Abstract
Asynchronous flowering of Brassica napus (canola) leads to seeds and siliques at varying stages of maturity as harvest approaches. This range of maturation can result in premature silique dehiscence (pod shattering), resulting in yield losses, which may be worsened by environmental stresses. Therefore, a goal for canola crop improvement is to reduce shattering in order to maximize yield. We performed a comprehensive transcriptome analysis on the dehiscence zone (DZ) and valve of Arabidopsis and Brassica siliques in shatter resistant and sensitive genotypes at several developmental stages. Among known Arabidopsis dehiscence genes, we confirmed that homologs of SHP1/2, FUL, ADPG1, NST1/3 and IND were associated with shattering in B. juncea and B. napus. We noted a correlation between reduced pectin degradation genes and shatter-resistance. Tension between lignified and non-lignified cells in the silique DZ plays a major role in dehiscence. Light microscopy revealed a smaller non-lignified separation layer in relatively shatter-resistant B. juncea relative to B. napus and this corresponded to increased expression of peroxidases involved in monolignol polymerization. Sustained repression of auxin biosynthesis, transport and signaling in B. juncea relative to B. napus may cause differences in dehiscence zone structure and cell wall constituents. Tension on the dehiscence zone is a consequence of shrinkage and loss of flexibility in the valves, which is caused by senescence and desiccation. Reduced shattering was generally associated with upregulation of ABA signaling and down-regulation of ethylene and jasmonate signaling, corresponding to more pronounced stress responses and reduced senescence and photosynthesis. Overall, we identified 124 cell wall related genes and 103 transcription factors potentially involved in silique dehiscence.
Keywords: Arabidopsis thaliana, Brassica juncea, Brassica napus, microarray, mutants, pod shatter, silique dehiscence
Abbreviations
- DZ
dehiscence zone
- Enb
endocarp b
- LL
lignified layer
- SL
separation layer
- SSP
seed storage protein
- TF
transcription factor
- VM
valve margin
- WT
wild type
Introduction
Canola is an economically successful oil crop because of its healthy fatty acid composition and high yield. However, the precise timing of crop harvest is challenging due to non-uniform seed maturity caused by asynchronous flowering. On the one hand, premature harvest results in poor oil quality due to the relatively high moisture and chlorophyll content of immature seeds. On the other hand, late harvesting results in yield losses of 8%-12%, and up to 50% under extreme conditions, since fully mature canola siliques are extremely sensitive to premature dehiscence, referred to as pod shatter.1,2,3 The timing challenge for harvest is exacerbated by water, wind and heat stresses; each of which can increase dehiscence. Attempts have been made to increase shatter resistance through conventional breeding since genetic variation for shattering occurs in the Brassica family. For example, B. juncea is relatively shatter resistant compared to canola.2 However, traditional breeding methods have so far been unsuccessful. Reduction of shattering has been accomplished in Arabidopsis by transgenic manipulation of upstream regulatory transcription factors (TFs) such as SHATTERPROOF1 and 2 (SHP1/2), FRUITFULL (FUL), ALCATRAZ (ALC) and INDEHISCENT (IND) as well as with downstream TFs or metabolic genes.4-8 However, the goal of reducing premature shattering while simultaneously maintaining facile seed release at maturity has proved challenging.
Dry fruit (capsule) development and subsequent dehiscence is a complex and highly organized process. The progressive development of fruits is coordinated with that of enclosed seeds and climaxes with capsule dehiscence (shattering) allowing seed dispersal. Crucifer capsules consist of 2 fused carpels divided by a false septum and are termed siliques. The fused carpels develop into valves that are joined with 2 medial repla which border the septum; seeds are attached to the septum by funicles. The valve and replum are separated by the valve margin (VM) consisting of cells that differentiate to become the dehiscence zone (DZ). The valves separate at the DZ during pod shattering. Development of Arabidopsis thaliana siliques has been studied extensively (for reviews see refs. 9, 10).
There are 2 cell types in the DZ. One type constitutes the non-lignified separation layer (SL) which is juxtaposed with the replum and allows the detachment of valves via cell-cell separation.11 The other VM cells are remote from the replum and form a rigid lignified layer (LL) which is continuous with the lignified endocarp b (enb) of the valve.12,13
The INDEHISCENT (IND) TF is responsible for the differentiation of the VM into the LL and SL by regulation of auxin transport14 whereas the TF ALCATRAZ (ALC) regulates the development of the SL.5,15 Lignification in the valve is controlled by IND and ALC, as well as SHP1, SHP2 and FUL.5 It has been postulated that during capsule desiccation tension builds up between rigid lignified and non-lignified layers to potentiate valve detachment.16 SHP1/2 promotes lignification of VM adjacent to the DZ. In the double mutant shp1/2, DZ formation and lignin deposition in the VM do not occur resulting in indehiscent siliques. FUL promotes valve differentiation and expansion (suppressing SHP and IND activity) and prevents VM lignification.5,17,18 Consequently, ful siliques are 80% smaller than WT siliques and there is ectopic lignification of all internal valve mesophyll cells and reduced shattering relative to WT. The association of reduced shattering and ectopic lignification in ful may be due to elimination of the tension between lignified and non-lignified cells. On the other hand, in siliques over-expressing FUL, both VM and replum cells become valve-like with no DZ or lignification resulting in completely indehiscent siliques.18 Siliques of alc acquire ectopically lignified cells in the inner VM that forms a continuous lignified bridge between lignified inner valve cell layers and the lignified replum vasculature and are indehiscent.15 Replum identity is specified by REPLUMLESS (RPL), which represses SHP1/2.10
After establishment of tissue identity, secondary wall deposition occurs in cells adjacent to the SL and in the valve endocarp. This deposition is required for dehiscence and is positively regulated by 2 plant-specific TFs NAC SECONDARY WALL THICKENING PROMOTING FACTOR 1 and 3 (NST1 and NST3) in a partially redundant manner and loss of function mutants of NST1/3 are indehiscent.7
A growing body of evidences supports the long-held hypothesis that the degradation of middle lamella and cell wall in the SL loosens cellular cohesion leading to valve detachment and that this engages proteins involved in cell wall loosening and hydrolysis such as cellulase (β-1,4 glucanase,), polygalacturonase (PG) and expansin.11,19-23 Direct evidence for this hypothesis came from studies on loss-of-function mutants of 2 closely related endo-PGs, ARABIDOPSIS DZ POLYGALACTURONASE1 (ADPG1) and ADPG2, which resulted in indehiscent siliques.8 ADPG1/2 act in a partially redundant fashion and normal expression of ADPG1 in the silique DZ depends on IND.
However, other factors in addition to cell wall composition may influence shattering. Magnetic resonance imaging of silique desiccation revealed that shatter sensitive B. napus (cv. Quantum) lost water from the inside of the valve in contrast with shatter resistant B. rapa (cv Parkland) which lost water from the outside of the valve.24 This suggested that the pattern of water loss contributed to a difference in tension on the SL and hence to a difference in susceptibility to shatter between the 2 cultivars.
A link between pod shatter TFs and hormonal regulation was revealed by the discovery that IND modulates auxin distribution by relocating the auxin efflux carrier PIN FORMED1 (PIN1) in the SL cells as they differentiate.14 In addition IND causes localized GA production leading to de-repression of ALC by DELLA proteins and hence (as noted earlier) to SL development.25 Reduced ethylene production in parthenocarpic (seedless) pods of canola was correlated with delayed pod dehiscence which was restored by application of the ethylene releasing chemical ethephon.26 On the other hand, siliques of loss-of-function mutants of ethylene receptors in Arabidopsis shatter at the normal time.26 Ethylene production was studied in shatter resistant B. juncea (mustard) and shatter sensitive canola and was very low in silique wall and high in seeds in both species.27,27 Interestingly, there was considerably higher ethylene inside mustard siliques than inside canola siliques and this interspecies difference became larger toward desiccation.27
The process of pod dehiscence in Arabidopsis is similar to that in canola based on comparative anatomical and physiological studies.16 Although several groups have identified regulators and metabolic genes involved in silique dehiscence that are described above, a complete description of shatter-related processes has not yet been obtained. In this paper, we present a comprehensive transcriptome analysis using: (1) shatter resistant and –sensitive Arabidopsis and Brassica genotypes and (2) selected developmental stages to identify genes that are differentially regulated between genotypes differing in shattering. Integration of transcriptome data from genotypic, interspecies and developmental comparisons has uncovered genes whose functions and pattern of expression suggest roles in pod development and dehiscence.
Results
Experimental design
The Combimatrix 90k Brassica array covers about 64% of the corresponding genes in the Arabidopsis genome and 78% of the ∼90000 contigs represented on the array were aligned with Arabidopsis genes. About 80% of Arabidopsis genes on the array corresponded to multiple Brassica probes (contigs).28 The effect of multiple probes for most Arabidopsis genes was to increase replication and hence the reliability of the Brassica transcriptome analysis. Arabidopsis microarray hybridizations (2 color) were performed between whole siliques of each of the shatter mutants – shp1xshp2, ful, and alc with that of WT and between manually peeled valve of B. juncea and B. napus at 3 developmental stages (3, 5 and 6) as described in the methods section (Table 1). Since Arabidopsis and Brassica share 86% DNA homology in protein coding regions, we initially hybridized Brassica onto Arabidopsis arrays. Such heterologous probing has been successfully employed in previous studies (e.g. see ref. 29). We later used homologous probing to the Brassica Combimatrix arrays described above.
Table 1.
Experiments and number of probed arrays used for data analysis
| Experiment I (Arabidopsis whole siliques; mutant vs. WT; Two color; Arabidopsis full genome oligo arrays) | |||||
|---|---|---|---|---|---|
|
Genotype |
stage 3 |
stage 5 |
stage 6 |
|
|
| shp1xshp2 vs. WT | 6 | 6 | 6 | ||
| alc vs. WT | 6 | 6 | 6 | ||
| ful vs. WT | 6 | 6 | 6 | ||
| Experiment II (Brassica tissues; 2 color; Arabidopsis full genome oligo arrays) | |||||
|
Genotype |
stage 2 |
stage 3 |
stage 5 |
stage 6 |
|
| DZ: B. juncea vs. B. napus | 2 | — | — | — | |
| Valve: B. juncea vs. B. napus | 2 | 4 | 5 | 6 | |
| Experiment III (Arabidopsis pod; 2 color; Arabidopsis full genome oligo arrays) | |||||
|
Genotype |
6 DPA |
11 DPA |
14 DPA |
|
|
| ful vs. WT | 4 | 4 | 4 | ||
| Experiment IV (Brassica tissues; single color; Combimatrix 90k Brassica oligo arrays) | |||||
|
Genotype |
stage 1 |
stage 2 |
stage 4 |
stage 5 |
stage 6 |
| B. juncea DZ | 6 | 6 | 6 | — | — |
| B. juncea valve | 6 | 6 | 6 | 5 | 6 |
| B. napus DZ | 5 | 6 | 5 | — | — |
| B. napus valve | 6 | 6 | 5 | 4 | 4 |
Numbers indicate probed arrays used for data analysis and obtained after quality control on samples in GeneSpring.
First, we compared the expression of known shatter-related genes such as ADPG1 in our microarray data with published results. Promoter-GUS analysis clearly demonstrated that ADPG1 is expressed in DZ but not in valve.8 In contrast, we detected moderate to high expression of ADPG1 in manually dissected valves. We concluded that valves contained residual DZ. In order to improve tissue uniformity and to explore gene expression at earlier stages, we dissected DZ and valve of B. napus and B. juncea microscopically at stages 1, 2 and 4 for definitive expression studies (Table 1). There were substantial differences in transcript profiles between B. juncea and B. napus as reflected by the high number of differentially expressed genes in both DZ and valve (Fig. 1A; Figs. S2A, S3). In contrast, there were fewer differences between the transcript profiles of DZ and valve within the same Brassica species (Fig. 1B). Therefore, we concluded that the effect of residual DZ on gene expression changes in manually dissected Brassica valve would be negligible. Consequently, we analyzed microarray data from all dissected Brassica tissues and Brassica analysis below refers to both experiments. In addition, a second set of gene expression comparisons in Arabidopsis pod tissues (with seeds removed for greater specificity) between ful and WT at earlier stages including 6 days post anthesis (DPA), 11 DPA and 14 DPA were performed. Since FUL is a negative regulator of dehiscence, genes associated with shattering will be overexpressed in valves in the ful background. Most of the Arabidopsis analyses refer to these second set of experiments.
Figure 1.
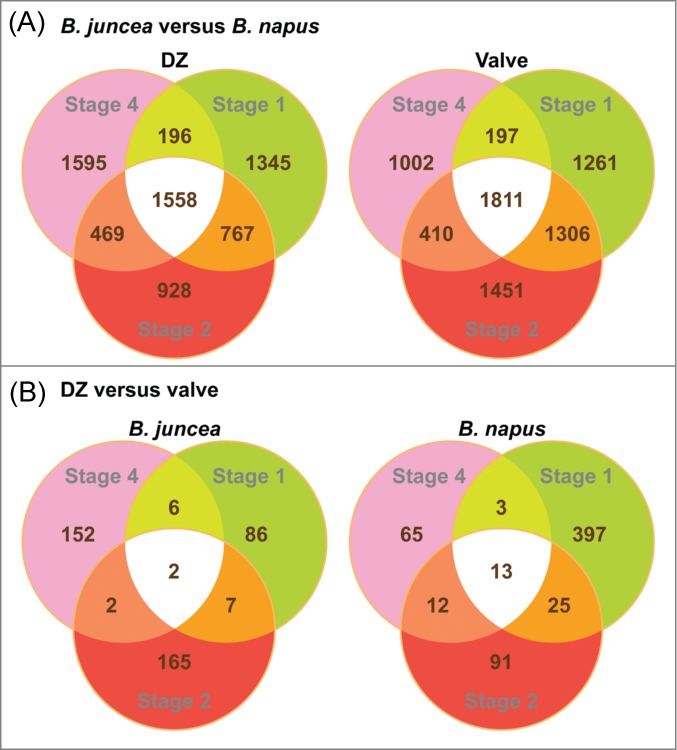
Venn diagrams to illustrate differences in transcript profiles of Brassica tissues between genotypes and at various stages.
Comparing Arabidopsis shatter mutants
The number of expressed genes as a percentage of the full Arabidopsis genome was calculated for each class of experiments. This percentage was 20 ± 2.6% for initial Arabidopsis comparisons and 74.7 ± 3.8% for Arabidopsis comparisons of ful vs. WT using the more sensitive RNA amplification protocol with seeds removed. The relatively low number of significantly expressed genes in whole siliques of Arabidopsis mutant versus WT comparisons (Table 2) could be partly due to the presence of highly abundant transcripts associated with seed nutrient reserve accumulation that may dilute out less abundant silique transcripts.
Table 2.
Number of significantly up- and downregulated genes (indicated by arrows) using a threshold change in expression of 1.5 fold and a p-value cut-off: ≤0.05
| Whole silique |
||||
|---|---|---|---|---|
| shp1 xshp2 vs. Col-0 | alc vs. Col-0 | ful vs. Col-0 | Pod ful vs. Col-0 | |
| Stage 3 | 189↑; 86↓ | 24↑; 233↓ | 28↑; 10↓ | — |
| Stage 5 | 55↑; 146↓ | 35↑; 94↓ | 157↑; 230↓ | — |
| Stage 6 | 78↑; 65↓ | 0↑; 56↓ | 60↑; 14↓ | v |
| 6 DPA | — | — | — | 479↑; 778↓ |
| 11 DPA | — | — | — | 576↑; 876↓ |
| 14 DPA | — | — | — | 245↑; 294↓ |
Genes encoding seed storage proteins
Accumulation of nutrient reserves during Arabidopsis and Brassica seed and Arabidopsis silique development has been extensively documented.30,31 Different classes of seed storage proteins (SSPs) accumulate at the mid maturation stage of seed development but follow distinct temporal expression patterns.30 Interestingly, SSP genes were the most highly upregulated genes in Arabidopsis pod starting from a very low level at 10 DPA but increasing almost 500 fold by 20 DPA.31 The expression of nutrient reserve transcripts including cruciferin, napin, oleosin, and late embryogenesis-associated (LEA) proteins in DZ of both B. juncea and B. napus, were highly upregulated at stage 4 (green-yellow tissue) with expression up to 170 fold above the basal level in B. juncea compared to 14-fold in B. napus (Fig. 2A; Table S3). The upregulation of storage product transcripts occurred at stage 2 of B. juncea valve. Accumulation of storage product transcripts in B. napus valve and DZ was very similar. Oddly enough, after a decline at stages 4 and 5, the transcripts were upregulated at stage 6 in valves of B. juncea.
Figure 2.
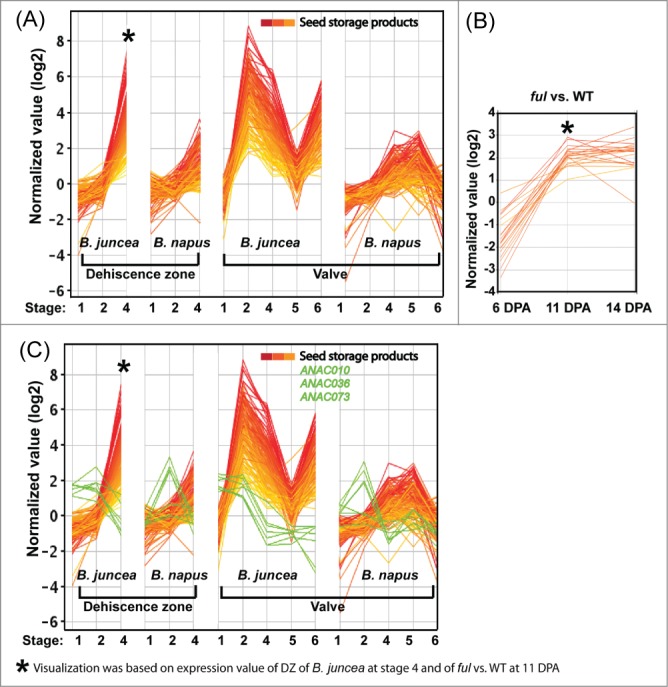
Comparison of transcript profiles of genes related to nutrient reserves between B. juncea and B. napus and between Arabidopsis ful and WT. Expression of individual contigs associated with Arabidopsis nutrient reserve was measured in various tissues of B. juncea and B. napus at different developmental stages using Brassica (Combimatrix, single color) microarray hybridization (A), (C). Expression of sequences corresponding to 3 Arabidopsis NAC TFs are overlaid on the SSP data in green (C). Expression profiles of nutrient reserve genes were compared in pod tissues of Arabidopsis ful vs. WT using Arabidopsis microarray (oligonucleotide probes, 2 color) hybridization (B).
In the Arabidopsis ful vs. WT (Fig. 2B; Table S3), the expression of SSPs was downregulated at 6 DPA but upregulated in 11 DPA and 14 DPA which indicates a possible role of FUL in regulating expression of storage proteins.
ABI3, FUS3, LEC1 and LEC2 are master regulators of SSP accumulation in seeds.32 However, we were unable to detect LEC2 and ABI3 transcripts in pods of Arabidopsis and Brassica. FUS3 and LEC1 were expressed in pods at very low levels and were not differentially expressed in ful vs. WT and B. juncea vs. B. napus comparisons. We performed SOM clustering among these nutrient reserve genes and differentially expressed significant TFs (fold change ≥4 and p-value cutoff ≤0.05) and identified 3 NAC TFs whose transcript profiles preceded those of the SSP transcripts (Fig. 2C). These TFs, NAC010 (At1g28470 [2 contigs]), NAC036 (At2g17040 [3 contigs]) and NAC073 (At4g28500 [2 contigs]), may be involved in the regulation of seed storage product accumulation in siliques.
The expression of known dehiscence genes
The Combimatrix microarray contained probes corresponding to SHP1, SHP2, FUL, ALC, ADPG2 and NST3, but none for IND, NST1, NST2 and ADPG1. Expression of both SHP1/2 genes showed similar profiles (Fig. 3A) and interestingly were higher in DZ than in valve at stages 1 and 2 in both B. juncea and B. napus. In contrast, the expression of FUL was lower in DZ than in valve at stages 1 and 2 of both Brassica cultivars. The expression of SHP1/2 were much higher than FUL in DZ of B. napus but the expression levels of these 3 TFs were similar in DZ of B. juncea. This is consistent with the fact that pods of B. napus are more shatter sensitive than B. juncea since we might expect expression of positive shatter regulators such as SHP1 and SHP2 to be relatively high, whereas expression of negative regulator FUL to be relatively low in DZ of siliques nearing dehiscence.
Figure 3.

Transcript profiling of known dehiscence genes in DZ and valve of B. juncea and B. napus using Brassica microarrays. Expression of TFs SHP1, SHP2, FUL and ALC (A). Expression of PIN3 and PID (B).
Intriguingly, the SHP1/2 expression levels in manually dissected valve at stages 5 and 6 (Fig. 3A), were higher in B. juncea than in B. napus and were verified by 2 color direct comparisons (Table S1).
FUL expression in DZ and valve at all stages studied was consistently 1.5-2 fold higher in B. juncea than in B. napus (Fig. 3A). The expression profile of ALC was opposite to that of SHP1/2 in DZ at stage 3 and in valve at all stages studied of both Brassica cultivars (Fig. 3A). Using heterologous Arabidopsis oligo arrays, no or low signal was detected for ALC, ADPG2, IND and NST2 (signal intensity of 200 for ALC and ADPG2 using RNA amplification system) in Brassica valves. It has been shown previously that FUL represses the expression of SHP1/2, IND and ALC.5,18 IND activates the expression of ADPG1 in the silique DZ.8
Dehiscence genes that were upregulated in pods of ful plants compared to that of WT included IND (12.93 fold), ADPG2 (11.08 fold), ADPG1 (4.95 fold), FIL (4.27 fold), SHP1 (3.26 fold), PID (3.13 fold), and SHP2 (2.72 fold) (Table 3). Indeed, in pods of ful, expression of all the positive regulators of dehiscence except ALC were upregulated compared to WT and therefore consistent with the role of FUL as a negative regulator of dehiscence. Observed expression patterns of ALC in pod are consistent with the previous findings that it has diffuse expression in valves of younger siliques (as reviewed in ref. 13). SHPs were shown to be specifically expressed in the VM starting before anthesis.33
Table 3.
Differential expression of known dehiscence genes from 2 color Arabidopsis array data
| Arabidopsis: ful vs WT |
||||
|---|---|---|---|---|
| Gene name | Locus ID | 6 DPA | 11 DPA | 14 DPA |
| IND (bHLH TF) | At4g00120 | 12.93 | 12.77 | 5.25 |
| ADPG2 (endo-polygalacturonase) | At2g41850 | 1.43 | 4.17 | 11.08* |
| ADPG1 (endo-polygalacturonase) | At3g57510 | 2.56 | 4.95* | 3.18* |
| FIL (YABBY1 TF) | At2g45190 | 2.43 | 4.27 | 1.21 |
| SHP1 (MADS TF) | At3g58780 | 3.26 | 3.10 | 2. 18 |
| PID (PINOID; protein kinase) | At2g34650 | 1.18 | 3.13 | 1.96 |
| SHP2 (MADS TF) | At2g42830 | 1.94 | 2.72 | 2.71 |
| FUL (bHLH TF) | At5g60910 | 0. 21 | 0.16 | 0.13 |
| ALC (bHLH TF) | At5g67110 | 0.94 | 1.05 | 0.92 |
| NST1 (NAM TF; ANAC043) | At2g46770 | 1.06 | 1.44 | 1.02 |
| NST2 (NAM TF; ANAC066) | At3g61910 | 1.04 | ||
| NST3 (NAM TF; ANAC012) | At1g32770 | 1.22 | 0.79 | 0.59 |
| PIN3 (auxin symporter) | At1g70940 | 0.65 | 1.77 | 1.08 |
| JAG (zinc finger C2H2 type TF) | At1g68480 | 0.68 | 0.83 | 0.74 |
| transducin family protein | At3g49400 | 1.29 | 1.13 | 0.96 |
| QRT2 (polygalacturonase) | At3g07970 | 1.32 | 1.22 | |
| YAB3 (YABBY3 TF) | At4g00180 | 1.01 | 1.05 | 1.04 |
| HEC3 (bHLH TF)) | At5g09750 | 1.08 | 1.07 | 0.84 |
*Expression exceeded the detection limit of intensity value of 65,000. Fold changes ≥2 are shaded and ≥5 are in bold text.
Promoter-GUS analysis revealed that ADPG1 and ADPG2 are expressed predominantly in silique DZs just prior to or at dehiscence but are absent in valve.8 The spot intensity of ADPG1 was 6270 (±1051) and 21,985 (±2884), respectively, in DZ of B. juncea and B. napus at stage 2 and was at the background level in valve at the same stage in both Brassica cultivars. Thus, ADPG1 was expressed 3.3-fold lower in stage 2 DZ (Table S5) of shatter resistant B. juncea than in shatter sensitive B. napus consistent with the direct involvement of ADPG1 in dehiscence. Expression of ADPG1 was also downregulated in silique of shp1xshp2 relative to WT at all stages (Table S11).
The consistency of the differential expression of dehiscence genes in pods of ful vs. WT with previous studies encouraged us to examine in detail other differentially expressed genes. We identified cell wall related genes and TFs using functional annotation based on http://cellwall.genomics.purdue.edu/ for cell wall genes and PlnTFDB (plant transcription factor database) for TFs34 (Table S5) that may be associated with the silique dehiscence. We selected these genes based on similarity to the expression patterns of IND, ADPG1, ADPG2, SHP1 and SHP2 in ful vs. WT and in DZ and valve of B. juncea vs. B. napus at stage 2.
Differences in silique lignin deposition and ultrastructure between B. juncea and B. napus
Previous studies on B. napus and B. juncea suggested that tensions generated between the non-lignified and rigid, lignified layers in the valve and VM precipitate silique dehiscence.16 Upstream positive regulators of pod shatter SHP1/2 promote lignification of a subset of VM cells.4 Furthermore, in the Arabidopsis alc mutant, ectopically lignified cells in the inner VM create a “lignified bridge” between the lignified inner valve cell layers and the lignified replum vasculature, rendering alc siliques resistant to shatter.15 We studied the progression of lignification and ultrastructural changes in silique development in both Brassica cultivars at stages 1, 2, and 4 and in dried mature siliques (Fig. 4). Circumferences (cross section) of siliques and sizes of most cell types of B. juncea were smaller than those of B. napus while the sizes of cells in enb, and septum (SE) were similar. There were 7-8 mesocarp (m) layer cells in both cultivars (Fig. 4A-F). At stage 1, M cells were rounded in shape in both cultivars but became flattened and elongated at stage 2 in both the inner and the outer layers of M in B. juncea but only in outer layer of B. napus.
Figure 4.

Degree of lignification and ultrastructural changes associated with the progression of Brassica silique dehiscence. Transverse sections were stained with toluidine blue O. Lignins stained blue and pectin and pectic substances stained purple. Sections of B. juncea silique at stage 1 (A), stage 2 (C), stage 4 (E) and mature-dried (G) and sections of B. napus silique at stage 1 (B), stage 2 (D), stage 4 (F) and mature-dried (H). Valves separated from replum of sections of mature-dried siliques during tissue preparation for microscopy. Abbreviations: ena, endocarp a; enb, endocarp b; ex, exocarp; m, mesocarp; r, replum; se, septum; sl, separation layer; v, vascular bundle; vm, valve margin. Scale bar, 100 μm. Stars depict remaining intact lignified cells after collapse of the ena layer at stage 4.
Toluidine blue staining revealed lignin in enb, M cells adjacent to SL and replum vasculature (v) in both cultivars as early as stage 1 (Fig. 4A, 4B). The degree of lignification in these cells, in addition to inner valve M cells, increased substantially at later stages of silique development. Nonetheless, the extent of lignification in these cells, specifically at stages 1 and 2 was lower in B. juncea than in B. napus (Fig. 4A-D) while conversely, lignin deposition was higher in stage 4 and mature-dry siliques of B. juncea than in B. napus (Fig. 4E-H). VM consists of lignified (LL) and non-lignified (SL) cells and there were 1-2 non-lignified cells in B. juncea and 3-5 in B. napus while the number of lignified cells was similar in both. Larger cell sizes and more cells in the VM of B. napus made the VM wider than that of B. juncea. The enb layer of B. napus, especially at stage 4, comprised 1-2 cell files whereas the enb of B. juncea consisted mostly of one cell file. Dissolution of the cells in the ena layer was observed at stage 4 leaving a few intact lignified cells close to both sides of the replum (indicated by asterisks in Fig. 4). In B. juncea, there were 1-4 very small surviving cells but there were 3-6 relatively large survivors in B. napus (Fig. 4E-H).
Careful examination of lignification at the inner VM cells in stage 4 revealed that the non-lignified cells extended a little beyond the ena toward the replum in B. napus while they ended, in most cases, just above the enb layer of B. juncea suggesting an alc-like lignified cell-bridge in B. juncea encompassing lignified enb, a few surviving-lignified cells of ena and lignified replum vasculature (Fig. 4E, F). This may be a significant factor in reducing shattering in B. juncea.
A previous study based on MRI revealed that during the early desiccation stage, shattering resistant B. rapa valves dried initially from the outside whereas B. napus valves dried initially from inside.24 It was suggested that this drying pattern contributed to the different susceptibilities to shattering in the 2 cultivars. The increase in tension on the DZ is a product of senescence and desiccation, which together result in loss of flexibility and contraction of the valves. The microscopy results are consistent with preferential drying of B. napus siliques from the inside (Fig. 4H) since most of the ena layer had collapsed at the mature, dry stage but the exocarp (EX) layer remained hydrated, although there is some shrinkage of cells within the M layer. In B. juncea, early collapse of the ena layer is apparent at stage 4 (Fig. 4E), but by the mature, dry stage (Fig. 4G) the EX layer had collapsed and there was substantial shrinkage of M cells, suggesting rapid drying from the outside. Therefore, the most obvious difference in silique drying between the 2 species is the relative resistance of the outer EX and M cells to water loss in B. napus. This asymmetric drying in B. napus may contribute toward additional tension on the DZ as described previously.24
Shikimate, phenylpropanoid and monolignol biosynthesis pathways are strongly downregulated in B. juncea relative to B. napus
The shikimate pathway links primary and secondary metabolism and products arising from the pathway include lignins (reviewed in ref. 35). As noted above, lignification plays an important role in silique dehiscence4,16 and we have described significant differences in silique lignin deposition between B. juncea and B. napus (Fig. 4). Moreover, lignification occurs in the whole inner valve of the ful mutant since the valves manifest replum-like characteristics, and as a result siliques fail to dehisce .17 Therefore, we compared genes of these pathways in Arabidopsis ful vs. WT and from DZ and valve of B. juncea vs. B. napus.
The shikimate pathway consists of 7 enzymes, and 9 genes coding for pathway enzymes were substantially downregulated in B. juncea compared to B. napus (Table S6). The final product of the shikimate pathway is chorismate, which is converted into multiple products, including lignin. Transcript downregulation was observed in B. juncea compared to B .napus for 5 genes involved in chorismate metabolism.
Next, we compared the expression of genes involved in monolignol biosynthesis in the Brassica cultivars. Expression of 27 genes involved in the pathway were detectable by microarray analysis and 16 of these were moderately to highly downregulated in valve and DZ of B. juncea compared to B. napus (Table 4). It was clear that monolignol (mainly coniferyl alcohol/G unit and coumaryl alcohol/H unit) biosynthetic genes were downregulated in B. juncea relative to B. napus. However, transcripts of sinapoyl glucose: malate sinapoyl transferase (SNG1) stayed upregulated up to stage 5 and transcripts of CCoAOMT6, O-methyltransferase Family 2 Protein, CAD5, and COMT-like1/2/3/4 were highly upregulated, especially in stages 5 and 6 in B. juncea relative to B. napus indicating enrichment of sinapoyl alcohol (S) units in lignin polymers and/or the formation of sinapoyl malate ester.36 Interestingly, only a few monolignol biosynthetic genes were differentially expressed in ful vs. WT comparisons (Table 4). However 10 of the 16 upregulated genes in the Brassicas were also downregulated in alc and shp1xshp2 relative to WT (Table S11).
Table 4.
Differential expression of genes encoding enzymes of phenylpropanoid pathway and biosynthesis of monolignol and sinapate ester (functional annotation based on http://cellwall.genomics.purdue.edu/) from 2-color Arabidopsis microarray data
| Stages: B. juncea vs. B. napus |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis: ful vs. WT |
DZ* |
valve* |
valve** |
|||||||
| Gene name | Locus ID | 6 DPA | 11 DPA | 14 DPA | 2 | 2 | 3 | 5 | 6 | Pathway |
| PAL1; phenylalanine ammonia-lyase 1 | At2g37040 | 0.99 | 0.90 | 0.33 | 0.97 | 1.13 | 0.72 | 0.22 | 0.16 | Phenylpropanoid |
| PAL2; phenylalanine ammonia-lyase 2 | At3g53260 | 1.46 | 1.26 | 0.49 | 0.31 | 0.24 | 0.45 | Phenylpropanoid | ||
| PAL4; phenylalanine ammonia-lyase 4 | At3g10340 | 0.80 | 1.20 | 1.86 | 0.57 | 0.13 | 0.03 | 0.05 | Phenylpropanoid | |
| C4H; Trans-cinnamate 4-hydroxylase; CYP73A5 | At2g30490 | 1.36 | 1.18 | 0.86 | 0.44 | 0.41 | 0.46 | 0.85 | 1.90 | Phenylpropanoid |
| 4CL1; 4-coumarate coenzyme A ligase 1 | At1g51680 | 0.83 | 1.22 | 1.01 | 0.32 | 0.39 | 0. 16 | 0.08 | 0.08 | Phenylpropanoid |
| 4CL3; 4-coumarate coenzyme A ligase 3 | At1g65060 | 1.03 | 0.80 | 0.34 | 1.48 | 1.21 | 1.32 | 0.24 | Phenylpropanoid | |
| 4CL4; 4-coumarate coenzyme A ligase 4 | At3g21230 | 0.95 | 0.90 | 1.14 | 0.57 | 0.56 | 0.48 | 0. 18 | 0.29 | Phenylpropanoid |
| 4CL-like1 | At1g20510 | 1.08 | 0.41 | 0.64 | 1.87 | 1.35 | 1.28 | 0.77 | 0.37 | Phenylpropanoid |
| HCT; Hydroxycinnamoyl-CoA:shikimate/quinate hydroxycinnamoyltransferase | At5g48930 | 1.64 | 1.56 | 1.25 | 1.55 | 1.58 | 0.74 | 0.36 | 0.37 | Phenylpropanoid |
| C3H1; p-coumarate-3-hydroxylase; CYP98A3 | At2g40890 | 1.04 | 1.07 | 0.92 | 0.51 | 0.53 | 0.32 | 0.27 | 0.58 | Phenylpropanoid |
| CCoAOMT1; caffeoyl-CoA 3-O-methyltransferase 1 | At4g34050 | 0.98 | 1.32 | 1.77 | 0.42 | 0.45 | 0.76 | 0. 21 | 0.55 | Phenylpropanoid |
| CCR-like5; cinnamoyl-CoA reductase-like5 | At5g58490 | 0.94 | 0.76 | 0.79 | 1.55 | 1.58 | 2.02 | 0.82 | 0.26 | Monolignol |
| F5H1; ferulate-5-hydroxylase 1; CYP84A1 | At4g36220 | 0. 23 | 0.58 | 0.37 | 0.44 | 0.50 | 0.35 | 0.88 | 3.19 | Monolignol |
| COMT; caffeic acid O-methyltransferase | At5g54160 | 0.67 | 1.16 | 1.13 | 0.34 | 0.40 | 0.41 | 0.16 | 0.21 | Monolignol |
| CAD4; cinnamyl alcohol dehydrogenase 4 | At4g37980 | 1.81 | 0.49 | 0.58 | 0.58 | 0.55 | 0.06 | 0.14 | Monolignol | |
| CAD6; cinnamyl alcohol dehydrogenase 6 | At4g34230 | 0.97 | 1.39 | 0.99 | 0.53 | 0.63 | 0.73 | 0.40 | 0.34 | Monolignol |
| CCoAOMT6;; caffeoyl-CoA 3-O-methyltransferase 6 | At1g67980 | 1.19 | 0.91 | 1.66 | 0.94 | 1.13 | 2.00 | 3.41 | 2.96 | Phenylpropanoid |
| O-methyltransferase Family 2 Protein | At4g35150 | 0.80 | 0.63 | 0.63 | 1.26 | 1.02 | 1.73 | 3.20 | ||
| O-methyltransferase Family 2 Protein | At4g35160 | 0.88 | 0.93 | 0.99 | 1.34 | 0.99 | 1.12 | 3.50 | ||
| CAD5; cinnamyl alcohol dehydrogenase 5 | At4g37990 | 9.62 | 3.61 | 1.24 | 1.09 | 0.74 | 1.32 | 6.06 | 4.99 | Monolignol |
| COMT-like1 | At1g21100 | 1.08 | 0.96 | 3.33 | 8.58 | 33.72 | Monolignol | |||
| COMT-like2 | At1g21110 | 0.68 | 0.51 | 0.43 | 1.11 | 0.96 | 2.76 | 9.94 | 22.64 | Monolignol |
| COMT-like3 | At1g21120 | 0.59 | 0.54 | 0.67 | 1.09 | 1.00 | 5.52 | 5.28 | Monolignol | |
| COMT-like4 | At1g21130 | 0.62 | 0.67 | 0.66 | 1.15 | 0.86 | 5.73 | 10.29 | 29.57 | Monolignol |
| sinapoylglucose:malate sinapoyltransferase (SNG1) | At2g22990 | 1.15 | 1.24 | 0.63 | 2.94 | 3.32 | 4.89 | 5.21 | 1.34 | Sinapate ester |
| peroxidase | At3g17070 | 0.38 | 0.43 | 0. 24 | 0.76 | 1.23 | 0.73 | 0.49 | 0.32 | |
| PER3 = Rcl3 | At1g05260 | 1.40 | 0.81 | 0. 20 | 0. 24 | 0.45 | 0.52 | |||
| peroxidase | At3g28200 | 1.28 | 0.87 | 0.68 | 2.00 | 2.10 | 2.14 | 1.34 | 2.79 | |
| PER17 | At2g22420 | 2.08 | 1.46 | 0.60 | 2.14 | 2.05 | 1.92 | 2.97 | 3.33 | |
| peroxidase | At5g14130 | 1.26 | 2.05 | 1.39 | 1.09 | 0.98 | 2.97 | 2.22 | 3.43 | |
| PER21 | At2g37130 | 1.28 | 0.35 | 0.58 | 3.10 | 9. 17 | 1.94 | 1.65 | 4.22 | |
| peroxidase | At5g64120 | 3.83 | 0.82 | 0.70 | 26.17 | 14.11 | ||||
| PER12 | At1g71695 | 0.37 | 1.92 | 1.65 | 2.17 | 1.73 | 1.88 | 9.71 | 27.56 | |
| peroxidase | At4g08770 | 0.85 | 0.79 | 0.97 | 2.26 | 1.69 | 5. 04 | 19.19 | 57.37 | |
| PERx34 | At3g49120 | 0.32 | 0.29 | 0.81 | 1.42 | 0.95 | 2.98 | 14.12 | 67.53 | |
| LAC15; Laccase | At5g48100 | 0.61 | 2.11 | 2.12 | 0.81 | 0.81 | 5.94 | 10.63 | ||
| LAC1; Laccase | At1g18140 | 0.94 | 0.90 | 0.93 | 0.93 | 0.96 | 0.33 | 0.25 | 0.44 | |
*DZ and valve dissected under microscope. **Manually dissected valve and septum. Fold change ≥5 are shaded and ≥10 have bold text.
After synthesis, monolignols are transported from cytoplasm to the cell wall and lignin is formed through polymerization of the monolignols involving peroxidases, laccases, polyphenol oxidases, and coniferyl alcohol oxidase.37 Peroxidases and laccases belong to large families in Arabidopsis and hence we only examined cell wall related peroxidases and laccases. Transcripts of 10 cell wall related peroxidases and one laccase were highly upregulated – especially in stages 5 and 6 in B. juncea relative to B. napus and this is correlated with our microscopy data (Fig. 4E-H) suggesting increased lignifications in stage 4 and dry-mature silique in B. juncea relative to B. napus.
Remodeling of cell wall components during silique dehiscence
Plant cell walls are composed of cellulose microfibrils coated by xyloglucans and embedded in a matrix of pectic polysaccharides.38 Pectins constitute a complex family of galacturonic acid (GalA)-rich (70% of pectin) polysaccharides. They are abundant in the primary cell wall and in the intercellular matrices of middle lamellae in which they form a gelling matrix for cell-cell adhesion. Homogalacturonans (HGs) are the most abundant pectins (65%) and are synthesized in the Golgi apparatus mostly as methylester precursors (60%). Methylated HGs are secreted into the apoplast, where they are deesterified by pectin methylesterase (PME) to form negatively charged pectate. Pectates are cross-linked with each other through calcium ions to form a gel.
Pectin modification enzymes
PMEs generate deesterified HGs, which are degraded preferentially by endo-PGs.39 Therefore, PMEs and PGs are thought to act together in the degradation of pectins and are involved in cell wall breakdown during silique dehiscence.8,40,41 While screening for cell-wall genes whose expression was reminiscent of ADPG1 and ADPG2 we identified homologs of 4 PMEs (At1g11580, At2g36710, At5g26810 & At3g59010) that were highly upregulated in ful relative to WT and were downregulated in DZ and valve of B. juncea relative to B. napus (Table S5). Conversely, 2 PMEs (At3g62170 & At1g53840) showed the opposite expression profiles in both ful vs. WT and Brassica cultivar comparisons. PME can be inactivated by PME inhibitors (PMEIs) and expression of PMEI1 (At1g48020) was downregulated in ful relative to WT, but was undetectable in Brassica. Overall, PMEs seemed to be associated with shattering.
Besides endo-PG ADPG1, 4 other PGs (At1g80170, At1g19170, At3g16850 and At5g41870) were highly downregulated in B. juncea relative to B. napus but unlike ADPG1, these genes were not upregulated in pods of ful vs. WT and therefore may not be directly involved in dehiscence or may not be regulated by IND. Transcript of PG - inhibiting protein (PGIP1) that inhibits fungal PGs,42 was highly upregulated in DZ and valve of B. juncea relative to B. napus.
Pectate can be degraded by pectate lyase (PL). Eight PLs were differentially regulated and 5 (At1g14420, At3g01270, At3g09540, At3g55250 & At5g15110) were downregulated and 3 (At1g04680, At4g24780 & At5g63180) were upregulated in both ful vs. WT and Brassica cultivar comparisons. Pectin acetylesterases (PAEs) deacetylate homogalacturonan polymers and thus solubilize pectin facilitating the access of PL to substrate. Two PAE genes were up- and 3 genes were downregulated in B. juncea compared to B. napus.
The enzyme β-galactosidase (BGAL) degrades structural pectins, xyloglucans or arabinogalactoproteins in plant cell walls and 3 BGALs (BGAL1/3/6) were upregulated by 2-6 fold in B. juncea relative to B. napus.
There is a unifying pattern in most of the pectin-related genes that can be related to pectin degradation and consequently to shattering – especially in the Brassica data. There is a tendency for reduced expression of PGs, PMEs and PLs associated with pectin breakdown in B. juncea vs B. napus comparisons (and for PGIP1 to increase). This could suggest stronger middle lamellae, contributing to reduced shattering. Alternatively, reduced pectin degradation might be a consequence of fewer SL cells in B. juncea than in B. napus (Fig. 4).
Xyloglucan endotransglucosylase/hydrolases (XTH)
XTHs have dual activities as endotransglucosylase (XET) and endohydrolase (XEH) and can strengthen or loosen cell walls under different circumstances. They belong to a large family of 33 genes43 and 7 XTHs (XTH3/6/7/8/10/16/29) were differentially expressed (Table S5). In our microarray analysis, expression levels of all 7 were very low in Brassica, and in Arabidopsis, expression of all except XTH3/6/10 were very low. In Arabidopsis, expression of XTH3 increased only in ful pod at 11 DPA and 14 DPA; XTH6 increased in WT pods at 6 DPA and of XTH10 increased only at 14 DPA in WT. In addition to low expression level, differential expression of XTHs exhibited noclear pattern in relation to pod dehiscence.
Enriched metabolic pathways
Expression data was mapped on to biological pathways using MapMan.44 We documented some key processes in silique development (Tables 6, 7). Genes in individual biochemical pathways were initially annotated according to the Plant Metabolic Pathway Database (http://www.plantcyc.org/), Cell Wall Genomics (http://cellwall.genomics.purdue.edu/) and MapMan and then updated from the published literature.
Table 6.
Significant pathways derived by Wilcoxon rank sum test (p-value cut-off: ≤ 0.05) in MapMan44 from Brassica (Combimatrix) microarray data
|
B. juncea |
B. napus |
B. juncea |
B. napus |
Genotype |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dehiscence zone | Valve | Tissue type | ||||||||||||||||
| Bin# | Assignment (MapMan) | 1 | 2 | 4 | 1 | 2 | 4 | 1 | 2 | 4 | 5 | 6 | 1 | 2 | 4 | 5 | 6 | Development stage |
| 35 | Not assigned | 235 | 269 | 268 | 159 | 544 | ||||||||||||
| 29 | Protein | 180 | 2 | 2 | 22 | 208 | 96 | 115 | 83 | 8 | 3 | 17 | 133 | |||||
| 34 | Transport | 4 | 4 | 50 | 4 | 11 | 5 | 61 | 114 | 55 | 3 | 88 | 159 | |||||
| 20 | Stress | 90 | 74 | 77 | 8 | 84 | 66 | 12 | 3 | 12 | 92 | 8 | 26 | 119 | ||||
| 26 | Miscellaneous | 36 | 20 | 6 | 38 | 6 | 15 | 34 | 18 | 17 | 44 | 40 | 45 | 8 | 10 | 40 | 37 | |
| 10 | Cell wall | 69 | 14 | 74 | 9 | 64 | 11 | 10 | 53 | 9 | 9 | 6 | 2 | |||||
| 33 | Development | 28 | 8 | 22 | 39 | 8 | 6 | 4 | 18 | 19 | 31 | 21 | 45 | |||||
| 11 | Lipid metabolism | 12 | 8 | 12 | 7 | 10 | 9 | 12 | 5 | 7 | 9 | 10 | 8 | 13 | ||||
| 27 | RNA | 5 | 45 | 5 | 3 | 15 | 3 | 6 | 16 | 3 | 2 | 11 | ||||||
| 1 | Photosynthesis | 8 | 2 | 3 | 22 | 19 | 2 | 2 | 24 | 7 | ||||||||
| 13 | Amino acid metabolism | 17 | 19 | 2 | 3 | 3 | 65 | |||||||||||
| 31 | Cell | 7 | 13 | 14 | 37 | 37 | ||||||||||||
| 17 | Hormone metabolism: auxin | 2 | 4 | 2 | 4 | 11 | 10 | 11 | 16 | |||||||||
| 18 | Hormone metabolism: ethylene | 4 | 3 | 4 | 5 | 6 | 5 | 6 | ||||||||||
| 16 | Secondary metabolism | 6 | 2 | 2 | 6 | 6 | 2 | 3 | 15 | |||||||||
| 30 | Signaling | 4 | 3 | 4 | 2 | 13 | 2 | |||||||||||
| 24 | Biodegradation of Xenobiotics | 5 | 6 | 5 | ||||||||||||||
| 21 | Redox | 3 | 4 | 6 | 6 | |||||||||||||
| 6 | Gluconeogenesis/glyoxylate cycle | 2 | 6 | 6 | 5 | |||||||||||||
| 18 | Hormone metabolism: salicylic acid | 3 | 2 | 3 | 3 | 3 | ||||||||||||
| 28 | DNA | 4 | 3 | |||||||||||||||
| 2 | Major CHO metabolism | 2 | 3 | 3 | ||||||||||||||
| 18 | Hormone metabolism: gibberelin | 4 | 3 | |||||||||||||||
| 3 | Minor CHO metabolism | 2 | 4 | |||||||||||||||
| 9 | mit. electron transport/ATP synthesis | 2 | ||||||||||||||||
| 18 | Hormone metabolism: jasmonate | 2 | ||||||||||||||||
| 4 | Glycolysis | 3 | ||||||||||||||||
| 23 | Nucleotide metabolism | |||||||||||||||||
| 8 | TCA | 2 | ||||||||||||||||
| 12 | N-metabolism | |||||||||||||||||
| 17 | Hormone metabolism: cytokinin | 2 | ||||||||||||||||
Table 7.
Transcript profiles of senescence related genes of Arabidopsis and Brassica cultivars in various tissues during silique development
| DZ*: B. juncea versus B. |
Valve*: B. juncea vs. B. napus |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene name | Locus ID | 1 | 2 | 4 | 1 | 2 | 4 | **5 | **6 | : Stages |
| APG6 | AT3g61710 | ↓8.49 | ↓7.87 | ↓8.04 | ↓10.56 | ↓9.66 | ↓16.2 | ↓18.68 | ↓48.93 | |
| APG8c | AT1g62040 | ↓3.95 | ↓5.22 | ↓12 | ↓5.54 | ↓21.78 | ↓5.84 | ↓6.03 | ↓14.73 | |
| APG8f | AT4g16520 | ↓10.71 | ↓16.66 | ↓9.01 | ↓20.12 | ↓57.44 | ↓19.28 | ↓60.2 | ↓115.04 | |
| APG12a | AT1g54210 | ↓2.36 | ↓3.97 | ↓2.82 | ↓2.55 | ↓3.53 | ↓7.57 | ↓6.52 | ↓42.78 | |
| APG12b | AT3g13970 | ↓6.9 | ↓7.49 | ↓5.71 | ↓17.37 | ↓11.07 | ↓12.35 | ↓43.51 | ↓24.88 | |
| PDF1.2a | AT5g44420 | ↓107.1 | ↓311.7 | ↓33.39 | ↓355 | ↓306.8 | ↓129.4 | ↓648.7 | ↓683.2 | |
| SAG12 | AT5g45890 | ↓1.94 | ↓8.63 | |||||||
| SAG21 | AT4g02380 | ↑4.09 | ↓2.03 | ↓6.12 | ↓2.36 | |||||
|
B. Juncea vs. B. napus |
||||||||||
|
Arabidopsis array data: |
Arabidopsis: ful vs. WT |
DZ* | valve* |
valve** |
: Stages | |||||
|
Gene name |
Locus ID |
6 DPA |
11 DPA |
14 DPA |
2 |
2 |
3 |
5 |
6 |
|
| APG8c | At1g62040 | ↓17.29 | ↓23.42 | ↓11.55 | ||||||
| PDF1.1 | At1g75830 | ↑9.62 | ↓2.31 | ↓2.22 | ↓1.68 | ↓5.68 | ↓4.29 | |||
| PDF1.2a | AT5g44420 | ↓4.88 | ↓5.13 | ↓3.73 | ↓21.71 | ↓24.93 | ||||
| PDF1.2b | At2g26020 | ↑1.9 | ↓4.79 | ↓3.73 | ↓4.94 | ↓3.85 | ||||
| PDF1.2c | At5g44430 | ↑2.75 | ↓3.15 | ↓3.31 | ↓5.57 | ↓3.3 | ||||
| PDF1.3 | At2g26010 | ↓2.88 | ↓2.68 | ↓3.85 | ↓2.96 | |||||
| PDF1.4 | At1g19610 | ↓3.41 | ↓2.27 | ↑5.2 | ↑8.58 | |||||
| PDF1.5 | At1g55010 | ↑2.39 | ↑1.96 | ↑1.8 | ↓14.49 | ↓18.57 | ||||
| PDF2.3 | At2g02130 | ↓3.64 | ↓4.41 | 5.29 | ||||||
↑ : fold upregulated. ↓ : fold downregulated. In Combimatrix data, fold changes of multiple contigs for the same gene are averaged. *DZ and valve dissected under microscope. **Manually dissected valve.
Photosynthesis
Both seed and silique lose chlorophyll and photosynthetic activity during development. Transcripts of 65 photosynthetic genes were downregulated in pod of ful compared to WT indicating that WT leaf-like valves became non-photosynthetic and VM-like in ful (Fig. 5). These genes were also downregulated in B. juncea vs. B. napus comparisons and in whole siliques of alc compared to WT but were not differentially regulated in whole siliques of the shp1xshp2 vs. WT comparison suggesting that changes in photosynthetic genes are not directly related to dehiscence.
Figure 5.

Heat map representation of expression of photosynthetic genes derived from Arabidopsis microarrays.
Senescence
Silique dehiscence shows similarities in gene expression and hormone signaling with senescence, wounding and defense.16,31 Genes such as SAG12, AUTOPHAGY 8 (APG8), APG12, PDF1.2, VSP2, and LOX2 that are correlated with cell death and organ senescence were upregulated in yellowing silique wall of Arabidopsis.31,45 Interestingly, genes of 6 plant defensin fusion proteins (PDF), PDF1.1/1.2a/1.2b/1.2c/1.3/2.3, were highly expressed in DZ and valve of B. napus and these PDFs along with PDF1.5 were downregulated strongly in B. juncea (Table 7). PDF1.1/1.2b/1.2c were also downregulated in alc but were upregulated in ful relative to WT (Table S1). Five autophagy genes APG6, APG8c, APG8f, APG12a and APG12b were also highly downregulated in B. juncea relative to B. napus in all tissues at all stages. Senescence marker genes SAG12 and SAG21 were downregulated in B. juncea relative to B. napus at early and later stages of silique development, respectively. Both SAG12/21 were highly downregulated in ful vs. WT but were upregulated in shp1 X shp2 vs. WT and alc vs. WT comparisons (Table S1) suggesting SAG12/21 are either unlinked to pod shattering and/or not regulated by SHPs and ALC. However, marker genes VSP1/2 of ethylene and jasmonate signaling pathways were downregulated in alc vs. WT and shp1xshp2 vs. WT comparisons (Table S1). Therefore, downregulation of PDFs, APGs, and SAGs suggests that delayed and/or reduced senescence may contribute toward reduced dehiscence in B. juncea.
Phytohormones
Auxin: Significant pathway analysis in MapMan revealed differentially regulated genes associated with auxin and ethylene (Tables 5, 6). Recent results have documented a local auxin minimum required for the specification of the VM SL.14 Therefore, we examined the expression of genes involved in auxin biosynthesis, transport and signaling. Several genes coding for auxin biosynthetic enzymes were strongly downregulated in B. juncea relative to B. napus (Table S7). IAA biosynthetic genes such as NIT1/2/3, IBR3, ILL6 and ILR1 were also downregulated in shp1xshp2, alc and ful in comparison to WT (Table S1). The majority of IAA in Arabidopsis is found conjugated as glycosyl esters or amide-linked IAA-conjugates, some of which could release free IAA upon hydrolysis.46,47 IAA-conjugates play an important role in IAA metabolism and homeostasis and in Arabidopsis, 90% of the conjugated IAAs are amide-linked and are formed by indole-3-acetic acid amido synthetases, which belong to the GH3 gene family of auxin primary response genes. Genes catalyzing the formation of IAA conjugates including several GH3s and UDP-GLUCOSE TRANSFERASE (UGT1/At1g05560) were highly upregulated in B. juncea relative to B. napus indicating the synthesis of storage forms of IAA (Table S7).46,48 In contrast, most of these auxin conjugating genes were downregulated in ful vs. WT.
Table 5.
Significant pathways derived by Wilcoxon rank sum Test (p-value cut-off: ≤0.05) in MapMan44 from Arabidopsis microarray data
|
B. juncea vs. B. napus |
ful vs. WT |
shp1 x shp2 vs. WT |
alc vs. WT |
Genotype |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| valve |
pod |
whole silique |
whole silique |
whole silique |
Tissue type |
||||||||||||
| Bin# | Assignment (MapMan) | 3 | 5 | 6 | 6 DPA | 11 DPA | 14 DPA | 3 | 5 | 6 | 3 | 5 | 6 | 3 | 5 | 6 | Development stage |
| 35 | Not assigned | 646 | 933 | 2 | 43 | ||||||||||||
| 27 | RNA | 8 | 19 | 56 | 117 | 139 | 6 | 6 | |||||||||
| 26 | Miscellaneous | 28 | 89 | 167 | 3 | 28 | 15 | 9 | 5 | 6 | |||||||
| 34 | Transport | 14 | 26 | 33 | 15 | 8 | 14 | 4 | |||||||||
| 20 | Stress | 21 | 6 | 26 | 45 | 18 | |||||||||||
| 16 | Secondary metabolism | 3 | 49 | 54 | 4 | 13 | 3 | ||||||||||
| 33 | Development | 5 | 11 | 11 | 7 | 6 | 2 | 3 | |||||||||
| 1 | Photosynthesis | 3 | 19 | 43 | 2 | 6 | 2 | ||||||||||
| 29 | Protein | 15 | 35 | 25 | 5 | 32 | 9 | 2 | |||||||||
| 11 | Lipid metabolism | 6 | 19 | 32 | 4 | 8 | 5 | ||||||||||
| 10 | Cell wall | 2 | 3 | 11 | 10 | 7 | 4 | ||||||||||
| 17 | Auxin | 20 | 31 | 12 | 2 | ||||||||||||
| 31 | Cell | 2 | |||||||||||||||
| 21 | Redox | 13 | 14 | 11 | 3 | ||||||||||||
| 2 | Major CHO metabolism | 5 | 24 | 9 | |||||||||||||
| 30 | Signaling | 4 | 20 | 8 | |||||||||||||
| 17 | Ethylene | 2 | 13 | 14 | |||||||||||||
| 13 | Amino acid metabolism | 10 | 2 | ||||||||||||||
| 15 | Metal handling | 14 | |||||||||||||||
| 17 | GA | ||||||||||||||||
| 12 | N-metabolism | 3 | 4 | 2 | |||||||||||||
| 18 | Co-factor and vitamine metabolism | 7 | |||||||||||||||
| 25 | C1-metabolism | 2 | 2 | ||||||||||||||
| 3 | Minor CHO metabolism | 3 | |||||||||||||||
| 14 | S-assimilation | 3 | |||||||||||||||
| 17 | CK | 3 | |||||||||||||||
| 9 | Electron transport | ||||||||||||||||
| 17 | ABA | 2 | |||||||||||||||
| 23 | Nucleotide metabolism | ||||||||||||||||
A proper auxin response is dependent on the delivery of IAA to target cells by polar transport involving influx and efflux carriers. The efflux carriers PIN3/4 as well as the influx carrier AUX1 and transporter PGP19 were downregulated whereas 2 auxin efflux carrier family proteins (At1g76520, At2g17500) were upregulated in B. juncea compared to B. napus (Table S7). The expression patterns of these transporter genes were reversed in ful vs. WT compared to B. juncea vs. B. napus, suggesting that their expression is linked to dehiscence.
Auxin responses involve ubiquitination and subsequent degradation of Aux/IAA transcriptional repressor proteins resulting in the release of auxin response factor (ARF) proteins which either activate or repress auxin-regulated genes.47 In the absence of auxin, ARFs are sequestered and inactivated by Aux/IAAs. There are 22 ARFs and 29 AUX/IAAs in Arabidopsis. No clear pattern of expression was apparent for ARFs (Table S7). Expression of IAAs including IAA9/10/12/13/17/18/26 were down regulated and IAA8/31 were up regulated in B. juncea relative to B. napus (Table S7). This suggests that, similar to auxin biosynthesis and transport, auxin signaling was predominantly repressed in B. juncea compared to B. napus.
Ethylene: Ethylene has a role in many processes including organ abscission, senescence and stress.49 Ethylene is synthesized from methionine (Met) by 3 enzymes: S-adenosyl-L-Met (AdoMet) synthetase, 1-amino-cyclopropane-1-carboxylate (ACC) synthase (ACS) and ACC oxidase (ACO). ACS catalyzes the first committed step and, in most instances, is rate limiting. Several ethylene biosynthetic genes were highly upregulated in all tissues and stages tested in B. juncea compared to B. napus including SAM1, 5 ACSs, ACO1, ACO2 and 4 others ACOs (Table S11). Ethylene biosynthesis is regulated, in part, by ETO1 and 2 ETO1-like genes EOL1 and EOL2 negatively regulate ACS activity and hence ethylene production.50 EOL1 was downregulated in B. juncea relative to B. napus (Table S8). Upregulation of several biosynthetic genes and downregulation of EOL indicates that ethylene biosynthesis is upregulated in shatter resistant B. juncea relative to shatter sensitive B. napus which is consistent with previous results that B. juncea siliques produced considerably higher ethylene inside siliques than B. napus throughout silique development.27
In the absence of ethylene, its receptors activate the Raf-like protein kinase CTR1 (MAPKKK), which is a negative regulator of ethylene action. Two among its 5 receptors, ETR1 and ETR2, were downregulated and CTR1 was upregulated in B. juncea compared to B. napus (Table S8). Among the downstream effectors, EIN2 and EIN3 are positive regulators of ethylene action and were downregulated in B. juncea more than in B. napus. EIN2 is upregulated in ful vs. WT. Taken together, the downregulation of 2 receptors ETR1 and ETR2 and 2 positive regulators EIN2 and EIN3 and upregulation of negative regulator CTR1 indicate that, in contrast to biosynthetic genes, ethylene signaling was downregulated in B. juncea more than in B. napus. Ethylene regulates its own biosynthesis through a positive feedback control.51 Therefore, down regulation of ethylene signaling may act as a positive feedback for the induction of ethylene biosynthesis. In Arabidopsis, ACO2 and a putative ACO were downregulated in shp1xshp2 and alc relative to WT suggesting reduced ethylene production in non-shattering genotypes (Table S11).
Jasmonic acid (JA): The bioactive form of JA (e.g., jasmonoyl-isoleucine, JA-Ile) plays an important role in wounding and pathogen invasion (for reviews, see refs. 52, 53). In our microarray data, downregulation of marker genes for JA signaling such as PDF1.2a/b/c in B. juncea relative to B. napus (Table 7) prompted us to look closely JA biosynthesis and signaling. JA is derived from α-linolenic acid (18:3) which can be generated by at least 3 enzymes FAD3, FAD7 and FAD8. FAD3 and several other genes coding for JA biosynthetic enzymes, including 4 lipoxygenases (LOX1/2/4), 3 allene oxide cyclases (AOC1/2/3), and 12-oxophytodienoate reductase (DDE1) were upregulated in B. juncea relative to B. napus (Table S9). In contrast to JA biosynthesis, genes JAR1 and JMT for the formation of JA-Ile and methyl jasmonate (MeJA), respectively, were downregulated in B. juncea relative to B. napus. Like auxin signaling, JA-Ile action results in the ubiquitination and subsequent degradation of JAZ repressors, which releases JA signaling. Positive regulators COI1 and MYC2 expressions were downregulated whereas negative regulators JAZ1/2/8/9/10 were upregulated in B. juncea relative to B. napus implying a suppression of JA signaling in B. juncea. This is consistent with the suppression of JA marker genes: PDFs and VSP2 in B. juncea, alc and shp1 x shp2 (Table 7; Table S11). Therefore, similar to ethylene, down regulation of JA signaling may in turn activate JA biosynthetic genes through positive feedback. JA biosynthetic genes FAD3, LOX1 and LOX2 were downregulated in shp1 x shp2 vs. WT (Table S9).
ABA: ABA is a key phytohormone in water stress responses. Several genes of ABA biosynthesis including NCED, AAO3 and ABA3 were induced and 2 genes coding for the catabolic enzyme ABA 8′-hydroxylase were repressed in B. juncea relative to B. napus and in ful vs. WT (Table S10).54 Induction of ABA biosynthesis correlates with the high expression of SSP, oleosin and LEAs and down regulation of photosynthetic genes in pods of B. juncea and ful as described above and similar effects of ABA have been described previously.55-57 ABA induces the expression of many genes that are important for adaptation to abiotic stresses.57 The expression of these stress related genes and of TFs involved in mediating ABA responses such as ABF2/3/4 are upregulated in B. juncea relative to B. napus and in alc, shp1xshp2 and ful relative to WT (Tables S10, S11) indicating stronger adaptive responses to desiccation in shatter-resistant genotypes.
Discussion
Expression of upstream regulators such as SHP1/2 and IND were upregulated in ful vs. WT comparisons (Table 3) whereas FUL and ALC were upregulated and SHP1/2 downregulated in DZ of B. juncea vs. B. napus comparison (Fig. 3A) (IND was absent from the Brassica array). SHP1/2 and FUL regulate silique dehiscence and their expression is consistent with the observed differences in shattering between the 2 species. In general, our microarray data showed that expression of dehiscence-related genes in ful vs. WT were opposite to that in B. juncea vs. B. napus. However, there were some exceptions to this relationship. Similarities between some differentially expressed genes in the B. juncea vs. B. napus and ful vs. WT comparisons might reflect the fact that ful siliques are also relatively resistant to dehiscence due to loss of VM identity and ectopic lignin deposition in the entire inner valve.
Among the known downstream shattering genes, ADPG1/2 were upregulated and NST3 was downregulated in ful relative to WT, whereas both ADPG1 and NST1/3 were downregulated in B. juncea vs. B. napus and shp1xshp2 vs. WT. Expression of these genes appears to be reduced in most shattering-resistant genotypes, consistent with their known functions. Although the ful genotype exhibits reduced shattering, dehiscence-related genes are expressed in the valve due to de-repression of native SHP. Therefore, expression of ADPG1/2 in the ful background appears to reflect expression of SHP-repressed genes, whereas NST3 expression is a manifestation of reduced shattering independent of SHP.
Similarities and differences between seed (embryo) and silique
Siliques and seeds follow distinct but coordinated developmental programs and have some similar morphogenetic and metabolic processes. For instance, both seeds and siliques desiccate during late maturation whereas siliques and endosperm senesce but embryos do not. Nutrient reserves in seeds account for 90% or more of the seed dry weight and in Brassica is comprised mostly of oils (triacyglycerols), and specialized SSPs.
Expression of genes coding for SSPs were highly upregulated in silique walls of Arabidopsis (Col-0).31 It is not clear why there were abundant transcripts for SSPs and oil body associated proteins in valves of Arabidopsis and Brassica even in the senescing stage. Moreover, these genes were upregulated in B. juncea relative to B. napus and in ful vs. WT comparison. High expressions of LEAs and stress/senescence/drought responsive genes (e.g. RAB18, DI21, SAG21, ERD14, LTI30, LTI65, KIN2 and P5CS1) in valve tissue are most likely related to cellular dehydration during drying of pods. Interestingly, LEAs related to cold stress responses such as COR15A, COR15B, and ERD10/LTI45 were downregulated in B. juncea relative to B. napus.
Previous studies have shown that LEAs are capable of stabilizing labile enzymes under stress conditions (reviewed in ref. 58) and they may be required to perform this function in siliques as well as in seeds. However, this is speculative since the extent to which LEA transcripts are translated in siliques is unknown. It is interesting that the expression of known master regulators of seed development, the TFs, ABI3, FUS3, LEC1/2, that regulate the accumulation of nutrient reserves in seed were either absent or barely detectable in the valves.
Differences in gene expression between the 2 Brassica species are related to dehiscence
Transcriptome comparisons between B. juncea and B. napus (Fig. 6; Table 6) revealed that large numbers of differentially regulated genes function in hormone metabolism and signaling, photosynthesis, cell wall and secondary metabolism.
The timing and amount of lignin deposition differed significantly in the 2 Brassica species as was evident from the light micrographs (Fig. 4) and microarray data (Table 4). However, overall there was more lignification in B. juncea and fewer non-lignified SL cells. Despite these observations, numerous genes in the phenylpropanoid and monolignol pathways were repressed in B. juncea relative to B. napus. However, higher expression of peroxidases in B. juncea may be the determinant of lignin deposition. Reduced shattering in B. juncea may be partly due to a combination of a smaller SL layer and increased lignin deposition. However, either an excess or a deficiency of lignin can be associated with silique dehiscence. For example, both ectopic lignification in the whole inner valve of ful siliques, and absence of lignification in cells adjacent to VM in plants overexpressing FUL rendered pods indehiscent.18 This suggests that the amount and localization of lignin deposition determines shattering rather than its overall presence or absence.
Lignin is not the only cell wall component that may influence shattering. The tendency of middle lamellae to rupture is likely increased by pectin degradation. Our observation that pectin degradation via PGs was downregulated in B. juncea relative to B. napus and in shp1 x shp2 vs. WT suggests an inverse relationship between shattering and pectin accumulation.
Siliques lose water prior to dehiscence and consequently suffer dehydration stress,24 as do embryos. It is well known that ABA induces the expression of many stress responsive genes.57 In this study, these stress responsive genes as well as ABA biosynthetic genes were induced, and ABA catabolic genes repressed in B. juncea relative to B. napus. These results suggest that B. juncea tissues may be better adapted to desiccation and this may be manifested in the greater resistance of vacuolated B. juncea cells to collapse during desiccation (Fig. 4F, H). Some ABA stress responsive genes were also induced in shp1xshp2 and alc relative to WT (Table S10), suggesting a possible association of enhanced abiotic stress responses with reduced silique dehiscence.
The significance of the down regulation of many genes encoding components of photosystem (PS) I and II, and Calvin cycle in B. juncea vs. B. napus, ful vs. WT and alc vs. WT is not clear. Degradation of photosynthetic components is an expected consequence of senescence but shatter resistance is associated with reduced senescence (as summarized below). Therefore, these differences may be a result of enhanced ABA effects in shattering-deficient genotypes. ABA strongly down-regulates chlorophyll biosynthesis and photosynthesis.57
Auxin biosynthesis, transport and signaling were repressed in B. juncea relative to B. napus. The consistent difference in auxin responses between the 2 species may result in differences in the VM layer. It has been shown previously that auxin is involved in VM specification,14 so the smaller size of the SL layer in B. juncea may be a consequence of sustained differences in auxin responses. Later in development, these consequences may include differences in lignin deposition and other cell wall constituents between B. juncea and B. napus that we have noted.
Desiccation and senescence in the silique lead to the imposition of tension on the DZ due to contraction and loss of flexibility in the valves. It has been observed previously that cell death and organ senescence related genes were induced in senescing Arabidopsis siliques.31,45 Gene expression during silique dehiscence confirms the importance of senescence, wounding and defense processes.16,31 Ethylene and JA signaling play important roles in these processes26,49,53 and ethylene and jasmonate signaling genes were repressed in B. juncea relative to B. napus. Results presented here suggest that the onset of senescence, wounding and defense responses were delayed or reduced in the shatter resistant genotypes. Reduced senescence may contribute toward greater resistance to pod shattering in B. juncea because less disruptive force is imposed on the SL.
Conclusions
Pod shattering in Brassicas depends on the presence of a layer of relatively fragile SL cells that are unlignified and with weakened cell walls due to pectin degradation. These SL cells are juxtaposed with rigid, lignified VM cells (the LL) and replum cells with reinforced secondary walls. Tension between these cell layers increases as valves shrink due to desiccation and senescence until a critical point is reached at which shattering can be triggered by a mechanical impact. The size of the SL layer and cell wall metabolism, including lignin deposition, are clearly crucial determinants of shattering. In addition, we noted that ABA signaling and stress responses were upregulated whereas ethylene- and jasmonate signaling pathways and senescence related genes were downregulated in B. juncea vs. B. napus, shp1xshp2 vs. WT and alc vs. WT. These relationships suggest that reduced dehiscence is associated with more pronounced adaptive response to water stress and a reduction in the degradative processes associated with senescence. The sustained effects of auxin on cell specification, cell wall formation and senescence may be a primary determinant of variations in dehiscence. We have summarized our results in a model of silique dehiscence (Fig. 7).
Figure 6.
Illustration of the metabolic pathways and genes that were differentially affected in siliques between B. juncea and B. napus. Unabbreviated gene names and AGI numbers are listed in respective pathway-Table 4 and Tables S6 to S11. Other abbreviations for each pathway are as follows. IAA biosynthesis: IAA, indole-3-acetic acid; IBA, indole-3-butyric acid. Shikimate pathway: PEP, phospho-enol-pyruvate; E4P, erythrose 4-phosphate. Isoprenoid biosynthesis: MVP, mevalonate pathway; IPP, isopentenyl diphosphate; MEP, methyl erythritol phosphate pathway; G3P, D-glyceraldehyde-3-phosphate; Pyr, pyruvate; GGP, geranylgeranyl diphosphate. Carotenoid biosynthesis: GGP, geranylgeranyl diphosphate. Jasmonates biosynthesis: 18:1-18:1-PC, 1,2-dioleoylphosphatidylcholine; 18:3-18:3-PC, 1,2-dilinolenoylphosphatidylcholine; JA, jasmonic acid; MeJA, methyl jasmonate; JA-Ile, JA-isoleucine. Ethylene biosynthesis: SAM, S-adenosyl methionine.
Figure 7.
Model summarizing the biological processes, hormones and regulatory proteins controlling silique dehiscence in Brassicas. A cross-section of a B. juncea silique from Fig. 4c is used to illustrate the location of effects.
A major objective of this study was to identify genes related to pod shatter. We have developed a list of pod shatter-related genes including 124 metabolic genes and 103 TFs (Table S8). Gene selection was based on expression patterns of known shatter genes or a link to cell wall, organ abscission or senescence. The genes listed here may serve as starting points for the functional analysis of specific aspects of dehiscence. Study of these genes will provide a better understanding of silique dehiscence and lead to new strategies for modulating pod shattering via transgenic alteration of selected target genes. As noted earlier, shattering is increased by stresses such as heat, drought and wind, therefore reduction of shattering is a useful approach to improving performance under abiotic stress.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana (Col-0 ecotype) wild type (WT) and mutant (shp1xshp2, ful, alc and ind) plants were grown under long-day conditions at 22°C and 40% humidity with 16 h of 150 μE light and 8 h dark cycles. Seeds of T-DNA insertion mutant lines for shp1xshp2, ful, alc and ind were obtained from the Arabidopsis Biological Research Center (ABRC) with accession numbers respectively: SALK_CS3844, SALK_033647, SALK_103763 and SALK_010533. Homozygous plants were selected by PCR (as described in ref. 59). Siliques from homozygous plants of shp1xshp2 and alc were indehiscent whereas that of ful was small as described previously.4,15,17 However, ind siliques were dehiscent like WT indicating the possible loss of T-DNA insert and hence were not studied further. Seeds of B. napus (cv. Nexera 715) and B. juncea (cv Vniimk405) were provided by DowAgroSciences. Growth conditions for Brassica plants were 22°C day and 18°C night with 16 h of 250-300 μE light and 8 h dark.
Developmental stages, tissue collection and RNA extraction
Tissue was collected from 6 developmental stages in Arabidopsis and Brassica. The stages were defined as follows: stage 1, embryos were approximately at heart stage and siliques were 70-90% elongated and 50% expanded with respect to diameter; stage 2, embryos were approximately at the bent cotyledon stage and occupied approximately 50-90% of the volume of the seed sac; stage 3, green silique color immediately prior to yellowing; stage 4, siliques were intermediate between green and yellow color; stage 5, siliques were yellow or green-yellow and stage 6, siliques were brown or yellow-brown. In a separate series of tissue collections, Arabidopsis flowers of WT and ful were tagged and green siliques were harvested at 6 and 11 days post anthesis (DPA) and green to yellow siliques were collected at 14 DPA.
In over 70% of samples, 6 independent biological replicates were collected of each tissue stage and in each replicate, tissues were harvested from no less than 10 (Arabidopsis) and 4 (Brassica) plants. DZ and valve tissues of Brassica were dissected under a microscope at stages 1, 2 and 4. Dissected tissues were frozen in dry ice and RNA was isolated using the RNAqueous RNA isolation kit (Ambion AM7021). Furthermore, whole siliques of Arabidopsis (with seeds) and Brassica (without seeds) were also collected in dry ice at stages 3, 5 and 6. For Brassica siliques (stages 3, 5 and 6), valve was peeled manually from septum-replum then frozen in liquid nitrogen and stored at −80°C until RNA was isolated (as described in ref. 60). In addition, Arabidopsis siliques of WT and ful plants were collected at 6 DPA, 11 DPA and 14 DPA in RNALater solution (Ambion AM1912) and seed, stigma, style and gynophore were removed from pods under a dissecting microscope and total RNA was isolated from these pod tissues using the RNAqueous kit as described above. The RNA quality was assessed in a Bioanalyzer (Agilent) prior to labeling and the RNA integrity numbers (RIN) obtained were in the range 7-9.
Microarray labeling, hybridization and data acquisition
Sample preparation and hybridization of Brassica and Arabidopsis samples on Arabidopsis microarrays without RNA amplification
Five micrograms of total RNA isolated from whole siliques of Arabidopsis and manually dissected valves of Brassica were used for cDNA synthesis using 3DNA Array 900 (Genisphere Inc.. cat# W500130 and W500140) and at least 6 independent hybridizations were performed for each type of sample. cDNA was labeled either with Cy3 or Cy5 followed by hybridization onto full genome Arabidopsis oligonucleotide microarray slides obtained from the University of Arizona (http://ag.arizona.edu/microarray/) and washed according to the manufacturer's instructions for 2 color hybridizations except for prehybridization. Immediately prior to use, slides were prehybridized for 20 mins at 65°C in a Coplin jar containing 3.5x SSC, 0.1% SDS and 10 mg/ml BSA. Subsequently, slides were washed for 1 min in autoclaved double distilled water (ddH2O) and for 1 min in isopropanol and then dried using a filtered air flow. Hybridization was performed in a hybridization cassette (ArrayIt cat#AHC). Each slide was scanned for Cy3 and Cy5 emissions at a resolution of 10 μm per pixel using a GenePix 4000B scanner (Molecular Devices).
RNA amplification
For microscopically dissected tissues of Brassica and Arabidopsis WT and ful siliques from which seeds were removed, there was limited tissue availability and a consequent low quantity of total RNA. In addition, Combimatrix slide chemistry is incompatible with the 3DNA Array 900 system. Therefore an aRNA amplification system (Allyl MessageAmp II aRNA amplification kit from Ambion: cat#AM1753) was adopted and 800 ng total RNA was used as a starting material to make fluorescent dye labeled aRNA.
Hybridization of Brassica samples to Combimatrix Brassica microarrays
For quantifying Brassica gene expression, we used custom-built Combimatrix 90k Brassica arrays. Array probes were developed using public EST sequences obtained from Brassica tissues.28 These ESTs were assembled into 95,000 unigenes. Unique 35-mer probe sequences were designed and synthesized in situ on slides using the Combimatrix technology (Combi Matrix diagnostics). Each full length individual Brassica contig/singleton was aligned against the A. thaliana unigene (TAIR8 cds database) and Uniprot plant sequence database and was scored by the BLAST similarity matrices. Combimatrix slide chemistry has inherent fluorescence at lower wavelengths; therefore, only Cy5 dye labeling and hence single color analyses were performed. Furthermore, as Combimatrix slides are not suitable for the autoPMT function in the GenePix scanner, 10 alien RNA spikes (Stratagene cat#252561-252570) were added to the 800 ng total RNA. Five micrograms of Cy5 labeled aRNA were fragmented using reagents from Ambion (cat#AM8740) prior to the hybridization step. Although aRNA amplification routinely produces around 60 μg aRNA from 800 ng total RNA, separate aRNA preparations were made for all hybridizations and at least 6 independent hybridizations were performed for each type of sample.
Combimatrix slide hybridization, washing, scanning and post scan stripping of labeled probes from slides and rehybridization were performed according to the manufacturer's instructions (CustomArrayTM 90K microarray; protocol#PTL020). GenePix scanner settings for Combimatrix slides were 5 μM pixel size and 130 μM focus position. During scanning, the spot intensity value of each RNA spike was kept approximately the same for all hybridized slides.
Hybridization of Arabidopsis samples to Arabidopsis microarrays using amplified RNA.
For 2 color hybridizations on Arabidopsis arrays (consisting of ful vs. WT comparisons with seeds removed), 4 independent hybridizations were performed and the above aRNA protocol was modified slightly as follows: No RNA spikes were added to the 800 ng starting total RNA and each aRNA prep was labeled with either Alexa647 or Alexa555 dyes. Slides were UV cross-linked and prehybridized as described above. Five micrograms of each Alexa647 and Alexa555 labeled aRNA were mixed together, fragmented and then added to 68 μl hybridization solution (SlideHyb #1 buffer; Ambion cat#AM8861). A 25X60 LifterSlip (Erie Scientific Company cat#25x60I-2-4789) was placed on top of the arrayed area and around 80 μl of the above labeled aRNA and hybridization solution mixture was added at one edge of the LifterSlip and the slide was placed inside an ArrayIt hybridization cassette. Hybridization was performed at 45°C overnight with gentle horizontal agitation at 40 rpm. After hybridization, the LifterSlip was removed in 2xSSC and 0.2% SDS solution prewarmed at 45°C. Slide washing was carried out with gentle agitation of 40 rpm as follows: 1) 10 min washing in 2xSSC and 0.2% SDS solution prewarmed at 45°C; 2) 10 min in 2xSSC at room temperature (RT) and 3) 10 min in 0.2xSSC at RT. Slides were scanned by a GenePix scanner.
Initial data extraction
Calculation of the signal intensity of spots and flagging of bad spots were carried out in GenePix Pro 6.1 software (Molecular Devices).
Quality control of data for subsequent analysis
All primary data, including images were uploaded into BioArray Software Environment (BASE) where background signal subtraction was performed.61 Subsequently, normalization (Print-tip loess), filtering bad spots and control spots and filtering minimum channel intensity (intensity for both channel should be <300 in most cases) were also carried out. Then calculation of correlation coefficients between replicates and visual spot quality inspection was also performed in BASE. Afterward, data were transferred to GeneSpring GX 10.0.2 (Agilent) for higher order data analysis. Single color Combimatrix microarray data was uploaded directly from GenePix Pro 6.1 into GeneSpring where the default normalization (each measurement was divided by the 75th percentile of all measurements in that sample and each gene intensity was divided by the median of its measurements in all samples), filtering of flagged spots and calculation of correlation coefficient among replicates were performed. For both 2 and single color hybridizations, quality control on sample data was performed in GeneSpring.
Determining differentially expressed genes and enrichment of biological pathways
A t-test against zero together with Benjamini-Hotchberg multiple testing correction with a 0.05 p-value cut-off was performed in GeneSpring to obtain statistically differentially expressed gene sets. Furthermore, a fold change ≥2.0 and ≥4.0, for 2 color and single color platforms, respectively, were employed as additional criteria of biological significance. Due to the large number of changes in expression, a conservative approach of 4-fold change was taken to reduce Brassica false positives. Afterward, log2 expression values were uploaded into MapMan ImageAnnotator version 3.0.0RC344 for pathway analysis. Microarray data from multiple contigs (probes) for the same gene were averaged before uploading to MapMan. To obtain statistically significant enriched biological pathways, a Wilcoxon rank sum t-test embedded in MapMan was performed with a p-value cut-off 0.05 and Benjamini Hochberg multiple testing correction. The heat map was generated using heatmap builder version 1.1 software.62 For multiple comparisons, 1-way ANOVA was performed in GeneSpring to obtain statistically differentially expressed gene lists. Additional quality control on gene lists for false positive / negative was performed in BASE using its spot visualization feature. Only genes that were differentially expressed in more than one comparison are reported.
Light microscopy
Canola siliques at various developmental stages were cut transversely by hand with a Teflon-coated razor blade under 3% (v/v) glutaraldehyde in 0.05 M PIPES buffer (pH 7.4) into thin ‘coins’ on a sheet of dental wax, then transferred to a scintillation vial containing this fixative for 2 h at room temperature with vacuum applied several times to remove air from the tissue. The specimens were then washed twice (15 min each) in cold PIPES buffer and dehydrated in an ethanol series of 25, 50 and 75% at 4°C, 2 hours each step, then 100% ethanol at −20°C for 24 hours. Samples were then infiltrated with increasing concentrations of LR White resin (25, 50, 75, and 100%) at −20°C for 24 h each step. The specimens were then placed on a rotator at room temperature for 24 hours and subsequently transferred individually to flat-bottomed BEEM capsules and polymerized at 55°C for 2 h. Sections for light microscopic examination were cut 0.5 μm thick on an ultramicrotome with a diamond knife, collected on slides, dried on a hotplate, and stained for 10–30 seconds with 1% (w/v) toluidine blue in 1% (w/v) sodium borate. Sections were then mounted with Polymount (Polysciences), observed on a Leica DM5000 microscope, and recorded with a Leica DCF500 digital camera.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Mr. Chad Matsalla for assistance with Bioinformatics, Dr. Van Ripley of Dow AgroSciences for providing B. juncea seeds, and Dr. Mark Thomson, Dr. Allan Feurtado and Dr. Daiqing Huang for helpful discussions. Dr. Yuejin Sun provided helpful input into microarray experiments and Dr. Tom Skokut provided valuable contributions during project initiation.
Funding
This project was funded by the National Research Council of Canada and Dow AgroSciences. This paper is NRCC number 55577.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. MacLeod J. Oilseed rape. Cambridge: Cambridge Agricultural, 1981; 107-19. [Google Scholar]
- 2. Kadkol GP, Macmillan RH, Burrow RP, Halloran GM. Evaluation of Brassica genotypes for resistance to shatter. I. Development of a laboratory test. Euphytica 1984; 33:63-73. [Google Scholar]
- 3. Child RD, Summers JE, Babij J, Farrent JW, Bruce DM. Increased resistance to pod shatter is associated with changes in the vascular structure in pods of a resynthesized Brassica napus line. J Exp Bot 2003; 54:1919-30; PMID:12837816 [DOI] [PubMed] [Google Scholar]
- 4. Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 2000; 404:766-70; PMID:10783890 [DOI] [PubMed] [Google Scholar]
- 5. Liljegren SJ, Roeder AHK, Kempin SA, Gremski K, Østergaard L, Guimil S, Reyes DK, Yanofsky MF. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 2004; 116:843-53; PMID:15035986 [DOI] [PubMed] [Google Scholar]
- 6. O/stergaard L, Kempin SA, Bies D, Klee HJ, Yanofsky MF. Pod shatter-resistant Brassica fruit produced by ectopic expression of the FRUITFULL gene. Plant Biotech J 2006; 4:45-51. [DOI] [PubMed] [Google Scholar]
- 7. Mitsuda N, Ohme-Takagi M. NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 2008; 56:768-78; PMID:18657234; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03633.x [DOI] [PubMed] [Google Scholar]
- 8. Ogawa M, Kay P, Wilson S, Swain SM. ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 2009; 21:216-33; PMID:19168715; http://dx.doi.org/ 10.1105/tpc.108.063768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lewis MW, Leslie ME, Liljegren SJ. Plant separation: 50 ways to leave your mother. Curr Opin Plant Biol 2006; 9:59-65; PMID:16337172 [DOI] [PubMed] [Google Scholar]
- 10. Girin T, Sorefan K, Østergaard L. Meristematic sculpting in fruit development. J Exp Bot 2009; 60:1493-502; PMID:19246597; http://dx.doi.org/ 10.1093/jxb/erp031 [DOI] [PubMed] [Google Scholar]
- 11. Peterson M, Sander L, Child R, van Onckelen H, Ulvskov P, Borkhardt B. Isolation and characterization of a pod dehiscence zone-specific polygalacturonase from Brassica napus. Plant Mol Biol 1996; 31:517-27; PMID:8790285 [DOI] [PubMed] [Google Scholar]
- 12. Dinneny JR, Weigel D, Yanofsky MF. A genetic framework for fruit patterning in Arabidopsis thaliana. Development 2005; 132:4687-96; PMID:16192305 [DOI] [PubMed] [Google Scholar]
- 13. Roeder AH, Yanofsky MF. Fruit development in Arabidopsis. Arabidopsis Book. Rockville, MD. 2006; 4:e0075. http://dx.doi.org/ 10.1199/tab.0075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galvan-Ampudia CS, Offringa R, Friml J, Yanofsky MF, Østergaard L. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 2009; 459:583-6; PMID:19478783; http://dx.doi.org/ 10.1038/nature07875 [DOI] [PubMed] [Google Scholar]
- 15. Rajani S, Sundaresan V. The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr Biol 2001; 11:1914-22; PMID:11747817 [DOI] [PubMed] [Google Scholar]
- 16. Spence J, Vercher Y, Gates P, Harris N. 'Pod shatter' in Arabidopsis thaliana, Brassica napus and B. juncea. J Microsc 1996; 181:195-203. [Google Scholar]
- 17. Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 1998; 125:1509-17; PMID:9502732 [DOI] [PubMed] [Google Scholar]
- 18. Ferrándiz C, Liljegren SJ, Yanofsky MF. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 2000; 289:436-8; PMID:10903201 [DOI] [PubMed] [Google Scholar]
- 19. Meakin PJ, Roberts JA. Dehiscence of fruit in oilseed rape (Brassica napus L.): II. The role of cell wall degrading enzymes and ethylene. J Exp Bot 1990; 41:1003-11. [Google Scholar]
- 20. Meakin PJ, Roberts JA. Anatomical and biochemical changes associated with the induction of oilseed rape (Brassica napus) pod dehiscence by Dasineura brassicae (Winn.). Ann Bot 1991; 67:193-7. [Google Scholar]
- 21. Bonghi ZC, Casadoro G, Ramina A, Rascio N. Abscission in leaf and fruit explants of Prunus persica (L.) Batsch. New Phytol 1993; 123:555-65. [DOI] [PubMed] [Google Scholar]
- 22. Cho H-T, Cosgrove DJ. Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 2000; 97:9783-8; PMID:10931949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sander L, Child R, Ulvskov P, Albrechtsen M, Borkhardt B. Analysis of a dehiscence zone endo-polygalacturonase in oilseed rape (Brassica napus) and Arabidopsis thaliana: evidence for roles in cell separation in dehiscence and abscission zones, and in stylar tissues during pollen tube growth. Plant Mol Biol 2001; 46:469-79; PMID:11485203 [DOI] [PubMed] [Google Scholar]
- 24. Squires TM, Gruwel MLH, Zhou R, Sokhansanj S, Abrams SR, Cutler AJ. Dehydration and dehiscence in siliques of Brassica napus and Brassica rapa. Can J Bot 2003; 81:248-54. [Google Scholar]
- 25. Arnaud N, Girin T, Sorefan K, Fuentes S, Wood TA, Lawrenson T, Sablowski R, Østergaard L. Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev 2010; 24:2127-32; PMID:20889713; http://dx.doi.org/ 10.1101/gad.593410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Child R, Chauvaux N, John K, Ulvskov P, van Onckelen H. Ethylene biosynthesis in oilseed rape pods in relation to pod shatter. J Exp Bot 1998; 49:829-38. [Google Scholar]
- 26. Ferrándiz C. Regulation of fruit dehiscence in Arabidopsis. J Exp Bot 2002; 53:2031-8; PMID:12324527 [DOI] [PubMed] [Google Scholar]
- 27. Johnson-Flanagan AM, Spencer MS. Ethylene production during development of mustard (Brassica juncea) and canola (Brassica napus) seed. Plant Physiol 1994; 106:601-6; PMID:12232353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang D, Koh C, Feurtado JA, Tsang E, Cutler AJ. MicroRNAs and their putative targets in Brassica napus seed maturation. BMC Genomics 2013; 14:140-64; PMID:23448243; http://dx.doi.org/ 10.1186/1471-2164-14-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fei H, Ferhatoglu Y, Tsang E, Huang D, Cutler AJ. Metabolic and hormonal processes associated with the induction of secondary dormancy in Brassica napus seeds. Botany 2009; 87:585-96. [Google Scholar]
- 30. Ruuska SA, Girke T, Benning C, Ohlrogge JB. Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 2002; 14:1191-206; PMID:12084821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carol W, Thomas JWY, Anthony DS, Vicky B-W, Jeremy AR. A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J 2009; 57:690-705; PMID:18980641; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03722.x [DOI] [PubMed] [Google Scholar]
- 32. Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L. Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. The Arabidosis Book. Rockville, MD. 2008; 6:e0113; PMID:22303238; http://dx.doi.org/ 10.1199/tab.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roeder AHK, Ferrándiz C, Yanofsky MF. The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr Biol 2003; 13:1630-5; PMID:13678595 [DOI] [PubMed] [Google Scholar]
- 34. Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LGG, Rensing SA, Kersten B, Mueller-Roeber B. PlnTFDB: updated content and new features of the plant transcription factor database. Nucl Acids Res 2010; 38:D822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Herrmann KM, Weaver LM. The shikimate pathway. Annu Rev Plant Physiol Plant Mol Biol 1999; 50:473-503; PMID:15012217 [DOI] [PubMed] [Google Scholar]
- 36. Ruegger M, Meyer K, Cusumano JC, Chapple C. Regulation of ferulate-5-hydroxylase expression in Arabidopsis in the context of sinapate ester biosynthesis. Plant Physiol 1999; 119:101-10; PMID:9880351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev of Plant Biol 2003; 54:519-46. [DOI] [PubMed] [Google Scholar]
- 38. Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 2009; 344:1879-900; PMID:19616198; http://dx.doi.org/ 10.1016/j.carres.2009.05.021 [DOI] [PubMed] [Google Scholar]
- 39. Benen JAE, Kester HCM, Visser J. Kinetic characterization of Aspergillus niger N400 endopolygalacturonases I, II and C. Eur J Biochem 1999; 259:577-85; PMID:10092840 [DOI] [PubMed] [Google Scholar]
- 40. Stolle-Smits T, Beekhuizen JG, Kok MTC, Pijnenburg M, Recourt K, Derksen J, Voragen AGJ. Changes in cell wall polysaccharides of green bean pods during development. Plant Physiol 1999; 121:363-72; PMID:10517827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ridley BL, O'Neill MA, Mohnen D. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001; 57:929-67; PMID:11423142 [DOI] [PubMed] [Google Scholar]
- 42. Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G. Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 2003; 15:93-106; PMID:12509524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rose JKC, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 2002; 43:1421-35; PMID:12514239 [DOI] [PubMed] [Google Scholar]
- 44. Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 2004; 37:914-39; PMID:14996223 [DOI] [PubMed] [Google Scholar]
- 45. Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 2005; 42:567-85; PMID:15860015 [DOI] [PubMed] [Google Scholar]
- 46. Jackson RG, Lim EK, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem 2001; 276:4350-6; PMID:11042207 [DOI] [PubMed] [Google Scholar]
- 47. Leyser O. Dynamic integration of auxin transport and signalling. Curr Biol 2006; 16:R424-33; PMID:16753558 [DOI] [PubMed] [Google Scholar]
- 48. Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005; 17:616-27; PMID:15659623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Binder BM, Walker JM, Gagne JM, Emborg TJ, Hemmann G, Bleecker AB, Vierstra RD. The Arabidopsis EIN3 binding F-box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 2007; 19:509-23; PMID:17307926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang KLC, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 2004; 428:945-950. [DOI] [PubMed] [Google Scholar]
- 51. Rzewuski G, Sauter M. Ethylene biosynthesis and signaling in rice. Plant Sci 2007; 175:32-42. [Google Scholar]
- 52. Chico JM, Chini A, Fonseca S, Solano R. JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol 2008; 11:486-94; PMID:18653378; http://dx.doi.org/ 10.1016/j.pbi.2008.06.003 [DOI] [PubMed] [Google Scholar]
- 53. Browse J. The power of mutants for investigating jasmonate biosynthesis and signaling. Phytochemistry 2009; 70:1539-46; PMID:19740496; http://dx.doi.org/ 10.1016/j.phytochem.2009.08.004 [DOI] [PubMed] [Google Scholar]
- 54. Nambara E, Marion-Poll A. Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 2005; 56:165-85; PMID:15862093 [DOI] [PubMed] [Google Scholar]
- 55. Zou J, Abrams GD, Barton DL, Taylor DC, Pomeroy MK, Abrams SR. Induction of lipid and oleosin biosynthesis by (+)-abscisic acid and its metabolites in microspore-derived embryos of Brassica napus L.cv Reston. Biological responses in the presence of 8'-hydroxyabscisic acid. Plant Physiol 1995; 108:563-71; PMID:12228493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiang L, Abrams SR, Kermode AR. Vicilin and napin storage-protein gene promoters are responsive to abscisic acid in developing transgenic tobacco seed but lose sensitivity following premature desiccation. Plant Physiol 1996; 110:1135-44; PMID:12226247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang D, Jaradat MR, Wu W, Ambrose SJ, Ross AR, Abrams SR, Cutler AJ. Structural analogs of ABA reveal novel features of ABA perception and signaling in Arabidopsis. Plant J 2007; 50:414-28; PMID:17376162 [DOI] [PubMed] [Google Scholar]
- 58. Hundertmark M, Hincha D. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 2008; 9:118-39; PMID:18318901; http://dx.doi.org/ 10.1186/1471-2164-9-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003; 301:653-7; PMID:12893945 [DOI] [PubMed] [Google Scholar]
- 60. Suzuki Y, Kawazu T, Koyama H. RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 2004; 37:542-4; PMID:15517963 [DOI] [PubMed] [Google Scholar]
- 61. Saal LH, Troein C, Vallon-Christersson J, Gruvberger S, Borg A, Peterson C. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol 2002; 3:software0003.1-software0003.6; PMID:12186655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. King JY, Ferrara R, Tabibiazar R, Spin JM, Chen MM, Kuchinsky A, Vailaya A, Kincaid R, Tsalenko A, Deng DX, et al. Pathway analysis of coronary atherosclerosis. Physiol Genomics 2005; 23:103-18; PMID:15942018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




