Abstract
We present an efficient method for the production of transgenic salt tolerant hexaploid wheat plants expressing the Arabidopsis AtNHX1 gene. Wheat mature zygotic embryos were isolated from two hexaploid bread wheat (Triticum aestivum) cultivars (namely: Gemmeiza 9 and Gemmeiza 10) and were transformed with the A. tumefaciens LBA4404 harboring the pBI-121 vector containing the AtNHX1 gene. Transgenic wheat lines that express the gus intron was obtained and used as control. The results confirmed that npt-II gene could be transmitted and expressed in the T2 following 3:1 Mendelian segregation while the control plant couldn't. The data indicate that, the AtNHX1 gene was integrated in a stable manner into the wheat genome and the corresponding transcripts were expressed. The transformation efficiency was 5.7 and 7.5% for cultivars Gemmeiza 10 and Gemmeiza 9, respectively. A greenhouse experiment was conducted to investigate the effect of AtNHX1 gene in wheat salt tolerance. The transgenic wheat lines could maintain high growth rate under salt stress condition (350 mM NaCl) while the control plant couldn’t. The results confirmed that Na+/H+ antiporter gene AtNHX1 increased salt tolerance by increasing Na+ accumulation and keeping K+/Na+ balance. Thus, transgenic plants showed high tolerance to salt stress and can be considered as a new genetic resource in breeding programs.
Keywords: Wheat plants, Agrobacterium, transformation, AtNHX1 gene expression, salt tolerance
Introduction
Environmental factors that impose water-deficit stress, such as drought, salinity and temperature extremes, place major limits on plant productivity worldwide.1 To overcome these limitations and production efficiency in the face of increasing world population, more stress tolerant crops must be developed.2 Traditional breeding strategies that have attempted to utilize genetic variation arising from varietal germplasm, interspesfic or intergeneric hybridization, induced mutations and somaclonal variations of cell and tissue cultures have met with only limited success; very few new plant introductions with improved stress resistance under field conditions have resulted.2,3
In Egypt wheat (Triticum aestivum L.) represents a major renewable resource for food, feed, and industrial raw materials, and is the most important grain crop. Egypt is an overpopulated country and there is a big gap between wheat production and consumption (www.fao.org). There are agricultural opportunities to increase wheat production by expanding into the new reclaimed regions. Wheat is classified as a semi tolerant crop to salinity. One way to alleviate the problem is the application of modern genetic techniques in breeding programs aiming to develop salt tolerant genotypes that perform better than current.4
Fortunately, plants have evolved several sophisticated systems to tolerate excessive salt. Among different salt-tolerance strategies plants use, sodium/proton antiporters (Na+/H+ exchangers, which can exclude Na+ from cytosol using proton gradient across membranes as driving force) which play crucial roles for plant’s survival under such condition.5 Therefore, the maintenance of ion homeostasis by the plants is essential to survive in these conditions. In order to avoid Na+ toxicity, the plant cell may either transport the ions outside the cell or store them inside the vacuole, processes mediated by specialized proteins. Some of them belong to the family of Na+/H+ exchangers, which can be located in the plasma membrane or vacuole.6
The AtNHX1 gene codes for the most abundant vacuolar Na+/H+ antiporter in Arabidopsis thaliana was expressed in wheat plants which improve grain yields under saline soils.7 Transgenic tomatoes overexpressing the AtNHX1 gene were able to grow, flower and produce seeds in the presence of 200 mM NaC1.8
The development of Agrobacterium-mediated transformation methods for wheat began in 19979 and for triticale eight years later.10 The main reason for applying this technique is its natural process of foreign DNA introduction into the plant genome, minimizing any rearrangements and multi-copy insertions.11 This was approved for wheat12 and barley.13 Since that time, several factors affecting the efficiency of transformation and the ability to transform new genotypes of wheat have been documented.14,15 These factors include the Agrobacterium strains, the binary vectors, the selectable genes and promoters, the inoculation method, and the in vitro culture conditions.15
Therefore, the present study was undertaken to transform two hexaploid wheat cultivars with AtNHX1 gene via Agrobacterium mediated gene transfer using an efficient and tissue culture-independent system, and to evaluate the salt tolerance in the transgenic plants in terms of biomass production.
Results and discussion
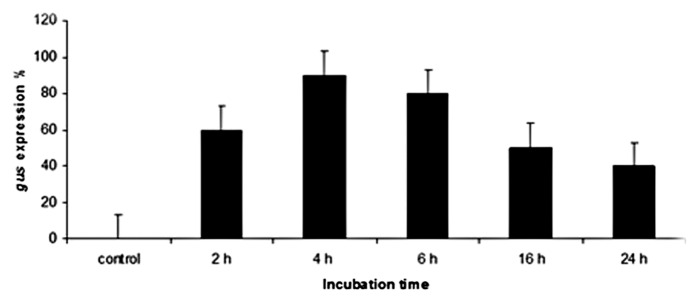
A potential limitation of plant transformation is the requirement for effective in vitro regeneration protocols, which are genotype-independent. Minimizing the role of tissue culture in the transformation procedure would therefore be advantageous under such circumstances.16,17 In the present study an efficient protocol for transformation which is quick, cultivar and tissue culture-independent in two hexaploid wheat cultivars was used. Mature zygotic embryo explants were inoculated with Agrobacterium strain LBA 4404 harboring the 35S-GUS-intron vector that carries the genes for β-glucuronidase (gus) and neomycin phosphotransferase (npt II). To determine the optimum co-cultivation time, the embryonic axes of mature seeds were immersed in a suspension of Agrobacterium in MS medium for 0, 2, 4, 6, 16 and 24 h respectively (Fig. 2). The data indicated that the gus gene expression increased with increasing incubation time from two to four hours, then it decreases with increasing the incubation period (Figs. 1 and 2A). The stable expression of gus gene was detected in transgenic T1 seeds and spikes by histochemical assay (Fig. 2B)

Figure 2. Histochemical gus assay showing gene expression (A) in transgenic plantlets while no expression can be detected in non-transgenic plant (C). (B) In transgenic T 1 seeds and spikes (B and C) while no expression can be detected in non-transgenic seeds and spikes (A).
Figure 1.Effect of co-cultivation time on transient GUS gene expression in wheat cultivar Gemmeiza 9.
Inheritance of thenpt-II gene in the T2 plants
The T2 plants derived from five transgenic lines from the cultivar Gimmaza-9 were analyzed for the inheritance of the npt-II gene. A 250 bp fragment equal to the molecular size of the npt-II gene was amplified from the total DNA of the transgenic plants. These DNA fragments were not detected in the DNA of non-transformed plants. The segregation patterns of 56 events are shown in Table 1. PCR positive and negative plants were clearly distinguishable by the presence of the specific bands of the npt-II gene. A 3:1 segregation ratio was observed for 21 out of 56 (37.5%) independent events (Table 1). An abnormal ratio 1:1 was observed for 35 out of 56 independent events (62.5%) which reflect its chimeric nature. These data confirmed the inheritance and integration of the npt-II gene in the T2 generation.
Table 1. Segregation analysis of npt-II gene among T2 progeny of transgenic events as revealed by PCR analysis.
| Wheat line | Total number of plants | Positive number | Negative number | Ratio | Calculated X2 |
|---|---|---|---|---|---|
| 1 2 3 4 5 |
10 11 18 9 8 |
7 9 10 5 4 |
3 2 8 4 4 |
3:1 3:1 1:1 1:1 1:1 |
0.16* 0.27 * 0.2 * 0.11 * 0.01* |
The improvement of plant salt-tolerance by the expression of a vacuolar Na+/H+ antiporter gene AtNHX1 from Arabidopsis has been reported in dicot species: A. thaliana,18 tomato19 and B. napus.20 In the present study the AtNHX1 gene was introduced to wheat plant genome through the Agrobacterium mediated gene delivery system.
Embryonic axes isolated from mature seeds of wheat cultivars Gemmeiza 9 and Gemmeiza 10 were incubated for four hours in a suspension culture of Agrobacterium harboring plasmid 35S AtNHX1 that contain the AtNHX1 gene under the genetic control of CaMV 35S promoter and nos terminator. Data for the different steps that had been undertaken to recover salt tolerant transgenic wheat plants are presented in Figure (3).

Figure 3. Recovery of fertile transgenic wheat plants expressing the AtNHX1 gene. (A) and (B) mature seeds, (C) embryonic excise, (D) inoculation step, (E-F) germination of transgenic plant, (G-I) acclimatization and maturation.
To confirm the stable integration of the T-DNA into the resulted wheat plantlets, genomic DNA samples isolated from T1 plants were subjected to PCR analysis using primers specific for the AtNHX1 gene and also for npt-II gene. The PCR analysis indicated that the majority of the tested plants showed a clear band corresponding to the relevant sequence of both of the npt-II gene (250 bp) and AtNHX1 gene (500 bp) (Fig. 4). The transformation percentages for the two tested wheat cultivars were 5.7% and 7.5% for the cultivars Gemmeiza 10 and Gemmeiza 9, respectively (Table 2).
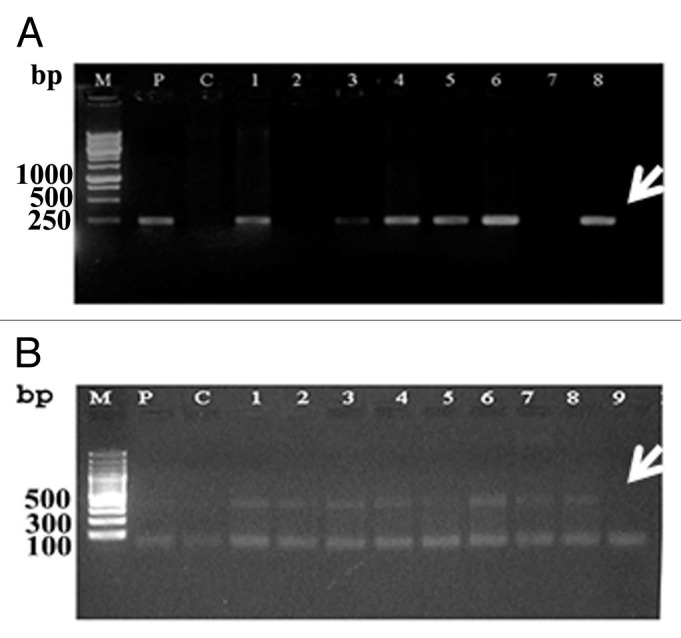
Figure 4. PCR analysis indicating the transformation status of wheat genome with the AtNHX1 gene. A and B PCR with AtNHX1 gene (500 bp) and npt-II (250bp), respectively. M, 100 bp DNA ladder; P, plasmid; C, control.
Table 2. Transformation percentages of two hexaploid wheat cultivars with ATNHX1.
|
Cultivar |
Number of explants used | Number of induced Shoots | PCR positive for npt-II | PCR positive for AtNHX1 | Trans. % |
|---|---|---|---|---|---|
| Gemmeiza 9 | 1900 | 1200 | 90 | 90 | 7.5 |
| Gemmeiza 10 | 963 | 1050 | 60 | 60 | 5.7 |
The T1 plants that show positive results with PCR analysis were self-pollinated to collect the T2 seeds. The inheritance of AtNHX1 gene into T2 plant genomes was confirmed by dot blot hybridization. Dot blot analysis indicated that AtNHX1 gene was transmitted into six out of the 11 transgenic lines tested (54.5%) (Fig. 5-a). Also the expression of the AtNHX1 gene in the transgenic wheat was confirmed by northern bloting analysis. The total mRNA was isolated from three transgenic lines and also from the non-transgenic plants (WT) and blot hybridized with the AtNHX1 probe. The data presented in Figure (5-b) indicate the presence of transcript that can hybridize specifically to the AtNHX1 gene accumulated in the T2 plants of the transgenic lines L-2 and L-3 confirming the correct integration and expression of the transgenes into their genomes. These two transgenic lines were selected for detailed physiological studies.
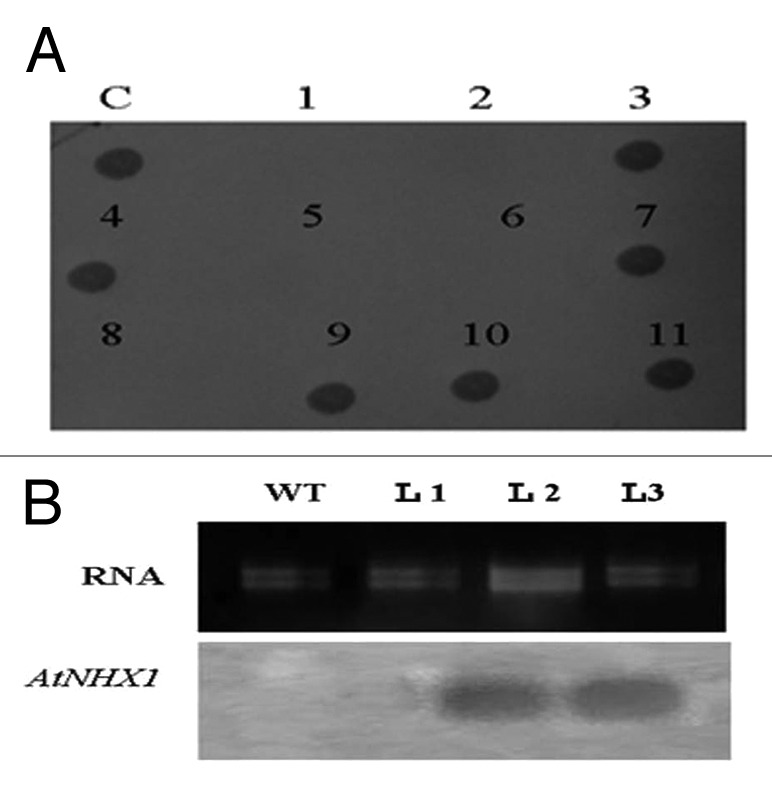
Figure 5. Dot blot analysis with AtNHX1 gene specific probe (A). Northern blot analysis confirming AtNHX1 gene expression in three transgenic lines of wheat. WT, non transgenic and T1-T3: transgenic lines (B).
AtNHX1-expressing wheat plants are more salt tolerant than wild-type
To test the salt tolerance of AtNHX1-expressing wheat plants, one month old seedlings of the two selected transgenic lines from the cultivars Gemmiza-9 were subjected to different concentrations of NaCl (0.0, 300, 350, 375 and 400 mM). The present data indicated that, with increasing salt concentration, the phenotypic differences between wild type and transgenic plants became apparent. When compared with wild-type controls, AtNHX1-expressing wheat plants displayed better growth under high salinity conditions (Fig. 6). For example the wild type plants showed growth retardation at 300 mM, and died at 350 mM while the transgenic plants could maintain higher growth rate under high salt concentrations (Fig. 6). The plant dry weight decreased with increasing NaCl concentration in the irrigation water but the decrease was more remarkable in the non-transgenic (WT) compared with the transgenic plants under the same salt concentration. The transgenic line-2 showed higher plant dry weight than the transgenic line-1 under high salt concentration (Fig. 7).

Figure 6. The effect of salt stress on growth rate of the wild type (WT) (upper panel) and AtNHX1 expressing wheat lines (lower panel).
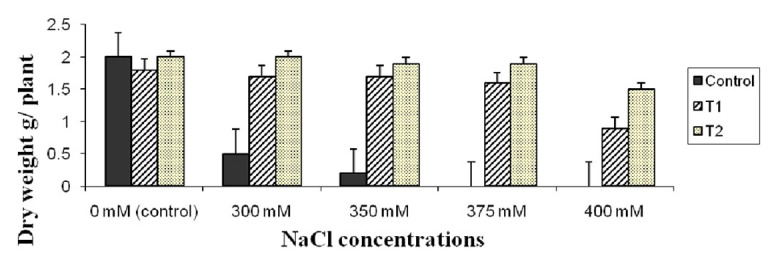
Figure 7. Plant dry weight of two wheat transgenic lines compared with the control (non-transgenic) under different NaCl concentrations. Data are averages of three replication ± SE
The data of the present study show that, by increasing salt concentration the AtNHX1 expressing wheat plants accumulate much higher Na+ concentration in their leaves compared with the wild type plants (Table 3), indicating that, the expression of AtNHX1 gene significantly improved salt tolerance in the transgenic lines. The transgenic and wild type plant leaves K+ concentration decrease by increasing NaCl concentration. There is a significant difference of K+ content between leaves of the wild type and transgenic plants under salinity stress condition (Table 5). According to Li et al. Twenty-six transgenic soybean plant cells could sequester efficiently the excessive sodium into vacuoles and decrease its cytosolic sodium concentration which could explain the observed higher growth rate in transgenic wheat plants under salt stress. Under high level of salt Na+ ions may displace K+ from its carrier binding sites and this competition results in impaired K+ uptake and lower cytosolic K+ concentrations. The results confirmed that AtNHX1 gene expression increased salt tolerance by increasing Na+ accumulation and keeping K+/Na+ balance. This agree with Banjara et al. Twenty-one who reported that, the accumulation of sodium in the vacuoles restores the correct osmolarity to the intracellular milieu, which favors water uptake by plant root cells and improves water retention in tissues under higher soil salinity. The results of the present study also agree with the previous reported data in which the transgenic plants that express AtNHX1, such as Arabidopsis18, tomato19, rapeseed20, cotton22, tobacco23, tall fescue24, Petunia hybrid25, soybean26, and peanut21 all displayed increased salt tolerance.
Table 3. Sodium, potassium and chloride concentrations in leaves of the transgenic wheat lines.
| Parameter | NaCl concentration (mM) | Control wheat | Transgenic wheat lines | |
|---|---|---|---|---|
| Line 1 | Line 2 | |||
| Sodium (mg g-1 DW) | 0.00 350 |
2.32 5.14 |
2.95 6.04 |
2.30 5.50 |
| Potassium (mg g-1 DW) | 0.00 350 |
1.50 1.12 |
1.71 1.13 |
1.56 1.26 |
| Chloride (mg g-1 DW) | 0.00 350 |
1.24 10.25 |
1.68 17.85 |
1.30 18.27 |
Materials and Methods
Plant material
Two cultivars of allohexaploid, spring wheat, Gemmeiza 9 and Gemmeiza 10 were used in the present study (Table 4).
Table 4. The pedigrees of the two wheat cultivars used.
| Cultivars | Pedigree |
|---|---|
| Gemmeiza 9 | ALD”S”//CMH74.A630/5X |
| Gemmeiza 10 | MAYA74”S”/ON//1160-147/3/Bb/4/CHAT”S”/5/CTOW |
Bacterial strains and vectors
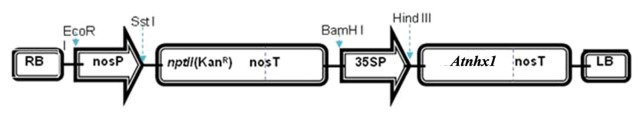
Agrobacterium strain LBA 4404 harboring the binary vector pBI-121 that carries the genes for β-glucuronidase (gus) and neomycin phosphotransferase (npt II) was used to generate transgenic line to be used as control. In order to develop transgenic salt tolerant wheat plants, the A. tumefacien strain LBA4404 harboring the binary vector pBI-121 that contains the At-NHX1 gene was used (Fig. 8). The strain harboring the previous constructs was kindly provided by Prof Dr. Ahmed E. Abo-Salha, Department of Genetics, Al-Minia University, Egypt.
Figure 8. Schematic representation of the transformation vector pBI-121 containing the ATNHX1 under the genetic control of 35S promoter.
Production of transgenic wheat plants
Wheat mature seeds were surface sterilized by dipping in 70% ethanol for one minute, followed by immersion in 20% sodium hypochlorite for 25min, and three rinses with sterile distilled water. To facilitate the isolation of wheat mature zygotic embryos, sterilized seeds were imbibed in sterile distilled water for overnight at 25 °C.
Wheat mature zygotic embryos were isolated under aseptic conditions using a sterile sewing needle and were dunked in the suspension of Agrobacterium in MS medium. The infection was performed by gentle agitation at 28 °C for 2, 4, 6, 16 and 24 h respectively. The explants were blot-dried, washed thoroughly with 500 mg l_1 of cefotaxime for 18 h and placed on MS medium for germination. After 6–10 d, the germinated seedlings were transferred to soilrite in pots and the seedlings were allowed to grow under growth room conditions for at least 10 d before they were transferred to the greenhouse. The pots were initially covered with polythene bags to maintain humidity. The growth chamber was maintained at 25 °C under a 14 h photoperiod with fluorescent light of intensity 35 mmol m_2 s_1. For each experiment 300 embryos were infected and the experiments were repeated three times.
Histochemical GUS assay
The histochemical assay to screen for the expression of β-glucurodinase (GUS) activity in the transgenic wheat plants was performed according to the method of Jefferson et al.27 For analysis, transgenic plant tissues were incubated in a reaction buffer containing 12.5 mM K3Fe (CN) 6, 12.5 mM K4Fe (CN) 6, 20% methanol, 1% Triton X-100 and 38.3 mM 5-bromo-4-chloro-3-indolyl glucuronide as a substrate for the enzyme. The tissues were incubated in a staining solution at 37 °C for 24 h and the developed blue spots were recorded.
Segregation analysis of the npt-II gene in the T2 transgenic wheat plants
In order to study the npt-II gene segregation in the next generation, seeds representing five transgenic lines obtained from the cultivar Gemmeiza 9 were used. Transgenic seeds from each line were germinated in plastic pots (3 L) each containing a mixture of sandy soil and peatmoss (1:1/ v: v). The temperature was 25 °C and the photosynthetically active radiation was 2743 μmole m−2s−1 (photosynthetic active radiation PAR). Three weeks after planting, the genomic DNA was isolated for PCR amplification. PCR reaction was conducted to ensure the presence of the npt-II gene in the genomic DNA of the T2 transgenic plants. The χ2 test was performed to test if segregation ratios were as expected.
PCR analysis
DNA was isolated from At-NHX1 transgenic plants (T2) and from control plants (gus expressing plants) according to the method described by Rogers and Bendich28 and were subjected to PCR analysis using specific primers for the At-NHX1 and npt-II genes. The sequences of the primers for the At-NHX1 gene detection were;
5′- TTTTGG CTT AAA TTC ATA TTC AA -3′ and 5′- GGC CGT TAA AGT GTC CATG -3′ respectively. The forward and reverse primers for the npt-II gene were 5′-CGC AGGTTC TCC GGCCGC TTG GTG G-3′ (position: 24–49) and 5′- TCG TCG GTC AGG GAAGGC GAA GTC-3′ (position: 254–277).
The reaction mixture (20 µl) contained 10 ng DNA, 200 mM dNTPs, 1 mM of each primer, 0.5 units of Red Hot Taq polymerase (ABgene Housse, UK) and 10-X Taq polymerase buffer (ABgene Housse, UK). The following profile was used for these reactions: 94 °C/1 min, followed by 30 cycles of 98 °C/20 s, 68 °C/ 1 min 30 s, and a final extension at 72 °C/10 min. PCR products were separated by (2%) agarose gel electrophoresis and visualized after ethidium bromide staining under UV light.
Dot bolt analysis
Total genomic DNA was denatured for 10 min., and spotted to a nylon membrane followed standard protocols. Labeling of the probes (AtNHX1 gene), hybridization and detection was performed using the Biotin Chromogenic Detection kit #K0661, #K0662 according to instructions provided by the manufacturer (Ferments Life Sciences, USA).
Northern blot analysis
Total RNA was isolated from leaf samples of both the transgenic and control plants using SV Total RNA Isolation System. (PROMEGA, cat. #Z3100 USA). RNA was separated by electrophoresis and transferred to Hybond NC nylon membrane (Amersham). Pre-hybridization and hybridization conditions were in strict accordance with the manufacturer’s recommendations. Labeling of the probes (AtNHX1 gene), hybridization and detection were performed using RPN 3450 Gene Image kit according to the manufacturer’s instructions (Amersham, Buckinghamshire, UK).
Evaluation of the salt tolerance enhancement in transgenic wheat plants
Salt-tolerance experiment
Control (gus transgenic plants) wheat seeds from the cultivar Gemmeiza 9and two independent AtNHX1 transgenic lines (T3) derived from the same cultivars were used. Fifteen seeds from each were planted in plastic pots (3 L) each containing a mixture of granite regosol, peat moss, and perlite (2:1:1, v:v:v), watered daily with 400 ml of 10% Hoagland solution and the soil water tension was maintained at ≤ 60 kP. At 21 d after planting, the plants were subjected to salt stress by the addition of 300, 350, 375 and 400 mM NaCl to the daily supply of Hoagland solution for 10 d. The temperature in the greenhouse was 25 °C, and the photosynthetically active radiation (PAR) was 2743 μmol m-2 s-1 (photosynthetic active radiation PAR). There were five replicates per treatment.
Measurement of plant dry weight
Plants were harvested 10 d after starting the salt treatment. The harvested plants were dried at 80 °C in an air-forced draft oven for more than three days, and then weighed.
Determination of ions concentration
Wheat leaves sodium, chloride and potassium contents were measured by HPLC analysis using a Waters Alliance system (Milford, MA) consisting of pump control separation module (Model 2690). A programmable photodiode array detector (Model 996), multi-wavelength UV absorbance detector (Model 490), an autosampler (Model 717). Samples were introduced via auto-injector with a 20 ml and all chromatographic separations were performed at ambient temperature. Data acquisition and analysis were performed on a computer using the Millenium 32 chromatography software (Waters Association), communicated with the HPLC equipment. Spectrophotometer UV2101- PC (Schimadzu, Japan) and centrifuge 5417 eppendorf were used for the analyses.
Statistical analysis
χ2 analysis was performed using Analyze-it software (Analyze-it, Leeds, UK) in accordance with the method of Maxwell and Delany.29
Acknowledgments
We would like to thank Prof Dr. Ahmed E. Abo-Salha, Department of Genetics, Al-Minia University, Egypt for providing the At-NHX1 gene.
References
- 1.Dixon J, Braun HJ, Crouch JH. Transitioning wheat research to serve the future needs of the developing world, in: J. Dixon, H.-J. Braum, P. Kosina, J.H. Crouch (Eds.), Wheat Facts and Futures, CIMMYT, Mexico DF, Mexico, 2009. pp. 1–25. [Google Scholar]
- 2.Flowers TJ. . Improving crop salt tolerance. J Exp Bot 2004; 55:307 - 19; http://dx.doi.org/ 10.1093/jxb/erh003; PMID: 14718494 [DOI] [PubMed] [Google Scholar]
- 3.Kebede B, Thiagarajah M, Zimmerli C, Rahman MH. . Improvement of open pollinated spring rapeseed (B. napus) through introgression of genetic diversity from winter rapeseed. Crop Sci 2010; 50:1236 - 43; http://dx.doi.org/ 10.2135/cropsci2009.06.0352 [DOI] [Google Scholar]
- 4.Moghaieb REA, Abdel-Hadi AA, Talaat NB. . Molecular markers associated with salt tolerance in Egyptian wheats. Afr J Biotechnol 2011; 10:18092 - 103 [Google Scholar]
- 5.Silva P, Gerós H. . Regulation by salt of vacuolar H+-ATPase and H+-pyrophosphatase activities and Na+/H+ exchange. Plant Signal Behav 2009; 4:718 - 26; http://dx.doi.org/ 10.4161/psb.4.8.9236; PMID: 19820346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horie T, Schroeder JI. . Sodium transporters in plants. Diverse genes and physiological functions. Plant Physiol 2004; 136:2457 - 62; http://dx.doi.org/ 10.1104/pp.104.046664; PMID: 15375202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue ZY, Zhi DY, Xue GP, Zhang H, Zhao YX, Xia GM. . Enhanced salt tolerance of transgenic wheat (Triticum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci 2004; 167:849 - 59; http://dx.doi.org/ 10.1016/j.plantsci.2004.05.034 [DOI] [Google Scholar]
- 8.Zhang HX, Blumwald E. . Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 2001; 19:765 - 8; http://dx.doi.org/ 10.1038/90824; PMID: 11479571 [DOI] [PubMed] [Google Scholar]
- 9.Cheng M, Fry JE, Pang S, Zhou H, Hironaka CM, Duncan DR, Conner TW, Wan Y. . Genetic transformation of wheat mediated by Agrobacterium tumefaciens.. Plant Physiol 1997; 115:971 - 80; PMID: 12223854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nadolska-Orczyk A, Przetakiewicz A, Kopera K, Binka A, Orczyk W. . Efficient method of Agrobacterium-mediated transformation for triticale (x Triticosecale Wittmack). J Plant Growth Regul 2005; 24:2 - 10; http://dx.doi.org/ 10.1007/s00344-004-0046-y [DOI] [Google Scholar]
- 11.Nadolska-Orczyk A, Orczyk W, Przetakiewicz A. . Agrobacterium mediated transformation of cereals—from technique development to its application. Acta Physiol Plant 2000; 22:77 - 88; http://dx.doi.org/ 10.1007/s11738-000-0011-8 [DOI] [Google Scholar]
- 12.Wu H, Sparks CA, Jones HD. . Characterisation of T-DNA loci and vector backbone sequences in transgenic wheat produced by Agrobacterium-mediated transformation. Mol Breed 2006; 18:195 - 208; http://dx.doi.org/ 10.1007/s11032-006-9027-0 [DOI] [Google Scholar]
- 13.Bartlett JG, Snape JW, Harwood WA. . Intron-mediated enhancement as a method for increasing transgene expression levels in barley. Plant Biotechnol J 2009; 7:856 - 66; http://dx.doi.org/ 10.1111/j.1467-7652.2009.00448.x; PMID: 19781005 [DOI] [PubMed] [Google Scholar]
- 14.Bhalla PL. . Genetic engineering of wheat--current challenges and opportunities. Trends Biotechnol 2006; 24:305 - 11; http://dx.doi.org/ 10.1016/j.tibtech.2006.04.008; PMID: 16682090 [DOI] [PubMed] [Google Scholar]
- 15.Shrawat AK, Lörz H. . Agrobacterium-mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnol J 2006; 4:575 - 603; http://dx.doi.org/ 10.1111/j.1467-7652.2006.00209.x; PMID: 17309731 [DOI] [PubMed] [Google Scholar]
- 16.Rohini VK, Rao KS. . Transformation of peanut (Arachis hypogaea L.): a non-tissue culture based approach for generating transgenic plants. Plant Sci 2000; 150:41 - 9; http://dx.doi.org/ 10.1016/S0168-9452(99)00160-0 [DOI] [Google Scholar]
- 17.Rakoczy-Trojanowska M. . The effects of growth regulators on somaclonal variation in rye (Secale cereale L.) and selection of somaclonal variants with increased agronomic traits. Cell Mol Biol Lett 2002; 7:1111 - 20; PMID: 12511978 [PubMed] [Google Scholar]
- 18.Apse MP, Aharon GS, Snedden WA, Blumwald E. . Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis.. Science 1999; 285:1256 - 8; http://dx.doi.org/ 10.1126/science.285.5431.1256; PMID: 10455050 [DOI] [PubMed] [Google Scholar]
- 19.Zhang HX, Blumwald E. . Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol 2001; 19:765 - 8; http://dx.doi.org/ 10.1038/90824; PMID: 11479571 [DOI] [PubMed] [Google Scholar]
- 20.Zhang HX, Hodson JN, Williams JP, Blumwald E. . Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc Natl Acad Sci U S A 2001; 98:12832 - 6; http://dx.doi.org/ 10.1073/pnas.231476498; PMID: 11606781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banjara M, Zhu L, Shen G, Payton P, Zhang H. . Expression of an Arabidopsis sodium/proton antiporter gene (AtNHX1) in peanut to improve salt tolerance. Plant Biotechnol. Rep. 2012; 6:59 - 67; http://dx.doi.org/ 10.1007/s11816-011-0200-5 [DOI] [Google Scholar]
- 22.He C, Yan J, Shen G, Fu L, Holaday AS, Auld D, Blumwald E, Zhang H. . Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant Cell Physiol 2005; 46:1848 - 54; http://dx.doi.org/ 10.1093/pcp/pci201; PMID: 16179357 [DOI] [PubMed] [Google Scholar]
- 23.Duan X, Song Y, Yang A, Zhang J. . The transgene pyramiding tobacco with betaine synthesis and heterologous expression of AtNHX1 is more tolerant to salt stress than either of the tobacco lines with betaine synthesis or AtNHX1. Physiol Plant 2009; 135:281 - 95; http://dx.doi.org/ 10.1111/j.1399-3054.2008.01194.x; PMID: 19236662 [DOI] [PubMed] [Google Scholar]
- 24.Tian L, Huang C, Yu R, Liang R, Li Z, Zhang L, Wang Y, Zhang X, Wu Z. . Overexpression of AtNHX1 confers salt-tolerance of transgenic tall fescue. Afr J Biotechnol 2006; 5:1041 - 4 [Google Scholar]
- 25.Xu K, Hong P, Luo L, Xia T. . Overexpression of AtNHX1, a vacuolar Na+/H+ antiporter from Arabidopsis thaliana, in Petunia hybrid enhances salt and drought tolerance. J Plant Biol 2009; 52:453 - 61; http://dx.doi.org/ 10.1007/s12374-009-9058-2 [DOI] [Google Scholar]
- 26.Li TX, Zhang Y, Liu H, Wu YT, Li WB, Zhang HX. . Stable expression of Arabidopsis vacuolar Na+/H+ antiporter gene AtNHX1, and salt tolerance in transgenic soybean for over six generations. Chin Sci Bull 2010; 55:1127 - 34; http://dx.doi.org/ 10.1007/s11434-010-0092-8 [DOI] [Google Scholar]
- 27.Jefferson RA, Kavanagh TA, Bevan MW. . GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 1987; 6:3901 - 7; PMID: 3327686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers SO, Bendich AJ. . Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol Biol 1985; 5:69 - 76; http://dx.doi.org/ 10.1007/BF00020088; PMID: 24306565 [DOI] [PubMed] [Google Scholar]
- 29.Maxwell SE, Delaney HD. Designing Experiments and Analyzing Data, 1989, pp. 241-260, Wadsworth, Belmont, CA, USA. [Google Scholar]