Abstract
A robust phenotypic plasticity to ward off adverse environmental conditions determines performance and productivity in crop plants. Flax (linseed), is an important cash crop produced for natural textile fiber (linen) or oilseed with many health promoting products. This crop is prone to drought stress and yield losses in many parts of the world. Despite recent advances in drought research in a number of important crops, related progress in flax is very limited. Since, response of this plant to drought stress has not been addressed at the molecular level; we conducted microarray analysis to capture transcriptome associated with induced drought in flax. This study identified 183 differentially expressed genes (DEGs) associated with diverse cellular, biophysical and metabolic programs in flax. The analysis also revealed especially the altered regulation of cellular and metabolic pathways governing photosynthesis. Additionally, comparative transcriptome analysis identified a plethora of genes that displayed differential regulation both spatially and temporally. These results revealed co-regulated expression of 26 genes in both shoot and root tissues with implications for drought stress response. Furthermore, the data also showed that more genes are upregulated in roots compared to shoots, suggesting that roots may play important and additional roles in response to drought in flax. With prolonged drought treatment, the number of DEGs increased in both tissue types. Differential expression of selected genes was confirmed by qRT-PCR, thus supporting the suggested functional association of these intrinsic genes in maintaining growth and homeostasis in response to imminent drought stress in flax. Together the present study has developed foundational and new transcriptome data sets for drought stress in flax.
Keywords: flax/linseed, drought, abiotic stress, microarray, gene expression profiling, transcriptomics
Introduction
Drought is a major global phenomenon that affects growth, accumulation of biomass, performance and productivity in crops. In addition to drought stress, crop plants also face, inter alia, other forms of abiotic stresses1 such as salt, temperature (low and high), low light irradiance, flood, metal toxicity (viz. heavy metal) during their life cycle that reduces yield potential up to 70%.2,3 Hydropenia, caused by drought, affects several key processes in plants that include stomatal opening and closure,4 decrease in photosynthetic carbon assimilation,5 rate of transpiration, water potential in tissues, and other physiological, biochemical and molecular parameters that ultimately lead to retardation of growth and development and death of the plant in severe cases.6 Although, hydropenia triggers a cascade of physiological and metabolic processes, plants circumvent this stress by three different adaptations: (1) drought escape, (2) drought avoidance and (3) drought tolerance.7,8 Drought stress occurs when soil moisture content and relative humidity in the air are low and the ambient temperature is high.9 In these conditions many plant species escape drought by accelerating flowering and complete their life cycles early though with compromised productivity. On the contrary, the predisposition of plants to maintain water homeostasis in their tissues during drought helps them in drought avoidance and this is accomplished by enhancing water absorption and/or reducing evapotranspiration. Drought tolerance refers to plants’ predisposition to survive and resist water deficiency below field capacity of soil by protoplasmic tolerance, de novo synthesis of osmolytes and/or compatible solutes.10,11 Both drought avoidance and drought tolerance traits in crops have a significant positive impact on agricultural production. Unlike, tolerance to biotic stresses which are mostly governed by monogenic traits, abiotic stresses are predominantly controlled by polygenic traits and hence are complex to study and more challenging to develop a comprehensive understanding of the underpinning processes.12
Because of its importance, drought has been investigated extensively and the advances and insights gained from these studies suggest that plants’ response to abiotic stress involves a plethora of genes that produce diverse responses at the biochemical, physiological and molecular levels.5,13 These genes are broadly classified into three main categories: (1) genes that function directly in the protection of membranes and proteins; water and ion uptake/transport, (2) regulatory genes involved in signaling cascades and transcriptional control, and (3) genes of unknown function.14 The first group consists of functional proteins which include chaperones, heat shock proteins, late embryogenesis abundant (LEA) proteins, osmotin, antifreeze proteins, mRNA-binding proteins; key enzymes for osmolyte biosynthesis such as proline, mannitol, glycine betaine, trehalose; water channel proteins and transporters, aquaporins; detoxification enzymes like catalases, peroxidases and various proteases.5,15-17 The second class comprises regulatory proteins that control activities of key enzymes of stress-signal transduction pathways or modulate the expression of stress responsive genes. These include transcription factors like18-22 DREB, AREB, MYC, MYB, bZIP, NAC, zinc finger, ERF and WRKY etc; protein kinases like mitogen-activated protein (MAP) kinases, calcium-dependent protein kinases (CDPKs), receptor-like kinases and histidine kinases, SNF1-related protein kinase (SnRK2); protein phosphatases like pp2c family; enzymes involved in phospholipid metabolism like PLD, PLC and other signaling molecules.23-25 These two classes of proteins along with unassigned putative proteins either work in coordination or may act independently to mitigate the effects of drought stress in plants.
Flax (Linum usitatissimum) is one of the ancient crops cultivated for dual purposes of fiber and oil. Other economic products such as animal feed-stock, industrial solvents and omega 3 fatty acids for human diet supplements are also obtained from flax. Because of these unique attributes, flax is grown in many parts of the world. Globally, top producers of flax/linseed are Canada, India, and China, with bulk of the world production and exports coming from Canada.26,27 In flax growing regions, this crop is vulnerable to drought and high temperature and the effects are pronounced at seedling, flowering and early seed development stages.28 While water deficit has been reported to reduce seed yield in flax toward the end of growing cycle29; irrigation, in dry periods, especially at flowering stage alone has been shown to increase yield.30 Thus, ideal agroclimatic conditions for cultivation of flax involves light precipitation, light cloud cover, and moderate air temperature (18–20◦C).26,31-33 These conditions positively influence optimal plant growth and development of proper anatomical structures of the stem that ensure the quantity and high quality of fiber and seed yield. Currently, in many regions of the world, one of the main factors limiting yield of flax is physiological drought.34,35 This situation, predicted to be the result of global climatic changes,36 significantly affects India, which ranks 2nd in global flax production. In central India, flax is extensively cultivated but relies on rain-fed and irrigated conditions. However, due to limited water supply the yields have been very low for flax in this region. To address this challenge, development of improved drought tolerant flax cultivars are required.
Because of its global importance in recent years, drought research has received significant attention using both model and crop species. Conventional approaches along with more recent genomic technology based tools are driving the advances in this field. While priority crops like corn,37 soybean,38 rice,39 wheat,40 and canola41,42 along with Arabidopsis model system43-46 have witnessed significant recent advances in drought research including the application of molecular breeding tools in new varietal development; so far very limited progress has been made in the flax crop. To address this gap, in the present study, we performed genome-wide gene expression analysis to identify genetic programs associated with drought in flax. Our study identified a large set of differentially expressed genes with implications to drought response and tolerance. These findings will contribute to advancing the basic understanding of drought in flax with potential value in breeding and development of new cultivars with improved tolerance to drought.
Results
Drought stress phenotypes in flax
Compared with other crops, flax is relatively more susceptible to drought. In the present study, we used “Flax variety T-397” that is known to be moderately tolerant to drought stress. First, we assessed the growth of this line under induced drought and controlled conditions in green-house. The seedlings were grown in nine pots (30 seedlings/pot) representing three treatments each of which was replicated three times. Two treatments were performed with seedling sets for four (4d) and five day (5d) stress and the third set of seedlings was used as control with no treatment. The control pots were irrigated from the bottom everyday while drought stressed pots were not irrigated and monitored for wilting symptoms. While there was no wilting in control plants during the entire period (Fig. 1A), 19 out of 30 seedlings (63%) showed wilting on 4th day and 24 seedlings (80%) showed aggravated form of wilting on the 5th day with drought stress (Fig. 1B and C). Relative water content (RWC) of wilted shoots as a measure of drought stress was 70% in 4d stress plants while by the end of 5d, RWC of leaves reduced to 60%. Further, on 6th and 7th day, severity of drought symptoms increased (data not shown) and these stressed plants on 8th day displayed complete wilting (Fig. 1D). Upon re-watering 100% recovery was obtained in 4d stress plants while only 50% showed recovery in 5d stress plants whereas plants exposed to 6 d stress did not revive, likely due to passing the critical recovery point. These observations suggest that at 4d and 5d the plants display both recovery and affected phenotypes. To capture the underlying gene expression programs, we selected these two drought treatment time points, along with corresponding controls, and isolated total RNAs from shoot and root tissues (Fig. 1E–H). Overall, the RNA yields from shoots were three times higher than the root samples and all samples were of good (A 280/ A 260 > 1.7) quality.
Figure 1. Drought stress phenotypes. (A) Control plants during the stress treatment period. (B) Four day stressed plants. (C) Five day stressed plants. (D) Eight day stressed plants. RNA from 4d stressed (E) Shoot and (F) Root and 5d stressed (G) Shoot and (H) Root. R1, R2, R3 represent three replications while C represents control for each treatment.
Gene expression in normal vs drought stress conditions
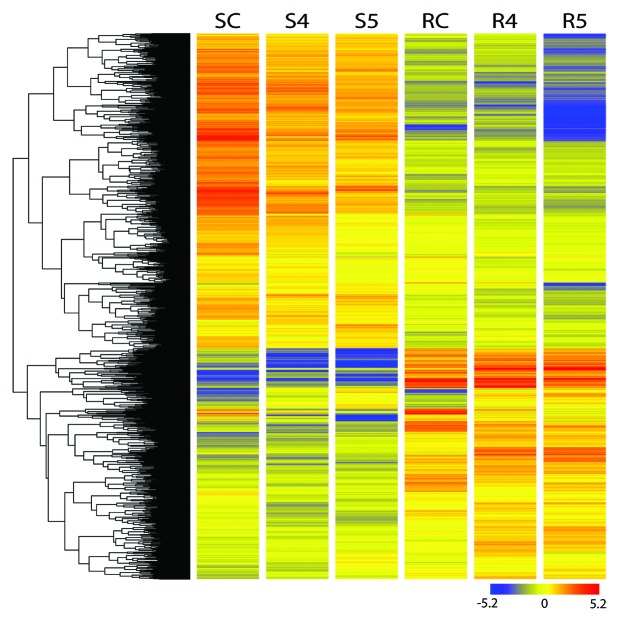
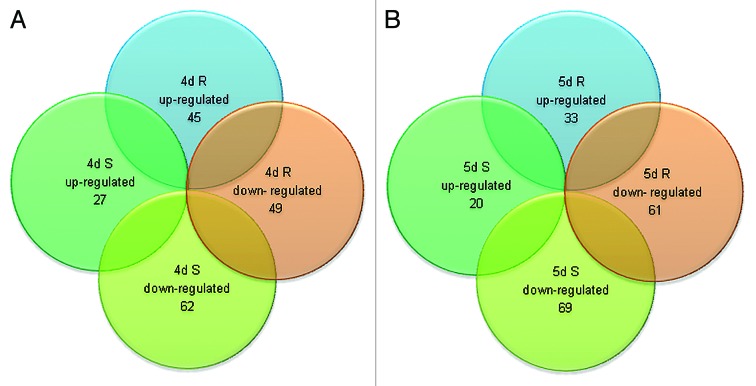
To obtain global gene expression profiles, we used a custom designed flax microarray based on the CombiMatrix system. This array contained probes for 48 904 predicted genes in flax.47 Equal amounts of RNA from flax root and shoot samples representing RWC 70% and 60% were used for microarray experiments. Analysis of results from these experiments identified genes that were differentially regulated by drought stress by comparing gene expression in two different tissues (root and shoot) over two time points along with the corresponding controls. The differentially expressed genes are presented in Figure 2 and additional details summarized in Fig. S1A and B and Tables S1 and S2. The results from these experiments showed that drought treatment significantly altered expressions of 183 target genes of which 72 genes displayed increased expression (≥2-fold upregulation) while 111 genes displayed reduced expression (≥2-fold downregulation) at 4d. Out of 72 highly expressed genes, 10 genes were identified with ≥4-fold upregulation. As expected, a high degree of overlap in gene expression profiles was observed between both tissue types and time points (Fig. 3A and B). In short, 27 and 20 genes were upregulated at 4d and 5d respectively while 45 and 33 genes were upregulated in root at these time points (Fig. S1A). Similarly, at 4d and 5d, 62, and 69 genes were downregulated in shoot while 49 and 61 genes were downregulated in root respectively (Fig. S1B).
Figure 2. Differentially expressed genes across two tissues at two time points of drought treatments. SC-Shoot control, S4 and S5 – shoot four day and five day after stress respectively, RC- root control, R4 and R5- root four day and five day after stress respectively.
Figure 3. Analysis of 183 drought stress inducible genes showing differential expression in shoot and root at two time points. (A) Up- or downregulated genes in shoot and root after 4 d drought stress. (B) Up- or downregulated genes in shoot and root after 5 d drought stress.
Our results showed LEA genes coding for late embryogenesis abundant protein LEA5 (7.0 fold increase), dehydrin (6.0 fold increase), brassinosteroid-regulated protein BRU1 precursor (4.6-fold increase), and calmodulin-binding heat-shock protein (2.2-fold increase) genes were highly induced under drought conditions in flax. Multiple genes encoding cytochrome P450 family proteins, six genes encoding serine/threonine protein kinases, and many lipid transfer protein 3 precursor genes were found to be highly induced in both root and shoot. A majority of the genes that were found to be significantly downregulated in drought treated samples correspond to unknown or hypothetical proteins. Among the known genes of this group, AP2/ERF domain-containing transcription factor (6-fold decrease), brassinosteroid insensitive 1-associated receptor kinase 1 (5-fold decrease), and histone h2b (4-fold decrease) were found to be downregulated. Together this microarray study identified differentially regulated genes associated with induced drought in shoot and root tissues of flax. Interestingly these include several gene candidates associated with drought in other plant species. Detection of many putative/predicted genes in flax tissues signifies that root/shoot microarray data are revealing new useful insights in this crop.
Identification of drought induced differentially expressed genes in root and shoot tissues
Since drought has different effects on different tissues of the plant, and the root is the first organ to be affected by soil moisture deficit, we investigated changes in gene expression in root and shoot tissues separately at the two selected drought stress treated time points. The results of our study showed that drought treatment produced some differential expression patterns in root and shoots. Strikingly, the number of genes upregulated in roots was 1.5-fold higher than the shoot (Fig. S2A and B), while the number of downregulated genes (120 ± 10) was comparable. Further, the comparative analysis revealed that the 57 genes were expressed at low level in shoot compared to root (Fig. S3A) mostly belonging to antioxidant and ABP transporter groups. Interestingly, 116 genes belonging to hormone biosynthesis, lipases and dehydratases involved in water homeostasis were highly expressed in shoot compared to root (Fig. S3B). Additionally, Plastocyanin A, Photosystem I reaction center subunit IV, LEA5 were specifically induced in roots and Ferredoxin 3, phospho-2-dehydro-aldolase 1 were specific to shoot. Similarly, microarray studies in Arabidopsis have reported induction of Plastocyanin A during drought and hypoxia stresses (Uniprot KB Gene investigator - P11490; https://www.genevestigator.com/gv/plant.jsp) and in our study we too found them to be downregulated (2–3-fold decrease). Since their exact role during drought in root tissue is still unknown, it is possible that these genes may be involved in the modulation of photosynthesis under drought stress conditions. Additionally, PSI in chloroplast thylakoids is a major site of ROS generation in higher plants. Since, water is in scarcity during drought, photolysis of H2O needs to be downregulated to prevent further generation of ROS. This downregulation of PSI and plastocyanin may be an efficient mechanism to reduce ROS generation and minimize cellular damage. Moreover, several genes corresponding to hypothetical/predicted proteins were also differentially regulated in both the tissues and some of these may be flax specific and were not identified earlier from previous studies in other plant species.
To address the genes that were differentially expressed at the two time points of the treatments in shoot and root, we analyzed the data sets accordingly and identified several gene targets. These include: (1) AP2/ERF domain containing transcription factor was highly downregulated in the shoot tissue at both the time points i.e., 4d (4.0 fold decrease) and 5d (6.9-fold decrease) but at varying level; (2) Lipid transfer protein3 precursor showed 2.8-fold increase in shoot tissue at 4d stress; (3) interestingly, brassinosteroid regulated protein BRU1 precursor was found to be highly expressed (4.7-fold increase) in shoot tissue at 5d stress but was found to be downregulated in shoot tissue of 4d stress compared with unstressed controls. Late embryogenesis abundant protein (LEA5) was found to be significantly upregulated in root tissue of 4d (7.0 fold increase) and 5d (7.2-fold increase) stress; (4) zinc/iron transporter protein (2.4-fold increase) and photosystem I subunit O (5.1 fold decrease) showed extremely low expression in root tissue of 4d and 5d stress respectively.
Drought associated biochemical pathways in flax
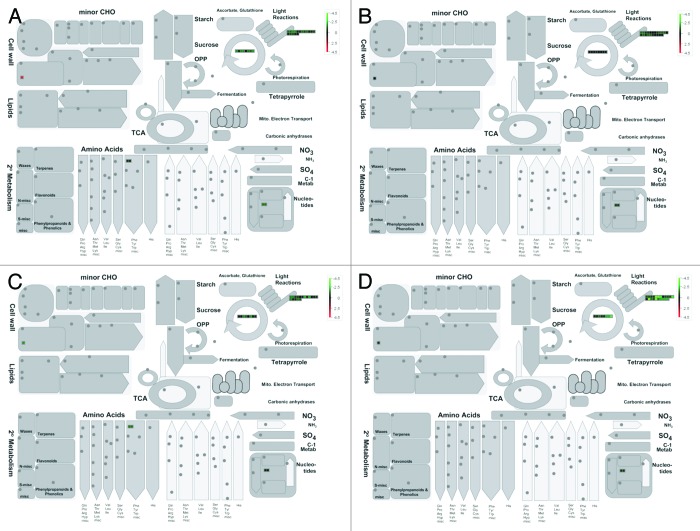
Flax metabolism as affected by drought stress in different tissues was visualized using MapMan software.48 For assigning flax transcripts to MapMan bins, L. usitatissimum_200 Phytozome V9.0 was used for classification. To define the effects of drought stress, temporal (two different time points) and spatial (two different tissues) expression profiles of relevant genes were compared and depicted in MapMan graph (Fig. 4). This provided general view of regulation of major biochemical pathways (Fig. 5A and B) and gene expression patterns (Fig. 5C) involved in drought stress in flax. Using this approach, we analyzed expression patterns of DEGs in root and shoot tissues for specific pathways. For root 85% of DEGs were mapped while 90% DEGs of shoot were mapped into several pathways. Photosystem bin created by MapMan represented the highest number of DEGs (30 in root and 25 in shoot) with most of them being downregulated (Table 1).
Figure 4. Differential expression of genes (log-scale) under drought treatment in two tissues at two different time points involved in flax metabolism grouped into different bins. Green indicates downregulated genes; red indicates upregulated genes. (A) Four day stressed shoot. (B) Four day stressed root. (C) Five day stressed shoot. (D) Five day stressed root.
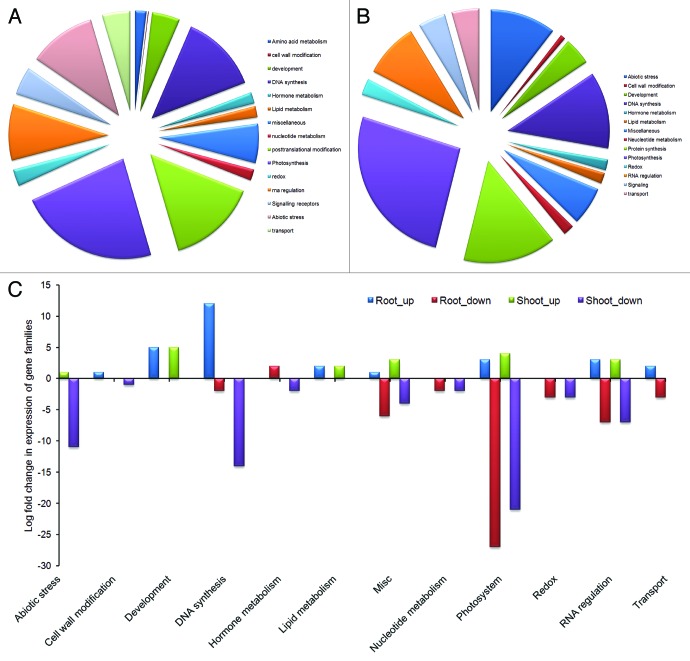
Figure 5. Classification of stress inducible gene families showing differential expression. (A) Families of up- and downregulated DEGs in shoot tissue after four and five day drought. (B) Families of up- and downregulated DEGs in root tissue after four and five day drought. (C) Families of genes showing differential expression in shoot and root with log –fold change values.
Table 1. Up/downregulation of important cellular genes under drought conditions in flax grouped into functional MapMan bins.
| MapMan Categories | Root | Shoot | ||
|---|---|---|---|---|
| Up | down | Up | down | |
| Abiotic stress | 0 | -12 | 1 | -11 |
| Cell wall modification | 1 | 0 | 0 | -1 |
| Development | 5 | 0 | 5 | 0 |
| DNA synthesis | 12 | -2 | 0 | -14 |
| Hormone metabolism | 0 | -2 | 0 | -2 |
| Lipid metabolism | 2 | 0 | 2 | 0 |
| Misc | 1 | -6 | 3 | -4 |
| Nucleotide metabolism | 0 | -2 | 0 | -2 |
| Photosystem | 3 | -27 | 4 | -21 |
| Redox | 0 | -3 | 0 | -3 |
| RNA regulation | 3 | -7 | 3 | -7 |
| Transport | 2 | -3 | 0 | -5 |
As shown in Table 1 under drought stress, downregulation of genes involved in photosynthetic light reactions is maximum (21 genes) followed by DNA synthesis (14 genes) and protein post-translational modification (13 genes). Among the 21 photosynthetic genes, phosphoribulokinase and ferrodoxin/iron-sulfur-cluster binding proteins were significantly downregulated, whereas photosystem bQ and photosystem subunit-O are moderately downregulated; Rubiscoactivase and photosystem subunit were minimally downregulated. With progression of stress, expression of genes such as AP2/ERF, photosystem I reaction center, histone acetyl transferase decreased 2-fold from 4d to 5d. Similarly, genes involved in vital plant processes viz. plastid specific 30S ribosomal protein and elongation factor Tu which were upregulated in 4 d stress exhibited rapid downregulation on 5th day. Reduction of plant recovery from 100% to 50% on fifth day of stress can be attributed to this decrease in expression of genes involved in primary plant functions. Surprisingly, Rubisco small subunit chain-1B and photosystem I subunit D were minimally upregulated. Additionally, five genes involved in plant development (viz. NAC) and three genes for lipid degradation were found to be upregulated. Among other bins that contained highly induced genes, protein post-translational modification bin contained 13 genes, whereas cell wall modification genes are represented by few followed by amino acid metabolism genes. Overall, the drought induced gene expression revealed an important photosystem bin which is likely involved in conferring some tolerance in flax cultivar T-397.
Drought induced co-regulated genes in flax
The analysis of our microarray data showed that there are 22 continuously downregulated (co-down) and four continuously upregulated (co-up) genes present in flax (Table 2). Prominent genes among these are associated with repression of photosynthesis, such as Rubisco, lipid transfer proteins and photosystem I reaction center were co-downregulated. In contrast, two of the genes belong to NAC domain proteins and one each to MYB transcription factor family and protein kinase APK1A were upregulated. Simultaneous up and downregulation of these genes in both the tissues and two time points suggest that several genes are temporally and spatially induced in response to drought stress.
Table 2. Continuously up/downregulated genes in drought stress in flax.
| Group | No. of genes downregulated | co-downregulation [examples] |
No. of genes upregulated | co-upregulation [examples] |
|---|---|---|---|---|
| 4d Leaf vs. Control | 62 | 22 (Ribulose biphosphate carboxylase/ oxygenase activase-2, Lipid transfer protein, Photosystem I reaction center, Elongation factor-TU, Cell wall synthesis genes etc.) |
27 | 4 (NAC-domain containing proteins (2), r2r3-MYB transcription factor (1) and Protein kinase APK1A (1)). |
| 5d Leaf vs. Control | 69 | 20 | ||
| 4d Root vs. Control | 49 | 45 | ||
| 5d Root vs. Control | 61 | 33 |
Validation of microarray data by quantitative qRT-PCR
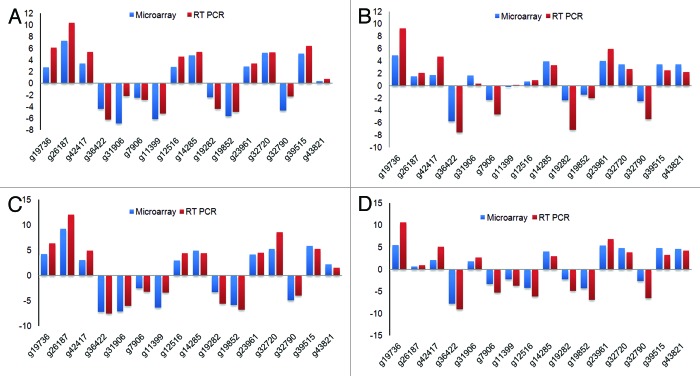
To validate microarray results, we used selected genes and performed qRT-PCR based analysis (Fig. 6). For this we used both up- and downregulated genes from the microarray experiments (Table 3). We selected 16 genes representing both upregulated (dehydrin and pp2c), downregulated (lipid transfer protein3) and few predicted proteins. Actin gene was used as control as its expression did not alter with treatments in our microarray data sets. The qPCR analysis of all the 16 selected gene targets showed similar expression patterns as observed in the microarray results. These confirmatory studies thus further support our microarray results and the derived conclusions.
Figure 6. qRT-PCR analysis of 16 genes compared with the expression levels observed in the microarray analysis. Y-axis values are log2 ratios. (A) log2-fold expression in shoot after four day stress. (B) log2-fold expression in root after four day stress. (C) log2-fold expression in shoot after five day stress. (D) log2-fold expression in root after five day stress.
Table 3. Validation of microarray results by qRT-PCR for 16 selected genes.
| Gene ID | 4d L log (Fc) | 5d L log (Fc) | 4d R log (Fc) | 5d R log (Fc) | Description | ||||
|---|---|---|---|---|---|---|---|---|---|
| Microarray | RT-PCR | Microarray | RT-PCR | Microarray | RT-PCR | Microarray | RT-PCR | ||
| g19736.t1|sl-582–617 | 2.77 | 6.12 | 4.30 | 6.35 | 4.90 | 9.24 | 5.46 | 10.60 | Dehydrin protein |
| g26187.t1|sl-1216–1256 | 7.33 | 10.37 | 9.31 | 12.08 | 1.49 | 2.03 | 0.68 | 0.97 | Cytochrome P450 |
| g42417.t1|sl-359–396 | 3.38 | 5.36 | 3.08 | 4.91 | 1.74 | 4.71 | 2.12 | 5.14 | Protein binding protein, putative |
| g36422.t1|sl-544–580 | –4.39 | –6.21 | –7.17 | –7.55 | –5.75 | –7.57 | –7.83 | –9.02 | PREDICTED: hypothetical protein |
| g31906.t1|sl-837–872 | –6.86 | –2.15 | –7.08 | –6.03 | 1.66 | 0.34 | 1.82 | 2.72 | H0306F03.8 |
| g7906.t1|sl-625–664 | –2.46 | –2.83 | –2.55 | –3.22 | –2.29 | –4.70 | –3.35 | –5.26 | PREDICTED: hypothetical protein |
| g11399.t1|sl-1167–1204 | –6.10 | –5.15 | –6.31 | –3.37 | –0.23 | 0.14 | –2.27 | –3.67 | Monooxygenase, putative |
| g12516.t1|sl-306–341 | 2.85 | 4.58 | 2.96 | 4.47 | 0.69 | 0.86 | –4.18 | –6.15 | Lipid transfer protein 3 precursor |
| g14285.t1|sl-2067–2102 | 4.84 | 5.42 | 4.98 | 4.46 | 3.96 | 3.33 | 4.02 | 2.94 | Unnamed protein product |
| g19282.t1|sl-845–885 | –2.37 | –4.36 | –3.30 | –5.56 | –2.36 | –7.14 | –2.26 | –4.87 | Predicted protein |
| g19852.t1|sl-3275–3312 | –5.58 | –4.87 | –5.79 | –6.71 | –1.49 | –2.04 | –4.33 | –6.95 | PREDICTED: hypothetical protein |
| g23961.t1|sl-1034–1069 | 2.87 | 3.41 | 4.21 | 4.52 | 3.98 | 5.94 | 5.38 | 6.82 | Protein phosphatase 2c, putative |
| g32720.t1|sl-2017–2052 | 5.25 | 5.32 | 5.26 | 8.63 | 3.47 | 2.71 | 4.82 | 3.81 | Unnamed protein product |
| g32790.t1|sl-1040–1077 | –4.66 | –2.25 | –4.87 | –3.93 | –2.53 | –5.41 | –2.63 | –6.56 | Predicted protein |
| g39515.t1|sl-675–710 | 5.12 | 6.38 | 5.86 | 5.28 | 3.44 | 2.48 | 4.78 | 3.28 | Hypothetical protein POPTRDRAFT_783792 |
| g43821.t1|sl-1315–1350 | 0.37 | 0.73 | 2.26 | 1.54 | 3.45 | 2.23 | 4.61 | 4.22 | Amino acid transporter, putative |
Discussion
Yield in temperate crops is predominantly governed by prevailing microclimate and edaphological conditions during their growth period. Nevertheless, abiotic stresses disturb both and are major impediments to achieve optimal yield potential in these crops. Drought is one of the most important yield affecting stresses and occurs when ambient temperature rises and soil moisture falls below field capacity. Synergism of both high temperature and low moisture results in physiological drought that has detrimental effect on yield of flax (fiber as well as seed). While drought during flowering and seed setting (terminal drought) affects seed production, seedling drought affects growth and development leading to reduction in linen production. For survival and productivity, temperate plants such as flax have developed robust molecular and phenotypic plasticity to counter adverse climatic conditions. Several genes inducible under drought stress have been isolated and characterized viz. glycine rich RNA binding protein from apple,49 RING domain containing E3 ubiquitin ligase from rice50 and Novel NAC gene family from leguminous chickpea.51 De novo induction of many cellular pathways and biochemical processes has also been reported to operate during drought stress. While metabolic pathways involved in sugar, amino acid and nitrogen metabolism, alkaloid and flavonoid biosynthesis are differentially regulated, gibberellin catabolism and signaling have been involved in controlling growth and adaptation in response to imminent adverse conditions.52
In the present study, we report a suite of gene regulation and metabolic changes in response to drought stress in flax using microarray based genome-wide gene expression analysis. Since response of plants to drought stress differs between tissues, duration and degree of stress, we performed the experiment in two different tissues, i.e., root and shoot; and at two different time points of four and five days after treatment. The relative water content of 70% and 60% observed in shoot at four and five days are comparable to long-term drought occurring under field conditions of central India and the simulated stress can be classified in between severe seedling drought to moderate terminal drought in long-term field trials. Since yield could not be assessed in a short-term experiment like the present study, we relied on a highly reproducible53 secondary parameter - shoot survival - as an indicator of drought stress. We found six days of stress as break-even point for drought tolerance in flax genotype T-397.
Hydropenia in experimental plants is induced by several methods such as PEG treatment,54,55 air-wilting, drydown and withholding water supply. While, methods such as PEG (induces anaerobic condition in roots), air wilting (induces sudden dehydration in plants in hours) and drying plants (takes too long in seedling to exhibit symptoms) are used by several researchers, we adopted the method of “withholding water”56 that simulated drought as encountered by plants in field condition resulted in diverse response in shoot and root tissues of flax. Altogether 183 genes were differentially expressed. Genes like protein kinase APK1A, leucine-rich repeat receptor, brassinosteroid insensitive1, and cytochrome P450 were differentially regulated in shoot and root (Fig. S3A). Similarly, genes like geranyl-geranyl reductase involved in chlorophyll biosynthesis, chlorophyll A/B binding protein involved in chlorophyll stabilization and trans-ketolase involved in oxidative/reductive pathway were highly expressed in shoot compared to root (Fig. S3B).
Simultaneous comparison of gene expression in different tissues of flax at different time points (Fig. 3A and B) revealed interesting commonalities in transcription responses. While, 90% of genes upregulated in shoot after 4d stress were the same as in root, 93% of the genes downregulated in root were identical to those identified to be downregulated in shoot. Additionally, comparison of the gene expression profiles between 4d and 5d shoot and root tissues revealed that 51% and 82% of genes were commonly upregulated and downregulated in shoot and 48% and 62% were up and downregulated in root respectively. This implies that simultaneous up/downregulation of genes in shoot and root is likely required to coordinate shoot-root ratio and water homeostasis.
Genes inducible under drought stress have been separated into three classes such as functional proteins, regulatory proteins14 and proteins of unknown function. Our microarray data identified genes belonging to all the three classes (Fig. 5A and B). Among the first class, prominent genes we identified belonged to late embryogenesis proteins,57 carbohydrate metabolism,58 amino acid metabolism,59 lipid transfer proteins, photosynthesis and chloroplastic proteins, heat shock proteins,60 and developmental proteins. In the second group, transcription factors like zinc finger protein, HAT, AP2/ERF domain, MYB and NAC; protein kinases like BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, MAPK2, APK1A and APK1B, and signaling protein like Bri1 were identified. We also identified several genes with unassigned/hypothetical functions. Analysis of nine such genes revealed that four (out of nine) genes are highly upregulated while five genes are downregulated by drought stress. It is possible that these predicted proteins are involved in yet to be defined functions associated with drought stress response.
Functional interpretation of genes of Linum usitatissimum using the Arabidopsis genome as reference revealed the gene expression changes in response to drought can be sorted by functional categories into different bins. Genes sorted into different bins were involved in cell wall modification, plant development, DNA/RNA synthesis, protein synthesis, hormone metabolism and photosynthesis. This suggests that core and essential developmental and biochemical processes were significantly affected in flax during drought and there is a massive transition of anabolic metabolism to catabolism. Although, drought induces cellular damage to vital organs such as cell membrane,61 tolerant plants exhibit immediate closure of stomata to avoid loss of water. But this consequently results in decreased photosynthetic efficiency due to reduced CO2 availability. In addition to stomatal closure, we found genes involved in photosynthetic light reaction were severely dampened during drought stress and our results are in agreement with similar findings in rice53,62 and barley.63,64
In root and shoot, five DEGs encoding transcription factors such as NAC012, NAC002, and NAC043 involved in developmental process65 were upregulated while two of them are continuously upregulated (co-up) (Table 2). NAC transcription factors are reported to impart drought stress tolerance by triggering a cascade of signaling pathways. In the signaling sub-group, out of five DEGs, we found two were upregulated in shoot while all were upregulated in root. Two of the DEGs were leucine-rich repeat trans-membrane protein kinases having higher expression level in root compared to shoot.
Post-translational protein modifications such as phosphorylation and dephosphorylation are important attributes that govern protein stability and function leading to plant growth and development, and modulation of ABA signaling by post translational modification.66-68 In flax, genes responsible for hormone biosynthesis were upregulated in 4d treatment. After drought treatment, genes involved in abiotic stress such as cell wall modification, photosynthesis, and transport were downregulated suggesting these genes and the associated processes influence the drought response and tolerance in flax. These are primary plant processes and are severely affected by water stress. Concomitant downregulation of genes involved in these cardinal processes exert a cumulative effect on plant response to drought.
It is reported that tolerant varieties display more changes in gene expression patterns than sensitive varieties69 to a particular stress. In susceptible varieties most of the affected genes are associated with stress induced damage and damage related responses70,71 such as upregulated enzymes for degradation of lipids and proteins. Most genes that were induced by drought stress in sensitive but not in tolerant cultivars are related to senescence rather than to stress tolerance mechanisms. Interestingly, we found few genes involved in lipid metabolism, functional proteins, transcription factors etc. were upregulated in T-397 suggesting that expression of these genes likely confer some tolerance to this flax genotype. This possibility is consistent with the observation that drought did not evoke damage-related responses in T-397 usually found in a susceptible cultivars. Similar observations were reported in rice where, a massive change in gene expression was observed in a sensitive cultivar compared with a tolerant cultivar.72,73 The downregulation of several genes involved in photosynthetic process suggests that it may be a regulatory response to limit further damage.
Drought and photosynthesis show a high level of connectivity where drought is the cause and reduction in photosynthesis is the effect. Our experiment indicates reduction of photosynthesis is an adaptive response rather than a regulatory mechanism to prevent photo-damage. Decreased CO2 diffusion from the atmosphere to the site of carboxylation is generally considered the main cause for decreased photosynthesis under mild to moderate water limitation. Decreased stomatal conductance coupled with sustained high irradiance of light is an effective defense mechanism in C3 plants to reduce generation of reactive oxygen species (ROS) that damages ATP synthase and Rubisco activase to slow down photosynthesis.74 Additionally, at molecular level, several TFs belonging to MYB family are involved in regulation of stomata number and size, and of metabolic components of the photosynthetic system to limit photosynthesis.75 Downregulation of photosynthetic genes in T-397 indicates an adaptive response to prevent photo-damage during times of reduced CO2 availability in the mesophyll when stomata are closed due to water shortage. Together, these factors likely contribute to minimal negative effect on photosynthesis and to the plant in the case of flax T-397 genotype.
Our result is the first report of global analysis of transcriptome associated with drought stress in flax. To validate these microarray based gene expression profiles, we performed real-time PCR with 16 selected differentially expressed gene targets. The expression results obtained by real-time PCR for all these genes viz., dehydrin, cytochome P450, lipid transfer proteins, amino acid transporters, and protein phosphatase 2c were in agreement with those obtained by microarray experiments and earlier reports. For example, dehydrin belonging to a class of late embryogenesis proteins were most abundant during water stress.76 Similarly, cytochrome P450 involved in the synthesis of fatty acids, secondary metabolites, suberin and protective tissue like cutin, may be involved in protecting water loss by forming a protective covering and these have been previously shown to be induced during drought.77 Lipid transfer proteins involved in the deposition of cuticular wax that are important in drought tolerance78 are also found to be upregulated in our study. It has been reported that mono-oxygenases are highly upregulated79,80 during drought stress, but our data for the expression of mono-oxygenase is different and shows that this gene is downregulated in both shoot and root. Since T-397 is moderately tolerant to drought, raising the possibility that other genes except mono-oxygenases are involved to impart drought tolerant attributes.
While most drought-related studies focused on genes that are highly upregulated, there are genes whose downregulation is equally important to sustain the plant life during stress. Therefore, profiling and analysis of downregulated genes is equally important to understand the molecular and biochemical basis of their functions under stress conditions. In our study, we regard those genes downregulated whose expression level is 2-fold less than the unstressed control tissue. Mostly, genes involved in photosynthesis like photosystem I reaction center subunit VI, oxygen-evolving enhancer protein 3–1; and chloroplast proteins like thylakoid membrane phosphoprotein 14 kDa, and chloroplast precursor were downregulated. This is consistent with earlier reports of drought-induced responses of photosynthesis and metabolism in higher plants.81,82
In conclusion, we have analyzed gene expression profiles associated with drought stress in flax, and identified 183 differentially expressed genes in shoot and root. These differentially regulated genes belong to 12 diverse functional categories and some of these display coordinated expression under stress conditions suggesting functional importance to drought responses in flax. Although, we did not see visible bleaching in drought stressed shoots, our findings suggest that photosynthetic activity is one of the main regulatory mechanisms affected by drought. Future studies could potentially use key findings of this study to advance critical knowledge base and also contribute to the development of drought tolerant flax cultivars.
Materials and Methods
Stress treatment
Seeds of flax variety (T-397) were obtained from project coordinating unit (Linseed), Kanpur, India. Seeds were sown in 10cm x 10cm plastic pots filled with soaked 1:1:1 mixture of Agro-coir peat, vermiculite and river sand and grown at a 12h photoperiod. The temperature regime was 24 °C /18 °C (day/night) and 75% relative humidity. Plants were grown by watering to field capacity every day till 20 d after germination, during this period plants attained 10–15cm height. Drought stress was standardized by with-holding irrigation over a period of 8 d from 21st to 28th day after germination; during which drought symptoms were monitored, visualized and recorded (data not shown). Initial symptoms were observed beginning 4th day after withholding irrigation and relative water content (RWC) of shoot was approx 70%. By end of 5th day RWC was 60% and plants showed distinct wilting symptoms. Thus, root and shoot samples were collected on 4th (4d) and 5th (5d) day. Further, on 6th, 7th, and 8th day intensity of drought symptoms increased. Plants exposed to 6th day stress partially revived after irrigation and were assumed to cross break-even point while drought stressed 8th day plants completely wilted. Control flax plants were grown normally by irrigating soil to field capacity and samples were collected on 4d and 5d along with stressed plants (Fig. 1). RWC of shoot was estimated using 100 mg tissue of flax following the method of Weatherly and Barr.83 After taking fresh weight, the tissue was floated on water for 4 h to achieve full turgidity and was weighed again to estimate turgid weight. The tissue was then oven-dried at 80 °C for 24 h and the dry weight was estimated. Relative water content was measured and expressed as percentage according to following equation:
RWC (%) = (Fresh weight – Dry weight) / Fresh weight x 100
RNA isolation
Total RNA was isolated by grinding 200 mg shoot and 400 mg root tissue of control and drought stressed samples (4th and 5th day) in liquid nitrogen using SpectrumTM plant total RNA kit (Sigma-Aldrich co.USA; Cat No: STRN250) according to manufacturer’s instructions with addition of an on-column DNase1 treatment (1:4 dilution of DNase 1 in DNase digestion buffer). Total RNA was checked for quality and quantity using a NanoDrop 1000 spectrophotometer (Thermo Scientific) and by denaturing agarose gel electrophoresis. RNA amplifications were performed with the MessageAmp™ aRNA Amplification Kit (Ambion, Cat. No.1750). Cy5 dye molecules (GE Healthcare) were coupled to the amplified RNA, and the dye-labeled RNA was fragmented before hybridization.
COMBIMATRIX Array
CombiMatrix 90K Array system was used in the design and synthesis of a set of 90K unique 35-mer flax oligo probes along with blanks and negative controls on re-useable slides. Probes were designed for 48 904 predicted genes (41 536 of the genes with 2 probes; 7368 of the genes with 1 probe; 90 controls) using the EST and the recent flax genome sequence information.47,84 The quality testing of these slides with cy5 labeled random 9 mers showed uniform spot morphology and efficient, consistent synthesis of probes across the slide.
Array hybridization, scanning, and image analysis
The high-density CombiMatrix 90K Flax oligonucleotide array was produced by the Plant Biotechnology Institute. A total of 90 527 probes (35–40 mer) were in situ synthesized using the CustomArray Synthesizer (CombiMatrix). Array hybridization, stripping and re-hybridization were performed following the CustomArray™ 90K Microarray: Hybridization, imaging protocol, and stripping and preparation of CombiMatrix 90K microarrays for re-hybridization protocols, as recommended by CombiMatrix (http://www.combimatrix.com). The arrays were first rehydrated at 65 °C for 10 min then prehybridized at 45 °C for 30 min before use. Fragmented Cy5-aRNAs (5 μg) were mixed with hybridization buffer (6 × SSPE, 0.05% Tween-20, 20 mM EDTA, 25% deionized formamide, 0.1 mg/mL sheared salmon sperm DNA and 0.04% SDS) and hybridized to the Combimatrix Brassica 90K arrays at 45 °C overnight in the dark. After hybridization, stringent washes were performed according to the manufacturer’s instructions, and the slides were immediately coated with an imaging solution (CombiMatrix) prior to scanning with a GenePix 4000B scanner (Axon Instruments) using 450 PMT, 5 μm resolution and 100% laser power. After scanning, the hybridized dye-labeled aRNA targets were stripped from the arrays using the CustomArray Stripping kit (CombiMatrix) and re-hybridized. Altogether, a single slide was used thrice for hybridization - de novo for the first time and twice by repeating the stripping followed by re-hybridization. In the present study for the 24 samples (12 controls and 12 drought stress treated plants) represented by 3 biological samples for each time point (4th and 5th day), tissue (root and shoot) for the control, and drought stress induced plants, 8 flax 90K CombiMatrix slides were used. After each stripping we ensured, complete removal of Cy5 signals from previous hybridization before rehybridization.
Image analysis was performed using GenePix Pro version 6.0 software (Axon Instruments). After automatic alignment, feature indicators were manually moved, resized and fine-tuned. The local background-corrected spot fluorescence intensities for the Cy5 channel were saved as GPR files.
Data processing and segregation into MapMan bins
GPR files were loaded in GeneSpring GX 12.6 (Agilent Technologies Inc. and Strand Life Sciences Pvt. Ltd) with percentile shift normalization and baseline to median of all samples. Signal intensity cut-off was set to 200. One-way ANOVA analysis was performed with the asymptotic P value computation method. Multiple testing correction method was performed using the Benjamini–Hochberg FDR.85 Only gene probes having an adjusted P value (FDR) < 0.05 and an absolute difference in expression fold-change higher than 2 were selected. Hierarchical clustering based on Euclidean distance was utilized to group gene probes in categories according to their expression profiles.
MapMan annotation of genomic positions of root and shoot transcripts of flax was determined by aligning transcripts against MapMan (ver.3.6.0RC1).The best hit was extracted. Genes whose array annotation and annotation of the best blast hit were identical were put in the MapMan bin of the blast hit.
qRT-PCR analysis
Microarray expression data were validated using real-time polymerase chain reactions in an optical 48-well plate with a StepOne™ Real-Time PCR System (Applied Biosystems), using SYBR® Green to monitor dsDNA synthesis. Reactions contained 5 µl 2 × SYBR® Green Master Mix reagent (Applied Biosystems), 1.0 ng cDNA and 200 nm of each gene-specific primer (Table S3) in a final volume of 10 µl. The following standard thermal profile was used for all PCRs: 50 °C for 2 min; 95 °C for 10 min; 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Actin 7 (g45138F, 5′-TTGCTGACCG TATGAGCAAG-3′; g45138R, 5′-ACCCTCCAAT CCAGACACTG-3′) gene was used as an endogenous reference. Data were analyzed using the StepOne™ Real-Time PCR System version 1.0 (Applied Biosystems).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We acknowledge ICAR project coordinating unit (linseed), Kanpur, for providing T-397 seeds for this work. This work was financially supported in the form of in-house project (RPP) to P.K.D. (Sanction no 7–5/2010-IC-IV) by Indian Council of Agricultural Research (ICAR), New Delhi, India. The work performed at National Research Council (R.D. and G.S.) was supported by Saskatchewan Agriculture Development Fund, Genome Canada and Genome Prairie TUFGEN projects. M.D. is supported by Genome Canada TUFGEN project. This is NRC publication number 56009.
Author Contribution
Concept, design, execution and coordination (P.K.D., P.A.K., R.D.); seed material procurement and multiplication (P.A.K., D.K.Y., P.K.D., P.G.); tissue sampling and RNA isolation (P.K.D., A.K.J., P.G.); Array experiments (Y.C., P.V., D.X., R.D.); interpretation of results and discussion (P.K.D., R.R., Y.C., P.G., P.V., R.D., R.S., N.T., M.Z.A., N.K.S.); new reagents, kits and arrays (P.A.K., M.D., J.S., G.S., R.D.); P.K.D., P.G., R.R., P.V., and R.D. wrote manuscript with input from Y.C., R.S., N.T., M.Z.A., M.D., J.S., N.K.S., and P.A.K..
Supplemental Material
Supplemental Material may be downloaded here: publisher's website
References
- 1.Begcy K, Mariano ED, Gentile A, Lembke CG, Zingaretti SM, Souza GM, Menossi M. . A novel stress-induced sugarcane gene confers tolerance to drought, salt and oxidative stress in transgenic tobacco plants. PLoS One 2012; 7:e44697; http://dx.doi.org/ 10.1371/journal.pone.0044697; PMID: 22984543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer JS. . Plant productivity and environment. Science 1982; 218:443 - 8; http://dx.doi.org/ 10.1126/science.218.4571.443; PMID: 17808529 [DOI] [PubMed] [Google Scholar]
- 3.Maybank J, Bonsai B, Jones K, Lawford R, O'brien E, Ripley E, Wheaton E. . Drought as a natural disaster. Atmos-ocean 1995; 33:195 - 222; http://dx.doi.org/ 10.1080/07055900.1995.9649532 [DOI] [Google Scholar]
- 4.Solari LI, Johnson S, DeJong TM. . Relationship of water status to vegetative growth and leaf gas exchange of peach (Prunus persica) trees on different rootstocks. Tree Physiol 2006; 26:1333 - 41; http://dx.doi.org/ 10.1093/treephys/26.10.1333; PMID: 16815835 [DOI] [PubMed] [Google Scholar]
- 5.Zhu JK. . Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 2002; 53:247 - 73; http://dx.doi.org/ 10.1146/annurev.arplant.53.091401.143329; PMID: 12221975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Affenzeller MJ, Darehshouri A, Andosch A, Lütz C, Lütz-Meindl U. . Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J Exp Bot 2009; 60:939 - 54; http://dx.doi.org/ 10.1093/jxb/ern348; PMID: 19213813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner NC, Wright GC, Siddique K. . Adaptation of grain legumes (pulses) to water-limited environments. Adv Agron 2001; 71:193 - 231; http://dx.doi.org/ 10.1016/S0065-2113(01)71015-2 [DOI] [Google Scholar]
- 8.Nguyen HT, Babu RC, Blum A. . Breeding for drought resistance in rice: physiology and molecular genetics considerations. Crop Sci 1997; 37:1426 - 34; http://dx.doi.org/ 10.2135/cropsci1997.0011183X003700050002x [DOI] [Google Scholar]
- 9.Lipiec J, Doussan C, Nosalewicz A, Kondracka K. . Effect of drought and heat stresses on plant growth and yield: a review. International Agrophysics 2013; 27:463 - 77; http://dx.doi.org/ 10.2478/intag-2013-0017 [DOI] [Google Scholar]
- 10.Blum A. . Drought resistance, water-use efficiency, and yield potential—are they compatible, dissonant, or mutually exclusive?. Crop Pasture Sci 2005; 56:1159 - 68; http://dx.doi.org/ 10.1071/AR05069 [DOI] [Google Scholar]
- 11.Vadez V, Kholova J, Choudhary S, Zindy P, Terrier M, Krishnamurthy L, Kumar PR. NC T. Responses to increased moisture stress and extremes: Whole plant response to drought under climate change. In: S.S. Yadav, R. Redden, J.L. Hatfield, H. Lotze-Campen, Hall AE, eds. Crop adaptation to climate change. Chichester: Wiley-Blackwell, 2011. [Google Scholar]
- 12.Vinocur B, Altman A. . Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol 2005; 16:123 - 32; http://dx.doi.org/ 10.1016/j.copbio.2005.02.001; PMID: 15831376 [DOI] [PubMed] [Google Scholar]
- 13.Wang W, Vinocur B, Altman A. . Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 2003; 218:1 - 14; http://dx.doi.org/ 10.1007/s00425-003-1105-5; PMID: 14513379 [DOI] [PubMed] [Google Scholar]
- 14.Shinozaki K, Yamaguchi-Shinozaki K, Seki M. . Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 2003; 6:410 - 7; http://dx.doi.org/ 10.1016/S1369-5266(03)00092-X; PMID: 12972040 [DOI] [PubMed] [Google Scholar]
- 15.Wani SH, Singh NB, Haribhushan A, Mir JI. . Compatible solute engineering in plants for abiotic stress tolerance - role of glycine betaine. Curr Genomics 2013; 14:157 - 65; http://dx.doi.org/ 10.2174/1389202911314030001; PMID: 24179438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neumann PM. . Coping mechanisms for crop plants in drought-prone environments. Ann Bot 2008; 101:901 - 7; http://dx.doi.org/ 10.1093/aob/mcn018; PMID: 18252764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. . Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol 2006; 17:113 - 22; http://dx.doi.org/ 10.1016/j.copbio.2006.02.002; PMID: 16495045 [DOI] [PubMed] [Google Scholar]
- 18.Mao X, Jia D, Li A, Zhang H, Tian S, Zhang X, Jia J, Jing R. . Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct Integr Genomics 2011; 11:445 - 65; http://dx.doi.org/ 10.1007/s10142-011-0218-3; PMID: 21472467 [DOI] [PubMed] [Google Scholar]
- 19.Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R. . TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot 2012; 63:2933 - 46; http://dx.doi.org/ 10.1093/jxb/err462; PMID: 22330896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, et al. . The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 2006; 60:107 - 24; http://dx.doi.org/ 10.1007/s11103-005-2910-y; PMID: 16463103 [DOI] [PubMed] [Google Scholar]
- 21.Mukhopadhyay A, Vij S, Tyagi AK. . Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci U S A 2004; 101:6309 - 14; http://dx.doi.org/ 10.1073/pnas.0401572101; PMID: 15079051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen H, Liu C, Zhang Y, Meng X, Zhou X, Chu C, Wang X. . OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol Biol 2012; 80:241 - 53; http://dx.doi.org/ 10.1007/s11103-012-9941-y; PMID: 22875749 [DOI] [PubMed] [Google Scholar]
- 23.Georges F, Das S, Ray H, Bock C, Nokhrina K, Kolla VA, Keller W. . Over-expression of Brassica napus phosphatidylinositol-phospholipase C2 in canola induces significant changes in gene expression and phytohormone distribution patterns, enhances drought tolerance and promotes early flowering and maturation. Plant Cell Environ 2009; 32:1664 - 81; http://dx.doi.org/ 10.1111/j.1365-3040.2009.02027.x; PMID: 19671099 [DOI] [PubMed] [Google Scholar]
- 24.Xiong L, Schumaker KS, Zhu JK. . Cell signaling during cold, drought, and salt stress. Plant Cell 2002; 14:Suppl S165 - 83; PMID: 12045276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marshall A, Aalen RB, Audenaert D, Beeckman T, Broadley MR, Butenko MA, Caño-Delgado AI, de Vries S, Dresselhaus T, Felix G, et al. . Tackling drought stress: receptor-like kinases present new approaches. Plant Cell 2012; 24:2262 - 78; http://dx.doi.org/ 10.1105/tpc.112.096677; PMID: 22693282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heller K, Woznica Z, Byczynska M, Wielgusz K, Baraniecki P. . The Efficacy of Salicylic and Acetylsalicylic Acids in the Protection of Fibrous Flax Against Drought Stress. J Nat Fibers 2013; 10:29 - 39; http://dx.doi.org/ 10.1080/15440478.2012.748298 [DOI] [Google Scholar]
- 27.FAO. FAO Statistical Yearbook 2009. http://wwwfaoorg/economic/ess/ess-publications/ess-yearbook/en/#UuTH_BC6bIU 2009.
- 28.Martin JH, Leonard WH. D.L. S. Principles of field crop production. N.Y.: Macmillan, 1976. [Google Scholar]
- 29.Casa R, Russell G, Lo Cascio B, Rossini F. . Environmental effects on linseed (Linum usitatissimum L.) yield and growth of flax at different stand densities. Eur J Agron 1999; 11:267 - 78; http://dx.doi.org/ 10.1016/S1161-0301(99)00037-4 [DOI] [Google Scholar]
- 30.Dutta HK, Rao RM. . H S. Response of linseed (Linum usitatissimum L.) to irrigation and nitrogen. Indian J Agron 1995; 40:130 - 1 [Google Scholar]
- 31.Heller K, Konczewicz W, Byczyńska M, Łukaszewska N, Praczyk M. The effect of fibre flax growing technologies on ontogenesis and cultivars yielding capacity. Conference on Flax and other Bast Plants 2008:315-25
- 32.Rolski S, Heller K. . Yielding capacity of different flax cultivars in varied environmental condition. Natural fibres 1998; 2:84 - 8 [Google Scholar]
- 33.Xinwen L. Analysis of ecological adaptation of flax in dry and cool areas in China. Proc of the Flax & Other Bast Plants Symposium 1997:43-8.
- 34.Widstoe JA. Dry farming for sustainable agriculture. Agrobios (India) 2007:361. [Google Scholar]
- 35.Gupta US. Physiology of stressed crops. Georgia, USA: Science Publishers, 2007. [Google Scholar]
- 36.IPCC. Contribution of working groups I, II and III to the fourth assessment report of the intergovernmental panel on climate change core writing team Geneva, Switzerland, 2007. [Google Scholar]
- 37.Hayano-Kanashiro C, Calderón-Vázquez C, Ibarra-Laclette E, Herrera-Estrella L, Simpson J. . Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS One 2009; 4:e7531; http://dx.doi.org/ 10.1371/journal.pone.0007531; PMID: 19888455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira SS, Guimarães FC, Carvalho JF, Stolf-Moreira R, Oliveira MC, Rolla AA, Farias JR, Neumaier N, Nepomuceno AL. . Transcription factors expressed in soybean roots under drought stress. Genet Mol Res 2011; 10:3689 - 701; http://dx.doi.org/ 10.4238/2011.October.21.5; PMID: 22033904 [DOI] [PubMed] [Google Scholar]
- 39.Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. . Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 2003; 133:1755 - 67; http://dx.doi.org/ 10.1104/pp.103.025742; PMID: 14645724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aprile A, Mastrangelo AM, De Leonardis AM, Galiba G, Roncaglia E, Ferrari F, De Bellis L, Turchi L, Giuliano G, Cattivelli L. . Transcriptional profiling in response to terminal drought stress reveals differential responses along the wheat genome. BMC Genomics 2009; 10:279; http://dx.doi.org/ 10.1186/1471-2164-10-279; PMID: 19552804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu S, Bahn SC, Qu G, Qin H, Hong Y, Xu Q, Zhou Y, Hong Y, Wang X. . Increased expression of phospholipase Dα1 in guard cells decreases water loss with improved seed production under drought in Brassica napus. Plant Biotechnol J 2013; 11:380 - 9; http://dx.doi.org/ 10.1111/pbi.12028; PMID: 23279050 [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Yang B, Liu WZ, Li H, Wang L, Wang B, Deng M, Liang W, Deyholos MK, Jiang YQ. . Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol 2014; 14:8; http://dx.doi.org/ 10.1186/1471-2229-14-8; PMID: 24397480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dean G, Cao Y, Xiang D, Provart NJ, Ramsay L, Ahad A, White R, Selvaraj G, Datla R, Haughn G. . Analysis of gene expression patterns during seed coat development in Arabidopsis. Mol Plant 2011; 4:1074 - 91; http://dx.doi.org/ 10.1093/mp/ssr040; PMID: 21653281 [DOI] [PubMed] [Google Scholar]
- 44.Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. . Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 2001; 13:61 - 72; http://dx.doi.org/ 10.1105/tpc.13.1.61; PMID: 11158529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu ZH, Wu WR. . Comparative and joint analyses of gene expression profiles under drought and rewatering in Arabidopsis. Genet Mol Res 2013; 12:3622 - 9; http://dx.doi.org/ 10.4238/2013.September.13.6; PMID: 24085426 [DOI] [PubMed] [Google Scholar]
- 46.Huang D, Wu W, Abrams SR, Cutler AJ. . The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J Exp Bot 2008; 59:2991 - 3007; http://dx.doi.org/ 10.1093/jxb/ern155; PMID: 18552355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Hobson N, Galindo L, Zhu S, Shi D, McDill J, Yang L, Hawkins S, Neutelings G, Datla R, et al. . The genome of flax (Linum usitatissimum) assembled de novo from short shotgun sequence reads. Plant J 2012; 72:461 - 73; http://dx.doi.org/ 10.1111/j.1365-313X.2012.05093.x; PMID: 22757964 [DOI] [PubMed] [Google Scholar]
- 48.Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. . MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 2004; 37:914 - 39; http://dx.doi.org/ 10.1111/j.1365-313X.2004.02016.x; PMID: 14996223 [DOI] [PubMed] [Google Scholar]
- 49.Wang S, Liang D, Shi S, Ma F, Shu H, Wang R. . Isolation and Characterization of a Novel Drought Responsive Gene Encoding a Glycine-rich RNA-binding Protein in Malus prunifolia (Willd.) Borkh. Plant Mol Biol Rep 2011; 29:125 - 34; http://dx.doi.org/ 10.1007/s11105-010-0221-1 [DOI] [Google Scholar]
- 50.Bae H, Kim SK, Cho SK, Kang BG, Kim WT. . Overexpression of OsRDCP1, a rice RING domain-containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.). Plant Sci 2011; 180:775 - 82; http://dx.doi.org/ 10.1016/j.plantsci.2011.02.008; PMID: 21497713 [DOI] [PubMed] [Google Scholar]
- 51.Peng H, Yu X, Cheng H, Shi Q, Zhang H, Li J, Ma H. . Cloning and characterization of a novel NAC family gene CarNAC1 from chickpea (Cicer arietinum L.). Mol Biotechnol 2010; 44:30 - 40; http://dx.doi.org/ 10.1007/s12033-009-9202-8; PMID: 19669952 [DOI] [PubMed] [Google Scholar]
- 52.Zawaski C, Busov VB. . Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short-day photoperiods in Populus trees. PLoS One 2014; 9:e86217; http://dx.doi.org/ 10.1371/journal.pone.0086217; PMID: 24465967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Degenkolbe T, Do PT, Zuther E, Repsilber D, Walther D, Hincha DK, Köhl KI. . Expression profiling of rice cultivars differing in their tolerance to long-term drought stress. Plant Mol Biol 2009; 69:133 - 53; http://dx.doi.org/ 10.1007/s11103-008-9412-7; PMID: 18931976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lata C, Bhutty S, Bahadur RP, Majee M, Prasad M. . Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setaria italica (L.)]. [Setaria italica (L.)] J Exp Bot 2011; 62:3387 - 401; http://dx.doi.org/ 10.1093/jxb/err016; PMID: 21414959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perdiguero P, Collada C, Barbero MdelC, García Casado G, Cervera MT, Soto A. . Identification of water stress genes in Pinus pinaster Ait. by controlled progressive stress and suppression-subtractive hybridization. Plant Physiol Biochem 2012; 50:44 - 53; http://dx.doi.org/ 10.1016/j.plaphy.2011.09.022; PMID: 22099518 [DOI] [PubMed] [Google Scholar]
- 56.Yao LM, Wang B, Cheng LJ, Wu TL. . Identification of key drought stress-related genes in the hyacinth bean. PLoS One 2013; 8:e58108; http://dx.doi.org/ 10.1371/journal.pone.0058108; PMID: 23472143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hundertmark M, Hincha DK. . LEA (late embryogenesis abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 2008; 9:118; http://dx.doi.org/ 10.1186/1471-2164-9-118; PMID: 18318901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kerepesi I, Galiba G. . Osmotic and Salt Stress-Induced Alteration in Soluble Carbohydrate Content in Wheat Seedlings. Crop Sci 2000; 40:482 - 7; http://dx.doi.org/ 10.2135/cropsci2000.402482x [DOI] [Google Scholar]
- 59.Serraj R, Sinclair TR. . Osmolyte accumulation: can it really help increase crop yield under drought conditions?. Plant Cell Environ 2002; 25:333 - 41; http://dx.doi.org/ 10.1046/j.1365-3040.2002.00754.x; PMID: 11841674 [DOI] [PubMed] [Google Scholar]
- 60.Rizhsky L, Liang H, Mittler R. . The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 2002; 130:1143 - 51; http://dx.doi.org/ 10.1104/pp.006858; PMID: 12427981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkinson S, Davies WJ. . Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ 2010; 33:510 - 25; http://dx.doi.org/ 10.1111/j.1365-3040.2009.02052.x; PMID: 19843256 [DOI] [PubMed] [Google Scholar]
- 62.Hazen SP, Pathan MS, Sanchez A, Baxter I, Dunn M, Estes B, Chang H-S, Zhu T, Kreps JA, Nguyen HT. . Expression profiling of rice segregating for drought tolerance QTLs using a rice genome array. Funct Integr Genomics 2005; 5:104 - 16; http://dx.doi.org/ 10.1007/s10142-004-0126-x; PMID: 15480887 [DOI] [PubMed] [Google Scholar]
- 63.Oztur ZN, Talamé V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ. . Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol 2002; 48:551 - 73; http://dx.doi.org/ 10.1023/A:1014875215580; PMID: 11999834 [DOI] [PubMed] [Google Scholar]
- 64.Talamè V, Ozturk NZ, Bohnert HJ, Tuberosa R. . Barley transcript profiles under dehydration shock and drought stress treatments: a comparative analysis. J Exp Bot 2007; 58:229 - 40; http://dx.doi.org/ 10.1093/jxb/erl163; PMID: 17110587 [DOI] [PubMed] [Google Scholar]
- 65.Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran L-SP. . Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 2011; 18:263 - 76; http://dx.doi.org/ 10.1093/dnares/dsr015; PMID: 21685489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Y, Liu Z-H, Feng L, Zheng Y, Li D-D, Li X-B. . Genome-wide functional analysis of cotton (Gossypium hirsutum) in response to drought. PLoS One 2013; 8:e80879; http://dx.doi.org/ 10.1371/journal.pone.0080879; PMID: 24260499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dai M, Xue Q, Mccray T, Margavage K, Chen F, Lee JH, Nezames CD, Guo L, Terzaghi W, Wan J, et al. . The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 2013; 25:517 - 34; http://dx.doi.org/ 10.1105/tpc.112.105767; PMID: 23404889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. . Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci U S A 2009; 106:5418 - 23; http://dx.doi.org/ 10.1073/pnas.0811088106; PMID: 19276109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hannah MA, Wiese D, Freund S, Fiehn O, Heyer AG, Hincha DK. . Natural genetic variation of freezing tolerance in Arabidopsis. Plant Physiol 2006; 142:98 - 112; http://dx.doi.org/ 10.1104/pp.106.081141; PMID: 16844837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gregersen PL, Holm PB. . Transcriptome analysis of senescence in the flag leaf of wheat (Triticum aestivum L.). Plant Biotechnol J 2007; 5:192 - 206; http://dx.doi.org/ 10.1111/j.1467-7652.2006.00232.x; PMID: 17207268 [DOI] [PubMed] [Google Scholar]
- 71.Lim PO, Kim HJ, Nam HG. . Leaf senescence. Annu Rev Plant Biol 2007; 58:115 - 36; http://dx.doi.org/ 10.1146/annurev.arplant.57.032905.105316; PMID: 17177638 [DOI] [PubMed] [Google Scholar]
- 72.Walia H, Wilson C, Zeng L, Ismail AM, Condamine P, Close TJ. . Genome-wide transcriptional analysis of salinity stressed japonica and indica rice genotypes during panicle initiation stage. Plant Mol Biol 2007; 63:609 - 23; http://dx.doi.org/ 10.1007/s11103-006-9112-0; PMID: 17160619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, et al. . Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiol 2005; 139:822 - 35; http://dx.doi.org/ 10.1104/pp.105.065961; PMID: 16183841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaves MM, Flexas J, Pinheiro C. . Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot 2009; 103:551 - 60; http://dx.doi.org/ 10.1093/aob/mcn125; PMID: 18662937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saibo NJM, Lourenço T, Oliveira MM. . Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Bot 2009; 103:609 - 23; http://dx.doi.org/ 10.1093/aob/mcn227; PMID: 19010801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cellier F, Conéjéro G, Breitler JC, Casse F. . Molecular and physiological responses to water deficit in drought-tolerant and drought-sensitive lines of sunflower. Accumulation of dehydrin transcripts correlates with tolerance. Plant Physiol 1998; 116:319 - 28; http://dx.doi.org/ 10.1104/pp.116.1.319; PMID: 9499218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Narusaka Y, Narusaka M, Seki M, Umezawa T, Ishida J, Nakajima M, Enju A, Shinozaki K. . Crosstalk in the responses to abiotic and biotic stresses in Arabidopsis: analysis of gene expression in cytochrome P450 gene superfamily by cDNA microarray. Plant Mol Biol 2004; 55:327 - 42; http://dx.doi.org/ 10.1007/s11103-004-0685-1; PMID: 15604685 [DOI] [PubMed] [Google Scholar]
- 78.Cameron KD, Teece MA, Smart LB. . Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant Physiol 2006; 140:176 - 83; http://dx.doi.org/ 10.1104/pp.105.069724; PMID: 16361524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang J, Tan W, Yang X-H, Zhang H-X. . Plastid-expressed choline monooxygenase gene improves salt and drought tolerance through accumulation of glycine betaine in tobacco. Plant Cell Rep 2008; 27:1113 - 24; http://dx.doi.org/ 10.1007/s00299-008-0549-2; PMID: 18437388 [DOI] [PubMed] [Google Scholar]
- 80.Lee M, Jung J-H, Han D-Y, Seo PJ, Park WJ, Park C-M. . Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 2012; 235:923 - 38; http://dx.doi.org/ 10.1007/s00425-011-1552-3; PMID: 22109847 [DOI] [PubMed] [Google Scholar]
- 81.Ramachandra Reddy A, Chaitanya KV, Vivekanandan M. . Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J Plant Physiol 2004; 161:1189 - 202; http://dx.doi.org/ 10.1016/j.jplph.2004.01.013; PMID: 15602811 [DOI] [PubMed] [Google Scholar]
- 82.Shanker AK, Maheswari M, Yadav SK, Desai S, Bhanu D, Attal NB, Venkateswarlu B. . Drought stress responses in crops. Funct Integr Genomics 2014; 14:11 - 22; http://dx.doi.org/ 10.1007/s10142-013-0356-x; PMID: 24408129 [DOI] [PubMed] [Google Scholar]
- 83.Barr HD, Weatherley PE. . A re-examination of the relative turgidity technique of estimating water deficit in leaves. Aust J Biol Sci 1962; 15:413 - 28 [Google Scholar]
- 84.Venglat P, Xiang D, Qiu S, Stone SL, Tibiche C, Cram D, Alting-Mees M, Nowak J, Cloutier S, Deyholos M, et al. . Gene expression analysis of flax seed development. BMC Plant Biol 2011; 11:74; http://dx.doi.org/ 10.1186/1471-2229-11-74; PMID: 21529361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benjamini Y, Hochberg Y. . Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc, B 1995; 57:289 - 300 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.