Abstract
Background
O6-methylguanine-DNA methyl-transferase (MGMT) gene, a DNA repair gene, plays a critical role in the repair of alkylated DNA adducts that form following exposure to genotoxic agents. MGMT is generally expressed in various tumors, and its function is frequently lost because of hypermethylation in the promoter. The promoter methylation of MGMT has been extensively investigated in head and neck squamous cell carcinoma (HNSCC). However, the association between the promoter methylation of MGMT and HNSCC risk remains inconclusive and inconsistent. Therefore, we performed a meta-analysis to better clarify the association between the promoter methylation of MGMT and HNSCC risk.
Methods
A systematical search was conducted in PubMed, Web of Science, EMBASE, and Ovid for studies on the association between MGMT promoter methylation and HNSCC. Odds ratio (ORs) and 95% confidence intervals (CI) were calculated to estimate association between MGMT promoter methylation and risk of HNSCC. The meta-regression and subgroup analysis were undertaken to explore the potential sources of heterogeneity.
Results
Twenty studies with 1,030 cases and 775 controls were finally included in this study. The frequency of MGMT promoter methylation was 46.70% in HNSCC group and 23.23% in the control group. The frequency of MGMT promoter methylation in HNSCC group was significantly higher than the control group (OR = 2.83, 95%CI = 2.25–3.56).
Conclusion
This meta-analysis indicates that aberrant methylation of MGMT promoter was significantly associated with the risk of HNSCC, and it may be a potential molecular marker for monitoring the disease and may provide new insights to the treatment of HNSCC.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common malignancy cancer worldwide and about 600,000 new cases each year [1]. Among them, approximately 500,000 HNSCC cases with high malignancy occur each year and the 5-year survival of patients was only 40–50% [2]. And most of HNSCC frequently occurred in the oral cavity, oropharynx, hypopharynx and larynx. At present, tobacco use and alcohol consumption both are well-established risk factors for the development of HNSCC [3]. Moreover, human papillomavirus (HPV) infection has recently been recognized as an independent etiologic factor in the development of HNSCC [4].
Hypermethylation of CpG islands in the promoter region of human genes often resulted in epigenetic inactivation, one of the most frequent events in human tumors. Gene-specific promoter methylation has been increasingly identified as a contributing factor to the development of HNSCC [5,6]. O6-methylguanine-DNAmethyl-transferase (MGMT) is a DNA repair gene that plays a crucial role in the mechanism of repair of DNA damage caused by alkylating agents [7]. MGMT is widely expressed in various tumors, and its function is frequently lost due to hypermethylation in the promoter. Some studies had found that methylation of MGMT gene promoter was closely related to poor prognosis, metastasis, and recurrence in HNSCC [8–10].
Up to now, many studies have explored the association between aberrant methylation of MGMT promoter and HNSCC risk. And most studies aimed to investigate the relationship by comparing the differences in the methylation frequencies of MGMT promoter between cancer and non-cancerous. However, the results remain inconclusive and inconsistent. Furthermore, some studies always have a small sample size and have different types of control. Therefore, we conducted a meta-analysis to better clarify the association between MGMT promoter methylation and risk of HNSCC.
Materials and Methods
As described in detail previously [11,12], the meta-analysis was performed according to the latest Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA).
Studies Identification
We searched the relevant studies in various online electronic databases (PubMed, Embase, Ovid, and Web of Science). The following search strategy was employed: (oropharyngeal or oral or oropharynx or tonsil or head and neck) and (squamous cell carcinoma or cancer) and (MGMT methylation). The search results were updated until May 20, 2016 with restricting to English language.
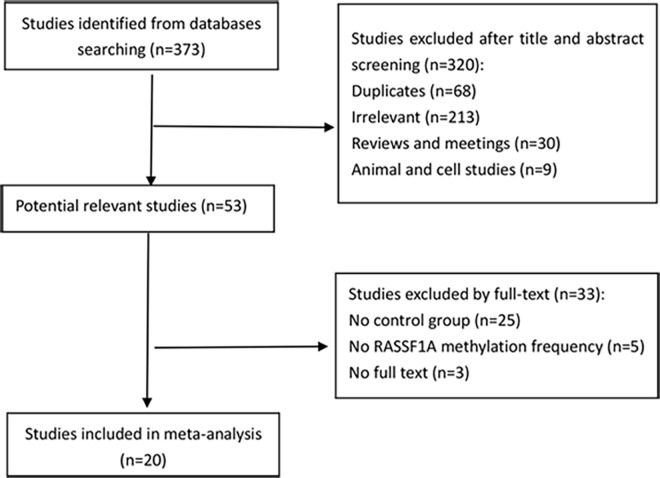
The inclusion criteria of the meta-analysis were: (1) articles studying with the association between MGMT promoter methylation and HNSCC, (2) case-control study and reporting the MGMT promoter methylation frequency in case and control groups, (3) specimens of HNSCC were limited to tissues. Firstly, we read the title and abstract of initial searching articles to assess whether it met the inclusion criteria. Then the potentially relevant articles were evaluated in full-text paper. Finally, a total of 20 articles which contain 1030 cases and 775 controls were included in the meta-analysis. The selection procedure of studies was illustrated in Fig 1.
Fig 1. Selection process of studies in the meta-analysis.
Data Extraction and Quality Assessment
Two reviewers (Fucheng Cai and Yi Zhong) independently reviewed the eligible studies. The following information was extracted from the eligible studies: first author’s name, publication year, study population, method for detecting the methylation status, sample type in case and control group, sample sizes (the number of people with MGMT methylation and the total people in the case and control groups). All the detailed information extracted from the eligible studies was checked by the third reviewer (Xiyue Xiao).
Statistical methods
The combined odds ratios (ORs) and its 95% confidence intervals (CIs) were calculated to evaluate the association between MGMT promoter methylation and HNSCC risk. The between-study heterogeneity was tested by the x2-based Cochran Q statistic test and I2 statistics [13]. The heterogeneity was considered significant (P<0.05 for the Q statistic or I2≥50%) and a random-effects model was used to calculate the pooled ORs. Otherwise, a fixed-effects model was applied to calculate the pooled ORs. Moreover, we performed the meta-regression and subgroup analysis to explore the source of heterogeneity. A sensitivity analysis was executed to investigate the influence of each individual study to the final results of the meta-analysis. The Begg’s funnel plot [14] and Egger’s test [15] were utilized to explore any possible publication bias. For all analyses, the two-sided P<0.05 was considered statistically significant. In the Meta package, the default is to add 0.5 to all zero counts when the individual studies have cells with zero counts. All statistical analyses were performed with the Meta package in R (version 3.2.3; http://www.r-project.org/).
Results
Study Characteristics
A total of 373 studies were identified by searching the electronic databases.
After eliminating duplicate articles and irrelevant studies (reviews, meeting reviews, and cell lines) by reviewing the titles and abstracts, 53 articles were identified. Finally, 20 studies were included in the meta-analysis by reviewing full-text removing some studies (no control group and no MGMT methylation frequency). The search and selection procedures of articles were shown in Fig 1. Among the 20 studies included in our meta-analysis, the control group consisted of HNSCC patients, benign disease patients, and healthy donors and the sample type of control group included tissue, saliva, serum, and buccal cells. The methylation detection methods of the studies included methylation-specific polymerase chain reaction (MSP), real-time quantitative MSP (QMSP), and pyrosequencing. Study characteristics are summarized in Table 1 [16–35].
Table 1. Characteristics of studies included in the study.
| Case | Control | Control source | Control sample | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Author | Year | Region | Age (case, years) | M | U | M | U | Method | ||
| Onerci Celebi[16] | 2016 | Turkey | mean = 56.6; range: 40–75 | 64 | 18 | 3 | 8 | Pyrosequencing | H | tissue |
| Yang[17] | 2015 | China | 27 | 50 | 12 | 50 | MSP | A | tissue | |
| Asokan[18] | 2014 | India | 4 | 6 | 0 | 5 | MSP | H | tissue | |
| Bhatia[19] | 2014 | India | mean = 53.0; sd: 12.97 | 58 | 18 | 34 | 36 | MSP | H | tissue |
| Rettori[20] | 2013 | Brazil | 14 | 53 | 2 | 55 | QMSP | H | saliva | |
| Koutsimpelas[21] | 2012 | Germany | mean = 62.0; range: 45–83 | 13 | 10 | 0 | 3 | MSP | H | tissue |
| Paluszczak[22] | 2011 | Poland | mean = 58.3; range: 41–75 | 22 | 19 | 15 | 26 | MSP | A | tissue |
| Weiss[23] | 2011 | Germany | mean = 63.7; sd: 11.8 | 13 | 39 | 3 | 28 | MSP | H | tissue |
| Su[24] | 2010 | Taiwan | mean = 54.94; range:37–82 | 7 | 23 | 3 | 27 | QMSP | A | tissue |
| 0 | 12 | H | buccal cell | |||||||
| Kordi[25] | 2010 | Iran | mean = 54.14 | 56 | 20 | 31 | 26 | MSP | H | tissue |
| Steinmann[26] | 2009 | Germany | mean = 57.0; range: 41–77 | 29 | 25 | 7 | 16 | MSP | A | tissue |
| De Schutter[27] | 2009 | Belgium | 17 | 23 | 0 | 5 | MSP | H | tissue | |
| Righini[28] | 2007 | France | median = 57.0; range: 33–7 | 20 | 70 | 0 | 30 | MSP | A | tissue |
| 0 | 30 | H | saliva | |||||||
| 13 | 47 | A | saliva | |||||||
| Martone[29] | 2007 | Italy | mean = 60.9; range: 27–89 | 10 | 10 | 5 | 6 | MSP | A | tissue |
| Kato[30] | 2006 | Japan | 27 | 24 | 0 | 18 | MSP | H | tissue | |
| 9 | 13 | A | tissue | |||||||
| Maruya[31] | 2004 | USA | mean = 58.3; range: 31–81 | 10 | 22 | 7 | 25 | MSP | A | tissue |
| 1 | 5 | MSP | H | tissue | ||||||
| Kulkarni[32] | 2004 | India | mean = 50.0; range:25–71 | 31 | 29 | 16 | 44 | MSP | A | tissue |
| 0 | 20 | MSP | H | buccal cell | ||||||
| Viswanathan[33] | 2003 | India | 21 | 30 | 0 | 25 | MSP | A | tissue | |
| Rosas[34] | 2001 | USA | 7 | 23 | 4 | 3 | MSP | A | saliva | |
| 1 | 29 | H | saliva | |||||||
| Sanchez[35] | 2000 | USA | 31 | 64 | 14 | 15 | MSP | A | serum | |
M: MGMT promoter methylated, U: MGMT promoter unmethylated
A: Autologous, H: Heterogeneous
sd: standard deviation
Meta-analysis
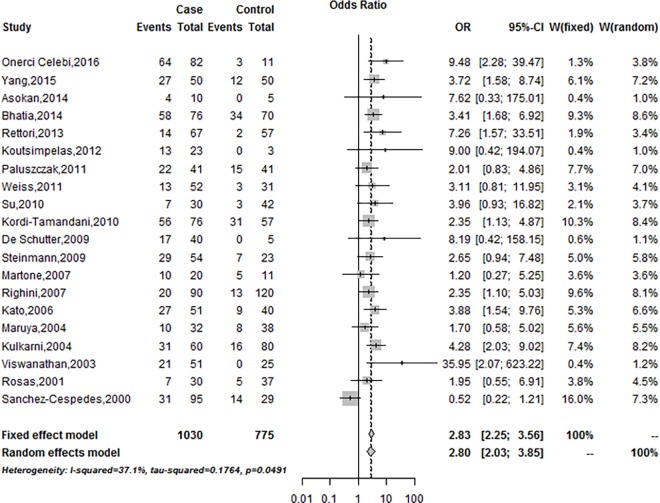
The combining result of the association of MGMT promoter methylation with HNSCC risk was shown in Fig 2. The fixed-effects model was employed due to the significant heterogeneity among the included studies (I2 = 37.1%, P = 0.05). MGMT promoter methylation frequency was significantly associated with an increased HNSCC risk based on the fixed effects model (Summary OR was 2.83, 95%CI = 2.25–3.56) (Fig 2).
Fig 2. Forest plot of MGMT promoter methylation associated with HNSCC risk under the fixed-effects model.
Meta-regression and Subgroup Analysis
Although the heterogeneity among the studies was found no significant (I2 = 37.1%, P = 0.05), we also conducted the meta-regression to find the potential sources of heterogeneity. The results of meta-regression showed that the potential source of the heterogeneity was only found in the control source (P = 0.02) (Table 2). We also performed the subgroup analysis to further evaluate the source of the heterogeneity according to race, method, control source, control sample type, and case sample size.
Table 2. Meta-regression analysis.
| 95%CI | ||||
|---|---|---|---|---|
| Heterogeneity sources | Coefficient | Lower | Upper | P |
| Publication year | 0.011 | -0.096 | 0.118 | 0.839 |
| Population | -0.297 | -1.106 | 0.511 | 0.471 |
| Method | 0.538 | -0.702 | 1.777 | 0.395 |
| Case sample size | 0.078 | -0.719 | 0.874 | 0.849 |
| Control source | 0.982 | 0.131 | 1.832 | 0.024 |
| Control sample | 0.450 | -0.610 | 1.510 | 0.406 |
In the race subgroup analysis, the OR in Asians group was 3.82 (95%CI = 2.75–5.30) under the fixed-effects model, and 2.28 (95%CI = 1.40–3.71) in Caucasians group under the random-effects model. Onerci Celebi [16] used pyrosequencing to detect MGMT promoter methylation and we have put the study classified as QMSP group in the methylation detection method. The OR for was 2.62 (95%CI = 2.07–3.33) in the MSP group under, and 6.48 (95%CI = 2.76–15.18) in the QMSP group under the fixed-effects model. With the control source, the OR in autologous group was 1.78 (95%CI = 1.12–2.81) under the random-effects model, and 5.18 (95%CI = 3.56–7.53) in the heterogeneous group under the fixed-effects model. In the control sample type group, we classified the studies into non-tissue group (control sample type: serum, saliva and buccal cell) and tissue group (control sample type: tissue).The OR for was 2.61 (95%CI = 0.81–8.41) in the non-tissue group, and 3.14 (95%CI = 2.03–4.86) in the tissue group under the random-effects model. The OR for was 2.62 (95%CI = 1.72–3.99) in case sample size < = 50 group under the fixed-effects model, and 3.05 (95%CI = 1.89–4.92) in case sample size >50 under the random-effects model. The results of subgroup analysis were summarized in Table 3.
Table 3. Summary of the subgroup analysis.
| Case | Control | M-H pooled ORƛ | D+L pooled ORǂ | Heterogeneity | |||||
|---|---|---|---|---|---|---|---|---|---|
| Group | M+ | N | M+ | N | OR (95%CI) | OR (95%CI) | I2 (%) | P | τ2 |
| Total | 481 | 1030 | 180 | 775 | 2.83 (2.25–3.56) | 2.80 (2.03–3.85) | 37.1 | 0.05 | 0.18 |
| Population subgroup | |||||||||
| Asians | 231 | 404 | 105 | 369 | 3.82 (2.75–5.30) | 3.57 (2.56–4.99) | 0 | 0.73 | 0 |
| Caucasians | 250 | 626 | 75 | 406 | 2.13 (1.55–2.94) | 2.28 (1.40–3.71) | 46 | 0.04 | 0.31 |
| Case sample size | |||||||||
| ≤50 | 117 | 276 | 48 | 232 | 2.62 (1.72–3.99) | 2.52 (1.64–3.88) | 0 | 0.78 | 0 |
| >50 | 364 | 754 | 132 | 543 | 2.92 (2.22–3.84) | 3.05 (1.89–4.92) | 60.4 | <0.01 | 0.36 |
| Control source | |||||||||
| Autologous | 242 | 604 | 105 | 420 | 1.93 (1.45–2.56) | 1.78 (1.12–2.81) | 54.1 | 0.01 | 0.33 |
| Heterogeneous | 341 | 719 | 75 | 355 | 5.18 (3.56–7.53) | 4.31 (2.88–6.46) | 2.2 | 0.43 | 0.01 |
| Control sample type$ | |||||||||
| Tissue | 429 | 838 | 146 | 530 | 2.91 (2.30–3.68) | 3.14 (2.03–4.86) | 58.2 | <0.01 | 0.52 |
| Non-tissue | 110 | 372 | 34 | 245 | 2.78 (1.56–4.96) | 2.61 (0.81–8.41) | 59.6 | 0.03 | 1.16 |
| Method | |||||||||
| MSP | 396 | 851 | 172 | 665 | 2.62 (2.07–3.33) | 2.52 (1.82–3.50) | 35.9 | 0.07 | 0.15 |
| QMSP | 85 | 179 | 8 | 110 | 6.48 (2.76–15.18) | 6.48 (2.78–15.11) | 0 | 0.69 | 0 |
ƛ: the fixed-effects model
ǂ: the random-effects model
$: Non-tissue: serum, saliva and buccal cell
Sensitivity Analysis
To evaluate the effects of each individual study on the overall effect, the sensitivity analysis was performed. The overall OR was changed from 2.68 (95%CI, 2.13–3.38) to 3.27 (95%CI, 2.57–4.16) under the fixed effects model by omitting each single study, which demonstrates that the pooled OR between the MGMT promoter methylation and risk of HNSCC was reliable and stable (Fig 3).
Fig 3. Sensitivity analysis of included studies under the fixed-effects model.
Publication Bias
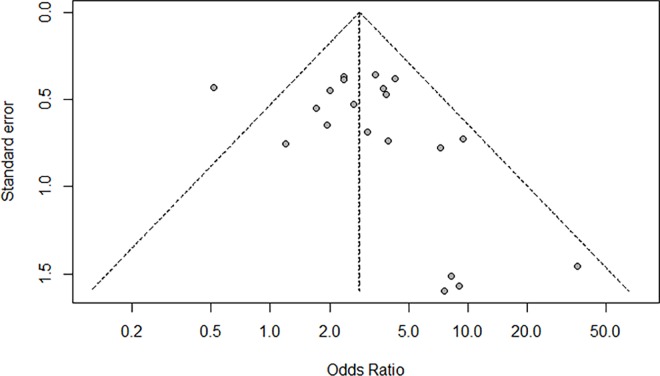
The Begg’s funnel plot and Egger’s test were employed to estimate the publication bias of the included studies. The Begg’s funnel plot of the pooled analysis in Fig 4 was quite symmetric and no publication bias was detected by Egger’s test (P = 0.31).
Fig 4. Funnel plot for assessment of publication bias in the meta-analysis.
Discussion
Epigenetic inactivation of the genes is common in human tumors. Hypermethylation of gene promoter is one of the important mechanisms for inactivation of tumor-suppressor genes involving apoptosis, DNA-repair, and cell cycle control [36]. The MGMT is a DNA repair gene, and the promoter methylation of MGMT plays an important role in the carcinogenic process and progression of cancer [37]. The protein MGMT also known as AGT (O6-alkylguanine transferase), is well known to play a crucial role in repairing O6-alkylguanine in DNA, a major premutagenic lesion produced by environmental and therapeutic alkylating agents [38]. Methylation of MGMT gene promoter can diminish MGMT protein expression in tumor tissues of various types of cancers, including lung cancer, gastric cancer, colorectal cancer, and breast cancer [39–43].
To our knowledge, this is the first meta-analysis on evaluating the association between MGMT promoter methylation and the risk of HNSCC. In this study, we found that 20 studies including 1030 cases and 775 controls were competent for inclusion criteria. The frequency of MGMT promoter methylation in tumor was 46.70% and 23.23% in control group. The result of the meta-analysis displayed that the methylation of MGMT promoter had an increased risk of HNSCC (OR = 2.83; 95%CI = 2.25–3.56).
Subgroup analysis by the methylation detection method showed the OR was 2.62 (95%CI = 2.07–3.33) in the MSP group and 6.48 (95%CI = 2.76–15.18) in the QMSP group under the fixed-effects model. In fact, QMSP is reported to be more specific and more sensitive, and able to detect much smaller magnitude (1/1000 methylated alleles) [44,45]. In contrast, the conventional MSP can only detect high concentrations of promoter methylation.
With the control style, the OR was 5.18 (95%CI = 3.56–7.53) in the heterogeneous control subgroup under the fixed-effects model and 1.78 (95%CI = 1.12–2.81) in autologous tissues subgroup under the random-effects model. The result suggested that the incidence of MGMT methylation in autologous control was higher than that in heterogeneous control. The result of the subgroup analysis is consistent with most previous studies [24,30,32]. In the control sample type subgroup, the OR was 3.14 (95%CI = 2.03–4.86) in the tissue group and 2.61 (95%CI = 0.81–8.41) in the non-tissue group under the fixed-effects model. In the subgroup analysis by sample size and race, significant associations were observed for all subgroups.
However, the present study had also several potential limitations. First, the search strategy was restricted to articles published in English language in this study. And then, some studies potentially suitable for inclusion that were published in other languages may be not included. Thus, some publication bias may exist. Second, we only study the association between MGMT promoter methylation and HNSCC in this study. We did not further investigate the association between MGMT promoter methylation and demographic (age and gender) and disease characteristics (stage, metastasis, and relapse) of HNSCC.
In conclusion, we found that hypermethylation of MGMT promoter was associated with an increased risk of HNSCC. The findings suggested that the promoter methylation of MGMT gene may play an important role in the carcinogenic process of HNSCC, and it may be a promising molecular marker for monitoring the disease and may provide new insights to the treatment of HNSCC. However, more studies with larger sample size must be performed to acquire a more precise and representative result.
Supporting Information
(DOC)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Nature Science Foundation of China (No. 81200745 and No. 81501297). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011; 61(2):69–90. 10.3322/caac.20107 [DOI] [PubMed] [Google Scholar]
- 2.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011; 11(1):9–22. 10.1038/nrc2982 [DOI] [PubMed] [Google Scholar]
- 3.Hashibe M, Brennan P, Chuang SC, Boccia S, Castellsague X, Chen C, et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev. 2009; 18(2):541–550. 10.1158/1055-9965.EPI-08-0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000; 92(9):709–720. [DOI] [PubMed] [Google Scholar]
- 5.Demokan S, Dalay N. Role of DNA methylation in head and neck cancer. Clin Epigenetics. 2011; 2(2):123–150. 10.1007/s13148-011-0045-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worsham MJ, Stephen JK, Chen KM, Havard S, Shah V, Gardner G, et al. Delineating an epigenetic continuum in head and neck cancer. Cancer Lett. 2014; 342(2):178–184. 10.1016/j.canlet.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burger PC, Green SB. Patient age, histologic features, and length of survival in patients with glioblastoma multiforme. Cancer. 1987; 59(9):1617–1625. [DOI] [PubMed] [Google Scholar]
- 8.Wong Y-K, Lee L-T, Liu C-J. Hypermethylation of MGMT and DAPK gene promoters is associated with tumorigenesis and metastasis in oral squamous cell carcinoma. Journal of Dental Sciences. 2011; 6(3):158–164. [Google Scholar]
- 9.Misawa K, Mochizuki D, Imai A, Endo S, Mima M, Misawa Y, et al. Prognostic value of aberrant promoter hypermethylation of tumor-related genes in early-stage head and neck cancer. Oncotarget. 2016;7(18):26087–98. 10.18632/oncotarget.8317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Celebi OO, Sener E, Hosal S, Gullu IH, Cengiz M, Sozeri AB, et al. 06-methylguanine-DNA-methyltransferase gene promoter region methylation pattern and its effect on survival, recurrence and chemosensitivity in laryngeal cancer. Virchows Archiv. 2013; 463(2):318–318. [Google Scholar]
- 11.Shi H, Li Y, Wang X, Lu C, Yang L, Gu C, et al. Association between RASSF1A promoter methylation and ovarian cancer: a meta-analysis. PLoS One. 2013; 8(10):e76787 10.1371/journal.pone.0076787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi H, Chen X, Lu C, Gu C, Jiang H, Meng R, et al. Association between P16INK4a promoter methylation and HNSCC: a meta-analysis of 21 published studies. PLoS One. 2015; 10(4):e0122302 10.1371/journal.pone.0122302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002; 21(11):1539–1558. [DOI] [PubMed] [Google Scholar]
- 14.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994; 50(4):1088–1101. [PubMed] [Google Scholar]
- 15.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997; 315(7109):629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onerci Celebi O, Tezel GG, Hosal AS, Cengiz M, Gullu IH, Hayran M. Detection of O6-methylguanine-DNA methyltransferase gene promoter region methylation pattern using pyrosequencing and the effect of methylation pattern on survival, recurrence, and chemotherapy sensitivity in patients with laryngeal cancer. Pathol Res Pract. 2016;212(5):456–62. 10.1016/j.prp.2016.02.022 [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Zhu X-B, He L-X, Gu Z-W, Jin M-Z, Ji W-Y. Clinical significance of epigenetic silencing and re-expression of O6-methylguanine-DNA methyltransferase using epigenetic agents in laryngeal carcinoma. Oncology Letters. 2015; 9(1):35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asokan GS, Jeelani S, Gnanasundaram N. Promoter hypermethylation profile of tumour suppressor genes in oral leukoplakia and oral squamous cell carcinoma. J Clin Diagn Res. 2014; 8(10):Zc09–12. 10.7860/JCDR/2014/9251.4949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatia V, Goel MM. Promoter region hypermethylation and mRNA expression of MGMT and p16 genes in tissue and blood samples of human premalignant oral lesions and oral squamous cell carcinoma. 2014; 2014:248419 10.1155/2014/248419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rettori MM, de Carvalho AC, Longo AL, de Oliveira CZ, Kowalski LP, Carvalho AL, et al. TIMP3 and CCNA1 hypermethylation in HNSCC is associated with an increased incidence of second primary tumors. J Transl Med. 2013; 11:316 10.1186/1479-5876-11-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutsimpelas D, Pongsapich W, Heinrich U, Mann S, Mann WJ, Brieger J. Promoter methylation of MGMT, MLH1 and RASSF1A tumor suppressor genes in head and neck squamous cell carcinoma: pharmacological genome demethylation reduces proliferation of head and neck squamous carcinoma cells. Oncol Rep. 2012; 27(4):1135–1141. 10.3892/or.2012.1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paluszczak J, Misiak P, Wierzbicka M, Wozniak A, Baer-Dubowska W. Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa. Oral Oncol. 2011; 47(2):104–107. 10.1016/j.oraloncology.2010.11.006 [DOI] [PubMed] [Google Scholar]
- 23.Weiss D, Basel T, Sachse F, Braeuninger A, Rudack C. Promoter methylation of cyclin A1 is associated with human papillomavirus 16 induced head and neck squamous cell carcinoma independently of p53 mutation. Mol Carcinog. 2011; 50(9):680–688. 10.1002/mc.20798 [DOI] [PubMed] [Google Scholar]
- 24.Su PF, Huang WL, Wu HT, Wu CH, Liu TY, Kao SY. p16(INK4A) promoter hypermethylation is associated with invasiveness and prognosis of oral squamous cell carcinoma in an age-dependent manner. Oral Oncol. 2010; 46(10):734–739. 10.1016/j.oraloncology.2010.07.002 [DOI] [PubMed] [Google Scholar]
- 25.Kordi-Tamandani DM, Moazeni-Roodi AK, Rigi-Ladiz MA, Hashemi M, Birjandian E, Torkamanzehi A. Promoter hypermethylation and expression profile of MGMT and CDH1 genes in oral cavity cancer. Arch Oral Biol. 2010; 55(10):809–814. 10.1016/j.archoralbio.2010.06.017 [DOI] [PubMed] [Google Scholar]
- 26.Steinmann K, Sandner A, Schagdarsurengin U, Dammann RH. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep. 2009; 22(6):1519–1526. [DOI] [PubMed] [Google Scholar]
- 27.De Schutter H, Geeraerts H, Verbeken E, Nuyts S. Promoter methylation of TIMP3 and CDH1 predicts better outcome in head and neck squamous cell carcinoma treated by radiotherapy only. Oncol Rep. 2009; 21(2):507–513. [PubMed] [Google Scholar]
- 28.Righini CA, de Fraipont F, Timsit JF, Faure C, Brambilla E, Reyt E, et al. Tumor-specific methylation in saliva: a promising biomarker for early detection of head and neck cancer recurrence. Clin Cancer Res. 2007; 13(4):1179–1185. [DOI] [PubMed] [Google Scholar]
- 29.Martone T, Gillio-Tos A, De Marco L, Fiano V, Maule M, Cavalot A, et al. Association between hypermethylated tumor and paired surgical margins in head and neck squamous cell carcinomas. Clin Cancer Res. 2007; 13(17):5089–5094. [DOI] [PubMed] [Google Scholar]
- 30.Kato K, Hara A, Kuno T, Mori H, Yamashita T, Toida M, et al. Aberrant promoter hypermethylation of p16 and MGMT genes in oral squamous cell carcinomas and the surrounding normal mucosa. J Cancer Res Clin Oncol. 2006; 132(11):735–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruya S, Issa JP, Weber RS, Rosenthal DI, Haviland JC, Lotan R, et al. Differential methylation status of tumor-associated genes in head and neck squamous carcinoma: incidence and potential implications. Clin Cancer Res. 2004; 10(11):3825–3830. [DOI] [PubMed] [Google Scholar]
- 32.Kulkarni V, Saranath D. Concurrent hypermethylation of multiple regulatory genes in chewing tobacco associated oral squamous cell carcinomas and adjacent normal tissues. Oral Oncol. 2004; 40(2):145–153. [DOI] [PubMed] [Google Scholar]
- 33.Viswanathan M, Tsuchida N, Shanmugam G. Promoter hypermethylation profile of tumor-associated genes p16, p15, hMLH1, MGMT and E-cadherin in oral squamous cell carcinoma. Int J Cancer. 2003; 105(1):41–46. [DOI] [PubMed] [Google Scholar]
- 34.Rosas SL, Koch W, da Costa Carvalho MG, Wu L, Califano J, Westra W, et al. Promoter hypermethylation patterns of p16, O6-methylguanine-DNA-methyltransferase, and death-associated protein kinase in tumors and saliva of head and neck cancer patients. Cancer Res. 2001; 61(3):939–942. [PubMed] [Google Scholar]
- 35.Sanchez-Cespedes M, Esteller M, Wu L, Nawroz-Danish H, Yoo GH, Koch WM, et al. Gene promoter hypermethylation in tumors and serum of head and neck cancer patients. Cancer Res. 2000; 60(4):892–895. [PubMed] [Google Scholar]
- 36.Scully C, Field JK, Tanzawa H. Genetic aberrations in oral or head and neck squamous cell carcinoma (SCCHN): 1. Carcinogen metabolism, DNA repair and cell cycle control. Oral Oncol. 2000; 36(3):256–263. [DOI] [PubMed] [Google Scholar]
- 37.Soejima H, Zhao W, Mukai T. Epigenetic silencing of the MGMT gene in cancer. Biochem Cell Biol. 2005; 83(4):429–437. [DOI] [PubMed] [Google Scholar]
- 38.Pegg AE. Mammalian O6-alkylguanine-DNA alkyltransferase: regulation and importance in response to alkylating carcinogenic and therapeutic agents. Cancer Res. 1990; 50(19):6119–6129. [PubMed] [Google Scholar]
- 39.Zuo C, Ai L, Ratliff P, Suen JY, Hanna E, Brent TP, et al. O6-methylguanine-DNA methyltransferase gene: epigenetic silencing and prognostic value in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2004; 13(6):967–975. [PubMed] [Google Scholar]
- 40.Krakowczyk L, Strzelczyk JK, Adamek B, Zalewska-Ziob M, Arendt J, Poltorak S, et al. Methylation of the MGMT and p16 genes in sporadic colorectal carcinoma and corresponding normal colonic mucosa. Med Sci Monit. 2008; 14(10):Br219–225. [PubMed] [Google Scholar]
- 41.Park TJ, Han SU, Cho YK, Paik WK, Kim YB, Lim IK. Methylation of O(6)-methylguanine-DNA methyltransferase gene is associated significantly with K-ras mutation, lymph node invasion, tumor staging, and disease free survival in patients with gastric carcinoma. Cancer. 2001; 92(11):2760–2768. [DOI] [PubMed] [Google Scholar]
- 42.Wu PF, Kuo KT, Kuo LT, Lin YT, Lee WC, Lu YS, et al. O(6)-Methylguanine-DNA methyltransferase expression and prognostic value in brain metastases of lung cancers. Lung Cancer. 2010; 68(3):484–490. 10.1016/j.lungcan.2009.08.010 [DOI] [PubMed] [Google Scholar]
- 43.Hu ZY, Tang LD, Zhou Q, Xiao L, Cao Y. Aberrant promoter hypermethylation of p16 gene in endometrial carcinoma. Tumour Biol. 2015; 36(3):1487–1491. 10.1007/s13277-014-2632-3 [DOI] [PubMed] [Google Scholar]
- 44.Yates DR, Rehman I, Meuth M, Cross SS, Hamdy FC, Catto JW. Methylational urinalysis: a prospective study of bladder cancer patients and age stratified benign controls. Oncogene. 2006; 25(13):1984–1988. [DOI] [PubMed] [Google Scholar]
- 45.Fackler MJ, Malone K, Zhang Z, Schilling E, Garrett-Mayer E, Swift-Scanlan T, et al. Quantitative multiplex methylation-specific PCR analysis doubles detection of tumor cells in breast ductal fluid. Clin Cancer Res. 2006; 12(11 Pt 1):3306–3310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.