Abstract
Background
Attention function filters and selects behaviorally relevant information. This capacity is impaired in some psychiatric disorders and has been proposed as an endophenotype for Attention-Deficit/Hyperactivity Disorder; however, its genetic basis remains largely unknown. This study aimed to identify single nucleotide polymorphism (SNPs) associated with attention function.
Materials and Methods
The discovery sample included 1655 children (7–12 years) and the replication sample included 546 children (5–8 years). Five attention outcomes were assessed using the computerized Attentional Network Test (ANT): alerting, orienting, executive attention, Hit Reaction time (HRT) and the standard error of HRT (HRTSE). A Genome-wide Association Study was conducted for each outcome. Gene set enrichment analyses were performed to detect biological pathways associated with attention outcomes. Additional neuroimaging analyses were conducted to test neural effects of detected SNPs of interest.
Results
Thirteen loci showed suggestive evidence of association with attention function (P<10−5) in the discovery sample. One of them, the rs4321351 located in the PID1 gene, was nominally significant in the replication sample although it did not survive multiple testing correction. Neuroimaging analysis revealed a significant association between this SNP and brain structure and function involving the frontal-basal ganglia circuits. The mTOR signaling and Alzheimer disease-amyloid secretase pathways were significantly enriched for alerting, orienting and HRT respectively (FDR<5%).
Conclusion
These results suggest for the first time the involvement of the PID1 gene, mTOR signaling and Alzheimer disease-amyloid secretase pathways, in attention function during childhood. These genes and pathways have been proposed to play a role in neuronal plasticity, memory and neurodegenerative disease.
Introduction
Attention is a cognitive function essential in daily life. Every day, our perceptual systems are exposed to a massive internal and external sensory input for which the relevant behavioural information is selected and prioritized [1, 2]. Attention function allows the selection and prioritization of stimuli by activating different neural systems that interact with each other in a complex manner. It has been proposed that three functionally and anatomically different networks are involved in this process: alerting, orienting and executive attention [3]. According to Posner and Rothbart (2009), ‘alerting is defined as achieving and maintaining a state of high sensitivity to incoming stimuli; orienting is the selection of information from sensory input; and executive attention involves mechanisms for monitoring and resolving conflict among thoughts, feelings, and responses’ [3].
From a developmental perspective, attention constitutes a precursor of other higher-level cognitive abilities such as learning and memory [4]. Therefore, attention function plays a key role in cognitive development. From a clinical perspective, attention function is impaired in neuropsychiatric disorders such as Attention-Deficit/Hyperactivity Disorder (ADHD) and schizophrenia [5–9], thus, research in attention may have etiological implications for these disorders. These facts highlight the relevance of investigating the sources of variation of attention function.
Although experiences with the caregivers, education and other exposures account for individual variation in cognitive functions, the development of attention is partly specified by genes [3]. Genetic effects on attention function variation have been found early in life [10]. The catechol-O-methyltransferase gene (COMT) and the dopamine D4 receptor gene (DRD4) are among the candidate genes for attention function development [1, 10, 11]. However, these studies found modest associations and they failed to identify consistent and replicable results. A twin study including only 26 pairs estimated the heritability of the abovementioned attention networks ranging from 0% for orienting to 72% for executive attention [12]. Other studies report low to moderate (28–38%) and high (79%) heritability estimates for attention function [13, 14]. Therefore, although we can expect genetic influences on attention function development, there is still scarce knowledge about the genetic basis of attention in general population. Furthermore, to our knowledge, there are no previous genome-wide association studies (GWAS) on attention function during childhood.
The main goal of the present study was to identify common genetic variants associated with attention at a genome-wide level. The steps followed include: i) identification of single nucleotide polymorphisms (SNPs) associated with attention, ii) replication of significant findings in an independent sample, iii) search for relevant biological pathways accumulating associated genetic variants using gene set enrichment analyses (GSEA) and iv) examination of potential association between relevant identified SNPs and variations in brain structure and function using neuroimaging tools.
Materials and Methods
Discovery sample
The discovery sample was obtained from the BRain dEvelopment and Air polluTion ultrafine particles in scHool childrEn (BREATHE) project aimed to analyze the association between air pollution and cognitive development of scholars [15]. From the total of 2897 children participating in this project, genotypic and neurocognitive data was available for 1655 individuals. All parents and legal guardians signed the informed consent approved by the Clinical Research Ethical Committee (No. 2010/41221/I) of the Institut Hospital del Mar d’Investigacions Mèdiques–Parc de Salut Mar, Barcelona, Spain.
Replication sample
The replication sample included 546 children from the INfancia y Medio Ambiente (INMA) multicenter birth cohort project recruited in the cities of Sabadell and Valencia (Spain) (INMA-SabVal) [16]. All parents and legal guardians signed the informed consent approved by the Clinical Research Ethical Committee of the Institut Hospital del Mar d’Investigacions Mèdiques–Parc de Salut Mar, Barcelona and institutional ethics committees in each region.
Measures
Attention function was assessed using the computerized Attentional Network Test [ANT; [17]] which assesses three attentional networks: alerting, orienting and executive attention. The computerized version of this test has been validated with brain imaging [17] and in the general population [18]. As part of the BREATHE project, a follow-up with four repeated measurements of the attention function were conducted in a period of a year. The outcomes analyzed herein correspond to the first administration of the test. Five attention outcomes were analyzed in the current study: alerting, orienting, executive attention, hit-reaction time (HRT) and the standard error of the HRT (HRTSE). Reaction time (RT) measures (i.e. time between the introduction of a stimulus and the reaction on the subject to that stimulus) were used to calculate alerting (RT for no cue minus RT for double cue trials), orienting (RT for central cue minus RT for spatial cue trials) and executive function (RT for incongruent minus RT for congruent trials) scores. HRT (Median RT for correct responses) and HRTSE (Standard error or RT for correct responses) were also analyzed as measures of variability. All the outcomes analyzed were continuous variables. Higher scores indicate worse performance. Children with >30% errors were excluded from the analysis. Further details can be found elsewhere [18].
Study Design, Genotyping and quality control
To identify novel loci associated with attention outcomes, we conducted a GWAS with follow-up of associations at suggestive evidence (P<10−5) in the replication sample.
In the discovery sample, DNA samples from 2492 children were obtained from saliva collected in Oragene DNA OG-500kit (DNA Genotek) following instructions of the manufacturer with minor modifications. DNA samples were quantified using Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies). A final subset of 1778 children was selected for genome-wide genotyping after applying a filtering criteria (low quality DNA, no neuropsychological data, non Caucasian descent origin and not born in Spain, parents born in Europe, and adopted children). Genome-wide genotyping for the discovery sample was performed using the HumanCore BeadChip WG-330-1101 (Illumina) at the Spanish National Genotyping Centre (CEGEN) coordinated by the Spanish National Cancer Research Centre CNIO). Genotype calling was done using the GeneTrain2.0 algorithm (with a default threshold of 0.15) based on HapMap clusters implemented in the GenomeStudio software. Twenty CEU HapMap duplicates and twenty BREATHE duplicates were included in the study and gave consistent results.
PLINK was used for the genotyping quality of the sample and SNPs [19]. Quality control procedures were samples with a minimum of 97% call rate (N = 3 exclusions) and a maximum of 4 SD heterozigosity were included (N = 5 exclusions). Further checking was conducted for gender discordance excluding mismatch information (N = 18 exclusions, representing 1% of the sample), sample relatedness excluding proportions of identity by-state above 0.185 (N = 80 exclusions: 1 twin, 32 siblings, 39 cousins, 8 incongruent sibling's couples) and population stratification. Five subjects were excluded due to mental disabilities. In total we excluded 111 subjects (6.26%) leaving 1667 individuals from who 1655 have data available for the attention outcomes considered in the present study.
Genetic variants were filtered by Hardy-Weinberg equilibrium (P<10−6), allele frequency (excluding minor allele frequency (MAF<1%) and SNP call rate with a minimum of 95%. In total, 58827 genetic variants (19.68%) were excluded. The final discovery genetic data set included 240103 SNPs.
The replication cohort was genotyped using the HumanOmni1-Quad v1.0 Beadchip (Illumina) at the CEGEN. Quality control procedures for the replication sample were also performed in PLINK. Samples with a minimum of 98% call rate and a maximum of 3 SD heterozigosity were included. Furthermore, gender discordance, sample relatedness (excluding proportions of identity by-state above 0.185) and population stratification were checked. Genetic variants were filtered using the same criteria as in the discovery sample. The final replication sample included 546 subjects.
Statistical Analysis
We used a two-sample t-test to check for differences in ANT outcomes and age, and a Pearson’s χ2 test to check for sex differences. Genome-wide association analyses were conducted using linear regression models in SNPtest [20]. Separate models were tested for each ANT outcome as dependent variables and genetic markers as predictors. Additive genetic models were assumed to assess the association of each SNP with each ANT outcome, adjusting for age, sex and school.
Quantile-quantile (Q-Q) and Manhattan plots were computed using the qqman package of R. Genome-wide significance was set at P<5x10-8, and suggestive evidence of association was set at P<10−5. These thresholds have been recommended by a simulation study taking into account linkage disequilibrium (LD) between SNPs [21]. SNPs showing an association with attention outcomes (at GWAS or suggestive significance) were taken forward for replication in the INMA-SabVal sample. In the replication sample, multiple linear regressions in SNPtest adjusting by age, sex and cohort were conducted. In order to be replicated, SNPs must be nominally significant (P<0.05) after multiple testing correction (FDR<0.05).
To further analyze the association signal, regions which include SNPs of potential interest for cognition were imputed using IMPUTE2 v2 [20] taking the 1000 Genomes project phase I integrated variant set (http:/www.1000genomes.org/) as a reference haplotype panel. Regional association plots were computed with LocusZoom [22].
In addition, potentially relevant SNPs detected were analyzed for associations with gene expression using the Brain expression quantitative trait loci (eQTL) Almanac (http://www.braineac.org/) [23]. BRAINEAC is a publicly accessible database which contains gene expression data (generated eQTL) analyzed in ten brain regions from postmortem human brains.
Gene set enrichment analyses (GSEA) were conducted using Meta-Analysis Gene-set Enrichment of variaNT Associations (MAGENTA) software (19) for each attention outcome. Data sources included Reactome, Panther, KEGG and Ingenuity. As described in detail previously [24], MAGENTA individually mapped genes in the genome to the lowest P-value single SNP within a 110kb upstream and 40kb downstream window. These P-values were adjusted for confounding factors (e.g. physical gene size, number of SNPs per kilobase for each gene and other genetic properties). Genes are then ranked according to these adjusted P-values, and the gene-set enrichment P-value for each biological pathway was calculated for a given significance threshold (95th percentile). To test whether genes were enriched in a pathway more than would be expected by chance, this value was compared with that generated with randomly permuted pathways of identical size. Individual pathways that reached FDR<0.05 were deemed significant and results for the 95th percentile cut-off analysis were reported.
Neuroimaging analyses
To further understand the role of SNPs of potential interest for cognition, its effects on brain structure and function were examined in a subsample of 185 children drawn from the BREATHE project who underwent neuroimaging studies with genetic and cognitive data available. More details in [25]. The imaging approach included whole-brain mapping of cortical thickness using high resolution 3D anatomic MRI, fractional anisotropy (FA) from diffusion tensor imaging (DTI) and resting-state functional connectivity in selected relevant large-scale networks [26–28]. Further details can be found in S1 Text.
MRI acquisition was performed using a 1.5 Tesla Signa Excite system (General Electric, Milwaukee, WI, USA) equipped with an eight-channel phased-array head coil and single-shot echo planar imaging (EPI) software was used (further details can be found in S1 Text.
Imaging data were analyzed using Statistical Parametric Mapping (SPM8) (http://www.fil.ion.ucl.ac.uk/spm, Wellcome Department of Cognitive Neurology, London, UK, 2008). Individual anatomical (cortical thickness), DTI and functional connectivity maps were included in second-level (group) analyses to map voxel-wise the correlation across-subjects between individual brain measurements and the SNP of interest. Results were considered significant with clusters of 1.032 ml (e.g., 129 voxels with a resolution of 2x2x2 mm) at a height threshold of p<0.005, which satisfied the family-wise error (FWE) rate correction of PFWE<0.05 according to recent Monte Carlo simulations [29]. Maps in figures are displayed at t>2.3.
Results
Descriptive results
Table 1 shows age, sex ratio and scores for the five attention outcomes of the discovery (BREATHE) and replication sample (INMA-SabVal). Within BREATHE sample, girls showed a better performance in executive attention [t(1493) = 4.21; P<0.001], but a worse performance in HRT [t(1493) = -8.39; P<0.001] and HRTSE [t(1493) = -5.63; P<0.001], compared to boys. Similar findings were observed within INMA-SabVal sample, in regard to sex differences for HRT [t(544) = 6.1; P<0.001] and HRTSE [t(544) = 4.6; P<0.001].
Table 1. Descriptive data for the variables of the study for the discovery (Breathe) and replication (INMA-SabVal) samples.
Percentage is indicated for categorical variables. Mean, SD and maximum and minim are indicated for continuous variables.
| Breathe (n = 1655) | INMA-SabVal (n = 546) | Comparison | |
|---|---|---|---|
| Sex, females (%) | 789 (47.7%) | 266 (48.7%) | Chi(1) = 0.3415; P = .599 |
| Age | 9.2 (.87) (7.5/11.6) | 7.12 (0.48) (5.36/8.55) | t(1738) = 71.62; P < .001 |
| Alerting | 48.3 (76.1) (-337.5/424) | 54.37 (92.9) (-450/448) | t(826) = -1.35; P = .175 |
| Orienting | 36.8 (74.3) (-297/403) | 33.31 (92.86) (-407.5/329) | t(813) = 0.78; P = .432 |
| Executive Attention | 62.2 (58.0) (-182/557) | 79.95 (84.63) (-185.5/646.5) | t(740) = -4.52; P < .001 |
| HRT | 803.9 (160.4) (438/1501) | 941.11 (186.84) (594.5/1724.5) | t(855) = -15.23; P < .001 |
| HRTSE | 266.3 (87.2) (76.5/528.5) | 317.12 (78) (97.25/507.99) | t(1074) = -12.63; P < .001 |
Genome-wide association study: Discovery sample
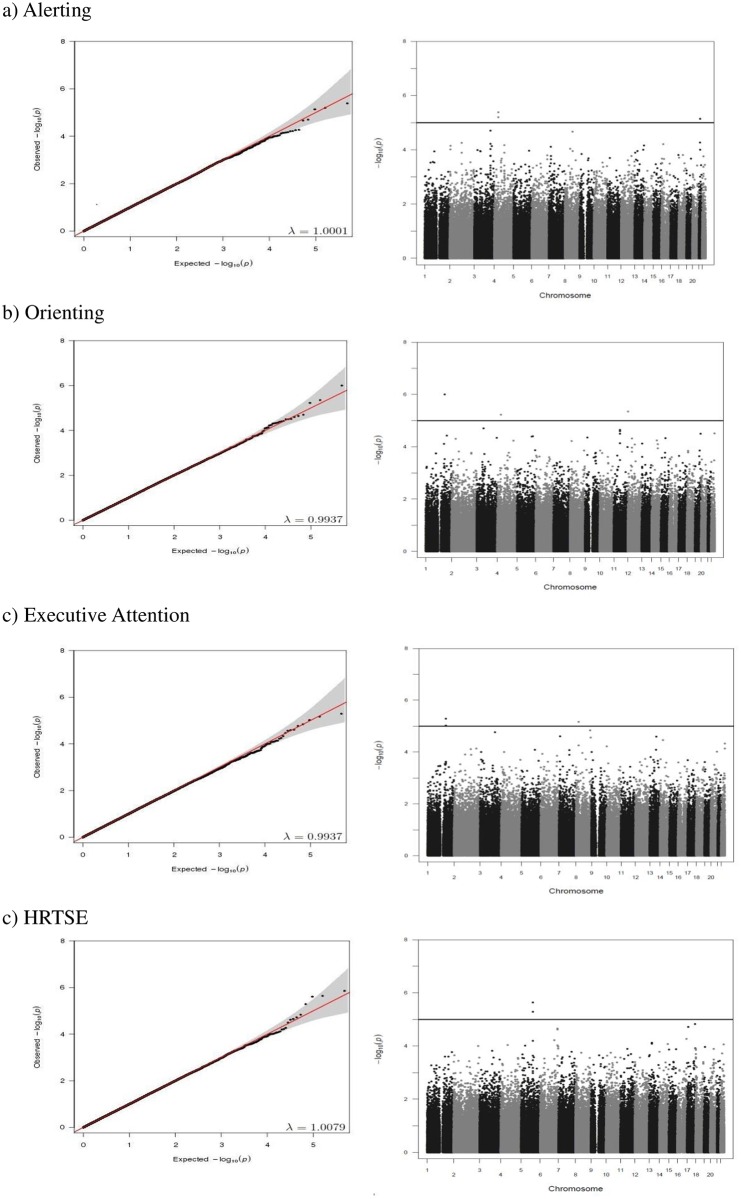
Q-Q plots of the observed versus expected P-values and Manhattan plots showing the distribution of negative log-transformed P-values for every attention outcome are provided in Figs 1 and 2a. The Q-Q plots showed no departure from the expected P-values distribution. Genomic control inflation factor (λ) is included in each Q-Q plot.
Fig 1. Quantile-quantile (Q-Q) plots (left side) and Manhattan plots (right side) of genome-wide association analyses for (a) alerting, (b) orienting, (c) executive attention, and (d) HRTSE attention outcomes in the discovery sample.
Genomic inflation factor (λ) is included in each Q-Q plot. The blue line in the Manhattan plots indicates the suggestive level of statistical significance (P<10−5).
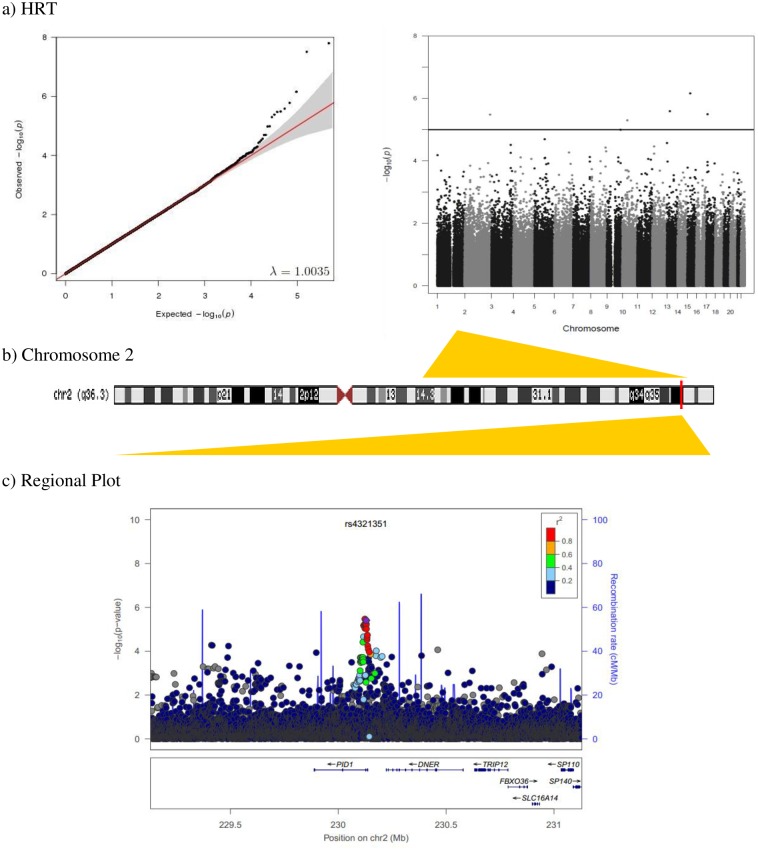
Fig 2.
a) Quantile-quantile and Manhattan plot of genome-wide association results for HRT. The blue line indicates the suggestive level of statistical significance (p<10−5). b) Diagram of the chromosome 2. The red line indicates the position of the rs4321351 (230,129,493 bp). c) Regional association plot of rs4321351 located in PID1 gene. The linkage disequilibrium (LD; r2) between the SNP in focus and its SNPs genotyped or imputed within 1Mb is showed in red (high LD) to blue (low LD). The recombination rate is plotted in blue according to HapMap (CEU).
No SNPs were genome-wide significant (P<10−8). Nevertheless, 13 loci showed suggestive evidence of association with attention outcomes (Table 2).
Table 2. SNPs associated with attention function outcomes at P<10−5 (ordered by significance).
| Attention outcome | SNP | CHR | position | Allelea | MAF | N | β | SE | P-value | Gene | Nearest gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alerting | rs10015679 | 4 | 40644376 | T/C | 0.319 | 1491 | -13.80 | 3.00 | 4.10 x 10−6 | Intergenic | RBM47 |
| rs13048083 | 21 | 28286853 | T/C | 0.247 | 1491 | -14.48 | 3.23 | 7.33 x 10−6 | Intergenic | ADAMTS5 | |
| Orienting | rs10911457 | 1 | 183843104 | T/C | 0.461 | 1492 | 13.14 | 2.69 | 9.99 x 10−7 | RGL1 | - |
| rs12579294 | 12 | 3289945 | T/C | 0.222 | 1492 | -14.84 | 3.23 | 4.46 x 10−6 | TSPAN9 | - | |
| rs4629469 | 4 | 36419047 | G/A | 0.357 | 1490 | 12.76 | 2.82 | 5.95 x 10−6 | Intergenic | DTHD1 | |
| Executive Attention | rs2207190 | 1 | 171415856 | G/A | 0.405 | 1493 | -9.84 | 2.16 | 5.12 x 10−6 | Intergenic | PRRC2C |
| rs2320783 | 8 | 25009089 | G/A | 0.134 | 1493 | -14.27 | 3.17 | 6.82 x 10−6 | Intergenic | DOCK5 | |
| HRT | rs4775379 | 15 | 46682794 | T/C | 0.216 | 1484 | 35.87 | 7.23 | 6.98 x 10−7 | Intergenic | SQRDL |
| rs951738 | 13 | 45479633 | G/A | 0.234 | 1442 | 32.85 | 6.99 | 2.60 x 10−6 | Intergenic | NUFIP1 | |
| rs757594 | 17 | 12000632 | G/A | 0.233 | 1493 | 33.09 | 7.11 | 3.25 x 10−6 | MAP2K4 | - | |
| rs4321351 | 2 | 230129493 | G/A | 0.318 | 1493 | -28.99 | 6.24 | 3.35 x 10−6 | PID1 | - | |
| rs6593376 | 10 | 44469148 | T/C | 0.215 | 1493 | 32.31 | 7.08 | 5.02 x 10−6 | Intergenic | LINC00841 | |
| HRTSE | rs1560054 | 5 | 111519018 | T/C | 0.282 | 1493 | -16.95 | 3.59 | 2.29 x 10−6 | EPB41L4A | - |
SNP, single nucleotide polymorphism; CHR, chromosome; MAF, minor allele frequency; β, regression coefficient; SE, standard error.
a Effect allele/Other allele
RBM47, RNA binding motif protein 47; ADAMTS5, ADAM metallopeptidase with thrombospondin type 1 motif, 5; RGL1, Ral guanine nucleotide dissociation stimulator-like 1; TSPAN9, tetraspanin 9; DTHD1, death domain containing 1; PRRC2C, proline-rich coiled-coil 2C; DOCK5, dedicator of cytokinesis 5; SQRDL; sulfide quinone reductase-like; NUFIP1, nuclear fragile X mental retardation protein interacting protein 1; MAP2K4, dual specificity mitogen-activated protein kinase kinase 4; PID1, phosphotyrosine interaction domain containing 1; LINC00841, long intergenic non-protein coding RNA 841; EPB41L4A, erythrocyte membrane protein band 4.1 like 4.
The SNP with the strongest association was the rs4775379 SNP (β = 35.9, P = 6.98 x 10−7) associated with HRT (Table 2). The nearest gene to this intergenic SNP located on chromosome 15 is the sulfide quinone reductase-like (SQRDL) gene. The second most significant SNP was the rs10911457, associated with orienting (β = 13.1, P = 10.0 x 10−7), located on chromosome 1 in the Ral guanine nucleotide dissociation stimulator-like 1 (RGL1) (Table 2).
Top five most significant SNPs associated with each attention outcome can be found in S1 Table. Full summary statistics for all SNPs tested in each attention outcome can be found in S1–S5 Files.
Genome-wide association study: Replication sample
The rs4321351 was nominally significant in the replication sample, although neither this SNP nor the others showing suggestive evidence of association with the attention outcomes remained significant after multiple testing correction (FDR<0.05).
The nominally significant SNP associated with HRT in the discovery sample (β = -29.0, P = 3.35 x 10−6) showed same direction of additive effect in INMA-SabVal sample (β = -27.7, P = 0.025) (Table 3). HRT scores decreased as a function of the G allele copies of the rs4321351. This SNP is located in an intronic region of the phosphotyrosine interaction domain containing 1 (PID1) gene. Regional association analysis within 1Mb of this loci (chr2:230129493) identified a linkage disequilibrium (LD) block of 18 SNPs (r2 > 0.8) yielding strong evidences of multiple association signals for HRT (Fig 2).
Table 3. SNPs replicated in INMA-SabVal sample.
| Attention outcome | SNP | CHR | position | Allelea | MAF | N | β | SE | P-value | FDR |
|---|---|---|---|---|---|---|---|---|---|---|
| Alerting | rs10015679 | 4 | 40644376 | T/C | 0.324 | 545 | 2.95 | 6.08 | 0.628 | 0.891 |
| rs13048083 | 21 | 28286853 | T/C | 0.247 | 545 | 6.60 | 6.45 | 0.307 | 0.891 | |
| Orienting | rs10911457 | 1 | 183843104 | C/T | 0.454 | 546 | 5.10 | 5.65 | 0.367 | 0.891 |
| rs12579294 | 12 | 3289945 | T/C | 0.221 | 546 | -10.09 | 6.98 | 0.148 | 0.858 | |
| rs4629469 | 4 | 36419047 | G/A | 0.376 | 546 | 3.69 | 5.71 | 0.517 | 0.891 | |
| Executive Attention | rs2207190 | 1 | 171415856 | G/A | 0.430 | 545 | -0.33 | 5.15 | 0.949 | 0.949 |
| rs2320783 | 8 | 25009089 | G/A | 0.146 | 545 | -3.15 | 7.21 | 0.662 | 0.891 | |
| HRT | rs4775379 | 15 | 46682794 | T/C | 0.253 | 546 | -4.46 | 12.74 | 0.726 | 0.891 |
| rs951738 | 13 | 45479633 | A/G | 0.241 | 546 | 5.65 | 12.69 | 0.656 | 0.891 | |
| rs757594 | 17 | 12000632 | A/G | 0.254 | 546 | -1.92 | 12.93 | 0.882 | 0.949 | |
| rs4321351 | 2 | 230129493 | G/A | 0.321 | 546 | -27.74 | 12.32 | 0.025 | 0.325 | |
| rs6593376 | 10 | 44469148 | T/C | 0.228 | 546 | -17.25 | 13.40 | 0.198 | 0.858 | |
| HRTSE | rs1560054 | 5 | 111519018 | C/T | 0.287 | 546 | 1.59 | 5.08 | 0.754 | 0.891 |
SNP, single nucleotide polymorphism; CHR, chromosome; MAF, minor allele frequency; β, regression coefficient; SE, standard error.
a Effect allele/Other allele
The eQTL analysis for rs4321351 indicated that PID1 and DNER genes were among the top ten most affected genes by this SNP. Moreover, exon-specific probesets in PID1 (ID: 2602738) and DNER (ID: 2602778) genes were expressed in putamen (p = 0.004 and p = 0.009, respectively) according to BRAINEAC database (S2 Table).
Gene set enrichment analysis results
Among the total of 195 functional pathways nominally associated with the attention outcomes (P<0.05), three remained significant after correcting for multiple testing (FDR<5%) (Table 4). The strongest enrichment was found for the alerting attention outcome involving the Alzheimer disease-amyloid secretase pathway (P = 9.40x10-5; FDR = 0.014) followed by the sex determination pathway associated with orienting (P = 6.00x10-4; FDR = 0.007). Also, a significant association was found for the mammalian target of rapamycin (mTOR) signalling pathway (P = 4.00x10-4; FDR = 0.043) for the HRT attention outcome.
Table 4. Gene set enrichment analysis (GSEA).
Pathways significantly associated with attention outcomes after applying multiple testing correction.
| Nominal 95th Percentile | |||||||
|---|---|---|---|---|---|---|---|
| Outcome | Data Base | Gene Set | Size (n° of genes) | Expected | Observed | P-value | FDR |
| Alerting | PB | Sex determination | 9 | 0 | 4 | 6.00 x 10−4 | 0.007 |
| Orienting | RE | mTOR signalling | 27 | 1 | 7 | 4.00 x 10−4 | 0.043 |
| HRT | PA | Alzheimer disease-amyloid secretase pathway | 23 | 1 | 7 | 9.40 x 10−5 | 0.014 |
RE, Reactome; PA, Panther; PB, Panther-Biological process
Top five most significant pathways associated with each attention function outcome can be found in S3 Table.
Neuroimaging results
Although none of the SNPs were replicated, we further explored the nominally significant SNP.
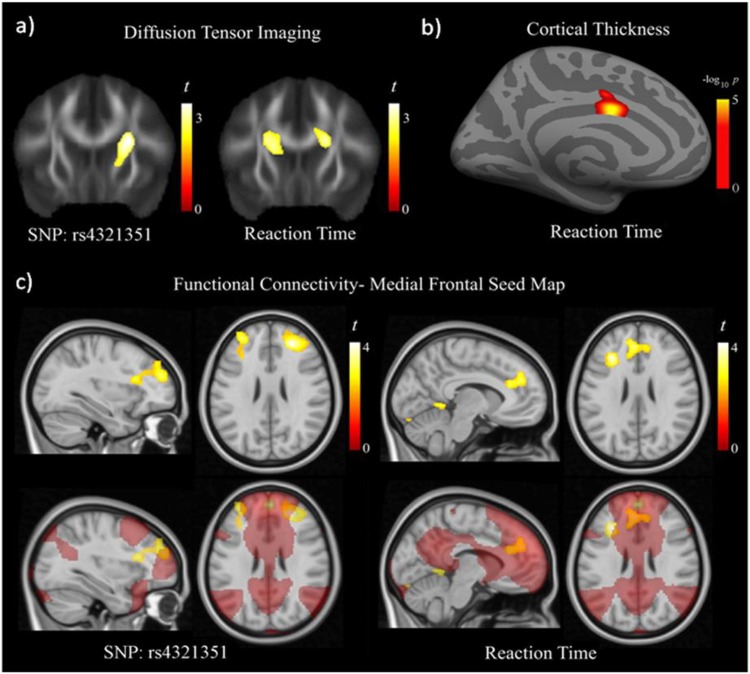
Significant associations were detected between the rs4321351 in PID1 gene and changes in both fractional anisotropy (FA) in DTI and functional connectivity. Specifically, carriers of the G allele of the rs4321351 presented lower FA values in the basal ganglia compared with homozygotes for the A allele (S4 Table and Fig 3a). Interestingly, lower FA in nearby white matter was associated with lower HRT (Fig 3a). Also, lower HRT was associated with thinner cortical thickness in the adjacent anterior cingulate cortex in the left hemisphere (Fig 3b). Functional results were also consistent with an effect of the SNP in frontal-basal ganglia circuits. In the medial frontal seed map, functional connectivity between the frontal medial cortex (selected seed region) and the prefrontal cortex bilaterally increased as a function of the G copies of the rs4321351 (S4 Table and Fig 3c). Increased functional connectivity in this map was associated with lower HRT involving both prefrontal cortex and anterior cingulate cortex (S4 Table and Fig 3c). Finally, in the frontal operculum seed map, carriers of the G allele presented higher functional connectivity between the frontal operculum and the basal ganglia at the putamen bilaterally (S1 Fig).
Fig 3. Diffusion tensor imaging results showed significantly lower fractional anisotropy (FA) in the region of the basal ganglia as a function of the G allele copies of the rs4321351 (a, left image).
Lower HRT scores correlated with lower FA in this region bilaterally (a, right image) and with thinner anterior cingulate cortex (b) (cortical area of 1.2 cm2 centered at MNI, x = -6 y = 2 z = 32, Pcorrected <0.05). Functional connectivity results from the medial frontal seed map (c) showing prefrontal regions with significantly higher functional connectivity as a function of the G allele copies of the rs4321351. Lower HRT scores correlated with higher functional connectivity also in the prefrontal cortex, and in the anterior cingulate cortex. T denotes statistics t value.–log10 p denotes log of the probability p value. The right side corresponds to the right hemisphere in coronal and axial images.
It is relevant to mention that the association between increased connectivity and lower HRT as a function of the G allele copies of the rs4321351, concerns almost selectively to the frontal-basal ganglia system; as posterior brain areas (i. e., parietal cortex), show the opposite association pattern (S4 Table).
Discussion
To our knowledge, this is the first GWAS on attention function assessed in children from the general population. No genome-wide significant results were detected but 13 loci were identified in the suggestive range of association (P<10−5) in the discovery sample. One of them, the rs4321351 located in the PID1 gene, was nominally significant in the replication sample although it did not survive multiple testing correction. This signal was further explored due to its potential relationship with the findings at the pathway level involving Alzheimer disease (AD).
The PID1 gene increases proliferation of preadipocytes without affecting adipocytic differentiation [30]. Studies examining PID1 has been mostly in the context of obesity and insulin resistance [30–33]. Overexpression of PID1 in human myoblasts results in reduced insulin signaling [31] which has been pointed out as a neuroprotective agent acting mainly against apoptosis, beta amyloid toxicity, oxidative stress, and ischemia [34]. Indeed, insulin signaling has been found to be impaired in brains of patients with AD [35]; and type 2 diabetes, characterized by insulin resistance or lack of insulin, has been proposed as a risk factor for AD [36, 37]. Furthermore, PID1 expression has been found to be significantly decreased in brains of patients with AD compared with controls [38]. These evidences suggest the involvement of PID1 gene in neuronal processes and neurodegenerative disease. Our findings add to these evidences relating this gene with attention function during childhood.
Although the rs4321351 SNP was located within the PID1 gene, fine-mapping results showed that the LD region was close to the Delta and Notch-like epidermal growth factor-related receptor (DNER) gene. Thus, we cannot discard that this SNP may be responsible for regulation of the DNER gene rather than PID1 gene. DNER is a neuron-specific Notch ligand required for cerebellar development [39–41]. Also, the DNER gene functions as an activator of the NOTCH1 pathway which has also been related to AD and postnatal myelination and adult plasticity [42, 43]. Furthermore, copy number variations in DNER have been associated with autism spectrum disorders [44].
Additional neuroimaging analyses revealed significant associations between the genetic variant rs4321351 located at PID1 gene and both brain structure and function with the most consistent findings involving the frontal-basal ganglia circuits. This is in accordance with models of attention consistently suggesting the interaction of cortical structures such as frontal cortex with subcortical structures such as basal ganglia to form a complex functional system implicated in sustained attention processes [45, 46]. Relevantly, individual measurement of structure and function in frontal-basal ganglia circuits showed, in turn, significant correlations with HRT where G allele carriers presented higher connectivity between these regions. The direction of the imaging results was in agreement with GWAS findings indicating that HRT scores decreased as a function of the G allele copies of the rs4321351. Furthermore, analyses based in BRAINEAC database showed that rs4321351 may act as eQTL in putamen, one of the structures comprising basal ganglia. Thus, the imaging and eQTL results reinforce the possibility that this SNP may play a role in neuronal structure and functioning related to HRT.
At the pathway level, three biological pathways were significantly associated with different attention outcomes. The sex determination pathway refers to any process that establishes and transmits the specification of sexual status of an individual organism. To our knowledge, neither the current findings nor previous research provide clear clues to link this pathway with alerting. Of note, no sex differences were detected in alerting scores. Thus, the possible role of this pathway in attention development requires replication. In contrast, the other two pathways identified, the mTOR singalling pathways and the Alzheimer disease-amyloid secretase pathway involve processes of interest for cognition which, again involve AD.
The mTOR is a ubiquitously expressed protein kinase that functions as a regulator of several cellular processes including metabolism, growth, proliferation and survival [47]. There is evidence supporting the role of mTOR signaling in synaptic plasticity and memory [48] and it has been suggested that dysregulation of mTOR signaling might be associated with neurodevelopmental, neurodegenerative and neuropsychiatric disorders [49–51]. Biological plausability for these evidences regards the modulating function of mTOR in autophagy since this signaling pathway receives inputs regarding the energetic state of the cell in order to trigger or stop the synthesis of proteins. Also, kinase mTOR is a downstream target of two other pathways, the phosphatidylinositol 3 kinase (PI3K) and kinase AKT (AKT) pathway, which together would downregulate autophagy while fostering cell growth, differentiation and survival. Therefore, activation of the PI3K/AKT/mTOR pathway would promote survival, neuronal protection, and inhibition of autophagy by mTOR activation. [50]. Interestingly, autophagy, which is partially modulated by mTOR as abovementioned, plays a critical role in multiple pathological lesions of AD, such as the formation of amyloid plaques [52] which is related to the second enriched pathway associated with attention function in our study. The AD amyloid secretase pathway refers to the role of the amyloid precursor protein (APP) in the formation of amyloid plaques in AD. However, APP is not only linked to this pathologic process, it has been suggested that APP is also involved in neurite outgrowth and synaptogenesis, neuronal protein trafficking along the axon, transmembrane signal transduction, cell adhesion, and calcium metabolism [53].
Other relevant findings include the nearest gene to the top hit SNP (rs4321351 associated with HRT at p = 6.98 x 10−7) the sulfide quinone reductase-like (SQRDL) gene, the rs10911457 (associated with orienting at p = 9.99 x 10−7) located in the Ral guanine nucleotide dissociation stimulator-like 1 (RGL1) gene, and the proline-rich coiled-coil 2C (PRRC2C) gene (nearest gene of the rs2207190 associated with executive attention at p = 5.12x10-6). The SQRDL is a protein-coding gene which product may function in mitochondria to catalyze the conversion of sulfide to persulfides, thereby decreasing toxic concencrations of sulfide. This gene has been related to ethylmalonic encephalopathy disease [54] and there is evidence indicating that SQRDL is expressed in neurons, oligodendrocytes, and endothelial cells [55]. The RGL1 gene is involved in Ras and Ral GTPase signaling pathways as a downstream effector protein. Interestingly, it has been suggested that the functions of the Ras and Ral signaling pathways also extend into neuronal differentiation and outgrowth [56]. Furthermore, the RGL1 gene has been associated with conduct problems in a GWAS of children with ADHD [57]. Of note, the SNP associated with conduct problems in the study of Anney and collaborators [57] (rs10797919) is in linkage disequilibrium (LD) (r2 = 0.60; D’ = 0.94) with the SNP within the RGL1 gene associated with orienting in the current study (rs10911457). It might be plausible that the RGL1 gene and its product may play a role in attention. Interestingly, the PRRC2C gene associated has been associated with cognitive decline in AD [58].
Of note, besides the eQTL results regarding rs4321351, the possible functionality of the genetic variants discussed above is currently unknown. To our knowledge, none of the loci were in linkage disequilibrium with any potential functional coding SNP.
It is worth mentioning that most of the relevant findings discussed above involved the HRT and HRTSE attention outcomes. Reaction Time (RT) variability is one of the most replicated deficits in ADHD [59] and previous research highlights RT as a promising cognitive target for molecular genetics investigation [60].
The current results should be interpreted considering its limitations and strengths. First, the main limitation of the study is the modest sample size which may increase type II error. Second, we examined multiple phenotypes under a massive univariate approach which may inflate type 1 error. Thus, further research and replication in larger samples are needed. That said, the strengths of the study include several aspects to overcome these limitations including, i) the use of quantitative traits and application of gene set enrichment analyses which helps increasing the power of the study, ii) the inclusion of a replication sample of a similar age and assessed with the same instrument, and iii) additional neuroimaging analyses using different techniques to get insight into the possible neural effects of the genetic variant replicated. Thus, while type II error may only be solved by increasing sample size, several genetic loci showed suggestive evidence for association with the attention outcomes analyzed. Although none of the loci was further replicated when adjusting by multiple testing, one SNP was nominally associated with the same outcome in an independent sample. Furthermore, this locus showed significant associations with different neuroimaging techniques assessing brain structure and function converging in frontal-basal ganglia connections, previously associated with attention and reaction time, as abovementioned. At pathway level, several interesting biological pathways were associated with the attention outcomes assessed underscoring proteins of interest for cognition such as mTOR and APP. Also, we used a computerized test to assess attention, the ANT, which provides homogeneous and reliable measures of attention function [17]. For these reasons, while we cannot discard that other potential genetic variants of interest would be detected in larger samples, we believe that it is unlikely that our results may be false positives since the loci, pathways and neuroimaging results obtained are likely to be biologically meaningful for attention function research.
To conclude, the current study has identified a new promising locus (rs4321351) which may be involved in attention function during childhood and is associated with brain structural and functional changes. Furthermore, to our knowledge, this is the first study suggesting that the PID1 and the DNER genes, the mTOR and the amyloid precursor pathways, proposed to be involved in the pathogenesis of AD, may play a role in the development of attention function during childhood. Evidences from previous studies also suggest that cognitive functions assessed in nondemented populations may share common genetic factors with neurodegenerative disorders such as AD. AD related pathways were associated with attention outcomes in adults affected by ADHD [24]. A marginal joint effect of established AD genes was found on memory in a population-based sample of nondemented middle-aged and elderly subjects [61]. Remarkably, a recent GWAS of cognitive functions and educational attainment in UK Biobank identified genomic regions previously associated with neurodegenerative disorders and AD [62]. Thus, further research is needed to elucidate whether AD and attention function development may share common genetic and biological pathways that can be detected early in life through GWAS methodologies.
Supporting Information
A bilateral portion of the putamen shows significantly higher functional connectivity with the seed region as a function of the G allele copies of the rs4321351. T denotes statistics t value. The right side corresponds to the right hemisphere in the coronal image. The sagittal image corresponds to the left hemisphere.
(DOCX)
(GZ)
(GZ)
(GZ)
(GZ)
(GZ)
(DOCX)
(DOCX)
Five top most significant associated pathways with attention outcomes.
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are acknowledged with all the children and their families participating into the BREATHE project for their altruism and particularly to the schools Antoni Brusi, Baloo, Betània–Patmos, Centre d’estudis Montseny, Col·legi Shalom, Costa i Llobera, El sagrer, Els Llorers, Escola Pia de Sarrià, Escola Pia Balmes, Escola concertada Ramon Llull, Escola Lourdes, Escola Tècnica Professional del Clot, Ferran i Clua, Francesc Macià, Frederic Mistral, Infant Jesús, Joan Maragall, Jovellanos, La Llacuna del Poblenou, Lloret, Menéndez Pidal, Nuestra Señora del Rosario, Miralletes, Ramon Llull, Rius i Taulet, Pau Vila, Pere Vila, Pi d'en Xandri, Projecte, Prosperitat, Sant Ramon Nonat—Sagrat Cor, Santa Anna, Sant Gregori, Sagrat Cor Diputació, Tres Pins, Tomàs Moro, Torrent d'en Melis, Virolai. The authors also would particularly like to thank all the participants of INMA project for their generous collaboration. Also, the authors are grateful to Silvia Fochs, Anna Sànchez, Maribel López, Nuria Pey, and Muriel Ferrer for their assistance in contacting the families and administering the questionnaires.
Data Availability
In order to protect participant’s privacy, data at individual level cannot be made publicly available. Full summary statistics can be found in the paper and supporting information. For further details, authors can be contacted at http://www.isglobal.org/ca/.
Funding Statement
The research leading to these results has received funding from the European Research Council under the ERC Grant Agreement number 268479 – the BREATHE project. The INMA project was funded by grants from Instituto de Salud Carlos III (Red INMA G03/176 and CB06/02/0041). The INMA-Sabadell cohort received funding from Instituto de Salud Carlos III (FIS-FEDER: PI041436 and PI081151), Generalitat de Catalunya-CIRIT 1999SGR 00241, and EU sixth framework project NEWGENERIS FP6-2003-Food-3-A-016320. The INMA-Valencia cohort received funding from UE (FP7-ENV-2011 cod 282957 and HEALTH.2010.2.4.5-1), and from Instituto de Salud Carlos III (FIS-FEDER: 03/1615, 04/1509, 04/1112, 04/1931, 05/1079, 05/1052, 06/1213, 07/0314, 09/02647, 11/0178, 11/01007, 11/02591, 11/02038, 13/1944, 13/2032, 14/00891, 14/01687 and Miguel Servet-FEDER MS15/0025) and the Conselleria de Sanitat, Generalitat Valenciana. SA is supported by a Sara Borrell grant from the Instituto de Salud Carlos III (CD14/00214). NV-T is funded by a pre-doctoral grant from the Agència de Gestió d’Ajuts Universitaris i de Recerca (2015 FI_B 00636) Generalitat de Catalunya – Fons Social Europeu. JJ is supported by a Miguel Servet (MS) Fellowship (MS14/00108) awarded by the Spanish Institute of Health Carlos III (Ministry of Economy and Competitiveness). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bellgrove MA, Mattingley JB. Molecular genetics of attention. Ann N Y Acad Sci. 2008;1129:200–12. 10.1196/annals.1417.013 [DOI] [PubMed] [Google Scholar]
- 2.Fan J, Gu X, Guise KG, Liu X, Fossella J, Wang H, et al. Testing the behavioral interaction and integration of attentional networks. Brain Cogn. 2009;70(2):209–20. 10.1016/j.bandc.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Posner MI, Rothbart MK. Toward a physical basis of attention and self regulation. Phys Life Rev. 2009;6(2):103–20. Epub 2010/02/18. 10.1016/j.plrev.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson P. Assessment and development of executive function (EF) during childhood. Child Neuropsychol. 2002;8(2):71–82. . [DOI] [PubMed] [Google Scholar]
- 5.Egeland J. Differentiating attention deficit in adult ADHD and schizophrenia. Arch Clin Neuropsychol. 2007;22(6):763–71. . [DOI] [PubMed] [Google Scholar]
- 6.Egeland J, Rund BR, Sundet K, Landro NI, Asbjornsen A, Lund A, et al. Attention profile in schizophrenia compared with depression: differential effects of processing speed, selective attention and vigilance. Acta Psychiatr Scand. 2003;108(4):276–84. . [DOI] [PubMed] [Google Scholar]
- 7.Gallagher R, Blader J. The diagnosis and neuropsychological assessment of adult attention deficit/hyperactivity disorder. Scientific study and practical guidelines. Ann N Y Acad Sci. 2001;931:148–71. . [DOI] [PubMed] [Google Scholar]
- 8.Mulet B, Valero J, Gutierrez-Zotes A, Montserrat C, Cortes MJ, Jariod M, et al. Sustained and selective attention deficits as vulnerability markers to psychosis. Eur Psychiatry. 2007;22(3):171–6. . [DOI] [PubMed] [Google Scholar]
- 9.Franke B, Faraone SV, Asherson P, Buitelaar J, Bau CH, Ramos-Quiroga JA, et al. The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol Psychiatry. 2012;17(10):960–87. 10.1038/mp.2011.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmboe K, Nemoda Z, Fearon RM, Csibra G, Sasvari-Szekely M, Johnson MH. Polymorphisms in dopamine system genes are associated with individual differences in attention in infancy. Dev Psychol. 2010;46(2):404–16. 10.1037/a0018180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefanis NC, van Os J, Avramopoulos D, Smyrnis N, Evdokimidis I, Stefanis CN. Effect of COMT Val158Met polymorphism on the Continuous Performance Test, Identical Pairs Version: tuning rather than improving performance. Am J Psychiatry. 2005;162(9):1752–4. . [DOI] [PubMed] [Google Scholar]
- 12.Fan J, Wu Y, Fossella JA, Posner MI. Assessing the heritability of attentional networks. BMC Neurosci. 2001;2:14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giubilei F, Medda E, Fagnani C, Bianchi V, De Carolis A, Salvetti M, et al. Heritability of neurocognitive functioning in the elderly: evidence from an Italian twin study. Age Ageing. 2008;37(6):640–6. 10.1093/ageing/afn132 [DOI] [PubMed] [Google Scholar]
- 14.Xu C, Sun J, Duan H, Ji F, Tian X, Zhai Y, et al. Gene, environment and cognitive function: a Chinese twin ageing study. Age Ageing. 2015;44(3):452–7. 10.1093/ageing/afv015 [DOI] [PubMed] [Google Scholar]
- 15.Sunyer J, Esnaola M, Alvarez-Pedrerol M, Forns J, Rivas I, Lopez-Vicente M, et al. Association between traffic-related air pollution in schools and cognitive development in primary school children: a prospective cohort study. PLoS Med. 2015;12(3):e1001792 10.1371/journal.pmed.1001792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guxens M, Ballester F, Espada M, Fernandez MF, Grimalt JO, Ibarluzea J, et al. Cohort Profile: the INMA—INfancia y Medio Ambiente—(Environment and Childhood) Project. Int J Epidemiol. 2011;41(4):930–40. . [DOI] [PubMed] [Google Scholar]
- 17.Rueda MR, Fan J, McCandliss BD, Halparin JD, Gruber DB, Lercari LP, et al. Development of attentional networks in childhood. Neuropsychologia. 2004;42(8):1029–40. . [DOI] [PubMed] [Google Scholar]
- 18.Forns J, Esnaola M, Lopez-Vicente M, Suades-Gonzalez E, Alvarez-Pedrerol M, Julvez J, et al. The n-back test and the attentional network task as measures of child neuropsychological development in epidemiological studies. Neuropsychology. 2014;28(4):519–29. 10.1037/neu0000085 [DOI] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–13. . [DOI] [PubMed] [Google Scholar]
- 21.Duggal P, Gillanders EM, Holmes TN, Bailey-Wilson JE. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics. 2008;9:516 10.1186/1471-2164-9-516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17(10):1418–28. 10.1038/nn.3801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alemany S, Ribases M, Vilor-Tejedor N, Bustamante M, Sanchez-Mora C, Bosch R, et al. New suggestive genetic loci and biological pathways for attention function in adult attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2015. . [DOI] [PubMed] [Google Scholar]
- 25.Pujol J, Martinez-Vilavella G, Macia D, Fenoll R, Alvarez-Pedrerol M, Rivas I, et al. Traffic pollution exposure is associated with altered brain connectivity in school children. Neuroimage. 2016;129:175–84. 10.1016/j.neuroimage.2016.01.036 [DOI] [PubMed] [Google Scholar]
- 26.Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102(27):9673–8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison BJ, Pujol J, Cardoner N, Deus J, Alonso P, Lopez-Sola M, et al. Brain corticostriatal systems and the major clinical symptom dimensions of obsessive-compulsive disorder. Biol Psychiatry. 2013;73(4):321–8. 10.1016/j.biopsych.2012.10.006 [DOI] [PubMed] [Google Scholar]
- 28.Pujol J, Del Hoyo L, Blanco-Hinojo L, de Sola S, Macia D, Martinez-Vilavella G, et al. Anomalous brain functional connectivity contributing to poor adaptive behavior in Down syndrome. Cortex. 2014;64:148–56. 10.1016/j.cortex.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 29.Pujol J, Macia D, Garcia-Fontanals A, Blanco-Hinojo L, Lopez-Sola M, Garcia-Blanco S, et al. The contribution of sensory system functional connectivity reduction to clinical pain in fibromyalgia. Pain. 2014;155(8):1492–503. 10.1016/j.pain.2014.04.028 [DOI] [PubMed] [Google Scholar]
- 30.Wang B, Zhang M, Ni YH, Liu F, Fan HQ, Fei L, et al. Identification and characterization of NYGGF4, a novel gene containing a phosphotyrosine-binding (PTB) domain that stimulates 3T3-L1 preadipocytes proliferation. Gene. 2006;379:132–40. . [DOI] [PubMed] [Google Scholar]
- 31.Bonala S, McFarlane C, Ang J, Lim R, Lee M, Chua H, et al. Pid1 induces insulin resistance in both human and mouse skeletal muscle during obesity. Mol Endocrinol. 2013;27(9):1518–35. 10.1210/me.2013-1048 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Wu WL, Gan WH, Tong ML, Li XL, Dai JZ, Zhang CM, et al. Over-expression of NYGGF4 (PID1) inhibits glucose transport in skeletal myotubes by blocking the IRS1/PI3K/AKT insulin pathway. Mol Genet Metab. 2011;102(3):374–7. 10.1016/j.ymgme.2010.11.165 [DOI] [PubMed] [Google Scholar]
- 33.Zhang CM, Chen XH, Wang B, Liu F, Chi X, Tong ML, et al. Over-expression of NYGGF4 inhibits glucose transport in 3T3-L1 adipocytes via attenuated phosphorylation of IRS-1 and Akt. Acta Pharmacol Sin. 2009;30(1):120–4. 10.1038/aps.2008.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blazquez E, Velazquez E, Hurtado-Carneiro V, Ruiz-Albusac JM. Insulin in the brain: its pathophysiological implications for States related with central insulin resistance, type 2 diabetes and Alzheimer's disease. Front Endocrinol (Lausanne). 2014;5:161 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hölscher C. Drugs developed for treatment of diabetes show protective effects in Alzheimer’s and Parkinson’s diseases. Acta Physiologica Sinica. 2014;66(5):497–510. [PubMed] [Google Scholar]
- 36.Vagelatos NT, Eslick GD. Type 2 Diabetes as a Risk Factor for Alzheimer's Disease: The Confounders, Interactions, and Neuropathology Associated With This Relationship. Epidemiol Rev. 2013. . [DOI] [PubMed] [Google Scholar]
- 37.Martins IJ, Hone E, Foster JK, Sunram-Lea SI, Gnjec A, Fuller SJ, et al. Apolipoprotein E, cholesterol metabolism, diabetes, and the convergence of risk factors for Alzheimer's disease and cardiovascular disease. Mol Psychiatry. 2006;11(8):721–36. . [DOI] [PubMed] [Google Scholar]
- 38.Kajiwara Y, Franciosi S, Takahashi N, Krug L, Schmeidler J, Taddei K, et al. Extensive proteomic screening identifies the obesity-related NYGGF4 protein as a novel LRP1-interactor, showing reduced expression in early Alzheimer's disease. Mol Neurodegener. 2010;5:1 10.1186/1750-1326-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, et al. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;8(7):873–80. . [DOI] [PubMed] [Google Scholar]
- 40.Saito SY, Takeshima H. DNER as key molecule for cerebellar maturation. Cerebellum. 2006;5(3):227–31. . [DOI] [PubMed] [Google Scholar]
- 41.Tohgo A, Eiraku M, Miyazaki T, Miura E, Kawaguchi SY, Nishi M, et al. Impaired cerebellar functions in mutant mice lacking DNER. Mol Cell Neurosci. 2006;31(2):326–33. . [DOI] [PubMed] [Google Scholar]
- 42.Liang H, Zhang Y, Shi X, Wei T, Lou J. Role of Notch-1 signaling pathway in PC12 cell apoptosis induced by amyloid beta-peptide (25–35). Neural Regen Res. 2014;9(13):1297–302. 10.4103/1673-5374.137577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodhoo A, Alonso MB, Droggiti A, Turmaine M, D'Antonio M, Parkinson DB, et al. Notch controls embryonic Schwann cell differentiation, postnatal myelination and adult plasticity. Nat Neurosci. 2009;12(7):839–47. 10.1038/nn.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griswold AJ, Ma D, Cukier HN, Nations LD, Schmidt MA, Chung RH, et al. Evaluation of copy number variations reveals novel candidate genes in autism spectrum disorder-associated pathways. Hum Mol Genet. 2012;21(15):3513–23. 10.1093/hmg/dds164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna BS, Young JW, Dawes SE, Asgaard GL, Eyler LT. Bridging the bench to bedside gap: validation of a reverse-translated rodent continuous performance test using functional magnetic resonance imaging. Psychiatry Res. 2013;212(3):183–91. 10.1016/j.pscychresns.2013.01.005 [DOI] [PubMed] [Google Scholar]
- 46.Riccio CA, Reynolds CR, Lowe P, Moore JJ. The continuous performance test: a window on the neural substrates for attention? Arch Clin Neuropsychol. 2002;17(3):235–72. . [PubMed] [Google Scholar]
- 47.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122(Pt 20):3589–94. 10.1242/jcs.051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33(2):67–75. 10.1016/j.tins.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costa-Mattioli M, Monteggia LM. mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat Neurosci. 2013;16(11):1537–43. 10.1038/nn.3546 [DOI] [PubMed] [Google Scholar]
- 50.Heras-Sandoval D, Perez-Rojas JM, Hernandez-Damian J, Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell Signal. 2014;26(12):2694–701. 10.1016/j.cellsig.2014.08.019 [DOI] [PubMed] [Google Scholar]
- 51.Oguro-Ando A, Rosensweig C, Herman E, Nishimura Y, Werling D, Bill BR, et al. Increased CYFIP1 dosage alters cellular and dendritic morphology and dysregulates mTOR. Mol Psychiatry. 2014. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai Z, Zhao B, Li K, Zhang L, Li C, Quazi SH, et al. Mammalian target of rapamycin: a valid therapeutic target through the autophagy pathway for Alzheimer's disease? J Neurosci Res. 2012;90(6):1105–18. 10.1002/jnr.23011 [DOI] [PubMed] [Google Scholar]
- 53.Zheng H, Koo EH. The amyloid precursor protein: beyond amyloid. Mol Neurodegener. 2006;1:5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palmfeldt J, Vang S, Stenbroen V, Pavlou E, Baycheva M, Buchal G, et al. Proteomics reveals that redox regulation is disrupted in patients with ethylmalonic encephalopathy. J Proteome Res. 2011;10(5):2389–96. 10.1021/pr101218d [DOI] [PubMed] [Google Scholar]
- 55.Ackermann M, Kubitza M, Maier K, Brawanski A, Hauska G, Pina AL. The vertebrate homolog of sulfide-quinone reductase is expressed in mitochondria of neuronal tissues. Neuroscience. 2011;199:1–12. 10.1016/j.neuroscience.2011.10.044 [DOI] [PubMed] [Google Scholar]
- 56.Goi T, Rusanescu G, Urano T, Feig LA. Ral-specific guanine nucleotide exchange factor activity opposes other Ras effectors in PC12 cells by inhibiting neurite outgrowth. Mol Cell Biol. 1999;19(3):1731–41. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anney RJ, Lasky-Su J, O'Dushlaine C, Kenny E, Neale BM, Mulligan A, et al. Conduct disorder and ADHD: evaluation of conduct problems as a categorical and quantitative trait in the international multicentre ADHD genetics study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(8):1369–78. 10.1002/ajmg.b.30871 [DOI] [PubMed] [Google Scholar]
- 58.Sherva R, Tripodis Y, Bennett DA, Chibnik LB, Crane PK, de Jager PL, et al. Genome-wide association study of the rate of cognitive decline in Alzheimer's disease. Alzheimers Dement. 2014;10(1):45–52. 10.1016/j.jalz.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Castellanos FX, Tannock R. Neuroscience of attention-deficit/hyperactivity disorder: the search for endophenotypes. Nat Rev Neurosci. 2002;3(8):617–28. . [DOI] [PubMed] [Google Scholar]
- 60.Kuntsi J, Wood AC, Rijsdijk F, Johnson KA, Andreou P, Albrecht B, et al. Separation of cognitive impairments in attention-deficit/hyperactivity disorder into 2 familial factors. Arch Gen Psychiatry. 2010;67(11):1159–67. 10.1001/archgenpsychiatry.2010.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verhaaren BF, Vernooij MW, Koudstaal PJ, Uitterlinden AG, van Duijn CM, Hofman A, et al. Alzheimer's disease genes and cognition in the nondemented general population. Biol Psychiatry. 2013;73(5):429–34. 10.1016/j.biopsych.2012.04.009 [DOI] [PubMed] [Google Scholar]
- 62.Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, et al. Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N = 112 151). Mol Psychiatry. 2016;21(6):758–67. 10.1038/mp.2016.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A bilateral portion of the putamen shows significantly higher functional connectivity with the seed region as a function of the G allele copies of the rs4321351. T denotes statistics t value. The right side corresponds to the right hemisphere in the coronal image. The sagittal image corresponds to the left hemisphere.
(DOCX)
(GZ)
(GZ)
(GZ)
(GZ)
(GZ)
(DOCX)
(DOCX)
Five top most significant associated pathways with attention outcomes.
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
In order to protect participant’s privacy, data at individual level cannot be made publicly available. Full summary statistics can be found in the paper and supporting information. For further details, authors can be contacted at http://www.isglobal.org/ca/.