Abstract
Uveal melanomas are molecularly distinct from cutaneous melanomas and lack mutations in BRAF, NRAS, KIT, and NF1. Instead, they are characterized by activating mutations in GNAQ and GNA11, two highly homologous α subunits of Gαq/11 heterotrimeric G proteins, and in PLCB4 (phospholipase C β4), the downstream effector of Gαq signaling 1–3. We analyzed genomics data from 136 uveal melanoma samples and found a recurrent mutation in CYSLTR2 (cysteinyl leukotriene receptor 2) encoding a p.Leu129Gln substitution in 4 of 9 samples that lacked mutations in GNAQ, GNA11, and PLCB4 but in 0 of 127 samples that harbored mutations in these genes. The Leu129Gln CysLT2R mutant protein constitutively activates endogenous Gαq and is unresponsive to stimulation by leukotriene. Expression of Leu129Gln CysLT2R in melanocytes enforces expression of a melanocyte-lineage signature, drives phorbol ester–independent growth in vitro, and promotes tumorigenesis in vivo. Our findings implicate CYSLTR2 as a uveal melanoma oncogene and highlight the critical role of Gαq signaling in uveal melanoma pathogenesis.
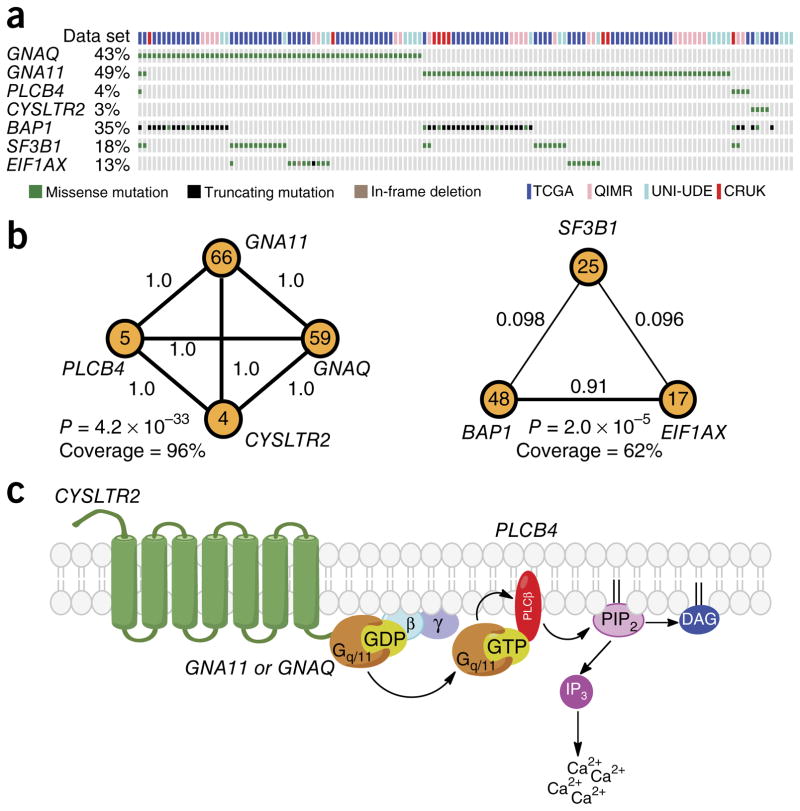
Uveal melanomas arise from melanocytes of the uveal tract and are the most prevalent tumors of the eye. To identify additional oncogenic drivers in uveal melanoma, we curated whole-genome or whole-exome sequencing data for 136 uveal melanomas from The Cancer Genome Atlas (TCGA, personal communication), Cancer Research UK (CRUK) 4, QIMR Berghofer Medical Research Institute (QIMR) 3, and University of Duisburg-Essen (UNI-UDE) 5 cohorts. We performed mutational analysis using an algorithm that is highly sensitive in detecting rare hotspot mutations found in oncogenes 6. We identified seven significantly mutated (q < 0.05) codons in six genes, including known mutations in GNAQ, GNA11, PLCB4, SF3B1, and EIF1AX. In addition, we found a new c.386T>A mutation in CYSLTR2 encoding a p.Leu129Gln substitution in four samples (q = 3.3 × 10−8) (Fig. 1a, Table 1, and Supplementary Fig. 1a). RNA-seq data validated the presence of the CYSLTR2 mutation encoding p.Leu129Gln and confirmed expression of the mutant allele (Supplementary Figs. 1a and 2c). For further validation, we performed Sanger sequencing on two Memorial Sloan Kettering Cancer Center (MSKCC) patient samples that lacked GNAQ and GNA11 mutations (2/28) and found one sample with a CYSLTR2 mutation encoding p.Leu129Gln (Supplementary Fig. 1b). Although CYSLTR2 is sporadically mutated in other cancer types sequenced by TCGA, these mutations do not occur at hotspots (except for mutations mapping to Arg136 (p.Arg136Cys or p.Arg136His) in three samples), the corresponding transcripts are not expressed at high levels, and the mutations are found in high-mutation-burden tumors, suggesting that CYSLTR2 might not be an oncogenic driver in other tumor types and that the CYSLTR2 hotspot mutation encoding p.Leu129Gln is unique to uveal melanoma (Supplementary Fig. 2).
Figure 1.
CYSLTR2 mutation encoding p.Leu129Gln is a hotspot mutation and is mutually exclusive with known drivers in uveal melanoma. (a) OncoPrint of a selection of frequently mutated genes in uveal melanomas from four data sets: TCGA (n = 80), UNI-UDE (n = 22), CRUK (n = 9), and QIMR (n = 25). (b) CoMEt plots of two distinct, mutually exclusive modules in uveal melanoma. Each node is a mutated gene, and the number inside each node is the number of samples with a mutation in that gene. The number on each edge between nodes is edge weight (δ), which represents the fraction of permutations where mutations in the two genes are significantly mutually exclusive. The P value, also known as Φ, represents the mid P value that the genes in the module are mutated in a mutually exclusive manner. Coverage is the percentage of samples with at least one mutation in the genes in the module. (c) A schematic of the pathway activated in uveal melanoma. GPCR (CYSLTR2) activation of Gαq/11 (GNAQ or GNA11) promotes the exchange of GDP for GTP and binding of Gαq/11 to phospholipase C β (PLCβ; PLCB4) to activate cleavage of phosphatidylinositol 4,5-bisphosphate (PIP2) to produce diacylglycerol (DAG) and inositol triphosphate (IP3), resulting in subsequent calcium release.
Table 1.
Recurrent hotspot mutations in uveal melanoma
| Gene | Codon | Alterationa | Mutations at codon | Mutations in gene | P value | q value |
|---|---|---|---|---|---|---|
| GNA11 | Gln209 | p.Gln209Leu (64) | 64 | 66 | 1.4 × 10−275 | 3.3 × 10−272 |
| GNAQ | Gln209 | p.Gln209Pro (40), p.Gln209Leu (15) | 55 | 59 | 1.3 × 10−211 | 3.0 × 10−208 |
| SF3B1 | Arg625 | p.Arg625His (10), p.Arg625Cys (8) | 18 | 25 | 9.5 × 10−37 | 9.6 × 10−33 |
| CYSLTR2 | Leu129 | p.Leu129Gln (4) | 4 | 4 | 3.4 × 10−11 | 7.5 × 10−8 |
| PLCB4 | Asp630 | p.Asp630Tyr (3), p.Asp630Phe (1), p.Asp630Asn (1) | 5 | 5 | 3.0 × 10−10 | 2.7 × 10−6 |
| SF3B1 | Lys666 | p.Lys666Thr (4) | 3 | 35 | 3.0 × 10−7 | 1.6 × 10−3 |
| EIF1AX | Gly6 | p.Gly6Asp (2), p.Gly6Val (1) | 3 | 16 | 3.0 × 10−6 | 2.4 × 10−3 |
The number of tumors in which an alteration was present is given in parentheses.
Because mutations that activate the same pathway or mutations that define molecular subtypes tend to be mutually exclusive, we used CoMEt to identify mutually exclusive mutational modules de novo in the data set with 136 uveal melanomas 7. We found that mutations in GNAQ, GNA11, PLCB4, and CYSLTR2 formed a highly significantly mutually exclusive module (P = 4.2 × 10−33), suggesting that these genes are in the same pathway. BAP1 (ref. 8), SF3B1, and EIF1AX formed a second mutually exclusive module (P = 2.0 × 10−5), consistent with the observation that BAP1 mutations primarily occur in tumors with monosomy 3 and SF3B1 and EIF1AX mutations primarily occur in tumors with disomy 3 (Fig. 1b) 5, 9.
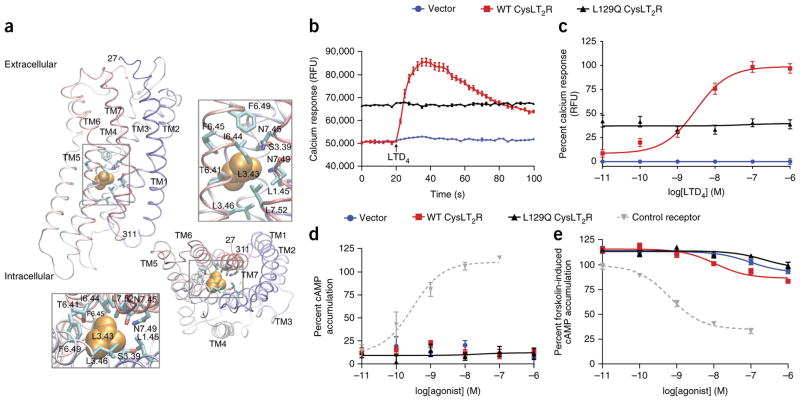
CYSLTR2 encodes cysteinyl leukotriene receptor 2 (CysLT2R), a seven-transmembrane G-protein-coupled receptor (GPCR), which functions in leukotriene-mediated signaling, including during inflammation and fibrosis. CysLT2R belongs to the rhodopsin-like family of GPCRs and is known to activate Gαq (Fig. 1c) 10, 11. Leu129 of CysLT2R is located in transmembrane helix 3 (TM3), a functional hub of the receptor that makes contact with the ligand extracellularly, associates with other transmembrane helices within the membrane, and interacts with the Gα subunit intracellularly (Fig. 2a). Leu129 resides in the highly conserved 3.43 position (Ballesteros–Weinstein numbering 12) that stabilizes the inactive configuration of GPCRs through intramolecular contacts. Alteration at the 3.43 position has been shown to confer constitutive activity in a number of GPCRs, including the β-adrenergic, luteinizing hormone, and thyroid-stimulating hormone receptors 13. To further understand the role of Leu129, we modeled the structure of CysLT2R on the basis of the known structure of its close homolog, protease-activated receptor 1 (PAR1) 14. Leu129 (3.43) resides in a hydrophobic core of the GPCR, which is highly enriched in hydrophobic residues (Leu80 (1.45), Ser125 (3.39), Leu132 (3.46), Thr252 (6.41), Ile255 (6.44), Phe256 (6.45), Phe260 (6.49), Asn297 (7.45), Asn301 (7.49), and Leu304 (7.52)) (Fig. 2a), suggesting that substitution with a more hydrophilic glutamine residue (p.Leu129Gln) might disrupt structural organization.
Figure 2.
Leu129Gln CysLT2R exhibits high basal coupling to Gαq. (a) Structural homology model of CysLT2R based on the structure for PAR1. Transmembrane (TM) segments are labeled. Leu129 (3.43) is shown in orange space fill. Boxed segments are expanded to show residues that interact with position 3.43. The bottom panels show the model rotated by 90° relative to the top panels; the intracellular surface faces outward. (b) Representative 100-s time course of calcium flux with 10 nM LTD4 added at 20 s (arrow) in HEK293T cells transfected with empty vector or expressing wild-type (WT) or Leu129Gln CysLT2R. Data are presented as relative fluorescence units (RFU). Error bars, s.e.m. from three biological replicates, each with three technical replicates. (c) LTD4 concentration-dependent calcium flux in HEK293T cells transfected with empty vector or expressing wild-type or Leu129Gln CysLT2R. The EC50 (half-maximal effective concentration) for wild-type CysLT2R is 3.07 nM. Expression of Leu129Gln CysLT2R causes sustained elevated calcium flux that is unaffected by LTD4. Data are presented as the percentage of relative fluorescence units at LTD4 concentrations and correspond to the means ± s.e.m. of seven independent experiments, each carried out in triplicate. (d) Dose-dependent Gαs activation by LTD4 in HEK293T cells cotransfected with vector encoding RLuc3-EPAC-GFP10 together with empty vector or vector encoding wild-type or Leu129Gln CysLT2R. An overlay shows activity for a control receptor (calcitonin receptor–like receptor (CLR); cotransfected with the RAMP2 accessory protein) in cells treated with adrenomedullin to cause dose-dependent production of cAMP. Data are expressed as the percentage of cAMP accumulation relative to that observed with forskolin induction and correspond to the means ± s.e.m. from two independent experiments, each carried out in triplicate. (e) Analysis of dose-dependent Gαi activation performed as in d with forskolin pretreatment. An overlay shows activity for a control receptor (C-C chemokine receptor 5 (CCR5)) in cells treated with RANTES to cause dose-dependent inhibition of forskolin-induced cAMP production. Data are expressed as the percentage of cAMP accumulation relative to that observed with forskolin induction and correspond to the means ± s.e.m. from four independent experiments, each carried out in triplicate.
To determine the ability of the wild-type and Leu129Gln CysLT2R proteins to couple with Gαq, we generated synthetic genes encoding wild-type CYSLTR2 and the mutant, transiently expressed them in HEK293 T cells, and measured calcium mobilization (Fig. 2b). Leukotriene D4 (LTD4), a potent CysLT2R agonist, mobilized calcium in cells expressing wild-type CysLT2R but not in those transfected with empty vector, in a concentration-dependent manner. Cells expressing Leu129Gln CysLT2R exhibited high basal calcium levels that were not augmented by increasing concentrations of LTD4, indicating that the receptor is constitutively active (Fig. 2b,c). Wild-type and Leu129Gln CysLT2R were expressed at similar levels, as determined by immunoblot analysis for the engineered N-terminal FLAG and C-terminal 1D4 tags, and the proteins demonstrated similar cellular localization by immunofluorescence and similar levels of cell surface expression by FACS analysis (for extracellular FLAG epitope), suggesting that the different functional behavior of the mutant is not due to different expression level or localization in comparison with the wild-type receptor (Supplementary Fig. 3a–c). To determine whether wild-type and Leu129Gln CysLT2R are coupled to the Gαs activating or Gαi inhibitory adenylyl cyclase pathway, we measured the ligand-mediated increase in cAMP levels, indicative of Gαs coupling, and the ligand-mediated decrease in forskolin-induced cAMP levels, indicative of Gαi coupling. The basal level of cAMP was not altered by expression of either wild-type or Leu129Gln CysLT2R; further, addition of LTD4 did not increase cAMP levels in cells expressing wild-type or Leu129Gln CysLT2R (Fig. 2d), arguing against coupling of Leu129Gln CysLT2R with Gαs. After stimulation with forskolin, LTD4 minimally inhibited the increase in cAMP levels in cells expressing wild-type CysLT2R in comparison to those transfected with vector control, but cAMP levels were unaffected by LTD4 in cells expressing Leu129Gln CysLT2R (Fig. 2e), indicating that Leu129Gln CysLT2R was not coupled with Gαi. These data, in combination with the calcium flux data, suggest that Leu129Gln CysLT2R is a gain-of-function mutant that primarily couples through Gαq.
To determine whether Leu129Gln CysLT2R possesses oncogenic properties, we stably expressed empty-vector control, wild-type CysLT2R, and Leu129Gln CysLT2R, as well as the Arg136His CysLT2R mutant receptor found in three independent TCGA samples (Supplementary Fig. 2b), in immortalized mouse NIH3T3 fibroblasts and subcutaneously grafted these cells into severe combined immunodeficient (SCID) mice. We found that only cells expressing Leu129Gln CysLT2R were able to form tumors in vivo (Supplementary Fig. 4a), indicating that Leu129Gln CysLT2R has oncogenic activities.
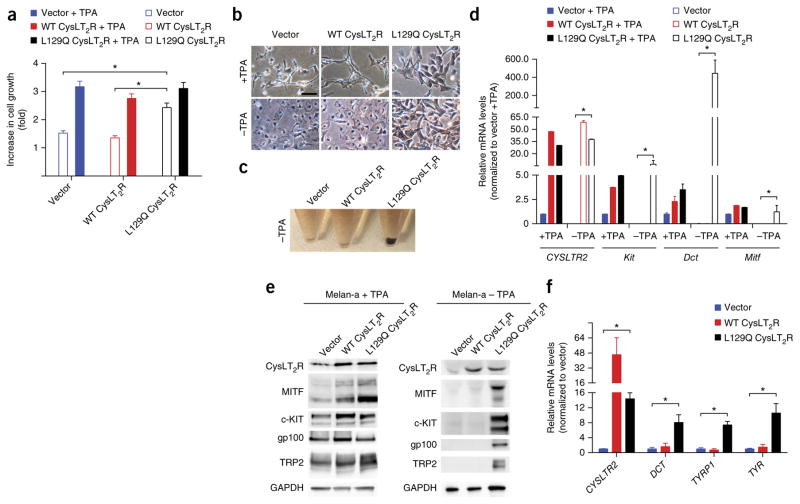
To determine the melanocyte-lineage-specific effects of the CysLT2R p.Leu129Gln substitution, we stably expressed the empty-vector control, wild-type CysLT2R, and Leu129Gln CysLT2R in melan-a cells, an immortalized mouse melanocytic cell line. Melan-a cells require phorbol esters such as 12-O-tetradecanoylphorbol-13-acetate (TPA) for growth 15. Leu129Gln but not wild-type CysLT2R conferred TPA-independent growth (Fig. 3a).
Figure 3.
Leu129Gln CysLT2R promotes TPA-independent growth in vitro and enforces a melanocyte-lineage-specific signature. (a) Cell growth of melan-a cells expressing empty-vector control, wild-type CysLT2R, or Leu129Gln CysLT2R and assayed by CellTiter-Glo in the presence or absence of 200 nM TPA for 3 d. The fold increase in growth relative to cell numbers at 0 d is shown and corresponds to means ± s.e.m. from six technical replicates. *P < 0.005. (b) Representative phase-contrast microscopy images of melan-a cells expressing empty-vector control, wild-type CysLT2R, or Leu129Gln CysLT2R and grown in the presence or absence of 200 nM TPA for 3 d. Scale bar, 50 μm. (c) Cellular pellets of melan-a cells expressing empty-vector control, wild-type CysLT2R, or Leu129Gln CysLT2R and grown in the absence of TPA for 18 d. (d) Relative mRNA levels of melanocyte-lineage-specific genes (Kit, Dct, and Mitf) in melan-a cells for each group in the presence or absence of 200 nM TPA for 18 d as assessed by RT–qPCR. Error bars, s.d. from three technical replicates. *P < 0.05. (e) Cropped immunoblots of CysLT2R and melanocyte-lineage markers in melan-a cells expressing empty- vector control, wild-type CysLT2R, or Leu129Gln CysLT2R and grown in the presence (left) or absence (right) of TPA for 18 d. Full-length immunoblots are presented in Supplementary Figure 5. (f) Relative mRNA expression of melanocyte-lineage-specific genes (DCT, TYRP1, and TYR) in MEL290 cells (a human melanoma cell line) expressing empty-vector control, wild-type CysLT2R, or Leu129Gln CysLT2R and grown for 14 d as assessed by RT–qPCR. Error bars, s.d. from three technical replicates. *P < 0.05.
We next sought to phenotypically characterize expression of Leu129Gln CysLT2R in the melanocyte lineage. In addition to being required for growth, supplemental TPA promotes melanocyte-lineage specification and pigmentation in both human and mouse melanocytes 16, 17. Notably, Leu129Gln CysLT2R expression dramatically augmented pigmentation in melan-a cells cultured in TPA-containing medium, with increased numbers of large pigmented melanosomes (Fig. 3b). When TPA was withdrawn, melan-a cells lost cytoplasmic branches and became rounded within 72 h and pigmentation was completely lost over 18 d (Fig. 3b,c and Supplementary Fig. 4b). Remarkably, Leu129Gln CysLT2R expression rescued both pigmentation and cell morphology (Fig. 3b,c). We next assayed for expression of melanocyte-lineage-specific genes (for example, Mitf, Kit, and Dct) and found that their expression was decreased upon TPA withdrawal. Leu129Gln CysLT2R expression modestly increased expression of these genes in the presence of TPA but dramatically increased their expression in the absence of TPA (Fig. 3d,e and Supplementary Fig. 5). To determine whether Leu129Gln CysLT2R also promotes melanocyte-lineage specification in the human context, we stably expressed the empty-vector control, wild-type CysLT2R, and Leu129Gln CysLT2R in MEL290 cells, a human uveal melanoma cell line that is wild type for GNA11 and GNAQ 18. Expression of Leu129Gln CysLT2R but not controls significantly increased expression of melanocyte-lineage-specific genes (for example, DCT, TYRP1, and TYR) (Fig. 3f).
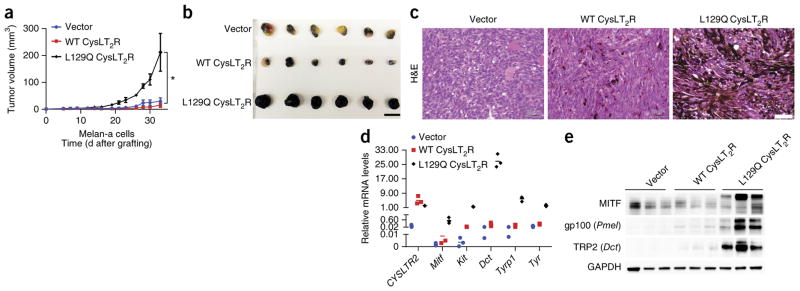
To determine the role of the CysLT2R p.Leu129Gln substitution in melanoma tumorigenesis in vivo, we grafted melan-a cells expressing empty-vector control, wild-type CysLT2R, and Leu129Gln CysLT2R into SCID mice and found that Leu129Gln CysLT2R significantly accelerated tumor formation in comparison to empty vector (P < 0.001; Fig. 4a). Examination of melan-a tumor grafts showed that tumors expressing Leu129Gln CysLT2R were highly pigmented and expressed higher levels of melanocyte-lineage-specific markers (for example, Mitf, Kit, Dct, Tyrp1, and Tyr) than grafts expressing empty-vector control and wild-type CysLT2R (Fig. 4b–e and Supplementary Fig. 5). These data indicate that, within the melanocyte lineage, Leu129Gln CysLT2R enforces a master melanocyte-lineage-specific transcriptional program and promotes transformation.
Figure 4.
Leu129Gln CysLT2R promotes tumorigenesis in vivo and enforces a melanocyte-lineage-specific signature. (a) Tumor volume over time of SCID mice subcutaneously injected with melan-a cells expressing empty-control vector, wild-type CysLT2R, or Leu129Gln CysLT2R (n = 8 mice per group). Error bars, s.e.m.*P < 0.001. (b) Photograph of six representative melan-a xenograft tumors per group explanted at 32 d after implantation. Scale bar, 1.51 cm. (c) Representative hematoxylin and eosin (H&E) images of melan-a xenograft tumors expressing empty-vector control, wild-type CysLT2R, or Leu129Gln CysLT2R at 32 d after implantation. Scale bar, 50 μm. (d) Relative mRNA levels of melanocyte-lineage-specific genes (Mitf, Kit, Dct, Tyrp1, and Tyr) assessed by RT–qPCR in three explanted xenograft tumors for each group. Each data point represents the mRNA level of a single tumor normalized to Rpl27. Statistical significance of P < 0.05 was observed for differences between tumors expressing Leu129Gln CysLT2R and tumors expressing empty-vector control or wild-type CysLT2R. (e) Cropped immunoblots of CysLT2R and melanocyte markers in three explanted melan-a xenograft tumors for each group. Full-length immunoblots are presented in Supplementary Figure 5.
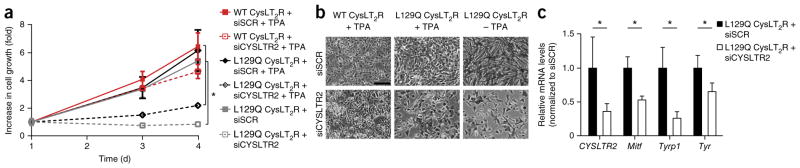
Because Leu129Gln CysLT2R conferred TPA-independent growth in melan-a cells, we asked whether melan-a cells expressing Leu129Gln CysLT2R had become dependent on it for proliferation. We performed small interfering RNA (siRNA)-mediated knockdown of the exogenous human CYSLTR2 in cells expressing wild-type CysLT2R and Leu129Gln CysLT2R. Knockdown of CYSLTR2 reduced growth in cells with Leu129Gln CysLT2R grown in the presence or absence of TPA and caused the cells to have a flattened morphology, but it did not affect the growth or morphology of cells with wild-type CysLT2R grown in the presence of TPA (Fig. 5a,b). Further, knockdown of CYSLTR2 in cells expressing Leu129Gln CysLT2R decreased the expression of melanocyte-lineage genes (Fig. 5c). These data suggest that inhibiting CysLT2R in cell lines with the CYSLTR2 mutation encoding p.Leu129Gln can have therapeutic potential.
Figure 5.
CYSLTR2 is required for the growth and maintenance of the melanocyte-lineage-specific signature in melan-a cells transformed to express Leu129Gln CysLT2R. (a) Growth curves of melan-a cells expressing wild-type or Leu129Gln CysLT2R transfected with either scrambled siRNA (siSCR) or siRNA targeting CYSLTR2 (siCYSLTR2) and assayed by CellTiter-Glo. Melan-a cells expressing wild-type CysLT2R were grown in the presence of 200 nM TPA, whereas cells expressing Leu129Gln CysLT2R were grown in the presence or absence of 200 nM TPA. All cells were grown for 3 d. The fold increase in growth is shown relative to cell numbers at 1 d and corresponds to the means ± s.e.m. from six technical replicates. *P < 0.05. (b) Representative phase-contrast microscopy images of melan-a cells expressing wild-type or Leu129Gln CysLT2R transfected with either scrambled siRNA or siRNA targeting CYSLTR2 at day 5 after transfection. Scale bar, 50 μm. (c) Relative mRNA levels of melanocyte-lineage-specific genes (Mitf, Tyrp1, and Tyr) in melan-a cells expressing Leu129Gln CysLT2R with scrambled siRNA or siRNA targeting CYSLTR2 grown in the presence of TPA. Error bars, s.d. from three technical replicates. *P < 0.05.
GPCR mutations drive several benign endocrine neoplasms, including mutations of thyroid-stimulating hormone receptor in thyroid adenoma 19 and luteinizing hormone receptor in testicular Leydig adenoma 20. In addition, many GPCRs can be oncogenic in experimental systems, and analysis of whole-exome studies has shown that GPCRs are among the most commonly mutated genes, mutated in approximately 20% of all cancers 21, 22. Despite the high mutation rate, to our knowledge, no single GPCR has been identified as a significantly mutated gene in any malignant tumor type 23, 24, possibly because GPCR genes generally lack hotspot mutations and mutations in these genes co-occur with mutations in other well-known driver oncogenes. We have defined a recurrent hotspot mutation of CYSLTR2 in uveal melanoma, which represents a bona fide mutated GPCR driver oncogene. Leu129Gln CysLT2R exhibits high basal activity and couples through Gαq, thereby activating a signaling pathway convergent with the one activated by GNAQ and GNA11 oncogenic mutations (Fig. 1c). We further demonstrated that Leu129Gln CysLT2R enforces expression of the melanocyte-lineage-specific transcriptional program, possibly having a role similar to that of MITF amplification in enforcing lineage in cutaneous melanoma 25.
GPCRs are attractive drug targets because of their location on the plasma membrane and the availability of high-throughput cell-based screens for drug discovery. Inhibitors of CysLT1R, such as montelukast, are broadly used in asthma 26. Unfortunately, montelukast has no activity against CysLT2R. Recently, several compounds have been discovered with purported activity against wild-type CysLT2R, including one that is undergoing phase II clinical trial 27. Transition of wild-type GPCR between the inactive and active conformations depends on the ligand environment, but mutations conferring constitutive activity strongly bias the protein toward the active conformation, either through destabilization of the inactive state or stabilization of the active state. To inhibit these mutants, high-affinity ‘inverse agonists’ that stabilize the inactive state will be required 28. Development of such drugs to inhibit Leu129Gln CysLT2R will have therapeutic potential in uveal melanoma with CYSLTR2 mutation encoding p.Leu129Gln, a disease that currently has no effective therapy.
URLs. 4Peaks, http://nucleobytes.com/4peaks; NIHTCGA server, https://tcga-data.nci.nih.gov/; hotspots algorithm, https://github.com/taylor-lab/hotspots; CoMEt algorithm, https://github.com/raphael-group/comet/releases; Cancer Genomics Hub, https://cghub.ucsc.edu/; Integrative Genomics Viewer, https://www.broadinstitute.org/igv/; MSKCC cBioPortal, http://www.cbioportal.org/; SAMtools, http://www.htslib.org/; Bowtie 2, http://bowtie-bio.sourceforge.net/bowtie2/.
ONLINE METHODS
Mutational analysis
Level 2 whole-exome mutational data (curated Broad Institute calls) and Level 3 RNA-seqV2 data from TCGA uveal melanomas (n = 80) were downloaded from the NIH TCGA server. Processed whole-exome sequencing data from the UNI-UDE uveal melanoma cohort (n = 22) and processed whole-genome sequencing data from the CRUK (n = 12) and QIMR (n = 28) cohorts were extracted from the supplementary tables of relevant publications 3–5. For downstream analysis, we merged the data for 3 duplicate samples present in both the CRUK and TCGA databases and removed 3 samples from the QIMR database that lacked any somatic mutations, leaving 136 samples. To identify significant hotspot mutations, we ran an algorithm that is highly sensitive in detecting rare hotspot mutations 6. To identify and visualize mutually exclusive mutations, we ran the CoMEt algorithm 7 using 10 million iterations and 100 permutations. Statistical significance is represented by mid P value (described as Φ in ref. 7). To examine raw DNA and RNA sequencing reads with CYSLTR2 mutation, we obtained TCGA BAM files from the Cancer Genomics Hub and UNI-UDE fastq files from the European Nucleotide Archive (study accession ERP003230) and mapped reads to the hg19 reference genome using Bowtie 2 (ref. 29). To determine the number of wild-type and mutant reads at CYSLTR2, we used SAMtools mpileup after filtering for base quality of at least 13 (ref. 30). Reads were visualized using the Integrative Genomics Viewer 31. The OncoPrint was generated using MSKCC cBioPortal 32.
For the MSKCC uveal melanoma cohort, all patients provided informed consent for tissue procurement and mutational testing, and the study was approved by the institutional review board (12-245A). Tumor samples were sequenced using MSK-IMPACT, a hybrid-capture deep sequencing assay of 341 key cancer-related genes. The MSK-IMPACT panel includes GNAQ and GNA11 but does not include CYSLTR2 (ref. 33). We obtained tumor and germline DNA from the MSK-IMPACT sequencing libraries of two patients who lacked mutations in GNAQ and GNA11 and performed Sanger sequencing of CYSLTR2.
For pan-TCGA analysis of CYSLTR2 mutations, we obtained curated data from MSKCC cBioPortal. We examined 24 disease cohorts (Supplementary Fig. 2a) and selected cases with both mutational data and RNA-seq-based gene expression data.
Sanger sequencing of CYSLTR2
Sequence analysis of codon 129 of CYSLTR2 by Sanger sequencing was performed by standard PCR using HotStarTaq Plus Master Mix (Qiagen) with CYSLTR2 Sanger sequencing primers (Supplementary Table 1). PCR products were purified using the E.Z.N.A. Cycle Pure kit (Omega) and sent for sequencing with the same primers used for PCR. Sequencing plots were created with 4Peaks.
Exogenous gene expression
For transient expression in HEK293T cells used in in vitro assays of calcium mobilization and cAMP accumulation, we generated vectors with synthetic CYSLTR2 encoding the protein fused to an N-terminal FLAG epitope (DYKDDDDK) and a C-terminal 1D4 epitope (TETSQVAPA) and subcloned this sequence into pcDNA3.1(+) (Life Technologies) using the restriction enzymes EcoRV and NotI (constructed by Genewiz). The human CYSLTR2 sequence was codon optimized for expression in mammalian cells by Genewiz. The construct encoding Leu129Gln CysLT2R mutant receptor was generated by QuikChange (Agilent Technologies) site-directed mutagenesis of CYSLTR2 to introduce a c.386T>A mutation (NM_020377.2) encoding the p.Leu129Gln substitution using synthetic CYSLTR2 L129Q QuikChange primers (Supplementary Table 1). HEK293T cells were transfected with the plasmids subcloned into pcDNA3.1(+) using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s recommendations. For stable expression in NIH3T3 and melan-a cells, the cDNA for wild-type human CYSLTR2 was obtained from Origene and cloned into the MSCV-IRES-GFP vector 34 (pMIGw; Addgene plasmid 12282). QuikChange site-directed mutagenesis was performed using CYSLTR2 L129Q QuikChange primers (Supplementary Table 1) on wild-type CYSLTR2 to introduce a c.386T>A mutation encoding the p.Leu129Gln substitution. In addition, site-directed mutagenesis was performed using CYSLTR2 R136H QuikChange primers (Supplementary Table 1) on wild-type CYSLTR2 to introduce a c.407G>A mutation encoding the p.Arg136His substitution. The sequences of all constructs were confirmed by sequencing.
Intracellular calcium flux mobilization assays (Gαq assays)
HEK293T cells were seeded at a density of 20,000 cells/well in 384-well plates (Corning) coated with poly-d-lysine hydrobromide (Sigma-Aldrich) and transiently transfected with vector encoding wild-type or Leu129Gln CysLT2R or with the empty vector (pcDNA3.1(+)). Twenty-four hours after transfection, cells were incubated for 1.5 h at 37 °C with 20 μl/well Ca4 dye (FLIPR Calcium 4 Assay Kit, Molecular Devices) with 2.5 mM probenecid (an inhibitor of the anion-exchange protein). The dye was dissolved in HBSS-H (Hank’s Balanced Salt Solution with 20 mM HEPES buffer, pH 7.4) supplemented with 0.4% BSA according to the manufacturer’s instructions. Subsequently, the plate was equilibrated for 15 min before the start of measurement on a FlexStation II 384 Plate Reader (Molecular Devices) preheated to 37 °C. Ligands were diluted in HBSS-H supplemented with 0.4% BSA. Fluorescence was monitored over a 100-s time course, with readings taken every 2.5 s, and 10 μl/well of ligand was added to the FlexStation in fluxo 20 s after the start of measurement. The excitation and emission wavelengths were set, respectively, to 488 and 530 nm. Relative fluorescence units were calculated from the mean of each set of triplicate experiments for each time point and were used to generate a representative time course curve (Fig. 2b) derived with GraphPad Prism. The sigmoid dose response curve (Fig. 2c) was generated using the absolute peak magnitude signal, from which the signal for cells transfected with empty vector was subtracted to correct for background fluorescence. Data were normalized to the maximal calcium response elicited by wild-type CysLT2R with 100 nM LTD4. Sigmoid dose response curves were calculated as the percentage of ((RFU)CysLT2R – (RFU)empty vector) versus ligand concentration with GraphPad Prism. EC50 values are means from seven independently repeated dose response curves.
Measurement of cAMP production (Gαi and Gαs biological assays)
cAMP production was monitored using the RLuc3-EPAC-GFP10 BRET2 biosensor as previously described 35. The plasmid encoding the EPAC biosensor was a kind gift of M. Bouvier (Université de Montréal). Briefly, the EPAC protein, which changes its conformation upon cAMP binding, is tagged on each extremity of the protein with a BRET donor (Renilla luciferase, Rluc3) and a BRET acceptor (GFP10) to monitor intramolecular BRET signal. Activation of the Gαs and Gαi pathways is determined by measuring the BRET ratio, that is, the GFP10 (530 nm)/Rluc3 (480 nm) ratio, resulting from structural rearrangements within EPAC 35. HEK293T cells were transiently cotransfected with empty vector (pcDNA3.1(+)) or vector encoding wild-type or Leu129Gln CysLT2R along with the RLuc3-EPAC-GFP10 cAMP sensor in 96-well plates. Twenty-four hours after transfection, cells were incubated with increasing concentrations of LTD4 agonist with (Gαi coupling assay) or without (Gαs coupling assay) 4 μM forskolin (which maximally activates adenylyl cyclase and produces intracellular cAMP) at room temperature for 5 min before BRET readings. The luciferase substrate Coelenterazine DeepBlue C (5 μM final concentration; Biotum) was added 10 min before readings. BRET2 signal was monitored on a BioTek Synergy NEO plate reader. Cells transfected with vector encoding control receptor coupling to Gαi or Gαs—CCR5 and CLR/RAMP2 receptors, respectively—and treated with increasing concentrations of RANTES or adrenomedullin agonist were used as controls in each experiment. Data are expressed as the percentage of cAMP accumulation relative to the maximal response obtained with forskolin induction and correspond to means ± s.e.m. from four (Gαi assay) and two (Gαs assay) independent experiments, in which each measurement was obtained in triplicate. Dose response curves were derived with GraphPad Prism.
Molecular modeling
Homology modeling of CysLT2R (UniProt, Q9NS75) was performed with the automodel routine of the software package MODELLER version 9.15 (ref. 36) using the crystal structure of human PAR1 (F2R/PAR1) 14 as a template (Protein Data Bank (PDB), 3VW7; UniProt, P25116). The sequence alignment provided as input for the automodel routine was generated with ClustalW 37 and manually refined in MacVector 14.0 (MacVector). Figures were generated using VMD 1.9.2 (ref. 38) to visualize residues 27–311 of the receptor, including the transmembrane helical bundle and the packing of the hydrophobic microdomain around Leu129 (3.43).
Flow cytometry
For FACS analysis of FLAG epitope on the cell surface, transiently transfected HEK293T cells were collected with PBS on ice, centrifuged at 350g and resuspended in FACS buffer (0.5 mM MgCl2 and 0.1% BSA in PBS). Cells were then incubated with a monoclonal antibody to FLAG conjugated to Alexa Fluor 647 (R&D System, clone 1042E; 3 μl antibody/1 × 106 cells) on ice for 45 min. Cells were then washed in PBS, fixed in 2% formaldehyde, and analyzed on an LSR-Fortessa (BD Biosciences). To determine the percentage of NIH3T3 and melan-a cells with stable GFP expression, cells were collected, centrifuged at 400g resuspended in FACS buffer (2% FBS, 5 mM EDTA, 25 mM HEPES, and 100 ng/ml DAPI), and analyzed on an LSR-Fortessa before subcutaneous injection into mice for xenograft experiments. Cells stably expressing GFP were sorted with a FACSAria II (BD Biosciences).
Cell lines and in vitro analysis
HEK293T cells were obtained from the American Type Culture Collection (ATCC) and were grown in DMEM (4.5 g/L glucose (Life Technologies)). Melan-a cells were provided by D. Bennett (St. George’s Hospital, University of London) 15 and were grown in RPMI with 200 nM TPA (Sigma-Aldrich) unless noted otherwise. NIH3T3 mouse embryonic fibroblasts (CRL-1658) were obtained from ATCC and cultured in DMEM. MEL290 cells (a gift from T. Wiesner) were grown in RPMI. All cell culture media contained 10% FBS, penicillin (100 U/ml), streptomycin (100 μg/ml), and l-glutamine (2 mM). All cell lines tested negative for mycoplasma contamination. Cell growth was assessed using CellTiter-Glo (Promega), and the fold change in growth at day 3 was calculated relative to the day 1 luminescence values. Results from a representative experiment are shown; cell growth was assessed in three independent experiments, each with quadruplicate measures.
Immunoblotting
Cell lysates for melan-a cells were prepared in SDS lysis buffer (1% SDS, 20 mM NaF, and 5 mM EDTA) supplemented with fresh proteinase and phosphatase inhibitors (PhosSTOP, Roche; Complete EDTA-Free, Roche). HEK293T cells were collected in PBS containing 1% PMSF (Sigma-Aldrich) and 1% aprotinin (Sigma-Aldrich). Cell pellets were lysed in lysis buffer (PBS containing 50 mM Tris, pH 6.8, 100 mM NaCl, 1 mM CaCl2, and 10% DM (dodecyl β-d-maltoside; Anatrace) supplemented with protease and phosphatase inhibitors (Complete EDTA-Free) for 1 h at 4 °C and subsequently centrifuged at 1000g for 15 min at 4 °C. Equal amounts of protein, as measured by BCA protein assay (Thermo Scientific) or Bradford protein assay (Bio-Rad), were resolved on NuPAGE Novex 4–12% Bis-Tris Protein Gels (Life Technologies) and transferred electrophoretically onto a 0.45-μm nitrocellulose membrane (Bio-Rad) or a PVDF membrane (Immobilon). Membranes were blocked for 1 h at room temperature in 5% milk in TBST or Odyssey blocking buffer (LI-COR) before being incubated overnight at 4 °C with primary antibodies diluted 1:1,000, unless otherwise noted, in either 5% milk in TBST or Odyssey blocking buffer. Antibodies to the following proteins were used: CysLT2R (Santa Cruz Biotechnology, E-20), MITF (Cell Signaling Technology, 12590), c-Kit (Cell Signaling Technology, 3074), gp100 (Abcam, ab137078), TRP2 (Abcam, ab74073), GFP (Abcam, ab6556), GAPDH (1: 5,000 dilution; Applied Biological Materials, G041), FLAG tag (1:3,000 dilution; Sigma-Aldrich, F7425), 1D4 tag (1:3,000 dilution; obtained from the National Cell Culture Center), and β-tubulin (1:500 dilution; Abcam, ab6046).
In vivo tumor formation
All mouse experiments were performed in accordance with a protocol approved by the MSKCC Institutional Animal Care and Use Committee (11-12-029). The size for each cohort was determined on the basis of previous experience without specific statistical methods. We resuspended 1 × 106 stably transduced melan-a or NIH3T3 cells in 100 μl of a 1:1 mix of medium and Matrigel (BD Biosciences) and subcutaneously and bilaterally injected the mix into the flanks of 7-week-old female CB17-SCID mice (Taconic). To assess tumor growth, five mice per group were injected for a total of ten tumors per group. No randomization or blinding was used in the analysis of tumor growth. A single tumor was excluded from the melan-a wild-type CysLT2R group because of deviation from the mean and erratic growth. Two tumors from the melan-a Leu129Gln CysLT2R group were excluded because of growth that differed from the mean. Melan-a in vivo tumor formation was assessed in two independent experiments, with the results of one experiment shown in Figure 2b. Tumors were measured with calipers every 2 or 3 d for up to 35 d. Growth curves were visualized with Prism GraphPad 6.0. Tumor volume was calculated using the formula
For hematoxylin and eosin staining, tumors were fixed at 4 °C overnight in 4% paraformaldehyde. Tissue embedding in paraffin, sectioning, and staining with hematoxylin and eosin were performed by Histoserv. Tumor protein lysates were generated by homogenization of flash-frozen tissue in cell lysis buffer (1% Triton X-100, 50 mM HEPES, pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 10 mM sodium pyrophosphate, 1 mM sodium orthovanadate, and 10% glycerol, containing freshly added protease and phosphatase inhibitors (PhosSTOP and Complete EDTA-Free)).
RNA isolation and RT–qPCR
For cultured melan-a cells, RNA was isolated using the E.Z.N.A. Total RNA kit (Omega). For tumor tissue, RNA was isolated by homogenizing the tumors in Ribozol (Amresco) followed by RNA purification with the RNeasy Plus Mini kit (Qiagen).
For RT–qPCR, RNA was reverse transcribed using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems) and PCR was run using Power SYBR Green PCR Master Mix (Applied Biosystems) on a QuantStudio 6 Flex system (Applied Biosystems). Primer sequences are listed in Supplementary Table 1. Expression was normalized to that of the ribosomal protein Rpl27. Relative mRNA expression was plotted as 2−ΔΔCt or 2−ΔCt. Each RT–qPCR was performed in at least triplicate and repeated in three different experiments.
siRNA
For siRNA experiments, the following pooled siRNAs were used: human CYSLTR2 (SMARTpool ON-TARGETplus CYSLTR2 siRNA, Dharmacon) and scrambled siRNA (ON-TARGETplus Non-Targeting Pool, Dharmacon). Transient transfection with siRNA was performed using Lipofectamine RNAiMAX reagent (Invitrogen), and siRNA was reverse transfected into cells according to the supplied protocol.
Immunofluorescence
Immunofluorescence images were acquired using an FSX100 Olympus inverted-base microscope with a 60× oil immersion objective. HEK293T cells were seeded at a density of 1 × 106 cells/well in six-well plates for 24 h before transfection. Twenty-four hours after transfection, cells were seeded and plated at a density of 300,000 cells/ml on coverslips in six-well plates. After 24 h, cells were fixed in cold methanol for 5 min, blocked in blocking buffer (CaCl2/MgCl2 (Life Technologies) and 0.5% BSA in PBS) for 30 min, and incubated with a mouse monoclonal antibody to 1D4 (1:2,000 dilution) and a rabbit polyclonal antibody to FLAG (1:1,000 dilution) for 1 h at room temperature. Cells were then washed (CaCl2/MgCl2 in PBS; 5 min, three times) and incubated with secondary antibody, either Alexa Fluor 488–conjugated anti-mouse antibody (1:500 dilution) (green) or Alexa Fluor 594–conjugated anti-rabbit antibody (1:500 dilution) (red) (Thermo Fisher, A-11029 and A-11012, respectively) for 30 min at room temperature. Non-specific secondary antibody signal was removed by washing with CaCl2/MgCl2 in PBS (5 min, three times), followed by a final incubation with Hoechst stain (1:15,000 dilution in CaCl2/MgCl2 in PBS) at room temperature for 5 min. Coverslips were mounted onto glass slides, which were stored at −20 °C before being visualized.
Statistics
All statistical comparisons between two groups in the qPCR and xenograft experiments were performed with GraphPad Prism 6.0 software and used a two-tailed parametric unpaired t test.
Acknowledgments
We gratefully acknowledge the members of the Molecular Diagnostics Service in the Department of Pathology and the Marie-Josée and Henry R. Kravis Center for Molecular Oncology. We thank T. Wiesner (MSKCC and Medical University of Graz) for the melan-a and MEL290 cell lines and intellectual input, D. Abramson for intellectual input, C. Kandoth and the TCGA for data sharing, M. Berger and N. Bouvier for obtaining patient DNA, Y.A. Berchiche for the flow cytometry analysis and scientific input, and M. Bouvier (Université de Montréal) for the kind gift of the EPAC construct. This work was supported in part by grants from the NIH to P.C. (P50CA140146, CDA; DP2CA174499; K08CA151660), the NIH to Y.C. (K08CA140946), the Sidney Kimmel Foundation to P.C. (Kimmel Scholar Award), the Cycle for Survival Fund to P.C. and Y.C., the Geoffrey Beene Award to P.C. and Y.C., the Prostate Cancer Foundation to Y.C., the STARR Cancer Consortium to P.C. and Y.C., and the François Wallace ç Monahan Fellowship to E.C.
Footnotes
AUTHOR CONTRIBUTIONS
Project planning and experimental design: P.C., Y.C., A.R.M., T.P.S., M.A.K., and E.C. Bioinformatic analysis of exome sequencing data: Y.C., A.R.M., M.T.C., and B.S.T. Clinical specimen acquisition, annotation, and analysis: A.N.S. Immunoblots, growth curves, qPCR, and all cellular assays: A.R.M., E.C., and Y.G. Xenograft assays: A.R.M., J.J.S., and E.G.W. Generation of the expression vectors: J.Q.Z., E.C., and A.R.M. Molecular modeling: T.H. Manuscript writing: A.R.M., P.C., Y.C., E.C., and T.P.S. All authors reviewed the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Van Raamsdonk CD, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Raamsdonk CD, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansson P, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furney SJ, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin M, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang MT, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leiserson MD, Wu HT, Vandin F, Raphael BJ. CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol. 2015;16:160. doi: 10.1186/s13059-015-0700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harbour JW, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbour JW, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet. 2013;45:133–135. doi: 10.1038/ng.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans JF. Cysteinyl leukotriene receptors. Prostaglandins Other Lipid Mediat. 2002;68–69:587–597. doi: 10.1016/s0090-6980(02)00057-6. [DOI] [PubMed] [Google Scholar]
- 11.Sakmar TP. Structure of rhodopsin and the superfamily of seven-helical receptors: the same and not the same. Curr Opin Cell Biol. 2002;14:189–195. doi: 10.1016/s0955-0674(02)00306-x. [DOI] [PubMed] [Google Scholar]
- 12.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein–coupled receptors. In: Stuart CS, editor. Methods in Neurosciences. Vol. 25. Academic Press; 1995. pp. 366–428. [Google Scholar]
- 13.Tao YX. Constitutive activation of G protein–coupled receptors and diseases: insights into mechanisms of activation and therapeutics. Pharmacol Ther. 2008;120:129–148. doi: 10.1016/j.pharmthera.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, et al. High-resolution crystal structure of human protease-activated receptor 1. Nature. 2012;492:387–392. doi: 10.1038/nature11701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett DC, Cooper PJ, Hart IR. A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int J Cancer. 1987;39:414–418. doi: 10.1002/ijc.2910390324. [DOI] [PubMed] [Google Scholar]
- 16.Prince S, Wiggins T, Hulley PA, Kidson SH. Stimulation of melanogenesis by tetradecanoylphorbol 13-acetate (TPA) in mouse melanocytes and neural crest cells. Pigment Cell Res. 2003;16:26–34. doi: 10.1034/j.1600-0749.2003.00008.x. [DOI] [PubMed] [Google Scholar]
- 17.Yaar M, Gilchrest BA. Human melanocyte growth and differentiation: a decade of new data. J Invest Dermatol. 1991;97:611–617. doi: 10.1111/1523-1747.ep12482985. [DOI] [PubMed] [Google Scholar]
- 18.Griewank KG, et al. Genetic and molecular characterization of uveal melanoma cell lines. Pigment Cell Melanoma Res. 2012;25:182–187. doi: 10.1111/j.1755-148X.2012.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parma J, et al. Somatic mutations in the thyrotropin receptor gene cause hyperfunctioning thyroid adenomas. Nature. 1993;365:649–651. doi: 10.1038/365649a0. [DOI] [PubMed] [Google Scholar]
- 20.Liu G, et al. Leydig-cell tumors caused by an activating mutation of the gene encoding the luteinizing hormone receptor. N Engl J Med. 1999;341:1731–1736. doi: 10.1056/NEJM199912023412304. [DOI] [PubMed] [Google Scholar]
- 21.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 22.Prickett TD, et al. Exon capture analysis of G protein–coupled receptors identifies activating mutations in GRM3 in melanoma. Nat Genet. 2011;43:1119–1126. doi: 10.1038/ng.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kandoth C, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence MG, et al. A preclinical xenograft model of prostate cancer using human tumors. Nat Protoc. 2013;8:836–848. doi: 10.1038/nprot.2013.043. [DOI] [PubMed] [Google Scholar]
- 25.Garraway LA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 26.Leff JA, et al. Montelukast, a leukotriene-receptor antagonist, for the treatment of mild asthma and exercise-induced bronchoconstriction. N Engl J Med. 1998;339:147–152. doi: 10.1056/NEJM199807163390302. [DOI] [PubMed] [Google Scholar]
- 27.Wunder F, et al. Pharmacological characterization of the first potent and selective antagonist at the cysteinyl leukotriene 2 (CysLT2) receptor. Br J Pharmacol. 2010;160:399–409. doi: 10.1111/j.1476-5381.2010.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bond RA, Ijzerman AP. Recent developments in constitutive receptor activity and inverse agonism, and their potential for GPCR drug discovery. Trends Pharmacol Sci. 2006;27:92–96. doi: 10.1016/j.tips.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 29.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng DT, et al. Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Refaeli Y, Van Parijs L, Alexander SI, Abbas AK. Interferon γ is required for activation-induced death of T lymphocytes. J Exp Med. 2002;196:999–1005. doi: 10.1084/jem.20020666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leduc M, et al. Functional selectivity of natural and synthetic prostaglandin EP4 receptor ligands. J Pharmacol Exp Ther. 2009;331:297–307. doi: 10.1124/jpet.109.156398. [DOI] [PubMed] [Google Scholar]
- 36.Martí-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 37.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]