Abstract
The gut microbiome plays a key role in human health. This community is dynamic during the first three years of life, before stabilizing to an adult-like state. However, relatively little is known about the impact of environmental factors on the developing human gut microbiome. Here we report a longitudinal study of the gut microbiome based on DNA sequence analysis of monthly stool samples and clinical information from 39 children, approximately half of whom received multiple courses of antibiotics during the first three years of life. While the gut microbiome of most vaginally born children was dominated by Bacteroides species, we found that all four children born by Cesarean section and approximately 20% of vaginally born children lacked Bacteroides in the first six to eighteen months. Our longitudinal sampling, coupled with whole-genome shotgun sequencing, allowed us to detect strain-level variation as well as the abundance of antibiotic resistance (AR) genes. The microbiota of antibiotic-treated children was less diverse at the level of both species and strains, with some species often dominated by single strains. In addition, we observed short-term composition changes between consecutive samples from children treated with antibiotics. AR genes carried on microbial chromosomes showed a strong peak in abundance after antibiotic treatment followed by a sharp decline, whereas some genes on mobile elements persisted longer after the end of antibiotic therapy. Our results highlight the value of dense longitudinal studies with high-resolution strain profiles in studying the establishment and response to perturbation of the infant gut microbiome.
Introduction
A growing number of studies have highlighted the critical role played by commensal bacteria in human health. These studies have largely focused on characterizing healthy adults (1) and finding commonalities among patients suffering from diseases such as inflammatory bowel disease (2, 3), type 2 diabetes (4), obesity (5-9), metabolic disorders (10-15), colorectal cancer (16, 17), liver cirrhosis (18, 19), rheumatoid arthritis (20), and others. Several longitudinal studies have focused on pediatric conditions, including malnutrition (21, 22) type 1 diabetes (23), and asthma (24). More recently, researchers have also begun to explore the impact of external factors such as antibiotic exposure, with studies in small samples of adults suggesting that antibiotic treatment decreases microbial diversity (25-27); similar results have been obtained in mice (28).
Fewer studies have sought to characterize the natural history of the gut microbiome in children. The majority of these studies have attempted to infer the natural history based on cross-sectional datasets comparing multiple children at a single time point. These cross-sectional observations have reported that birth mode (vaginal vs. Cesarean section) affects gut microbiome composition in the first six months of life (29-31) and that the gut microbiome of children matures by the age of three years (32). More recently, a longitudinal study analyzing four time points during the first year of life reported that birth mode and nutrition influence gut microbiome composition at the level of bacterial species (33). However, no longitudinal studies have analyzed the developing gut microbiome with dense sampling or detailed analysis at the level of strains within species. In addition, despite widespread use of antibiotics in children, the effect of antibiotic exposure on the developing infant gut microbiome remains unexplored. The high prevalence of antibiotic use and concurrent increase in antibiotic-resistant bacteria, coupled with our growing appreciation of the microbiota role in human health, highlights a critical need to understand the short and long-term effects of repeated antibiotic treatments on the gut microbiome.
We therefore undertook a natural history study of the infant gut microbiome with unique strengths arising from the combination of dense sampling, multiple clinical variables (including birth mode and antibiotic usage), and strain-level analysis. We collected and analyzed samples from 39 children with an average of 28 samples per child for the first three years of life. During this time period, 20 of the children received 9-15 antibiotic treatments, while the remaining 19 children never received antibiotics. We performed both 16S rRNA gene and whole-genome shotgun sequencing to analyze microbial diversity at all taxonomic levels, including genus, species, and strain. In addition to gaining insight on how the gut microbiome develops when unperturbed by antibiotics, metagenomic sequencing of samples before and after antibiotic exposure allowed us to explore changes in the abundance of antibiotic resistance genes. We found decreased microbial diversity and increased short-term composition changes in the gut microbiomes of antibiotic-treated children. Furthermore, we observed an increased abundance of antibiotic resistance genes following treatment, along with concurrent increases in specific bacteria likely harboring these genes.
Results
Study design
To study the development of the infant gut microbiome and the effect of multiple antibiotic treatments, we collected monthly stool samples from 39 Finnish children aged 2 - 36 months (average of 28 samples per child, total of 1,069 samples). We collected multiple clinical metadata variables per child (Materials and Methods) and focused on a subset of variables (mode of delivery, breastfeeding, infant diet) that have been reported in previous studies to affect the infant gut microbiome (29, 30, 33). Of the 39 children tested, 19 received no antibiotics (Abx− children) and the remaining 20 children received 9-15 antibiotic courses in their first three years of life (Abx+ children; Fig. 1A). All courses consisted of systemic antibiotics given orally, most commonly to treat otitis media (ear infections, 91% of courses) and respiratory infections (6%). To analyze the composition of the microbial communities in this cohort, we isolated DNA from stool samples and amplified and sequenced the V4 region of the 16S rRNA gene. Sequences were sorted into operational taxonomic units (OTUs) (Materials and Methods) and the results were integrated to construct genus-level composition maps for all 1,069 samples. We identified 142 genera across the samples, 36 of which were found at a relative abundance greater than 1% in at least 25 samples. We selected 240 samples for whole-genome shotgun (WGS) sequencing (triangles in Fig. 1A) based on two criteria: (1) samples from all children at ages 2, 12, 24, and 36 months, and (2) at least four samples before and after selected antibiotic treatments (with minimal additional treatments in this time period). WGS sequencing allowed us to identify bacterial species within the genera (1–25 species per genus). In addition, we could often use the WGS data to infer the strain composition within those species (1–10 strains per species) by mapping the reads to the species reference sequences and analyzing the frequencies of multiple polymorphism sites across time for each child. Finally, we used the WGS data to determine the presence and quantify the abundance of specific genes, including antibiotic resistance genes.
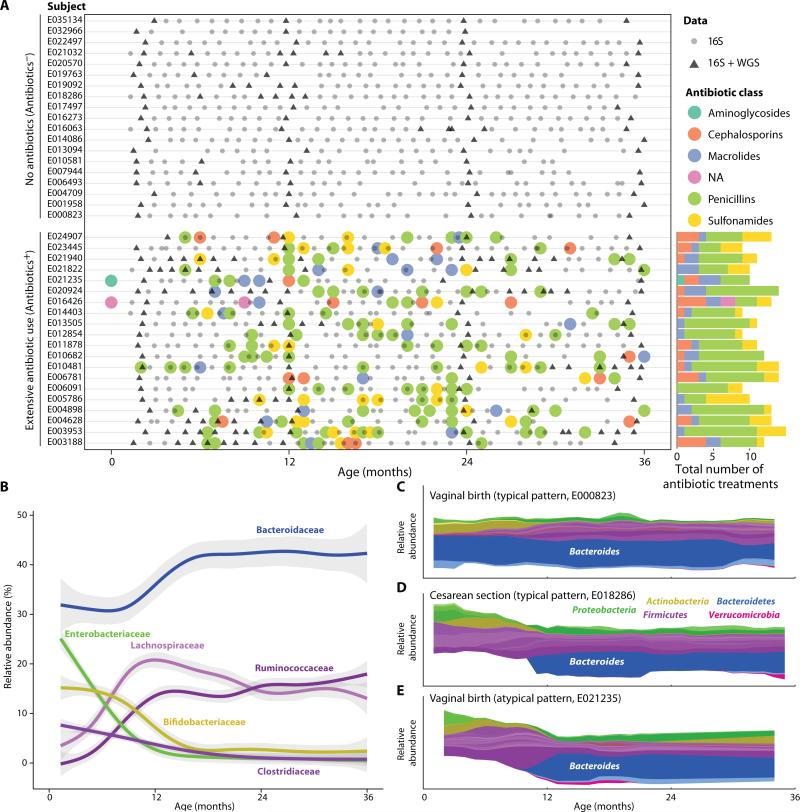
Fig. 1.
Study design and common features of infant gut microbiota. (A) Study design showing 1,069 samples (gray circles and triangles) collected from 39 children (y-axis) over 36 months (x-axis), together with information regarding the time and type of antibiotic treatments (colored circles). 16S rRNA gene sequencing was performed for all samples (gray circles and triangles) and 240 samples were selected for metagenomic sequencing (triangles). (B) Common features of the developing infant gut microbiota. Graph shows relative abundance (y-axis) of the most common bacterial families over time (x-axis), averaged across all 39 children. Shaded regions indicate 95% confidence intervals. See figure S1 for genus-level abundance plots. (C to E) Stream plots (57) of individual microbial trajectories of three children over time (x-axis), where each genus is color-coded by its phylum. Shown are typical trajectories for a vaginally born child (C), where species of the Bacteroides genus are present from an early age; a child born by Cesarean section (D), in which members of the Bacteroides genus are undetectable in the first 10 months; and an atypical trajectory for a vaginally born child (E), which appears similar to the Cesarean section signature.
Infant gut microbiome development shares common features across individuals
We first determined the change in microbial composition over time for each child (Fig. 1B). The microbial trajectory (namely, the succession of bacterial populations in the gut communities) showed multiple similarities across all infants, several of which have been observed in previous studies (21, 23). For example, at the family level, nearly all infants (87%) had significant levels of Enterobacteriaceae (average relative abundance 25%), Bifidobacteriaceae (average 15%), and Clostridiaceae (average 8%) at age two months, with each family steadily decreasing to an average relative abundance of 1%, 3%, and 2.5%, respectively, by the age of 18 months. In contrast, members of the Lachnospiraceae and Ruminococcaceae families were barely present at two months (average relative abundance of 4% and 0%, respectively) and increased in the first 12 months before stabilizing at 12–20% of the microbial community (Fig. 1B). In family-level measurements, a single genus often accounted for most of their abundance (fig. S1). For example, members of the Bifidobacterium and Clostridium genera were present in most children in the first few months (average relative abundance of 16% and 7%, respectively) and decreased over time to nearly zero by 18 months of age (fig. S1). The opposite pattern was observed for two notable beneficial commensal bacteria (34, 35). Akkermansia muciniphila (a mucin-degrading bacterium) and Faecalibacterium prausnitzii were absent at age two months but appeared in most children by months 24 and 12, stabilizing at 2% and 3% abundance, respectively (fig. S1).
We also found several clear differences from previous studies, particularly regarding the abundance of species from the Bacteroides and Bifidobacterium genera. Cross-sectional comparisons of breastfed and formula-fed children have reported a positive association between the total abundance of various Bifidobacterium species and the length of breastfeeding (29). All of the children in our study were breastfed for some period of time. On average, the breastfeeding period correlated with higher abundance of Bifidobacterium species (fig. S2); however, this longitudinal cohort of Finnish children did include some infants who had low abundance of these species even during the breastfeeding period (fig. S2). In addition, previous studies by Ley and others (36) (reviewed in (37)) have reported that the Bacteroides genus is absent prior to the introduction of solid food. However, many individuals in our cohort showed a significant Bacteroides species presence at the earliest time points, prior to the introduction of solid food (fig. S3). Specifically, the median relative abundance of the Bacteroides genus prior to introduction of solid food was 47%.
Finally, we examined the degree of consistency in the developing gut microbiome by querying whether the gut microbiome of a child at a given age is more similar to itself at an earlier time point, or to unrelated children at the same age. For this purpose, we compared the microbial composition of all sample pairs from the same child by calculating the Jaccard index (Materials and Methods). Simply stated, the Jaccard index measures the fraction of shared OTUs between two samples, such that higher values indicate greater similarity. We found that the infant gut microbiota developed continuously and the composition of a child at any given age was more similar to adjacent samples from the same child versus age-matched samples from unrelated children (fig. S4). However, adjacent samples of the same child in the first 6 months of age were not as similar as adjacent samples in older ages, indicating that the community developed most rapidly in the first six months of life (fig. S4).
A distinct microbial signature of low Bacteroides in the first 6 months is found in all Cesarean section-born and a subset of vaginally born children
Despite the commonalties among all infants, we observed one particularly striking difference in a significant portion of the cohort. Previous studies have reported that children born by Cesarean section initially lack members of the Bacteroides genus in their gut microbiome, in contrast to vaginally born children (30, 38, 39). We did indeed observe that members of the Bacteroides genus were undetectable in the guts of children born by Cesarean section (4/39) , and that this genus was detected for the first time only between 6 and 18 months of age (dark blue, Fig. 1C-E and fig. S5A). However, we unexpectedly found that a substantial proportion of vaginally born children (7/35) also exhibited this “low-Bacteroides” signature (Fig. 1E and fig. S5A), an observation that has not been previously reported. The low-Bacteroides signature was not associated with any clinical variables surrounding the delivery of the child, including antibiotic treatments of the mothers, gestational age, duration of delivery, use of enemas prior to delivery, or time spent at the hospital. We denote the group of 11 children (4 born by Cesarean section, and 7 vaginally born with a Cesarean section signature) as the low-Bacteroides group (see Materials and Methods for exact criteria).
Previous studies have also reported a decreased abundance of the Bifidobacterium genus in the gut microbiomes of children born by Cesarean section (29, 39); however, we observed the opposite pattern. On average, children from the low-Bacteroides group had a higher abundance of Bifidobacterium species in the first 6 months of life compared to all other individuals (fig. S5B). It is possible that the Bifidobacterium species occupy the niche left open by the lack of Bacteroides. Functionally, Bacteroides are major contributors to the breakdown of human milk oligosaccharides (HMO) (40). Using our metagenomic data, we quantified the relative contribution of each species to this function, revealing that Bifidobacterium were the main contributors to HMO breakdown in the low-Bacteroides group, whereas the Bacteroides species were the dominant contributors for all other children (fig. S5C).
Antibiotic-treated children have a less diverse gut microbiota
We next sought to investigate the effects of multiple antibiotic treatments in first three years of life. Specifically, we set out to measure the diversity and richness of the microbiota, first at the level of species, and then at the level of strains. We measured richness (alpha diversity) by calculating the Chao1 metric (Materials and Methods) for each of the 1,069 samples. On average, the Abx− children had a richer microbial community compared to the Abx+ children; however, the difference was relatively modest and was evident only after the first year of life (fig. S6A). Additionally, the low-Bacteroides signature was associated with a further decrease in alpha diversity, which was evident by six months of age (fig. S6). Previous studies have suggested that children born by Cesarean section have less diverse gut microbiomes (30, 38); however, we found decreased alpha diversity in low-Bacteroides children irrespective of mode of delivery.
One virtue of having multiple longitudinal samples from each individual is the ability to infer the strain composition of the abundant species. Using the 240 metagenomes that we obtained at both regular intervals and surrounding antibiotic treatments, we evaluated diversity at the strain level using ConStrains (41). In brief, this approach recruits metagenomic reads on a set of unique genes per species (42) and infers the strain composition and their relative abundance from detected single nucleotide polymorphism sites (SNPs) (commonly 1-10 strains per species). At the strain level, the difference in diversity of the Abx+ children's gut microbiomes became more evident. Within each species, we measured the strain diversity by calculating the diversity index using the probability of observing each strain within the species (Materials and Methods), and defined a species as dominated by a single strain if this score was lower than 0.1. The microbiota of the Abx+ children had significantly more species dominated by a single strain compared to the Abx− children (P = 1.38e-9; Fig. 2A-C).
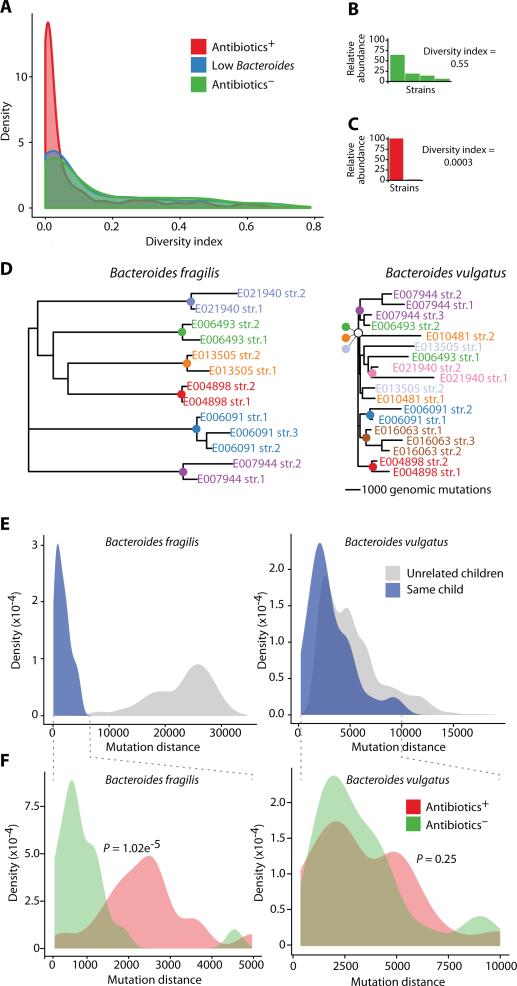
Fig. 2.
Diversity and strain similarities of the infant gut microbiota. (A) Diversity index of the strain distribution within each species shown for all metagenomic samples. Samples are colored according to three groups: children who received antibiotics (red), children with low Bacteroides (blue), and children who received no antibiotics (green). (B-C) Strain distributions of two selected samples (y-axis is relative abundance of each strain), and their calculated diversity indices. The selected samples were E032966, month 24 (B) and E006091, month 23 (C). (D) Partial phylogenetic trees based on the mutation distance between all strains of B. fragilis (left) and B. vulgatus (right). Strains are colored by the child in whom they were detected, and colored nodes represent the most recent common ancestor of the strains found in that child (scale bars of 1,000 mutations are shown per tree; Full trees appear in fig. S8). (E) Distributions of the mutation distance for all pairwise comparisons of B. fragilis (left) and B. vulgatus (right) strains, within (blue) or across (gray) individuals. (F) Distributions of mutation distances within individuals, colored as Antibiotics− (green), or Antibiotics+ (red), with P values for the separation of these two distributions (Kolmogorov-Smirnov (KS)-test) for B. fragilis (left) and B. vulgatus (right) strains.
Bacterial strains evolve within the gut (43) and are also introduced from the external environment (e.g., diet, household animals) (36, 37, 44). To attempt to determine the origin of the dominant strains in the Abx+ children, we investigated the differences among strains in each individual. For each species, we measured the difference between all of its strains both within and across all children by calculating the number of common SNPs between pairs of strains. Using this mutation-distance matrix, we constructed a phylogenetic tree of all the strains of each species, where the mutation distances are represented by the branch lengths (Fig. 2D), and compared the distance of strains within and across all children (Fig. 2E). To quantify the difference among the various strains in a species within a given individual, we calculated the median branch length of the strains to their most recent common ancestor (MRCA, colored nodes, Fig. 2D). To quantify the difference between strains of unrelated individuals, we calculated the median branch length from the MRCA to the MRCA of all other individuals (Materials and Methods; fig. S7). Using these phylogenetic strain metrics, we observed two patterns in strain-similarity measures. For some species, the strains within an individual were much more closely related to one another than they were to strains in other individuals. This pattern is consistent with the introduction of a single species, followed by evolution within the individual. Note that the single-colonization pattern may reflect multiple exposures, but only a single colonization event. In these cases, children were likely exposed to various strains of this species, but only a single strain robustly colonized their gut. Species with this pattern (referred to as ‘single-colonization species’) included Escherichia coli, Faecalibacterium prausnitzii, Bacteroides fragilis, and Haemophilus parainfluenzae (Fig. 2D-E, left and fig. S7, S8). In contrast, for other species such as Bacteroides vulgatus, some strains within an individual were more closely related to strains in other individuals (Fig. 2D-E, right). This pattern is referred to as ‘multiple-colonization species’ and is consistent with the introduction and colonization of multiple distinct strains into an individual, potentially over time.
We hypothesized that antibiotic treatments would affect the diversity of strains in the gut. We therefore expected that repeated antibiotic treatments would impact the strain similarity of ‘single-colonization species’ to a greater degree than ‘multiple-colonization species’. Focusing on the examples above, we examined diversity of B. fragilis as a representative ‘single-colonization species’ and B. vulgatus as a representative ‘multiple-colonization species’. For these analyses, we compared within an individual, and stratified the results according to antibiotic treatment history. Consistent with our hypothesis, we found that B. fragilis strains were less similar within Abx+ children compared to the strains within Abx− children (Fig. 2F, left); correspondingly, in the case of B. vulgatus, in which strains within individuals and strains in unrelated children were nearly inseparable, we could not differentiate the Abx− children from the Abx+ children (Fig. 2F, right).
Finally, the sampling and sequencing capabilities of our study offered a unique opportunity to examine the differential abundance of various taxa between Abx− and Abx+ children, at both the species and strain levels. Recent population-wide studies have identified members of the Clostridium clusters 4 and 14a as inducers of T regulatory immune cells (45), and we focused on this group of species as an illustrative example. We found that at the age of three, the total abundance of species from these clusters differed significantly between the Abx− and Abx+ children (P = 0.037, fig. S8). This difference was largely due to Eubacterium rectale, one of the most abundant members of this group (P = 0.031, fig. S8). Furthermore, when we examined the E. rectale strain-level differences within each individual, we found that strains within Abx+ children were as different as strains from unrelated individuals (fig. S8). One potential explanation for these observations is that E. rectale is a ‘single-colonization species’ for Abx− children; however, E. rectale was likely introduced multiple times but did not persist in at least some Abx+ children.
Antibiotic-treated children have less stable communities
The above analyses considered the number of different taxa (species or strains) identified at each time point. We next examined how the microbial community changes over time (i.e., the stability of the community). Taking advantage of the longitudinal nature of our study, we compared the microbial composition at all pairs of time points for each child by calculating the Jaccard index, in which higher values indicate similar microbial composition (Fig. 3A-C and fig. S9). We omitted the low-Bacteroides children from our analysis because the eventual appearance of Bacteroides greatly affected the Jaccard index (see *, Fig. 3B).
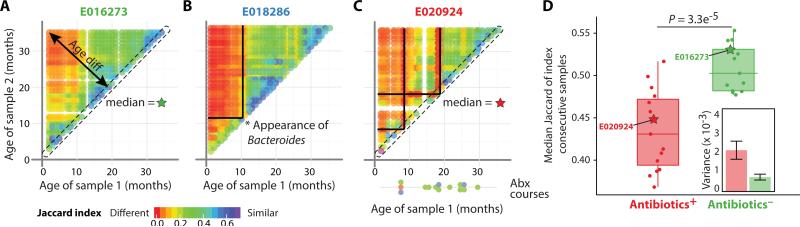
Fig. 3.
Stability of the infant gut microbiota. (A to C) Individual plots depicting the stability of the gut microbiome over time for three children (see figure S9 for plots for all children). All sample pairs from the same child were compared using the Jaccard index and plotted as a function of their age. Child identifiers are colored as Abx− (green, A), low Bacteroides (blue, B), or Abx+ (red, C). (D) Median of stability was calculated for all consecutive samples and plotted for Abx+ and Abx− children. Box boundaries are the 25th and 75th percentiles, and the median is highlighted. P = 3.3 × 10−5, KS-test. Inset shows the estimated variance of measurements for each group (mean + standard error). The median values for subjects presented in panels (A) and (C) are shown as green and red stars, respectively.
We observed that the Jaccard index of consecutive samples of Abx− individuals was consistently high, indicating the overall stability of the community, with many OTUs shared between these time points (points on the diagonal, Fig. 3A). In contrast, we observed decreased short-term stability around the time of the antibiotic treatment (red lines in Fig. 3C). We defined the overall stability index of each child as the median of the Jaccard indices for consecutive time points. Using this stability index, we found that, in contrast to the Abx− children, the Abx+ children had significantly less stable microbial communities (P = 3.3e-5; Fig. 3D and fig. S9; Materials and Methods). Furthermore, the variation in the stability indices across all Abx+ children was much larger compared to the Abx− children, indicating variability in the stability response per child (Fig. 3D inset).
Antibiotic resistance genes are detected following treatment
Next, we explored whether the use of antibiotics promotes the expansion of antibiotic resistance (AR) genes in the gut of these children, perhaps due to increased growth of bacteria harboring AR genes or increased mobilization of AR genes by other mobile elements (46). We determined the presence and abundance of antibiotic resistance genes by mapping the whole-genome shotgun sequence reads from all samples to a database of known antibiotic resistance genes. We focused on genes that are not normally present in the core genome of a species and whose presence, either chromosomally or episomally (either on a plasmid, or on a transposon), confers resistance to a specific type of antibiotics (47).
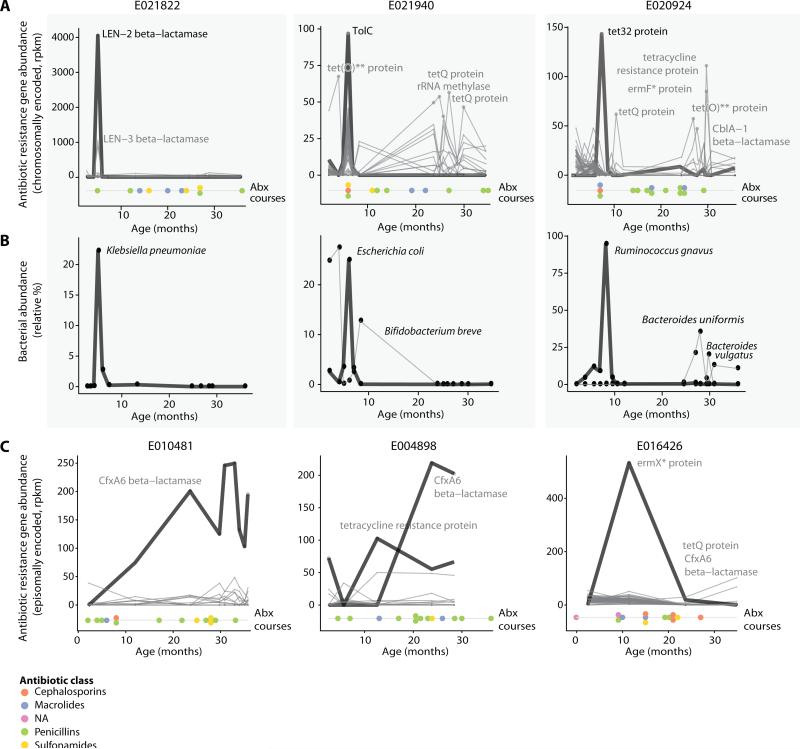
For chromosomally encoded genes, we found a number of cases in which the abundance of the antibiotic resistance gene rose extremely rapidly during antibiotic treatment and then decreased swiftly after antibiotic withdrawal (Fig. 4A). In each case, we also identified a bacterial species that showed a strongly correlated change in abundance, which was likely to harbor the resistance gene (Fig. 4B). A notable example followed penicillin treatment in subject E021822 at 5 months (Fig. 4A, left panel), where we detected an increase in a Klebsiella pneumoniae beta-lactamase resistance gene. In this example, the sharp increase of the beta-lactamase gene (from 0 to 4000 rpkm) correlated perfectly with an increase in the relative abundance of K. pneumoniae (from 0% to 22%; Fig. 4B, left panel); both the gene and species decreased to nearly zero abundance in the sample from the following month. Another example is the increase of a tolC antibiotic efflux gene (from 0 to 100 rpkm) in subject E021940 at age 6 months (Fig. 4A, middle panel) that is tightly correlated with an increase of E. coli, which is annotated as the host of this gene (from 4% to 24%, Fig. 4B, middle panel). A third example (subject E020924 at 8 months) involves the tet32 resistance gene, which is annotated as belonging to an unclassified Clostridiaceae bacterium (K10). Indeed, we observed that the relative abundance of Ruminococcus gnavus (a Clostridiaceae species) increased to a remarkable 97% at that time point, consistent with the tet32 gene being associated with this species (Fig. 4B, right panel).
Fig. 4.
Antibiotic resistance gene profiles. (A) Abundance of antibiotic resistance (AR) gene products (mostly chromosomally encoded, rpkm values) in three children over time, together with the timing of individual antibiotic courses (colored dots; see figure S10 for plots for all children). (B) Relative abundance of species most correlating with the resistance gene profiles of panel (A). (C) Examples as in (A) of AR genes found on mobile elements and that are present in the gut for longer time periods. In (A) and (C), the number and order of antibiotic courses are shown with each antibiotic class indicated by color.
For some antibiotic resistance genes that are encoded episomally, we observed a somewhat different pattern. Specifically, their abundance increased upon antibiotic treatment, but did not decrease sharply upon withdrawal of the antibiotic. Instead, the presence of the antibiotic resistance genes continued for much longer periods of time (Fig. 4C). This might be explained by the fact that episomally encoded elements can be broadly distributed across multiple species, whereas chromosomally encoded elements are non-mobile and could impose a significant fitness cost on the single host species.
Interestingly, some children (11/39) harbored antibiotic resistance genes as early as two months of age, prior to any antibiotic treatments (yellow plots; fig. S10). The reason for the increased abundance of these genes is unclear. Possible explanations could include (1) response to antibiotics passed on to the child from the environment (e.g., mothers’ breast milk); (2) a response to a natural antibiotic molecule produced by other gut bacteria; or (3) pre-existing presence of the antibiotic resistance gene in the genomes of some early inhabitants of the gut (48). The last explanation would be consistent with the spread of antibiotic resistance through natural populations due to widespread use of antibiotics. Additional studies will be needed to determine the explanation for the early presence of antibiotic resistance genes.
Discussion
We followed the development of the gut microbiome in 39 infants by densely sampling stool over a three-year period and investigating correlations with factors such as mode of birth (vaginal vs. Cesarean), infant nutrition (breast milk vs. formula), and exposure to antibiotics (33). The dense sampling per child, together with deep metagenomic sequencing that facilitated the recognition of distinct strains within species, allowed us to characterize the developing infant gut microbiome and the effect of repeated antibiotic exposure at unprecedented resolution.
Infants in the first six months of life showed one of two microbial signatures based on the abundance of Bacteroides species. There have been contradictory reports in the literature regarding the typical microbial profile in infants. Multiple studies have shown that children born by Cesarean section have a low-Bacteroides signature, whereas children born vaginally have higher levels of Bacteroides (30, 38, 39). Furthermore, one study (36) and a review (37) have described the low-Bacteroides signature as the typical profile for all children. Our study clearly demonstrates that the low-Bacteroides signature is not confined solely to children born by Cesarean section. While the low-Bacteroides signature was seen in all infants in our cohort born by Cesarean section, it was also observed in 7 of 35 (20%) children delivered vaginally. The differences cannot be explained by technical variation because the signal is extremely strong and long lasting: the abundance of Bacteroides species is exceptionally high (average 45%) and we observe it across multiple time points per child. We searched extensively for clinical variables that might explain the low-Bacteroides signature in vaginally born children (including exposure of mothers, length of delivery, and mother's use of enema prior to delivery), but with only seven children, we were unable to identify any variables that were statistically significantly correlated with the difference. Larger sample sizes may reveal the explanatory variable. Previous studies have shown that Cesarean section delivery is associated with decreased microbial diversity, even at two years of age (38). Our results critically expand this finding by demonstrating that the decrease in microbial diversity is seen across children with the low-Bacteroides signature regardless of their mode of birth. Interestingly, the decreased microbial diversity persisted, even at 36 months, when Bacteroides had become highly abundant in the low-Bacteroides children. The low-Bacteroides signature merits further study to identify its cause and to understand the implications of the associated decrease in microbial diversity.
Using our extensive metagenomic data, we identified organisms at the species level and often at the strain level, offering us the opportunity to study community diversity at a much higher resolution. The use of antibiotics was associated with a decreased microbial diversity at three years of age, especially at the strain level. It has been suggested that decreased community diversity can limit the education of the immune system, including its ability to recognize commensal bacteria, and can result in less robust microbial communities (reviewed in (49)).
Our results also shed light on the resilience of the gut microbiome, which relates to the ability of an ecosystem to return to equilibrium after perturbations (50). In ecology, repeated perturbations to an ecosystem are recognized as especially harmful when they recur before the ecosystem has had time to recover from an initial insult (51). In adults, the gut microbiome has been reported to recover from a single antibiotic exposure within approximately two weeks, but multiple treatments can cause this time frame to expand substantially (26, 50). We found that antibiotic-treated children had less stable gut microbiomes compared to untreated children. While the gut microbiome largely appeared to return to baseline within one month based on our taxonomic classification, we cannot exclude longer-term effects that are not captured by our measures. Sub-therapeutic antibiotic doses lead to increased body mass in both farm animals (52) and laboratory mice (53), however despite the repeated antibiotic treatment in children examined here, we did not observe any increase in body weight in the Abx+ children (median BMI at 36 months for the Abx+ and Abx− children was 16.2 kg/m2 and 16.0 kg/m2, respectively; P = 0.69). More detailed analysis of both gut microbiome and host will be required to understand long-term effects of antibiotic exposure.
Antibiotic treatment corresponded with short-term microbial community instability; however, we did not observe reproducible bacterial composition changes following antibiotic treatments, even when considering each antibiotic class separately. Because responses to antibiotics may occur over just a few weeks, it is possible that denser sampling would reveal more common short-term responses to antibiotic treatment. Alternatively, it is possible that the variation in responses reflects unique factors in each individual, based on the previous microbial state, host genetics, and/or overall disease state.
One important consequence of antibiotic usage is the spread of AR genes, which is a serious public health issue. Following antibiotic exposures, we observed two alternative patterns for AR gene abundance depending on their genomic context: (1) genes carried on the microbial chromosome tended to show a strong peak in abundance following treatment, followed by a sharp decline, and (2) genes on mobile elements tended to persist much longer after antibiotic withdrawal. Where these genes originated remains unclear, indeed, we detected the presence of AR genes in 2-month-old infants, prior to any antibiotic exposure. Previous studies have also reported that some newborn infants harbor AR genes (54, 55), potentially acquired from their mothers (54). Moreover, AR genes have been found in the guts of Amerindian adults (48) as well as a South American mummy (56), even in the absence of any known exposure to commercial antibiotics. Regardless of their origin, our data suggest that the genomic context of the AR gene may influence its spread and persistence upon exposure to antibiotics. Given the growing prevalence of antibiotic treatments and the frequency at which children exchange microbiota, it is particularly important to understand the effects of childhood antibiotic exposure on the presence and spread of AR genes in the community.
The pediatric gut microbiome field is still new, with relatively few large studies that include dense longitudinal sampling and multiple clinical covariates. Our work contributes many unexpected features to the field, including the highly individualized microbial profiles of infants and the variable responses of those microbial communities to antibiotics. Follow-up studies examining events closer to delivery will be critical for understanding the origin of these individualized microbial profiles as well as for determining potential sources of AR genes. Fortunately, improved methods for sequencing and analysis make it increasingly feasible to fully elucidate the natural history of the infant gut microbial community, including the long-term effects of important perturbations such as antibiotics and Cesarean delivery.
Materials and Methods
Classification of the low-Bacteroides children based on 16S rRNA gene sequencing data
The median relative abundance of the Bacteroides genus was calculated per child for samples collected in the first 6 months of age. Children with median abundance of less than 0.1% were classified as low-Bacteroides children.
Density plots
Multiple density plots appear throughout the manuscript. Similar to a histogram, the density plots shown here display the distribution of the observed data; unlike a histogram, the density function reflects the estimated underlying continuous probability from which the observed data has been sampled (i.e., the area under the curve sums to 1).
Inferring strain profiles using ConStrains
WGS samples were additionally analyzed using ConStrains (41) (https://bitbucket.org/luochengwei/constrains), which conducts within-species strain haplotyping by clustering SNP patterns detected from mapping reads to species pangenomes across samples. Using the longitudinal nature of our cohort, we analyzed the strain composition for all samples of the same child simultaneously, allowing us to follow within-subject strain profile evolution in. ConStrains requires 10x coverage of a given pangenome to reliably identify strains within its corresponding species.
We used the strain-level abundance profiles to compute species-specific haplotype diversity scores for each sample, defined as H = 1 - , where pi denotes the abundance of strain i. This measure was motivated by the concept of heterozygosity in population genetics, and is bounded by [0,1]. A higher value is consistent with a nearly uniform distribution of all strains within the species, and values closer to zero indicate a single dominant strain.
Supplementary Material
One Sentence Summary.
A longitudinal study with strain-level analysis of the infant gut microbiome reveals effects of repeated antibiotic treatments, including decreased diversity and stability, as well as transient increases in antibiotic resistance genes following treatments.
Acknowledgments
We thank Tiffany Poon and Scott Steelman (Broad Institute) for help in sequence production and sample management, Katariina Koski and Matti Koski (University of Helsinki) for the coordination and database work in the DIABIMMUNE study, Chengwei Luo (Broad Institute) for help in the strain analysis, Lawrence David (Duke University) for scripts, Roby Bhattacharyya, Jonathan Livny and Schragi Schwarts (Broad Institute) for helpful discussions, and Natalia Nedelsky (Massachusetts General Hospital) for editorial help and figure generation.
Funding: M.Y. was supported by the Klarman Family Foundation. E.S.L. was supported by the National Human Genome Research Institute grant 2U54HG003067-10. The DIABIMMUNE study (M.K.) was supported by the European Union Seventh Framework Programme FP7/2007-2013 under grant agreement number 202063, and the Academy of Finland Centre of Excellence in Molecular Systems Immunology and Physiology Research grant Decision number 250114, 2012–2017. R.J.X. was supported by funding from JDRF, National Institutes of Health (NIH) grants U54 DK102557, R01 DK092405, and P30 DK043351, the Leona M. and Harry B. Helmsley Charitable Trust, and the Crohn's and Colitis Foundation of America.
Footnotes
Author contributions: M.Y. performed 16S and metagenomic data analysis. M.Y, H.V., E.S.L., and R.J.X. assembled and wrote the paper. H.S., A.H., T.H., S.J.R., and M.K. designed the cohort study and collected clinical samples. M.Y., and D.G. designed DNA sequencing experiments and sample management pipelines. M.Y., T.V., E.A.F., C.H., H.V., D.G. and E.S.L. led method and research development. E.S.L., M.K., and R.J.X. served as principal investigators.
Competing interests: The authors declare no competing financial interests.
Data and materials availability: The NCBI BioProject ID for these data is PRJNA290381.
References and Notes
- 1.Human Microbiome Project Consortium, Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, Gonzalez A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 5.Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS ONE. 2014;9:e84689. doi: 10.1371/journal.pone.0084689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE. Obesity and the human microbiome. Curr. Opin. Gastroenterol. 2010;26:5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 7.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 9.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 10.Tilg H, Adolph TE. Influence of the human intestinal microbiome on obesity and metabolic dysfunction. Curr. Opin. Pediatr. 2015;26:496–501. doi: 10.1097/MOP.0000000000000234. [DOI] [PubMed] [Google Scholar]
- 11.Parekh PJ, Balart LA, Johnson DA. The Influence of the Gut Microbiome on Obesity, Metabolic Syndrome and Gastrointestinal Disease. Clin Transl Gastroenterol. 2015;6:e91. doi: 10.1038/ctg.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanz Y, Olivares M, Moya-Perez A, Agostoni C. Understanding the role of gut microbiome in metabolic disease risk. Pediatr. Res. 2015;77:236–244. doi: 10.1038/pr.2014.170. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda S, Ohno H. Gut microbiome and metabolic diseases. Semin Immunopathol. 2014;36:103–114. doi: 10.1007/s00281-013-0399-z. [DOI] [PubMed] [Google Scholar]
- 14.Kovatcheva-Datchary P, Arora T. Nutrition, the gut microbiome and the metabolic syndrome. Best Pract. Res. Clin. Gastroenterol. 2013;27:59–72. doi: 10.1016/j.bpg.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Tilg H, Kaser A. Gut microbiome, obesity, and metabolic dysfunction. J. Clin. Invest. 2011;121:2126–2132. doi: 10.1172/JCI58109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, Kostic AD, Giannakis M, Watanabe H, Bullman S, Milner DA, Harris CC, Giovannucci E, Garraway LA, Freeman GJ, Dranoff G, Chan AT, Garrett WS, Huttenhower C, Fuchs CS, Ogino S. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 19.Roderburg C, Luedde T. The role of the gut microbiome in the development and progression of liver cirrhosis and hepatocellular carcinoma. Gut Microbes. 2014;5:441–445. doi: 10.4161/gmic.29599. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, Lan Z, Chen B, Li Y, Zhong H, Xie H, Jie Z, Chen W, Tang S, Xu X, Wang X, Cai X, Liu S, Xia Y, Li J, Qiao X, Al-Aama JY, Chen H, Wang L, Wu QJ, Zhang F, Zheng W, Li Y, Zhang M, Luo G, Xue W, Xiao L, Li J, Chen W, Xu X, Yin Y, Yang H, Wang J, Kristiansen K, Liu L, Li T, Huang Q, Li Y, Wang J. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 21.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA, Jr., Ahmed T, Gordon JI. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, Liu J, Houpt E, Li JV, Holmes E, Nicholson J, Knights D, Ursell LK, Knight R, Gordon JI. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, Peet A, Tillmann V, Poho P, Mattila I, Lahdesmaki H, Franzosa EA, Vaarala O, de Goffau M, Harmsen H, Ilonen J, Virtanen SM, Clish CB, Oresic M, Huttenhower C, Knip M, Group DS, Xavier RJ. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Investigators CS, Mohn WW, Turvey SE, Brett Finlay B. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 25.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, Chung J, Sohn J, Barber CM, Goldfarb DS, Raju K, Abubucker S, Zhou Y, Ruiz VE, Li H, Mitreva M, Alekseyenko AV, Weinstock GM, Sodergren E, Blaser MJ. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 30.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Jun W. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 34.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L, Andersson AF. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 39.Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010;86(Suppl 1):13–15. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 40.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo C, Knight R, Siljander H, Knip M, Xavier RJ, Gevers D. ConStrains identifies microbial strains in metagenomic datasets. Nat. Biotechnol. 2015;33:1045–1052. doi: 10.1038/nbt.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods. 2012;9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, Metcalf JL, Ursell LK, Vazquez- Baeza Y, Van Treuren W, Hasan NA, Gibson MK, Colwell R, Dantas G, Knight R, Gilbert JA. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 46.Waters JL, Salyers AA. Regulation of CTnDOT conjugative transfer is a complex and highly coordinated series of events. MBio. 2013;4:e00569–00513. doi: 10.1128/mBio.00569-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJ, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcon O, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1 doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Relman DA. The human microbiome: ecosystem resilience and health. Nutr. Rev. 2012;70(Suppl 1):S2–9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paine RT, Tegner MJ, Johnson EA. Compounded Perturbations Yield Ecological Surprises. Ecosystems. 1998;1:535–545. [Google Scholar]
- 52.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 2009;7:887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zarate Rodriguez JG, Rogers AB, Robine N, Loke P, Blaser MJ. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alicea-Serrano AM, Contreras M, Magris M, Hidalgo G, Dominguez-Bello MG. Tetracycline resistance genes acquired at birth. Arch. Microbiol. 2013;195:447–451. doi: 10.1007/s00203-012-0864-4. [DOI] [PubMed] [Google Scholar]
- 55.Fouhy F, Ogilvie LA, Jones BV, Ross RP, Ryan AC, Dempsey EM, Fitzgerald GF, Stanton C, Cotter PD. Identification of aminoglycoside and beta-lactam resistance genes from within an infant gut functional metagenomic library. PLoS ONE. 2014;9:e108016. doi: 10.1371/journal.pone.0108016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santiago-Rodriguez TM, Fornaciari G, Luciani S, Dowd SE, Toranzos GA, Marota I, Cano RJ. Gut Microbiome of an 11th Century A.D. Pre-Columbian Andean Mummy. PLoS ONE. 2015;10:e0138135. doi: 10.1371/journal.pone.0138135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.David LA, Materna AC, Friedman J, Campos-Baptista MI, Blackburn MC, Perrotta A, Erdman SE, Alm EJ. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.