To explore whether this is an early disease-relevant molecular event in A152T neurons, we started by examining tau protein levels and PTMs. To this end, we utilized three NPC lines derived from the FTD19 A152T carrier (19-L3-RC3, 19-L5-RC6, and 19-L5-RC7), two NPC lines from the Tau6 A152T carrier (Tau6-1-RC1 and Tau6-1-RC2), and three control NPC lines (8330-8, 8330-8-RC1, and CTR2-L17-RC2) (Table S1).
We first performed a time-course analysis of tau levels by western blot between proliferative state (week 0) and 5 weeks of differentiation, by employing a panel of seven tau and P-tau antibodies. By comparing averaged tau levels between controls and A152T lines, we detected a time-dependent increase in total tau levels (TAU5 and DA9 antibodies), with a more rapid tau upregulation in A152T neurons relative to control cells at 5 weeks of differentiation (Figures 2A–2C). These antibodies report on total protein independently of PTMs, which, together with alternative splicing isoforms, give rise to distinct molecular weight bands (Figure 2A, blue arrows). Recombinant human tau, consisting of a ladder of six tau isoforms without PTMs, was also included in the analysis to guide interpretation of band pattern, even though equivalent migration does not report on a particular endogenous tau isoform, given endogenous tau PTMs. TAU1 antibody that recognizes a form of non-P-tau (non-P-S199/S202/T205) corroborated overall tau upregulation in A152T neurons (Figures 2A and 2D). Further individual analysis of all cell lines at 5 weeks of differentiation revealed that, whereas FTD19-derived A152T neurons showed an upregulation of total tau, Tau6-1-derived neurons’ total tau levels were more comparable with control after the same period of differentiation (Figures S4A and S4B).
Figure 2.
Time-Dependent Upregulation of Tau in A152T Neurons
(A and E) Total tau (A) and P-tau (E) expression by western blot analysis of up to 5-week differentiated neurons in control and A152T cell lines. Far-right panels show recombinant tau ladder (six tau isoforms, no PTMs); GAPDH is the loading control (n ≥ 3 independent experiments). Arrows indicate bands quantified: blue for bands with corresponding migration to the tau ladder, green for HMW bands. Samples ran in same blot as far as possible.
(B–D and F–I) Semi-quantitative analysis of western blots in (A) and (E). Band intensities in a.u. relative to GAPDH, averaged (Avrg) across genotype (controls/gray, A152T/black) ± SEM. Two-way ANOVA and Bonferroni post test, n ≥ 3 independent experiments: ns, not significant (p > 0.05); ∗p < 0.05, ∗∗p < 0.01.
See also Figure S4.
Next, P-tau was examined with the antibodies AT8 (P-Ser202/Thr205), AT180 (P-Thr231/Ser235), PHF1 (P-Ser396/Ser404), and S396 (P-Ser396), which recognize phospho-epitopes associated with autosomal dominant forms of tauopathy (Davies, 2000, Goedert et al., 1995). The increase in P-tau levels was also time dependent and, whether analyzed between averaged controls versus A152T lines (Figures 2E–2I and S4B inset) or individually (Figures S4A and S4B), all A152T neurons showed an upregulation of P-tau relative to controls. In particular, AT8 and AT180 tau were barely detected in control neurons (Figures 2F and 2G). Only PHF1 and S396 antibodies revealed tau species of high molecular weight (HMW; >250 kDa) with specificity to A152T neurons (green arrows in Figures 2E and S4A), indicating the presence of oligomeric, hyper-P-tau at the Ser396/Ser404 epitope. Even with some variability among clonal lines, our findings suggest tau upregulation in A152T neurons, particularly in the form of P-tau (Figures 2E–2I and S4B). Semi-quantitative RT-PCR analysis revealed that MAPT expression was constant between control and A152T neurons (total MAPT, Figures S4C and S4D), and that 3R- and 4R-tau expression was also constant across cell lines, with higher 3R-tau levels, consistent with the immaturity of these 5-week neurons (Figures S4C and S4E) (Stein et al., 2014). These results suggest that upregulation of tau is a result of protein accumulation rather than altered gene expression. Subsequent studies are shown at 5 weeks of differentiation, unless noted otherwise, but all phenotypes were also validated up to 20 weeks of differentiation (not shown).
We next utilized IF to examine the subcellular distribution of tau and P-tau. A152T neurons, in particular FTD19-derived neurons, showed a stronger signal for total tau K9JA antibody in the cell body and neuronal processes relative to control (red in Figures 3I–3V). The distinction between A152T and control neurons was even more accentuated for P-tau PHF1 staining, which showed a weaker signal in control neurons, being almost absent in cell bodies, whereas A152T neurons showed strong PHF1 staining in cell body and neuronal processes (red in Figures 3VI–3X). Upon co-staining of tau (K9JA) and P-tau (PHF1), we observed a higher PHF1-to-K9JA (green-to-red) relative intensity in A152T neurons, with a stronger PHF1 signal in the cell body and neuronal processes of A152T neurons relative to control (Figures 3XI–3XV). We did not, however, detect foci-like tau staining, reminiscent of tau aggregates or tangles, and neurons were Thioflavin-S negative (not shown). Due to the high density of neuronal processes in these cultures, we were unable to use automated microscopy to accurately quantify subcellular tau levels.
Figure 3.
Upregulation of Tau in A152T Neurons by IF Analysis
Total tau (K9JA), P-tau (PHF1), and MAP2 by IF in control and A152T 5-week neurons. Dotted insets correspond to zoomed-in images shown on the right. Scale bars, 50 μm.
Collectively, these results suggest that an early marker of A152T pathogenesis is an early and time-dependent accumulation of tau, predominantly in the form of P-tau and oligomeric P-tau. IF corroborated the accumulation of tau/P-tau in A152T neurons, further revealing increased P-tau in neuronal processes and cell body of A152T neurons, consistent with somatodendritic redistribution, both important pathophysiological features of tauopathy, detected here as an early phenotype in patient-derived neurons in vitro (de Calignon et al., 2012, Fong et al., 2013, Li et al., 2011, Sydow et al., 2016).
Quantitative Analysis of Tau in Human Neurons by Mass Spectrometry
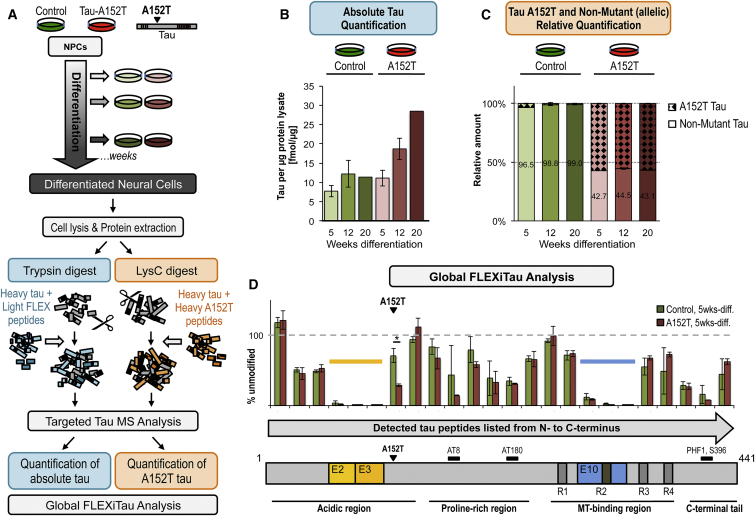
To corroborate our antibody-based results we employed FLEXITau, a mass spectrometry (MS)-based targeted assay for absolute quantification of tau protein levels and PTMs (Mair et al., 2016). A schematic of this methodology is shown in Figure 4A, and in-depth details have been described (Mair et al., 2016, Singh et al., 2009). FLEXITau absolute quantification of tau in control (8330-8-RC1) and A152T (19-L5-RC6) neurons at 5, 12, and 20 weeks of differentiation revealed upregulation of tau in A152T neurons, which was more pronounced at 20 weeks of differentiation (2.5-fold relative to control, average 28.5 fmol/μg and 11.4 fmol/μg protein lysate, respectively) (Figure 4B). Notably, the levels of tau in control neurons reached a plateau after 12 weeks, whereas A152T neurons showed a linear increase in tau levels up to 20 weeks of differentiation (Figure 4B, R2 = 0.999; not shown), again indicative of aberrant tau accumulation in A152T neurons.
Figure 4.
Tau Profile by MS Analysis
(A) Workflow for preparation of control (8330-8-RC1) and A152T (19-L5-RC6) samples for MS analysis.
(B) Quantification of total endogenous tau by FLEXITau. Shown is average tau in fmol/μg of protein lysate ± SD (n = 3, except n = 2 for week 20, see Experimental Procedures).
(C) Relative amount of endogenous non-mutant tau and A152T tau in control and A152T neurons (mean ± SD, n = 3, except n = 2 for week 20, see Experimental Procedures).
(D) Global quantitative peptide profile by FLEXITau, relative to full-length 2N4R-tau. Shown is the normalized amount of unmodified peptides relative to heavy tau protein standard (mean ± SD, n = 3 independent experiments). ∗p < 0.05, Student’s t test.
See also Figure S5.
Differential Expression of Non-mutant and A152T Tau
Given the lack of variant-specific antibodies to date, all studies of expression of endogenous mutant tau have relied on the inference that the mutated allele is expressed. However, this hindrance precludes gaining an understanding of the ratio between non-mutant and mutant protein levels and misinterpretation of total protein levels, because gene mutations can alter protein stability and biochemical properties. Therefore, we used a targeted MS approach to measure non-mutant and variant tau protein levels in control and A152T neurons (LysC digest, Figure 4A). Shotgun tandem MS (MS/MS) analysis from LysC-digested control and A152T neurons identified the peptides 151IATPRGAAPPGQK163 (non-mutant) and 151ITTPRGAAPPGQK163 (A152T) by specific peptide fragment spectra (Figure S5). For quantification of these peptides, we developed a targeted selected reaction monitoring (SRM) assay (Supplemental Experimental Procedures), which showed that in A152T neurons the levels of A152T tau are higher (56.6%) than non-mutant tau (43.4%), and this ratio stayed constant over the time points analyzed (Figure 4C), even though overall levels of total tau increased (Figure 4B). This is the first demonstration and quantification of allele-specific expression of a tau variant protein in a human iPSC-derived neuronal model.
Tau PTMs and Isoforms by Mass Spectrometry
FLEXITau MS analysis also enables the characterization of MAPT splicing and tau PTMs (Mair et al., 2016). This strategy relies on the quantification of unmodified tau peptides in a cell lysate relative to spiked-in stable isotope-labeled unmodified peptides derived from heavy-labeled tau protein. A reduction in cell lysate-measured light peptides is evidence of a “modification” in the endogenous tau peptide, and this modification can be a PTM, alternative splicing, or a mutation. We employed global FLEXITau analysis on 5-week neurons, using both trypsin and LysC samples to maximize sequence coverage (Figure 4A). The regions with the highest “modifications” were the exons that undergo alternative splicing (E2, E3, E10; Figure 4D). Within the sensitivity of our MS assay, the peptides corresponding to E2 and E3 were not detected, whereas E10 (4R-tau) was detected by one peptide (Figure 4D), corroborating gene-expression analysis (Figures S4C and S4E) and predominance of the 0N3R isoform. FLEXITau also detected “modified” peptides within the proline-rich region and the C-terminal tail, probably corresponding to PTMs. The peptides harboring Ser202/Thr205 (AT8), Thr231/Ser235 (AT180), and Ser396/Ser404 (PHF1) epitopes showed a decrease in unmodified peptides, i.e., an increase in “modified” epitopes in A152T neurons relative to control (p = 0.013), consistent with increased P-tau (Figure 4D). As expected, in A152T neurons the non-mutant (unmodified) peptide that covers the A152 site (151IATPRGAAPPGQK163) was only 47.5% of the corresponding peptide in control neurons, as the remaining “modified” peptide harbors to A152T variant (Figure 4D).
One possible mechanism for the variant A152T to alter tau function (Derisbourg et al., 2015) would be by creating a new threonine (Thr/T) P-site. To test this, we attempted to quantify the P-peptide 151IpTTPRGAAPPGQK163 by SRM analysis, using a synthetic heavy P-peptide as a reference. We were unable to detect the endogenous light counterpart within the sensitivity range of our SRM assay (as low as 2.5 fmol/μL of endogenous tau, not shown), suggesting that in human iPSC-derived neurons, the variant A152T is unlikely to create a new phosphorylation site or, if it does, it is phosphorylated to less than 2.5% occupancy.
Reduced Tau Solubility in A152T Neurons
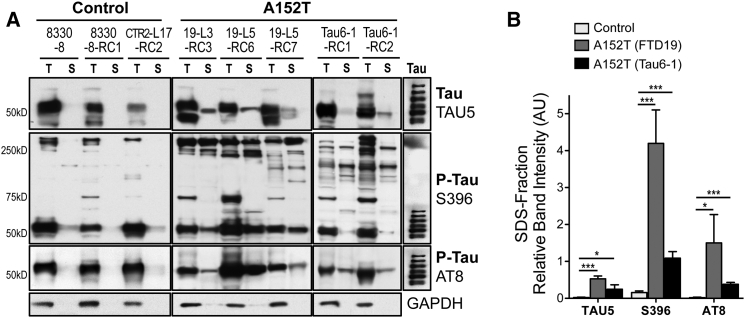
To determine whether accumulation of P-tau led to the formation of protein species of reduced solubility, a hallmark of tauopathy, we tested tau protein differential solubility to Triton X-100 and SDS detergents (Guo and Lee, 2011, Guo and Lee, 2013, Kfoury et al., 2012). A152T neurons showed significantly more accumulation of Triton-insoluble total tau (TAU5) and P-tau (AT8, S396), i.e., SDS-fraction tau (S, Figure 5A). The HMW species (>250 kDa) detected by the P-tau S396 antibody showed a mixture of detergent-soluble and -insoluble tau in A152T neurons, but only Triton-soluble tau in control neurons. When comparing only the insoluble fractions (S), A152T neurons showed a preferential accumulation of P-tau, which was particularly accentuated in FTD19-derived neurons (Figure 5B). These results suggest that accumulation of tau in A152T neurons leads to the formation of species of reduced solubility, which is likely to negatively affect cellular function.
Figure 5.
Detection of Insoluble Tau in A152T Neurons
(A) Western blot analysis of total tau (TAU5) and P-tau (S396, AT8) in Triton X (T)-soluble and Triton-X-insoluble SDS (S) fractions from 5-week neurons. GAPDH is the loading control.
(B) Semi-quantitative analysis of S-fraction band intensities from (A), relative to GAPDH in T-fraction (AU, arbitrary units). Values averaged between control, FTD19, and Tau6 lines (±SEM, n ≥ 3 independent experiments; ∗p ≤ 0.05, ∗∗∗p ≤ 0.001, Student’s t test).
Affected Proteostasis in FTD Neurons
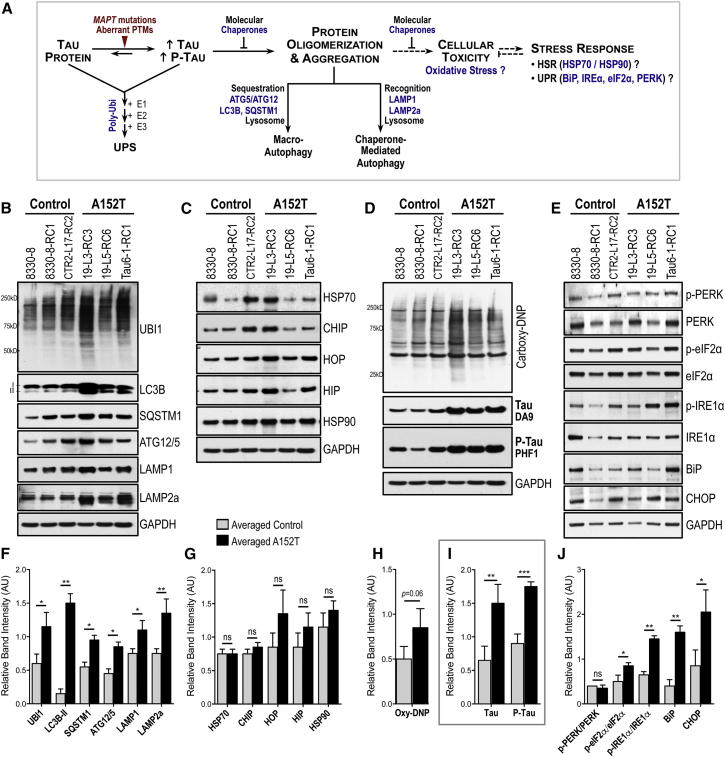
We examined whether upregulation of tau in A152T neurons affected proteostasis, which is kept by the concerted function of multiple pathways (Figure 6A), including the heat-shock response (HSR) and the ER and mitochondrial unfolded protein responses (UPR) that upregulate expression of molecular chaperones and other factors to prevent aberrant accumulation of proteins, and the ubiquitin proteasome system (UPS) and autophagy-lysosomal pathways responsible for clearance of these proteins (Balch et al., 2008, Wong and Cuervo, 2010). We asked whether these pathways were affected in A152T neurons by measuring pathway-specific components (Figure 6A, in blue). We examined 8-week differentiated neurons to take advantage of time-dependent accumulation of tau.
Figure 6.
Enhanced Stress Markers in A152T Neurons
(A) Simplified representation of proteostasis pathways responsive to proteotoxic stress (blue denotes components measured).
(B–J) Western blot and semi-quantitative analysis, in 8-week control and A152T neurons, of: (B and F) polyubiquitinated (UBI1), macro-autophagy (LC3B, SQSTM1/p62, ATG12-ATG5), and chaperone-mediated autophagy (LAMP1, LAMP2a) proteins; (C and G) molecular chaperones (HSP70, HSP90) and HSP70 co-chaperones (CHIP, HIP, HOP) of the HSR; (D and H) polycarbonylated proteins (DNP-derived); (E and J) UPR activation of PERK, eIF2α, and IRE1α, and downstream targets CHOP and BiP; (D and I) tau (DA9) and P-tau (PHF1). Shown is mean band intensity (±SD) relative to GAPDH; n = 3 independent experiments. Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant.
Relative to control, A152T neurons showed an increase in polyubiquitinated proteins (UBI1), which accumulate when UPS is disrupted, as well as an upregulation of autophagy markers (LC3B-II, ATG12/5, SQSTM1/p62, LAMP1, and LAMP2a), revealing activation of autophagy in A152T neurons (Figures 6B, 6F, 6D, and 6I). In contrast, there was no significant HSP70 chaperone family (HSP70, CHIP, HIP, and HOP) or HSP90 upregulation, suggesting no HSR induction (Figures 6C and 6G). To examine oxidative stress, we measured the levels of carbonylated proteins that accumulate in the cell by excessive protein oxidation (Figure 6D) (Dalle-Donne et al., 2003). A152T neurons showed a modest increase in stable 2,4-dinitrophenylhydrazone (DNP) proteins, suggesting disruption of proteostasis leading to oxidative stress (p = 0.06, Figure 6H). Finally, we examined UPR by measuring the ratios between the phospho-active forms and total levels of PERK, eIF2α, and IRE1α (Stutzbach et al., 2013), and the downstream targets CHOP and ER-HSP70 BiP (Figure 6A). Our results showed P-eIF2α/eIF2α and P-IRE1α/IRE1α upregulation in A152T neurons, with further increase in BiP and CHOP levels (Figure 6J). Even though we did not detect upregulation of P-PERK/PERK, CHOP is a downstream target of P-PERK and P-eIF2α, and transcriptional regulation cannot be excluded. Altogether, these results suggest that protein clearance and UPR are the most affected proteostasis pathways at the early stage of tau accumulation in an A152T iPSC-derived neuronal model.
Selective Vulnerability of A152T Neurons to Stress
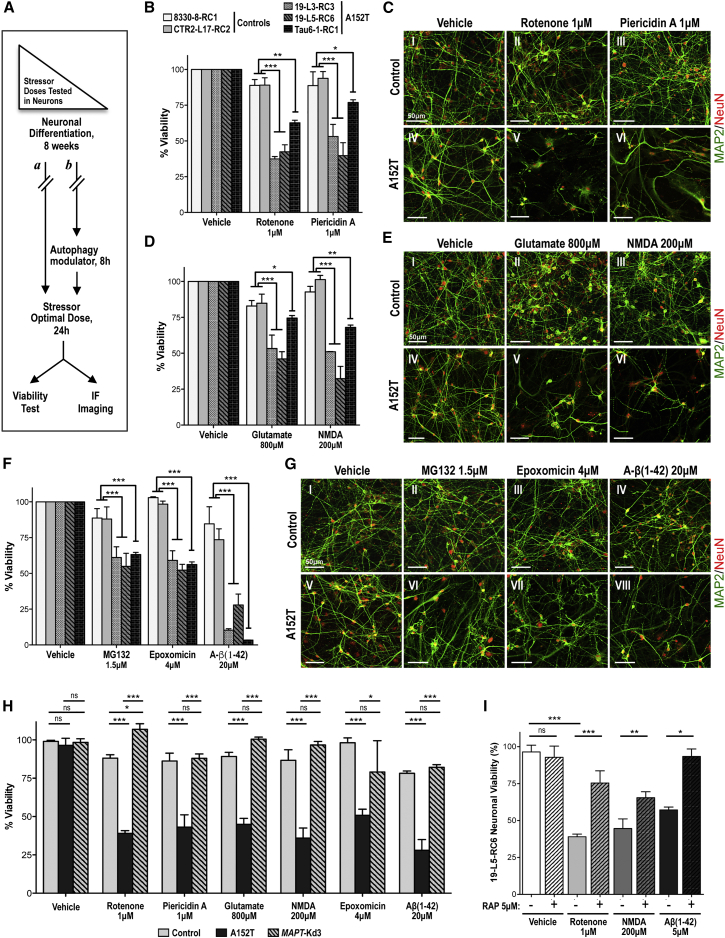
Based upon the premise that tauopathy results from time-dependent cumulative cell damage due to buildup of toxic proteins, we asked whether accumulation of tau in A152T neurons was deleterious. If so, by “adding insult to injury” neurons affected by tau accumulation should have lower capacity to survive additional stress (Frost et al., 2015, Gidalevitz et al., 2010). With the goal of also identifying pathways that may be compromised by tau pathology, we treated 8-week neurons with pathway-specific chemical stressors and compared cell viability between A152T and control neurons (Figure 7A). These “stressors” included environmental toxins that inhibit the mitochondrial electron transport chain (ETC) complex I (rotenone and piericidin A) (Hoglinger et al., 2005, Hollerhage et al., 2014); excitotoxic agonists of the glutamatergic AMPA and/or NMDA receptors (glutamate and NMDA) (Dong et al., 2009, Huey et al., 2006); and proteotoxic proteasome inhibitors (MG132 and epoxomicin) and A-β(1–42) amyloid peptide (a predominant component of amyloid plaques in Alzheimer's disease) (Roberson et al., 2007). To identify conditions whereby control and A152T neurons showed different survival, we started by performing a dose-dependent study in which compounds were added to neuronal cultures at different concentrations for 24 hr and cell viability was measured (Figures S6A–S6G). At the concentrations shown in Figure 7, stressor compounds had minimum impact in control neurons but caused a significant reduction in viability (<50% viable) of A152T neurons (Figures 7B, 7D, and 7F). These effects were also detected by IF of MAP2 and NeuN as markers of neuronal integrity, where A152T neurons incubated with mitochondrial ETC inhibitors (Figure 7C), excitotoxins (Figure 7E), or proteotoxins (Figure 7G) showed selective cell loss. These results revealed a phenotype of cellular toxicity suggesting that tau/P-tau accumulation in A152T neurons is associated with increased stress vulnerability, consistent with the observation of altered stress-inducible markers (Figure 6).
Figure 7.
Increased Vulnerability to Stress in A152T Neurons Is Tau Dependent
(A) Overview of the stress vulnerability tests done in 8-week neurons (a) using high doses of stress, (b) pre-treating neurons with the autophagy inducer rapamycin, or by testing genetically engineered MAPT-Kd neurons.
(B, D, and F) Cell viability of 8-week neurons, treated with compounds for 24 hr. Values represent percent viability (±SEM) relative to vehicle-treated neurons. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, two-way ANOVA and Bonferroni post test (n ≥ 3 independent experiments).
(C, E, and G) IF analysis of neuronal integrity of control (CTR2-L17-RC2) and A152T (19-L3-RC3) neurons upon 24-hr treatment, based on MAP2/NeuN staining. Scale bars 50 μm.
(H) Cell viability of 8-week neurons upon 24-hr treatment. The genetically engineered line MAPT-Kd3 (19-L5-RC6;MAPT-Kd3) showed rescue of vulnerability to stress relative to A152T neurons. Percent viability (±SEM) is relative to vehicle-treated cells (100%). Two-way ANOVA and Bonferroni post test: ∗p < 0.05, ∗∗∗p < 0.001; ns, not significant (p > 0.05) (n ≥ 3 independent experiments).
(I) Rescue of A152T neuronal (19-L5-RC6) vulnerability to stress [24 hr of rotenone, NMDA, and Aβ(1–42)] by 8-hr pre-treatment of neuronal cultures with rapamycin (RAP, 5 μM). Percent viability (±SEM) is relative to vehicle-treated cells (100%). Student’s t test: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p ≤ 0.001; ns, not significant (p > 0.05) (n ≥ 3 independent experiments).
See also Figure S6.
Vulnerability of A152T Neurons to Stress Is Tau Dependent
To test the direct correlation between upregulation of tau in A152T neurons and increased vulnerability to stress, we examined whether downregulation of the “toxic” tau would be sufficient to rescue viability. First, we genomically engineered the A152T cell line 19-L5-RC6 with CRISPR/Cas9 constructs for targeting and disrupting expression of the MAPT gene (Hsu et al., 2014, Shalem et al., 2014). The resulting cell lines (19-L5-RC6;MAPT-Kd) revealed a sharp downregulation (≥80%) of tau protein upon differentiation (Figures S6K and S6L), without detectable effect on neuronal integrity (Figures S6M and S2A) or cellular viability (Figure 7H). Remarkably, upon exposure to the cellular stressors that revealed vulnerability of the parental tau-A152T neurons, we observed a rescue of stress vulnerability in 8-week 19-L5-RC6;MAPT-Kd3 neurons, with viability increase to 90%–100%, similar to control (Figure 7H).
Second, we utilized the mTOR (mammalian target of rapamycin) inhibitor and autophagy activator rapamycin (Berger et al., 2006, Ozcelik et al., 2013). In control neurons, rapamycin downregulated tau by ≥50% via activation of the autophagy pathway, as shown by upregulation of the autophagosome marker LC3B-II (Figures S7N–S7P). We then pre-treated 8-week differentiated A152T neurons with rapamycin (8 hr) before exposure to stressor compounds (Figure 7A). Rapamycin-pre-treated A152T neurons showed a significant increase in cell viability relative to untreated neurons (Figure 7I). Consistent with the level of tau downregulation, rapamycin caused a milder but consistent rescue of viability compared with genetic knockdown (50% versus >80%, respectively).
Stress vulnerability tau dependence was further corroborated by testing the same stressors in 2-week neurons (Figures S6H–S6J), before tau accumulation, without a difference in survival between A152T and control neurons.
Altogether, these findings unequivocally demonstrate that accumulation of tau in A152T neurons is coupled to disruption of proteostasis and increased vulnerability to stress, early molecular events leading to neurodegeneration in tauopathy. Our study also demonstrates that endogenous tau-mediated cellular stress can be rescued by compounds that mediate tau clearance, which is of crucial relevance for the use of this cellular model for the development of therapeutics.
Discussion
Human iPSC-derived neurons allow measurement of disease-relevant cellular and molecular phenotypes in a physiologically and genomically relevant context, potentially recapitulating the early stages of disease etiology, and allow direct testing of therapeutic targets and small molecules in a human neuronal environment (Almeida et al., 2012, Bilican et al., 2012, Ehrlich et al., 2015, Fong et al., 2013, Haggarty et al., 2016, Iovino et al., 2015, Wren et al., 2015). We took advantage of this system to investigate the early events of tau-A152T pathology that may be causal in FTD. Our findings contribute to mounting evidence, across studies of different tau mutations and across model systems, of tau-mediated molecular events associated with neurodegeneration, toward the identification of relevant therapeutic targets (Ehrlich et al., 2015, Fong et al., 2013, Iovino et al., 2015, Maeda et al., 2016, Decker et al., 2016, Pir et al., 2016, Sydow et al., 2016, Wren et al., 2015). We biochemically profiled neurons derived from control and A152T carriers, and report on a phenotyping platform for tau regarding protein levels, PTMs, and solubility. We demonstrate accumulation of endogenous tau/P-tau in A152T human neurons by multiple powerful methods. By western blot we found a significant upregulation of P-tau in all A152T neurons, whereas total tau upregulation was specific to the FTD19-derived neurons, consistent with previous reports (Fong et al., 2013, Sydow et al., 2016). This result might be a consequence of variability among individuals of the same MAPT genotype, and possibly the clinical outcome, but only future analysis of larger cohorts of cases could allow us to draw accurate conclusions. By MS analysis we verified upregulation of absolute levels of endogenous tau in A152T neurons that became accentuated with time. By targeted MS, we demonstrated differential levels of tau A152T versus the non-mutant form in A152T neurons, which is relevant to the identification of “the toxic species.” Even though this ratio (∼60% versus 40%, respectively) was constant during the period of time tested (Figure 4C), the gradual increase in total tau (Figure 4B), suggests that this amount of tau-A152T is sufficient to cause some imbalance that leads to overall accumulation of tau.
By IF we detected an overall increase in tau in A152T neurons, with particular accumulation of P-tau in the cell body, consistent with somatodendritic redistribution of tau, as described for tauopathies and Alzheimer’s disease (de Calignon et al., 2012, Fong et al., 2013, Li et al., 2011, Sydow et al., 2016). We did not detect A152T-specific changes in cell morphology, as reported in another A152T model (Fong et al., 2013), which may be due to differences in methodologies and line-specific variability. Nonetheless, the overall conclusion of tau accumulation, somatodendritic redistribution, and tau-mediated neuronal toxicity in A152T cells was a common observation, unmasked either by additional stress (Figure 7) or by genetic manipulation (A152T homozygous line in Fong et al., 2013). Taken together, these two studies are complementary by demonstrating that even with technical variability, key aspects in pre-clinical disease events were corroborated.
We also reveal here, for an FTD A152T proband-derived neuronal culture, a decrease in tau solubility, again most accentuated for P-tau. Although we did not detect tau puncta-like aggregates per se, Triton X-insoluble tau and the HMW species detected are consistent with oligomeric species of reduced solubility. Based on our findings we propose a model in which accumulation of P-tau and oligomeric tau of reduced solubility are early disease-causal phenotypes, consistent with other A152T animal models demonstrating tau-mediated toxicity independent of aggregation (Maeda et al., 2016, Pir et al., 2016).
To explore the mechanisms of tau-mediated toxicity, we focused on specific proteostasis pathways shown previously to be involved in tauopathy, including the HSP70/HSP90 chaperone machinery (Blair et al., 2013), UPR markers (Stutzbach et al., 2013), mitochondrial function and oxidative stress (Schulz et al., 2012), and autophagy (Wang and Mandelkow, 2012). We report on disruption of the UPS and autophagy pathways, in addition to UPR and oxidative stress induction, in A152T neurons. Disruption of protein clearance is a strong model for tau-mediated toxicity in neurodegeneration, and is consistent with the observed accumulation of total tau protein. Also consistent with our findings, a recent study identified alterations of the endosomal-lysosomal pathway associated with another tau mutation (Wren et al., 2015); yet another study identified LAMP1 and ubiquitin as cerebrospinal fluid markers of neurodegeneration, reinforcing the value of our system in the identification of tauopathy biomarkers (Heywood et al., 2015). In turn, mounting evidence points to UPR activation “in close connection with early stages of tau pathology” (Abisambra et al., 2013, Stoveken, 2013), and to mitochondrial dysfunction and vulnerability to oxidative stress as common aspects in iPSC-derived models of tauopathy (Ehrlich et al., 2015, Iovino et al., 2015).
Tau-mediated disruption of proteostasis was further revealed by increased sensitivity of A152T neurons to exogenous “stressors,” unveiling toxicity phenotypes and pathways that may be affected in the early stages of tau pathology, offering a powerful method for phenotypic profiling of patient-derived neurons. Not all stressor compounds tested affected neuronal viability (not shown), revealing pathway-specific vulnerability. These phenotypes were tau dependent as shown by CRISPR/Cas9-mediated downregulation of MAPT expression in A152T neurons, which rescued neuronal stress vulnerability without affecting cell viability or integrity (Figures 7H and S6M). Most importantly, we demonstrate that stress vulnerability can be rescued pharmacologically by pre-treating neurons with the autophagy activator rapamycin, highlighting the value of this model for small-molecule screening. This is the first time, in a human neuronal cell context and with endogenous tau expression, that a compound has been shown to clear tau and consequently rescue toxicity. This result also further corroborates a model of tauopathy associated with disruption or inefficiency of protein clearance.
The results presented here are consistent with the proposition that the onset of cellular pathology in FTD occurs earlier than clinical manifestations, which has been corroborated by biomarker studies in Alzheimer’s disease (Jack et al., 2013). By implementing a panel of biochemical and cellular assays, and a toolbox of compounds that reveal pathways affected by tau toxicity, we demonstrated the power of phenotypic characterization of patient-derived neuronal cells at the pre-clinical level, which is now suitable for mechanistic studies across MAPT genotypes. Finally, we show that endogenous tau phenotypes can be pharmacologically rescued, proving the suitability of our patient iPSC-derived neuronal model for screening therapeutic targets and disease-modifying agents relevant to FTD and other tauopathies.
Experimental Procedures
iPSC, NPC, and Neuronal Cultures
Reprogramming, characterization, and maintenance of iPSCs, and NPC derivation have been described elsewhere (see Supplemental Experimental Procedures) (Zhao et al., 2012). NPCs were cultured on polyornithine and laminin (POL)-coated plates, with DMEM/F12-B27 media supplemented with EGF, FGF, and heparin, and passaged with TrypLE (Life Technologies). Approval for work with human subjects' materials and iPSCs was obtained under IRB-approved protocols at Massachusetts General Hospital/Partners Healthcare (2009P002730) and University of California, San Francisco (10-00234). Neural differentiation was achieved by plating NPCs (passages 30–35) at a density of ∼50,000 cells/cm2 on POL plates, with DMEM/F12-B27 media only (no growth factors), and with ½ medium change every 3–4 days.
Protein Analysis by Western Blot
Western blot analysis was performed using the Novex NuPAGE SDS-PAGE Gel System (Invitrogen) and standard immunoblotting techniques (see Supplemental Experimental Procedures).
LC-MS/MS Analysis
Absolute abundance of endogenous tau was determined using the FLEX peptide light-to heavy (L/H) ratio as recently described (Mair et al., 2016, Singh et al., 2009). The number of independent experiments was three; however, due to inherent challenges with long-term human neuronal differentiated cultures, there was a punctual necessity to pool samples for enough protein material at 20 weeks, for which n = 2 (Figures 4B–4D). See Supplemental Experimental Procedures for Expression and Purification of Heavy FLEXITau Standard, FLEXITau Sample Preparation and analysis, spectra analysis, data processing, and FLEXITau liquid chromatography (LC)-SRM measurements.
Analysis of Protein Solubility by Detergent Fractionation
The analysis was performed according to published protocols (see Supplemental Experimental Procedures).
MAPT Knockdown by CRISPR/Cas9 Genomic Engineering
The MAPT gene locus was targeted by the Tau CRISPR/Cas9 KO Plasmid (h) and Tau HDR Plasmid (h) (Santa Cruz Biotechnology sc-400136/sc-400136-HDR) in 19-L5-RC6 NPCs, according to the manufacturer's instructions (see Supplemental Experimental Procedures).
Compound Treatment and Viability Assays
Compound or vehicle alone was added directly onto the cellular medium. Viability was measured with the Alamar Blue Cell viability reagent (Life Technologies), according to the manufacturer's instructions. Readings were done in the EnVision Multilabel Plate Reader (PerkinElmer). For IF, fixed neurons image acquisition was done with the IN Cell Analyzer 6000 Cell Imaging System (GE Healthcare Life Sciences). For further details see Supplemental Experimental Procedures.
Author Contributions
Conceptualization, M.C.S., C.C., W.M., B.L.M., K.S.K., J.A.S., and S.J.H. Methodology, M.C.S., C.C., W.M., S.A., H.F., and S.J.H. Investigation, M.C.S., C.C., W.M., S.A., H.F., and S.J.H. Writing – Original Draft, M.C.S., W.M., S.T., D.H.G., J.A.S., and S.J.H. Writing – Review & Editing, M.C.S., C.C., S.A., Y.H., S.T., G.C., D.H.G., F.B.G., J.A.S., and S.J.H. Funding Acquisition, M.C.S., C.C., B.L.M., K.S.K., J.A.S., and S.J.H. Resources, S.A., H.F., H.U.B., Z.Z., Y.H., S.T., A.K., B.L.M., K.S.K., F.B.G., J.A.S., and S.J.H. Supervision, J.A.S. and S.J.H.
Acknowledgments
We thank the Tau Consortium for funding, continued support, and helpful discussions; members of the Haggarty laboratory for technical assistance and feedback on the manuscript content; Drs. Eva-Maria and Eckhard Mandelkow for discussions around the neurobiology and biochemistry of tau; Dr. Peter Davies for the generous contribution of antibodies; and the Stanley Medical Research Institute for brain tissue samples. Additional funding support came from AFTD, 1R21NS085487, Bluefield Project to Cure Frontotemporal Dementia, P50AG023501, and P01AG019724.
Published: September 1, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.08.001.
Supplemental Information
References
- Abisambra J.F., Jinwal U.K., Blair L.J., O'Leary J.C., III, Li Q., Brady S., Wang L., Guidi C.E., Zhang B., Nordhues B.A. Tau accumulation activates the unfolded protein response by impairing endoplasmic reticulum-associated degradation. J. Neurosci. 2013;33:9498–9507. doi: 10.1523/JNEUROSCI.5397-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida S., Zhang Z., Coppola G., Mao W., Futai K., Karydas A., Geschwind M.D., Tartaglia M.C., Gao F., Gianni D. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep. 2012;2:789–798. doi: 10.1016/j.celrep.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Berger Z., Ravikumar B., Menzies F.M., Oroz L.G., Underwood B.R., Pangalos M.N., Schmitt I., Wullner U., Evert B.O., O'Kane C.J. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum. Mol. Genet. 2006;15:433–442. doi: 10.1093/hmg/ddi458. [DOI] [PubMed] [Google Scholar]
- Bilican B., Serio A., Barmada S.J., Nishimura A.L., Sullivan G.J., Carrasco M., Phatnani H.P., Puddifoot C.A., Story D., Fletcher J. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc. Natl. Acad. Sci. USA. 2012;109:5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas M.H.U., Almeida S., Lopez-Gonzalez R., Mao W., Zhang Z., Karydas A., Geschwind M.D., Biernat J., Mandelkow E.-M., Futai K., Miller B.L., Gao F.-B. MMP-9 and MMP-2 contribute to neuronal cell death in iPSC models of frontotemporal dementia with MAPT mutations. Stem Cell Rep. 2016;7 doi: 10.1016/j.stemcr.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair L.J., Zhang B., Dickey C.A. Potential synergy between tau aggregation inhibitors and tau chaperone modulators. Alzheimers Res. Ther. 2013;5:41. doi: 10.1186/alzrt207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola G., Chinnathambi S., Lee J.J., Dombroski B.A., Baker M.C., Soto-Ortolaza A.I., Lee S.E., Klein E., Huang A.Y., Sears R. Evidence for a role of the rare p.A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer's diseases. Hum. Mol. Genet. 2012;21:3500–3512. doi: 10.1093/hmg/dds161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalle-Donne I., Rossi R., Giustarini D., Milzani A., Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta. 2003;329:23–38. doi: 10.1016/s0009-8981(03)00003-2. [DOI] [PubMed] [Google Scholar]
- Davies P. Characterization and use of monoclonal antibodies to tau and paired helical filament tau. Methods Mol. Med. 2000;32:361–373. doi: 10.1385/1-59259-195-7:361. [DOI] [PubMed] [Google Scholar]
- de Calignon A., Polydoro M., Suarez-Calvet M., William C., Adamowicz D.H., Kopeikina K.J., Pitstick R., Sahara N., Ashe K.H., Carlson G.A. Propagation of tau pathology in a model of early Alzheimer's disease. Neuron. 2012;73:685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker J.M., Krüger L., Sydow A., Dennissen F.J., Siskova Z., Mandelkow E., Mandelkow E.M. The Tau/A152T mutation, a risk factor for frontotemporal-spectrum disorders, leads to NR2B receptor-mediated excitotoxicity. EMBO Rep. 2016;17:552–569. doi: 10.15252/embr.201541439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derisbourg M., Leghay C., Chiappetta G., Fernandez-Gomez F.J., Laurent C., Demeyer D., Carrier S., Buee-Scherrer V., Blum D., Vinh J. Role of the Tau N-terminal region in microtubule stabilization revealed by new endogenous truncated forms. Sci. Rep. 2015;5:9659. doi: 10.1038/srep09659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X.X., Wang Y., Qin Z.H. Molecular mechanisms of excitotoxicity and their relevance to pathogenesis of neurodegenerative diseases. Acta Pharmacol. Sin. 2009;30:379–387. doi: 10.1038/aps.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Hallmann A.L., Reinhardt P., Arauzo-Bravo M.J., Korr S., Ropke A., Psathaki O.E., Ehling P., Meuth S.G., Oblak A.L. Distinct neurodegenerative changes in an induced pluripotent stem cell model of frontotemporal dementia linked to mutant TAU protein. Stem Cell Rep. 2015;5:83–96. doi: 10.1016/j.stemcr.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong H., Wang C., Knoferle J., Walker D., Balestra M.E., Tong L.M., Leung L., Ring K.L., Seeley W.W., Karydas A. Genetic correction of tauopathy phenotypes in neurons derived from human induced pluripotent stem cells. Stem Cell Rep. 2013;1:226–234. doi: 10.1016/j.stemcr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B., Gotz J., Feany M.B. Connecting the dots between tau dysfunction and neurodegeneration. Trends Cell Biol. 2015;25:46–53. doi: 10.1016/j.tcb.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson J.E., Castillo-Carranza D.L., Kayed R. Advances in therapeutics for neurodegenerative tauopathies: moving toward the specific targeting of the most toxic tau species. ACS Chem. Neurosci. 2014;5:752–769. doi: 10.1021/cn500143n. [DOI] [PubMed] [Google Scholar]
- Gidalevitz T., Kikis E.A., Morimoto R.I. A cellular perspective on conformational disease: the role of genetic background and proteostasis networks. Curr. Opin. Struct. Biol. 2010;20:23–32. doi: 10.1016/j.sbi.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M., Jakes R., Vanmechelen E. Monoclonal antibody AT8 recognises tau protein phosphorylated at both serine 202 and threonine 205. Neurosci. Lett. 1995;189:167–169. doi: 10.1016/0304-3940(95)11484-e. [DOI] [PubMed] [Google Scholar]
- Guo J.L., Lee V.M. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J. Biol. Chem. 2011;286:15317–15331. doi: 10.1074/jbc.M110.209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.L., Lee V.M. Neurofibrillary tangle-like tau pathology induced by synthetic tau fibrils in primary neurons over-expressing mutant tau. FEBS Lett. 2013;587:717–723. doi: 10.1016/j.febslet.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggarty S.J., Silva M.C., Cross A., Brandon N.J., Perlis R.H. Advancing drug discovery for neuropsychiatric disorders using patient-specific stem cell models. Mol. Cell Neurosci. 2016;73:104–115. doi: 10.1016/j.mcn.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heywood W.E., Galimberti D., Bliss E., Sirka E., Paterson R.W., Magdalinou N.K., Carecchio M., Reid E., Heslegrave A., Fenoglio C. Identification of novel CSF biomarkers for neurodegeneration and their validation by a high-throughput multiplexed targeted proteomic assay. Mol. Neurodegener. 2015;10:64. doi: 10.1186/s13024-015-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoglinger G.U., Lannuzel A., Khondiker M.E., Michel P.P., Duyckaerts C., Feger J., Champy P., Prigent A., Medja F., Lombes A. The mitochondrial complex I inhibitor rotenone triggers a cerebral tauopathy. J. Neurochem. 2005;95:930–939. doi: 10.1111/j.1471-4159.2005.03493.x. [DOI] [PubMed] [Google Scholar]
- Hollerhage M., Deck R., De Andrade A., Respondek G., Xu H., Rosler T.W., Salama M., Carlsson T., Yamada E.S., Gad El Hak S.A. Piericidin A aggravates Tau pathology in P301S transgenic mice. PLoS One. 2014;9:e113557. doi: 10.1371/journal.pone.0113557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey E.D., Putnam K.T., Grafman J. A systematic review of neurotransmitter deficits and treatments in frontotemporal dementia. Neurology. 2006;66:17–22. doi: 10.1212/01.wnl.0000191304.55196.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovino M., Agathou S., Gonzalez-Rueda A., Del Castillo Velasco-Herrera M., Borroni B., Alberici A., Lynch T., O'Dowd S., Geti I., Gaffney D. Early maturation and distinct tau pathology in induced pluripotent stem cell-derived neurons from patients with MAPT mutations. Brain. 2015;138:3345–3359. doi: 10.1093/brain/awv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol. Med. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- Jack C.R., Jr., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S., Shaw L.M., Vemuri P., Wiste H.J., Weigand S.D. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson G.V., Stoothoff W.H. Tau phosphorylation in neuronal cell function and dysfunction. J. Cell Sci. 2004;117:5721–5729. doi: 10.1242/jcs.01558. [DOI] [PubMed] [Google Scholar]
- Kara E., Ling H., Pittman A.M., Shaw K., de Silva R., Simone R., Holton J.L., Warren J.D., Rohrer J.D., Xiromerisiou G. The MAPT p.A152T variant is a risk factor associated with tauopathies with atypical clinical and neuropathological features. Neurobiol. Aging. 2012;33:2231.e7–2231.e14. doi: 10.1016/j.neurobiolaging.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgiou E., Miller B.L. Frontotemporal lobar degeneration: a clinical approach. Semin. Neurol. 2014;34:189–201. doi: 10.1055/s-0034-1381735. [DOI] [PubMed] [Google Scholar]
- Kfoury N., Holmes B.B., Jiang H., Holtzman D.M., Diamond M.I. Trans-cellular propagation of Tau aggregation by fibrillar species. J. Biol. Chem. 2012;287:19440–19451. doi: 10.1074/jbc.M112.346072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik K.S., Orecchio L.D., Bakalis S., Neve R.L. Developmentally regulated expression of specific tau sequences. Neuron. 1989;2:1389–1397. doi: 10.1016/0896-6273(89)90077-9. [DOI] [PubMed] [Google Scholar]
- Labbe C., Ogaki K., Lorenzo-Betancor O., Soto-Ortolaza A.I., Walton R.L., Rayaprolu S., Fujioka S., Murray M.E., Heckman M.G., Puschmann A. Role for the microtubule-associated protein tau variant p.A152T in risk of alpha-synucleinopathies. Neurology. 2015;85:1680–1686. doi: 10.1212/WNL.0000000000001946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Tartaglia M.C., Yener G., Genc S., Seeley W.W., Sanchez-Juan P., Moreno F., Mendez M.F., Klein E., Rademakers R. Neurodegenerative disease phenotypes in carriers of MAPT p.A152T, a risk factor for frontotemporal dementia spectrum disorders and Alzheimer disease. Alzheimer Dis. Assoc. Disord. 2013;27:302–309. doi: 10.1097/WAD.0b013e31828cc357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Kumar Y., Zempel H., Mandelkow E.M., Biernat J., Mandelkow E. Novel diffusion barrier for axonal retention of Tau in neurons and its failure in neurodegeneration. EMBO J. 2011;30:4825–4837. doi: 10.1038/emboj.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S., Djukic B., Taneja P., Yu G.Q., Lo I., Davis A., Craft R., Guo W., Wang X., Kim D. Expression of A152T human tau causes age-dependent neuronal dysfunction and loss in transgenic mice. EMBO Rep. 2016;17:530–551. doi: 10.15252/embr.201541438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair W., Muntel J., Tepper K., Tang S., Biernat J., Seeley W.W., Kosik K.S., Mandelkow E., Steen H., Steen J.A. FLEXITau: quantifying post-translational modifications of tau protein in vitro and in human disease. Anal. Chem. 2016;88:3704–3714. doi: 10.1021/acs.analchem.5b04509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S.W., Cho S.H., Zhou Y., Schroeder S., Haroutunian V., Seeley W.W., Huang E.J., Shen Y., Masliah E., Mukherjee C. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris M., Maeda S., Vossel K., Mucke L. The many faces of tau. Neuron. 2011;70:410–426. doi: 10.1016/j.neuron.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemati S., Hatami M., Kiani S., Hemmesi K., Gourabi H., Masoudi N., Alaei S., Baharvand H. Long-term self-renewable feeder-free human induced pluripotent stem cell-derived neural progenitors. Stem Cells Dev. 2011;20:503–514. doi: 10.1089/scd.2010.0143. [DOI] [PubMed] [Google Scholar]
- Neumann M., Kovacs G.G., Mackenzie I.R.A. Neuropathology of frontotemporal dementia and related disorders. In: Dickerson B.C., editor. Hodges' Frontotemporal Dementia. Cambridge University Press; 2015. pp. 165–184. [Google Scholar]
- Ozcelik S., Fraser G., Castets P., Schaeffer V., Skachokova Z., Breu K., Clavaguera F., Sinnreich M., Kappos L., Goedert M. Rapamycin attenuates the progression of tau pathology in P301S tau transgenic mice. PLoS One. 2013;8:e62459. doi: 10.1371/journal.pone.0062459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pir G.J., Choudhary B., Mandelkow E., Mandelkow E.M. Tau mutant A152T, a risk factor for FTD/PSP, induces neuronal dysfunction and reduced lifespan independently of aggregation in a C. elegans tauopathy model. Mol. Neurodegener. 2016;11:33. doi: 10.1186/s13024-016-0096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson E.D., Scearce-Levie K., Palop J.J., Yan F., Cheng I.H., Wu T., Gerstein H., Yu G.Q., Mucke L. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- Schulz K.L., Eckert A., Rhein V., Mai S., Haase W., Reichert A.S., Jendrach M., Muller W.E., Leuner K. A new link to mitochondrial impairment in tauopathies. Mol. Neurobiol. 2012;46:205–216. doi: 10.1007/s12035-012-8308-3. [DOI] [PubMed] [Google Scholar]
- Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan S.D., Theriault K.M., Reis S.A., Zhou F., Madison J.M., Daheron L., Loring J.F., Haggarty S.J. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS One. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Springer M., Steen J., Kirschner M.W., Steen H. FLEXIQuant: a novel tool for the absolute quantification of proteins, and the simultaneous identification and quantification of potentially modified peptides. J. Proteome Res. 2009;8:2201–2210. doi: 10.1021/pr800654s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J.L., de la Torre-Ubieta L., Tian Y., Parikshak N.N., Hernandez I.A., Marchetto M.C., Baker D.K., Lu D., Hinman C.R., Lowe J.K. A quantitative framework to evaluate modeling of cortical development by neural stem cells. Neuron. 2014;83:69–86. doi: 10.1016/j.neuron.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoveken B.J. Tau pathology as a cause and consequence of the UPR. J. Neurosci. 2013;33:14285–14287. doi: 10.1523/JNEUROSCI.2961-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutzbach L.D., Xie S.X., Naj A.C., Albin R., Gilman S., PSP Genetics Study Group, Lee V.M., Trojanowski J.Q., Devlin B., Schellenberg G.D. The unfolded protein response is activated in disease-affected brain regions in progressive supranuclear palsy and Alzheimer's disease. Acta Neuropathol. Commun. 2013;1:31. doi: 10.1186/2051-5960-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sydow A., Hochgrafe K., Konen S., Cadinu D., Matenia D., Petrova O., Joseph M., Dennissen F.J., Mandelkow E.M. Age-dependent neuroinflammation and cognitive decline in a novel Ala152Thr-Tau transgenic mouse model of PSP and AD. Acta Neuropathol. Commun. 2016;4:17. doi: 10.1186/s40478-016-0281-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungrin M.D., Joshi C., Nica A., Bauwens C., Zandstra P.W. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS One. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Mandelkow E. Degradation of tau protein by autophagy and proteasomal pathways. Biochem. Soc. Trans. 2012;40:644–652. doi: 10.1042/BST20120071. [DOI] [PubMed] [Google Scholar]
- Wang Y., Martinez-Vicente M., Kruger U., Kaushik S., Wong E., Mandelkow E.M., Cuervo A.M., Mandelkow E. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum. Mol. Genet. 2009;18:4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E., Cuervo A.M. Integration of clearance mechanisms: the proteasome and autophagy. Cold Spring Harb. Perspect. Biol. 2010;2:a006734. doi: 10.1101/cshperspect.a006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren M.C., Zhao J., Liu C.C., Murray M.E., Atagi Y., Davis M.D., Fu Y., Okano H.J., Ogaki K., Strongosky A.J. Frontotemporal dementia-associated N279K tau mutant disrupts subcellular vesicle trafficking and induces cellular stress in iPSC-derived neural stem cells. Mol. Neurodegener. 2015;10:46. doi: 10.1186/s13024-015-0042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S.H., Martin J., Elia J., Flippin J., Paramban R.I., Hefferan M.P., Vidal J.G., Mu Y., Killian R.L., Israel M.A. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6:e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W.N., Cheng C., Theriault K.M., Sheridan S.D., Tsai L.H., Haggarty S.J. A high-throughput screen for Wnt/beta-catenin signaling pathway modulators in human iPSC-derived neural progenitors. J. Biomol. Screen. 2012;17:1252–1263. doi: 10.1177/1087057112456876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.