Abstract
In China, species of Atraphaxis (Polygonaceae) primarily inhabit arid zones across temperate steppe and desert regions. The complex geologic history (e.g., expansion of deserts) and extreme climate shifts of the region appear to have played an important role in shaping the phylogeography of Atraphaxis. The present study focuses on species-level phylogeographic patterns of Atraphaxis in China, with the goal of determining the impact of past environmental changes, in northern China, on the evolutionary history of the genus. Five hundred and sixty-four individuals distributed among 71 populations of 11 species of Atraphaxis from across the geographic range of the genus were studied using sequence data from two plastid spacers, psbK-psbI and psbB-psbH. The results demonstrate that most chloroplast haplotypes are species-specific, except for some present among widespread species. The phylogeny of Atraphaxis was well structured, and molecular dating analyses suggest that the main divergence events occurred during the late Pliocene and Pleistocene (5.73–0.03 million years ago). The statistical dispersal-vicariance analysis (S-DIVA) results provide evidence that phylogeographic patterns for the genus were characterized by both vicariance events and regional dispersal. The presented data suggest that the rapid expansion of deserts and climatic changes in northern China during the late Pliocene and Pleistocene have driven the diversification and spread of Atraphaxis in the region. The expansion of the Tengger Desert provided appropriate conditions for the origin of A. bracteata. Additionally, a contact zone in the north of the Hexi Corridor was identified as having played a significant role as a migratory route for species in adjacent areas.
Introduction
The Pleistocene was characterized by extensive oscillations of the global climate that strongly affected the evolution and geographic distributions of organisms and communities [1,2]. The effects of these Pleistocene climate oscillations on phylogeographic patterns throughout temperate alpine and high-latitude regions have been well supported [3]. However, the impact of these oscillations remains poorly explored on the phylogeographic patterns of species in arid areas. The limited number of phylogeographic studies suggest that Pleistocene climatic cycles associated with habitat fragmentation are important environmental factors that shaped the dynamic distribution and genetic patterns of drought-resistant desert plants in the northern hemisphere [4–6]. The few studies that have been conducted on the impact of Pleistocene glaciations in arid regions provide evidence that desert plants suffered from increased aridity and lower temperatures [2,7], and, as a result, suitable habitat for these plants was reduced and fragmented, which promoted allopatric divergence among isolated populations and, in some instances, drove speciation [7]. Throughout periods of glaciation, glacial refugia have served as important reservoirs for biological diversity [8,9], and during interglacial periods when the climate became warmer and more humid, many lineages left glacial refugia and recolonized and diversified into newly available habitats [10]. Thus, the climatic oscillations during the Pleistocene helped drive species diversification and migration, processes that can be critically examined using phylogeographic analyses to better understand the patterns of spatial arrangements and genetic history of species [11,12]. Phylogeographic studies describe historical changes at spatial and temporal scales that impact patterns of gene flow, isolation, and migration within a species or group of closely related species. These studies can provide evidence for the location of glacial refugia and routes of colonization and range expansion after glacial periods [13–17].
Northern China, part of arid central Asia, has regional variation in aridity, ranging from the typical arid region of northwestern China (annual precipitation ≤200 mm) to the eastern semi-arid sandlands (annual precipitation ≤400 mm) [18]. The arid conditions prevailing over much of the region date back to the early Miocene, and these conditions were strongly influenced by both the uplift of the Qinghai-Tibet Plateau (QTP) and the westward regression of the Palaeo-Mediterranean [19–21]. The uplift of the QTP obstructed sea breezes, from the Indian Ocean, from reaching northern China, and this resulted in the East Asian monsoons from the Pacific Ocean restricting precipitation far inland. Consequently, the decreased precipitation accelerated the formation of ancient deserts, such as the Taklimakan, Gurbantunggut and Badain Jaran-Tengger deserts [22]. During the Pleistocene, these deserts increased in aridity and expanded dramatically [23]. Computer simulations provided additional evidence that the expansion of deserts in China during the Quaternary glacial periods arose via a decrease in the strength of the horizontal winds, enhanced atmospheric subsidence, and reduced moisture transport [22]. Additionally, rapid orogenic movements in the Tianshan and other mountain ranges during the Pleistocene caused local rain shadows and promoted extreme local aridity in some areas [22]. Thus, the extensive perturbations of the climate coupled with geological movements during the Pleistocene sharply increased aridity in northwestern China [24,25].
Despite the dry conditions, arid northern China exhibits outstanding floristic diversity, with many species endemic to the area [26,27]. Plant diversity in the arid core of northern China is largely restricted to the alpine melt ecosystems and areas of sporadic seasonal precipitation. From the perspective of phytogeographical studies, arid northwestern China belongs to the “Irano-Turanian Region” [28]. Except for some ancient relict species (e.g., Potaninia, Tetraena) [18], evidence suggests most species (e.g., Salsola, Tamarix, Zygophyllum) originated and migrated from central Asia. The eastern sandlands of this region are part of the “Mongolia Flora” and species that inhabit the sandlands are hypothesized to have primarily migrated from Mongolia and East Asia [18].
Both increased aridification and range contraction and isolation during dry/cold episodes in glacial cycles might have played a major role in promoting population divergence and speciation in arid northern China. Genetic evidence of population fragmentation is available for Tetraena mongolica Maxim., Gymnocarpos przewalskii Maxim., Lagochilus Bunge ex Benth., and Atraphaxis frutescens (L.) K. Koch [22,29–31], and these arid plants currently have limited distributions in Taklimakan Desert, Hexi Corridor, and Alashan desert. Very little is known concerning the response to past environmental changes of arid species that have continuous geographic distributions across northern China. Given the small amount of available data, it can be postulated that these plant species were affected by climate changes during glacial cycles, and these plants followed migratory routes in response to changes in the climate; however, these postulations have yet to be tested.
To better understand the impact of these environmental changes on the diversification of arid northern China, we examined the evolutionary history of Atraphaxis L. (Polygonaceae). The genus includes approximately 25 species primarily distributed throughout northern Africa and western and central Asia [32,33]. In arid northern China, Atraphaxis is one of the most representative and diverse plant genera, with ca. 11 species (A. laetevirem, A. jrtyschensis, A. pungens, A. pyrifolia, A.decipiens, A. frutescens, A. bracteata, A. compacta, A. canescens, A. spinosa, A. manshurica) [32] distributed in the deserts of northwestern China as well as extending eastwards into the semi-arid monsoon region of eastern China. Previous studies [32,33] have suggested that Atraphaxis originated in central Asia, with a few species expanding to northern China. In China, Atraphaxis primarily occurs in northern China, including ten species in the arid northwestern part of the country and one species, A. manshurica, distributed in the semi-arid Horqin sandlands [32]. Three species in the genus present in northern China, A. jrtyschensis, A. bracteata and A. manshurica, are endemic to the country.
Atraphaxis is highly drought-tolerant and inhabits areas along the foothills of mountains and edges of deserts [33]. Recent studies of phylogenetic relationships within the genus, using chloroplast DNA (cpDNA) and nuclear ribosomal DNA regions [34,35], suggest that Atraphaxis is monophyletic, but the patterns of temporal and geographical diversification remain poorly understood. The present study aims to investigate these patterns, with the goal of providing a more comprehensive historical perspective on both the biota and geological evolution of species in the arid northern China.
To tackle this project, we employed cpDNA sequence data to infer genetic patterns and population responses of species of Atraphaxis to past environmental changes throughout arid northern China. The palaeoclimatic scenario proposed for the region by Meng and Zhang [22] will allow for an interpretation of phylogeographic patterns, derived from molecular markers, in a specific environmental context. Together, this information will help determine the impact of past environmental changes, in northern China, on species of Atraphaxis.
Material and Methods
Sample collection
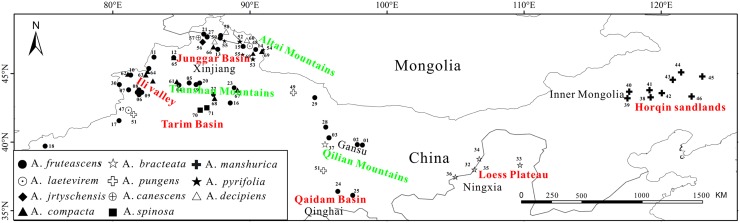
A total of 564 individuals representing 11 species of Atraphaxis were sampled from 71 populations throughout northern China (Table 1, Fig 1). The sampling included: A. jrtyschensis from Burqin County in Xinjiang Province; A. bracteata collected in sandy areas of Inner Mongolia, Ningxia, Shaanxi and Gansu Province; A. manshurica from the Horqin sandy and its adjacent regions; the dominant species, A. frutescens, sampled from throughout Xinjiang, Gansu, Qinghai Province; and seven species (A. canescens, A. compacta, A.decipiens, A. laetevirem, A. pungens, A. pyrifolia, and A. spinosa)_sampled in and around the Tianshan and Altai Mountains. This sampling scheme encompassed most of the natural geographic distribution of each species in China. In previous phylogenetic analyses of Atraphaxideae (Polygonaceae) based on five cpDNA regions, species of Atraphaxis in China were resolved as a well-supported monophyletic group [36]. Sample size per population ranged from two to 18 individuals, according to population density. The latitude and longitude of each locality were recorded using a global positioning system (GPS). Silica-dried tissues (leaves and/or flowers from each individual) were collected for DNA extraction. Voucher specimens for all 71 populations are deposited in the Herbarium of the Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences (XJBI). Our study did not concern Human Subject Research or Animal Research. We can confirm that the leaf materials did not come from conservation parks, and none of the samples involved endangered or protected species
Table 1. Details of Atraphaxis populations in study, sample sizes and cpDNA haplotypes observed.
| Species | n | h(±SD) | π(±SD) (×10−3) | mean number of pairwise differences |
|---|---|---|---|---|
| A. frutescens | 256 | 0.8365±0.0080 | 105.211±55.310 | 6.4178±3.050 |
| A. bracteata | 40 | 0.3576±0.0911 | 23.992±16.387 | 1.5115±0.9292 |
| A. manshurica | 104 | 0.0752±0.0354 | 2.567±3.809 | 0.1720±0.2304 |
| A. laetevirem | 13 | 0.6154±0.0782 | 11.349±10.310 | 0.6923±0.5595 |
| A. pungens | 22 | 0.6277±0.0602 | 29.806±19.897 | 1.8182±1.0878 |
| A. pyrifolia | 25 | 0.5267±0.0836 | 80.098±44.464 | 5.4467±2.7132 |
| A. jrtyschensis | 5 | 0 | 0 | 0 |
| A. canescens | 2 | 0 | 0 | 0 |
| A. decipiens | 16 | 0.5417±0.0985 | 54.645±33.148 | 3.3333±1.8052 |
| A. compacta | 66 | 0.4103±0.0709 | 39.213±23.797 | 2.4704±1.3522 |
| A. spinosa | 15 | 0 | 0 | 0 |
| Total | 564 | 0.8969±0.0069 | 7.234±3.748 | 7.3712±3.4531 |
| West region | 357 | 0.9138±0.0055 | 7.054±3.666 | 7.1883±3.3775 |
| Central region | 103 | 0.5877±0.0368 | 127.597±65.858 | 8.5490±3.9840 |
| East region | 104 | 0.0390±0.0267 | 2.049±3.368 | 0.1373±0.2038 |
Fig 1. Sampling locations and distributions of populations in 11 species of Atraphaxis in arid northern China.
Population codes are consistent with population names in Table 1.
DNA extraction, amplification and sequencing
Total genomic DNA was extracted, using a modified CTAB method [37], from silica-dried tissues of the 564 individuals of Atraphaxis. To assess phylogeographic patterns, we amplified and sequenced two cpDNA intergenic spacers, psbK-psbI [38] and psbB-psbH [39]. Each PCR amplification was implemented in a 30 μl volume including 2 μl of 10×PCR reaction buffer, 2 μl of 25mM MgCl2, 1.8 μl of each primer at 50 ng/μl, 2.0 μl of 2.5mM dNTP, 0.5 μl of Taq DNA-polymerase, and 1μl of genomic DNA at 60 ng/μl. The cycling procedures of the reaction were as follows: initial denaturation at 95°C for 4 minutes (min), followed by 32 cycles of 30 seconds (s) at 94°C, 30 s at 52°C, 60 s at 72°C, and a final extension step of 10 min at 72°C. PCR products were visualized on agarose gels and purified using an ethanol precipitation prior to sequencing [40].
Sequencing was carried out by Sangon Biotech (Shanghai) Co., Ltd., China using an ABI 3730XL automatic sequencer (Applied Biosystems, Foster City, CA, USA). Sample sequences were edited in Seqman (Lasergene, DNASTAR Inc., Madison, Wisconsin, USA), aligned using CLUSTAL_W [41], and then adjusted manually to ensure accurate allelic sequences. All obtained sequences are deposited in GenBank with accession numbers KM452785-KM452792, KX236017-KX236033, KR183867, KR183868, KR183870 for psbK-psbI; and KM452793-KM452796, KR183863, KR183864, KR183866, KX236034-KX236045 for psbB-psbH (S2 Table).
Analysis of genetic diversity
The psbK-psbI and psbB-psbH sequences were concatenated for analysis. DnaSP v. 5.0 [42] was utilized to estimate nucleotide polymorphism at all sites in a sequence. For the sequence data set, we calculated haplotype diversities (h) and nucleotide diversities (π) using ARLEQUIN 3.5 [43]. These measurements of molecular diversity reflect both the overall number of haplotypes and their molecular divergence [9]. We subdivided the distribution range into three approximately equal geographical regions based on mean humid conditions, and these closely corresponded to longitudinal regions: West (80–95°E, includes the Tarim and Junggar Basin; extremely arid conditions), Central (95–115°E, includes the Hexi Corridor, Alashan mountains, and Muus desert; arid conditions), and East (115–125°E, includes the Horqin sandlands; semi-arid conditions). Genetic variation in different regions and species was estimated using AMOVA, as implemented in ARLEQUIN 3.5, with significance tested via 10,000 permutations [44]. Permut version 1.0 [45] was employed to estimate within-population diversity (GST, NST) and total gene diversity (HS, HT), using 10,000 permutations. The GST and NST coefficients estimate the ratio between the mean within-population genetic diversity and total genetic diversity. While the GST index makes use only of the allelic frequencies, NST also takes into account the genetic distances among haplotypes. A higher NST signifies that more closely related haplotypes occur in the same population, and this indicates the presence of phylogeographic structure [45].
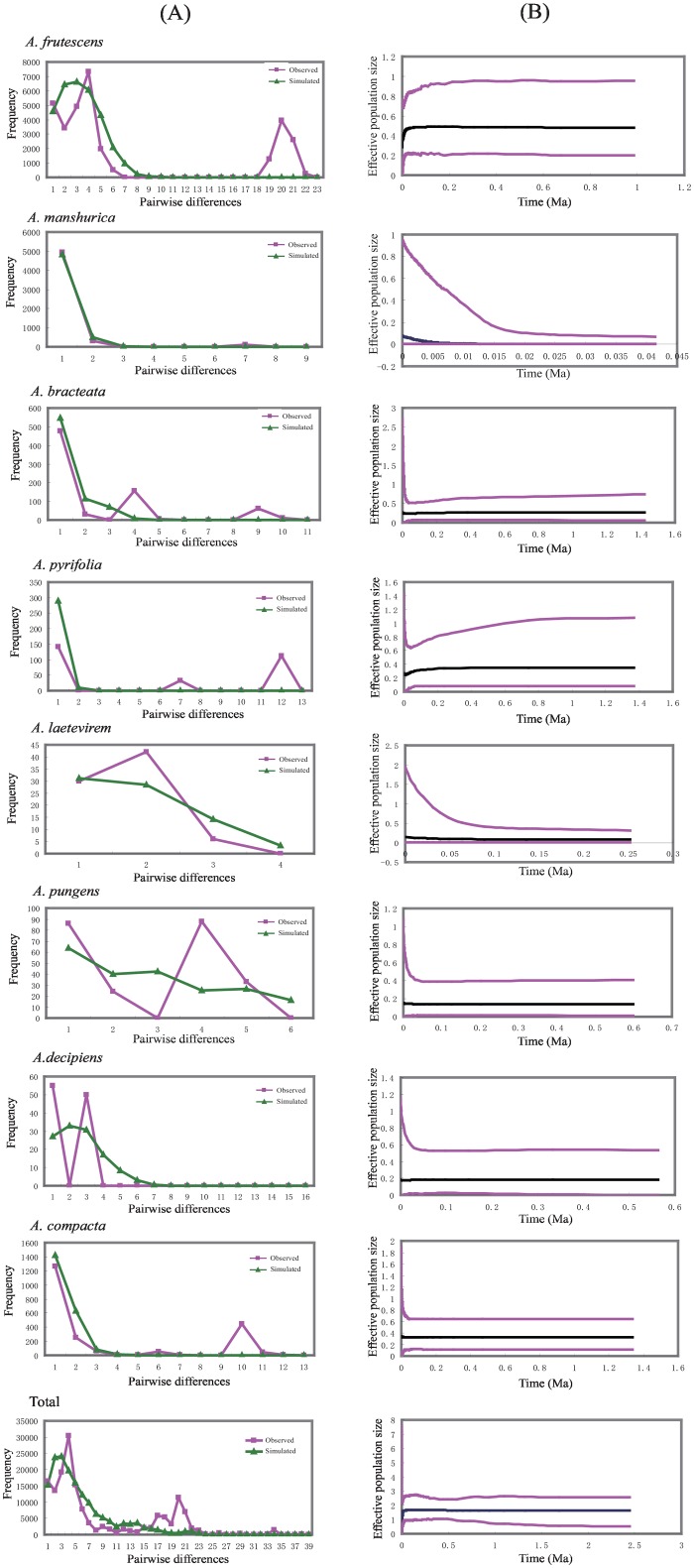
To test for demographic history, the mismatch distribution and neutrality tests (Tajima’s D and Fu’s Fs) were performed using ARLEQUIN 3.5. The significance of these tests was evaluated as the proportion of random statistics less than or equal to the observed value. For the mismatch distribution, a “ragged” multimodal distribution generally indicates that a population is in demographic equilibrium, whereas a smooth unimodal distribution is associated with recent demographic or range expansion [46]. We tested hypotheses of expansion for all populations of Atraphaxis as well as for populations for each species. In addition, in order to take full advantage of historical signals within DNA sequences, the estimates of changes in demographic growth over the history of major areas and the historical demographic dynamics of Atraphaxis were inferred via Extended Bayesian skyline plot (EBSP) analyses using BEAST version 1.5.4 [47]. The EBSP analyses are useful because two chloroplast sequences are employed to estimate effective population size through time. Linear and stepwise models were explored using an uncorrelated lognormal relaxed clock. Runs consisted of 10,000,000 generations, with trees sampled every 1,000 generations. The EBSP was visualized in the program Tracer version 1.5.
Bayesian divergence time estimations and network representation
Approximate divergence times among haplotypes were estimated using a Bayesian log-normal relaxed clock method, as implemented in BEAST version 1.5.4 [47]. We used the Bayesian Information Criterion (BIC) to estimate the best substitution model for the BEAST analyses, which was determined via Modeltest version 3.7 [48]. We used a coalescent model, a GTR substitution model with a Gamma site heterogeneity model, and the uncorrelated log-normal relaxed clock. Due to the lack of fossil data for the genus, along with no specific substitution rates available, we used a range (1.0–10.0×10−3 s/s/million) of nucleotide evolution for the genus to estimate divergence times [49,50]. The MCMC simulation was run for 10,000,000 generations, with trees sampled every 1,000 generations and the first 20% of generations were discarded as burnin. Two independent runs achieved the same results. The results were viewed in Tracer version 1.5 to check for convergence to a stationary distribution and for an effective sample size (ESSs) >200. Logs and tree files were combined in Logcombiner version 1.5.4 and summarized in Treeanotator 1.5.4 [47]. Final trees were viewed and edited in FigTree version 1.3.1 (http://tree.bio.ed.as.uk/software/figtree/). The phylogenetic tree was reconstructed without the inclusion of some individuals from species (populations FYD, QKE, SRBP, ALT) that had overlapping distributions as well as widespread species (species A. frutescens and A. compacta; see S3 Table). This approach was undertaken to reduce the impact of introgression on the resulting phylogeny [18].
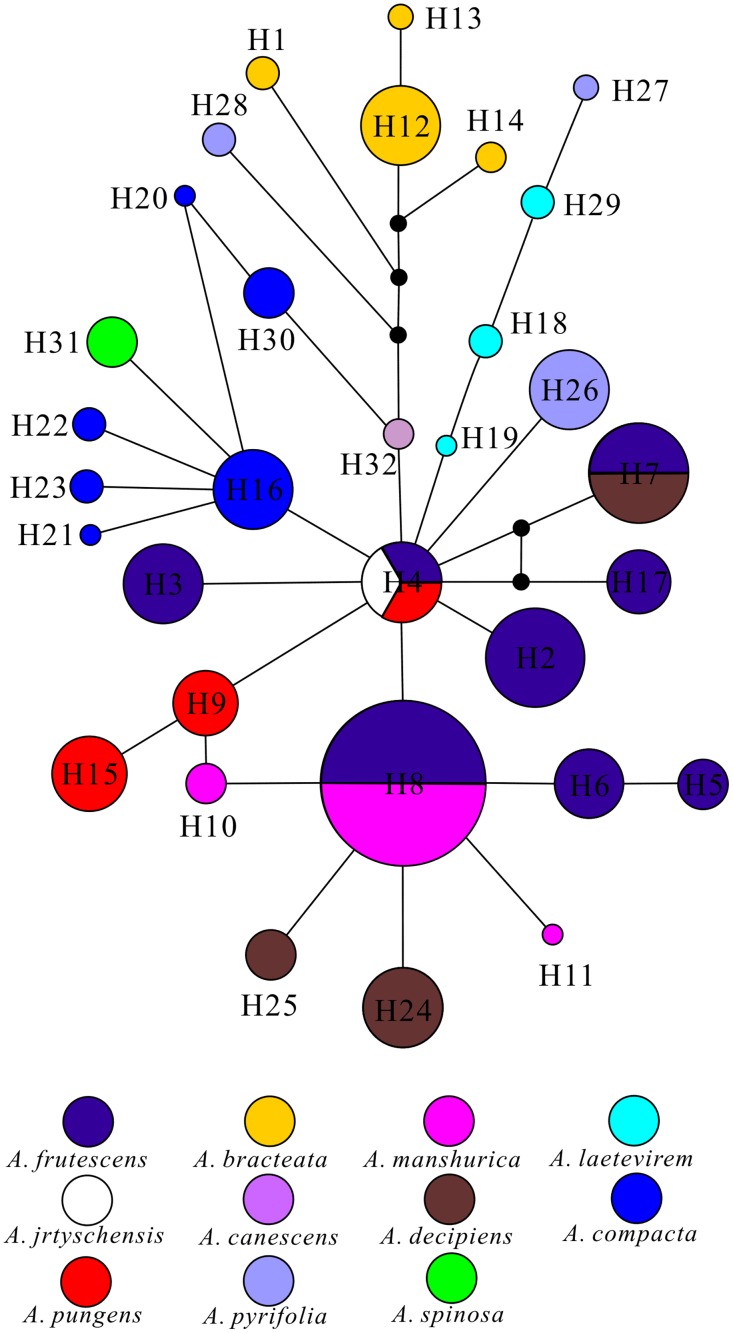
To investigate relatedness among haplotypes, we resolved a median-joining (MJ) network using Network version 4.6.0.0 [51]. This network can display genealogical relationships and reticulation among haplotypes that tree-building methods may not detect (Fig 2).
Fig 2. Median-joining network of 32 haplotypes of Atraphaxis sampled in northern China.
Each circle (H1-H32) represents a unique haplotype, with circle size reflecting haplotype frequencies. Black circles indicate potentially unsampled or extinct haplotypes. The colors within each species are consistent in all Figures.
Ancestral area optimization
Although many biogeographic analyses have used dispersal-vicariance analysis (DIVA) [52] to infer ancestral distributions [53,54], DIVA requires fully bifurcated trees [55]. Because our Bayesian trees were not fully resolved, we performed statistical dispersal-vicariance analysis (S-DIVA) to reconstruct the geographic diversification of Atraphaxis, as implemented in RASP version 2.0 (http://mnh.scu.edu.cn/soft/blog/RASP), which allows for uncertainties in phylogenetic trees [56]. Six geographic regions were identified for species of Atraphaxis in China: (A) Ili valley; (B) Loess Plateau; (C) Horqin sandlands; (D) Junggar Basin; (E) Tarim Basin; (F) and Qaidam Basin (Fig 1). We used 9,000 trees from the Bayesian phylogenetic analyses as input for S-DIVA to compute the condensed tree, and subsequently the ancestral areas and potential vicariance and dispersal events were inferred with RASP.
Results
Characteristics of cpDNA regions
The total aligned lengths of the psbK-psbI and psbB-psbH intergenic spacers for all species of Atraphaxis are 424 bp and 595 bp, respectively, with a total of 70 variable characters, including 31 structural characters (i.e., gaps) coded using simple gap coding [57]. The combined chloroplast intergenic spacers are A/T rich (64.98%), and this nucleotide composition is similar to that of most noncoding spacers and pseudogenes because of low functional constraints. From all of the variable sites, a total of 32 haplotypes (H1-H32) were identified, and the distribution and frequency of each haplotype is presented in Table 1, S1 Table, Fig 1 and S1 Fig.
Thirteen of the 32 haplotypes were identified in more than two populations, with haplotype frequencies ranging from less than 1% to 24% (Table 1). Nineteen haplotypes were very rare and highly localized, as they occurred in only one or two closely associated localities (Table 1, Fig 1 and S1 Fig). These haplotypes were concentrated in the western region (Xinjiang province), with four in the central region and one in the eastern region of the Horqin sandlands. The most widely distributed haplotypes were H8, H4 and H3. Indeed, 16 populations throughout the western and eastern regions of the genus in northern China share haplotype H8. Haplotype H4 was present in seven populations, primarily occurring in the Ili valley and Junggar Basin. Haplotype H3 was distributed among nine populations present in the lower montane zones of the central and east Tianshan range. Some haplotypes were present in only a single species, while other haplotypes were represented among widespread species (e.g., A. frutescens and A.compacta). In some instances, several species that are closely related phylogenetically or geographically share the same haplotype (Table 1 and S1 Fig). For example, haplotype H4 was shared by A. frutescens, A. jrtyschensis, and A. pungens; H7 was shared by A. frutescens and A. decipiens; and haplotype H8 was shared by A. frutescens and A. manshurica.
Phylogeographic patterns in Atraphaxis
The cpDNA sequences showed considerable variation in Atraphaxis. For the genus, data on haplotype diversity, as measured by two different indices, gene diversity (H) and molecular diversity (π), are presented in Table 1. Total genetic diversity (hT) was 0.919 (±0.0152), and within-population diversity (hS) was 0.113 (±0.0242).
In the three defined geographic regions, genetic diversity in the western region was much greater than that in the others (Table 1). Total genetic variation was partitioned by species and by geographic region (Table 2). In analyses based on species, the results demonstrated that 47.25% of the total genetic diversity was attributed to differences among species while a similar amount, 46.36%, was due to differences among populations. Only a small quantity, 6.38%, was attributed to differences within populations. In analyses based on geographic regions, variation among populations occupied the largest quantity, 72.11%, with variation among regions accounting for a smaller amount, 21.34%. As with total genetic diversity by species, within population variation was quite small, 6.55% (Table 2). Permut calculated a high NST (0.909), and this was substantially greater than GST (0.877), indicating obvious population differentiation and suggesting the existence of phylogeographic structure for all populations.
Table 2. Analysis of molecular variance of Atraphaxis populations, partitioned by species and geographic region.
| Partitioning | Source of variation | d.f | Sum of squares | Variance components | Percentage of variation |
|---|---|---|---|---|---|
| By species | Among species | 10 | 919.467 | 1.901 | 47.25 |
| Among populations | 60 | 908.493 | 1.865 | 46.36 | |
| Within populations | 488 | 125.286 | 0.257 | 6.38 | |
| Total | 558 | 1953.247 | 4.022 | ||
| By region | Among regions | 2 | 315.719 | 0.836 | 21.34 |
| Among populations | 68 | 1512.242 | 2.826 | 72.11 | |
| Within populations | 488 | 125.286 | 0.257 | 6.55 | |
| Total | 558 | 1953.247 | 3.919 |
The observed mismatch distribution analysis for the total samples revealed similar results with those of the Extended Bayesian Skyline Plots (EBSP) analyses. The mismatch distribution for all samples was multimodal (Fig 3). For the more conservative neutrality estimates, Tajima’s D (D = -1.563, P<0.05) was significant and Fu’s Fs was not (Fs = 0.308, Table 3). The mismatch distribution analysis performed for each species resulted in multimodal distributions. Combined with the non-significant distribution of the EBSP analysis and unapparent neutrality estimates (Fig 3), the results indicated that the populations for each species did not depart from equilibrium.
Fig 3. Historical demography for each species and total samples inferred from cpDNA sequences.
(A) Mismatch distribution analysis for each species and total samples. (B) Extended Bayesian skyline plot (EBSP) analyses for each species and total samples, showing effective population size as a function of time.
Table 3. Results of Tajima’s D and Fu’s Fs tests and mismatch analyses for 11 species of Atraphaxis.
| Tajima’s D test | Fu’s Fs test | Mismatch distribution | |||||
|---|---|---|---|---|---|---|---|
| Species | D | P | Fs | P | τ | θ0 | θ1 |
| A. frutescens | 0.835 | 0.819 | 13.119 | 0.983 | 3.176 | 0.000 | 5.933 |
| A. manshurica | -0.802 | 0.205 | -1.331 | 0.159 | 3.000 | 0.000 | 0.073 |
| A. bracteata | -1.485 | 0.055 | 1.852 | 0.845 | 3.000 | 0.000 | 0.430 |
| A. pyrifolia | 0.168 | 0.622 | 9.349 | 0.999 | 0.000 | 0.000 | 99999.000 |
| A. laetevirem | 0.208 | 0.662 | 0.049 | 0.438 | 0.895 | 0.000 | 99999.000 |
| A. jrtyschensis | 0.000 | 1.000 | - | - | - | - | - |
| A. pungens | 1.845 | 0.972 | 3.043 | 0.924 | 4.227 | 0.000 | 2.967 |
| A. decipiens | 0.007 | 0.546 | 4.747 | 0.978 | 2.047 | 0.000 | 3.600 |
| A. compacta | -0.384 | 0.400 | 2.412 | 0.844 | 3.000 | 0.000 | 0.546 |
| A. canescens | 0.000 | 1.000 | - | - | - | - | - |
| A. spinosa | 0.000 | 1.000 | - | - | - | - | - |
| Total | -1.563 | 0.016 | 0.308 | 0.608 | 1.551 | 3.579 | 12.471 |
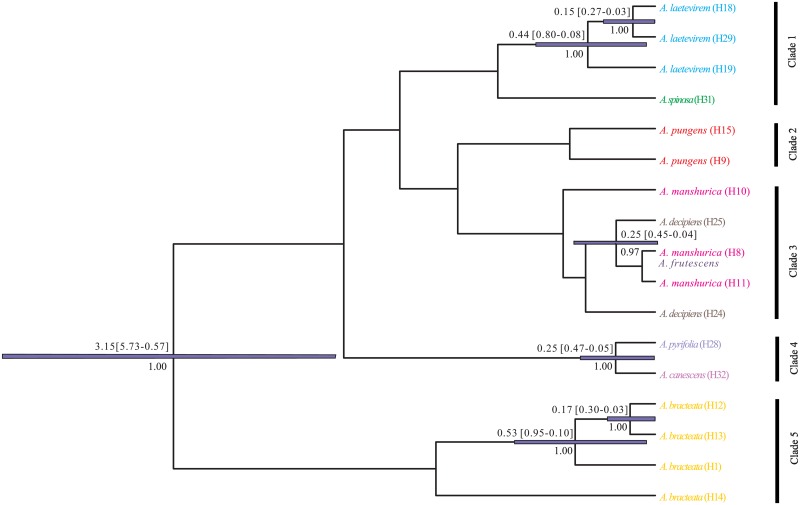
Phylogenetic relationships and divergence time
Based on the Bayesian log-normal relaxed clock method, we obtained an evolutionary tree and divergence time for all of the haplotypes (S2 Fig). However, this tree did not resolve each of the 11 species as reciprocally monophyletic. Some species are sympatric in particular regions (e.g., A. frutescens and A. laetevirem are both distributed in Xiaer Lake and in Aretuobie country), and this could induce sympatric introgression among neighboring individuals of different species. For this reason, the phylogenetic relationships of the haplotypes of Atraphaxis were well resolved, with the exception of a limited number of haplotypes from these sympatric individuals and two widespread species, A. frutescens and A. compacta (S3 Table). Based on the phylogenetic tree, five main haplotype clades can be recognized (Fig 4). Clade 5 comprised all haplotypes of A. bracteata, a species restricted to the Loess Plateau and separated from other Atraphaxis species. Clade 1 contained the haplotypes of A. laetevirem and A. spinosa. Clade 3 included haplotypes of two species, A. decipiens and A. manshurica. These two species occur in the north of Junggar Basin and the Horqin sandlands. Whereas, haplotype H8 was shared in the species of A. frutescens and A. manshurica, suggesting that A. manshurica has a close relationship with A. frutescens and may have originated north of Junggar Basin. Clade 4 included the haplotypes of A. canescens and A. pyrifolia, which are primarily distributed in the Junggar Basin. Clade 2 contained the haplotypes of A. pungens.
Fig 4. Bayesian phylogenetic tree for Atraphaxis based on combined cpDNA matrix.
Numbers above branches indicate range of divergence time in millions of years (Ma), and posterior probabilities larger than 0.95 are shown. Clades 1–5 denote five main clades based on identified haplotypes. Haplotype numbers refer to those in Table 1. Colors of each species are consistent with Fig 2.
In the BEAST analysis, the ESS values were over 200 for all nodes. Divergence time among the different haplotypes was estimated between 5.73–0.03 million years ago (Ma). Detailed information concerning relationships among each of the haplotypes separated is depicted in Fig 4.
Ancestral area reconstructions
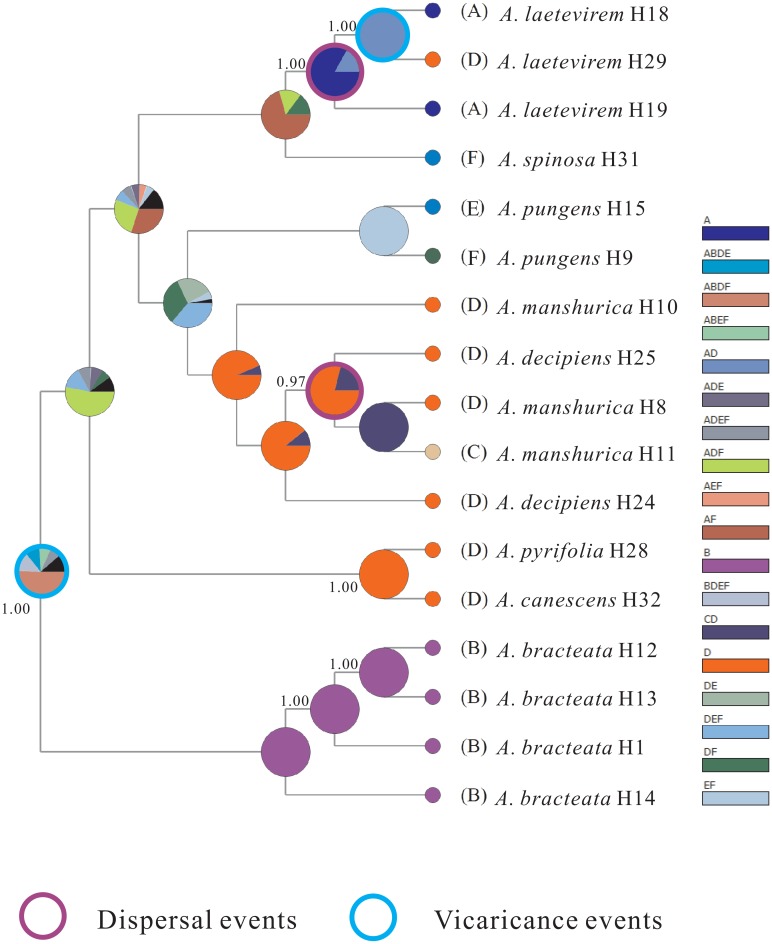
Results of the statistical dispersal-vicariance analysis (S-DIVA) suggest a complex phylogeographic history of the species of Atraphaxis in China. Pie charts at nodes indicate the relative frequencies of ancestral area optimization across the entire ancestral area reconstruction tree shown in Fig 5. The Ili valley, Junggar basin, Tarim Basin, and Loess plateau (ABDF, 47.2%) are together resolved to be the most probable ancestral area(s) for Atraphaxis in China. Collectively, these regions encompass the entire Western region and some areas of the Central region. Additionally, two dispersal events and two vicariance events are hypothesized based on the S-DIVA analyses, and these events are depicted in Fig 5. The most likely putative dispersal event, recovered by S-DIVA, in Atraphaxis is from Junggar Basin (D) to the Horqin sandlands (C), and this was followed by species diversification in these two regions.
Fig 5. Results of statistical dispersal-vicariance analysis (S-DIVA).
Coloured circles correspond to hypothetical ancestral area(s). Letters before names of terminal taxa represent current geographic distributions. Distribution regions: (A) Ili river valley; (B) Loess Plateau; (C) Horqin sandlands; (D) Junggar Basin; (E) Tarim Basin; (F) Qaidam Basin (see Fig 1).
Discussion
Past climatic changes in northern China have resulted in modified ecosystems and species distributions [22]. Indeed, these have affected population contraction and recolonization, range contractions and expansions, and the formation of contact zones between species, all of which have shaped current regional phylogeographic patterns [58]. The phylogeographic patterns of Atraphaxis are congruent with those previously described for other arid shrubs and small herbs, although previous studies primarily focused on a single species and a particular geographic area in the arid regions of northern China (e.g., northwestern China, Alashan desert, Horqin sandlands) [7,59,60]. In the present study, we provide a phylogeographic analysis of a species-rich genus throughout its entire geographic range in an arid region.
Genetic diversity of Atraphaxis
The use of cpDNA markers in combination with phylogeographic analyses provide detailed insights into the evolution of species during the late Pliocene and Pleistocene climate shifts. The results obtained from the present study demonstrate phylogenetic relationship and highly structured phylogeographic history in populations and species of Atraphaxis (Table 1 and Fig 4). The extreme climate changes that developed during the Pleistocene, and have also occurred since [11], likely played important roles in the diversification of the genus. Cooling sharply increased aridity, in the Chinese deserts, since the late Pliocene [24,25]. Subsequently, geographic isolation and genetic drift with local adaptions for populations (e.g., regarding climate tolerance) may have led to the high levels of genetic differentiation in the species of Atraphaxis in northern China. This amount of genetic diversity is important for the maintenance of biodiversity within the genus [61].
As many species inhabit similar ecosystems, admixed species (i.e., species with haplotypes also found in other species) are present in some populations, especially in the Junggar Basin. H4 is shared by three species (A. frutescens, A. jrtyschensis, and A. pungens), and H8 is shared by two species (A. frutescens and A. manshurica) (Table 1). This phenomenon can be explained as either retained ancestral polymorphism or introgression. Given the young age and overlapping geographic ranges of the species of the genus, it is likely that both retained ancestral polymorphisms and introgression have played roles in the observed admixture of haplotypes among species (Fig 1). The two processes would function in similar manners. The young age would allow for the observed retention of putative ancestral polymorphisms and the lack of reciprocal monophyly among species, while the sympatric geographic ranges would provide opportunities for introgression, which can lead to genetic admixture. Taking both processes together, it is unsurprising that some widespread haplotypes are present in multiple species.
Western populations within Atraphaxis show the largest number of haplotypes and harbour a higher genetic and nucleotide diversity than other areas (Table 1, S1 Table), suggesting that the species in the western region may persist for a longer period of time than species in other areas, such as through multiple glacial cycles. Additionally, the western region includes one of the widespread haplotypes (H4). In contrast, our data demonstrate that A. manshurica in the Horqin sandlands has the lowest genetic diversity of the three areas, and a small number of haplotypes are present at low frequency (Table 1), with H8 being most common. Interestingly, although H8 is quite common in A. manshurica, the most eastward species in China, this haplotype is also present in multiple populations throughout the western range of the Chinese species in the genus (A. frutescens and A. decipiens), but none in the central part of the distribution. Consequently, it seems likely that A. manshurica arose via dispersal from western species of Atraphaxis. This dispersal event may have occurred via migration along the mountain front and lower montane zones of the slope of Altai Mountains range in China and Mongolia and through the Mongolian Plateau.
Rapid genetic differentiation in relation to Quaternary climatic events
Based on the BEAST analyses, divergence time between haplotypes of Atraphaxis in China is estimated to have occurred 5.73–0.03 Ma (late Pliocene and Pleistocene). This diversification is much more recent than the origin of Atraphaxis [33], and this short range of time is likely due, in part, to the inclusion of only Chinese species in the present study. Populations over a limited geographic range may more easily introgress than those from across a larger area, and this can lead to homogenization of the populations and a decreased divergence time. However, the time estimates from BEAST are within the late Pliocene and Pleistocene and fit well with a time when climate changes and aridification began to develop and expand in northwestern and northern China. During the Pleistocene, there were intermittent colder periods as well as aridification in connection with glacial periods [62,63]. Since the early Pleistocene, the climate cooled, and there were several episodes of rapid aridification occurring across northwestern China [64]. The rapid drying of the region and the expansion of desert habitats during the Pleistocene may have acted as an effective stimulus to promote allopatric or parapatric divergence, leading to speciation and intraspecific differentiation within Atraphaxis. The consistency between the estimated timeframe of speciation and the glacial and arid expansion history through the Quaternary suggest that mixed and complex climate effects forced rapid speciation, local adaptation and intraspecific differentiation of Atraphaxis.
Ancestral area reconstruction and recent dispersal event
The results of S-DIVA suggest that the ancestor of the species of Atraphaxis in China primarily inhabited the Western and Central regions (including the Ili valley, Junggar basin, Tarim Basin, and Loess plateau). The identification of the ancestral area for species of Atraphaxis included in the present study coincides with previous hypotheses concerning the origin of the genus. Kransnov considered that the Tertiary mesic montane flora of the Tianshan and other mountains was the area of origin for Atraphaxis [65]. Molecular evidence by Zhang et al. [33] indicated that the area of origin for the genus consisted of the Junggar Basin and the uplands of the Pamir-Tianshan-Alatau-Altai mountain chains. The results of these studies are congruent with the central region in the current biogeographic analyses.
It is apparent that Atraphaxis in China experienced geographical isolation between the six biogeographic areas (A-F), followed by the detected inter-regional dispersal events (Fig 5). We suggest that the expansion of aridification may have caused these vicariance events. In northern China, Atraphaxis grows in areas with extreme drought conditions, such as deserts and semidesert grasslands. In these environments, unfavourable conditions during reproduction would restrict pollination and dispersal capacity of species. This would obstruct gene flow among populations, yet retain gene flow within populations and communities, which can include multiple species of Atraphaxis.
According to S-DIVA, a unique dispersal event has been identified from the Junggar Basin (D) to the Horqin sandlands (C). This dispersal event may have occurred via migration along the mountain front and lower montane zones of the Altai Mountains in China and Mongolia and through the Mongolian Plateau. The complexity of topology and melted snows from the peaks of the Altai Mountains may have played an important role in facilitating this migration event. Melted snow in the Altai Mountains would have been an integral source of water in these usually dry areas. During the interglacial period, the climate became warmer, and a greater amount of run-off from melting snow and glacial ice would have infused the lowland areas, making habitats moister [66]. These more favourable environments would have provided better conditions for species survival and range expansion. When populations migrated to semi-arid northeastern China, the altered environment would have provided appropriate conditions for rapid species divergence, including the origin of A. manshurica. New species resulting from dispersal events may suffer from founder effects, and this seems likely in the case of A. manshurica. This species is composed of a small number of haplotypes that are also only present in the eastern range of the genus in China.
Atraphaxis frutescens has the greatest number of haplotypes, eight (Table 1), and these occur throughout its continuous geographic distribution in the front and lower montane zones of the Tianshan and Altai Mountains, Hexi Corridor, and Qaidam Basin (Table 1, Fig 1). Despite the genetic differentiation of some populations of A. frutescens, such as those in the east Tianshan Mountains (H3) and the Hexi Corridor (H2), one haplotype dominates. This uniformity probably reflects recent colonization events or the purging of deleterious haplotypes over time. Two of the widespread haplotypes (H3 and H2) also dominate two disjunct populations of A. frutescens. Between the two regions, we identified a contact zone (LY) in the north of the Hexi Corridor in which mixed haplotypes were present in populations (Table 1 and Fig 1). This contact zone possessed haplotypes (H2 and H3) present in individuals from both regions (i.e., the Tianshan Mountains and Hexi Corridor). Because of the mixed haplotypes, populations in the contact zone display high levels of genetic diversity compared with other populations inhabiting adjacent areas. This contact zone (LY) is located in the transition between the temperate and the warm temperate zones as well as at the intersection of arid and extremely arid regions [67]. This particular geographic position and climatic conditions make this region a significant migratory route for species in adjacent areas.
Origin and evolution of A. bracteata in the Loess plateau
From the species included in the BEAST analyses, A. bracteata is recognized as sister to all other studied species in the genus, and the estimated divergence time for this split is 5.73–0.57 Ma. This is close to the range of the formation of the Tengger Desert, as determined by paleomagnetic data, at approximately 1.8 Ma, which is in the background of a glacial cycle and the uplift of the QTP [68]. Thus, we speculate that Atraphaxis existed in the Loess plateau prior to the speciation of A. bracteata. Subsequently, the expansion of the Tengger Desert split the continuous habitat into two isolated groups. Geographic isolation limited gene flow, and climatic changes may have acted as an effective stimulus to promote divergence between the two groups, leading to the origin of A. bracteata. The only population in the north of the Hexi Corridor (AX see in Table 1 and Fig 1) consists only of a widely distributed haplotype (H12) of A. bracteata, and this likely reflects a population expansion event during interglacial cycles of the recent Holocene. During this period, the climate warmed and became wetter, and more snow melted from the Qilian Mountains resulting in large amounts of run-off. This made habitats more suitable for the expansion of populations of A. bracteata. These findings highlight the predominant role of geographic vicariance in shaping the distribution of A. bracteata.
Supporting Information
Pie charts reflect frequency of haplotypes in each population. Haplotype colours correspond to those in panel.
(TIF)
(TIF)
(DOC)
(DOC)
(DOC)
Acknowledgments
We thank Hong-Xiang Zhang, Xiao-Jun Shi and Shu-Wen Jia for discussion of methodological design. We thank Jian Zhang for assistance with collecting samples and investigating in field.
Data Availability
All obtained sequences are deposited in GenBank with accession numbers KM452785-KM452792, KX236017-KX236033, KR183867, KR183868, KR183870 for psbK-psbI; and KM452793-KM452796, KR183863, KR183864, KR183866, KX236034-KX236045 for psbB-psbH.
Funding Statement
This work was supported by the China National Basic Research Program (2014CB954201) and Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences.
References
- 1.Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000; 405(6789): 907–913. [DOI] [PubMed] [Google Scholar]
- 2.Willis KJ, Niklas KJ. The role of Quaternary environmental change in plant macroevolution: the exception or the rule? Philos T R Soc B. 2004; 359(1442): 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taberlet P, Fumagalli L, Wust A, Cosson J. Comparative phylogeography and postglacial colonization routes in Europe. Mol Ecol. 1998; 7(4): 453–464. [DOI] [PubMed] [Google Scholar]
- 4.Nason JD, Hamrick J, Fleming TH. Historical vicariance and postglacial colonization effects on the evolution of genetic structure in Lophocereus, a Sonoran Desert columnar cactus. Evolution. 2002; 56(11): 2214–2226. [DOI] [PubMed] [Google Scholar]
- 5.Garrick R, Nason J, Meadows C, Dyer R. Not just vicariance: phylogeography of a Sonoran Desert euphorb indicates a major role of range expansion along the Baja peninsula. Mol Ecol. 2009; 18(9): 1916–1931. 10.1111/j.1365-294X.2009.04148.x [DOI] [PubMed] [Google Scholar]
- 6.Ossa PG, Pérez F, Armesto JJ. Phylogeography of two closely related species of Nolana from the coastal Atacama Desert of Chile: post-glacial population expansions in response to climate fluctuations. J Biogeogr. 2013; 40(11): 2191–2203. [Google Scholar]
- 7.Wang Q, Abbott RJ, Yu QS, Lin K, Liu JQ. Pleistocene climate change and the origin of two desert plant species, Pugionium cornutum and Pugionium dolabratum (Brassicaceae), in northwest China. New Phytol. 2013; 199(1): 277–287. 10.1111/nph.12241 [DOI] [PubMed] [Google Scholar]
- 8.Bennett KD, Tzedakis PC, Willis KJ. Quaternary refugia of north European trees. J Biogeogr. 1991; 18: 103–115. [Google Scholar]
- 9.Guest HJ, Allen GA. Geographical origins of North American Rhodiola (Crassulaceae) and phylogeography of the western roseroot, Rhodiola integrifolia. J Biogeogr. 2014; 41(6): 1070–1080. [Google Scholar]
- 10.Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philos T R Soc B. 2004; 359(1442): 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avise JC. Phylogeography: the history and formation of species. Harvard University Press; 2000. [Google Scholar]
- 12.Hardy ME, Grady JM, Routman EJ. Intraspecific phylogeography of the slender madtom: the complex evolutionary history of the Central Highlands of the United States. Mol Ecol. 2002; 11(11): 2393–2403. [DOI] [PubMed] [Google Scholar]
- 13.Hewitt GM. Speciation, hybrid zones and phylogeography—or seeing genes in space and time. Mol Ecol. 2001; 10(3): 537–549. [DOI] [PubMed] [Google Scholar]
- 14.Liu YF, Wang Y, Huang HW. Species-level phylogeographical history of Myricaria plants in the mountain ranges of western China and the origin of M. laxiflora in the Three Gorges mountain region. Mol Ecol. 2009; 18(12): 2700–2712. 10.1111/j.1365-294X.2009.04214.x [DOI] [PubMed] [Google Scholar]
- 15.Schaal BA, Olsen KM. Gene genealogies and population variation in plants. P Natl Acad Sci. 2000; 97(13): 7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petit RJ, Aguinagalde I, de Beaulieu J-L, Bittkau C, Brewer S, Cheddadi R, et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003; 300(5625): 1563–1565. [DOI] [PubMed] [Google Scholar]
- 17.McLachlan JS, Clark JS, Manos PS. Molecular indicators of tree migration capacity under rapid climate change. Ecology. 2005; 86(8): 2088–2098. [Google Scholar]
- 18.Liu YX. A study on origin and formation of the Chinese desert floras. Acta Phytotaxon Sin. 1995; 33(2): 133–143. [Google Scholar]
- 19.Wang CS, Zhao XX, Liu ZF, Lippert PC, Graham SA, Coe R S, et al. Constraints on the early uplift history of the Tibetan Plateau. P Natl Acad Sci. 2008; 105(13): 4987–4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu ZH, Barosh PJ, Wu ZH, Hu DG, Zhao X, Ye PS. Vast early Miocene lakes of the central Tibetan Plateau. Geol Soc Am Bull. 2008; 120(9–10): 1326–1337. [Google Scholar]
- 21.Guo ZT, Sun B, Zhang ZS, Peng SZ, Xiao GQ, Ge JY, et al. A major reorganization of Asian climate by the early Miocene. Clim Past. 2008; 4(3): 153–174. [Google Scholar]
- 22.Meng HH, Zhang ML. Diversification of plant species in arid Northwest China: Species-level phylogeographical history of Lagochilus Bunge ex Bentham (Lamiaceae). Mol Phylogenet Evol. 2013; 68(3): 398–409. 10.1016/j.ympev.2013.04.012 [DOI] [PubMed] [Google Scholar]
- 23.Yang XP, Rost KT, Lehmkuhl F, Zhu ZD, Dodson J. The evolution of dry lands in northern China and in the Republic of Mongolia since the Last Glacial Maximum. Quatern Int. 2004; 118: 69–85. [Google Scholar]
- 24.Ding ZL, Derbyshire E, Yang SL, Sun JM, Liu TS. Stepwise expansion of desert environment across northern China in the past 3.5 Ma and implications for monsoon evolution. Earth Planet Sc Lett. 2005; 237(1): 45–55. [Google Scholar]
- 25.Fang XM, Lu LQ, Yang SL, Li JJ, An ZS, Jiang PA, Chen XL. Loess in Kunlun Mountains and its implications on desert development and Tibetan Plateau uplift in west China. Science in China Series D: Earth Sciences. 2002; 45(4): 289–299. [Google Scholar]
- 26.Dang RL, Pan XL. The Chinese endemic plant analysis in west-north desert of China. Bulletin of Botanical Research. 2001; 4: 520–526. [Google Scholar]
- 27.Zhao YZ. Endemic genera and their basic characteristics of the Mongolian Planteau plants Acta Scientiarum Naturalium Universitatis Neimongol. 1997; 4. [Google Scholar]
- 28.Takhtajan A. Floristic regions of the world. Berkeley, etc:(Transl by TJ Crovello) Univ Calif Press; 1986. [Google Scholar]
- 29.Ge XJ, Hwang CC, Liu ZH, Huang CC, Huang WH, Hung KH, et al. Conservation genetics and phylogeography of endangered and endemic shrub Tetraena mongolica (Zygophyllaceae) in Inner Mongolia, China. BMC Genet. 2011; 12(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma SM, Zhang ML, Sanderson SC. Phylogeography and conservation genetics of the relic Gymnocarpos przewalskii (Caryophyllaceae) restricted to northwestern China. Conserv Genet. 2012; 13(6): 1531–1541. [Google Scholar]
- 31.Xu Z, Zhang ML. Phylogeography of the arid shrub Atraphaxis frutescens (Polygonaceae) in northwestern China, evidence from cpDNA sequences. J Hered. 2015; 106(2): 184–195. 10.1093/jhered/esu078 [DOI] [PubMed] [Google Scholar]
- 32.Bao BJ, Li AR. A study of the genus Atraphaxis in China and the system of Atraphaxideae (Polygonaceae). Acta Phytotaxon Sin. 1993; 31(2): 127–139. [Google Scholar]
- 33.Zhang ML, Sanderson SC, Sun YX, Vyacheslav BV, Hao XL. Tertiary montane origin of the Central Asian flora, evidence inferred from cpDNA sequences of Atraphaxis. J Integr Plant Biol. 2014; 56(12): 1125–1135. 10.1111/jipb.12226 [DOI] [PubMed] [Google Scholar]
- 34.Sun YX, Zhang ML. Molecular phylogeny of tribe Atraphaxideae (Polygonaceae) evidenced from five cpDNA genes. J Arid Land. 2012; 4(2): 180–190. [Google Scholar]
- 35.Tavakkoli S, Osaloo SK, Mozaffarian V, Maassoumi AA. Molecular phylogeny of Atraphaxis and the woody Polygonum species (Polygonaceae): taxonomic implications based on molecular and morphological evidence. Plant Syst Evol. 2015; 301(4): 1157–1170. [Google Scholar]
- 36.Sun YanXia Zhang MingLi. Molecular phylogeny of tribe Atraphaxideae (Polygonaceae) evidenced from five cpDNA genes. Journal of Arid Land. 2012; 4(2): 180−190. [Google Scholar]
- 37.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochemical Bulletin. 1987; 19: 11–15. [Google Scholar]
- 38.Sang T, Crawford D, Stuessy T. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am J Bot. 1997; 84(8): 1120–1120. [PubMed] [Google Scholar]
- 39.Xu D, Abe J, Sakai M, Kanazawa A, Shimamoto Y. Sequence variation of non-coding regions of chloroplast DNA of soybean and related wild species and its implications for the evolution of different chloroplast haplotypes. Theor Appl Genet. 2000; 101(5–6): 724–732. [Google Scholar]
- 40.Nishizawa T, Kurabayashi A, Kunihara T, Sano N, Fujii T, Sumida M. Mitochondrial DNA diversification, molecular phylogeny, and biogeography of the primitive rhacophorid genus Buergeria in East Asia. Molecular Phylogenetics & Evolution. 2011; 59(1): 139–147. [DOI] [PubMed] [Google Scholar]
- 41.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22(22): 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25(11): 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 43.Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol bioinform. 2005; 1: 47. [PMC free article] [PubMed] [Google Scholar]
- 44.Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992; 131(2): 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pons O, Petit R. Measwring and testing genetic differentiation with ordered versus unordered alleles. Genetics. 1996; 144(3): 1237–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grant W S. Problems and cautions with sequence mismatch analysis and Bayesian skyline plots to infer historical demography. Journal of Heredity. 2015; 106(4): 333–346. 10.1093/jhered/esv020 [DOI] [PubMed] [Google Scholar]
- 47.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007; 7(1): 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998; 14(9): 817–818. [DOI] [PubMed] [Google Scholar]
- 49.Alsos I G, Engelskjøn T, Gielly L, Taberlet P, Brochmann C. Impact of ice ages on circumpolar molecular diversity: insights from an ecological key species. Molecular Ecology. 2005; 14(9): 2739–2753. [DOI] [PubMed] [Google Scholar]
- 50.Ho SY, Lanfear R, Bromham L, Phillips MJ, Soubrier J, Rodrigo AG, Cooper A. Time dependent rates of molecular evolution. Molecular ecology. 2011; 20 (15), 3087–3101. 10.1111/j.1365-294X.2011.05178.x [DOI] [PubMed] [Google Scholar]
- 51.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999; 16(1): 37–48. [DOI] [PubMed] [Google Scholar]
- 52.Ronquist F. Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Systematic Biol. 1997; 46(1): 195–203. [Google Scholar]
- 53.Baird KE, Funk VA, Wen J, Weeks A. Molecular phylogenetic analysis of Leibnitzia Cass.(Asteraceae: Mutisieae: Gerbera-complex), an Asian-North American disjunct genus. J Syst Evol. 2010; 48(3): 161–174. [Google Scholar]
- 54.Harris A, Wen J, Xiang QYJ. Inferring the biogeographic origins of inter-continental disjunct endemics using a Bayes-DIVA approach. J Syst Evol. 2013; 51(2): 117–133. [Google Scholar]
- 55.Gao XY, Meng HH, Zhang ML. Diversification and vicariance of desert plants: Evidence inferred from chloroplast DNA sequence variation of Lagochilus ilicifolius (Lamiaceae). Biochem Syst Ecol. 2014; 55: 93–100. [Google Scholar]
- 56.Ali SS, Yu Y, Pfosser M, Wetschnig W. Inferences of biogeographical histories within subfamily Hyacinthoideae using S-DIVA and Bayesian binary MCMC analysis implemented in RASP (Reconstruct Ancestral State in Phylogenies). Ann Bot. 2012; 109(1): 95–107. 10.1093/aob/mcr274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biol. 2000; 49(2): 369–381. [PubMed] [Google Scholar]
- 58.Zhang HX, Zhang ML. Identifying a contact zone between two phylogeographic lineages of Clematis sibirica (Ranunculeae) in the Tianshan and Altai Mountains. J Syst Evol. 2012; 50(4): 295–304. [Google Scholar]
- 59.Su ZH, Zhang ML. Evolutionary response to Quaternary climate aridification and oscillations in north-western China revealed by chloroplast phylogeography of the desert shrub Nitraria sphaerocarpa (Nitrariaceae). Biol J Linn Soc. 2013; 109(4): 757–770. [Google Scholar]
- 60.Xu Z, Zhang ML. The effect of past climatic oscillations on spatial genetic structure of Atraphaxis manshurica (Polygonoideae) in the Horqin sandlands, northern China. Biochem Syst Ecol. 2015; 60: 88–94. [Google Scholar]
- 61.Shafer A, Côté SD, Coltman DW. Hot spots of genetic diversity descended from multiple Pleistocene refugia in an alpine ungulate. Evolution. 2011; 65(1): 125–138. 10.1111/j.1558-5646.2010.01109.x [DOI] [PubMed] [Google Scholar]
- 62.Shi YF, Cui ZJ, Su Z. The Quaternary glaciations and environmental variations in China. Hebei Science and Technology Publishing House. 2005. [In Chinese with English abstract]. [Google Scholar]
- 63.Xu X, Kleidon A, Miller L, Wang S, Wang L, Dong G. Late Quaternary glaciation in the Tianshan and implications for palaeoclimatic change: a review. Boreas. 2010; 39(2): 215–232. [Google Scholar]
- 64.Guo ZT, Peng SZ, Hao QZ, Chen XH, Liu TS. Late Tertiary development of aridification in northwestern China: link with the arctic ice-sheet formation and Tibetan uplifts. Quaternary Sci. 1999; 6: 556–567. [Google Scholar]
- 65.Wulff EV. Historical plant geography: history of the world flora. Waltham, MA: Chronica Botanica; 1944. [Google Scholar]
- 66.Williams MA. Quaternary environments. London: Edward Arnold; 1993. [Google Scholar]
- 67.Xu XY. First class national protected animal in Anxi protection zone: Capra ibex. China Biosphere Reserve. 1995; 2: 37–38. [Google Scholar]
- 68.Dong Y, Fang XM, Dong GR, Peng ZC, Li JJ. Aeolian deposit evidence for formation and evolution of the Tengger Desert in the north of China since early Pleistocene. Marine Geology and Quaternary Geology. 2006; 26 (1): 93. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pie charts reflect frequency of haplotypes in each population. Haplotype colours correspond to those in panel.
(TIF)
(TIF)
(DOC)
(DOC)
(DOC)
Data Availability Statement
All obtained sequences are deposited in GenBank with accession numbers KM452785-KM452792, KX236017-KX236033, KR183867, KR183868, KR183870 for psbK-psbI; and KM452793-KM452796, KR183863, KR183864, KR183866, KX236034-KX236045 for psbB-psbH.