Abstract
Gene-environment (G×E) interactions play key roles in many complex diseases. An increasing number of epidemiological studies have shown the combined effect of multiple environmental exposures on disease risk. However, no appropriate statistical models have been developed to conduct a rigorous assessment of such combined effects when G×E interactions are considered. In this paper, we propose a partial linear varying multi-index coefficient model (PLVMICM) to assess how multiple environmental factors act jointly to modify individual genetic risk on complex disease. Our model includes the varying-index coefficient model as a special case, where discrete variables are admitted as the linear part. Thus PLVMICM allows one to study nonlinear interaction effects between genes and continuous environments as well as linear interactions between genes and discrete environments, simultaneously. We derive a profile method to estimate parametric parameters and a B-spline backfitted kernel method to estimate nonlinear interaction functions. Consistency and asymptotic normality of the parametric and nonparametric estimates are established under some regularity conditions. Hypothesis testing for the parametric coefficients and nonparametric functions are conducted. Results show that the statistics for testing the parametric coefficients and the non-parametric functions asymptotically follow a χ2-distribution with different degrees of freedom. The utility of the method is demonstrated through extensive simulations and a case study.
Key words and phrases: Association study, Backfitting, B-spline, Single index model, Varying coefficient model
1. Introduction
There has been great interest in identifying gene-environment (G×E) interaction in the scientific literature. G×E interaction is defined as how genotypes influence phenotypes differently under different environmental conditions (Falconer (1952)), a phenomenon also termed as genetic sensitivity to environmental stimulus. A growing number of reports have confirmed the role of G×E interaction in many diseases, such as Parkinson disease (Ross and Smith (2007)) and type 2 diabetes (Zimmet et al. (2001)). G×E interaction has traditionally been pursued based on a single environment exposure model. Evidence from epidemiological studies has clearly indicated that disease risk can be modified by simultaneous exposure to multiple environmental factors, higher than what would be expected from simple addition of the effects of factors acting alone (Carpenter et al. (2002); Sexton and Hattis (2007)). Thus, assessing the combined effect of environmental mixtures and the mechanism in which they, as a whole, interact with genes to affect disease risk could shed novel insight into disease etiology. Suppose that Y is the trait response of primary interest. In many genetic studies, one collects a p-dimensional continuous covariate vector X, and a q-dimensional discrete covariate vector Z. Motivated by an empirical analysis to study G×E interaction, see Section 5, we propose a partial linear varying multi-index coefficient model (PLVMICM):
| (1.1) |
where Gl, l = 1, ⋯,L are genetic variables (e.g., single nucleotide polymorphisms (SNPs)) of interest, ε is an error term with mean 0 and finite variance; ml(·), l = 0, 1, ⋯,L are unknown index functions; α0, ⋯, αL and β0, ⋯, βL are parameters of interest, where the index coefficients βl are the index loadings or the loading parameters. The SNP variable Gl can be coded as 2, 1, and 0 for genotype AA, Aa, and aa, assuming an additive model. Note that the main genetic effect for each Gl is captured by the function . Thus we do not need to have a separate term to model the main genetic effect for each SNP. Model (1.1) provides a unified model framework for many existing models used for studying G × E interaction. Specifically, the model proposed in Ma et al. (2011) can be viewed as a special case with p = 1 (the dimension of βl), q = 0 (the dimension of αl), and L = 1. Model (1.1) also include the semiparametric varying-index coefficient model proposed by Ma and Xu (2015), studying G×E interaction, as a special case with β0 = βl = β, l = 1, ⋯,L, i.e., assuming the same index loading parameter. Our empirical analysis in the data example in Section 5 clearly shows that this assumption is not realistic, making it necessary to allow different loading parameters in the model.
Model (1.1) also includes many other existing models as special cases. It reduces to the partial linear single-index model (Carroll et al. (1997); Xia and Li (1999); Xia and Hardle (2006); Liang et al. (2010); Cui et al. (2011)), in which the discrete variable in the linear part is admitted if all Gl = 0; it reduces to VICM proposed by Ma and Song (2015) if Z = 0.
This paper aims to develop a set of statistical estimation and hypothesis procedures for model (1.1). We employ a B-spline backfitted kernel smoothing (BSBK) procedure to estimate the parametric parameters and the nonparametric functions (Wang and Yang (2007)). We first develop a profile least squares method to estimate the index coefficients βl and the linear coefficients αl by approximating unknown function ml(·) with B-spline basis functions. The parametric estimates can be shown to be n1/2-consistent and asymptotically normal. We also obtain uniformly consistent estimators of the nonparametric functions. Given the n1/2-consistent parametric estimators and the consistent estimators of the nonparametric functions, the kernel estimators of nonparametric functions can be obtained from which we establish the asymptotic normality.
Under model (1.1), it is natural to ask whether there is an interaction between discrete/continuous environments and genes, and whether the interaction with the combined environmental exposures is linear or nonlinear. Cai et al. (2000) studied the nonparametric testing problem for varying coefficient models based on the generalized likelihood ratio test. Nonparametric inferences for additive models were previously discussed by employing the generalized likelihood ratio (GLR) statistic (Fan and Jiang (2005)). We propose a parametric likelihood ratio test to test for the linear interaction term and a nonparametric GLR test to test for the nonparametric interaction functions (Fan et al. (2001)). We further show that the proposed nonparametric GLR statistic is asymptotically χ2. We conduct rigorous theoretical evaluation of the proposed estimators and test statistics and show the utility of the model through extensive simulations and a case study.
The paper is organized as follows. In Section 2.2, we formulate the model and describe the BSBK procedure and the parametric estimators for the continuous and discrete parts based on a profile least squares method. The nonparametric kernel estimators for index functions are given in Section 2.3. The consistency and normality of parametric and nonparametric estimators are given in Section 2.4. Section 3 gives the parametric likelihood ratio statistic and several nonparametric GLR statistics, as well as their theoretical properties. In Section 4, we report on simulation studies that illustrate the finite sample performance of the proposed estimators and test statistics. In Section 5 we show the utility of the method by applying it to a baby birthweight data set. Some concluding remarks are given in Section 6. The proofs of the main results are relegated to the Appendix.
2. Estimation Procedures
2.1. Estimation Procedures
We focus on the situation with L = 1 for ease of presentation, and rewrite (1.1) as
| (2.1) |
The proposed procedure for model (2.1) can be easily extend to model (1.1) with multiple G’s (i.e., multiple SNPs), and it is still more general than the existing ones used for G×E interaction. It is motivated by a recent genome-wide association study to identify genetic risk factors interacting with maternal uterine environments to increase the risk of low and high birth weight (HAPO Study Cooperative Research Group (2009)). The underlying hypothesis is that the variation of birth weight can be explained by complex G×E interactions in the context of the maternal-fetal unit. As a fetus resides inside its mother’s womb, there is intensive signalling and chemical exchanges between the two. The effects of fetal genes could be modified by simultaneous exposure to multiple stimuli from the mother’s side such as mother’s glucose level and blood pressure. For continuously measured environmental variables, we propose to model the joint effect of environment variables as a whole through an unknown index function m(·). The index function can be linear or nonlinear. That is determined by the data, with flexibility to capture the underlying mechanism of environmental mixtures modifying genetic influences on disease risk. For such discrete environmental variables as smoking status and family disease history, their interaction effects with genes can be modeled through a parametric function.
The motivation for assessing nonlinear G×E interaction in complex disease has been discussed extensively in Ma et al. (2011) and Wu and Cui (2013). The model for testing nonlinear G×E interactions in Ma et al. (2011) can be viewed as a special case of (2.1) with p = 1 (the dimension of βl) and q = 0 (the dimension of αl). We assume the index loading parameters β0 and β1 to be different; this differs from the single index model assuming common loading parameters for different index functions proposed by Xia and Li (1999). Li et al. (2010) studied the generalized functional linear models with semi-parametric single index interaction, but did not allow dissimilar loading parameters in different index functions. Although the varying-index coefficient model (VICM) proposed by Ma and Song (2015) could consider the joint interaction of multiple environments with genes, it does not admit discrete variables Z. Such discrete environmental variables are common in G×E studies and the inclusion of these variables is crucial to assess the discrete G×E interactions, as implemented in most partial linear single index models (Carroll et al. (1997); Xia and Li (1999); Xia and Hardle (2006);Liang et al. (2010)). Nevertheless, including both parametric and nonparametric terms into the same model poses computational and theoretical challenges. As discussed earlier, our model differs from that proposed by Ma and Xu (2015) in which they assumed the same loading parameters for different index functions. This assumption is too strong in reality, the modulation effect of environmental variables may differ from gene to gene. Our data analysis results in Section 5 indicate that such an assumption is invalid there. Theoretical and practical considerations thus motivate us to consider a more flexible model that can incorporate both linear and nonlinear interactions, and without too many assumptions on the model parameters, as in (2.1).
2.2. Parameter estimation
Consider the PLVMICM model given in (2.1). Let θ = (αT, βT)T, where and . Let Vi = (Xi,Zi,Gi), i = 1, ⋯, n, be the observations, and Θα and Θβ be the parameter spaces for α and β, respectively. In this section, we derive the detailed estimation procedure employing the BSBK method proposed by Wang and Yang (2007). Let ℱn be the space of B-spline basis functions of order r (r ≥ 2) (de Boor (2001)) with the B-spline basis Br(u) = (Bs,r(u) : 1 ≤ s ≤ Jn)T, u ∈ [a, b], where Jn = N + r and N = Nn is the number of interior knots for a knot sequence ξ1 = ⋯= 0 = ξr < ξr+1 < ⋯< ξr+Nn < 1 = ξr+Nn+1 = ⋯= ξNn+2r in which Nn increases along with the sample size n. Then ml(ul) with , l = 0, 1, can be approximated by a spline function,
where λl(β) = (λs,l(β), 1 ≤ s ≤ Jn)T and λ(β) = (λ0(β)T, λ1(β)T). For given β, the B-spline coefficients λ(β) and α can be estimated as
where . Let , where and . Let D(Z̃, β) = (D1(Z̃1, β), ⋯, Dn(Z̃n, β))T, an n × 2(q + Jn) matrix, and Y = (Y1, ⋯, Yn)T, where Z̃ = (Z̃1, ⋯, Z̃n)T is an n × 2q matrix. Then the least squares estimators of α and λ(β) is
| (2.2) |
Once the B-spline coefficients λ(β) are estimated, we can obtain the first derivative of the spline approximation of the nonparametric function as , where is the first derivative of Br(ul). Given the estimator λ̂l(β) in (2.2), we can estimate the loading parameters β by
Let λ̂l(β̂) be the estimators of the spline coefficients obtained by replacing D(Z̃, β) with D(Z̃, β̂) in (2.2). Based on the parametric estimator θ̂, it is easy to obtain the estimator of the nonparametric function ml(ul) as
| (2.3) |
A detailed estimation algorithm is given in Supplementary Materials.
2.3. Kernel estimator of nonparametric functions
To obtain the asymptotic normality of the spline estimators for the nonparametric functions ml(ul), l = 0, 1, as in Wang and Yang (2007), we use the BSBK estimator to establish their asymptotic normality. Define Ỹl = (Ỹ1l, ⋯, Ỹnl)T as the new pseudo-responses, and their corresponding “oracle” responses as , l = 0, 1. By using the B-spline estimators m̃l(·) and the parametric estimators of Section 2.2, we have
Similarly, Ỹi0 and can be defined. In the “oracle” responses, the functions ml(·) are assumed to be known.
Based on the new responses Ỹ1, we can obtain the BSBK estimator of m1(u1) as m̂1(u1, β̂) = â +b̂u1 by local linear fitting, in which
where Kh(t) = K(t/h)/h and K(·) is a kernel function and h is a bandwidth. By minimizing the weighted least squares, the estimator m̂1(u1, β̂) has a closed form
| (2.4) |
where
Similarly, we can also obtain the “oracle” kernel estimator of m1(u1) as based on new data by local linear fitting
| (2.5) |
An outline of the algorithm can be found in Supplementary Materials. We use the BIC criterion to select the number of interior knots, while fixing the order of basis function as cubic to approximate the unknown functions, as described in Ma and Song (2015). The positions of interior knots are chosen as the uniform quantiles of in the (k + 1)-th step (l = 0, 1, ⋯,L). Thus they change at each step while the number of knots remain fixed. This, however, does not affect the convergence of the algorithm in practice. To prove convergence of the algorithm with changes in knots is beyond the scope of this work. The BSBK estimator m̂l(ul, θ̂) is sensitive to the choice of bandwidth hl, l = 0, 1. Bandwidth selection has been intensively studied, see Sepanski et al. (1994) and Ruppert et al. (1995) for good discussions. To avoid the estimation of high order derivatives, we employ a bandwidth selector based on the mean squared error (MSE) criterion, called empirical bias bandwidth selection (EBBS) (Ruppert (1997); Carroll et al. (1998); Liu et al. (2014)). The details of EBBS are provided in Supplementary Materials.
Remark 1
Cui et al. (2011) and Ma and Song (2015) relaxed the constraints ||βl||2 = 1 to ||βl,−1|| < 1 with β l,−1 = (βl2, ⋯, βlp)T, l = 0, 1. We work directly on the equality constraints ||βl||2 = 1 which allows us to easily develop a Newton-Raphson algorithm. We can then test H0 : βlk = 0 for all k = 1, ⋯, p (see Section 5 for a demonstration). In addition, the Newton-Raphson algorithm is faster than the nonlinear optimization method adopted in Ma and Song (2015), especially under nonlinear constraints.
2.4. Theoretical results
We need some additional notation to show the asymptotic normality of the estimator. Let θ0 = ((α0)T, (β0)T)T be the true parameter θ, where and . Let the space ℳ be a collection of functions with finite L2 norm on [a0, b0]×[a1, b1]×ℛ with ℳ= {g(u) = g0(u0) + g1(u1)G, Egl(ul)2 ≤ ∞}, where u = (u0, u1)T. For 1 ≤ k ≤ q, let be a maximizer in ℳ for the optimization problem,
where . Let and P(Z) = (P1(Z1), ⋯, Pq(Zq))T. We take P(X) = (P1(X1), ⋯, Pp(Xp))T with . Let Ẑ = Z − P(Z), X̂ = X − P(X) and ϕ(V,β0) = (ϕ1(V,β0)T,ϕ2(V,β0)T)T, where ϕ1(V,β0) = (ẐT, ẐT G)T and . Define the covariance matrix of θ0 as
where ζ⊗2 = ζζT for any vector ζ. Σ can be simplified as if the error variance σ(V) is a constant .
Theorem 1
If assumptions (A.1)–(A.4) in the Appendix hold, and nN4 → ∞ and nN−2r−2 → 0, then ||θ̂−θ0||2 = Op(n−1/2), and as n → ∞, .
Theorem 2
If assumptions (A.1)–(A.4) in the Appendix hold, and nN4 → ∞ and nN−2r−2 → 0, then for l = 0, 1,
where m̃l(ul, β̂) is given in (2.3), and ml(·) is the true function.
Next we show that the order of the asymptotic uniform magnitude of the difference between the BSBK estimator m̂l(ul, β̂) and its “oracle” version is op(n−2/5), so m̂l(ul, β̂) and share the same asymptotic distribution.
Theorem 3
If assumptions (A.1)–(A.6) in the Appendix hold, and nN4 → ∞ and nN−δ → 0 with δ = min(2r + 2, 5r/2), then for l = 0, 1,
Set μk = ∫tkK(t)dt, νk = ∫tkK2(t)dt. The consistency and asymptotic normality of the unknown functions m0(·) and m1(·) now follow.
Theorem 4
If assumptions (A.1)–(A.6) in the Appendix hold, and nN4 → ∞ and nN−2r−2 → 0, then, for l = 0, 1,
where , l = 0, 1, , and .
If Equation , the variance vl(ul) can be simplified as for l = 0, 1.
3. Hypothesis tests
3.1. Testing for nonparametric components
Our model can assess the interaction of the combined effect of multiple environmental exposures with genes. This can be achieved by testing the nonparametric component m1(·) to discover the change trend of the interaction of the combined environmental effect. We consider a test to detect whether m1(u1) is a linear function ,
| (3.1) |
via a generalized likelihood ratio (GLR) test (Fan et al. (2001); Liang et al. (2010); Ma and Song (2015)). Rejecting H0 indicates statistical evidence of nonlinear interaction between G and multiple environmental mixtures. If we fail to reject H0, we can further assess whether there exists a genetic effect as well as linear interaction effect between a gene and multiple environmental exposures by fitting a parametric linear interaction model.
Remark 2
In addition to the linear hypothesis, we are interested in testing H0 : m1(·) = 0 or H0 : m1(·) = c where c is a constant. Testing for zero or constant effect can be done under the varying-coefficient model proposed in Ma et al. (2011), this cannot be done in the current model setup due to the fact that the loading parameters β1 are not identifiable under the above nulls. If we fail to reject the null in hypotheses (3.1), we can fit a linear interaction model as , where no constraints on β1 are imposed. Then one can proceed to test to assess the overall effect of G on Y. One can continue to assess the marginal effect of G on Y and the interaction effect between G and X or Z if is rejected.
Consider (3.1). Let θ̂ be the BSBK estimate of θ proposed in Section 2.2. Let m̂l,H0(ul) and m̂l,H1 (ul) be the estimators under H0 and H1, respectively. Let the residual sums of squares under H0 and H1 in (3.1) be and , where Ŷi = Yi − α̂TZ̃i. We define the generalized likelihood ratio (GLR) test statistic as
| (3.2) |
Let aK = {K(0) − 1/2 ∫ K2(u)du} [∫{K(u) − 1/2K * K(u)}du]−1, where K * K(u) denotes the convolution of K. Denote by Ωl the support of , and by |Ωl| the length of Ωl, l = 0, 1.
Theorem 5
If assumptions (A.1)–(A.6) in the Appendix hold, and nN4 → ∞ and nN−2r−2 → 0, then under H0 in (3.1), when is a linear function of u1,
where , and . Furthermore, , where d1 = aKμ1n.
When assessing the linear form of the function, RSS1(H0) and RSS1(H1) can be calculated by first getting the estimators of m0(·) and m1(·) using the B-spline method under the null and alternative hypotheses. The B-spline estimators under H0 are given by and m̂1,H0(u1) = δ̂0 +δ̂1u1, where δ̂0, δ̂1, and λ̂0 are the ordinary least squares estimators of δ0, δ1, and λ0. Then, we can obtain the kernel estimator m̂0,H0(u0) based on the new data (ŶH0,X,Z,G) and , using the arguments in Section 2.3, where ŶH0 = (Ŷ1,H0, ⋯, Ŷn,H0)T and . Here m̂0,H1(·) and m̂1,H1(.) are the BSBK estimators which can be obtained as in (2.4).
To illustrate the testing for the case with l > 1, we consider a model with two genetic variables G1 and G2,
| (3.3) |
One can simultaneously test m1(·) and m2(·), for example, testing
| (3.4) |
where and are linear functions. Similarly, we can construct the corresponding GLR test statistic
| (3.5) |
where , and . Note that , l = 0, 1, 2, are different from those in T1, but the estimation is similar.
Theorem 6
If assumptions (A.1)–(A.6) in the Appendix hold, nN4 → ∞ and nN−2r−2 → 0, then under H0 in (3.4), when and are linear functions,
where , μ2n = bn{K(0) − 1/2 ∫ K2(u)du} and bn = Σl=1,2 |Ωl|/hl. Furthermore, , where with .
Remark 3
The formulation of asymptotic normality in Theorem 6 is that in Fan and Jiang (2005). Theorem 6 can be generalized to cases where three or more genetic variables can be fitted and tested (l ≥ 3). One can apply Theorem 6 for simultaneous inference on the functions of some components of varying index coefficients. While the asymptotic results for T1 and T2 are available, they may not perform well when sample sizes are small. We recommend the conditional bootstrap method (Cai et al. (2000); Fan et al. (2001)) in applications.
3.2. Testing parametric components
We are also interested in assessing the interaction effects of genes with discrete environments. This can be addressed via parametric hypothesis testing. Furthermore, if there is G×E interaction, one may be interested in testing which index coefficients contribute to the joint effect. This results in another parametric hypothesis testing problem. We consider a class of general hypothesis testing problems with
| (3.6) |
where A is a known k × (q + s) full-rank matrix, s is the number of elements in S ⊂ {1, ⋯, p}, with βS = (βj1, ⋯, βjs)T, jl ∈ S, and γ is a k-dimensional vector. For a special case, we can detect whether α1 and βS are zeros by taking
| (3.7) |
Let be the parameters corresponding to θ under H0 in (3.7) and be the counterparts under H1. Define the residual sums of squares under H0 and H1 as
where θ̂H0 and θ̂H1 are the estimators of θ under H0 and H1 proposed in Section 2.2, and m̂l,H0(·) and m̂l,H1 are estimators of ml(·) proposed in (2.4) under H0 and H1, l = 0, 1, respectively. We take the test statistic
| (3.8) |
Theorem 7
If assumptions (A.1)–(A.6) in the Appendix hold, nN4 → ∞ and nN−2r−2 → 0, then when σ(V) is a constant ,
4. Monte Carlo simulation
The finite sample performance of the proposed method was evaluated by simulation studies. Under model (2.1), we generated continuous X variables X1, X2, X3 as independent uniform U(0, 1) and discrete Z variables Z1, Z2 as independent Bernoulli Ber(1, 0.5). The genetic variable G was coded as (2, 1, 0) corresponding to genotypes (AA, Aa, aa). We set the minor allele frequency (MAF) pA = (0.1, 0.3, 0.5) and assumed Hardy-Weinberg equilibrium. SNP genotypes AA, Aa, and aa were simulated from a multinomial distribution with frequencies , 2pA(1 − pA) and (1 − pA)2, respectively. The error term ε was normal N(0, 0.1).
We set m0(u) = cos(πu) and m1(u) = sin{π(u − A)/(B − A)} with and , and , α0 = (0.5, 0.5)T, and α1 = (0.3, 0.3)T. We drew 1000 data sets with sample size n = 200, 500. The Epanechnikov kernel K(t) = 0.75(1 − t2)+ was chosen to localize the unknown functions m0(·) and m1(·). The suitable smoothing bandwidths for estimating both functions were selected using the EBBS method described in Section 2.3. The number of interior knots Nk was selected by the BIC method.
4.1. Performance of estimation
Table 1 summarizes the average bias of the estimators (Bias), the standard deviation of the 1000 estimators (SD), the average of the estimated standard errors (SE) based on the theoretical calculation, and the estimated coverage probability (CP) at the nominal 95% confidence level for the parameters. In general, the coverage probability for all the parameters was close to 95% and reasonably controlled. As the sample size increased, the performance of the parameter estimators improved. We observed consistently smaller SD and SE when n increased from 200 to 500. The same trend was observed when n increased to 1000 (see Supplementary Materials for more details). The parameter estimators for the interaction effects (β1, α1) improved as MAF increased. For example, the SD of β̂11 went from 0.028 to 0.012 when MAF increased from 0.1 to 0.5 under a fixed sample size n = 200. However, the estimators for the main effects (β0,α0) showed an opposite direction due to limited data information to estimate these parameters when MAF increased. This is due to the fact that the amount of data used to estimate these parameters is proportional to (1 − pA)2.
Table 1.
Simulation results for pA = 0.1, 0.3, 0.5 with sample size n = 200, 500.
| n | Param | True | pA = 0.1
|
pA = 0.3
|
pA = 0.5
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bias | SD | SE | CP | Bias | SD | SE | CP | Bias | SD | SE | CP | |||
| 200 | α01 | 0.500 | 4.4E-04 | 0.016 | 0.016 | 95.2 | 3.1E-04 | 0.020 | 0.020 | 95.2 | 9.9E-04 | 0.026 | 0.026 | 95.1 |
| α02 | 0.500 | −1.6E-04 | 0.016 | 0.016 | 95.3 | 4.1E-04 | 0.020 | 0.020 | 95.3 | 5.6E-04 | 0.026 | 0.026 | 95.8 | |
| α11 | 0.300 | 9.4E-05 | 0.040 | 0.039 | 94.1 | 6.0E-04 | 0.024 | 0.024 | 94.1 | 6.7E-05 | 0.022 | 0.022 | 95.2 | |
| α12 | 0.300 | −1.1E-03 | 0.040 | 0.039 | 95.0 | −1.1E-03 | 0.023 | 0.024 | 95.9 | −4.4E-04 | 0.021 | 0.022 | 96.3 | |
| β01 | 0.620 | −3.7E-04 | 0.011 | 0.011 | 94.7 | −1.7E-03 | 0.012 | 0.013 | 94.8 | −2.1E-03 | 0.014 | 0.014 | 94.5 | |
| β02 | 0.555 | 3.3E-04 | 0.012 | 0.012 | 95.3 | 1.0E-03 | 0.013 | 0.013 | 96.4 | 1.5E-03 | 0.014 | 0.015 | 96.6 | |
| β03 | 0.555 | −2.7E-04 | 0.012 | 0.012 | 94.0 | 4.2E-04 | 0.013 | 0.013 | 95.3 | 3.1E-04 | 0.015 | 0.015 | 95.4 | |
| β11 | 0.577 | 1.4E-03 | 0.028 | 0.027 | 92.9 | −4.0E-04 | 0.015 | 0.015 | 95.5 | −7.5E-05 | 0.012 | 0.012 | 95.1 | |
| β12 | 0.577 | −3.4E-04 | 0.029 | 0.028 | 93.5 | 9.5E-05 | 0.015 | 0.015 | 95.3 | 2.9E-04 | 0.011 | 0.012 | 96.2 | |
| β13 | 0.577 | −3.2E-03 | 0.028 | 0.027 | 94.3 | −2.6E-04 | 0.015 | 0.015 | 96.1 | −5.7E-04 | 0.012 | 0.012 | 96.0 | |
| 500 | α01 | 0.500 | −3.2E-04 | 0.010 | 0.010 | 95.8 | −5.5E-04 | 0.012 | 0.012 | 95.2 | −4.0E-04 | 0.016 | 0.016 | 96.1 |
| α02 | 0.500 | 1.9E-04 | 0.010 | 0.010 | 94.1 | 2.0E-04 | 0.013 | 0.012 | 94.2 | 3.8E-04 | 0.016 | 0.016 | 94.6 | |
| α11 | 0.300 | 5.6E-04 | 0.023 | 0.022 | 93.7 | 9.9E-04 | 0.015 | 0.014 | 93.8 | 6.5E-04 | 0.013 | 0.013 | 94.5 | |
| α12 | 0.300 | 1.2E-05 | 0.023 | 0.022 | 94.0 | 2.6E-04 | 0.015 | 0.014 | 93.8 | 2.0E-04 | 0.013 | 0.013 | 94.1 | |
| β01 | 0.620 | −4.6E-04 | 0.007 | 0.007 | 95.2 | −1.0E-03 | 0.008 | 0.008 | 95.7 | −1.2E-03 | 0.009 | 0.009 | 94.9 | |
| β02 | 0.555 | 1.2E-04 | 0.007 | 0.007 | 95.5 | 4.3E-04 | 0.008 | 0.008 | 95.1 | 5.5E-04 | 0.009 | 0.009 | 95.1 | |
| β03 | 0.555 | 2.6E-04 | 0.007 | 0.007 | 94.2 | 5.2E-04 | 0.008 | 0.008 | 94.1 | 5.2E-04 | 0.009 | 0.009 | 94.4 | |
| β11 | 0.577 | 5.2E-04 | 0.015 | 0.016 | 95.0 | 3.0E-05 | 0.009 | 0.009 | 96.6 | −8.5E-06 | 0.007 | 0.007 | 95.9 | |
| β12 | 0.577 | −3.4E-04 | 0.016 | 0.016 | 94.0 | −8.0E-06 | 0.009 | 0.009 | 95.6 | 1.0E-04 | 0.007 | 0.007 | 96.3 | |
| β13 | 0.577 | −8.3E-04 | 0.016 | 0.016 | 94.5 | −2.3E-04 | 0.009 | 0.009 | 95.2 | −2.3E-04 | 0.007 | 0.007 | 94.8 | |
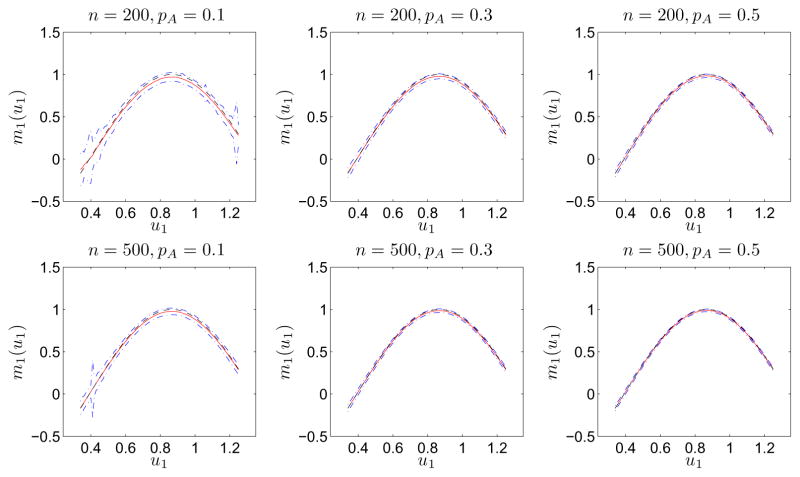
Figure 1 shows the plot of the estimators of m1(u1), and its corresponding confidence bands under different sample sizes and MAFs in the interval of u1 from 0.25 to 1.25. It can be there seen that the estimated curves almost overlap with the corresponding true curves, and the confidence bands are very tight, especially under large MAF and sample size. We also plotted the estimate of m0(·), see the Supplementary Materials.
Figure 1.
The estimation of function m1(·) under different MAFs and sample sizes. The estimated and true functions are denoted by the solid and dashed lines, respectively. The 95% confidence band is denoted by the dotted-dash line.
4.2. Performance of hypothesis tests
We first evaluated the performance of the test for the nonparametric function under the hypothesis , where , and δ0 and δ1 are some constants. Power was evaluated under a sequence of alternative models indexed by τ, . When τ = 0, the test results provide the false positive rates. The null model corresponds to a linear G×E effect.
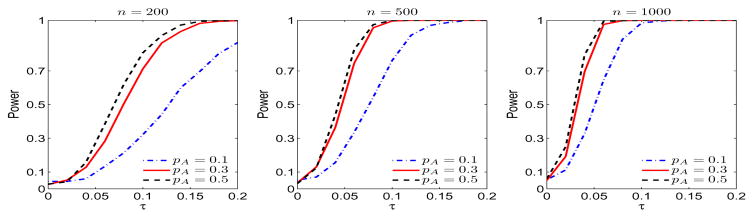
Figure 2 shows the size (τ = 0) and power function (τ > 0) at significance level 0.05 based on 500 Monte Carlo simulations each with 500 bootstrap samples under sample sizes n = 200, 500, 1000. The empirical type I errors under the three scenarios are very close to the nominal level 0.05. We observed dramatic power increase when MAF increased from 0.1 to 0.3 in all scenarios. The results indicate that our method can reasonably control the false positives and has appropriate power to detect genetic difference. We also considered the PLVMICM model in (3.3) with two genetic components and tested if both m1(·) and m2(·) are simultaneously linear, following Theorem 6. The results are in the Supplemental Materials.
Figure 2.
The empirical size and power function of testing nonparametric function m1(·) under different sample sizes and MAFs.
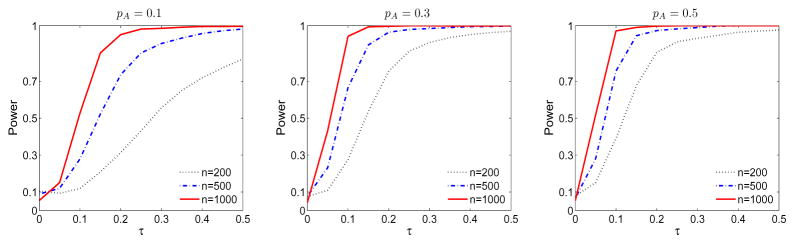
To check the performance of the interaction test between G and discrete variable Z, under model (2.1), we considered the hypothesis H0 : α1 = 0. The power of the test was evaluated under a sequence of alternatives indexed by τ, . Data were simulated as in the previous section. Figure 3 depicts the empirical size (τ = 0) and power functions (τ > 0) under different sample sizes and MAFs at the 0.05 significance level. As expected, the power and size improve as MAF and sample size increase. Under low MAF (pA = 0.1), the size is a little inflated when n is small (200 and 500), but is well controlled when n increases to 1000. As tith the nonparametric test, dramatic power improvement is observed when MAF increases from 0.1 to 0.3. The power difference between MAF=0.3 and MAF=0.5 is small indicating good performance of the test.
Figure 3.
The empirical size and power functions of testing H0 : α1 = 0 under differen sample sizes and MAFs.
5. A case study
We applied the proposed PLVMICM model to a data set from the Gene Environment Association Studies initiative (GENEVA, http://www.genevastudy.org) funded by the trans-NIH Genes, Environment, and Health Initiative (GEI), to show the utility of the method. Low and high birth weights are not only the major causes of neonatal morbidity and mortality, but are also related to increased risk of metabolic diseases later in life. Fetal growth is determined by fetal genes as well as complex interactions between fetal genes and the maternal uterine environment. We focused on the Thai population with 1126 subjects genotyped with the Omni1-Quad v1-0 B platform after removing outliers. After regressing the baby’s body weight on twelve environmental variables, including nine continuous and three discrete variables, five continuous variables and one discrete variable remained significant at the 0.0001 significance level. Three of the five continuous variables were chosen, including mother’s mean OGTT diastolic blood pressure (denoted as X1), mother’s one hour OGTT glucose level (denoted as X2), and mother’s mean OGTT systolic blood pressure (denoted as X3). The discrete variable, denoted as Z, is baby’s gender. To show the utility of the method, we picked one candidate gene CDKAL1 for a demonstration. The gene is located on chromosome 6 and contains 192 SNPs after removing those with MAF< 0.05. Low birth weight has been shown to be associated with high risk in type 2 diabetes later in life. Evidence of genetic studies on type 2 diabetes loci suggests that this gene is associated with reduced birth weight in Caucasian populations (Zhao et al. (2009); Andersson et al. (2010)). Our goal is to evaluate whether this gene also functions in the Thai population and, if so, how SNPs in the gene interact with mother’s condition (considered as environment) to affect birth weight and further determine the interaction mechanism.
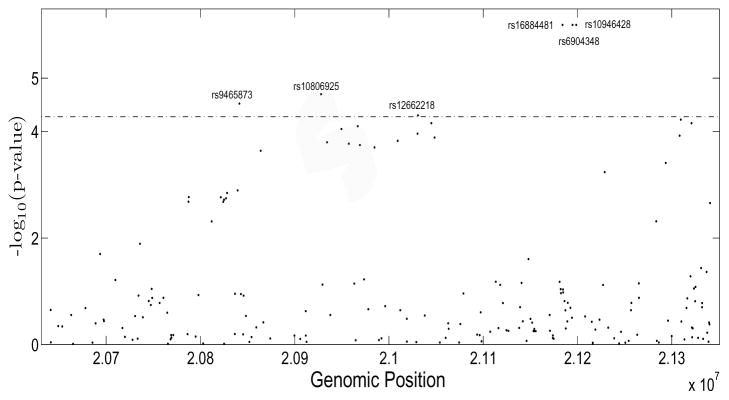
We first tested whether any SNP is associated with birth weight based on the nonparametric test of H0 : m1(u1) = δ0 + δ1u1 with p-value denoted by pm1. Since we tested each SNP individually, we applied a simple multiple testing correction method. We first calculated the effective number of tests E0 by using the Cheverud estimation method, given by , where L = 192 is the total number of SNPs and rij are the pairwise correlation coefficients of SNPs (Cheverud (2001)). The estimated E0 = 188.09, which yields a gene-wide significance level of α = 0.01/E0 = 5.3 × 10−5. Figure 4 depicts the −log10(p-values). Clearly, six SNPs rs16884481 rs10946428, rs6904348, rs10806925, rs9465873, and rs12662218 passed the significance level based on 105 bootstrap samples.
Figure 4.
Plot of −log10(p-value) for SNPs within gene CDKAL1.
The testing results for the six SNPs are reported in Table 2. We report SNP ID, MAF, allele information with bold font letter as the minor allele, p-values for the nonparametric test (described in Section 4.2). We also report the p-value of the test H0 : β0 = β1 v.s. H1 : β0 ≠ β1 in the column labeled by pβ as opposed to the model by Ma and Xu (2015) based on the generalized likelihood ratio test in Section 3.2. The p-value of the parametric test H0 : α1 = 0 is reported in the column labeled by pα1 following the procedure described in Section 4.2. To compare the goodness of fit for PLVMICM with an additive varying-coefficient model (AVCM), , and to see the relative gain by the integrative analysis, we calculated the MSEs of both models; they are given in the last two columns of Table 2. The p-values for testing H0 : m11(X1) = m12(X2) = m13(X3) = 0 when fitting the AVCM model are reported in the column labeled by pAVCM.
Table 2.
List of SNPs with MAF, allele, p-values under different hypothesis testing and MSE
| SNP ID | MAF | Alleles | p-value
|
MSE
|
||||
|---|---|---|---|---|---|---|---|---|
| pm1 | pβ | pα1 | pAVCM | PLVMICM | AVCM | |||
| rs16884481 | 0.1960 | C/T | ≤1.0E-05 | 5.1E-04 | 0.2517 | 0.1799 | 0.1342 | 0.1402 |
| rs10946428 | 0.2744 | A/G | ≤1.0E-05 | 1.5E-05 | 0.0960 | 0.1227 | 0.1333 | 0.1399 |
| rs6904348 | 0.2766 | A/C | ≤1.0E-05 | 1.9E-05 | 0.0869 | 0.1358 | 0.1334 | 0.1399 |
| rs10806925 | 0.4761 | C/T | 2.0E-05 | 2.2E-06 | 0.3671 | 0.2733 | 0.1340 | 0.1405 |
| rs9465873 | 0.4503 | A/G | 3.0E-05 | 6.5E-06 | 0.4911 | 0.2562 | 0.1340 | 0.1403 |
| rs12662218 | 0.2719 | A/G | 5.0E-05 | 5.4E-06 | 0.2802 | 0.4616 | 0.1345 | 0.1408 |
The p-values in column pβ for the comparison of different model assumptions clearly show that the loading parameters are different for different index functions, indicating the necessity of the proposed model vs the one proposed by Ma and Xu (2015). The p-values in column pα1 indicate that SNP×gender interactions are not significant for these six SNPs. The goodness of fit measure in the last two columns shows that the PLVMICM model fits the data better than the AVCM model, indicating the potential benefit of integrative G×E analysis. Furthermore, the testing p-values for the AVCM model do not show significance. The results imply that the genetic effects of these six SNPs are modified by the mixture effect of the three X variables, rather than separately, which further indicate the power of the integrative analysis.
For the 186 SNPs that were rejected, we fitted the model assuming m1(u1) = δ0 + δ1u1, assuming linear G×E interaction, then testing H0 : δ0 = δ1 = 0. No SNPs showed signs of significance at the 5.3E-05 significant level. The most significant SNP was rs12209806 with a p-value of 6.72E-05. This indicates that there is no linear interaction between these SNPs and the three environmental variables. However, there are four SNPs, rs12196595, rs6908425, rs6917599, and rs7773189 showing interactions with gender based on pα1 for the 186 SNPs; the p-values were 6.12E-08, 1.89E-07, 3.69E-07, and 1.61E-05, respectively.
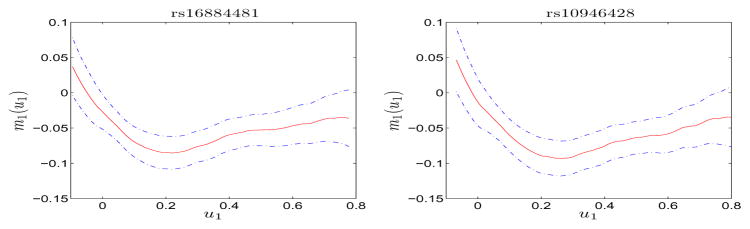
We tested the significance of the individual X variable that contributes to the joint effect following the procedure given in Section 3.2. The results showed that X1 and X2 contribute significantly to the joint effect in these six SNPs, but not X3 (see Table S2 in Supplementary Materials). The estimators of the nonparametric function m1(u1) for the first two SNPs, rs16884481 and rs10946428 along with their 95% confidence band are given in Figure 5. The estimators for the other four SNPs are shown in Section 3 in Supplementary Materials due to space limit. The estimated function shows a decreasing pattern then slightly increases as the index value u1 increases. Our model clearly reveals the nonlinear modulating effect of environmental mixtures on genetic effect of birth weight. Such dynamic effects can be helpful in designing prevention strategy when the model is applied to other complex diseases such as diabetes.
Figure 5.
Plot of the estimate (solid curve) of the nonparametric function m1(u1) for SNPs rs16884481 and rs10946428 along with their 95% confidence band (dash-dotted line).
6. Discussion
G×E interaction has been studied intensively in the literature and many statistical methods have been proposed. In this paper, we developed a partially linear varying multi-index coefficient model (PLVMICM) to conduct a rigorous assessment of the combined effect of multiple environmental exposures on the risk of disease under the paradigm of G×E interaction. Our model can be interpreted as a systems genetics approach to modeling the joint effect of environmental mixtures as a whole, then assessing how the integrative effect modifies genetic influence on disease risk. Our model is biologically attractive in that it addresses a long-term question on G×E interaction from a systems genetics perspective and is well supported by epidemiological studies (Carpenter et al. (2002); Monosson (2005); Powers et al. (2008)); and it has the flexibility to detect nonlinear interactions, and therefore, is more powerful when genetic effects are nonlinearly modified by simultaneous exposure to multiple environments.
From a statistical point of view, the index coefficient function treats multiple environmental variables X as a single index variable, and therefore can reduce multiple testing burden when interactions between the X variables and G are modelled separately. In addition, when there exist interactions between the X variables, our model has the flexibility to incorporate such interactions by adding interaction terms to the index function. PLVMICM is flexible and includes several existing models as special cases, for example, the partially linear single-index model (Carroll et al. (1997); Xia and Li (1999); Xia and Hardle (2006); Liang et al. (2010); Cui et al. (2011)) and the nonparametric additive model discussed by Fan and Jiang (2005).
In a typical G×E study, there are usually a large number of genetic variables (e.g., SNPs), and it is important to fit multiple SNPs in a single model and to select important players that interact with environmental mixtures to affect disease risk in a high dimensional model setup. In addition, many human diseases are measured on a binary scale. It is natural to extend the current PLVMICM model to a generalized PLVMICM model framework. This will be considered in a future investigation.
Supplementary Material
Acknowledgments
This work was partially supported by grants from NSF (IOS-1237969, DMS-1209112 and DMS-1512422), from NIDA/NIH (P50 DA10075 and P50 DA039838), and from NSFC (31371336). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF, the NIDA/NIH and the NSFC. The authors thank an associate editor and two reviewers for their constructive and helpful comments. Funding support for the GWA mapping: Maternal Metabolism-Birth Weight Interactions study was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01HG004415). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession number phs000096.v4.p1. Code for implementing the method was written in Matlab and C, and is available for free download at http://www.stt.msu.edu/~cui/software.html.
Appendix: Proofs
Notations
For any vector ξ = (ξ1, ⋯, ξs)T ∈ ℛs, let ||ξ||∞ = max1≤l≤s |ξl|. For any nonzero matrix As×s, denotes its Lr norm as . For any matrix , let . Let C(p)[a, b] = {ψ : ψ(p) ∈ C[a, b]} be the space of the pth-order smooth functions. Denote the space of Lipschitz continuous functions for any fixed constant c0 as Lib([a, b], c0) = {ψ : | ψ(x1) − ψ(x2)| ≤ c0|x1−x2|, ∀x1, x2 ∈ [a, b]}. The following assumptions are required.
-
A.1
For each l = 0, 1, the density function fU(βl)(·) of random variable is bounded away from 0 on Ωl, and there exists a constant 0 < c0 < ∞ such that fU(βl)(·) ∈ Lib([a, b], c0) for βl in the neighborhood of , where and 𝒳 is a compact support of X.
-
A.2
The nonparametric function ml ∈ C(r)[al, bl], l = 0, 1.
-
A.3
The noise ε satisfies E(ε|V) = 0, E(|ε|4) < ∞ and σ(v) = var(ε|V = v) < c1 for some 0 < c1 < ∞.
-
A.4
There exist constants 0 < cz ≤ Cz < ∞ such that cz ≤ Q(x) = E(Z̃Z̃T |X = x) ≤ Cz for all x ∈ 𝒳.
-
A.5
The kernel function K(·) is a symmetric density function with compact support [−1, 1] and K ∈ Lib([a, b], cK) for some constant cK. The bandwidth hl = O(n−1/5), l = 0, 1.
-
A.6
The function u3K(u) and u3K′(u) are bounded and ∫ u4K(u)du < ∞.
Let , Yz = (Yz,1, ⋯, Yz,n)T, e = (ε1, ⋯, εn)T, 𝕏 = (X1, ⋯, Xn)T, ℤ = (Z1, ⋯, Zn)T, ℤ̃ = (1n, ℤ), and G = (G1, ⋯, Gn)T. Define
| (A.1) |
where Di(β) = (Di,sl(βl), 1 ≤ s ≤ Jn, l = 0, 1)T and D(β) = (D1(β), ⋯, Dn(β))T, an n × 2Jn matrix.
Proof of Theorem 1
This is a straightforward result of Lemma S.6 in the Supplementary Materials.
Proof of Theorem 2
For simplicity, we assume [al, bl] = [a, b] for l = 0, 1. Since for any ul ∈ [al, bl], Bs,l(ul), s = 1, ⋯, Jn, l = 0, 1, have bounded first derivatives, by Lemmas S.4 and S.5 in the Supplementary Materials and Theorem 1, we have for any ul ∈ [a, b],
Then, combined with Lemma S.4, we have
This completes the proof of Theorem 2.
Proof of Theorem 4
As nh5 = O(1), we have (nhl)1/2n−2/5 = o(1). By Theorem 3, we have
Thus Theorem 4 can be shown straightforwardly following Lemma S.7 in the Supplementary Materials.
Proof of Theorem 7
This proof is similar to that of Liang et al. (2010). Accordingly, we only provide a sketch of the proof here, more details can be found in the Supplementary Materials. We first prove n−1R(H1) = E{σ(V)}+op(1). Let m̂(X, β) = m̂0(XTβ0, β) + m̂1(XTβ1, β)G and, correspondingly, . By Theorem 3 and Lemma S.7 in the Supplementary Materials, n−1R(H1) can be decomposed as
where , and . It is easy to see by the Law of Large Numbers that 𝕀1 = E{σ(V)} + Op(n−1/2). By Theorem 2.6 in Li and Racine (2007), we have maxi | m̂O(Xi, β0) − m(Xi, β0)| = Op((log(n)/(nh))1/2), which results in 𝕀2 = Op((log(n)/(n2h))1/2) and 𝕀3 = Op(log(n)/(nh)). This leads to n−1R(H1) = E{σ(V)} + op(1).
The difference R(H0) − R(H1) can be decomposed as
Under the null, we have , and under the alternative σ−2𝕀4 asymptotically follows a noncentral Chi-squared distribution with k degrees of freedom and noncentrality parameter ϕ. It remains to show that 𝕀5 = op(1). This can be shown along the same lines as 𝕀4. This completes the proof of Theorem 7.
The proofs of Theorem 3, 5, and 6 are in the Supplementary Materials.
Footnotes
Supplementary Materials Proofs of theorems and lemmas, additional simulation, and data analysis results can be found in the Supplementary Materials.
References
- Andersson EA, Pilgaard K, Pisinger C, Harder MN, Grarup N, Faerch K, Poulsen P, Witte DR, Jrgensen T, Vaag A, Hansen T, Pedersen O. Type 2 diabetes risk alleles near ADCY5, CDKAL1 and HHEX-IDE are associated with reduced birthweight. Diabetologia. 2010;53:1908–1916. doi: 10.1007/s00125-010-1790-0. [DOI] [PubMed] [Google Scholar]
- Cai Z, Fan J, Li R. Efficient estimation and inferences for varying-coefficient models. J Am Stat Assoc. 2000;95:888–902. [Google Scholar]
- Carroll RJ, Fan J, Gijbels I, Wand MP. Generalized partially linear single-index models. J Am Stat Assoc. 1997;92:477–489. [Google Scholar]
- Carroll RJ, Ruppert D, Welsh AH. Local estimating equations. J Am Stat Assoc. 1998;93:214–227. [Google Scholar]
- Carpenter DO, Arcaro K, Spink DC. Understanding the human health effects of chemical mixtures. Environ Health Perspect. 2002;110(suppl 1):25–42. doi: 10.1289/ehp.02110s125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87:52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- Cui X, Härdle W, Zhu L. The EFM approach for single-index models. Ann Stat. 2011;39:1658–1688. [Google Scholar]
- de Boor C. A Practical Guide to Splines. Springer; New York: 2001. [Google Scholar]
- Falconer DS. The problem of environment and selection. Am Natural. 1952;86:293–299. [Google Scholar]
- Fan J, Jiang J. Nonparametric inferences for additive models. J Am Stat Assoc. 2005;100:890–907. [Google Scholar]
- Fan J, Zhang C, Zhang J. Generalized likelihood ratio statistics and Wilks phenomenon. Ann Stat. 2001;29:153–193. [Google Scholar]
- HAPO Study Cooperative Research Group. Hyperglycemia and Adverse Pregnancy Outcome (HAPO) Study: associations with neonatal anthropometrics. Diabetes. 2009;58:453–459. doi: 10.2337/db08-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Racine RS. Nonparametric Econometrics: Theory and Practice. Princeton University Press; Princeton, N. J: 2007. [Google Scholar]
- Li Y, Wang N, Carroll RJ. Generalized functional linear models with semi- parametric single-index interactions. J Am Stat Assoc. 2010;105:621–633. doi: 10.1198/jasa.2010.tm09313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Liu X, Li R, Tsai CL. Estimation and testing for partially linear single index models. Ann Stat. 2010;38:3811–3836. doi: 10.1214/10-AOS835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang H, Zhou Y. Local empirical likelihood inference for varying-coefficient density-ratio models based on case-control data. J Am Stat Assoc. 2014;109:635–646. [Google Scholar]
- Ma S, Song PX. Varying index coefficient models. J Am Stat Assoc. 2015;110:341–356. [Google Scholar]
- Ma S, Xu S. Semiparametric nonlinear regression for detecting gene and environment interactions. J Stat Plan Inf. 2015;156:31–47. [Google Scholar]
- Ma S, Yang L, Romero R, Cui Y. Varying coefficient model for gene-environment interaction: a non-linear look. Bioinformatics. 2011;27:2119–2126. doi: 10.1093/bioinformatics/btr318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosson E. Chemical mixtures: considering the evolution of toxicology and chemical assessment. Environ Health Perspect. 2005;113:383–390. doi: 10.1289/ehp.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert D. Empirical-bias bandwidths for lcoal polynomial nonparametric regression and density estimation. J Am Stat Assoc. 1997;92:1049–1062. [Google Scholar]
- Ruppert D, Sheathers SJ, Wand MP. An effective bandwidth selector for local least squares regression. J Am Stat Assoc. 1995;90:1257–1270. [Google Scholar]
- Ross CA, Smith WW. Gene environment interactions in Parkinson’s disease. Parkins Rel Dis. 2007;13:S309–S315. doi: 10.1016/S1353-8020(08)70022-1. [DOI] [PubMed] [Google Scholar]
- Powers KM, Kay DM, Factor SA, Zabetian CP, Higgins DS, Samii A, Nutt JG, Griffith A, Leis B, Roberts JW, Martinez ED, Montimurro JS, Checkoway H, Payami H. Combined effects of smoking, coffee, and NSAIDs on Parkinson’s disease risk. Mov Disord. 2008;23:88–95. doi: 10.1002/mds.21782. [DOI] [PubMed] [Google Scholar]
- Sepanski JH, Knickerbocker R, Carroll RJ. A semiparametric correction for attenuation. J Am Stat Assoc. 1994;89:1366–1373. [Google Scholar]
- Sexton K, Hattis D. Assessing cumulative health risks from exposure to environmental mixtures - three fundamental questions. Environ Health Perspect. 2007;115:825–832. doi: 10.1289/ehp.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang L. Spline-backfitted kernel smoothing of nonlinear additive autoregression model. Ann Stat. 2007;35:2474–2503. [Google Scholar]
- Wu C, Cui Y. A novel method for identifying nonlinear gene-environment inter- actions in case-control association studies. Hum Genet. 2013;132:1413–1425. doi: 10.1007/s00439-013-1350-z. [DOI] [PubMed] [Google Scholar]
- Xia Y, Härdle W. Semi-parametric estimation of partially linear single-index models. J Multiv Anal. 2006;97:1162–1184. [Google Scholar]
- Xia YC, Li WK. On single-index coefficient regression models. J Am Stat Assoc. 1999;94:1275–1285. [Google Scholar]
- Zhao J, Li M, Bradfield JP, Wang K, Zhang H, Sleiman P, Kim CE, Annaiah K, Glaberson W, Glessner JT, Otieno FG, Thomas KA, Garris M, Hou C, Frackelton EC, Chiavacci RM, Berkowitz RI, Hakonarson H, Grant SF. Examination of type 2 diabetes loci implicates CDKAL1 as a birth weight gene. Diabetes. 2009;58:2414–8. doi: 10.2337/db09-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmet P, Alberti K, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.