Abstract
Cyclin-dependent kinases 12 and 13 (CDK12 and 13) play critical roles in the regulation of gene transcription. However, the absence of CDK12 and 13 inhibitors has hindered the ability to investigate the consequences of their inhibition in healthy cells and cancer cells. Here we describe the rational design of a first-in-class CDK12 and 13 covalent inhibitor, THZ531. Co-crystallization with CDK12-cyclin K indicates that THZ531 irreversibly targets a cysteine located outside the kinase domain. THZ531 causes a loss of gene expression with concurrent loss of elongating and hyperphosphorylated RNA polymerase II. In particular, THZ531 substantially decreases the expression of DNA damage response genes and key super–enhancer–associated transcription factor genes. Coincident with transcriptional perturbation, THZ531 dramatically induced apoptotic cell death. Small molecules capable of specifically targeting CDK12 and 13 may thus help identify cancer subtypes that are particularly dependent on their kinase activities.
Introduction
RNA polymerase II (Pol II) is responsible for the transcription of protein–coding genes in eukaryotic cells.1 The process of transcription itself can be divided into discrete stages including initiation, elongation, and termination. Pol II activity throughout the transcription cycle is controlled by coordinated, reversible, post-translational modification of residues in the heptad (YSPTSPS) amino acid repeats found in its C-terminal domain (CTD).2–4 Phosphorylation of serine at position 5 (Ser5) of the CTD is required for proper transcriptional initiation from gene promoters, while Ser2 phosphorylation promotes elongation of Pol II through the gene body and the production of mature mRNA transcript.5 In mammalian cells, Ser2 phosphorylation has, until recently, been attributed solely to the activity of cyclin –dependent kinase 9 (CDK9), the kinase component of the positive transcription elongation factor b (P-TEFb).6,7 Research in both yeast and metazoans demonstrates that CDK12 and CDK13 may also play important roles in Ser2 phosphorylation and gene transcription, particularly elongation, though their exact roles in these processes remain unclear.8–10
Complexes containing CDK12 and 13 regulate transcriptional elongation and processes occurring co-transcriptionally, including mRNA splicing and 3’ end RNA processing.11–13 CDK12 and 13 aid in regulating RNA processing both directly by physical interaction with RNA-processing factors and indirectly by phosphorylation of the CTD, which recruits these processing factors.13–17 Because of their roles in regulating these processes, loss of CDK12 and 13, or their associated cofactor cyclin K, impedes both Pol II processivity and RNA processing. For example, CDK12 binds in exon junction complexes with other arginine-serine (RS) domain–containing splicing factors including SRSF1, and its loss leads to mRNA splicing defects.13,16 Factors involved in 3’ end cleavage and polyadenylation of RNA transcripts, including CstF64 and CstF77, are recruited to 3’ ends coincident with CTD Ser2 phosphorylation, which is dependent on CDK12 function. Depletion of CDK12 leads to simultaneous loss of Ser2 phosphorylation, recruitment of these factors, and subsequent 3’ processing defects.14,15,17 Lastly, CDK12 lacking N-terminal RS domains also exhibits 3’ end processing defects, suggesting that dominant negative mutant forms of CDK12 that disrupt structure and physical interactions may also impact transcription.14
CDK12-cyclin K and CDK13-cyclin K complexes exhibit both distinct and overlapping regulation of Pol II –mediated gene expression. Genetic depletion of CDK12 or CDK13 demonstrated that both complexes similarly regulate the expression of roughly 1,000 genes including RNA processing genes13, while separately regulating distinct classes of genes.13,18 In particular, loss of CDK13, but not CDK12, decreases the expression of genes encoding proteins that regulate protein translation.13 Conversely depletion of CDK12, but not CDK13, reduces the expression of core members of the DNA damage response (DDR), leading to a marginal increase in unrepaired double -strand breaks and increased susceptibility to treatment with DNA damaging agents.13,18–21 Interestingly, breast and ovarian cancers harboring inactivating mutations in BRCA1/2, two members of the homologous recombination DDR pathway, demonstrate synthetic lethality with pharmacological inhibition of PARP1, which repairs single –strand breaks.22 Consistent with these findings, genetic screens have indicated that loss of CDK12 potentiates response to PARP1 inhibition.23,24 These data suggest that CDK12 and 13 inhibitors alone, or in combination treatment, may represent an effective strategy for targeting cancers demonstrating extreme dependencies on the expression of CDK12 and 13 – regulated genes.
While genetic depletion of CDK12 and 13 indicate that the proteins themselves are required for gene transcription, it remains unclear how CDK12 and 13 kinase activities regulate these processes. Mutational studies using CDK12 variants that are either devoid of N-terminal RS domains or that cannot complex with cyclin K indicate that loss of function mutants may elicit phenotypes comparable to deletion.14,21 Recently, CRISPR/Cas9 –mediated gene editing was used to generate cells expressing a CDK12 mutant that is selectively inhibitable by a cell-permeable adenine analog.25 While this approach permits investigation of CDK12 kinase activity, its utility is limited to a particular genetic background. Therefore, the identification of inhibitors of wild-type CDK12 and 13 function is necessary to understand the effects of pharmacologically inhibiting CDK12 and 13 in normal and disease states.
Inspection of the recently published CDK12-cyclin K and CDK13-cyclin K co-crystal structures reveals that CDK12 Cys-1039 and CDK13 Cys-1017 occupy similar spatial positions as Cys-312 in CDK7.26–28 Therefore we surmise that we could use THZ1, which covalently targets Cys-312 of CDK7, as starting material for developing selective CDK12 and 13 agents.29 Here we report the development of THZ531 as a new covalent inhibitor of CDK12 and 13 and present a 2.7 Å co-crystal structure with CDK12-cyclin K, which shows THZ531 bound to the ATP pocket and connected to Cys-1039 via its flexible linker. Treatment of Jurkat cells with THZ531 resulted in diminished global Ser2 phosphorylation (pSer2). Closer inspection indicated that loss of pSer2, and resulting loss of elongating Pol II, occurs specifically at THZ531 –responsive genes indicating that loss of transcription elongation following CDK12/13 inhibition contributes to loss of gene expression. Analysis of the genes whose expression was particularly sensitive to treatment with THZ531 showed two classes of genes: at low doses of THZ531, genes encoding DDR factors were exquisitely sensitive, whereas complete inhibition of CDK12 and 13 with a high dose of THZ531 led to significant downregulation of key Jurkat transcription factors. Concomitant with this extreme transcriptional stress, we observed potent induction of apoptosis. Thus, inhibition of CDK12 and 13 -mediated transcription in tumors by targeting CDK12 and 13 kinase activity may be a potent means of affecting oncogenic cell survival.
Results
THZ531 covalently inhibits CDK12 and 13
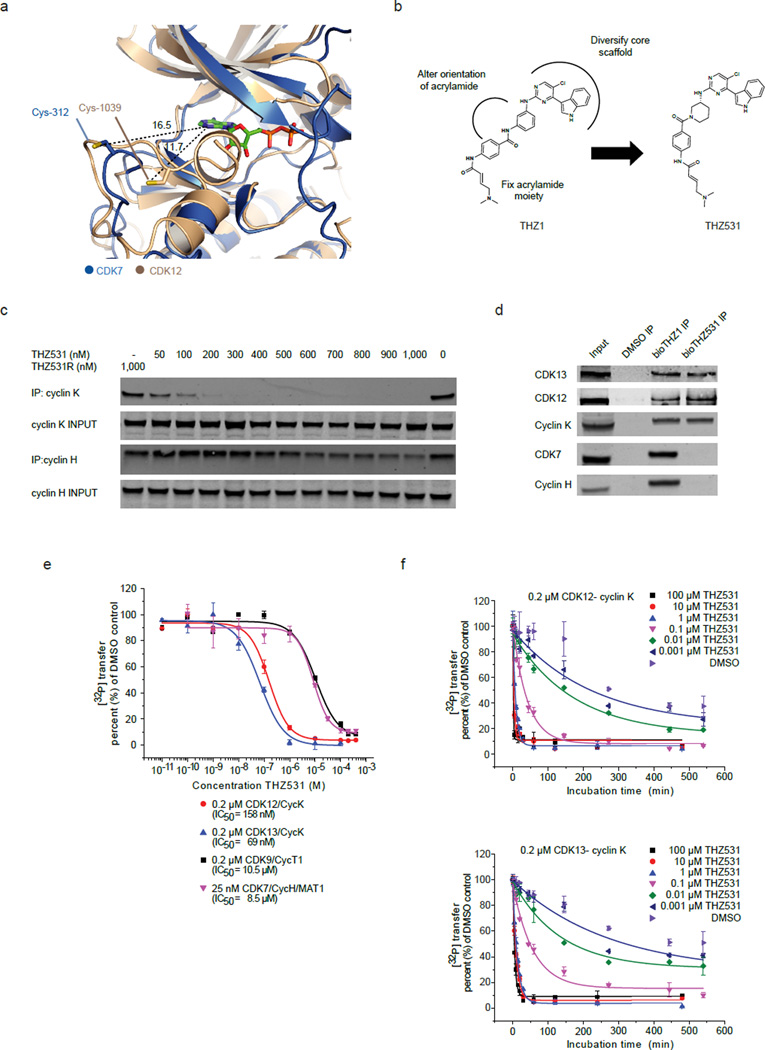
We previously described the characterization of THZ1, a covalent inhibitor of CDK7, which has the unprecedented ability to covalently modify Cys-312 located on a C-terminal extension of the kinase domain that traverses the ATP–binding site. Affinity chromatography with a biotin-modified version of THZ1 revealed that it is additionally capable of covalently binding to CDK12 and 13, both of which were also predicted by sequence alignment to have C-terminal cysteines.29 Comparison of the CDK7 and CDK12 crystal structures revealed that indeed both kinases have similarly positioned, yet spatially distinct, cysteines in their C-terminal domains (Fig. 1a).26,28 We reasoned that modification of the substituent bearing the acrylamide could serve to alter the orientation of the acrylamide warhead to selectively target Cys-1039 of CDK12. To discriminate amongst CDK7, 12, and 13 binders we developed a ‘target-engagement’ assay where the ability of new compounds to protect these kinases from affinity purification by biotinylated THZ1 (bioTHZ1) was assessed by western blot (see detailed protocol in Supplementary Results, Supplementary Fig. 1a). Following the iterative synthesis and testing of a variety of analogs (to be reported elsewhere), we discovered THZ531 (1), which possesses a piperidine ring in the acrylamide ‘arm’ (Fig. 1b). We discovered that this configuration resulted in more selective binding of THZ531 to CDK12-cyclin K and CDK13-cyclin K relative to CDK7-cyclin H and CDK9-cyclin T (Fig. 1c, Supplementary Fig. 1b). Mass spectrometry of recombinant CDK12-cyclin K complex incubated with the inhibitor demonstrated covalent modification of Cys-1039 of CDK12 (Supplementary Fig. 1c,d). To corroborate covalent bond formation in cell lysates, we synthesized a biotinylated analog of THZ531 (bioTHZ531, 2) that could affinity purify covalently bound targets with streptavidin. Inspection of precipitated proteins confirmed CDK12-cyclin K and CDK13-cyclin K complexes as the main targets of THZ531 (Fig. 1d, Supplementary Data Set 1).
Figure 1. THZ531 covalently inhibits CDK12 and 13.
a, Overlay of CDK7 and CDK12 crystal structures reveals similarly positioned cysteines in the C-terminal extension segments of the two kinase domains. Cys-312 was modeled into the crystal structure of CDK7 as previously described.29 PDB codes: 1UA2 (CDK7)28 and 4NST (CDK12).26 b, Structures of THZ1 and THZ531 and rational design strategy to synthesize THZ531 from THZ1. c, THZ531 binds intracellular CDK12-cyclin K complexes with higher affinity than CDK7-cyclin H complexes out competing lysate –introduced bioTHZ1 in target engagement assay. d, Biotinylated THZ531 (bioTHZ531, 1 µM) pulls down CDK12-cyclin K and CDK13-cyclin K complexes, but not CDK7-cyclin H complexes, whereas bioTHZ1 (1 µM) pulls down CDK7, CDK12, and 13 complexes. e, THZ531 potently inhibits CDK12 and CDK13. IC50 values for CDK12 and 13 were 158 and 69 nM, respectively, compared to 10.5 µM for CDK9 and 8.5 µM for CDK7. Measurements were performed in triplicate and data represent mean values ± S.D. f, THZ531 inhibition of CDK12 and 13 is time –dependent. In vitro kinase activity assay of CDK12-cyclin K (top) and CDK13-cyclin K (bottom) with different concentrations of THZ531 and varying preincubation times. For all incubation time series, the counts per minute of the kinase activity measurements were normalized to the relative [32P] transfer. Measurements were performed in triplicate and data represent the mean values ± S.D. Uncut western blots are in Supplementary Fig. 10.
To confirm that THZ531 inhibits the enzymatic activity of CDK12 and 13, we performed a radiometric kinase assay measuring the ability of recombinant CDK12 and 13 to phosphorylate a Pol II CTD-peptide substrate.26 In fixed- end point kinase assays, THZ531 potently inhibited CDK12 and 13 with IC50s of 158 nM and 69 nM, respectively; whereas inhibition of CDK7 and CDK9 was more than 50-fold weaker with IC50s of 8.5 and 10.5 µM, respectively (Fig. 1e). In contrast both THZ531R (3), where the electrophilic acrylamide is replaced with a propyl amide incapable of covalent reaction and THZ532 (4), the enantiomer of THZ531 (Supplementary Fig. 1e), were both 50–100 fold less active on CDK12 and 13, but exhibited similarly–low–inhibition activity on CDK7 and CDK9, implying that THZ531 inhibits the enzymatic functions of CDK12 and 13 and that covalent binding was important for this activity (Supplementary Fig. 2a–c). As an additional control, THZ531 showed no appreciable inhibitory effect on ERK1, using this kinase as example for the MAP kinase family of the CMGC group (Supplementary Fig. 2d). Similar to THZ1, increasing incubation time augmented THZ531 potency, further supporting the notion that covalent binding of THZ531, which occurs with slower kinetics than reversible binding is critical for activity (Fig. 1f, Supplementary Fig. 2e,f).
To broadly survey kinase selectivity we performed in vitro profiling using the Ambit™ binding technology and intracellular profiling using KiNativ™ profiling, which measures the ability of a compound to block nucleotide-dependent enzymes from biotinylation with a reactive ATP-desthiobiotin probe.30 Ambit™ profiling indicated that CDK13 was among the top scoring kinases (Supplementary Data Set 2). KiNativ™ profiling confirmed CDK12 and 13 as the primary targets of THZ531 and demonstrated the excellent selectivity of the inhibitors – none of the other 211 kinases were labeled to greater than 55% percent (Supplementary Table 1, Supplementary Fig. 2g).
THZ531 co-crystal structure with CDK12-cyclin K
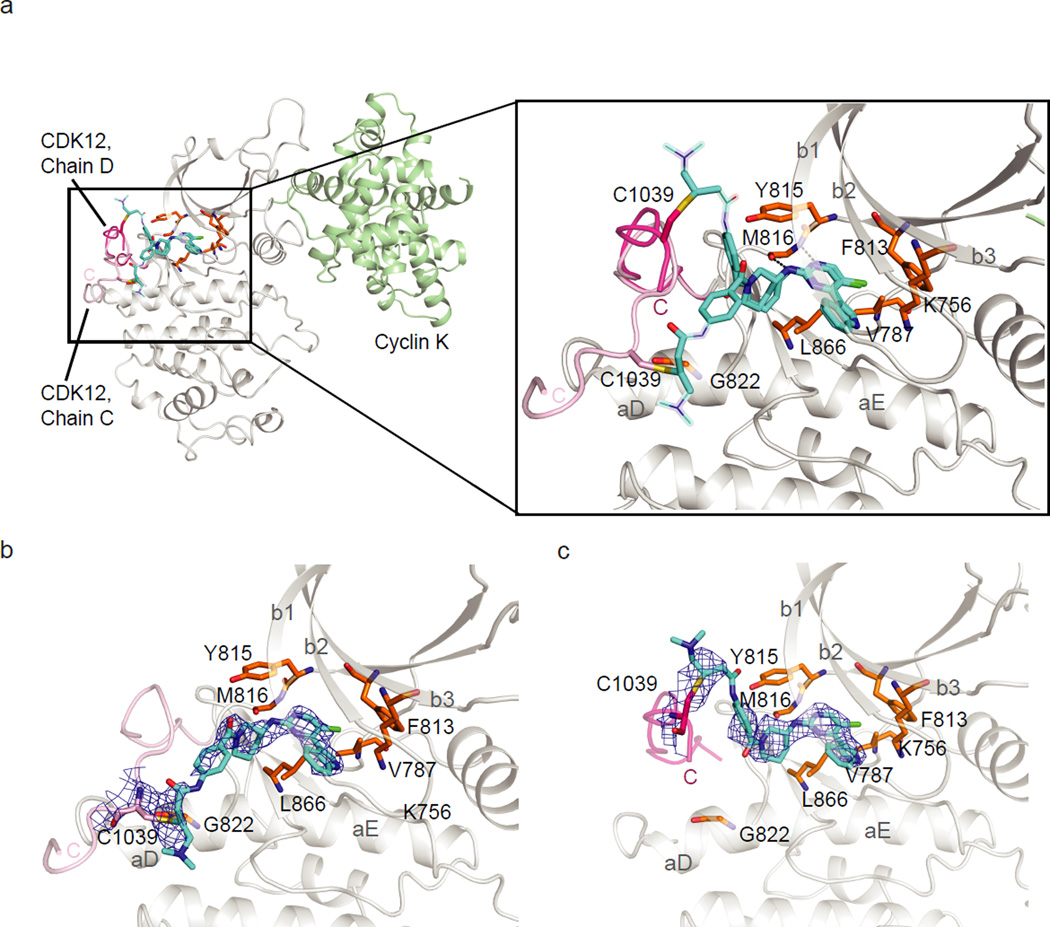
To understand how THZ531 selectively targets Cys-1039 in CDK12, we determined its co-crystal structure with CDK12-cyclin K (see Supplementary Table 2 for diffraction data collection and refinement statistics). Two CDK12-cyclin K complexes were found in the asymmetric unit, each binding to THZ531 in a different rotamer (Fig. 2a–c). The 2.7 Å structure revealed that the labile αK helix can be displaced from CDK12 allowing Cys-1039 to reorient towards the ATP –binding pocket for cross-linking. The aminopyrimidine bound to the kinase hinge region forming two hydrogen bonds to the backbone of M816, while the pendant 3-indolyl and piperazine groups mediated further hydrophobic interactions. Different conformations of the secondary amide were observed in the two THZ531 complexes causing the solvent-exposed groups to pack either against the N-lobe β1 strand or C-lobe αD helix. These exposed sites were only poorly defined, except for the positions of Cys-1039, which were clearly visible in the electron density maps (Supplementary Fig. 3). Notably, the equivalent sulfhydryl in CDK7, Cys-312, was at a more distant site (over 12 Å from CDK12 Cys-1039), suggesting a basis for the weaker binding of THZ531 to CDK7.
Figure 2. THZ531 co-crystal structure with CDK12-cyclin K.
a, THZ531 binds to M816 in the kinase hinge region and connects to Cys-1039 in two conformations via the compound’s flexible linker. Solvent-exposed regions of THZ531 with poor electron density are represented by thin sticks. b. Omit map contoured at 2.5σ for THZ531 bound to CDK12 chain C. c. Omit map contoured at 2.5σ for THZ531 bound to CDK12 chain D.
THZ531 induces apoptosis in Jurkat cells
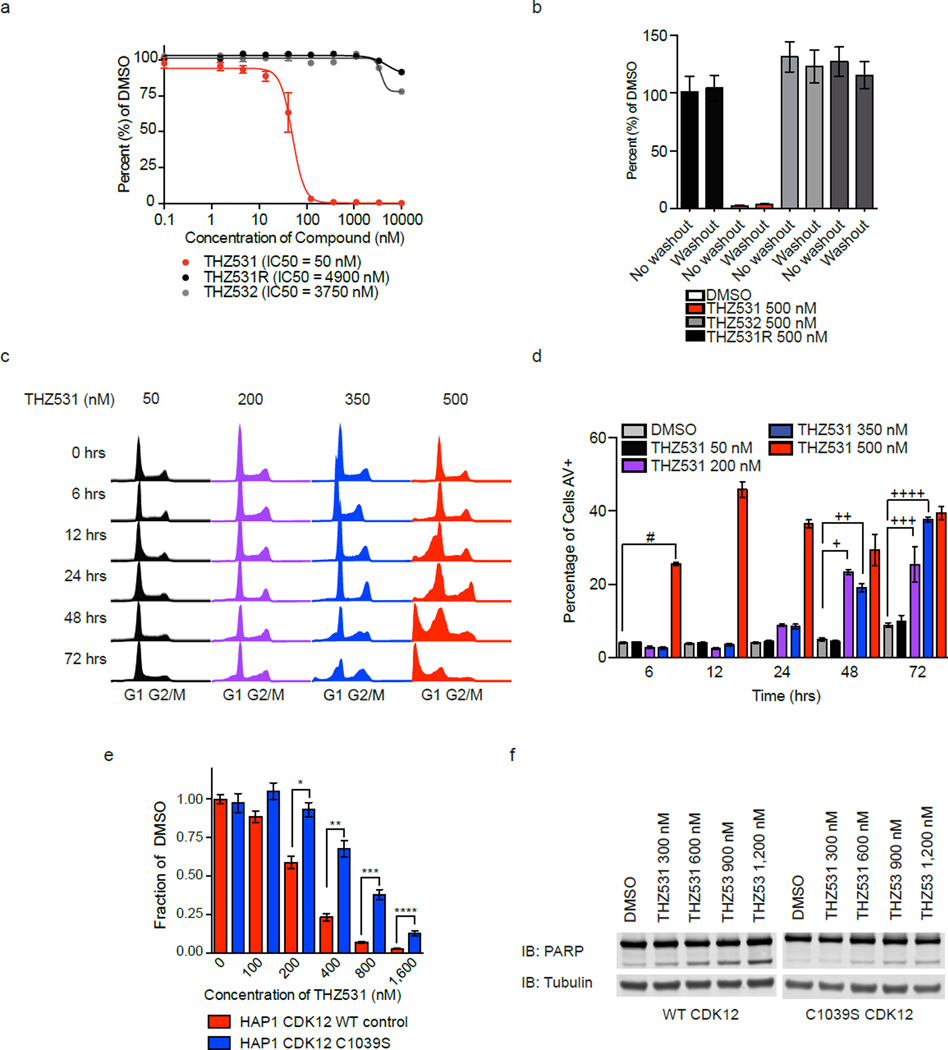
THZ1, the predecessor to THZ531, displays strong anti-proliferative activity in models of various cancer types.29,31,32 To evaluate if THZ531 exhibits similar anti-cancer activity, we treated Jurkat cells with THZ531 in escalating doses for 72 hours. THZ531 treatment led to a dramatic and irreversible decrease in Jurkat cell proliferation with an IC50 of 50 nM (Fig. 3a,b), which correlates with the IC50 concentration required to achieve 50% covalent CDK12 and 13 binding (Fig. 1c). In agreement with their reduced anti-CDK12 and 13 activity, THZ531R and THZ531’s enantiomer, THZ532, displayed minimal antiproliferative activity in Jurkat cells (Fig. 3a,b). FACS cell cycle analysis following treatment with escalating doses of THZ531 displayed a dose and time-dependent increase in the number of cells exhibiting sub-G1 content, which is suggestive of cell death (Fig. 3c). To investigate this further, we used flow cytometry to assess annexin V (AV) and propidium iodide (PI) staining to identify apoptotic and dead cells, respectively. At 50 nM THZ531, no increase in the percentage of apoptotic cells was observed over DMSO control for the time course of the experiment (Fig. 3d). However, higher doses of THZ531 led to pronounced Annexin V signal with 30–40% annexin V–positively stained cells by 72 hrs (Fig. 3d, Supplementary Fig. 4a). Similarly, PARP cleavage further indicated that cell death as a result of apoptosis occurred following THZ531 administration (Supplementary Fig. 4b). Thus, THZ531 induces apoptosis in a dose- and time-dependent manner with low doses (<350 nM) leading to slow onset of apoptosis over several cell doublings, while doses exceeding 350 nM induce rapid apoptosis which may result from a combination of on- and off- target effects.
Figure 3. THZ531 induces apoptosis in Jurkat cells.
a. THZ531, but not THZ531R or THZ532, exhibits strong antiproliferative effect in Jurkat T-cell acute lymphoblastic leukemia cells. Cells were treated with the indicated compounds for 72 hrs and assessed for antiproliferative effect using CellTiter Glo. b, THZ531 demonstrates irreversible suppression of Jurkat cell proliferation. Jurkat cells were treated with the indicated compounds for 6 hrs. Cell growth medium containing inhibitors was then removed (washout) and cells were allowed to grow for the remainder of the 72 hr proliferation assay without inhibitors. CellTiter Glo results were compared to cellular treatments whereby cells were treated with inhibitors for the full 72 hrs (no washout). c. THZ531 increases sub-G1 population. Jurkat T-ALL cells were treated with THZ531 for the indicated time periods. Cell cycle progression was assessed using FACS cell cycle analysis. d. Treatment with THZ531 induces apoptosis. Jurkat cells were treated with THZ531 for the indicated times. Cells were stained with Annexin V and propidium iodide. All proliferation and apoptosis assays were performed in biological triplicate and error bars are +/− SD. #p-value=3.33e-07, +p-value=1.04e-04, ++p-value=1.35e-04, +++p-value=9.94e-04, ++++p-value=1.53e-07. e, 72-hour antiproliferation assay using WT and C1039S HAP1 cells. Cells were analyzed using Cell Titer Glo. Experiment was performed in biological triplicate and error bars are +/− SD. *p-value=5.84e-04, **p-value=1.76e-04, ***p-value=9.09e-05, ****p-value=4.25e-04. f, Expression of C1039S CDK12 partially reduces PARP cleavage in HAP1 cells. Western blot analysis of indicated proteins in HAP1 CDK12 WT and C1039S cells. Uncut western blots are in Supplementary Fig. 10. P-values were determined with a two-tailed Student’s T test.
To assess whether mutation of CDK12 Cys-1039 to a less nucleophilic serine residue could rescue THZ531 –mediated effects we used CRISPR/Cas9 to mutate the CDK12 gene locus (Supplementary 5a, b). This substitution was sufficient to make CDK12 refractory to covalent affinity pull down and led to partial restoration of cellular proliferation and reduced apoptosis (Fig. 3e,f; Supplementary Fig. 5c). For these experiments the chronic myeloid leukemia cell line HAP1 was used, as technical problems precluded the use of CRISPR/Cas9 in Jurkat cells. Additionally, we believe the partial rescue may result, in part, from the continued presence of WT CDK13 or from an unknown off-target.
CDK12 binds regulatory and coding regions of active genes
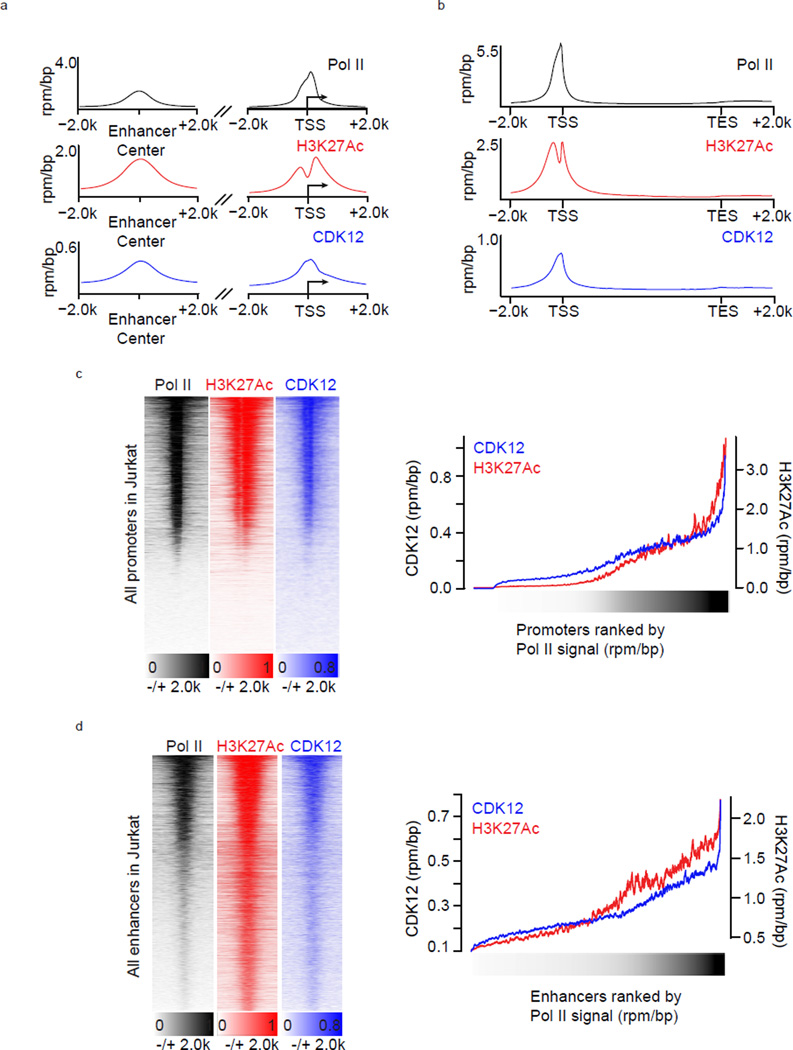
Given the role CDK12 and 13 play in transcriptional regulation we next sought to explore the potential transcriptional underpinnings of this anti-proliferative effect. CDK12 is believed to regulate Pol II –mediated transcription. However, with the exception of Drosophila, where CDK12 was shown to co-localize with hyperphosphorylated Pol II (II0) on larval polytene chromosomes and to ChIP mainly to gene interiors on cultured cell chromosomes, little is known about how CDK12 interacts with the genome.9 To address this question, we performed chromatin immunoprecipitation coupled to massively parallel DNA sequencing (ChIP-seq) using an antibody against CDK12.16 Genome –wide analysis of CDK12 ChIP-seq signal indicated that CDK12 bound to both protein -coding genes and enhancer regions (Fig. 4a). At enhancers, CDK12 ChIP-seq signal coincided with Pol II and H3K27Ac, marks associated with active enhancers and the production of enhancer RNAs, suggesting that CDK12 binds to active enhancers (Fig. 4a). At protein –coding genes, CDK12 ChIP-seq signal was found at promoters and gene bodies, where it largely overlapped with Pol II signal, indicating that it may travel with elongating Pol II (Fig. 4a, 4b). Furthermore, the intensity of CDK12 ChIP-seq signal at both enhancers and promoters increased commensurate with Pol II and H3K27Ac signals implying that CDK12 levels correlate with transcriptional activity (Fig. 4c, 4d). Therefore, these data suggest that CDK12 is present at actively transcribed regions of the genome.
Figure 4. CDK12 binds regulatory and coding regions of active genes.
a. Pol II, H3K27Ac, and CDK12 co-occupy active enhancers and promoters. Metagene representation of average Pol II, H3K27Ac, and CDK12 occupancy at H3K27ac-defined enhancers (left) and 37974 RefSeq promoters (right). b. Pol II, H3K27Ac, and CDK12 bind at promoters and across gene bodies. Metagene representation of global Pol II, H3K27Ac, and CDK12 occupancy at promoters and gene bodies. Average ChIP-seq signal in 13906 RefSeq genes expressed in 6h DMSO conditions. c. CDK12, Pol II, and H3K27Ac co-occupy active gene promoters. Left: Heatmaps show the density of Pol II- (left), H3K27Ac- (center), and CDK12- (right) ChIP-Seq reads relative to RefSeq promoters and comprising the region +/−2kb from RefSeq transcription start sites. Right: ChIP-seq read densities for CDK12 and H3K27ac at RefSeq promoters of 4kb centered on transcription start sites. d. CDK12, Pol II, and H3K27Ac co-occupy active enhancers. Left: Heatmaps show the density of Pol II- (left), H3K27Ac- (center), and CDK12- (right) ChIP-Seq reads relative to 32855 H3K27ac –enriched enhancer sites. For each H3K27ac-bound site (y axis), the densities of H3K27Ac (red) and CDK12 (blue) are displayed within a 2 kb window centered on the center of the H3K27ac-bound site. Right: ChIP-seq read densities for CDK12 and H3K27ac at enhancers of 4kb centered on the middle of H3K27ac peaks. For all heatmaps, promoters and enhancers were ranked by Pol II density (black) and a smoothing function was applied. All metagenes are presented in units of rpm/bp.
THZ531 inhibits transcription elongation and gene expression
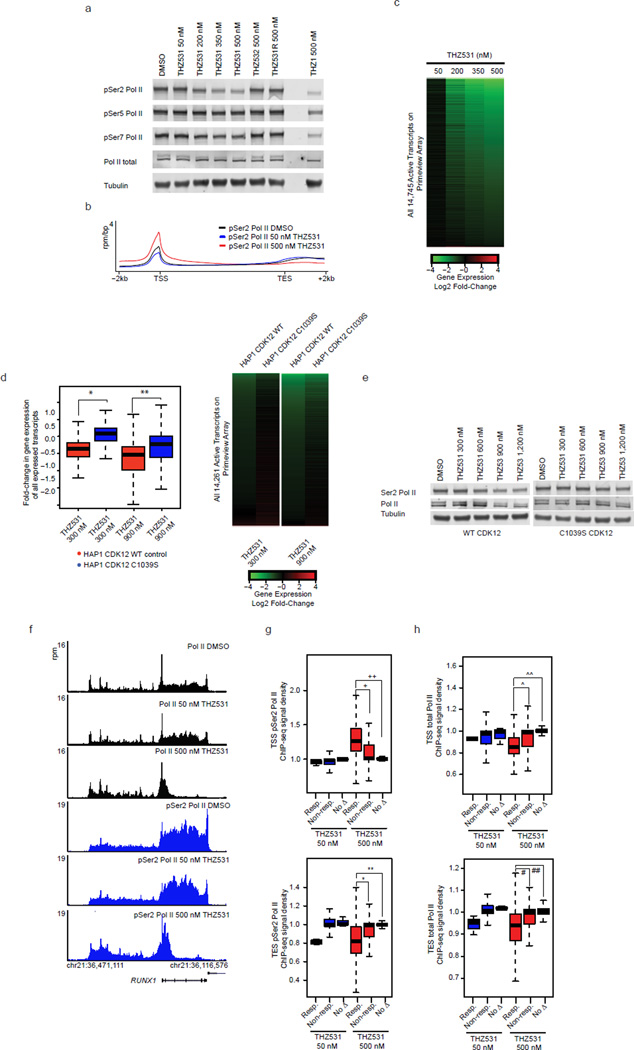
CDK12 is implicated in the control of elongating Pol II, in part, through phosphorylation of the Pol II CTD. To assess if CDK12 and 13 kinase activity is required for Pol II CTD phosphorylation, we treated Jurkat cells with THZ531 at doses across the effective concentration range of the inhibitor (Fig. 1c). In contrast to THZ1, which causes complete loss of Pol II CTD phosphorylation29, THZ531 selectively reduced Ser2 phosphorylation levels without appreciable effect on CTD pSer5/pSer7 levels (Fig. 5a). To understand how these findings extend to gene –associated pSer2 signal we conducted genome-wide analysis of pSer2 ChIP-seq signal. In comparison to total Pol II ChIP-seq signal that displays enrichment primarily at the TSS, pSer2 ChIP-seq signal shows additional pronounced signal in the gene body, particularly near the 3’ ends of genes, consistent with pSer2 being an established mark of elongating Pol II (Fig. 5b). While 50 nM THZ531 treatment did not appreciably change pSer2 ChIP-seq signal, 500 nM THZ531 led to a dramatic decrease in 3’ end –associated pSer2 ChIP-seq signal with a reciprocal accumulation of 5’ end –associated pSer2 signal (Fig. 5b).
Figure 5. THZ531 inhibits transcription elongation and gene expression.
a, THZ531 reduces Pol II CTD Ser2 phosphorylation (pSer2) in a concentration-dependent manner. b, THZ531 reduces 3’ pSer2 Pol II signal density. Metagene representation of global pSer2 Pol II occupancy at gene bodies. Average ChIP-seq signal (rpm/bp) across 13906 genes expressed in DMSO. c, THZ531 downregulates mRNA levels in a concentration-dependent manner. Heatmaps display the Log2 fold-change in gene expression vs. DMSO for 14,745 transcripts expressed in DMSO. Heatmap is ranked by 500 nM THZ531. d, C1039S CDK12 expression partially restores gene expression in HAP1 cells. Box plots (left) and heatmaps (right) displaying expression of 14,261 expressed transcripts. *p-value < 10−16 and **p-value < 10−16. Heatmaps are ranked by WT HAP1 fold-change. e, C1039S CDK12 expression partially restores pSer2 in HAP1 cells. f, THZ531 reduces elongating Pol II at THZ531-response genes. g and h, THZ531 reduces 3’ pSer2 Pol II (g) and Pol II (h) ChIP-seq signal at THZ531-responsive genes. Box plots of pSer2 Pol II and Pol II signal density at transcriptional start sites (TSS, top) and termination sites (TES, bottom) at THZ531–responsive genes (Resp.) and equal numbers of randomly selected non-responsive genes (Non-resp.) or genes exhibiting no change (no Δ). 7 and 3801 genes were sensitive to THZ531 at 50 and 500 nM THZ531, respectively. +p-value = 1.64e-252, ++p-value = 0, *p-value = 1.31e-136, **p-value = 9.00e-180. ^p-value = 1.17e-231, ^^p-value = 0, #p-value = 4.43e-101, ##p-value = 1.21e-155. Sensitive genes are defined by > 2 log2 fold-change in gene expression. Uncut western blots are in Supplementary Fig. 10. P-values were determined with a two-tailed Student’s T test.
Reduction of Pol II CTD Ser2 phosphorylation, a mark of active transcriptional elongation, led us to investigate whether THZ531 affects gene expression. To address this, we treated Jurkat cells for 6 hours with increasing doses of THZ531 and examined the effect on steady-state mRNA levels. Treatment with 50 nM THZ531, a concentration that leads to minimal loss of CDK12 activity and pSer2 levels, resulted in the loss of expression of a small subset of genes (Fig. 5c, Supplementary Data Set 3). Further dose escalation of THZ531 using concentrations that result in the complete targeting of CDK12 and greater reduction of pSer2 levels, led to gradual erosion of mRNA output with 25% and 46% of transcripts displaying at least 2- and 1.5- log2 fold reduction (4- and 2.25 linear fold-change) in expression following 500 nM THZ531 treatment, respectively (Fig. 5c). CRISPR/Cas9 –mediated replacement of CDK12 WT for CDK12 C1039S was sufficient to partly restore Ser2 CTD phosphorylation and gene expression (Fig. 5d,e, Supplementary Fig. 6a). We believe the partial restoration of gene expression may result from continued CDK13 activity or from an unknown off-target. Consistent with a lack of anti-CDK12 activity, THZ531R and THZ532 decreased neither Ser2 phosphorylation nor steady-state mRNA levels (Fig. 5a, Supplementary Fig. 6b). Gene expression analysis following THZ1 treatment revealed a similar, but not identical, transcriptional downregulation, consistent with the general role of CDK7 in mediating the transcription initiation-to-elongation step33 (Supplementary Fig. 6c).
THZ531 treatment did not completely abolish Ser2 phosphorylation levels, suggesting that loss of Ser2 phosphorylation may occur in a gene –specific manner. To understand if THZ531 –responsive genes preferentially suffer loss of Ser2 phosphorylation upon THZ531 treatment, thereby contributing to their sensitivity, we first analyzed pSer2 Pol II ChIP-seq signal at genes whose expression is reduced with THZ531 treatment. Coincident with loss of gene expression, we observed reduced pSer2 ChIP-seq signal near the 3’ ends of THZ531 –responsive genes (Fig. 5f, Supplementary Fig. 7a). Global analysis of pSer2 ChIP-seq density at all THZ531 –responsive genes (>2 log2 fold reduction in gene expression) demonstrated a significant decrease in 3’ end -associated and an increase in 5’ end –associated pSer2 ChIP-seq signal density compared to non –responsive genes, suggestive of a loss of Pol II processivity at THZ531 –responsive genes (Fig. 5g).
In agreement with a marked decrease in gene body -associated pSer2 ChIP-seq signal, we observed that THZ531 led to a dramatic depletion of elongating Pol II at these same genes (Fig. 5f, h; Supplementary Fig. 7a). Importantly, the Pol II elongation defect observed following THZ531 was significantly different from that obtained following inhibition of CDK9 by Flavopiridol treatment. Whereas Flavopiridol causes an arrest proximal to transcription start sites (TSS), THZ531 treatments results in reduced TSS – associated Pol II and reduced Pol II processivity across the gene body (Supplementary Fig. 7b, c). These results suggest that THZ531 downregulates gene expression, in part, through the loss of Pol II processivity.
THZ531 inhibits DDR and transcription factor gene expression
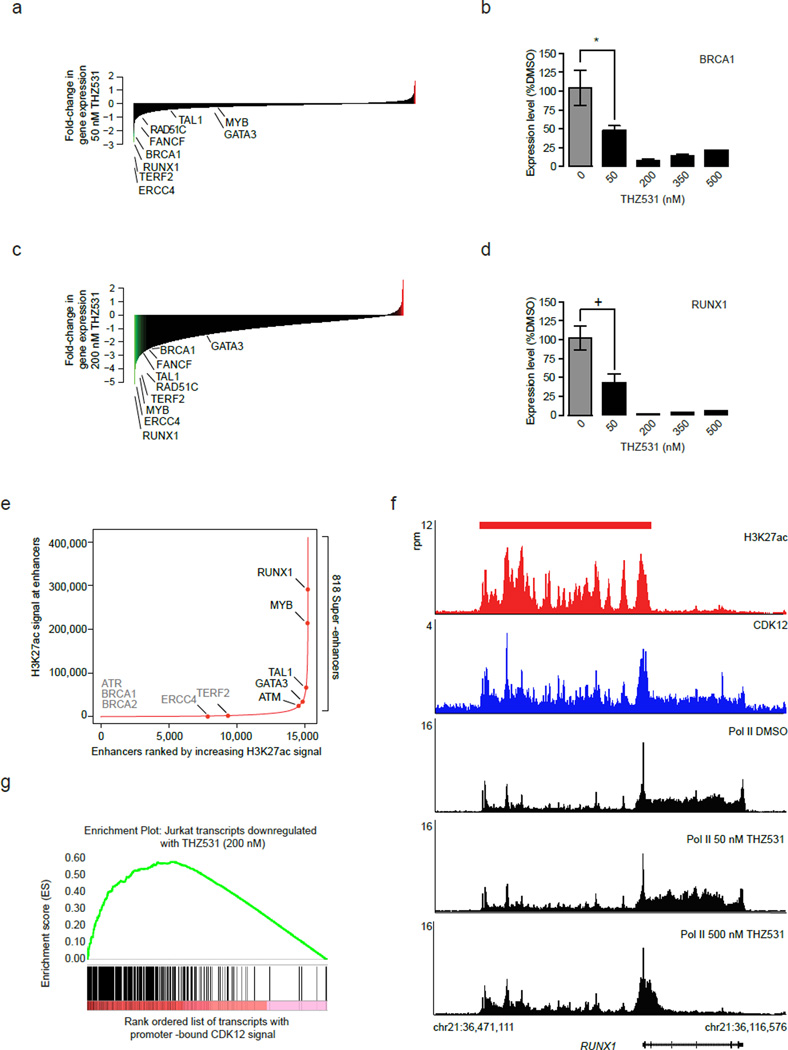
We asked whether the loss of expression of certain genes or gene sets could contribute to the sensitivity of Jurkat cells to THZ531. To address this we subjected the set of gene transcripts sensitive to THZ531 to gene ontology (GO) term enrichment analysis and identified two distinct gene signatures. At 50 nM THZ531 we observed enrichment for DNA damage response (DDR) pathway genes among those genes most sensitive to THZ531 (Fig. 6a; Supplementary Fig. 8a). Core DDR genes including BRCA1, FANCF, and ERCC4 all showed substantial loss of expression with 50 nM THZ531, an effect that was further exacerbated at higher doses (Fig. 6b; Supplementary Fig. 8b). Loss of DDR gene expression following CDK12-cyclin K knockdown is believed to result, in part, from reduced active transcription across these genes.18 Indeed we also observed reduced Pol II occupancy at these genes commensurate with the loss of gene expression (Fig. 5; Supplementary Fig. 7).
Figure 6. THZ531 inhibits DDR and transcription factor gene expression.
a, DDR genes are sensitive to 50 nM THZ531. b, RT-qPCR of BRCA1 gene expression following THZ531 treatment. *p-value=9.42e-05, c, Transcription factor genes are sensitive to 200 nM THZ531. d, RT-qPCR of RUNX1 gene expression following THZ531 treatment. +p-value=5.33e-06. e, Total H3K27Ac ChIP-seq signal in enhancer regions for all stitched enhancers ranked by increasing H3K27Ac ChIP-seq signal. Gene names in gray either have no associated enhancer or associated typical enhancers. f, RUNX1 gene contains exceptional CDK12 and H3K27Ac signal and suffers profound loss of elongating Pol II following THZ531 treatment. CDK12 (blue), H3K27Ac (red) and Pol II (black) ChIP-seq occupancy at the promoter and across the gene body of RUNX1 locus following THZ531 treatment. The red bar indicates the genomic coordinates of a super –enhancer. g, Transcripts downregulated by 200 nM THZ531 are enriched for transcripts whose gene promoters contain high levels of CDK12. Gene set enrichment analysis of top 500 genes downregulated following THZ531 treatment in comparison to CDK12 promoter-bound ChIP-seq signal of these transcripts, GSEA-supplied p-value < 0.001. Heatmaps (a and c) display the log2 fold-change in gene expression vs. DMSO for the 14,745 transcripts expressed in Jurkat cells. RT qPCRs were performed in biological triplicate and error bars are +/− SD. P-values were determined with a two-tailed Student’s T test.
Interestingly, while the transcripts most sensitive to 50 nM THZ531 were enriched for regulators of DDR pathway, the transcripts that were most sensitive to higher doses THZ531 (200 nM or greater) were enriched for transcripts that encode for transcription factors including those known to be drivers of Jurkat T-cell acute lymphoblastic leukemia (Fig. 6c; Supplementary Fig. 8c). Among these transcription factors are RUNX1, MYB, TAL1, and GATA3, which comprise the Jurkat transcriptional autoregulatory loop (Fig. 6d; Supplementary Fig. 8d).34,35 All of these transcription factor genes are associated with super –enhancers that were previously shown to contain disproportionate amounts of transcriptional apparatus, including CDK7 (Fig. 6e, Supplementary Fig. 8e and Data Set 4).29 Indeed, super-enhancer—associated genes were collectively more downregulated upon THZ531 treatment than those associated with typical enhancers (Supplementary Fig. 8f). Analyzing genome-wide CDK12 ChIP-seq signal revealed that these genes, in particular RUNX1, MYB, TAL1, and GATA3, as well as their associated super–enhancers contained large amounts of CDK12 ChIP-seq signal as well as Pol II (Fig. 6f, Supplementary Fig. 9a, 9b). Interestingly, we observed a dramatic reduction in elongating Pol II following THZ531 treatment, in contrast to a reduction in overall Pol II occupancy following THZ1 treatment (Supplementary Fig. 9a). As CDK12 has been implicated in the phosphorylation of Ser2, a mark of transcriptional elongation, these results are consistent with CDK12 inhibition. Therefore, we asked whether the profound loss of gene expression following 200 nM THZ531 treatment correlated with the levels of CDK12 (i.e –THZ531 target). Gene set enrichment analysis of the top 500 transcripts downregulated by 200 nM THZ531 revealed that the most THZ531 -responsive genes tended to be particularly enriched in promoter-proximal and enhancer –bound CDK12 signal (Fig. 6g, Supplementary Fig. 9c). This indicated that at saturating doses of THZ531, CDK12 target occupancy may contribute to gene expression sensitivity to THZ531.
Discussion
Here we have reported the rational design and characterization of the first covalent inhibitor of CDK12 and 13 enzymatic activities. The CDK12 crystal structure revealed that it contains an extended C-terminal domain that traverses the kinase domain, positioning a cysteine residue proximal to the ATP –binding site in a manner analogous to CDK7. Therefore, we reasoned that THZ1, a recently reported CDK7 inhibitor that targets the CDK7 C-terminal cysteine, would make a suitable synthetic starting point. Indeed we show that simple chemical modification to alter the orientation of the electrophilic acrylamide permitted us to selectively target CDK12 and CDK13. We demonstrated that covalent binding to CDK12 and 13 is critical for the potency and selectivity of THZ531 as its reversible analog is more than 100-fold less active. Therefore, while high sequence and structural similarity within ATP–binding pockets poses a significant challenge to developing selective ATP–competitive inhibitors of CDKs; targeting unique cysteine nucleophilic hotspots, particularly those lying outside the conserved kinase domain, may circumvent these confounding factors. It remains to be seen whether this strategy can be readily deployed to target other cysteine – targetable members of the kinome (or proteome). The identification of starting chemical matter that can be modified to deliver an electrophilic warhead proximal to these cysteines presents the first hurdle.
We found that pharmacological inhibition of CDK12 and 13 –cyclin K complexes with THZ531 led to diminished pSer2 and elongating Pol II at THZ531 –target genes. This post-translation modification has previously been ascribed to CDK9 kinase activity, largely from studies using pharmacological agents. However, recent work has shown that Flavopiridol inhibits CDK12 at concentrations that decrease Ser2 phosphorylation, suggesting that CDK12 may influence the pharmacology of this inhibitor.16,26 Furthermore, using several unbiased and orthogonal approaches we found no evidence that THZ531 targets CDK9, which lacks a C-terminal cysteine. Lastly, Pol II occupancy at THZ531 –responsive genes following drug treatment is distinct from that of Flavopiridol. Whereas Pol II undergoes a pronounced pause proximal to gene promoters following Flavopiridol treatment, Pol II entered the gene body following THZ531 treatment but experienced reduced processivity, suggesting a defect in elongation proficiency or termination steps. These data would seem to support existing models of Pol II regulation in other organisms, including Saccharomyces cerevisiae, where regulation of transcriptional elongation is sequentially and differentially regulated by BUR1 and CTK1.36
Coincident with THZ531 decreasing the elongating Pol II population, we found that THZ531 downregulated the expression of certain genes in Jurkat T-ALL. We found two general signatures of genes that were particularly sensitive to THZ531 treatment. At concentrations leading to complete CDK12 and 13 inhibition, genes encoding factors that regulate transcription were exceptionally sensitive to THZ531. At this concentration, the loss of expression of the most sensitive genes positively correlated with the amount of promoter and enhancer–bound CDK12 with super-enhancer—associated genes being especially sensitive. These data suggest that inhibition of CDK12, like that of CDK7, may provide another means of targeting super –enhancer –associated oncogene expression in various cancers. Additionally, we found that genes involved in DDR were exquisitely sensitive to low doses of THZ531 and continue to lose expression with escalating doses of THZ531. A similar set of DDR genes was recently shown to be sensitive to loss of CDK12 and 13 cyclin K complexes.13,18 In contrast to THZ531 treatment, neither depletion of CDK12 and 13 nor mutations that disrupt complex integrity leads to gross disruption of the larger gene expression program. This discrepancy could be explained by numerous factors. We favor the explanation that THZ531 treatment effectively creates an inactive or dominant negative CDK12 and 13 complexes, which elicits fundamentally different effects on transcription. However, the modest recovery of wild-type function following expression of CDK12 C1039S suggests that remaining CDK13 activity, or off-target activity on other transcription-associated kinases such as JNK1/2/3 (identified in Supplementary Table 1 and Data Set 2) or other unknown off-targets may also account for this discrepancy.
Recent research in both yeast and mammalian systems has identified genetic and physical interactions between the CDK12-cyclin K complex and the DDR pathway. Genetic depletion of the complex reduces the expression of a core set of DDR genes.13,18–21 Therefore, while the CDK12-cyclin K complex exerts control over Pol II – mediated transcription, its loss appears to have pleiotropic consequences for both transcription and the DNA damage response (DDR) pathway. THZ531 does indeed downregulate core DDR pathway genes, which may have very important mechanistic and therapeutic implications for CDK12 and 13 inhibition. Mechanistically, this suggests a link between the integrity of DNA damage signaling and transcription that is specific to CDK12, as other CDKs that were probed in previous research did not affect DDR gene expression.13,18
From a therapeutic standpoint, it is attractive to think that an inhibitor, such as THZ531, could target both aberrant transcription and genomic instability, two enabling characteristics that facilitate the acquisition of cancer hallmark capabilities. While both of these characteristics are common features of all cancers, certain cancers may be particularly susceptible to CDK12 and 13 inhibition. T-cell acute lymphoblastic leukemia (T-ALL) exploits a variety of genetic perturbations including chromosomal translocations, deletions, and insertions to create super –enhancers that drive aberrant expression of oncogenic transcription factors, including TAL1.34,37,38 As a single agent, THZ531 would appear to induce significant transcriptional stress that primes cells for apoptotic cell death. In Jurkat T-ALL, the loss of transcription factors that form the transcriptional regulatory core is sufficient to initiate apoptotic signaling.29,34,35 Breast and ovarian cancers harboring BRCA1 and 2 mutations are deficient in homologous recombination (HR), yet they show excellent response to PARP1 inhibitors resulting from a synthetic lethal relationship between the combined loss of HR and loss of single strand repair. The loss of BRCA expression following CDK12 inhibition would suggest a similar relationship to hold for CDK12 and PARP1, allowing treatment with potentially low doses of THZ531 in combination with recently clinically –approved PARP1 inhibitors. Indeed recent work in ovarian cancers that harbor CDK12 inactivating mutations, shows that attenuation of CDK12 signaling is sufficient to sensitize cells to PARP1 inhibition.23,24 CDK12 and 13 inhibition therefore may hold promise for the treatment of cancers that utilize aberrant transcription or genomic instability to gain a selective growth advantage.
Online Methods
Chemistry
All solvents and reagents were used as obtained. 1H NMR spectra were recorded with a Varian Inova 600 NMR spectrometer and referenced to dimethylsulfoxide. Chemical shifts are expressed in ppm. Mass spectra were measured with Waters Micromass ZQ using an ESI source coupled to a Waters 2525 HPLC system operating in reverse mode with a Waters Sunfire C18 5 µm, 4.6 mm × 50 mm column. Purification of compounds was performed with either a Teledyne ISCO CombiFlash Rf system or a Waters Micromass ZQ preparative system. The purity was analyzed on an above-mentioned Waters LC-MS Symmetry (C18 column,4.6 mm × 50 mm, 5 µM) using a gradient of 5–95% methanol in water containing 0.05% trifluoroacetic acid (TFA). Detailed synthetic schemes and characterization data are presented below and elsewhere.1
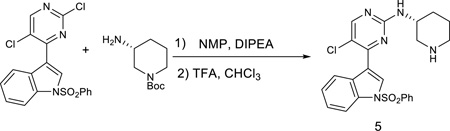
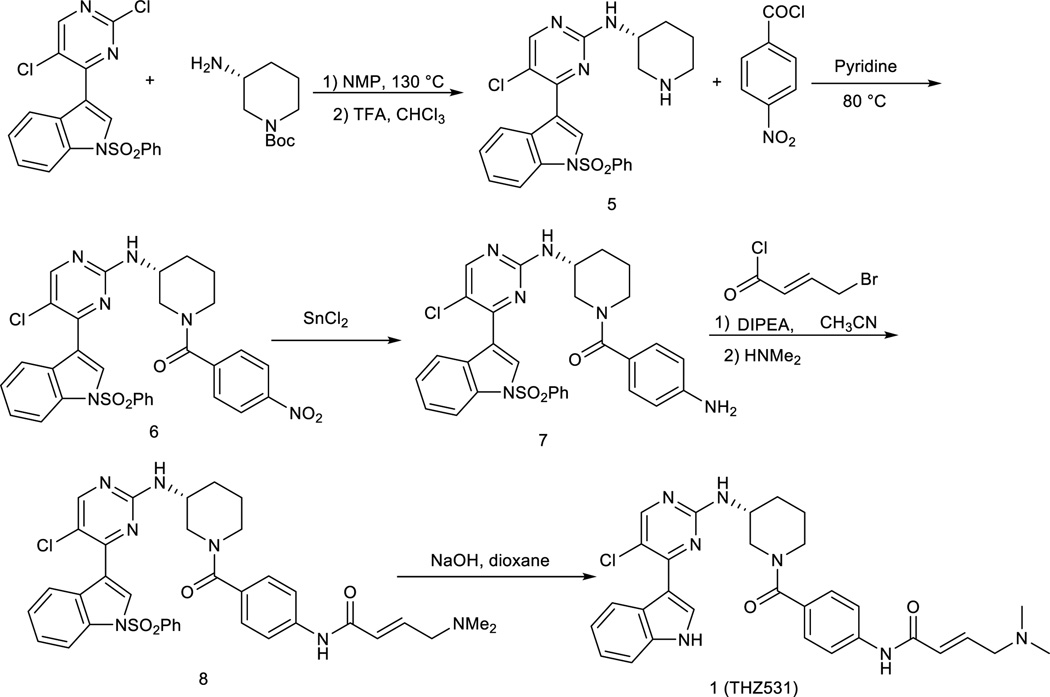
(R)-5-chloro-4-(1-(phenylsulfonyl)-1H-indol-3-yl)-N-(piperidin-3-yl)pyrimidin-2-amine
To a solution of 3-(2, 5-dichloropyrimidin-4-yl)-1-(phenylsulfonyl)-1H-indole (402mg) in NMP (5.0mL) was added tert-butyl (R)-3-aminopiperidine-1-carboxylate (200 mg, 1.0 equiv) and diisopropylethylamine (129 mg, 1.0 equiv). The solution was heated for 2 h at 130 °C. The cooled solution was diluted with 100 mL of CHCl3 and i-PrOH (4:1, 100 mL)) and then washed with water. After removing solvent, the crude was dissolved in CHCl3 (10 mL) and then was treated with TFA (5 mL). After 30 min stirring at room temperature, the solvent was then removed. The separation by silica gel chromatography column with CH2Cl2/methanol (10/1) to give the product 5 (350 mg, 76%, two steps). LC-MS: m/z (M+H) 468.1H NMR (500 MHz, DMSO) δ 8.70 (s, 2H), 8.54 (s, 1H), 8.48 (s, 1H), 8.29 (d, J = 8.2 Hz, 1H), 8.07 – 8.03 (m, 2H), 8.01 (d, J = 8.4 Hz, 1H), 7.73 (t, J = 7.5 Hz, 1H), 7.63 (t, J = 7.9 Hz, 2H), 7.45 (dd, J = 11.3, 4.1 Hz, 2H), 7.37 (t, J = 7.6 Hz, 1H), 4.18 (td, J = 13.5, 6.9 Hz, 1H), 3.41 (s, 1H), 3.22 – 3.17 (m, 1H), 2.91 (dd, J = 21.6, 11.2 Hz, 2H), 2.04 (dd, J = 12.6, 3.8 Hz, 1H), 1.92 (d, J = 4.4 Hz, 1H), 1.80 – 1.56 (m, 2H).13C NMR (125 MHz, DMSO) 13C NMR (126 MHz, DMSO) δ 160.19, 158.72, 156.61, 137.33, 135.47, 134.50, 130.47 (2C), 129.85, 128.99, 127.35 (2C), 126.03, 124.61, 123.56, 117.82, 116.04, 113.51, 47.06, 45.65, 43.76, 28.57, 20.97.
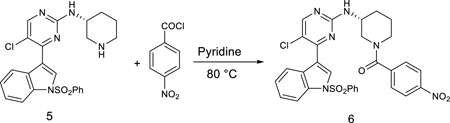
(R)-(3-((5-chloro-4-(1-(phenylsulfonyl)-1H-indol-3-yl)pyrimidin-2-yl)amino)piperidin-1-yl)(4-nitrophenyl)methanone
To a stirred solution of the compound 5 (467 mg) in pyridine (10 mL) was added 4-nitrobenzoyl chloride (180 mg, 1.0 equiv) and then was heated to 80 °C. The reaction mixture was stirred for 2 hours and then was concentrated under reduced pressure. The resulting crude was purified by silica gel column with CH2Cl2/methanol (10/1) to give the product 6 (450 mg, 73%). LC-MS: m/z (M+H) 617. 1H NMR (500 MHz, DMSO) δ 8.49 (s, 1H), 8.31 (d, J = 41.7 Hz, 2H), 8.05 (d, J = 7.7 Hz, 2H), 8.00 (d, J = 8.3 Hz, 1H), 7.73 (t, J = 7.4 Hz, 1H), 7.63 (t, J = 7.8 Hz, 2H), 7.55 (s, 1H), 7.44 (t, J = 7.7 Hz, 1H), 7.37 (dd, J = 16.3, 8.5 Hz, 2H), 3.90 (s, 1H), 3.52 (s, 2H), 3.28 (s, 2H), 2.10 – 2.00 (m, 1H), 1.91 (s, 1H), 1.75 (s, 1H), 1.58 (s, 1H).13C NMR (125 MHz, DMSO) δ 168.01, 160.29, 158.51, 156.27, 148.12, 143.03, 137.36, 135.43, 134.48, 130.45 (2C), 129.71, 128.95, 128.49 (2C), 127.31 (2C), 126.00, 124.57, 123.62 (2C), 117.85, 115.58, 113.45, 50.52, 48.04, 42.61, 29.88, 22.67.
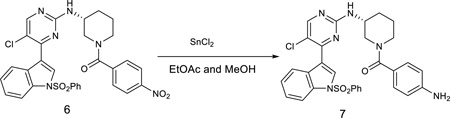
(R)-(4-aminophenyl)(3-((5-chloro-4-(1-(phenylsulfonyl)-1H-indol-3-yl)pyrimidin-2-yl)amino)piperidin-1-yl)methanone
The nitro compound 6 (617 mg) was suspended in ethyl acetate/methanol (5:1, 30 mL) and was treated with SnCl2 (380 mg, 2.0 equiv). After stirring for 2 hours at 80 °C, the reaction mixture was cooled down to room temperature and poured onto saturated aqueous NaHCO3. The mixture was stirred for 10 min followed by extraction with chloroform and 2-propanol (4:1, 100 mL). The organic layer was washed with water and brine, dried over sodium sulfate, filtered through a pad of celite and concentrated under reduced pressure. The resulting crude was purified by flash column with CH2Cl2/methanol (10/1) to provide the compound 7 (290 mg, 50%). LC-MS: m/z (M+H) 587. 1H NMR (500 MHz, DMSO) δ 8.54 (s, 1H), 8.40 (s, 1H), 8.32 (d, J = 7.9 Hz, 1H), 8.09 – 8.03 (m, 2H), 8.00 (d, J = 8.3 Hz, 1H), 7.72 (t, J = 7.5 Hz, 1H), 7.62 (t, J = 7.9 Hz, 2H), 7.47 – 7.40 (m, 1H), 7.34 (t, J = 7.3 Hz, 1H), 7.20 (d, J = 8.2 Hz, 2H), 6.88 (s, 4H), 6.70 (d, J = 7.9 Hz, 2H), 4.07 (d, J = 11.7 Hz, 1H), 3.92 (s, 1H), 3.80 (d, J = 12.5 Hz, 1H), 3.10 (dt, J = 22.1, 7.5 Hz, 2H), 2.12 – 1.96 (m, 1H), 1.82 (dd, J = 9.0, 4.4 Hz, 1H), 1.67 (ddd, J = 16.5, 11.9, 3.8 Hz, 1H), 1.59 – 1.45 (m, 1H).13C NMR (125 MHz, DMSO) δ 170.41, 160.37, 158.53, 156.41, 146.65, 137.37, 135.40, 134.49, 130.44 (2C), 129.78, 129.20 (2C), 129.05, 127.33 (2C), 126.32, 125.95, 124.54, 123.72, 117.94, 115.57 (2C), 115.44, 113.41, 49.66, 48.26, 45.39, 30.52, 23.96.
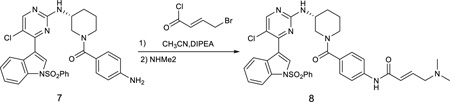
(R,E)-N-(4-(3-((5-chloro-4-(1-(phenylsulfonyl)-1H-indol-3-yl)pyrimidin-2-yl)amino)piperidine-1-carbonyl)phenyl)-4-(dimethylamino)but-2-enamide
To the solution of the compound 7 (60 mg) in acetonitrile (10 mL) was added diisopropylethylamine (13 mg, 1.0 equiv). The reaction mixture was cooled down to 0 °C and then treated with 4-chlorobut-2-enoyl chloride (54 mg, 3.0 equiv) in CH2Cl2. The reaction mixture was stirred for 10 min at 0 °C followed by adding dimethylamine in THF (1M, 2.0 mL). The reaction mixture was then warmed up to room temperature, stirred for 1 hour and then concentrated under reduced pressure. The resulting crude was purified by preparative HPLC to give the product 8 (38 mg, 55%) LC-MS: m/z (M+H) 698. 1H NMR (500 MHz, DMSO) δ 10.38 (s, 1H), 8.53 (s, 1H), 8.37 (s, 1H), 8.30 (d, J = 6.9 Hz, 1H), 8.08 – 8.02 (m, 2H), 7.99 (d, J = 8.3 Hz, 1H), 7.72 (t, J = 7.5 Hz, 1H), 7.70 – 7.58 (m, 4H), 7.47 – 7.40 (m, 1H), 7.39 – 7.27 (m, 4H), 6.88 – 6.80 (m, 1H), 6.54 (d, J = 15.4 Hz, 1H), 5.95 (s, 1H), 3.95 (d, J = 6.6 Hz, 2H), 3.91 (s, 1H), 3.67 (d, J = 25.4 Hz, 1H), 3.29 – 3.09 (m, 2H), 2.81 (s, 6H), 2.56 (d, J = 8.7 Hz, 1H), 2.11 – 1.98 (m, 1H), 1.85 (s, 1H), 1.70 (dd, J = 9.4, 3.5 Hz, 1H), 1.54 (dd, J = 9.7, 3.8 Hz, 1H). 13C NMR (125 MHz, DMSO) δ 169.74, 162.73, 160.33, 158.51, 156.38, 140.22, 137.35, 135.41, 134.47, 132.74, 131.95, 131.80, 130.44 (2C), 129.81, 129.04, 128.15 (2C), 127.32 (2C), 125.94, 124.55, 123.68, 119.42 (2C), 117.89, 115.50, 113.41, 57.35, 49.48, 48.17, 44.88, 42.54 (2C), 30.27, 23.65
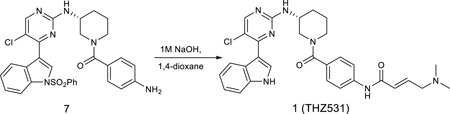
(R,E)-N-(4-(3-((5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl)amino)piperidine-1-carbonyl)phenyl)-4-(dimethylamino)but-2-enamide
The compound 8 (70 mg) was dissolved in 1,4-dioxane (5 mL) and aqueous NaOH (1.0 M, 5 mL). The solution was allowed to stir at room temperature for 2 hours followed by neutralization with HCl (1M, 5 mL). The solution was then extracted with chloroform and 2-propanol (4:1, 30 mL) and was washed with water. The removal of solvent provided the crude which was purified by HPLC to give the final product (25 mg, 43%). LC-MS 558 (M+1), 1H NMR (500 MHz, DMSO-d6) 11.83 (s, 1H), 10.94 (s, 1H), 10.41 (s, 1H), 8.53 (s, 1H), 8.44 (s, 1H), 8.25 (s, 1H), 7.67 (d, J = 7.0 Hz, 1H), 7.52 (d, J = 8.0 Hz, 1H), 7.36 (d, J = 8.5 Hz, 2H), 7.22 (t, J = 8.0 Hz, 1H), 7.16 (d, J = 8.0 Hz, 1H), 6.83 (m, 1H), 6.52 (d, J = 15.0 Hz, 1H), 4.02 (br, 2H), 3.93-3.80 (m, 3H), 3.20 (br, 2H), 2.80 (s, 6H), 2.10 (m, 1H), 1.86 (m, 1H), 1.71 (m, 1H), 1.61 (m, 1H). 13C NMR (126 MHz, DMSO) δ 169.89, 162.78, 160.36, 157.54, 153.27, 140.36, 136.78, 132.99, 132.70, 131.99, 131.65, 128.11 (2C), 126.74, 123.20, 122.96, 121.69, 119.42 (2C), 113.66, 112.59, 111.53, 57.19, 49.12, 48.30, 45.07, 42.31 (2C), 30.21, 23.51.
Same synthetic method as compound 1 (THZ531)
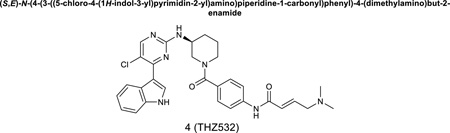
(S,E)-N-(4-(3-((5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl)amino)piperidine-1-carbonyl)phenyl)-4-(dimethylamino)but-2-enamide
LC-MS 558 (M+1), 1H NMR (500 MHz, DMSO-d6) 11.83 (s, 1H), 10.94 (s, 1H), 10.41 (s, 1H), 8.53 (s, 1H), 8.44 (s, 1H), 8.25 (s, 1H), 7.67 (d, J = 7.0 Hz, 1H), 7.52 (d, J = 8.0 Hz, 1H), 7.36 (d, J = 8.5 Hz, 2H), 7.22 (t, J = 8.0 Hz, 1H), 7.16 (d, J = 8.0 Hz, 1H), 6.83 (m, 1H), 6.52 (d, J = 15.0 Hz, 1H), 4.02 (br, 2H), 3.93-3.80 (m, 3H), 3.20 (br, 2H), 2.80 (s, 6H), 2.10 (m, 1H), 1.86 (m, 1H), 1.71 (m, 1H), 1.61 (m, 1H). 13C NMR (126 MHz, DMSO) δ 169.89, 162.78, 160.36, 157.54, 153.27, 140.36, 136.78, 132.99, 132.70, 131.99, 131.65, 128.11 (2C), 126.74, 123.20, 122.96, 121.69, 119.42 (2C), 113.66, 112.59, 111.53, 57.19, 49.12, 48.30, 45.07, 42.31 (2C), 30.21, 23.51.
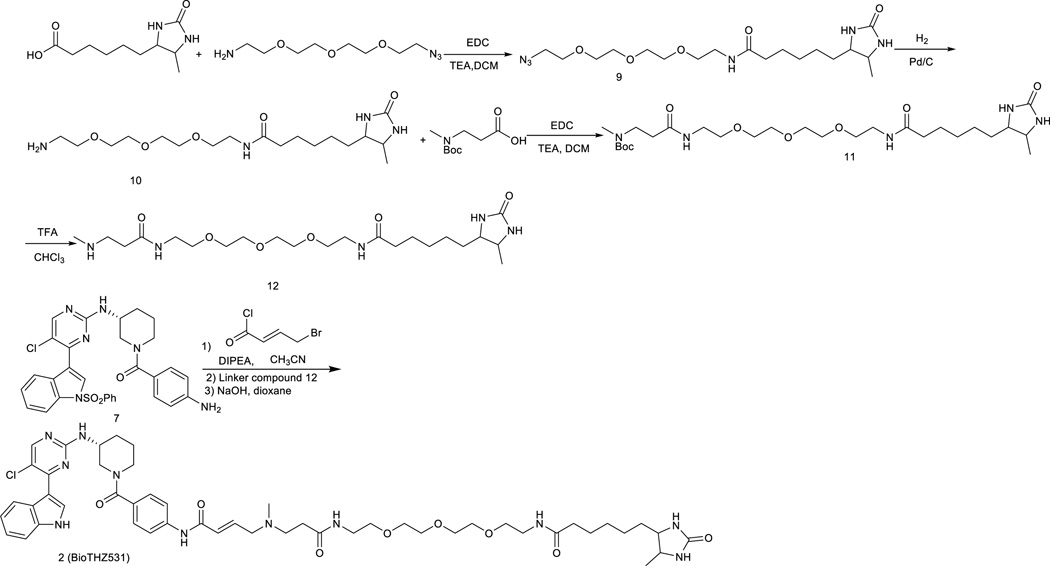
N-(2-(2-(2-(2-azidoethoxy)ethoxy)ethoxy)ethyl)-6-(5-methyl-2- oxoimidazolidin-4-yl)hexanamide
Desthiobiotin (430 mg, 2.0 mmol), 11-Azido-3,6,9-trioxaundecan-1-amine (435 mg, 2.0 mmol) and 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (576 mg, 3.0 mmol), were dissolved in methylene chloride (10 mL) and trimethylamine (380 uL, 3.0 mmol) was added at 0°C. The mixture was stirred at room temperature for 6 hours. The reaction solution was added with water and extracted with chloroform and isopropanol (4:1). The organic layer was dried with sodium sulfate and the solvent was removed under the reduced pressure to give a crude product which was used directly in the next step.
N-(2-(2-(2-(2-aminoethoxy)ethoxy)ethoxy)ethyl)-6-(5-methyl-2-oxoimidazolidin-4-yl)hexanamide
the crude from above reaction was dissolved in methanol (10 mL) and then 10% Pd on carbon (50 mg) was added. The mixture solution was stirred for 3 hours with hydrogen balloon at room temperature. The solution was filtered to remove catalyst and solvent was removed under reduced pressure. The product was purified by silica gel column with CH2Cl2/methanol (10/1) to provide the product (233 mg, 30%). LC-MS: m/z (M+H) 389. 1H NMR (500 MHz, MeOD) δ 3.85 (dd, J = 7.8, 6.5 Hz, 1H), 3.76 – 3.70 (m, 1H), 3.71 – 3.64 (m, 8H), 3.62 – 3.50 (m, 4H), 3.40 (d, J = 5.5 Hz, 1H), 3.34 (dt, J = 3.3, 1.6 Hz, 1H), 2.81 (dd, J = 7.2, 3.4 Hz, 2H), 2.25 (dd, J = 15.2, 7.9 Hz, 2H), 1.72 – 1.60 (m, 2H), 1.59 – 1.50 (m, 2H), 1.50 – 1.27 (m, 4H), 1.14 (d, J = 6.5 Hz, 3H).
tert-butyl methyl(22-(5-methyl-2-oxoimidazolidin-4-yl)-3,17-dioxo-7,10,13-trioxa-4,16-diazadocosyl)carbamate
The compound 10 (388 mg, 1.0 mmol) 3-((tert-butoxycarbonyl)(methyl)amino)propanoic acid (260 mg, 1.3 mmol) and 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (370 mg, 2.0 mmol) in methylene chloride (15 mL) and then trimethylamine (270 uL, 2.0 mmol) was added at 0°C. The mixture solution was stirred for 6 hours. The solvent was removed under the reduced pressure and the residue was purified by purified by silica gel column with CH2Cl2/methanol (10/1) to provide the product (120mg, 21%) LC-MS: m/z (M+H) 574.37 1H NMR (500 MHz, MeOD) δ 3.85 (dd, J = 7.9, 6.5 Hz, 1H), 3.76 – 3.62 (m, 8H), 3.59 – 3.50 (m, 5H), 3.40 (d, J = 5.5 Hz, 3H), 3.34 (dt, J = 3.3, 1.6 Hz, 1H), 3.21 – 3.12 (m, 1H), 2.89 (s, 3H), 2.45 (t, J = 6.9 Hz, 2H), 2.36 (s, 2H), 2.24 (t, J = 7.5 Hz, 2H), 1.69 (dq, J = 29.6, 7.5 Hz, 2H), 1.53 (ddd, J = 7.5, 5.2, 2.7 Hz, 2H), 1.49 (s, 9H), 1.45 – 1.30 (m, 3H), 1.15 – 1.10 (m, 3H).
6-(5-methyl-2-oxoimidazolidin-4-yl)-N-(5-oxo-9,12,15-trioxa-2,6-diazaheptadecan-17-yl)hexanamide
Crude compound 11 was dissolved in chloroform (10 mL) and then trifluoroacetic acid (5 mL) was added. The solution was stirred at room temperature for 1 hour and then solvent was removed under the reduced pressure. The residue was purified by preparative LC-MS with methanol and water (1% TFA) as eluent to provide compound 11 (165 mg, 35% for two steps). LC-MS: m/z (M+H) 474.
N-((E)-20-((4-((R)-3-((5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl)amino)piperidine-1-carbonyl)phenyl)amino)-16-methyl-13,20-dioxo-3,6,9-trioxa-12,16-diazaicos-18-en-1-yl)-6-(5-methyl-2-oxoimidazolidin-4-yl)hexanamide
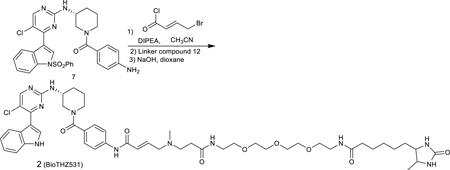
To the solution of the compound 7 (60 mg) in acetonitrile (10 mL) was added diisopropylethylamine (13 mg, 1.0 equiv). The reaction mixture was cooled down to 0° and then treated with 4-chlorobut-2-enoyl chloride (54 mg, 3.0 equiv) in CH2Cl2. The reaction mixture was stirred for 10 min at 0 °C and propanol (4:1, 30 mL) and was washed with water. The removal of solvent provided the crude which was used then dissolved in THF (3.0 mL). To this solution, the free amino compound 12 (94.0 mg, 0.2 mmol) was added and the solution were then heated for 2 hours at 50°. The reaction mixture was then cooled down to room temperature. The solvent was removed under reduced pressure and the residue was dissolved again in 1,4-dioxane (2.0 mL) followed by adding 1M NaOH solution (2.0 mL). The solution was stirred at room temperature for 2 hours and then was quenched with 1M HCl solution (2.0mL), extracted with chloroform/2-propanol (4/1, vol/vol, 20 mL). The organic layer was washed with water, brine and dried over sodium sulfate. The crude after removing the solvent was purified by HPLC to provide BioTHZ531 as TFA salt (15.0 mg, 15%) LC-MS: m/z (M+H) 987,1H NMR (500 MHz, DMSO-d6) 11.67 (s, 1H), 10.23 (s, 1H), 8.54 (s, 1H), 8.39 (s, 1H), 8.21 (s, 1H), 8.05 (s, 1H), 7.65 (d, J = 7.0 Hz, 2H), 7.55 (br, 1H), 7.51 (d, J = 8.0 Hz, 1H), 7.38 (d, J = 8.0 Hz, 2H), 7.22 (t, J = 8.0 Hz, 1H), 7.16 (t, J = 8.0 Hz, 1H), 7.06 (br, 1H), 6.83 (m, 1H), 6.52 (d, J = 15.0 Hz, 1H), 4.20-3.20 (m, 20H), 2.80 (s, 3H), 2.65 (t, J = 7.0 Hz, 2H), 2.08 (m, 3H), 1.86 (m, 1H), 1.71 (m, 1H), 1.58 (m, 1H), 1.51 (m, 2H), 1.40-1.22 (m, 4H), 1.00 (d, J = 6.0 Hz, 3H). 13C NMR (125 MHz, DMSO-d6) δ 173.76, 170.70, 169.77, 163.10, 160.22, 158.68, 158.38, 157.41, 140.41, 136.63, 131.44, 131.08, 128.15, 126.81, 123.27, 122.76, 121.01, 119.18, 114.07, 112.24, 111.66, 55.75, 55.54, 55.20, 50.81, 48.27, 35.77, 34.38, 33.41, 30.63, 29.99, 29.21, 25.96, 25.30, 24.00, 19.75, 15.90.
(R)-N-(4-(3-((5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl)amino)piperidine-1-carbonyl)phenyl)-4-(dimethylamino)butanamide
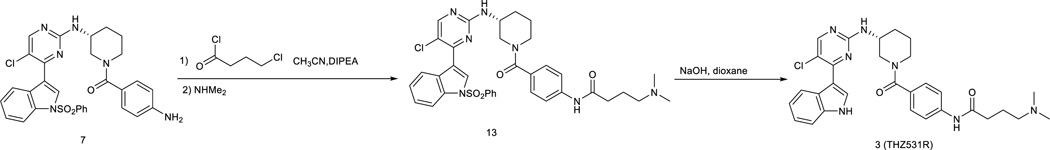
The free amino compound 7 (60mg) was dissolved in acetonitrile (2.0 mL) was added diisopropylethylamine (20.0 uL) and 4-chlorobutanoyl chloride (30 mg, 2.0 equiv) in dichloromethane (1.0 mL). After stirring for 5 min, dimethylamine (1M, 2.0 mL)) in THF was added and the reaction mixture was then heated to 50 °C for 2 hours. After removal of solvent, the crude was then treated with 1,4-dioxane (2.0 mL) and 1 M NaOH solution (2.0 mL) and then the solution was stirred at room temperature for 2 hours. After quench with 1M HCl (2.0 mL), the solution was extracted with chloroform/2-propanol (4:1, 20 mL). The organic layer was washed with water, brine and dried with MgSO4.The concentration under reduced pressure to give the crude produce which was purified by HPLC to provide THZ531R as TFA salt (16.0 mg, 60%) LC-MS: m/z (M+H) 560 1H NMR (500 MHz, DMSO-d6) 11.67 (s, 1H), 9.95 (s, 1H), 9.34(br, 1H), 8.53 (s, 1H), 8.40 (s, 1H), 8.21 (s, 1H), 7.67 (d, J = 7.0 Hz, 1H), 7.52 (d, J = 8.0 Hz, 1H), 7.36 (d, J = 8.5 Hz, 2H), 7.22 (t, J = 8.0 Hz, 1H), 7.16 (d, J = 8.0 Hz, 1H), 7.04 (br, 1H), 3.99 (br, 2H), 3.13 (m, 5H), 2.80 (s, 6H), 2.44 (t, J =7.0 Hz), 2.10 (m, 1H), 1.98 (m, 2H), 1.85 (m, 1H), 1.71 (m, 1H), 1.58 (m, 1H). 13C NMR (126 MHz, DMSO) δ 170.66, 169.81, 160.24, 158.66, 157.46, 140.40, 136.62, 131.43, 131.07, 128.14 (2C), 126.80, 123.25, 122.78, 121.03, 119.18 (2C), 114.09, 112.24, 111.65, 57.14, 49.40, 48.27, 45.06, 42.99 (2C), 33.36, 30.61, 23.97, 20.17.
Cell culture conditions
Jurkat cell lines (T. Look Laboratory, DFCI) were grown in RPMI-1640, Glutamax (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies).. HAP1 cells (Horizon Discovery Group, cat # C859) were grown in IMDM medium supplemented (Life Technologies) with 10% fetal bovine serum and 100 U/ml penicillin, 100 mg/ml (Life Technologies). All cell lines were cultured at 37°C in a humidified chamber in the presence of 5% CO2, unless otherwise noted. All cells were mycoplasma tested upon initial receipt.
Reagents and Antibodies
The following antibodies were used for immunoblots: Pol II CTD Ser-2 (cat# 04-1571, 1:1000), Ser-5 (cat# 04-1572, 1:5000), and Ser-7 (cat# 04-1570, 1:1000) phosphoantibodies (Millipore); Total Pol II (Santa Cruz cat# sc-899, 1:500); CDK7 (Santa Cruz cat# sc-723, 1:500); CDK9 (Bethyl cat# A303-493A, 1:1000); CDK12 and CDK13 (Arno Greenleaf, 1:1000); cyclin K (Bethyl cat# A301-939A, 1:500); cyclin H (Bethyl cat# A301-674A, 1:1000); PARP (Cell Signaling cat# 9542, 1:1000); and α-Tubulin DM1α (Sigma cat# T9026, 1:5000). The following ChIP-grade antibodies were used for ChIP-seq (10 µg each): Pol II (Santa Cruz cat# sc-899); CDK12 (gift of Arno Greenleaf); Pol II CTD Ser-2 (Millipore cat# 04-1571); and H3K27Ac (Abcam cat# AB4729).
Constructs
For structural studies, human CDK12 residues 715-1052 (Uniprot Q9NYV4) and human cyclin K residues 11-267 (Uniprot O75909) were cloned into the baculoviral transfer vector pFB-LIC-Bse by ligation-independent cloning. The vector encodes an N-terminal hexahistidine tag and a Tobacco Etch Virus Protease A (TEV) cleavage site. A pFBHTb expression vector containing Cdk-activating kinase (CAK1) from Saccharomyces cerevisiae (Uniprot P43568) was purchased from the MRC Protein and Phosphorylation Unit (University of Dundee, UK). For CRISPR studies, guide RNAs (sgRNA) targeting CDK12 were cloned into pX330 (Addgene, cat# 42230). pUC57-AMP was used as storage vector for CDK12 genome reference sequence.
Genome Editing
The CRISPR/Cas9 system was used to mutate the endogenous CDK12 WT locus to encode for CDK12 C1039S, a THZ531 -refractory mutant of CDK12. Target-specific oligonucleotides were cloned into pX330 (Addgene, cat# 42230), which carries a codon-optimized version of Cas9 and was further modified to express GFP for identifying transfectants. Cells were co-transfected (X-tremeGENE 9 (Roche) with 1) pX330 expressing Cas9 and CDK12 targeting sgRNA, and 2) a pUC57-AMP construct bearing 1500 bp of modified CDK12 reference genome (RefSeq Accession NM_016507). that is centered around the CRISPR targeting site in CDK12. Two days after transfection, cells were sorted using GFP as a marker of transfected cells and cells were re-plated for five days. Cells were then re-plated at low density to facilitate the isolation of individual clones. Individual clones were isolated, expanded, and PCR genotyped using mutant specific PCR primers. Following initial PCR screening, individual clones were Sanger sequenced to confirm the presence of the desired mutation. Western blot confirmed the presence of intact CDK12 kinase. Subsequent experiments were conducted using a CDK12 C1039S clone and a WT control clone that was carried through the entirety of the CRISPR protocol but that was verified by Sanger sequencing to be WT CDK12.
The genomic sequence complementary to the CDK12 -directed guide RNA that was cloned into pX330 and used in the genome editing experiments is: GGCAGGATTGCCATGAGTTG.
The modified genomic sequence that was cloned into pUC57-AMP by Genewiz and used as the repair template for genome editing is found in Supplementary Fig. 11.
The primers used for PCR –based diagnostic sequencing were the following:
GGATTGCCATGAGTTGTGGAGTA (specific to WT CDK12 allele)
AGACTCCCACGAGCTCTGGAGTA (specific to C1039S mutant allele)
TGCTTTCGCTTAAGAAATACAGG (anchored in the genome and amplifies both WT and mutant alleles)
The primers used for PCR amplification and Sanger sequencing were the following:
GCTTTAATGTCAAATTTTGGAGGTC
CTGGCACTGACCTATTGCAT
GGCTTGTTAATGTATACAGAAGGTCG
GATGCAATAGGTCAGTGCCAGAATGG
TGGCAGATGGTAGTCTGAGGA
In vitro kinase assays
Recombinant kinases CDK9-cyclin T1, CDK12-cyclin K and CDK13-cyclin K were prepared from baculo virus-infected insect cells similary as described.2,3 Radioactive kinase activity measurements were performed at a concentration of 0.2 µM CDK/cyclin complex.. Recombinant CDK7-cyclin H-MAT1 was purchased from EMD Millipore (cat#14-476). Typically, 35 µl reaction volumes of 0.2 µM active kinase were equilibrated in kinase buffer (40 mM Hepes (pH 7.6), 34 mM NaCl, 34 mM KCl, 10 mM MgCl2, 5% glycerol, and 1× PhosSTOP (Roche). Cold ATP to a final concentration of 200 µM and 3 µCi [32P]-γ-ATP (Perkin-Elmer) and 50 µM of a substrate peptide containing five phosphorylation sites were added and the reaction mixture incubated for 30 min at 30°C at 350 r.p.m. in a thermomixer (Eppendorf). Reactions were stopped by adding EDTA to a final concentration of 50 mM. Aliquots of 15 µl each were spotted onto paper squares using the Optitran BA-S85 reinforced membrane. Paper squares were washed three times for 5 min with 0.75% (v/v) phosphoric acid, with at least 5 ml washing solution per paper. Radioactivity was counted in a Beckman Scintillation Counter (Beckman-Coulter) for 1 min. Measurements were performed in triplicate and are represented as mean with standard deviation. For experiments studying the effect of an upstream pre-incubation time, compounds THZ531 or THZ531R were added at varying concentrations to 0.2 µM CDK/cyclin complex and incubated for varying times from 1 to 540 min at 30°C and 350 r.p.m. before ATP and substrate was added to the reaction mix.
Inhibitor treatment experiments
Cells were treated with THZ531, THZ531R, or THZ532 for 6 hrs at the indicated concentrations unless otherwise noted. For inhibitor washout experiments (Figure 3b) cells were treated with compounds for 6 hrs. Medium containing inhibitors was subsequently removed to effectively ‘washout’ the compound and the cells were allowed to grow in the absence of inhibitor for 66hrs.
Immunoblotting
Whole cell lysates for immunoblotting were prepared by pelleting cells from each cell line at 4°C (1,200 rpm) for 5 min using a Sorvall Legend centrifuge (ThermoFisher). The resulting cell pellets were washed 1× with ice-cold 1× PBS and then resuspended in lysis buffer containing 50 mM TrisHCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% SDS, 5 mM EDTA with protease (Roche) and phosphatase inhibitors (Roche). Whole cell lysates were collected and snap-frozen in liquid nitrogen before being stored at −80°C. Protein concentrations were determined by using the Biorad DC protein assay kit (Bio-Rad) or Pierce BCA protein assay kit (Life Technologies). Whole cell lysates were loaded into Bolt 4–12% or 8% bis-tris gels (Invitrogen) and separated by electrophoresis at 90 V for 2 hr. The gels were then transferred onto nitrocellulose membrane (Biorad) and blocked by incubation with 5% dry milk or BSA in TBST (TBS with 0.2% Tween-20). Membranes were probed using antibodies raised against the indicated proteins. Appropriate IR-labeled secondary antibodies were used and Licor Odyssey CLx was used for detection.
Pull down/ Immunoprecipitation (IP) experiments
Cells were treated with THZ531, THZ531R, or DMSO for 6 hrs. Following treatment cells were washed 2-fold with cold PBS and then lysed in the following lysis buffer: 50 mM Hepes pH7.4, 150 mM NaCl, 1% Nonidet P40 substitute, 5 mM EDTA, 1mM DTT, and protease/phosphatase cocktails. Following clearance, lysates were treated with bio-THZ1 or bio-TH531 for pulldown overnight at 4 °C. Lysates were further incubated at room temperature for 3 hrs to increase the efficiency of covalent bond formation. Lysates were then incubated with streptavidin agarose (Thermo scientific, cat. #20349) for pulldown for an additional 2–3 hrs at 4°C. Agarose beads were washed 5 times with lysis buffer and then boiled in 2× LDS for 10 minutes at 95 °C. SDS-page resolved precipitated proteins were probed for the indicated proteins.
Proliferation Assays
Proliferation assays were conducted using Cell Titer Glo assay kit (Promega cat# G7571). Jurkat cells were plated in 96-well plates at 20,000 cells/well in fresh media and treated with THZ531, THZ531R, THZ532, or DMSO at the indicated concentrations for 72 hours. HAP1 cells were seeded in 96-well plates at 12,000 cells/well in fresh media and 24 hours later were treated with THZ531 at the indicated concentrations for 72 hours. Anti-proliferative effects of compounds were assessed using Cell Titer Glo as described in product manual by luminescence measurements on a Tecan Safire plate reader. To assess the effect of inhibitor washout on anti-proliferation of Jurkat cells, cells were treated with THZ531, THZ531R, THZ532, or DMSO for 6 hrs. Inhibitor-containing medium was then removed and incubated with or without inhibitors for 66 hrs. Anti-proliferative effects of compounds were assessed using Cell Titer Glo as described in product manual by luminescence measurements on an Envision 2104 Multilabel Reader. All proliferation assays were performed in biological triplicate and error bars are +/− SD. IC50s were determined using GraphPad Prism 6 non-linear regression curve fit.
Fluorescence-Activated Cell Sorting Analysis (FACS)
For cell cycle analysis, cells were treated with inhibitors for indicated time periods. Cells were collected by centrifugation, washed once in ice-cold phosphate-buffered saline (PBS), and fixed overnight at −20 °C with 80% ethanol in PBS. Cells were washed three times with PBS. Finally, cells were resuspended in PBS containing 0.1% Triton X-100, 25 µg· ml−1 propidium iodide (PI, Molecular Probes), and 0.2 mg·ml−1 RNase A (Sigma) and incubated for 45 minutes at 37 °C. For discrimination of apoptotic vs. non-apoptotic cells by Annexin V/ PI staining, cells were treated with compound for indicated periods of time. Cells were collected by centrifugation, washed once with PBS, and processed according to manufacturers protocol (Invitrogen cat# V13242). All FACS samples (cell cycle distribution or PI/ Annexin V double staining) were analyzed on a FACS Canto II (BD Biosciences) instrument and processed on FlowJo (Treestar). All PI/ Annexin V experiments were performed in biological triplicate. Error bars are +/− SD.
HPLC/Mass Spectrometry
Recombinant CDK12-cyclin K complex was incubated with a 5-fold excess of THZ531 or DMSO for 1 hour at room temperature and analyzed by LC/ESI-MS essentially as described.4 In each analysis, 5 µg protein was injected onto a self-packed reversed phase column (1/32” O.D. × 500 um I.D., 5 cm of POROS 10R2 resin). After desalting for four minutes, protein was eluted with an HPLC gradient (0–100% B in 4 minutes, A=0.2M acetic acid in water, B=0.2 M acetic acid in acetonitrile, flow rate = 10 µL/min) into an LTQ ion trap mass spectrometer (ThermoFisher). Mass spectra were deconvoluted using MagTran1.03b2 software.5 To determine the site of modification, proteins were reduced with TCEP (10 mM final concentration), alkylated with iodoacetamide (22.5 mM final concentration) and digested with glu-c (37 °C, overnight) and analyzed by nanoLC-MS.6 Digested peptides were injected onto the precolumn (4 cm POROS 10R2, Applied Biosystems) and eluted with an HPLC gradient (NanoAcquity UPLC system, Waters, Milford, MA; 10–70% B in 60 minutes; A=0.1 M acetic acid in water, B=0.1M acetic acid in acetonitrile). Peptides were resolved on a self-packed analytical column (50 cm Monitor C18, Column Engineering, Ontario, CA) and introduced to the mass spectrometer (LTQ Orbitrap XL) at a flow rate of ~30 nL/min (ESI spray voltage = 3.2 kV). The mass spectrometer was programmed to perform data-dependent MS/MS on the five most abundant precursors (35% collision energy) in each MS1 scan (image current detection, 30K resolution, m/z 300–2000). MS/MS spectra were matched to peptide sequences using Mascot (version 2.2.1) after conversion of raw data to .mgf using multiplierz scripts.7 Search parameters specified glu-c digestion with up to 2 missed cleavages, fixed carbamidomethylation of cysteine residues, as well as variable oxidation of methionine and THZ531 modification of cysteine (i.e. cysteine residues are either carbamidomethylated or THZ531 modified). Precursor and peptide ion mass tolerances were 10 ppm and 0.6 Da, respectively.
Protein expression and purification
Bacmid DNA was prepared in Escherichia coli strain DH10Bac and used to generate baculovirus in Sf9 insect cells. Baculoviruses for CDK12, cyclin K and yeast CAK1 were used to co-infect Sf9 cells grown in suspension to a density of 2×106 cells/mL. Cells were lysed by sonication in binding buffer (50 mM HEPES pH 7.5, 500 mM NaCl, 5% glycerol, 5 mM Imidazole) supplemented with protease inhibitor cocktail set III (Calbiochem). Polyethylenimine (PEI) was then added to a final concentration of 0.5% to precipitate DNA. The lysate was clarified by centrifugation at 21,000 RPM for 1 hour at 4°C. Recombinant proteins were purified using nickel-sepharose resin (GE Healthcare) and eluted stepwise with imidazole. Fractions containing the phosphorylated CDK12- cyclin K complex were treated with TEV protease for cleavage of the N-terminal hexahistidine tags. The cleaved proteins were then exchanged into a buffer containing 50 mM MES pH 6.5, 0.5 mM TCEP and loaded onto a 5 mL HiTrap SP column (GE Healthcare) equilibrated in the same buffer. A linear elution gradient was run from 0 to 1 M NaCl. Fractions containing the CDK12 complex were pooled and the buffer adjusted to 50 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM TCEP during concentration in a 10 kD MWCO Amicon Ultra concentrator.
Crystallization
The purified CDK12- cyclin K complex at 17 µM concentration was incubated overnight at 4°C with 25 µM THZ531. Near complete covalent binding was shown by a mass shift of 559 Da using mass spectrometry. Excess free compound was removed by size-exclusion chromatography using a HiLoad Superdex S75 26/60 column (GE Healthcare). The protein-inhibitor complex was concentrated to 5.5 mg/mL buffered in 50 mM HEPES pH 7.5, 300 mM NaCl, 0.5 mM TCEP. Crystals were grown at 20°C in 195 nL sitting drops mixing 75 nL protein solution and 20 nL crystal seed stock with 100 nL of a reservoir solution comprising 0.1 M HEPES pH 7.0, 10% PEG8000, 0.2 M magnesium chloride. Crystals were cryo-protected with mother liquor supplemented with an additional 15% ethylene glycol and vitrified in liquid nitrogen after mounting.
Structure determination
Diffraction data were collected at a wavelength of 0.97625 Å at 100 K from a single crystal on Diamond Light Source beamline I03. Data were indexed and integrated using XDS8 and scaled using AIMLESS9 in the CCP4 suite of programs.10 Phases were found using molecular replacement in PHASER.11 The structure of the CDK12715–1038/cyclin K11–267 complex (PDB 4UN0) was used as a search model, with input of CDK12 and cyclin K done separately. A PDB file for the inhibitor, THZ531, was created using PRODRG in the CCP4 suite. Models were built initially using COOT12 and then refined and modified using alternate rounds of REFMAC513 and COOT. The refined structures were validated with MolProbity14 and the atomic coordinate files deposited in the Protein Data Bank. Ramachandran plot indicates good stereochemistry of the three-dimensional structure with all residues falling within the favored (96%) and allowed (4%) regions. Structure figures were prepared with PyMOL.15
Mass spectrometry for crystallography
Protein masses were determined using an Agilent LC/MSD TOF system with reversed-phase high-performance liquid chromatography coupled to electrospray ionization and an orthogonal time-of-flight mass analyzer. Proteins were desalted prior to mass spectrometry by rapid elution off a C3 column with a gradient of 5–95% isopropanol in water with 0.1% formic acid. Spectra were analyzed using the MassHunter software (Agilent).
RNA Extraction and Synthetic RNA Spike-In
Total RNA and sample preparation was performed as previously described.16 Briefly, Jurkat cells were incubated in media containing THZ531, THZ531R, or THZ532 at the indicated concentrations or with DMSO for the specified duration of time. Cell numbers were determined by manually counting cells using C-Chip disposable hemocytometers (Digital Bio, DHC-N01) prior to lysis and RNA extraction. Total RNA from biological replicates (equivalent to 5 million cells per replicate) was subsequently isolated using RNeasy Plus Mini kit (Qiagen) following the manufacturer’s instructions and resuspended in 50 µL nuclease-free water (Ambion). Total RNA was spiked-in with ERCC RNA Spike-In Mix (Ambion, cat# 4456740) and analyzed on Agilent 2100 Bioanalyzer for integrity. RNA with the RNA Integrity Number (RIN) above 9.8 was hybridized to GeneChip PrimeView Human Gene Expression Arrays (Affymetrix).
cDNA Preparation and TaqMan Expression Analysis
RNA utilized for RT-PCR was extracted as outlined above. One microgram of purified RNA was reverse transcribed using Superscript III First-Strand (Invitrogen) with oligo dT primers to prime first-strand synthesis according to the manufacturer's protocol. qPCR was carried out on the 7000 ABI Detection System using the following Taqman probes according to the manufacturer's protocol (Applied Biosystems). All experiments shown were performed in biological triplicate. Each individual biological sample was qPCR-amplified in technical duplicate. Error bars are +/− SD. Expression was normalized to GAPDH, and fold change in expression was calculated relative to the indicated conditions. The following Taqman probes from Life Technologies were used for qPCR-based gene expression analysis: RUNX1 - Hs00231079_m1; TAL1 - Hs01097987_m1; GATA3 - Hs00231122_m1; MYB - Hs00920556_m1; GAPDH - Hs02758991_g1; BRCA1 - Hs01556193_m1; ERCC4 - Hs00193342_m1; TERF2 - Hs00194619_m1; FANCF - Hs00256030_s1; PARP9 - Hs00967084_m1; MDC1 - Hs00206182_m1; RAG2 - Hs01851142_s1; RAD51C - Hs00427442_m1; and ATR - Hs00992123_m1.
Microarray Sample Preparation and Analysis
For microarray analysis, 100 ng of total RNA containing ERCC RNA Spike-In Mix (see above) was used to prepare biotinylated aRNA (cRNA) according to the manufacturer’s protocol (30 IVT Express Kit, Affymetrix 901228). Briefly, total RNA undergoes T7 oligo(dT)-primed reverse transcription to synthesize first-strand cDNA containing a T7 promoter sequence. This cDNA is then converted into a double-stranded DNA template for transcription using DNA Polymerase and RNase H to simultaneously degrade the RNA and synthesize second strand cDNA. In vitro transcription synthesizes aRNA and incorporates a biotin-conjugated nucleotide. The aRNA is then purified to remove unincorporated NTPs, salts, enzymes, and inorganic phosphate. Fragmentation of the biotin-labeled aRNA prepares the sample for hybridization onto GeneChip 3’ expression arrays. Samples were prepared for hybridization using 10 µg of biotinylated aRNA in a 1× hybridization cocktail according the Affymetrix hybridization manual. Additional hybridization cocktail components were provided in the Affymetrix GeneChip Hybridization, Wash and Stain Kit. GeneChip arrays (Human PrimeView, Affymetrix 901837) were hybridized in a GeneChip Hybridization Oven at 45°C for 16 hrs at 60 RPM. Washing was done using a GeneChip Fluidics Station 450 according to the manufacturer’s instructions, using the buffers provided in the Affymetrix GeneChip Hybridization, Wash and Stain Kit. Images were extracted with Affymetrix GeneChip Command Console (AGCC) and analyzed using GeneChip Expression Console. A Primeview CDF that included probe information for the ERCC controls (GPL16043), provided by Affymetrix, was used to generate .CEL files.
Batches of microarray experiments were normalized within themselves to ERCC spike-in probes as described in.1,16 Briefly, probes were collapsed into probesets using expresso with mas5 normalization. Probeset expression profiles were normalized using loess from the affy R package to equilibrate ERCC spike-in probes across microarrays in a given batch. Where possible, log2 values of biological replicates were averaged. Fold-changes were taken by subtracting average log2 DMSO signal from average log2 treatment signal. Expressed genes were those with log2(expression) > log2(100) in the corresponding DMSO sample. Sensitive genes were defined as those that decreased >= 2 log2-fold (4 linear fold-change). Expression heatmaps (Figure 5c,d; Supplementary Figure 6b) were created using the fold-change of expressed genes and plotted using heatmap.2 ranked by the values noted in the legend. Expression barplots (a.k.a. waterfall plots; Figure 6a, 6c) were created by ranking expressed transcripts by their fold change and plotting the fold change of each as a bar plot.
PrimeView microarrays were submitted in the following batches used for normalization:
HAP1 –WT DMSO, WT THZ531 300nM, WT THZ531 900nM, C1039S CDK12 - DMSO, C1039S CDK12 - THZ531 300nM, C1039S CDK12 - THZ531 900nM
Jurkat – 6h DMSO (2 replicates), 6h THZ531 50nM (2 replicates), 6h THZ531 200nM (2 replicates), 6h THZ531 350nM (2 replicates), 6h THZ531 500nM (2 replicates), THZ531R 500nM (2 replicates), THZ532 500nM (2 replicates)
Gene Set Enrichment Analysis (Figure 6g, Supplementary Figure 9c)
Gene Set Enrichment Analysis17 was performed using a gene list pre-ranked by CDK12 ChIP-seq signal at promoters or enhancers. The top 500 genes expressed in 6h DMSO from the same microarray batch as 6h 200 nM THZ531 and most downregulated by 6h 200 nM THZ531 treatment was used as the gene set.
Functional enrichment analysis using DAVID (Supplementary Figure 8a, c)
Gene transcripts that were sensitive to THZ531 (50 or 200 nM) were used as input in the functional analysis tool at http://david.abcc.ncifcrf.gov/.18 GO terms enrichment was performed using the default settings of the DAVID tool V6.7.
Chromatin Immunoprecipitation
Cells were crosslinked for 20 min at room temperature by the addition of one-tenth of the volume of 11% formaldehyde solution (11% formaldehyde, 50mM HEPES pH 7.3, 100mM NaCl, 1mM EDTA pH 8.0, 0.5mM EGTA pH 8.0) to the growth media followed by 5 min quenching with 100 mM glycine. Cells were washed twice with PBS, then the supernatant was aspirated and the cell pellet was flash frozen in liquid nitrogen. Frozen crosslinked cells were stored at −80°C. 50 µL of Dyna magnetic beads (Sigma) were blocked with 0.5% BSA (w/v) in PBS. Magnetic beads were bound with 10 µg of the indicated antibody. For RNA polymerase II-occupied genomic regions, we performed ChIP-Seq experiments using a SantaCruz Biotechnology (sc-899, lot C1413) antibody. The affinity-purified antibody was raised in rabbit against an epitope mapping to the N-terminus of murine RBP1, the largest subunit of RNA Pol II. For CDK12 occupied genomic regions, we performed ChIP-Seq experiments using an antibody shared with us from Arno Greenleaf.19 For pSer2 Pol II occupied genomic regions, we performed ChIP-seq experiments using a Millipore (04-1571) antibody. For H3K27Ac-occupied genomic regions, we performed ChIP-Seq experiments using an Abcam (AB4729A) antibody. The affinity-purified antibody was raised in rabbit against an epitope corresponding to amino acids 1–100 of human Histone H3 that is acetylated at K27. For Jurkat, crosslinked cells were lysed with lysis buffer 1 (50mM HEPES pH 7.5, 140mM NaCl, 1mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100), pelleted and resuspended in lysis buffer 2 (10 mM TrisHCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). The subsequent pellet was resuspended in and sonicated in sonication buffer (50 mM HEPES pH 7.5, 140 mM NaCl, 1 mM EDTA pH 8.0, 1 mM EGTA, 0.1% Na-deoxycholate, 0.1% SDS, and 1% Triton X-100). Cells were sonicated for 10 cycles at 30 s each on ice (18–24 W) with 60 s on ice between cycles. Sonicated lysates were cleared and incubated overnight at 4°C with magnetic beads bound with antibody to enrich for DNA fragments bound by the indicated factor. Beads were washed two times with sonication buffer, one time with sonication buffer with 500 mM NaCl, one time with LiCl wash buffer (10 mM TrisHCl pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate) and one time with TE. DNA was eluted in elution buffer (50 mM TrisHCl pH 8.0, 10 mM EDTA, 1% SDS). Cross-links were reversed overnight. RNA and protein were digested using RNase A and Proteinase K, respectively and DNA was purified with phenol chloroform extraction and ethanol precipitation.
ChIP-Seq Analysis
Illumina sequencing libraries generated and data was processed according to.20 In brief, libraries were generated for ChIP samples following the Illumina TruSeq™ DNA Sample Preparation v2 kit protocol with minor changes. All ChIP-Seq data sets were aligned using bowtie21 to build NCBI37/hg19 of the human genome with parameters −p 4 −−best −k 2 −m 2 −−sam −l 40. Wiggle files for gene tracks were created using Macs 1.4.222 with options −w −S −space=50 −shiftsize=200 –nomodel to count extended reads in 50bp bins, were divided by the number of considered reads to normalize to mapped-reads-per-million, and were displayed in the UCSC genome browser (Fig. 5f, 6f, Supplementary Fig. 5a, 7a, 7b, 9a).
Enhancer Identification
Enhancers (Fig. 4a, 4d, 6e, 6f, Supplementary Fig. 8e, 8f, 9a, 9b, 9c,) were identified as having a significant enrichment in ChIP-Seq reads for H3K27ac as determined by MACS 1.4.222 with input control and parameters −keep-dup=auto −p 1e-9.
Metagene Analysis (Fig. 4a, 4b, 5b; Supplementary Fig. 7b)
Metagenes were created to summarize the binding activities of proteins at particular genomic region types. For promoter/enhancer metagenes, the RPM-normalized (−r) ChIP-Seq read counts in 200 equally sized bins (−m 150) was calculated using bamToGFF (https://github.com/BradnerLab/pipeline), and the mean for each bin across all regions was plotted. Enhancer metagenes (Figure 4a) used a 4kb window centered on the center of MACS-defined H3K27ac peaks. Promoter metagenes (Fig. 4a) used a 4kb window centered on the transcription start sites defined by RefSeq.23 Gene body metagenes (Fig. 4b, 5b, Supplementary Fig. 7b) were created in three parts: 2kb upstream of the transcription start site (TSS) to TSS; TSS to transcription end site (TES); TES to 2kb downstream of the TES. 50 bins divided the gene-flanking regions; 150 bins divided the genic regions. Strandedness was preserved. Only genes whose transcript was considered expressed in the DMSO condition calculated in the THZ531 treatment batch were considered.
ChIP-Seq Heatmap Analysis (Fig. 4c, 4d)
Heatmaps were created to show per-region occupancy of proteins. The RPM-normalized (−r) ChIP-Seq read counts (−d) in 200 equally sized bins (−m 150) dividing 4 kb regions (+/− 2 kb from transcription start sites or +/− 2kb from the center of an H3K27ac peak) was calculated using bamToGFF (https://github.com/BradnerLab/pipeline) and plotted using heatmap.2 in R.
Correlation Lineplots (Fig. 4c, 4d)
To correlate the presence of CDK12, H3K27ac, and RNA Pol II at enhancers and promoters, the RPM-normalized read counts from ChIP-Seq experiments targeting these factors in these regions was calculated using bamToGFF with one window for the entire region (−m 1). Promoters and enhancers were separately ranked by RNA Pol II read counts and CDK12 and H3K27ac read counts were displayed using rollmean from the zoo package in R with a width of 200 values. The Pol II heatmap was constructed using heatmap.2 on the Pol II read counts.
Region Boxplots (Fig. 5g, 5h)
Boxplots were created to show the effects of different transcriptional inhibitors on Pol II distributions across gene bodies. Promoter boxplots refer to a 4kb window centered on the transcription start site; TES boxplots refer to a 4kb window centered on the transcription end site, both defined by RefSeq. The RPM-normalized (−r) read counts from ChIP-Seq experiments targeting RNA Pol II in these regions was calculated using bamToGFF with one window for the entire region (−m 1). Boxplots display these normalized read counts across all regions.
Super-Enhancer Identification (Fig. 6e, 6f, Supplementary Fig. 8e, 8f, 9a, 9b)
Super-enhancers were identified as described in24–26 with a stitching distance of 12.5kb. Briefly, Macs peaks of H3K27ac were used to identify constituent enhancers. H3K27ac signal (less input control) was used to rank enhancers by their enrichment. In Jurkat 818 super-enhancers were separated from typical enhancers by finding the point at which the line y=x is tangent to the curve. Super-enhancers were assigned to the expressed gene whose TSS falls nearest to the center of the super-enhancer. Expressed genes for enhancer assignment were determined by interrogating a 1kb region centered on RefSeq transcription start sites for RPM-normalized (−r) H3K27ac read density (−d) using bamToGFF with one bin (−m 1). The top 2/3 of genes were considered expressed.
Super-Enhancer Analysis (Supplementary Fig. 9b)
To determine if super-enhancers and their associated genes are particularly enriched in CDK12, we interrogated the set of stitched enhancer regions for CDK12 ChIP-Seq read counts and 37973 4 kb promoters centered on RefSeq transcription start sites. bamToGFF was used with the −t option to count reads per region.
Data Presentation and Statistics
All the results presented in graphics are reported as the mean ±SD unless otherwise noted. All statistical analyses herein represent comparison of continuous variables performed using an unpaired (two-tailed) T test unless otherwise noted. All graphics and statistics were generated using GraphPad Prism, Microsoft Excel, or R.
Supplementary Material
Scheme 1.
The synthesis of THZ531
Scheme 2.
The synthesis of BioTHZ531
Scheme 3.
The synthesis of THZ531R
Acknowledgments
We thank R. George, K. Wong, P. Hammerman, J. Bradner and members of the Gray and Young laboratories for helpful discussions. We thank P. Wisniewski and C. Zollo from the Whitehead FACS facility for help with FACS instruments. We thank J. Love, T. Volkert, and S. Gupta from the Whitehead Genome Core for help with genomics experiments. The authors would like to thank Diamond Light Source for beamtime (proposal mx8421), and the staff of beamline I03 for assistance with data collection. We thank W. Massefski from the Dana-Farber NMR lab for help in collecting NMR data for the manuscript. M.G. is a member of the DFG excellence cluster ImmunoSensation. This work was supported by the National Institutes of Health (HG002668 and CA109901 - R.A.Y. and 5 R01 CA179483-02 C – N.S.G.), the Koch Institute and Dana-Farber/Harvard Cancer Center Bridge Grant (N.K., N.S.G., R.A.Y.), the DFG (GE 976/9-1 – M.G.), the Hope Funds for Cancer Research Grillo-Marxuach Family Fellowship (B.J.A.), and a NDM Prize Studentship, funded in part by the Medical Research Council (S.E.D.C.). The SGC is a registered charity (no. 1097737) that receives funds from AbbVie, Bayer, Boehringer Ingelheim, Genome Canada through Ontario Genomics Institute Grant OGI-055, GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and Wellcome Trust Grant 092809/Z/10/Z.
N.S.G, T.Z., N.K. are inventors on patent applications covering THZ531 (WO 2015/058126, WO/2014/063068, WO 2015/058140), which are licensed to a company co-founded by N.S.G and R.A.Y.
Footnotes
Accession Numbers
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession number PDB ID: 5ACB. Sequencing and expression data were deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE72031 (Supplementary Table 3). CDK12 RefSeq Accession NM_016507 was used as repair template in CRISPR studies.
Author Contributions. N.S.G., R.A.Y., N.K., and T.Z. conceived the project. N.S.G. and T.Z. conceived and directed chemistry effort with input from Y.L. Chemical synthesis and small molecule structure determination was performed by both T.Z. and T.M. R.A.Y., N.S.G., B.J.A. and N.K. conceived genomics effort. N.K. and C.M.O. designed and executed cellular biological experimental research with input from N.S.G. and R.A.Y. S.E.D.C. solved co-crystal structure of CDK12- cyclin K with THZ531 with guidance from J.M.O. and A.N.B. A.K.G. designed and executed CDK in vitro kinase assays with input from M.G. S.B.F. designed and performed protein mass spectrometry on THZ531-CDK12 adducts with input from J.A.M. N.M.H. providing cloning expertise. B.B. and A.L.G. developed and provided CDK12 and CDK13 antibodies. B.J.A. designed and performed genomics data analyses. N.K., T.Z., N.S.G. and R.A.Y. co-wrote the paper. All authors edited the manuscript.
Competing financial interests. The authors declare the following competing interests.
References
- 1.Young RA. RNA polymerase II. Annual review of biochemistry. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- 2.Buratowski S. Progression through the RNA polymerase II CTD cycle. Molecular cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz MJ, de la Mata M, Kornblihtt AR. The carboxy terminal domain of RNA polymerase II and alternative splicing. Trends in biochemical sciences. 2010;35:497–504. doi: 10.1016/j.tibs.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Sanso M, Fisher RP. Pause, play, repeat: CDKs push RNAP II's buttons. Transcription. 2013;4:146–152. doi: 10.4161/trns.25146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Current opinion in cell biology. 2008;20:334–340. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes & development. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartkowiak B, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes & development. 2010;24:2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng SW, et al. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Molecular and cellular biology. 2012;32:4691–4704. doi: 10.1128/MCB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HH, Wang YC, Fann MJ. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Molecular and cellular biology. 2006;26:2736–2745. doi: 10.1128/MCB.26.7.2736-2745.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen HH, Wong YH, Geneviere AM, Fann MJ. CDK13/CDC2L5 interacts with L-type cyclins and regulates alternative splicing. Biochemical and biophysical research communications. 2007;354:735–740. doi: 10.1016/j.bbrc.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 13.Liang K, et al. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Molecular and cellular biology. 2015;35:928–938. doi: 10.1128/MCB.01426-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eifler TT, et al. Cyclin-dependent kinase 12 increases 3' end processing of growth factor-induced c-FOS transcripts. Molecular and cellular biology. 2015;35:468–478. doi: 10.1128/MCB.01157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson L, Muniz L, West S. 3' end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes & development. 2014;28:342–356. doi: 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartkowiak B, Greenleaf AL. Expression, purification, and identification of associated proteins of the full-length hCDK12/CyclinK complex. The Journal of biological chemistry. 2015;290:1786–1795. doi: 10.1074/jbc.M114.612226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3' end processing. Molecular cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- 18.Blazek D, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes & development. 2011;25:2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winsor TS, Bartkowiak B, Bennett CB, Greenleaf AL. A DNA damage response system associated with the phosphoCTD of elongating RNA polymerase II. PloS one. 2013;8:e60909. doi: 10.1371/journal.pone.0060909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bennett CB, et al. Yeast screens identify the RNA polymerase II CTD and SPT5 as relevant targets of BRCA1 interaction. PloS one. 2008;3:e1448. doi: 10.1371/journal.pone.0001448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekumi KM, et al. Ovarian carcinoma CDK12 mutations misregulate expression of DNA repair genes via deficient formation and function of the Cdk12/CycK complex. Nucleic acids research. 2015;43:2575–2589. doi: 10.1093/nar/gkv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rottenberg S, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajrami I, et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer research. 2014;74:287–297. doi: 10.1158/0008-5472.CAN-13-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi PM, Sutor SL, Huntoon CJ, Karnitz LM. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. The Journal of biological chemistry. 2014;289:9247–9253. doi: 10.1074/jbc.M114.551143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartkowiak B, Yan C, Greenleaf AL. Engineering an analog-sensitive CDK12 cell line using CRISPR/Cas. Biochimica et biophysica acta. 2015;1849:1179–1187. doi: 10.1016/j.bbagrm.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosken CA, et al. The structure and substrate specificity of human Cdk12/Cyclin K. Nature communications. 2014;5:3505. doi: 10.1038/ncomms4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greifenberg AK, et al. Structural and Functional Analysis of the Cdk13/Cyclin K Complex. Cell reports. 2016;14:320–331. doi: 10.1016/j.celrep.2015.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Lolli G, Lowe ED, Brown NR, Johnson LN. The crystal structure of human CDK7 and its protein recognition properties. Structure. 2004;12:2067–2079. doi: 10.1016/j.str.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Kwiatkowski N, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patricelli MP, et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chemistry & biology. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christensen CL, et al. Targeting transcriptional addictions in small cell lung cancer with a covalent CDK7 inhibitor. Cancer cell. 2014;26:909–922. doi: 10.1016/j.ccell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larochelle S, et al. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nature structural & molecular biology. 2012;19:1108–1115. doi: 10.1038/nsmb.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanda T, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer cell. 2012;22:209–221. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowman EA, Kelly WG. RNA polymerase II transcription elongation and Pol II CTD Ser2 phosphorylation: A tail of two kinases. Nucleus. 2014;5:224–236. doi: 10.4161/nucl.29347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 38.Brown L, et al. Site-specific recombination of the tal-1 gene is a common occurrence in human T cell leukemia. The EMBO journal. 1990;9:3343–3351. doi: 10.1002/j.1460-2075.1990.tb07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 1.Kwiatkowski N, et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature. 2014;511:616–620. doi: 10.1038/nature13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosken CA, et al. The structure and substrate specificity of human Cdk12/Cyclin K. Nature communications. 2014;5:3505. doi: 10.1038/ncomms4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nature communications. 2012;3:842. doi: 10.1038/ncomms1846. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T, et al. Discovery of potent and selective covalent inhibitors of JNK. Chemistry & biology. 2012;19:140–154. doi: 10.1016/j.chembiol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z, Marshall AGA. universal algorithm for fast and automated charge state deconvolution of electrospray mass-to-charge ratio spectra. Journal of the American Society for Mass Spectrometry. 1998;9:225–233. doi: 10.1016/S1044-0305(97)00284-5. [DOI] [PubMed] [Google Scholar]
- 6.Ficarro SB, et al. Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Analytical chemistry. 2009;81:3440–3447. doi: 10.1021/ac802720e. [DOI] [PubMed] [Google Scholar]
- 7.Askenazi M, Parikh JR, Marto JA. mzAPI: a new strategy for efficiently sharing mass spectrometry data. Nature methods. 2009;6:240–241. doi: 10.1038/nmeth0409-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta crystallographica. Section D, Biological crystallography. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The PyMOL Molecular Graphics System (Version 1.2r3pre) [Google Scholar]
- 16.Loven J, et al. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 18.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartkowiak B, Greenleaf AL. Expression, purification, and identification of associated proteins of the full-length hCDK12/CyclinK complex. The Journal of biological chemistry. 2015;290:1786–1795. doi: 10.1074/jbc.M114.612226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruitt KD, et al. RefSeq: an update on mammalian reference sequences. Nucleic acids research. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.