Abstract
Dendritic cells (DCs) are uniquely potent in orchestrating T cell immune response, thus they are indispensable immune sentinels. They originate from progenitors in the bone marrow through hematopoiesis, a highly regulated developmental process involving multiple cellular and molecular events. This review highlights studies of DC development—from the discovery of DCs as glass-adherent antigen presenting cells to the debate and resolution of their origin and lineage map. In particular, we summarize the roles of lineage-specific cytokines, the placement of distinct hematopoietic progenitors within the DC lineage and transcriptional programs governing DC development, which together have allowed us to diagram the current view of DC hematopoiesis. Important open questions and debates on the DC development and relevant models are also discussed.
Keywords: Dendritic cell, hematopoiesis, mononuclear phagocyte system, lineage, progenitor, MDP, CDP, pre-cDC, cytokine, transcription factor
Introduction
Dendritic cells (DCs) are uniquely potent in orchestrating T cell immune response, thus they are indispensable immune sentinels. They originate from progenitors in the bone marrow through hematopoiesis, a highly regulated developmental process involving multiple cellular and molecular events. This review highlights studies of DC development—from the discovery of DCs as glass-adherent antigen presenting cells to the debate and resolution of their origin and lineage map. In particular, we summarize the roles of lineage-specific cytokines, the placement of distinct hematopoietic progenitors within the DC lineage and transcriptional programs governing DC development, which together have allowed us to diagram the current view of DC hematopoiesis. Important open questions and debates on the DC development and relevant models are also discussed.
1. Identification of the DC – a mononuclear phagocyte uniquely potent in initiating T cell immunity
1.1. Discovery of DCs
The discovery of DCs resulted from the effort to understand the cellular initiator of adaptive immune response. Macrophages, monocytes and their bone marrow precursors, comprising the mononuclear phagocyte system (MPS), were long appreciated for their capacity to adhere to, engulf and degrade infectious pathogens (reviewed in [1]. Importantly, some MPS cells retain antigens that are not entirely degraded and present them on the cell surface in the context of the MHC-peptide complex, resulting in T cell activation [2, 3]. MPS cells adhere to glass readily, thus glass-adherence was recognized as a characteristic feature of MPS cells and widely used for their isolation. In the 1960’s it was recognized that glass-adhering cells from mouse spleen could prime adaptive immune response, i.e., induce antibody production from naïve lymphocytes in vitro [4]. It was speculated that antigen-presenting cells (APCs) initiated the response by activating T cells, which subsequently stimulated antibody production from B cells, thereby bridging innate immunity and adaptive immunity. However, the exact identity of such immunity-initiating APC within the MPS was unclear. In 1973, Ralph Steinman and Zan Cohn, during microscopic study of the glass-adhering mouse splenocytes, discovered a small population of cells with unique stellate morphology, and named them dendritic cells (DCs) [5]. It was soon found that DCs express high level of MHC II and are uniquely potent in stimulating T cell proliferation—orders of magnitude more potent than macrophages and other APCs examined [6]. Thus DCs were discovered as the T cell-activating member of MPS.
The early studies of DC origin and in vivo dynamics utilized cell surface markers, bone marrow chimeras, 3H-thymidine injection, GM-CSF culture and colony formation assays (reviewed in [7]). Glass-adhering DCs in mouse spleen express characteristic myeloid markers such as CD33 and CD11b and are replenished by rapidly proliferating bone marrow progenitors. These progenitors produce DCs together with granulocytes, monocytes and macrophages. Based on these studies, DCs were considered to be of myeloid origin.
1.2 DCs, macrophages and monocytes are separate lineages
Because monocytes were originally considered to be the migratory bone marrow precursor to all MPS cells, scientists understandably reasoned DCs also descend from monocytes. This reasoning was supported by the fact that both human and mouse monocytes can acquire typical DC features. When cultured with GM-CSF in vitro, monocytes up-regulate MHC II and accessory molecules, develop dendrites and become immune-stimulatory to T cells [8, 9]. Similarly, in vivo, monocytes can develop DC features during inflammation [10, 11]. However, it is now clear that neither DCs nor tissue resident macrophages descend from monocytes in the steady state (reviewed in [12]). As demonstrated from genetic fate mapping and adoptive transfer experiments, tissue resident macrophages derive from a yolk-sac-derived erythroid-myeloid progenitor ([13] [14]), whereas DCs descend from progenitors distinct from monocytes (discussed in section 3). Thus, despite belonging to the same MPS, DCs, macrophage and monocytes have distinct origins.
2. DC subsets
DCs control many aspects of immunity. Broadly, they initiate T cell immunity and tolerance [15]. Specifically they can regulate differentiation of T cells toward Th1, Th2, Th17 or Treg subsets. DCs can also indirectly skew T cell subset differentiation by activating innate lymphocytes to produce conditioning cytokines for T cell subset specification [16]. Such diverse functions of DCs are unlikely executed by one type of cell. Indeed, multiple DC subsets have been identified, each displaying distinct phenotype, carrying out distinct functions, and collaboratively enabling adaptive T cell immune response (reviewed in [17]). As growing numbers of DC subsets were identified, one question arose: do these DC subsets share the same origin? This question has generated heated and ongoing debate, tantalizing hypotheses, and surprising findings that provide deeper insights and more questions about DC development and hematopoiesis as a whole.
2.1. From glass-adhering DCs to CD11c+ DCs
Although the morphologic and functional differences between DCs and other leukocytes were striking, few immunology laboratories in the 70’s were equipped to perform the purification procedures required to study DCs until monoclonal antibodies to DC markers became available. A series of mouse DC-restricted monoclonal antibodies were produced by Steinman and others starting in the 1980s, including 33D1 (specific for the cell surface receptor DCIR2/Clec4a4), NLDC-145 (specific for the adsorptive endocytosis receptor, DEC205/cluster of differentiation (CD205)), and N418 (specific for the integrin, CD11c). These monoclonal antibodies were used to establish the unique functions of DCs within heterogeneous mixtures of leukocytes and to identify DCs in situ. The ligand of N418, CD11c, is highly expressed on all DCs [18]. CD11c and MHC II expression soon became the criteria for identification of DCs, replacing old adherence and morphologic criteria. The new criteria then allowed in-depth study of DC diversity. It is worth noting that “DCs” identified using this criteria included some tissue macrophages that have been shown to also express CD11c and MHC class II (reviewed in [12]).
2.2 Identification of DC subsets with lymphoid features
The idea of “lymphoid DC” was formed when DC subsets expressing lymphoid markers were identified. Shortman and colleagues found that CD11c+MHC II+ DCs in multiple lymphoid organs could be divided into two subsets [19]. One subset expresses the CD11b and CD4 (CD11b+ or CD4+ DCs), while the other lacks CD11b and CD4 but expresses CD8a (CD8a+ DCs). Both CD4 and CD8 are markers typically found on T cells. In mice, CD11b+ DCs and CD8a+ DCs differ from plasmacytoid DCs (discussed below) that lack MHC II expression, and are classified as conventional DCs (cDCs). Other lymphoid markers, including CD2 and CD25, were also found on thymic DCs. An additional DC subset, the plasmacytoid dendritic cell (pDC), demonstrates even more dramatic lymphoid features. pDCs are uniquely capable of producing large amount of type I IFN upon detecting infection and correspondingly exhibit the morphology of a secretory lymphocyte. These cells were first found in human blood and tonsils [20]. Later their equivalents were identified in the mice as CD11c+B220+ cells [21]. When freshly isolated, pDCs do not have dendrites, but display plasma cell morphology, which was responsible for their name. When cultured with IL-3, they acquire DC features, developing long dendrites, up-regulating CD11c, MHC II and co-stimulatory molecules, and becoming immunostimulatory to T cells. For this reason, the pDC was once called pre-DC2 because it was considered to be a precursor of an IL-3-dependent DC lineage, as opposed to the monocyte (pre-DC1), which was considered the precursor of GM-CSF-dependent myeloid DCs. In addition to B220 and CD4, pDCs express several other genes typical of lymphoid lineage, including RAG1, rearranged immunoglobulin heavy chain, and pre-T cell receptor, suggesting a lymphoid past (reviewed in [22]). Developmentally, a mouse thymic T cell precursor and human bone marrow progenitor that lacked myeloid potential were shown to give rise to DCs [23] [24]. Given DCs’ diverse function, their expression of genes typical of lymphoid cells, and development relationship to a T cell precursor, it was proposed that different DC subsets descend separately from myeloid and lymphoid origins.
2.3 Both myeloid and lymphoid progenitors produce all DC subsets
The hypothesis that both lymphoid and myeloid progenitors give rise to DCs was tested in 2000. At the time, common myeloid progenitors (CMPs) and common lymphoid progenitors (CLPs) had been identified in the mouse bone marrow [25, 26]. CMPs and CLPs have committed to myeloid and lymphoid lineages, respectively, representing progenitors of two independently operating lineages that are mutually exclusive [27]. Their potential to generate DCs was immediately tested in mice [28]. Both CMPs and CLPs produced all DC subsets. Importantly, DC subsets descending from CMPs and their counterparts descending from CLPs are indistinguishable in phenotype, function and gene expression [29]. For instance, pDCs derived from CMPs equally express RAG1, and pre-TCR as their CLP-derived counterparts [30]. Therefore, DCs can arise from both myeloid and lymphoid origin. The “dual” origin of DCs is difficult to fit into the conventional bifurcation model of mutually exclusive myeloid and lymphoid lineages, and prompted the search of a separate DC lineage (see next section).
3. A specific lineage of DC
The key elements defining a hematopoietic lineage are lineage-specific cytokines, progenitors and transcription factors. Work over the past two decades has accumulated substantial knowledge allowing a better understanding of the DC lineage. For example, several cytokines have proved essential for normal DC development and homeostasis, sequential DC progenitors have been identified in mouse and humans indicating how DC lineage develop through increasing commitment, and multiple transcription factors form regulatory network controlling DC development and subset specification.
3.1 Cytokines of DC lineage
Although whether cytokines instruct lineage differentiation is debated, it is generally accepted that specific cytokines are essential for proliferation and survival of each hematopoietic lineages. DC development and homeostasis depend on Flt3L, GM-CSF, M-CSF and Lymphotoxin-β.
3.1.1 Flt3 ligand
Fms-like tyrosine kinase 3 (Flt3) is a tyrosine kinase receptor first noted on stem cells and committed lymphoid precursors [31]. The ligand, Flt3L was then cloned and shown to be an active proliferative stimulus for stem cells and developing dendritic cells [32, 33]. Flt3L culture of mouse bone marrow cells can produce large numbers of pDCs and cDC subsets, equivalent in composition to their in vivo counterparts [34]. Deficiency of Flt3L or its receptor results in dramatic loss of pDCs and cDCs in the spleen and lymph nodes, as well as in the peripheral tissues [35]. In these mice, monocytes and granulocytes are unaffected [36]. Conversely, injection of Flt3L or increased serum level of Flt3L, resulting from malaria infection, for example, stimulates expansion of DCs in vivo [33]. Consistent with dependence on and responsiveness to Flt3L, most DC progenitors express the receptor Flt3. Two groups simultaneously found that CLPs and CMPs are heterogeneous and only Flt3+ fractions in these progenitors are able to produce DCs [37] [38]. Notably, ectopic expression of Flt3 in MEP can confer DC potential to these progenitors that otherwise would not produce DCs [39]. Therefore, Flt3L is a cytokine necessary and sufficient for development of DC lineage. Intriguingly, monocyte and granulocytes are independent of this program.
3.1.2 GM-CSF
Granulocyte-macrophage colony stimulating factor (GM-CSF) is a cytokine discovered for its ability to stimulate the formation of myeloid cell colonies from bone marrow progenitors [40, 41]. The role of GM-CSF in DC development was first inferred by in vitro studies. In these studies, GM-CSF was sufficient to promote the differentiation of hematopoietic progenitors and circulating monocytes into cells that resemble DCs in morphology, cell surface marker expression and capacity to stimulate T cells [9] [8] [42]. However, GM-CSF’s role in vivo is limited to certain tissues. In mice deficient in GM-CSF or its receptor, spleen and lymph node cDCs develop normally [43] but the number of resident cDCs in the peripheral non-lymphoid tissues is significantly reduced [44]. In these mice, DCs are able to differentiate but their survival is compromised because of apoptosis induced by increased mitochondria fission [44]. These mice also failed to mount T cell response to antigens inoculated in the lung [44]. Thus, although GM-CSF has no effect on spleen and lymph node DCs, nor on differentiation of DC progenitors, it regulates homeostasis and function of resident DCs in non-lymphoid tissues. Correspondingly, a prolonged culture combining Flt3L and GM-CSF has been established to produce large number of CD103+ DCs equivalent to the tissue resident DCs [45]. In addition, although GM-CSF-generated cells have been shown to be transcriptionally and developmentally different from the splenic DCs [46], they are potent in stimulating T cells. Therefore, GM-CSF remains a key cytokine to generate DC-based vaccines for clinical use [47].
3.1.3 Other cytokines regulate DC development and homeostasis
In addition to the well-studied Flt3L and GM-CSF, several other cytokines have been shown to participate in DC development and homeostasis. First, in the non-inflammatory state, DCs in lymphoid and non-lymphoid tissues undergo several divisions in situ before they die [48] [49]. Their division is regulated by Flt3L [36] and Lymphotoxin-β [50]. The effects of LTα1β2 appear to be restricted to CD11b+ spleen DCs as mice deficient in the receptor (LTβR) showed smaller population of CD11b+ DCs with fewer dividing cells [50]. Second, during inflammation, monocytes can differentiate to DCs (mo-DCs) [11]. In contrast to the in vitro differentiation of mo-DC that depends on GM-CSF, in vivo, mo-DC only depend on M-CSF and its receptor (CD115), but not GM-CSF [44].
3.2 DC progenitors
In classic hematopoiesis model, isolation of prospective progenitors is prerequisite to defining a cellular and molecular pathway of a given hematopoietic lineage. The aforementioned heterogeneity of CLPs and CMPs based on Flt3 expression suggested that DC progenitors could be further purified. Indeed, a series of DC progenitors have been identified in mouse and humans. Their differentiation potential and inter-progenitor relationships have begun to outline the map of DC lineage development.
3.2.1 DC progenitors in mouse
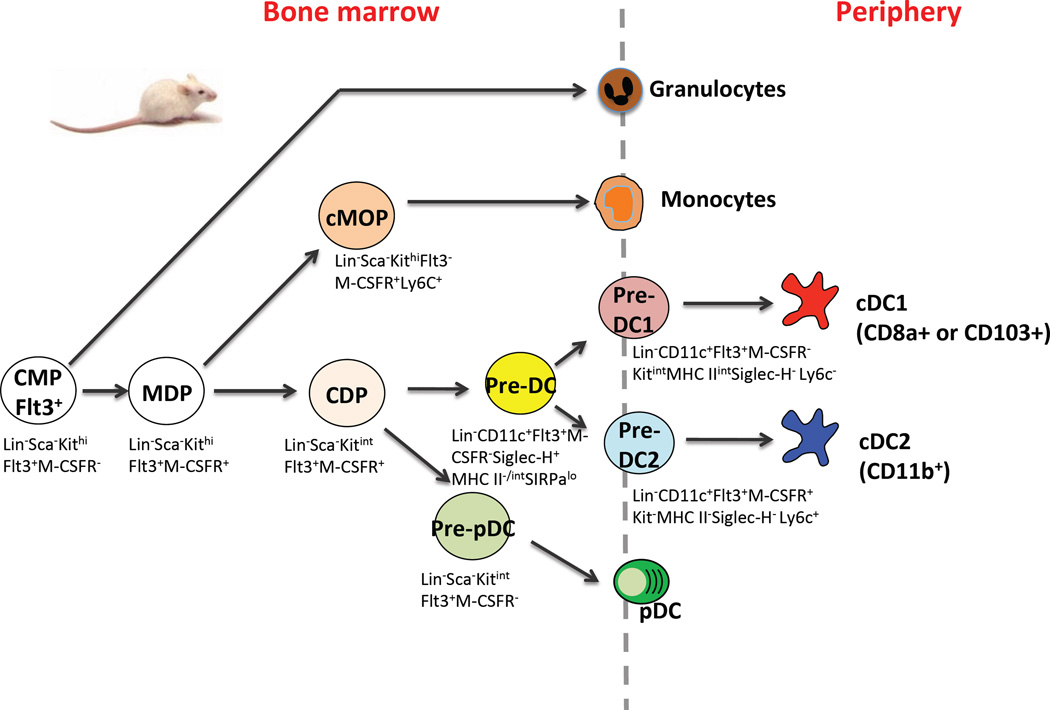
In mice, identification of macrophage-dendritic cell progenitor (MDP) that has lost granulocyte potential was a milestone [51]. MDPs are Lin−Sca−KithiFlt3+CD115+CX3CR1+ cells that lack lymphoid, megakaryocyte, and granulocyte potential but can produce monocytes, macrophages and DCs. As such, MDPs represent a key bifurcation point in DC hematopoiesis, i.e. a specialized progenitor dedicated to producing MPS cells that bridge the innate and adaptive immune response. From MDP, two distinct pathways branch to monocytes and DCs, respectively. Along the monocyte pathway, MDPs give rise to a common monocyte progenitor (cMOP), which lacks Flt3, thus loses DC potential and produces only monocytes [52]. Along the DC pathway, MDPs give rise to a common dendritic progenitor (CDP) [53]; CDPs retain Flt3 and M-CSFR (CD115), but have lost monocyte potential, and subsequently diverge to two pathways to produce pDC or cDC, respectively [39] [54]. On the pathway from CDPs to pDCs, CDPs give rise to the pre-pDC, a progenitor that retains Flt3 but lacks M-CSFR and produces pDCs [55]. On the pathway from CDPs to cDCs, CDPs appear to differentiate in the bone marrow, through a SiglecH+ stage, after which they give rise to SiglecH− pre-CD4/CD11b DCs and pre-CD8 DCs [56] [57] (Fig. 1). Pre-CD4/CD11b and pre-CD8 DCs emigrate from the bone marrow, travel through the blood, and enter the spleen and other tissues [58], where they terminally differentiate to CD11b+ cDCs and CD8+ cDCs [56, 57]. Thus cDC commitment and terminal differentiation are spatially separated and take place in the bone marrow and peripheral tissues, respectively.
Figure 1.
Phenotype and relationship of DC progenitors in mouse
3.2.1 DC progenitors in human
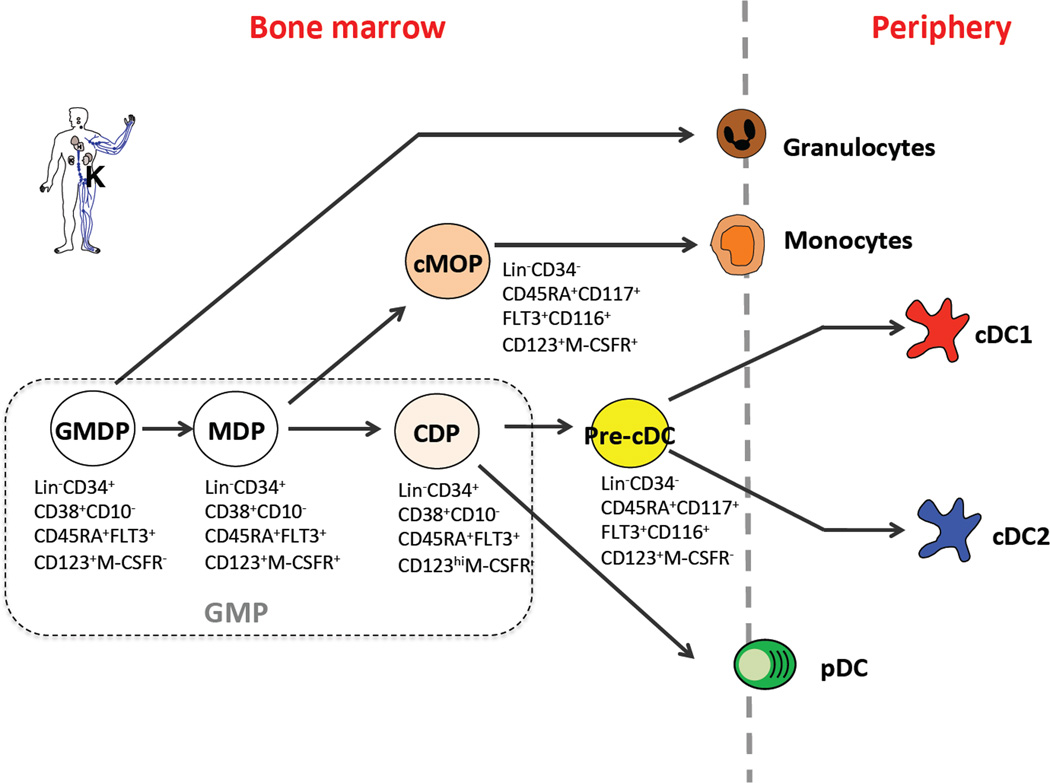
DC development in humans resembles that of mice. Previously, human granulocyte macrophage progenitors (GMPs) have been shown to possess granulocyte, macrophage and DC potential as a bulk population [59] [60] [61]. Recently, a culture system was established to simultaneously assess differentiation of human granulocytes, monocytes, DCs and lymphoid cells at the population and single cell level [62] [63]. When clonal analysis of GMPs was performed using this culture, heterogeneity of clonal potential was revealed: some GMPs appear to have lost granulocyte and monocyte potential [62], prompting further fractionation of the GMP population. Using cytokine receptor expression, Lee et al. isolated three distinct sub-populations within GMPs with DC potential and sequential progenitor-progeny relationship [63] (Fig. 2): one population having granulocyte, monocyte and DC potential (hGMDP, the equivalent to the Flt3+ CMP in mice); downstream of hGMDPs, a population that produces only monocyte and DCs and lacks granulocyte potential (hMDP); arising from hMDP is a third population that produces only DCs and lacks monocyte and granulocyte potential (hCDP). Further, hCDPs give rise to a committed precursor of cDCs (hpre-cDCs) [64], which is present in the bone marrow, blood and peripheral lymphoid organs. This suggests that pre-cDCs emigrate from the bone marrow, travel through the blood to enter peripheral lymphoid organs such as tonsils, where they terminally differentiate to cDCs (Fig. 2).
Figure 2.
Phenotype and relationship of DC progenitors in human
Therefore, in both mouse and human, development of DC lineage is a process of increasing commitment through sequential loss of potential to granulocyte, monocyte and other DC subsets. This degree of conservation is an indication of the relative importance of this pathway during vertebrate evolution.
3.3 Transcription regulation of DC development
It is widely accepted that in hematopoiesis, cytokines only play a permissive role to sustain cell viability and proliferation, whereas the “instructive” role is executed by nuclear transcription regulatory factors (TFs). Through restricted expression within particular lineages, these factors establish gene expression programs intrinsic to cell diversification [65]. Work over the past two decades has revealed important transcription factors that collaboratively regulate DC development and subset specification. The identification of these regulatory networks is made possible by large databases, knowledge of DC progenitors, along with genetic models relevant to the TFs.
3.3.1 Distinct transcriptional programs drive DC and macrophage development
Transcriptional comparison between mouse and human DC subsets initially revealed evolutionarily conserved molecular pathways governing development and functions of DCs [66]. The Immunological Genome Project further provides the expression database of more than 50 mouse macrophage, monocyte, DC subsets, and hematopoietic progenitors enabling an unbiased assessment of genetic relationships among cell types and along development [67] [68] [69]. Such analyses identified a core set of DC signature genes that differentiate DCs from macrophages, including Flt3, CCR7, Zbtb46, Kit, Btla (encoding CD272) and Dpp (encoding CD26), many of which are consistent with the morphological, behavioral and developmental distinction between DCs and macrophages [67, 69]. These analyses imply that a transcriptional program, orchestrated by TFs, drives the DC lineage.
3.3.2 Approach of studying TFs controlling DC development
Prior to the identification of DC progenitors, only differentiated DCs were compared in genetic models that lacked, overexpressed or reported the expression of transcription factors (TFs). These studies revealed that multiple TFs contribute to DC development and subset specification. DC development, a process of increasing commitment and sequential loss of potential, is marked by distinct stages of progenitors. Now, using these genetic models to compare newly defined, sequential DC progenitors has revealed how multiple transcription factors interact in cis and in trans, spatially and temporally, to drive the DC development and specification of DC subsets (Table 1).
Table 1.
Change of DC and progenitor population caused by transcription factor deficiency
| Classical DC | Tissue DC | |||||||
|---|---|---|---|---|---|---|---|---|
| MDP | CDP | pre- cDC |
pDC | CD8+ | CD8− | CD103+ | CD103− | |
| PU.1−/− | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ | ↓ |
| Irf8−/− | ↑ | ↓ | ↓ | ↓ | ↓ | - | ↓ | nd |
| Irf4−/− | nd | nd | nd | - | - | ↓ | nd | nd |
| ID2−/− | nd | nd | nd | - | ↓ | - | ↓ | nd |
| E2-2−/− | nd | nd | nd | ↓ | - | - | nd | nd |
| Stat3−/− | nd | nd | nd | ↓ | ↓ | ↓ | - | - |
| Xbp1−/− | nd | nd | nd | ↓ | ↓ | ↓ | nd | nd |
| Batf3−/− | - | - | - | - | ↓ | - | ↓ | - |
| Mtg16−/− | ↓ | - | ↑ | ↓ | ↑ | - | nd | nd |
↓ decrease
↑ increase
- no change
nd not determined
3.3.3 Flt3-STAT3-PU.1 feedback loop controls DC development
PU.1 plays important role several hematopoietic lineages in a dose dependent manner. High dose of PU.1 is required for monocytes and DCs, whereas low dose of PU.1 is required for lymphoid lineage development (reviewed in [70] [71]). PU.1 expression is controlled by STAT3, downstream of many tyrosine kinase receptors, but a positive feedback loop between Flt3-STAT3-PU.1 is unique to the DC lineage (reviewed in [72]). Flt3 signaling activates STAT3, which drives PU.1 expression [39]; PU.1 protein binds the Flt3 promoter and regulates Flt3 expression in a dose-dependent manner. This positive feedback loop reinforces continuous Flt3 signaling, continuous expression and accumulation of PU.1 to a high concentration, which is required for DC development [73] (see below).
3.3.4 IRF, Batf and PU.1 in regulating DC subset specification
Two IRF family members, IRF8 and IRF4, regulate DC subset specification. Unlike other IRF family members, IRF4 and IRF8 have low affinity to interferon response elements, thus they must be recruited to Ets-IRF (EICE) or AP-1–IRF (AICE) composite motifs through interactions with PU.1 (an Est family protein) and BATF (AP-1), respectively [74]. Many immunocytes, including DCs, use PU.1 to recruit IRF4 and IRF8. As a result, PU.1 deficiency results in loss of DCs and abnormal development of multiple lineages [70].
IRF not only requires protein interaction with PU.1 to exert its DNA binding function, but it also requires PU.1 for its transcription. In early DC progenitors, PU.1 drives transcription of IRF8, which is required for establishing transcription identity of DC lineage. As mentioned above, a Flt3-STAT3-PU.1 positive feedback loop leads to accumulation of PU.1. High concentration of PU.1 is required for continuous expression of IRF8 through a mechanism involving a cell-specific enhancer and chromatin remodeling. A model has been proposed [75]: in MDPs, PU.1 at high concentration binds an enhancer at −50kb of transcription starting site (TSS) of Irf8, and loops the enhancer into physical proximity to Irf8 promoter to drive Irf8 transcription. Importantly, such chromatin remodeling is cell-stage specific, as it was observed only in MDPs, not in the upstream myeloid progenitors [75]. Continuous IRF8 expression is essential for maintaining DC transcriptional identity and drives differentiation from MDP to CDP [75, 76]. In IRF8−/− mice, the MDP population is increased, while CDPs are reduced dramatically [76]. Correspondingly, CD8+ DCs and pDCs are lacking in IRF8−/− mice, leading to the inference that IRF8 deficiency blocks the transition from MDP to CDP [77]. Similarly, IRF8 mutations in humans appear to block a transition between hGMDP to hMDP stage. As a result, monocytes and DCs fail to develop in these patients [78] [63].
As work from Murphy’s group has demonstrated, IRF8 works closely with BATF in specification of DC subsets [56, 79]. IRF8 expression starts in MDPs and remains high in CDPs. Apparent in the CDP stage, IRF8 uses auto-activation to regulate its own expression [56]. This is evident from dramatic reduction of IRF8 protein level in IRF8+/− CDPs, and from the fact that ectopically transduced IRF8 induces endogenous IRF8 expression. Interestingly, auto-activation of IRF8 becomes dependent on Batf3 in the stage of pre-cDCs and such dependence regulates subsequent differentiation to CD8+ cDCs. The major role of Batf3 is to maintain IRF8 expression for CD8+ DC differentiation [56]. BATF3 is only expressed in pre-CD8 DCs, not pre-CD4/CD11b DCs. BATF3 can form a heterodimer with IRF8 and maintain IRF8 expression through binding to a super-enhancer region in IRF8 locus, −40kb from the TSS. Importantly, this super-enhancer is also activated in a cell-specific manner, only in CD8+ DCs but not CD11b+ DCs. When Batf3 is deficient, IRF8 decays and pre-CD8 DCs fail to differentiate to CD8+ cDCs. Instead, they start to gain features of CD11b+ cDCs. In pre-CD4/CD11b DCs, absence of Batf3 is associated, and perhaps responsible, for the gradual loss of IRF8. During differentiation from pre-CD4/CD11b DCs to CD11b+ cDCs, IRF8 is lost and IRF4 expression starts. It is possible that in CD11b+ cDCs, IRF4 instead of IRF8 serves as the binding partner with PU.1 to turn on the DC transcriptional program, including expression of CD80, CD86 and CCR7 [80]. However, IRF4 is not completely redundant to IRF8; IRF4 ‘preferentially’ activates genes encoding accessory molecules required for efficient generation of peptide–MHC class II complexes, an activation achieved through binding of IRF4-PU.1 complex to Ets-IRF (EICE) site in the Ciita locus [80]. As a result, IRF4’s role in CD11b DC development cannot be replaced by IRF8.
Thus IRF8 appears to be an important transcription factor with “high maintenance”. Its continuous expression requires transcriptional regulation in cis and in trans, auto-activation, and modulation by different TFs and enhancers at different stages. Yet when its expression is established and sustained, it is powerful in driving the differentiation and specification of DCs.
3.3.5 E2-2 regulates pDC development
E2-2, a basic helix-loop-helix transcription factor, is the master regulator of pDCs. E2-2 simultaneously guides development toward pDCs and inhibits development of cDCs. E2-2 drives expression of pDC-specific genes, such as ILT7 and PACSINI [81]. E2-2 also represses genes marking cDC fate, such as ITGAX (CD11c) and ID2 [81, 82]. The combined up-regulation of E2-2 and low expression of E protein inhibitor, ID2, serve as a key fate determinants in pDC development. Suppression of Id2 depends on MTG16, an ETO family protein, which forms a complex with E2-2 and is subsequently recruited to Id2 locus, suppressing its expression [83]. Together, deficiency of E2-2 in hematopoietic stem cells blocks differentiation to pDCs, whereas deficiency of E2-2 or Mtg16 in mature pDCs prompts spontaneous differentiation of pDCs into cDC-like cells [82, 83]. Therefore, continuous expression of E2-2 is not only essential for commitment to the pDC lineage, but also required to maintain the lineage and prevent their spontaneous differentiation into cDC-like cells.
In summary, DC development not only employs multiple transcription regulators, but also takes advantage of their dose effect, lineage-specific- and cell-stage-specific expression as well as cell-stage-specific enhancers. Such multi-dimensional coordination allows the reuse of the same set of transcription factors to establish distinct lineage-specific programs..
4. Debates and open questions
The increasing knowledge of DC development also generates new questions and reveals significant debates that need to be resolved.
4.1 Does DC lineage start at LMPP, MDP or CDP?
The first question is: at which stage does the DC lineage start? MDPs in both mouse and humans have been shown to have lost granulocyte activity, thus representing a dedicated progenitor of MPS [51] [36] [63]. However, developmental potential of murine MDPs has been challenged by a recent study arguing that MDPs still possess granulocyte potential and have low potential to produce DCs, in particular, pDCs [84]. According to this study, the CDP but not the MDP is the formal DC progenitor. One other study by Naik et al. proposed a much earlier start of DC lineage [85]. By labeling MPPs or LMPPs with bar-coded viruses to trace the origin of single cells in vivo, Naik et al found that DCs and lymphoid cells or DC and myeloid cells rarely descend from the same LMPP progenitors, thus suggesting that DC lineage is imprinted and separated from lymphoid and myeloid lineages at the LMPP stage. However, whether such “imprinting” is a result of intrinsic commitment or is directed by external signaling is unclear. Thus whether the DC lineage is committed at the LMPP, MDP or CDP stage, and whether there is a difference between mice and humans must be clarified.
4.2 Pattern of progenitor-progeny relationship
The second issue that has arisen from identification of prospective progenitors is whether progenitor-progeny relationship is linear. Repeatedly it is found that one type of progeny can descend from different or even mutually exclusive progenitors. For example, indistinguishable DCs of all subsets descend from CMPs and CLPs that are mutually exclusive and lack linear relationship; although murine pre-pDCs are downstream of CDPs [55], which descend from Flt3+ CMPs [86], they can also directly descend from LMPPs after a single division [55]; similarly, murine cMOPs can descend from Flt3+ MDPs [52], but also can descend from Flt3− GMPs [87]. In addition, some studies argue that most DCs descend from myeloid progenitors [88], while others suggest that DCs descend from lymphoid primed progenitors [61]. These conflicts are difficult to resolve using the classic model, which emphasizes homogeneous progenitors, bifurcation and linear development.
4.3 Cellular stage of DC subset commitment
The third unresolved issue is that precursors committed to DC subsets are found within different progenitor populations. Although subset-committed pre-CD4/CD11b DCs and pre-CD8 DCs have been identified [56] [57], committed precursors of specific DC subsets appear to emerge before pre-cDC stage. Single cells that have committed to one DC subset can be identified in mouse CDP, and pre-pDC populations [57][86][55], as well as in human CDP, and even MDP, populations [63]. These single cell analyses reveal heterogeneity of progenitors and indicate that committed precursors of DC subsets can exist over several different progenitor stages. This phenomenon could result from two possibilities: first, development is a continuous process that transitions through sequential cellular stages [57], starting with early “imprinting” [85]. Alternatively, the final commitment can take place at different stages in an asynchronous manner, e.g., LMPP can directly produce pre-pDC within one cell cycle, by-passing the sequential MDP and CDP stages [55]. These two possibilities are not mutually exclusive. More experiments are required to differentiate and determine the contribution from each scenario.
5. Conclusion
In conclusion, work from the past two decades has tremendously improved our understanding of DC development. The DC lineage is comprised of sequential progenitors that proliferate in response to Flt3L, utilize multiple transcriptional factors and enhancers through temporal and spatial coordination to achieve increased commitment and subset specification. However, important questions remain to be resolved. Because the DC lineage development does not fit perfectly in the classic bifurcation model of hematopoiesis, a new model is in need to explain and reconcile seemingly conflicting data of DC lineage development, and guide further study of transcription regulation. In order to construct a new model of hematopoiesis and coherent DC-development scheme, progenitor-progeny relationship between lymphoid progenitors, myeloid progenitors, and DC progenitors should be revisited at a population and clonal level.
Highlights.
History of Dendritic cell discovery as a member of mononuclear phagocyte system.
Recent discoveries of dendritic cell development and its transcriptional regulation.
Debate and open questions about dendritic cell development.
Acknowledgments
This work is in supported by NYSTEM (to K.L.), and NIH grants AI101251 (to K.L.) and NS084776 (to S.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van Furth R, et al. Mononuclear phagocytic system: new classification of macrophages, monocytes and of their cell line. Bull World Health Organ. 1972;47(5):651–658. [PMC free article] [PubMed] [Google Scholar]
- 2.Unanue ER, Cerottini JC. The immunogenicity of antigen bound to the plasma membrane of macrophages. J Exp Med. 1970;131(4):711–725. doi: 10.1084/jem.131.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Unanue ER, Askonas BA. Persistence of immunogenicity of antigen after uptake by macrophages. J Exp Med. 1968;127(5):915–926. doi: 10.1084/jem.127.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishell RI, Dutton RW. Immunization of dissociated spleen cell cultures from normal mice. J Exp Med. 1967;126(3):423–442. doi: 10.1084/jem.126.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steinman RM, Cohn ZA. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J Exp Med. 1973;137(5):1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steinman RM, Witmer MD. Lymphoid dendritic cells are potent stimulators of the primary mixed leukocyte reaction in mice. Proc Natl Acad Sci U S A. 1978;75(10):5132–5136. doi: 10.1073/pnas.75.10.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young JW, Steinman RM. The hematopoietic development of dendritic cells: a distinct pathway for myeloid differentiation. Stem Cells. 1996;14(4):376–387. doi: 10.1002/stem.140376. [DOI] [PubMed] [Google Scholar]
- 8.Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med. 1994;179(4):1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inaba K, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176(6):1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randolph GJ, et al. Differentiation of phagocytic monocytes into lymph node dendritic cells in vivo. Immunity. 1999;11(6):753–761. doi: 10.1016/s1074-7613(00)80149-1. [DOI] [PubMed] [Google Scholar]
- 11.Cheong C, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143(3):416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guilliams M, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14(8):571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336(6077):86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 14.Gomez Perdiguero E, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518(7540):547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392(6673):245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 16.Briseno CG, Murphy TL, Murphy KM. Complementary diversification of dendritic cells and innate lymphoid cells. Curr Opin Immunol. 2014;29:69–78. doi: 10.1016/j.coi.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shortman K, Liu YJ. Mouse and human dendritic cell subtypes. Nat Rev Immunol. 2002;2(3):151–161. doi: 10.1038/nri746. [DOI] [PubMed] [Google Scholar]
- 18.Metlay JP, et al. The distinct leukocyte integrins of mouse spleen dendritic cells as identified with new hamster monoclonal antibodies. J Exp Med. 1990;171(5):1753–1771. doi: 10.1084/jem.171.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vremec D, et al. CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen. J Immunol. 2000;164(6):2978–2986. doi: 10.4049/jimmunol.164.6.2978. [DOI] [PubMed] [Google Scholar]
- 20.Grouard G, et al. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J Exp Med. 1997;185(6):1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Keeffe M, et al. Dendritic cell precursor populations of mouse blood: identification of the murine homologues of human blood plasmacytoid pre-DC2 and CD11c+ DC1 precursors. Blood. 2003;101(4):1453–1459. doi: 10.1182/blood-2002-03-0974. [DOI] [PubMed] [Google Scholar]
- 22.Liu YJ. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu Rev Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- 23.Wu L, Li CL, Shortman K. Thymic dendritic cell precursors: relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J Exp Med. 1996;184(3):903–911. doi: 10.1084/jem.184.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galy A, et al. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3(4):459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 25.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91(5):661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 26.Akashi K, et al. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404(6774):193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 27.Weissman IL. Translating stem and progenitor cell biology to the clinic: barriers and opportunities. Science. 2000;287(5457):1442–1446. doi: 10.1126/science.287.5457.1442. [DOI] [PubMed] [Google Scholar]
- 28.Traver D, et al. Development of CD8alpha-positive dendritic cells from a common myeloid progenitor. Science. 2000;290(5499):2152–2154. doi: 10.1126/science.290.5499.2152. [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa F, et al. The developmental program of human dendritic cells is operated independently of conventional myeloid and lymphoid pathways. Blood. 2007;110(10):3591–3660. doi: 10.1182/blood-2007-02-071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shigematsu H, et al. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21(1):43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 31.Matthews W, et al. A receptor tyrosine kinase specific to hematopoietic stem and progenitor cell-enriched populations. Cell. 1991;65(7):1143–1152. doi: 10.1016/0092-8674(91)90010-v. [DOI] [PubMed] [Google Scholar]
- 32.Lyman SD, et al. Molecular cloning of a ligand for the flt3/flk-2 tyrosine kinase receptor: a proliferative factor for primitive hematopoietic cells. Cell. 1993;75(6):1157–1167. doi: 10.1016/0092-8674(93)90325-k. [DOI] [PubMed] [Google Scholar]
- 33.Maraskovsky E, et al. Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J Exp Med. 1996;184(5):1953–1962. doi: 10.1084/jem.184.5.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naik SH, et al. Cutting edge: generation of splenic CD8+ and CD8− dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J Immunol. 2005;174(11):6592–6597. doi: 10.4049/jimmunol.174.11.6592. [DOI] [PubMed] [Google Scholar]
- 35.McKenna HJ, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95(11):3489–3497. [PubMed] [Google Scholar]
- 36.Waskow C, et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol. 2008;9(6):676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karsunky H, et al. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med. 2003;198(2):305–313. doi: 10.1084/jem.20030323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Amico A, Wu L. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt3. J Exp Med. 2003;198(2):293–303. doi: 10.1084/jem.20030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Onai N, et al. Flt3 in regulation of type I interferon-producing cell and dendritic cell development. Ann N Y Acad Sci. 2007;1106:253–261. doi: 10.1196/annals.1392.015. [DOI] [PubMed] [Google Scholar]
- 40.Burgess AW, Camakaris J, Metcalf D. Purification and properties of colony-stimulating factor from mouse lung-conditioned medium. J Biol Chem. 1977;252(6):1998–2003. [PubMed] [Google Scholar]
- 41.Sheridan JW, Metcalf D. A low molecular weight factor in lung-conditioned medium stimulating granulocyte and monocyte colony formation in vitro. J Cell Physiol. 1973;81(1):11–23. doi: 10.1002/jcp.1040810103. [DOI] [PubMed] [Google Scholar]
- 42.Caux C, et al. CD34+ hematopoietic progenitors from human cord blood differentiate along two independent dendritic cell pathways in response to GM-CSF+ TNF alpha. J Exp Med. 1996;184(2):695–706. doi: 10.1084/jem.184.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Witmer-Pack MD, et al. Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J Cell Sci. 1993;104(Pt 4):1021–1029. doi: 10.1242/jcs.104.4.1021. [DOI] [PubMed] [Google Scholar]
- 44.Greter M, et al. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 2012;36(6):1031–1046. doi: 10.1016/j.immuni.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayer CT, et al. Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow. Blood. 2014;124(20):3081–3091. doi: 10.1182/blood-2013-12-545772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helft J, et al. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity. 2015;42(6):1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 47.Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nat Rev Cancer. 2012;12(4):265–277. doi: 10.1038/nrc3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu K, et al. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat Immunol. 2007;8(6):578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- 49.Liu K, Nussenzweig MC. Development and homeostasis of dendritic cells. Eur J Immunol. 2010;40(8):2099–2102. doi: 10.1002/eji.201040501. [DOI] [PubMed] [Google Scholar]
- 50.Kabashima K, et al. Intrinsic lymphotoxin-beta receptor requirement for homeostasis of lymphoid tissue dendritic cells. Immunity. 2005;22(4):439–450. doi: 10.1016/j.immuni.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Fogg DK, et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311(5757):83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- 52.Hettinger J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013;14(8):821–830. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- 53.Liu K, et al. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324(5925):392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naik SH, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8(11):1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 55.Onai N, et al. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity. 2013;38(5):943–957. doi: 10.1016/j.immuni.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 56.Grajales-Reyes GE, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat Immunol. 2015;16(7):708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlitzer A, et al. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat Immunol. 2015;16(7):718–728. doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- 58.Liu K, Nussenzweig MC. Origin and development of dendritic cells. Immunol Rev. 2010;234(1):45–54. doi: 10.1111/j.0105-2896.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- 59.Manz MG, et al. Prospective isolation of human clonogenic common myeloid progenitors. Proc Natl Acad Sci U S A. 2002;99(18):11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chicha L, Jarrossay D, Manz MG. Clonal type I interferon-producing and dendritic cell precursors are contained in both human lymphoid and myeloid progenitor populations. J Exp Med. 2004;200(11):1519–1524. doi: 10.1084/jem.20040809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11(7):585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 62.Lee J, et al. Clonal analysis of human dendritic cell progenitor using a stromal cell culture. J Immunol Methods. 2015 doi: 10.1016/j.jim.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J, et al. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J Exp Med. 2015;212(3):385–399. doi: 10.1084/jem.20141442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Breton G, et al. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J Exp Med. 2015;212(3):401–413. doi: 10.1084/jem.20141441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Orkin SH, Zon LI. Hematopoiesis and stem cells: plasticity versus developmental heterogeneity. Nat Immunol. 2002;3(4):323–328. doi: 10.1038/ni0402-323. [DOI] [PubMed] [Google Scholar]
- 66.Robbins SH, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008;9(1):R17. doi: 10.1186/gb-2008-9-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller JC, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13(9):888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heng TS, Painter MW C. Immunological Genome Project. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 69.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13(11):1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carotta S, Wu L, Nutt SL. Surprising new roles for PU.1 in the adaptive immune response. Immunol Rev. 2010;238(1):63–75. doi: 10.1111/j.1600-065X.2010.00955.x. [DOI] [PubMed] [Google Scholar]
- 71.Belz GT, Nutt SL. Transcriptional programming of the dendritic cell network. Nat Rev Immunol. 2012;12(2):101–113. doi: 10.1038/nri3149. [DOI] [PubMed] [Google Scholar]
- 72.Merad M. PU.1 takes control of the dendritic cell lineage. Immunity. 2010;32(5):583–585. doi: 10.1016/j.immuni.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 73.Carotta S, et al. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32(5):628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 74.Glasmacher E, et al. A genomic regulatory element that directs assembly and function of immune-specific AP-1-IRF complexes. Science. 2012;338(6109):975–980. doi: 10.1126/science.1228309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schonheit J, et al. PU.1 level-directed chromatin structure remodeling at the Irf8 gene drives dendritic cell commitment. Cell Rep. 2013;3(5):1617–1628. doi: 10.1016/j.celrep.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 76.Becker AM, et al. IRF-8 extinguishes neutrophil production and promotes dendritic cell lineage commitment in both myeloid and lymphoid mouse progenitors. Blood. 2012;119(9):2003–2012. doi: 10.1182/blood-2011-06-364976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aliberti J, et al. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 2003;101(1):305–310. doi: 10.1182/blood-2002-04-1088. [DOI] [PubMed] [Google Scholar]
- 78.Hambleton S, et al. IRF8 mutations and human dendritic-cell immunodeficiency. N Engl J Med. 2011;365(2):127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murphy KM. Transcriptional control of dendritic cell development. Adv Immunol. 2013;120:239–267. doi: 10.1016/B978-0-12-417028-5.00009-0. [DOI] [PubMed] [Google Scholar]
- 80.Vander Lugt B, et al. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat Immunol. 2014;15(2):161–167. doi: 10.1038/ni.2795. [DOI] [PubMed] [Google Scholar]
- 81.Cisse B, et al. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135(1):37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ghosh HS, et al. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33(6):905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghosh HS, et al. ETO family protein Mtg16 regulates the balance of dendritic cell subsets by repressing Id2. J Exp Med. 2014;211(8):1623–1635. doi: 10.1084/jem.20132121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sathe P, et al. Lymphoid tissue and plasmacytoid dendritic cells and macrophages do not share a common macrophage-dendritic cell-restricted progenitor. Immunity. 2014;41(1):104–115. doi: 10.1016/j.immuni.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 85.Naik SH, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496(7444):229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 86.Onai N, et al. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat Immunol. 2007;8(11):1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- 87.Yanez A, et al. IRF8 acts in lineage-committed rather than oligopotent progenitors to control neutrophil vs monocyte production. Blood. 2015;125(9):1452–1459. doi: 10.1182/blood-2014-09-600833. [DOI] [PubMed] [Google Scholar]
- 88.Manz MG, et al. Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood. 2001;97(11):3333–3341. doi: 10.1182/blood.v97.11.3333. [DOI] [PubMed] [Google Scholar]