Abstract
From January 2014 to December 2014, 972 consecutive non-replicate carbapenemase-producing Enterobacteriaceae isolates from colonised or infected patients were collected at the Associated French National Reference Centre as part of the French national survey on antimicrobial resistance. It included 577 Klebsiella spp. (59%), 236 Escherichia coli (24%), 108 Enterobacter spp. (11%), 50 Citrobacter spp. (5%), and a single Salmonella spp. isolate (0.1%). Of 561 K. pneumoniae isolates, 35 were found to be resistant to colistin (6.2%). PFGE analysis revealed a clonal outbreak involving 15 K. pneumoniae isolates belonging to sequence type ST11, recovered in a single hospital in the Picardie region in northern France. Those clonally related isolates showed variable levels of resistance to colistin, ranging from 4 to 64 mg/L. They harboured the blaOXA-48 carbapenemase gene and the blaCTX-M-15 extended-spectrum beta-lactamase gene. Among the 91 Enterobacter cloacae isolates, seven were resistant to colistin and produced different types of carbapenemases. Surprisingly, none of the E. coli and Citrobacter spp. isolates showed resistance to colistin. This national survey including carbapenemase-producing isolates recovered in 2014 reported a high rate of colistin resistance in K. pneumoniae and E. cloacae (6.2% and 7.7%, respectively) in France.
Keywords: colistin, polymyxins, resistance, Klebsiella pneumoniae, Enterobacter cloacae, outbreak
Introduction
Carbapenemase-producing Enterobacteriaceae (CPE) resistant to colistin are increasingly reported. They represent an additional link in the development of pan-drug resistance. However, the epidemiology of colistin resistance among enterobacterial isolates is currently almost unknown in most parts of the world. In Italy, an increase in carbapenemase-producing Enterobacteriaceae has been noted in the past years, but the situation remains unknown in France [1]. The lack of information about the prevalence of colistin resistance among multidrug-resistant enterobacterial isolates derives from several reasons: (i) so far, there has been limited interest in that field, (ii) methods used for determination of colistin susceptibility are not adequate, and (iii) the lack of well-defined breakpoints does not allow precise determination of prevalence. However, the recent identification of a plasmid-borne polymyxin resistance determinant (MCR-1) raised a very serious concern in that resistance to colistin might widely disseminate [2].
The aim of this study was to evaluate retrospectively the prevalence of colistin resistance among a collection of CPE strains recovered in France during a period of one year and to analyse the phenotypic, genotypic features and clonality of the colistin-resistant isolates.
Methods
Carbapenemase-producing Enterobacteriaceae isolates
From January to December 2014, 972 consecutive non-duplicate isolates of carbapenemase-producing Enterobacteriaceae were isolated in private laboratories and hospitals in France either by screening for colonisation or by analysing clinical samples in the context of infections. They were recovered from rectal swabs or stools (n = 625), urine samples (n = 250), respiratory tract samples (n = 35), blood samples (n = 22), wounds (n = 24), catheter (n = 7), vaginal swabs (n = 3) and other specimens (n = 6). Those isolates were sent to the Associated French National Reference Centre for characterisation of resistance mechanisms to carbapenems as part of the French antibiotic resistance survey. The 972 carbapenemase-producing Enterobacteriaceae isolates included 577 isolates of Klebsiella spp. (59%), 236 isolates of Escherichia coli (24%), 108 isolates of Enterobacter spp. (11%), 50 isolates of Citrobacter spp. (5%), and a single isolate of Salmonella spp. (0.1%). Species that are naturally resistant to colistin (Proteus spp., Morganella morganii, Providencia spp., and Serratia spp.) had been excluded before the initiation of this study. Only a single isolate per patient was included in the study. All isolates were identified using the Microflex bench-top MALDI-TOF mass spectrometer (Bruker, Champs-sur-Marne, France).
Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MIC) of colistin (CS) were determined using broth microdilution method according to the guidelines of the Clinical Laboratory Standards Institute (CLSI) [3]. As recommended, E. coli ATCC 25922 was used as quality control strain.
For the colistin-resistant isolates, susceptibility to other classes of antibiotics was also tested. Susceptibility to imipenem, ertapenem, and tigecycline was tested by broth microdilution method according to CLSI guidelines, whereas susceptibility to the other antibiotics was tested by the standardised agar disk diffusion method according to the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [4]. The antibiotics tested using disk diffusion method were: amoxicillin (AMX), amoxicillin/clavulanic acid (AMC), cefotaxime (CTX), cefoxitin (FOX), ceftazidime (CAZ), cefepime (FEP), temocillin (TEM), ciprofloxacin (CIP), gentamicin (GM), amikacin (AK), trimethoprim-sulfamethoxazole (SXT) and fosfomycin (FOS).
The MIC results for colistin and the disk diffusion diameters were interpreted according to susceptibility breakpoints of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [4].
Molecular characterisation
The mgrB genes of K. pneumoniae and Enterobacter spp. isolates were amplified using specific primers (Table 1), knowing that the MgrB protein is a negative regulator of the PhoPQ two-component system and that alterations in the mgrB gene are commonly involved in acquisition of colistin resistance in K. pneumoniae [5-7]. The plasmid-mediated mcr-1 gene encoding colistin resistance was sought as described previously [2]. Detection of extended-spectrum beta-lactamases (ESBL) and carbapenemases genes was performed with specific primers as described previously [8]. Both strands of the amplification products obtained were sequenced with an ABI 3100 sequencer (Applied Biosystems, Foster City, US). The nucleotide and deduced protein sequences were analysed at the National Centre for Biotechnology Information website (www.ncbi.nlm.nih.gov) by the Basic Local Alignment Search Tool (BLAST) programme.
Table 1. Oligonucleotides used as primers in this study, France, January–December 2014 .
| Oligonucleotides | Sequence (5’-3’) | Reference |
|---|---|---|
| Kpn mgrB ext F | TTA AGA AGG CCG TGC TAT CC | [7] |
| Kpn mgrB ext R | AAG GCG TTC ATT CTA CCA CC | [7] |
| Kpn mgrB int F | CGG TGG GTT TTA CTG ATA GTC | This study |
| Kpn mgrB int R | GAA CAT CCT GGT CGC ACA TT | This study |
| Ent mgrB ext F | CGG TTT ACT CTA TGA AAC AAG TGC | This study |
| Ent mgrB ext R | GCG AAG GAA GGA AAT CAC CT | This study |
Genotyping
Genotyping was performed to evaluate the clonal relationship of the colistin-resistant K. pneumoniae and E. cloacae isolates by pulsed-field gel elctrophoresis (PFGE) with XbaI-digested genomic DNA and interpreted according to Tenover criteria [9]. Multilocus sequence typing (MLST) for K. pneumoniae was performed using the simplified protocol at the Institut Pasteur website (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html) [10].
Results
Klebsiella pneumoniae
Of 561 K. pneumoniae isolates, 35 were found to be resistant to colistin (6%). Fifteen of the 35 colistin-resistant K. pneumoniae isolates were recovered from a single hospital in the Picardie region, northern France (Figure 1). We could not obtain the exact dates of their isolations due to the retrospective nature of the study. These isolates had mostly been recovered from rectal swab specimens, but also from a catheter, a urinary sample, a wound exudate and a respiratory specimen (isolates 1 to 15, Table 2). PFGE analysis revealed that the 15 isolates were clonally related (Figure 2, Table 2). The clone was of the ST11 type, and was susceptible only to cefoxitin, amikacin and fosfomycin (Table 2). A single isolate among these 15 was susceptible to tigecycline. The 15 isolates harboured both the blaOXA-48 carbapenemase gene, and the blaCTX-M-15 extended-spectrum beta-lactamase (ESBL) gene, and the MICs for colistin ranged from 4 to 64 mg/L (Table 2).
Figure 1.
Geographic distribution of colistin-resistant Enterobacteriaceae isolates, France, January–December 2014 (n = 43)
Easb: Enterobacter asburiae; Ecl: Enterobacter cloacae; Kpn: Klebsiella pneumoniae.
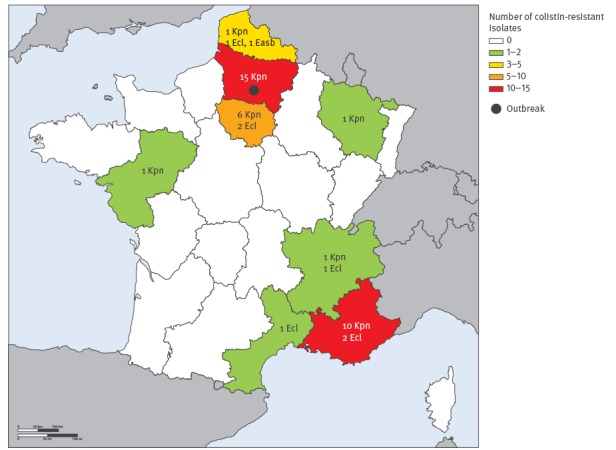
Table 2. Characteristics of the colistin-resistant Klebsiella pneumoniae and Enterobacter spp. clinical isolates, France, January–December 2014 (n = 43) .
| Isolate | Site of isolation |
Origin | MIC CSa | mgrB genotype | Carbapenemase | Associated beta-lactamase | Co-resistancesb | ST | PFGE |
|---|---|---|---|---|---|---|---|---|---|
| 1 | ? | Picardie | 32 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT | 11 | A |
| 2 | Catheter | Picardie | 8 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 3 | Rectal swab | Picardie | 4 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 4 | Rectal swab | Picardie | 4 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 5 | Rectal swab | Picardie | 64 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 6 | Urine | Picardie | 32 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 7 | Rectal swab | Picardie | 64 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 8 | Wound | Picardie | 4 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 9 | Respiratory | Picardie | 4 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 10 | Rectal swab | Picardie | 4 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 11 | Rectal swab | Picardie | 8 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 12 | Rectal swab | Picardie | 64 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 13 | Rectal swab | Picardie | 8 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 14 | Rectal swab | Picardie | 4 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 15 | Rectal swab | Picardie | 4 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 11 | A |
| 16 | Rectal swab | Nord-Pas-de-Calais | 128 | IS1R in promoter region (between nt −45 and −46) | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 147 | B |
| 17 | Rectal swab | Ile-de-France | 128 | mgrB WT | NDM | CTX-M-15 | CIP GM SXT TIG | 147 | C |
| 18 | Rectal swab | PACA | >128 | ISKpn26-like in coding region (between nt +74 and +75) | KPC | - | CIP AK SXT TIG | 258 | D |
| 19 | Rectal swab | Rhône-Alpes | 128 | MgrB truncated (27 amino acids) | KPC | - | CIP AK SXT TIG | 258 | E |
| 20 | Blood | PACA | 16 | Full gene deletionc | KPC | - | CIP AK SXT TIG | 258 | F |
| 21 | Rectal swab | Lorraine | 64 | Single nucleotide deletion (nt 74) | OXA-48 | CTX-M-15 | CIP GM | 101 | G |
| 22 | Rectal swab | Ile-de-France | 32 | Single nucleotide deletion (nt 23) | OXA-48 | CTX-M-15 | CIP GM | 101 | H |
| 23 | Rectal swab | Ile-de-France | 64 | IS1R in promoter region (between nt −36 and −37) | OXA-48 + NDM | CTX-M-15 | CIP GM | 101 | I |
| 24 | Urine | PACA | 64 | IS1R in promoter region (between nt −45 and −46) | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 101 | J |
| 25 | Abcess | Ile-de-France | 32 | MgrB M27K | OXA-48 | CTX-M-15 | CIP GM | 101 | K |
| 26 | Urine | Pays de la Loire | 128 | Duplication of 19 nucleotides | OXA-48 | CTX-M-15 | CIP SXT FOS | 101 | L |
| 27 | Urine | PACA | 64 | IS1R in coding region (between nt +21 and +22) | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 307 | M |
| 28 | Rectal swab | PACA | 64 | mgrB WT | OXA-48 | CTX-M-15 | CIP GM SXT TIG FOS | 307 | M |
| 29 | Rectal swab | PACA | 64 | IS5-like in coding region (between nt +74 and +75) | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 307 | M |
| 30 | Urine | PACA | >128 | Full gene deletionb | OXA-48 | - | CIP GM AK SXT TIG FOS | 611 | N |
| 31 | Rectal swab | Ile-de-France | 32 | ISKpn14-like in promoter region (between nt −45 and −46) | OXA-48 | - | CIP GM SXT TIG | 23 | O |
| 32 | Rectal swab | PACA | 32 | IS102-like in coding region (between nt +36 and +37) | OXA-48 | CTX-M-15 | CIP GM SXT TIG | 20 | P |
| 33 | Respiratory | Ile-de-France | 32 | IS1R in promoter region (between nt −61 and −62) | OXA-48 | CTX-M-15 | CIP SXT TIG FOS | Q | |
| 34 | Blood | PACA | >128 | mgrB WT | OXA-48 | CTX-M-15 | CIP SXT TIG | 39 | R |
| 35 | Rectal swab | PACA | 32 | MgrB truncated (32 amino acids) | OXA-48 | - | FOS | 13 | S |
| 36 | Rectal swab | Nord-Pas-de-Calais | 64 | mgrB WT | OXA-48 + VIM | CTX-M-15 | CIP GM SXT | NA | T |
| 37 | Stools | Languedoc-Roussillon | 64 | mgrB WT | OXA-48 | CTX-M-15 | GM AK | NA | U |
| 38 | Respiratory | Ile-de-France | 32 | mgrB WT | VIM | - | SXT TIG | NA | V |
| 39 | Rectal swab | PACA | >128 | mgrB WT | OXA-48 | - | CIP GM SXT TIG | NA | W |
| 40 | Rectal swab | Ile-de-France | 16 | mgrB WT | OXA-48 | - | FOS | NA | X |
| 41 | Rectal swab | PACA | >128 | mgrB WT | IMP | CTX-M-2 | No | NA | Y |
| 42 | Respiratory | Rhône-Alpes | >128 | mgrB WT | OXA-48 | - | TIG | NA | Z |
| 43 | Urine | Nord-Pas-de-Calais | >128 | mgrB WT | VIM-1 | - | CIP TIG | NA | α |
Isolates 1-35: Klebsiella pneumoniae; 36–42: Enterobacter cloacae; 43: Enterobacter asburiae.
AK: amikacin; CIP: ciprofloxacin; CS: colistin; FOS: fosfomycin; GM: gentamicin; MIC: minimum inhibitory concentration; NA: not applicable; nt: nucleotide; PACA: Provence-Alpes-Côte-d’Azur; PFGE: pulsed-field gel electrophoresis; ST: sequence type; SXT: trimethoprim-sulfamethoxazole; TIG: tigecycline; WT: wildtype.
a MIC of colistin determined by broth microdilution method.
b Resistant or intermediate susceptibility to antibiotic.
c Full gene deletion: no PCR product was detected with external or internal primers.
Figure 2.
PFGE patterns of XbaI-digested chromosomal DNA of colistin-resistant Klebsiella pneumoniae isolates, France, January–December 2014 (n = 35)
The numbers correspond to the isolates from Table 2 and the letters indicate the PFGE type.
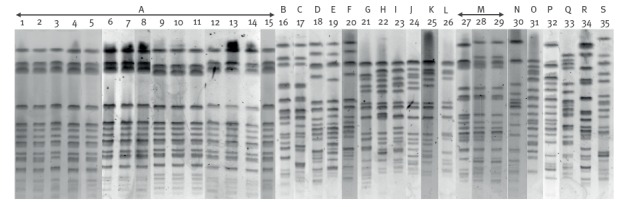
The other 20 colistin-resistant K. pneumoniae strains were mostly recovered from the regions Ile-de-France (n = 6) and Provence-Alpes-Côte d’Azur (n = 10) (Figure 1). These strains presented high MIC values for colistin ranging from 16 to >128 mg/L (isolates 16 to 35, Table 2). They produced either the carbapenemases OXA-48 (15/20), KPC-2 (3/20), NDM-1 (1/20), or both OXA-48 and NDM-1 together (1/20) (Table 2). Overall, 14 of the 20 isolates produced the ESBL CTX-M-15. PFGE analysis identified 18 clonal patterns among the 20 isolates (n = 3 for clone M) (Figure 2, Table 2), and MLST assigned the isolates to eight sequence types (STs) (Table 2).
Sequencing of the mgrB gene of those K. pneumoniae isolates revealed various mgrB alterations and none of the strains harboured the plasmid-encoded mcr-1 gene.
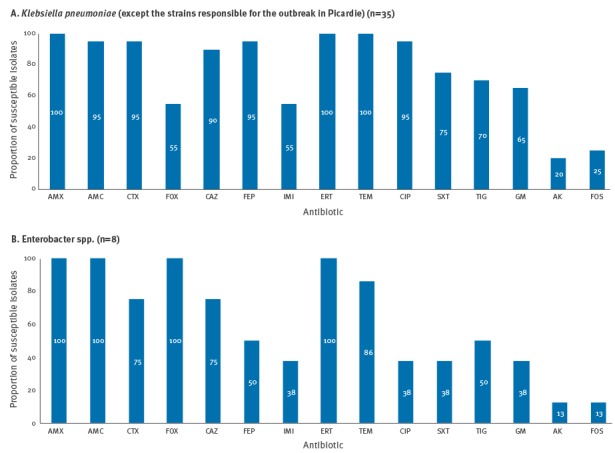
Antimicrobial susceptibility data for the colistin-resistant K. pneumoniae isolates not involved in the outbreak revealed that most isolates (19/20) were non-susceptible to third- and fourth-generation cephalosporins (Figure 3A). They were also frequently resistant to ciprofloxacin (19/20), trimethoprim-sulfamethoxazole (15/20) and tigecycline (14/20). They were less often resistant to gentamicin and cefoxitin (13/20 and 11/20, respectively). Amikacin and fosfomycin remained the most active agents against colistin-resistant K. pneumoniae (16/20 and 15/20 were susceptible, respectively) (Figure 3A).
Figure 3.
Antimicrobial non-susceptibilities among colistin-resistant isolates, France, January–December 2014
AK: amikacin; AMC: amoxicillin/clavulanic acid; AMX: amoxicillin; CAZ: ceftazidime; CIP: ciprofloxacin; CTX: cefotaxime; ERT: ertapenem; FEP: cefepime; FOS: fosfomycin; FOX: cefoxitin; GM: gentamicin; IMI: imipenem; SXT: trimethoprim-sulfamethoxazole; TEM: temocillin; TIG: tigecycline.
Enterobacter spp.
Among the 91 Enterobacter cloacae isolates, seven were resistant to colistin (7.7%). They showed high MIC values for colistin (ranging from 16 to >128 mg/L) (isolates 36 to 42, Table 2). They produced the carbapenemases OXA-48 (4/7), VIM-1 (1/7), IMP-1 (1/7), or both OXA-48 and VIM-1 together (1/7) (Table 2). In total, three of seven strains were CTX-M producers, with two isolates producing CTX-M-15 and a single isolate producing CTX-M-2. The colistin-resistant E. cloacae isolates were recovered in different geographical regions in France (Figure 1, Table 2), and results of the PFGE analysis revealed that they were not clonally related (data not shown).
The single carbapenem-resistant E. asburiae strain was resistant to colistin. It had an MIC of colistin above 128 mg/L and produced the VIM-1 carbapenemase (isolate 43, Table 2).
All Enterobacter spp. isolates had a wild-type mgrB gene, leaving unexplained the colistin resistance mechanism (E. cloacae and E. asburiae) (Table 2).
Of the eight colistin-resistant Enterobacter spp. isolates, four were non-susceptible to cefepime and tigecycline, and three were non-susceptible to ciprofloxacin, trimethoprim-sulfamethoxazole and gentamicin (Figure 3B). Amikacin and fosfomycin were the most active agents against colistin-resistant E. cloacae (all seven isolates were susceptible) (Figure 3B).
Other species
None of the E. coli (n = 236) and Citrobacter spp. (n = 50) isolates were resistant to colistin.
Discussion
We describe here a clonal outbreak involving 15 K. pneumoniae isolates recovered from a single hospital in the Picardie region in northern France. This outbreak was caused by a colistin-resistant OXA-48 and CTX-M-15-producing K. pneumoniae of ST11 type that was susceptible only to cefoxitin, amikacin and fosfomycin. Surprisingly, those clonally related isolates had variable MIC values for colistin ranging from 4 to 64 mg/L. An ST11 clone co-producing OXA-48 and CTX-M-15 was responsible for a large outbreak involving 44 patients in a hospital in Madrid, Spain, from 2009 to 2014 but only 3.4% of the isolates were resistant to colistin [11].
Several outbreaks of colistin-resistant KPC-producing K. pneumoniae (mainly attributed to the international epidemic clone type ST258) have been reported across Europe, in Greece [12,13], Hungary [14], Italy [15-17] and the Netherlands [18]. A single outbreak of colistin-resistant VIM-1-producing K. pneumoniae has also been described in Spain [19].
We report also 20 colistin-resistant K. pneumoniae strains recovered from the regions Ile-de-France and Provence-Alpes-Côte d’Azur. These strains belonged to 10 sequence types (n = 2 ST147, n = 3 ST258, n = 6 ST101, n = 3 ST307) and PFGE analysis identified 18 patterns among the 20 isolates. All three KPC-producing K. pneumoniae isolates belonged to ST258, the most common clone for KPC-producing isolates [20]. The OXA-48-producing K. pneumoniae isolates belonged to nine sequence types with six strains that were ST101, the most common clone identified among OXA-48-positive K. pneumoniae [21].
Sequencing of the mgrB gene revealed mgrB alterations which are likely to be responsible for colistin resistance as described previously [5-7]. Interestingly, the three strains belonging to the single clone M recovered in the Provence-Alpes Côtes-d’Azur region had different mechanisms of mgrB inactivation (Table 2). The occurrence of such different mechanisms of colistin resistance among clonally related isolates indicates that it is not the product of clonal dissemination of a single colistin-resistant K. pneumoniae strain, but rather clonal dissemination of a carbapenemase-producing isolate, which has acquired colistin resistance thereafter.
The rates of colistin resistance among the carbapenemase-producing isolates were 7.7% for Enterobacter spp. and 3.6% for K. pneumoniae isolates (excluding the isolates responsible for the outbreak in the Picardie region). The resistance rate observed among the carbapenemase-producing K. pneumoniae isolates was much lower than the high rates reported in the neighbouring countries of southern Europe such as Spain (20%) [19] and Italy (43%) [1].
None of the 236 carbapenemase-producing E. coli isolates were colistin-resistant or carried the mcr-1 gene. This is surprising considering that a recent report of the French antimicrobial resistance Resapath surveillance network identified the plasmid-borne mcr-1 gene in 21% of ESBL-producing E. coli isolates recovered from faeces of veal calves in France between 2005 and mid-2014 [22]. The plasmid-borne mcr-1 colistin resistance gene has also been found in many neighbouring countries of France, for example among ESBL-producing Enterobacteriaceae isolates recovered from river water and imported vegetable samples in Switzerland [23], in E. coli isolates recovered from calves and piglets in Belgium [24], in swine and human wound infections in Germany [25], and in food and human bloodstream infections in Denmark [26]. The mcr-1 gene was also detected in Salmonella enterica from food samples in Portugal [27] and France [28]. An E. coli isolate co-harbouring the blaVIM-1 carbapenemase gene and the mcr-1 gene was described in Switzerland [29] and an isolate co-producing NDM-9 and MCR-1 was reported from China [30]. We believe that the plasmid carrying the mcr-1 gene might be currently more prevalent among ESBL-producing isolates than among carbapenemase-producing isolates in human samples, which would explain why we did not identify this gene in our collection of carbapenemase-producing isolates.
Amikacin and fosfomycin were most effective against the colistin and carbapenem-resistant K. pneumoniae (susceptibility rates of 80% and 75%, respectively) and E. cloacae isolates (susceptibility rates of 87%). The rate of tigecycline non-susceptibility was high (70% for K. pneumoniae and 50% for Enterobacter spp.), probably because of a strong selective pressure by this last-line antibiotic.
Conclusion
This national survey on carbapenemase-producing isolates recovered in 2014 discovered a high rate of colistin resistance in K. pneumoniae and E. cloacae (6.2% and 7.7%, respectively) in France. These resistance rates remain much lower than those observed in other European countries such as Greece, Italy and Spain. No plasmid-encoded mcr-1 gene was identified here. Therefore it seems that it is still possible to control the spread of those multidrug-resistant isolates based on accurate identification of colistin resistance and isolation of plasmid-encoded MCR-1 producers. Amikacin and fosfomycin remained the antibiotic agents most effective against those isolates which were resistant to polymyxins and produced a carbapenemase.
Acknowledgements
This work was supported by the University of Fribourg and partially funded by the Institut de Veille Sanitaire (InVS).
Conflict of interest: None declared.
Authors’ contributions: AJ, LP, and PN contributed to the design of the study. AJ performed the experiments. AJ, LP, and PN analysed the data. AJ, LP, LD, and PN contributed to the writing of the manuscript.
References
- 1. Monaco M, Giani T, Raffone M, Arena F, Garcia-Fernandez A, Pollini S, et al. Colistin resistance superimposed to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly evolving problem in Italy, November 2013 to April 2014. Euro Surveill. 2014;19(42). 10.2807/1560-7917.ES2014.19.42.20939 [DOI] [PubMed] [Google Scholar]
- 2. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute (CLSI). Methods for dilution of antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 9th ed. CLSI document M07-A10. Wayne: CLSI; 2015. [Google Scholar]
- 4.European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoints tables for interpretation of MICs and zone diameters, Version 5.0. Växjö: EUCAST. 2015. [Google Scholar]
- 5. Cannatelli A, Giani T, D’Andrea MM, Di Pilato V, Arena F, Conte V, et al. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. 2014;58(10):5696-703. 10.1128/AAC.03110-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng YH, Lin TL, Pan YJ, Wang YP, Lin YT, Wang JT. Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob Agents Chemother. 2015;59(5):2909-13. 10.1128/AAC.04763-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Türkoglu S, et al. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70(1):75-80. 10.1093/jac/dku323 [DOI] [PubMed] [Google Scholar]
- 8. Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791-8. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brañas P, Villa J, Viedma E, Mingorance J, Orellana MA, Chaves F. Molecular epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital in Madrid: Successful establishment of an OXA-48 ST11 clone. Int J Antimicrob Agents. 2015;46(1):111-6. 10.1016/j.ijantimicag.2015.02.019 [DOI] [PubMed] [Google Scholar]
- 12. Antoniadou A, Kontopidou F, Poulakou G, Koratzanis E, Galani I, Papadomichelakis E, et al. Colistin-resistant isolates of Klebsiella pneumoniae emerging in intensive care unit patients: first report of a multiclonal cluster. J Antimicrob Chemother. 2007;59(4):786-90. 10.1093/jac/dkl562 [DOI] [PubMed] [Google Scholar]
- 13. Kontopoulou K, Protonotariou E, Vasilakos K, Kriti M, Koteli A, Antoniadou E, et al. Hospital outbreak caused by Klebsiella pneumoniae producing KPC-2 beta-lactamase resistant to colistin. J Hosp Infect. 2010;76(1):70-3. 10.1016/j.jhin.2010.03.021 [DOI] [PubMed] [Google Scholar]
- 14. Tóth A, Damjanova I, Puskás E, Jánvári L, Farkas M, Dobák A, et al. Emergence of a colistin-resistant KPC-2-producing Klebsiella pneumoniae ST258 clone in Hungary. Eur J Clin Microbiol Infect Dis. 2010;29(7):765-9. 10.1007/s10096-010-0921-3 [DOI] [PubMed] [Google Scholar]
- 15. Giani T, Arena F, Vaggelli G, Conte V, Chiarelli A, Henrici De Angelis L, et al. Large nosocomial outbreak of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae traced to clonal expansion of an mgrB deletion mutant. J Clin Microbiol. 2015;53(10):3341-4. 10.1128/JCM.01017-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mammina C, Bonura C, Di Bernardo F, Aleo A, Fasciana T, Sodano C, et al. Ongoing spread of colistin-resistant Klebsiella pneumoniae in different wards of an acute general hospital, Italy, June to December 2011. Euro Surveill. 2012;17(33):17. [PubMed] [Google Scholar]
- 17. Mezzatesta ML, Gona F, Caio C, Petrolito V, Sciortino D, Sciacca A, et al. Outbreak of KPC-3-producing, and colistin-resistant, Klebsiella pneumoniae infections in two Sicilian hospitals. Clin Microbiol Infect. 2011;17(9):1444-7. 10.1111/j.1469-0691.2011.03572.x [DOI] [PubMed] [Google Scholar]
- 18. Weterings V, Zhou K, Rossen JW, van Stenis D, Thewessen E, Kluytmans J, et al. An outbreak of colistin-resistant Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae in the Netherlands (July to December 2013), with inter-institutional spread. Eur J Clin Microbiol Infect Dis. 2015;34(8):1647-55. 10.1007/s10096-015-2401-2 [DOI] [PubMed] [Google Scholar]
- 19. Pena I, Picazo JJ, Rodríguez-Avial C, Rodríguez-Avial I. Carbapenemase-producing Enterobacteriaceae in a tertiary hospital in Madrid, Spain: high percentage of colistin resistance among VIM-1-producing Klebsiella pneumoniae ST11 isolates. Int J Antimicrob Agents. 2014;43(5):460-4. 10.1016/j.ijantimicag.2014.01.021 [DOI] [PubMed] [Google Scholar]
- 20. Nordmann P. Carbapenemase-producing Enterobacteriaceae: overview of a major public health challenge. Med Mal Infect. 2014;44(2):51-6. 10.1016/j.medmal.2013.11.007 [DOI] [PubMed] [Google Scholar]
- 21. Potron A, Poirel L, Rondinaud E, Nordmann P. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill. 2013;18(31). 10.2807/1560-7917.ES2013.18.31.20549 [DOI] [PubMed] [Google Scholar]
- 22. Haenni M, Poirel L, Kieffer N, Châtre P, Saras E, Métayer V, et al. Co-occurrence of extended spectrum beta lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis. 2016;16(3):281-2. 10.1016/S1473-3099(16)00007-4 [DOI] [PubMed] [Google Scholar]
- 23.Zurfuh K, Poirel L, Nordmann P, Nüesch-Inderbinen M, Hächler H, Stephan R. Occurrence of the plasmid-borne mcr-1 colistin resistance gene in ESBL-producing Enterobacteriacae in river water and imported vegetable samples in Switzerland. Antimicrob Agents Chemother. 2016; 60(4):2594-5. [DOI] [PMC free article] [PubMed]
- 24. Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Butaye P, Goossens H. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis. 2016;16(3):283-4. 10.1016/S1473-3099(16)00012-8 [DOI] [PubMed] [Google Scholar]
- 25. Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, et al. Colistin resistance gene mcr-1 in extended-spectrum beta-lactamase-producing and carbapenemase-producing Gram-negative bacteria in Germany. Lancet Infect Dis. 2016;16(3):282-3. 10.1016/S1473-3099(16)00009-8 [DOI] [PubMed] [Google Scholar]
- 26. Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, et al. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill. 2015;20(49). 10.2807/1560-7917.ES.2015.20.49.30085 [DOI] [PubMed] [Google Scholar]
- 27. Tse H, Yuen KY. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):145-6. 10.1016/S1473-3099(15)00532-0 [DOI] [PubMed] [Google Scholar]
- 28. Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC, Nightingale KK, et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(2):144-5. 10.1016/S1473-3099(15)00538-1 [DOI] [PubMed] [Google Scholar]
- 29. Poirel L, Kieffer N, Liassine N, Thanh D, Nordmann P. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis. 2016;16(3):281. 10.1016/S1473-3099(16)00006-2 [DOI] [PubMed] [Google Scholar]
- 30. Yao X, Doi Y, Zeng L, Lv L, Liu JH. Carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-9 and MCR-1. Lancet Infect Dis. 2016;16(3):288-9. 10.1016/S1473-3099(16)00057-8 [DOI] [PubMed] [Google Scholar]


