Abstract
This study explores the use of human embryonic stem cells (hESCs) for assessing nanotoxicology, specifically, the effect of gold nanoparticles (AuNPs) of different core sizes (1.5 nm, 4 nm, and 14 nm) on the viability, pluripotency, neuronal differentiation, and DNA methylation of hESCs. The hESCs exposed to 1.5 nm thiolate-capped AuNPs exhibited loss of cohesiveness and detachment suggesting ongoing cell death at concentrations as low as 0.1 µg/mL. The cells exposed to 1.5 nm AuNPs at this concentration did not form embryoid bodies but rather disintegrated into single cells within 48 hours. Cell death caused by 1.5 nm AuNPs also occurred in hESC-derived neural progenitor cells. None of the other nanoparticles exhibited toxic effects on the hESCs at concentrations as high as 10 µg/mL during a 19 day neural differentiation period. Thiolate-capped 4 nm AuNPs at 10 µg/mL caused a dramatic decrease in global DNA methylation (5mC) and a corresponding increase in global DNA hydroxymethylation (5hmC) of the hESC’s DNA in only 24 hours. This work identifies a type of AuNPs highly toxic to hESCs and demonstrates the potential of hESCs in predicting nanotoxicity and characterizing their ability to alter the DNA methylation and hydroxymethylation patterns in the cells.
Keywords: gold, nanoparticles, assays, toxicology, human embryonic stem cell
1. Introduction
Nanotoxicology is an emerging discipline studying the interference of engineered nanomaterials with the functions of cellular and extracellular nanomachineries.[1] The accelerated pace of clinical trials and commercialization of nanomaterials places increased demands on toxicological evaluations of engineered nanomaterials.[2] There is an urgent need to develop predictive and validated nanotoxicological assessment methods for nanomaterials due to their unique size range, surface chemistry, and interactions with biological systems.[3] Nanomaterials have the potential to cross the placental and the blood-brain barriers.[4, 5] Neurotoxicity of engineering nanomaterials will become more relevant with the development of central nervous system-targeting nanomedicine.
Among the diverse nanomaterials currently being explored for nanomedicine, gold nanoparticles (AuNPs) are particularly attractive due to their versatile surface chemistry, ease of imaging, and tunability for targeted drug delivery.[5, 6] AuNPs are being widely investigated for targeted delivery of drugs, genetic materials, antigens, and diagnostic agents to the brain.[7] For example, in combination with radiation, AuNPs have proven successful at dissolving deposits of beta-amyloid, the protein involved in Alzheimer’s disease.[8–11] Because of the wide biomedical applications of AuNPs, human exposure to AuNPs will be increasingly likely, which warrants careful evaluation of their toxicological effects. AuNPs have been described by most studies as nontoxic,[12–15] but some studies have found them to be toxic.[16–22] AuNPs with 1.4–1.5 nm in diameter (Au55) have been found to be highly toxic to cells.[18, 23] A few studies have shown that chronic exposure to AuNPs might interfere with brain function. For example, exposure to high concentrations of 20 nm AuNPs was found to increase the proliferation of human neurosphere-derived neural precursor/pregenitor cells (NPCs)[24] and induce oxidative stress in a mouse immortalized NPC line.[25] In rodent models, intraperitoneal injections of 17–20 nm AuNPs resulted in learning and memory deficits associated with elevated dopamine levels and decreased serotonin levels,[26] as well as increased brain expression levels of markers of oxidative stress, apoptosis, and inflammation.[27] More recently, cultured mouse hippocampal CA1 neurons were shown to be more excitable when exposed to 5–40 nm AuNPs.[28] Taken together, these data demonstrate that AuNPs as tools for neuromedicine should be carefully evaluated since they may adversely alter neuronal differentiation, synapse formation, and functional plasticity (which affect memory and learning) in a size-dependent manner.
Currently, there are no systematic methods to predict toxicological effects of AuNPs on human health. A majority of neurotoxicological assessments are conducted in vitro using immortalized cell lines with standard toxicological assays such as the MTT (measurement of mitochondrial enzymatic activity) and LDH (measurement of cell membrane disruption) assays with short exposure time (usually limited to within 48 h). One shortcoming of standard in vitro toxicity assays, however, is that the potential interference of nanoparticles with embryonic development or stem cell differentiation cannot be clearly addressed. On the other hand, it is often difficult to correlate outcomes from animals to humans based on in vivo tests. Human embryonic stem cells (hESCs)[29] and their subsequent differentiation into NPCs and neurons have enabled researchers to establish pluripotent cell-based models to study developmental neurotoxicology. This process mimics aspects of the early stages of human brain development, giving us a unique opportunity to identify the effects of exposure to nanomaterials on neural specification Though mouse ESCs were recently used to evaluate the neurotoxicity of AuNPs[30, 31], hESCs have yet to be applied for nanotoxicological testing.
This paper explores the use of hESCs for neurotoxicity tesing of AuNPs and dendrimers. It describes the morphological and epigenetic effects of AuNPs of three core sizes, 1.5 nm, 4 nm, and 14 nm, on the neuronal differentiation of hESCs. The 1.5 nm AuNP (AuNP1.5) was chosen because AuNPs of this size have been found to be highly toxic to cells.[18, 23] AuNP4 and AuNP14 are within the size range widely used for targeted drug delivery applications including those targeting the brain.[5, 6] The nanoparticle size and surface chemistry were characterized by UV-vis, TEM, and zetasizer. We show that AuNPs have size-dependent differential effects on the viability, pluripotency, and neuronal differentiation potentials of hESCs, AuNP1.5 being highly toxic. In addition, we show that exposure to certain sized AuNPs modifies the DNA methylation and hydroxymethylation profiles of hESCs.
2. Results
2.1. AuNP Synthesis and Characterization
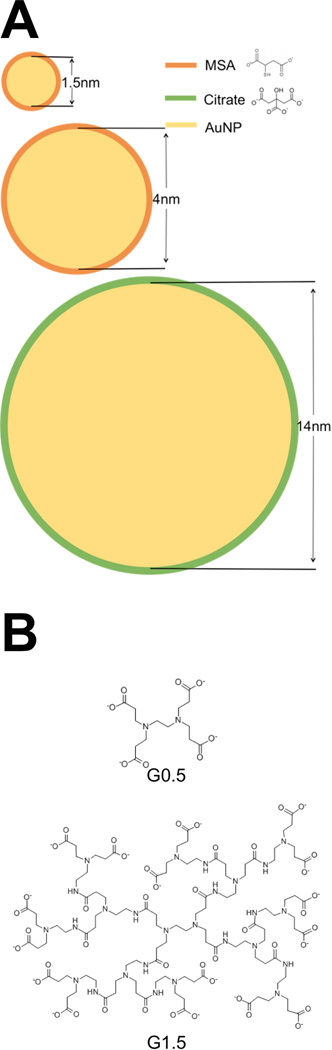
All the nanoparticles used in this study are given in Scheme 1. AuNPs were synthesized according to established procedures in the literature. The detailed procedures are described in the Experimental Section and only briefly summarized here. Ultrasmall AuNP1.5 was synthesized by modifying a literature procedure using a mild reductant tert-butylamine-borane to slow-down the reduction rate of AuCl(PPh3) precursor.[32] AuNP1.5 was capped by mercaptosuccinic acid (MSA) following the ligand exchange reaction.[33] AuNP4 capped by MSA was synthesized by following a literature procedure.[34] Citrate-capped AuNP14 was synthesized by using the Turkevich method.[35–38] For comparison with the AuNPs, two half-generation dendrimers with carboxyl termination G0.5 and G1.5 were used.
Scheme 1.
Nanoparticle chemical structures: (A) AuNP1.5, AuNP4, and AuNP14 and (B) dendrimer G0.5 and G1.5.
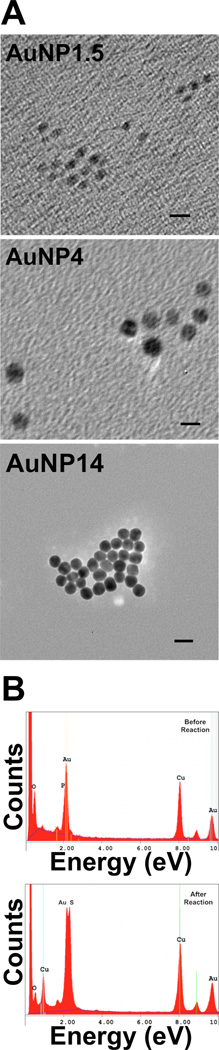
The nanoparticles were characterized by TEM with energy dispersive spectroscopy (EDAX), zetasizer, and UV-vis spectroscopy. Figure 1A shows the TEM images of synthesized AuNPs. Their diameters were determined to be 1.5 ± 0.1 nm (AuNP1.5), 4.2 ± 0.5 nm (AuNP4), and 13.6 ± 0.4 nm (AuNP14), respectively. The various AuNPs were thus identified by their TEM measured diameters. Figure 1B shows the TEM EDAX spectra of AuNP1.5 before and after the ligand exchange reaction. The phosphorus peak at 2.013 eV was replaced by the sulfur peak at 2.307 eV confirming the exchange reaction. The gold peak was unchanged at 2.120 eV.
Figure 1.
(A) TEM images of AuNPs of average core diameters 1.5 nm, 4 nm, and 14 nm. The scale bar length is 5 nm for AuNP1.5 and AuNP4 images and 20 nm for the image of AuNP14. (B) EDAX spectra of AuNP1.5 before (left) and after replacing PPh3 with MSA.
When AuNP4 and AuNP14 were added to the hESC culture medium, we observed aggregation of the nanoparticles from the disappearance of the red color associated with stabilized AuNPs. To prevent this, we incubated the nanoparticles with bovine serum albumin (BSA) prior to their addition to the culture medium as BSA has been shown to be effective in preventing nanoparticle aggregation in culture medium.[39, 40] BSA coating was proven to be effective in preventing AuNPs from aggregation in the hESC culture medium (see Section 2.2). The nanoparticle size measurements from dynamic light scattering (DLS) (Figure S1, Supporting Information (SI)) are consistent with the expected sizes of the AuNPs and dendrimers. After applying the BSA, an increase in particle hydrodynamic radius of 4–6 nm was observed for AuNP4 and AuNP14 indicating the formation of the corona (BSA has the following molecular dimensions 3 nm by 3 nm by 8 nm[39]). The particles retained their negative charges after being coated with BSA. We were not able to determine the size of AuNP1.5, G0.5, and G1.5 after incubating with BSA due to their extreme small sizes. The UV-vis spectra of the AuNPs after coating with BSA showed no shift in the absorption peak position in the range of 500–520 nm dispersed either in deionized water or PBS buffer further supporting the stabilization effect of BSA (Figure S2, SI). In contrast to AuNP4 and AuNP14, AuNP1.5 does not absorb light in the 500–520 nm range (Figure S2(a), SI) because ultrasmall AuNPs of 1–2 nm are no longer metallic but exhibit discrete electron excitations.[41]
2.2. AuNP Size Effect on hESC Viability
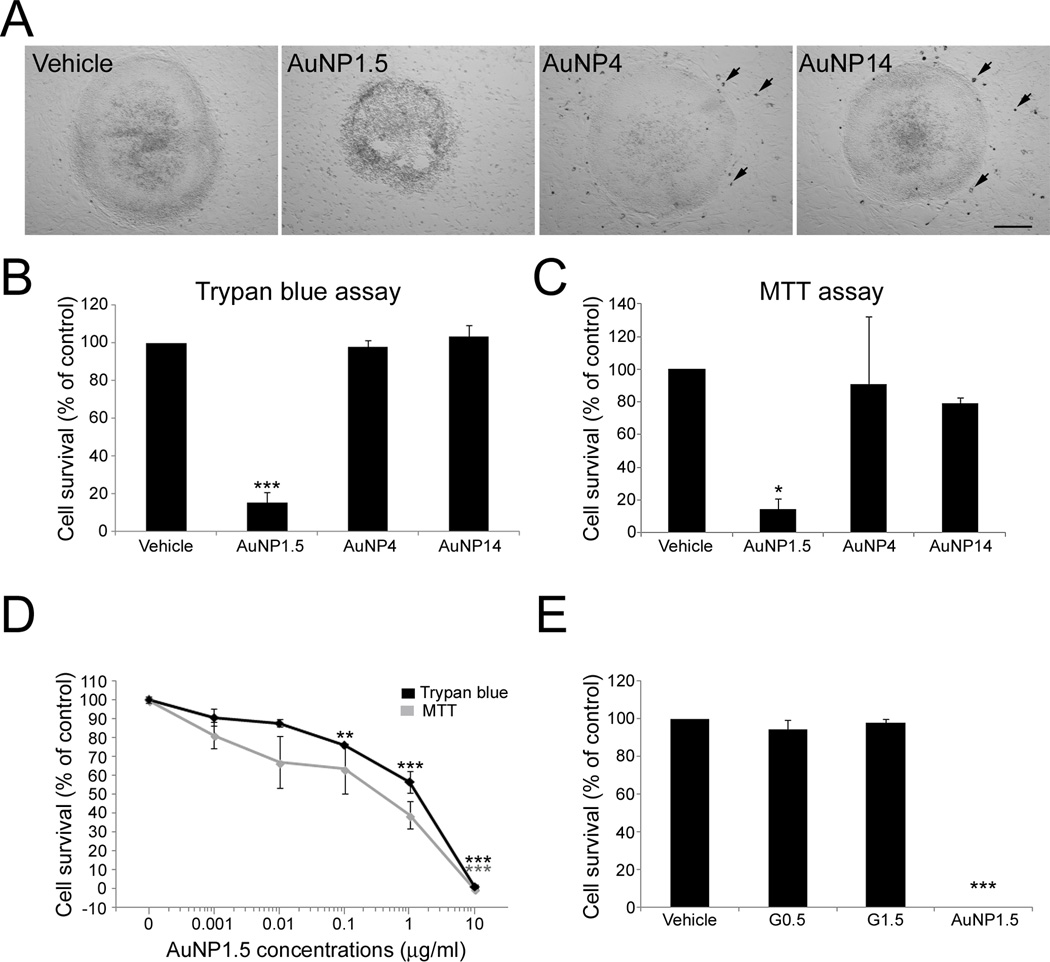
Here, we sought to determine the influence that AuNPs of various sizes may have on hESCs and their neuronal specification. We started by examining the effects that AuNP exposure has on the viability of hESCs. We first verified the efficacy of the BSA-coating procedure at preventing AuNP aggregation in the hESC culture medium. The culture medium maintained its red color when BSA-coated AuNPs were introduced indicating the AuNP colloidal stability. Figure 2A shows light microscopic images of WA09 hESC cell colonies on a mouse embryonic fibroblast (MEF) feeder layer following 24 h exposure to vehicle, or 10 µg/mL of AuNP1.5, AuNP4, or AuNP14. We chose to express particle dosage in mass concentration (e.g., µg/mL) as in a majority of publications. However, considering the importance of particle surface area concentration (e.g., cm2/mL) and dose per cell number,[42] we also listed the corresponding dosages in Table 1 for various sized AuNPs. The cell density of 100,000 cells per mL was used in the calculations. Colonies exposed to AuNP1.5 exhibited altered morphology suggestive of cell death. As shown in Figure 2A, no significant AuNP aggregates were visible for any size of BSA-coated AuNPs tested, confirming earlier reports.[39, 40] To identify the potential AuNP toxicity, hESCs were exposed to 10 µg/mL AuNPs, largely above the nontoxic range[5] for 24 h. At the end of exposure with AuNP4 and AuNP14, no remarkable differences in the growth and morphology of hESC colonies were observed compared to control cultures (Figure 2A). In contrast, cells in AuNP1.5-treated hESC colonies exhibited loss of cohesiveness, rounding up, and detachment, suggesting ongoing cell death (Figure 2A). Similar time-course and severity of AuNP1.5-induced cell loss was observed when hESCs were cultured on gelatin-coated plates instead of MEFs, suggesting a direct toxic effect of AuNP1.5 on hESCs (data not shown). Trypan blue assays, which stains dead cells, at the end of the 24-h treatment revealed statistically significant differences in the viability of hESCs between the different experimental groups (p < 0.001) (Figure 2B). Thus, whereas no change in viability was observed among control, AuNP4- (p = 0.79), and AuNP14- (p = 0.89) treated cultures, a statistically significant 88% decrease in the number of viable hESCs was observed following treatment with AuNP1.5 (p < 0.001) (Figure 2B). Assessment of hESC viability using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) viability assay, a measure of mitochondrial function, provided comparable results with a statistically significant 85% decrease (p = 0.028) in cell viability in AuNP1.5-treated hESCs compared to controls (Figure 2C).
Figure 2.
Exposure of hESCs to AuNPs. (A) WA09 hESC cell colonies on MEF feeder layer following 24 h exposure to vehicle, or 10 µg/mL of AuNP1.5, AuNP4, and AuNP14. Only colonies exposed to AuNP1.5 exhibited altered morphology suggestive of cell death. Arrows indicate MEFs with visible intracellular uptake of AuNPs in the case of AuNP4 and AuNP14. AuNP1.5 was too small to be seen. The scale bar is 160 µm in the bright field microscopic images. (B) Cell viability analysis of hESCs exposed for 24 h to 10 µg/mL of the different sized AuNPs using Trypan blue assay. The percentage of viable cells [(total number of cells - number of non-viable dark-blue stained cells) / (total number of cells) ×100] was significantly decreased following exposure to AuNP1.5. Histogram values are means ± SEM. Statistical significance compared to the vehicle control group is designated as (***) (p < 0.001; n = 3, one-way ANOVA). (C) Measurement of mitochondrial activity in hESCs exposed for 24 h to 10 µg/mL of the different sized AuNPs by the MTT assay. A decrease in mitochondrial enzymatic activity was observed between the vehicle and the AuNP1.5 experimental groups. Histogram values are means ± SEM. Statistical significance compared to the vehicle control group is designated as (*) (p = 0.028; n = 3, one-way ANOVA). (D) Dose-response curves for the survival of hESCs exposed to AuNP1.5 for 4 days using Trypan blue and MTT assays. Statistical significances compared to the vehicle group are noted as (***) (p < 0.001; n = 3) and as (**) (p < 0.01; n = 3). (E) Cell viability analysis of hESCs exposed for 24 h to 10 µg/mL of AuNP1.5 and dendrimers G0.5 and G1.5 using Trypan blue assay. Histogram values are means ± SEM. Statistical significance compared to the vehicle control group is designated as (***) (p < 0.001; n = 3, one-way ANOVA).
Table 1.
AuNP dosage expressed in various units.
| AuNP1.5 D = 1.5 nm Conc. = 10 µg/mL |
AuNP1.5 D = 1.5 nm Conc. = 0.6 µg/mL |
AuNP4 D = 4 nm Conc. = 10 µg/mL |
AuNP14 D = 14 nm Conc. = 10 µg/mL |
|
|---|---|---|---|---|
|
Number dose (1/ml) |
2.93×1014 | 1.76×1013 | 1.55×1013 | 3.61×1011 |
|
Number dose (µM) |
0.487 | 0.0292 | 0.0257 | 5.99×10−4 |
|
Surface dose (cm2/mL) |
20.7 | 1.24 | 7.79 | 2.22 |
| Particle No./cell | 2.93×109 | 1.76×108 | 1.55×108 | 3.61×106 |
We then analyzed the sub-lethal range of exposure to AuNP1.5 by assessing the viability of hESCs treated for 4 days with six different concentrations ranging from 0.001 to 10 µg/mL (Figure 2D). The Trypan blue exclusion assay revealed a dose-dependent decrease in hESC viability, with concentrations of 0.1 and 1 µg/mL resulting in a significant 24% (p = 0.004) and 43% (p < 0.001) reduction, respectively (Figure 2D). Almost no hESCs survived following exposure to 10 µg/mL AuNP1.5 (p < 0.001) (Figure 2D). We found that the lowest AuNP1.5 concentrations of 0.001 and 0.01 µg/mL slightly reduced hESC viability compared to control cultures, but these differences did not reach statistical significance (p > 0.05). The IC50 value, i.e., the concentration necessary to kill 50% of the cells, measured in this 4-day exposure paradigm was 1.45 µg/mL (or 0.0706 µM). MTT viability was also significantly reduced after exposure to 10 µg/mL (p = 0.007) (Figure 2D). A non-significant trend (p = 0.11) toward reduced cell viability at the 1 µg/mL concentration was also observed with the MTT assay for which the IC50 value was 1.03 µg/mL.
In comparison, viability of hESCs was tested against BSA-treated G0.5 and G1.5 dendrimers with the same carboxyl surface group and in a close hydrodynamic size range as AuNP1.5 (Figure S1, SI). We compared the viability of hESCs exposed for 24 h to AuNP1.5 or G0.5 and G1.5 dendrimers at a concentration of 10 µg/mL. As shown in Figure 2E using the Trypan blue assay, the percentage of viable hESCs after exposure to dendrimers of both sizes was not different from that observed in control cultures. However, as previously observed, no hESCs survived treatment with AuNP1.5 (p < 0.001), suggesting that it is the chemical nature of gold that likely plays a role in AuNP1.5-induced cytotoxicity.
2.3. AuNP Cell Uptake by hESCs
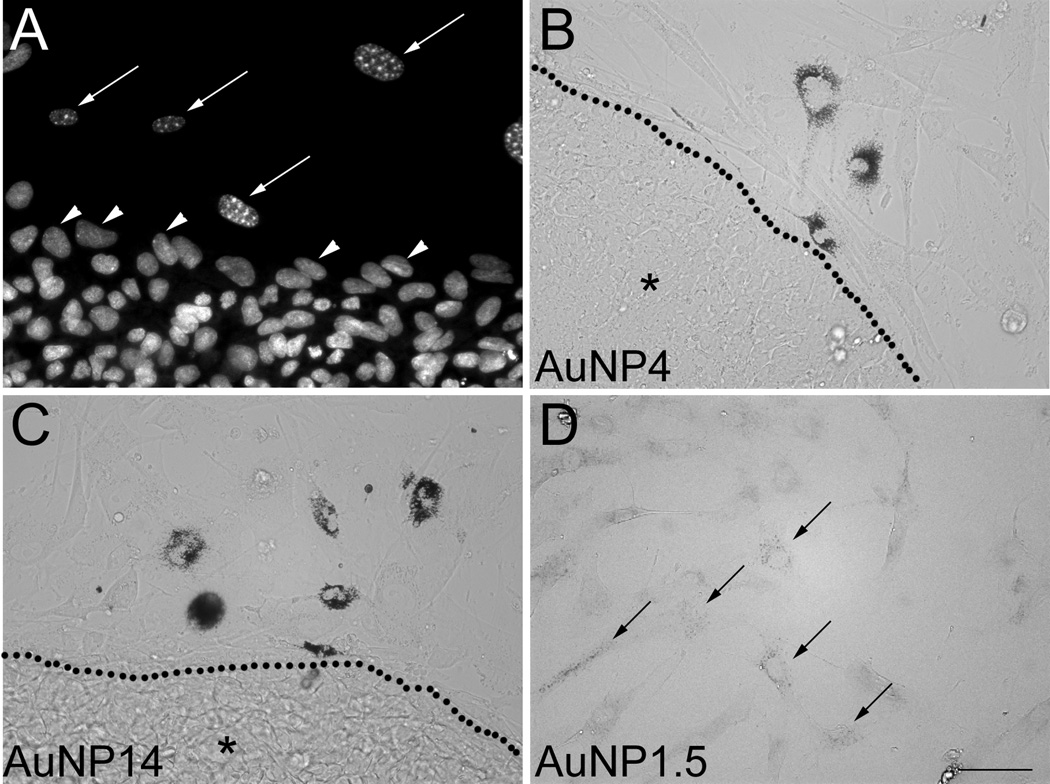
Size, shape, surface chemistry, and dosage are factors that may affect the uptake of AuNPs by cells.[5] The main route for AuNP cellular entry is through endocytosis and it depends on the size, shape, and surface chemistry of AuNP.[13, 43] As shown in Figure 2A, intracellular accumulation of AuNP4 and AuNP14 could clearly be detected by optical microscopy in cultures of hESCs grown on MEFs after a 24-h exposure to 10 µg/mL concentration. Using DAPI (4’,6-diamidino-2-phenylindole) staining to visualize the differences in nuclear chromatin patterns that exist between MEFs and hESCs,[44, 45] we observed that the nuclei of AuNP-positive cells displayed the bright fluorescent heterochromatin foci, characteristic of both proliferating and senescent MEFs[45] (Figure 3A). In MEFs, AuNPs appeared as black dots that distributed throughout the cellular cytoplasm, preferentially aggregating around the nucleus (Figure 3B–C). Because no clear uptake of AuNP1.5 could be observed in the cultures, we used the GoldEnhanceTM (Nanoprobes, Inc.) amplification method, which revealed the presence of labeled MEFs (Figure 3D). In contrast to the MEF case, no accumulation of AuNP1.5 (after GoldEnhance treatment) (data not shown), AuNP4, or AuNP14 could be observed in hESCs (Figure 3B–C). While we cannot exclude that cell death being a factor preventing active transport of AuNP1.5 at 10 µg/ml, we observed no visible intracellular entry of AuNP1.5 at sub-lethal dosages.
Figure 3.
Uptake of AuNPs by MEFs but not hESCs. (A) Cell cultures stained with the nuclear marker DAPI showing the bright heterochromatin foci characteristic of MEFs (long arrows), which allow to distinguish them from hESCs (arrowheads). (B, C) Uptake of AuNP4 (B) and AuNP14 (C) was clearly visible in MEFs, but not in hESC colonies (region marked by *) delineated by dotted lines. (D) Uptake of AuNP1.5 by MEFs was only visible following intensification with GoldEnhance™ (arrows). The scale bar length in (D) valid for (A–C) is 50 µm.
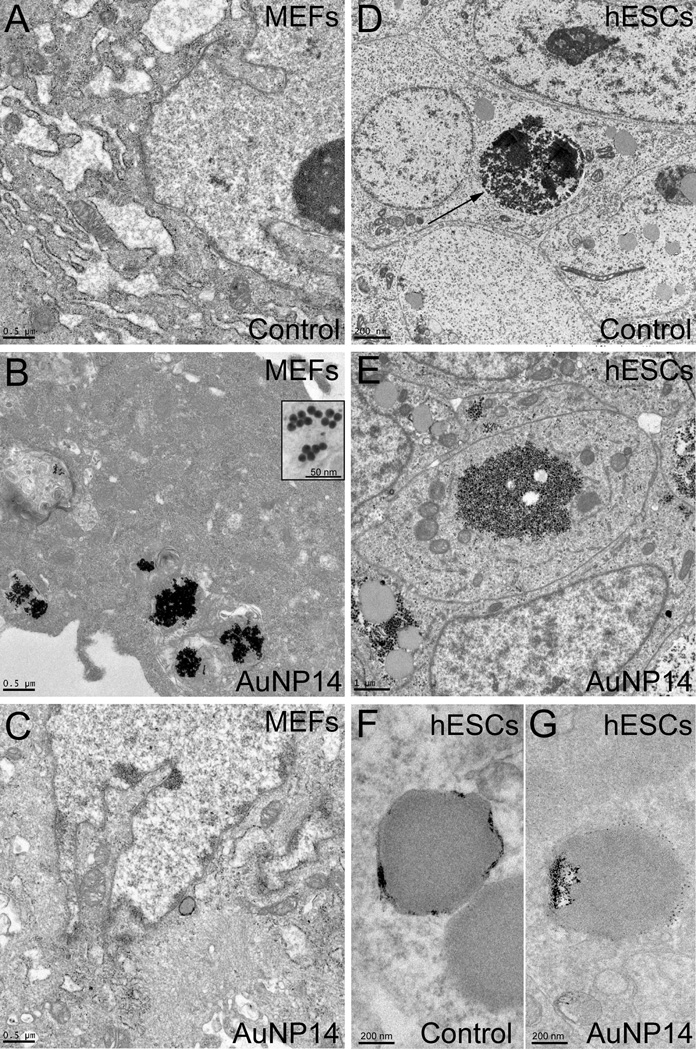
This peculiar observation prompted us to further examine the uptake efficiency of AuNPs by MEFs and hESCs. To this end, we treated MEFs and hESCs with either vehicle (control) or 10 µg/mL AuNP14 for 48 h and then looked for the presence of AuNP14 using TEM. Compared to control MEFs (Figure 4A), nanoparticle clusters could clearly be observed in MEFs exposed to AuNP14 (Figure 4B). Clusters were mostly found in the cytoplasm, and very rarely localized to the nucleus (Figure 4C). As shown in Figure 4D, hESCs appeared as tightly arranged cells with high nuclear to cytoplasmic ratio. Clusters of dark small sized glycogen granules (< 50 nm) were frequently observed in the cellular cytoplasm (Figure 4D, arrow) as previously observed in hESCs.[46–49] AuNP14-exposed hESCs exhibited a cellular ultrastructure similar to that observed in control cells, and no AuNP14 could be clearly observed (Figure 4E), suggesting that no intracellular AuNP uptake had occurred in hESCs. Because staining of the sections may make it difficult to discriminate the AuNP from the electron-dense glycogen granules, we also analyzed unstained sections by TEM. No AuNP could clearly be observed in AuNP-exposed versus control hESCs (Figure 4F–G). Combining the data from optical and electron microcopies we conclude that AuNP14 was taken up readily by MEFs but not by hESCs.
Figure 4.
TEM analysis of AuNP14 uptake in irradiated MEFs and hESCs. (A) TEM analysis of MEFs exposed to vehicle for 48 h allows for the identification of cellular organelles. (B–C) After 48 h-exposure, AuNP14 can clearly be observed in irradiated MEFs (see inset in B), where they mostly distribute in the cytoplasm (B) but are mostly absent from the nucleus (C). (D) Cytoarchitecture of vehicle-exposed hESCs was clearly visible using TEM, including clusters of glycogen granules (arrows). (E) hESCs exposed to AuNP14 for 48 h showed an ultrastructure similar to that of control cells. (F–G) No visible AuNP14 could be detected in unstained sections of exposed hESCs (G) compared to unstained sections of control hESCs (F). The scale bar length is 500 nm for (A), (B), and (C), 50 nm for the inset in (B), 200 nm for (D), (F), and (G), and 1 µm for (E).
2.4. AuNP Effect on hESC Pluripotency
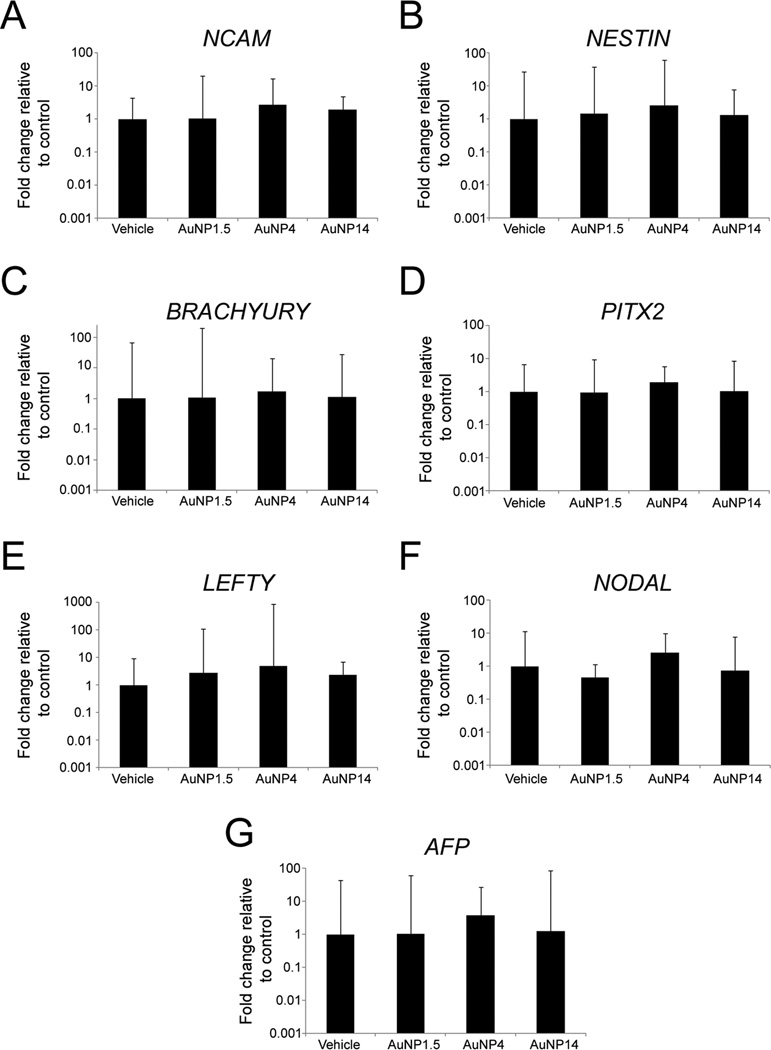
Next, we sought to determine if AuNP exposure affects the capabilities of hESCs to generate ectodermal, mesodermal, and endodermal derivatives. The pluripotency of hESCs enables them to differentiate into multiple tissue types. To this end, we passively differentiated hESC-derived embryoid bodies (EBs) for 2 weeks in basic fibroblast growth factor (bFGF)-free stem cell medium, while exposing them to either AuNP or vehicle. The concentration for AuNP4 and AuNP14 was 10 µg/mL, while a sub-lethal dosage of 0.6 µg/mL was used for AuNP1.5. As shown in Figure 5, quantification of marker mRNAs indicated that there were no statistically significant differences in the expression levels of NCAM (Figure 5A), NESTIN (Figure 5B), BRACHYURY (Figure 5C), PITX2 (Figure 5D), LEFTY (Figure 5E), NODAL (Figure 5F), and AFP (Figure 5G). These gene expression results suggest that, at the concentrations tested, exposure to AuNPs did not markedly alter the in vitro differentiation potentials of hESCs.
Figure 5.
Characterization of the effects that 14 day-exposure to AuNP1.5 (0.6 µg/mL), AuNP4 (10 µg/mL), and AuNP14 (10 µg/mL) has on the capability of hESCs to passively differentiate into cells of the three germ layers. (A, B) Quantitative RT-PCR expression analysis for the ectodermal markers (NCAM) (A), and (NESTIN) (B). (C–E) Quantitative RT-PCR expression analysis for the mesodermal markers (BRACHYURY) (C), (PITX2) (D), and (LEFTY) (E). (F, G) Quantitative RT-PCR expression analysis for the endodermal markers (NODAL) (F) and (AFP) (G). At the concentrations examined, exposure to AuNPs did not affect the in vitro pluripotency capabilities of hESCs. Histogram values represent means and error bars represent 95% interval confidences (p > 0.05; n = 3, one-way ANOVA and Kruskal Wallis tests).
2.5. AuNP Effect on the Neuronal Differentiation of hESCs
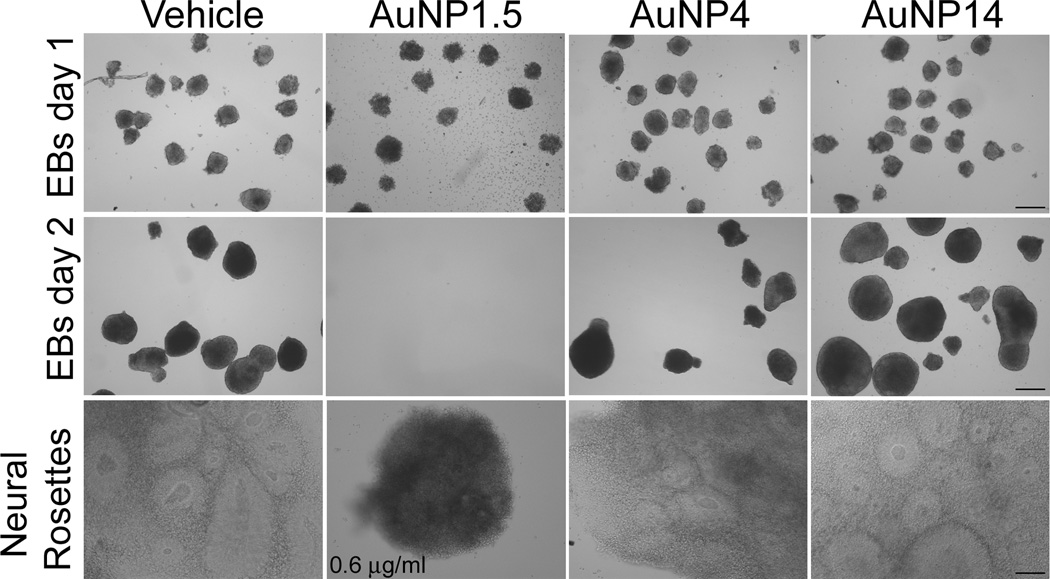
To examine the effects of exposure to various sized AuNPs on the neural differentiation of hESCs, we generated EBs and maintained them throughout the entire neural differentiation protocol (19 days) in the presence of vehicle or 10 µg/mL AuNP1.5, AuNP4, or AuNP14. The presence of AuNP4 and AuNP14 in the medium did not prevent the formation of EBs, which appeared morphologically similar to those generated from vehicle-exposed EBs (Figure 6). In contrast, AuNP1.5-exposed hESCs did not form EBs but rather rapidly disintegrated into single cells within 48 h of treatment, so that by day 3, no EBs survived (Figure 6). Therefore, following experiments were carried out using a 0.6 µg/mL (EC30) concentration for AuNP1.5, which allowed the formation of EBs. During neural differentiation, both control and AuNP1.5-treated EBs were able to generate neural rosettes (Figure 6). However, the neural rosettes formed by AuNP1.5-exposed EBs were small, exhibiting a dark aspect (Figure 6), and did not survive. The data suggest that even at sub-lethal dosage AuNP1.5 causes adverse effect on neuronal differentiation of hESCs.
Figure 6.
Formation of embryoid bodies and neural rosettes by control and AuNP-exposed hESCs. Exposure to 10 µg/ml AuNP4 or AuNP14 did not affect the capability of hESCs to generate EBs and neural rosettes, compared to control cells (vehicle). In contrast, cells exposed to 10 µg/ml AuNP1.5 were not able to form EBs and died within 2 days of exposure. When treated with a lower concentration of 0.6 µg/ml AuNP1.5, hESCs were able to form EBs, but exhibited a decrease in growth, which resulted in the formation of small sized neural rosettes compared to control cells. Bars: 150 µm (EBs day 1), 100 µm (in EBs day 2 and neural rosettes).
Even though NPCs generated from AuNP4- and AuNP14-treated hESCs appeared similar to those obtained from control hESCs, we further verified their capability to differentiate into neurons by using previously published protocol.[50] By day 21, about 15.1 ± 2.3% of MAP2-positive neurons were observed in control cultures. No statistically significant differences (p = 0.912) in the percentage of MAP2-positive neurons were observed between NPC cultures derived from AuNP4- (16.3 ± 3%) or AuNP14- (14.7 ± 2.6%) hESCs. We then analyzed the complexity of the differentiated neurons by quantitatively measuring several morphological parameters. Sholl analysis (Figure S3, SI) indicated no remarkable differences in the average sum of crossings (p = 0.21) (Figures S3A–B, SI), average neuritic length (p = 0.45) (Figure S3C, SI) or average number of branchings (p = 0.25) (Figure S3D, SI) between control hESCs and AuNP-treated hESCs. Taken together, these data show that exposure to AuNP4 or AuNP14 of hESCs does not alter the neuronal differentiation capabilities of their derived NPCs.
2.6. AuNP Effect on hESC-derived NPCs
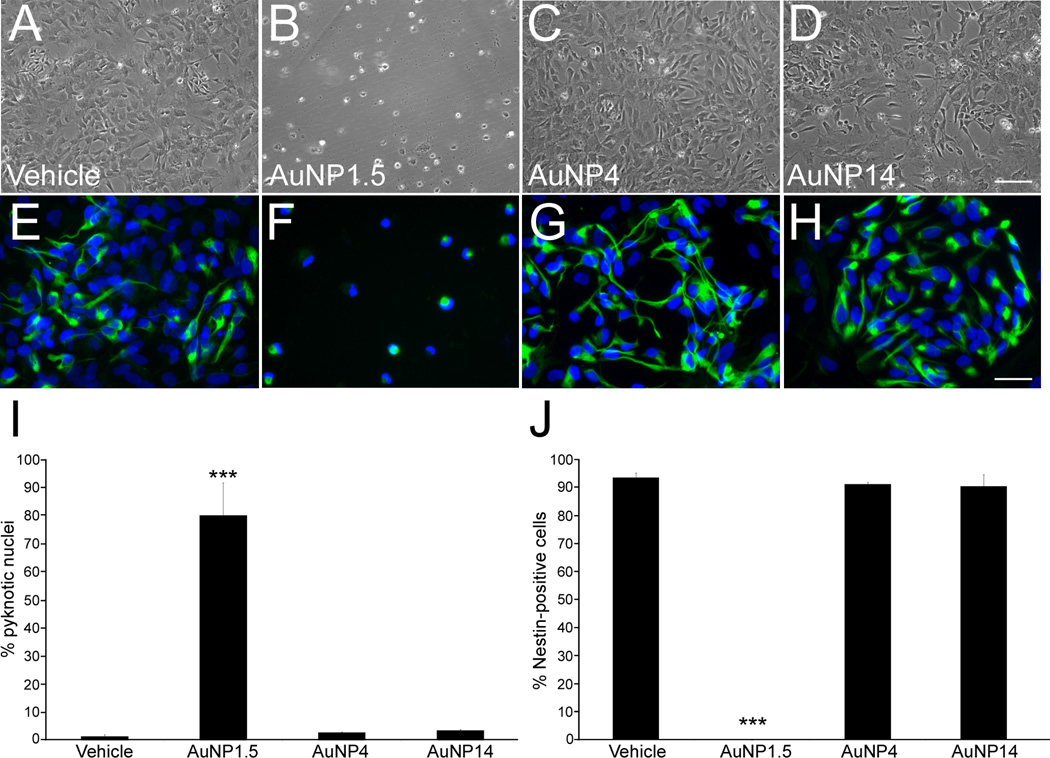
To better understand the neurotoxic effects of AuNP1.5 on the formation of neural rosettes, we examined the direct effects of AuNP1.5 on the survival of NPCs. To this end, proliferating hESC-derived NPCs were exposed for 3 days to vehicle or 10 µg/mL of AuNPs, and immunostained for the neural marker Nestin and the DNA marker DAPI. While cultures treated with vehicle, AuNP4, and AuNP14 exhibited numerous NPCs, those exposed to AuNP1.5 were almost completely depleted of cells (Figure 7A–H). There was a significant 54-fold increase (p = 0.02) in the number of pyknotic nuclei (condensed chromatin state that precedes apoptosis) in AuNP1.5-treated wells (Figure 7I), which also exhibited loss of Nestin-positive cells (p < 0.001), compared to other experimental exposures (Figure 7J).
Figure 7.
Examination of the effects of AuNP exposure on the survival of hESC-derived NPCs. NPCs were treated with vehicle or 10 µg/mL of AuNP1.5, AuNP4, and AuNP14 for 72 h, processed for immunofluorescence detection of Nestin, and then stained with the nuclear marker DAPI. (A–H) hESC-derived NPCs exposed for 72 h to vehicle (A, E), AuNP1.5 (B, F), AuNP4 (C, G), and AuNP14 (D, H). Whereas exposure to AuNP1.5 resulted in cell death (B, F), NPCs treated with AuNP4 (C, G) and AuNP14 (D, H) displayed a viability and a Nestin immunostaining (E–H) similar to those observed in control conditions (A, E). (I) Quantitative analysis of NPCs harboring pyknotic nuclei showed ongoing cell death in cells exposed to 10 µg/mL AuNP1.5. (J) No Nestin-positive NPCs survived following 72-h exposure to AuNP1.5. Histogram values are means ± SEM. Statistical significance compared to the vehicle control group is designated as (***) (p < 0.001; n = 3, one-way ANOVA). The scale bar length is 100 µm for (A–D) and 30 µm for (E–H).
2.7. AuNP Effect on hESC Epigenetics
We hypothesized that some of the physiological effects of AuNP exposure can be mediated by changes in the methylcytosine (5mC) profile of the genome. Therefore, we investigated the global 5mC status of the genome using a 5mC-immunoprecipitation-based colorimetric assay in hESC exposed to sub-lethal dosage of 1 µg/mL AuNP1.5, 10 µg/mL AuNP4, or 10 µg/mL AuNP14 for 24 hours. We chose 10 µg/mL also because it was used on one of few studies using embryonic stem cells.[51] This concentration avoids exposing cells to an excessive amount of buffer. We observed a marked reduction of 5mC levels 24 h after AuNP exposure irrespective of AuNP size (Figure 8A). Interestingly, exposure to AuNP4 showed the most dramatic decrease in 5mC levels (Figure 8A). This decrease was coupled with an increase in hydroxymethylcytosine (5hmC), which is consistent with 5hmC being the first step in the demethylation process of DNA (Figure 8B; see Discussion). We performed a two-sample t-test, assuming equal variances, to determine if the reported differences were statistically significant. For 5mC, the p-value associated with a decrease in global 5mC levels in AuNP4 treated cells was 0.062. For 5hmC, the p-value associated with an increase in global 5hmC levels in AuNP4 treated cells was 0.093. Even though the p-values for the colorimetric assays did not reach the significant cut-off of 0.05, the data suggested a trend toward AuNP4-induced global demethylation of the genome. To further explore this result, we looked at the locus specific methylation profile of hESCs exposed to AuNP1.5, AuNP4, and AuNP14 using the HM450K array, which measures the DNA methylation levels at over 480,000 CpG sites in the human genome (see Methods).
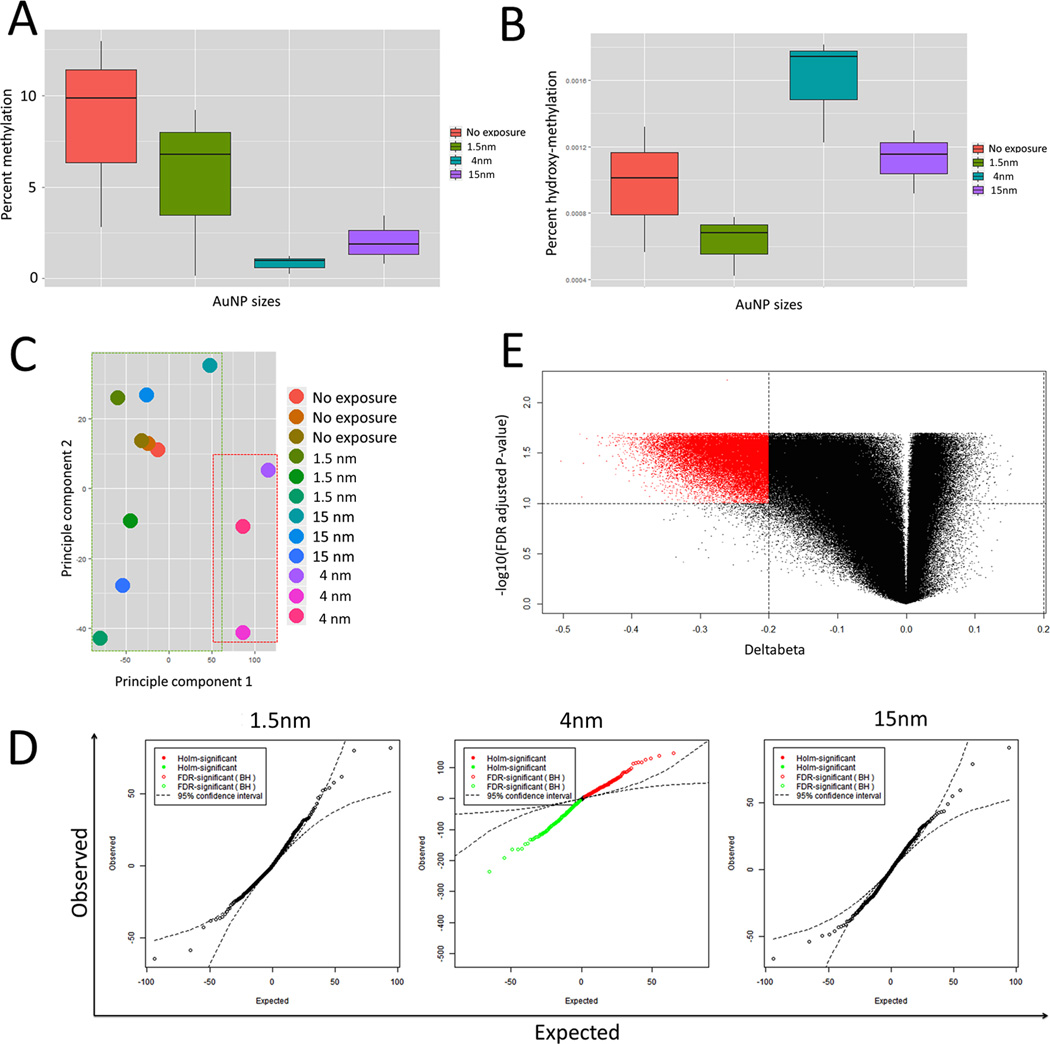
Figure 8.
Exposure to 10 µg/ml AuNP4 for 24 h causes global demethylation of genomic DNA. (A) Global 5mC levels measured using MethylFlash™ Methylated DNA Quantification Kit. Genomic DNA from AuNP4 treated hESCs shows a observable decrease in 5mC, though the p-value from 2 sample t.test assuming equal variances is 0.062. (B) Global 5hmC levels measured using MethylFlash™ Hydroxymethylated DNA Quantification Kit. Genomic DNA from AuNP4 treated hESCs shows a observable increase in 5hmC, though the p-value from 2 sample t.test assuming equal variances is 0.093. (C) PCA on normalized beta matrix indicating that the AuNP4 cluster seperately from the other treatments (AuNP1.5 and AuNP14) and the non-treated controls. (D) q-q plot for genome wide association analysis using fixed effect model for AuNP1.5, AuNP4, AuNP14. The results show a large number of CpG sites (red = increase, green = decrease) where called to be significant differentially methylated at a FDR corrected p-value cut-off < 0.05. (E) Further filtering by effect size (or deltabeta) ≥ abs (0.20 or 20%) result in 31,311 CpG sites all showing decrease in methylation on treatment with AuNP4.
Principle component analysis (PCA) of the matrix of normalized beta values for all sample indicated the hESCs treated with AuNP4 clustered separately along principle component 1 compared to the non-treated hESCs, and hESCs treated with AuNP1.5 and AuNP14 (Figure 8C). All of the samples also showed separation along Principle component 2, suggesting large variability between replicates (Figure 8C). In agreement with the PCA, we did not observe any significant association of 5mC levels for single CpG sites (at an FDR corrected cutoff of p < 0.05) with exposure to either 1 µg/mL AuNP1.5 or 10 µg/mL AuNP14 (Figure 8D). However hESCs treated with 10 µg/mL AuNP4 showed a large number of exposure-dependent genome-wide associations between 5mC levels for single CpG sites, as illustrated by the q-q plot (Figure 8D). A q-q plot is a plot of the quantiles of the first data set against the quantiles of the second data set. By a quantile, we mean the fraction (or percent) of points below the given value. That is, the 0.2 (or 20%) quantile is the point at which 20% percent of the data fall below and 80% fall above that value. Previous studies have indicated that the HM450K array is able to detect 0.20 or 20% difference in CpG methylation (Deltabeta) with a confidence of ~95%. At an exposure effect size ≥ |0.20| or 20%, FDR corrected p-value ≤ 0.05, 31,311 single CpG sites showed a decrease in beta values (negative Deltabeta) on exposure to AuNP4 (Figure 8E). This result confirms our previous observation that treatment of hESCs with AuNP4 cause global DNA demethylation.
3. Discussion
This is one of the first studies to demonstrate the use of hESCs for assessing nanotoxicology, specifically, the effect of AuNPs on the viability, pluripotency, neuronal differentiation, and DNA methylation of hESCs. Some significant advantages in using hESCs compared to other (e.g., animal-derived) systems are that 1) they have unlimited self-renewal capabilities and can be grown indefinitely in the laboratory; 2) they can differentiate into a variety of specialized cell types, such as neurons over a longer period of time; 3) they preclude interspecies variability, making them a better model to predict human response to toxins; and 4) subtle changes in cell functionality rather than cell death can be tested on nanomaterials of low toxicity such as majority of AuNPs. This study contributes a hESC-based assay to the study of human developmental neurotoxicology. The differentiation of hESCs into neurons mimics aspects of the early stages of human brain development. Cytotoxicity to hESCs caused by AuNP1.5 therefore implies that such ultrasmall AuNPs may be toxic to human (especially considering the size effect on tissue distribution and ability to cross the blood-brain barrier) and their use be further regulated.
Of the three AuNP sizes studied, only AuNP1.5 shows toxicity to hESC viability and differentiation into EBs. The IC50 value measured during 4-day exposure of AuNP1.5 is 0.0706 µM. In comparison, the IC50 (3 day test) for a common chemotherapeutic agent, cisplatin, is 0.45 µM tested on fibroblast CCD-919Sk cells, 7.93 µM on cervical cancer HeLa cells, and 26.10 µM on bone MCT3T3-E1 cells.[18] It shows that AuNP1.5 can be as or more toxic than chemotherapeutic drugs. Indeed, some have suggested that such AuNPs could be used for cancer treatment if toxicity to normal cells can be controlled.[1, 52]
Our study also found that dendrimers of generation 0.5 and 1.5 with similar size and carboxylic acid termination to AuNP1.5 do not exhibit cytotoxicity to hESCs. However, one must consider differences beyond a simple size comparison. Dendrimers of such early generations (G0.5 and G1.5) behave more like an open prolate ellipsoid than a hard sphere.[53, 54] In addition, the interaction between amphiphilic dendrimers with hydrophobic core and hydrophilic surface may interact with cell membranes differently from hydrophilic AuNPs with fully protected surface.
Though our study is not the first one to show cytotoxicity of ultrasmall AuNPs of the cluster structure Au55, we want to point out some key new findings based on our results. First we briefly summarize previous studies on size-dependent cytotoxicity of AuNPs both in vitro (e.g., using melanoma cells, cervix carcinoma cells, mouse fibroblasts, and mouse macrophages)[18, 23, 55] and in vivo (using zebrafish embryos).[56] Au55 cluster with a diameter of 1.4 nm capped by triphenylphosphine monosulfonate (TPPMS), denoted as Au1.4MS, was found highly toxic to a number of human cancer and healthy cell lines.[18] For example, the IC50 value of Au55 was determined to be 0.62 µM for CCD-919Sk fibroblast cells measured at 24 h. Au1.4MS was found to cause necrosis by oxidative stress and mitochondrial damage while AuNPs of 15 nm in size are nontoxic.[23] The same study also showed that when the phosphine ligand was replaced with a thiol ligand, glutathione, Au55 cytotoxicity was greatly reduced.[23] The authors attributed the toxicity of Au1.4MS to weak phosphine/metal binding that allows highly electronegative gold atoms to interact directly with biological targets. However, in our study, we found that even thiolate-capped Au55 is toxic to both MEFs and hESCs. In addition, the low IC50 of hESCs suggests that hESCs may be more sensitive to Au55 than other cell types.
The second key finding of this study is hESC uptake of AuNPs is much more limited than other cell types reported in the literature.[13, 43, 57] We found little evidence of intracellular uptake of AuNP4 and AuNP14 by hESCs while at the same time these nanoparticles were located readily in the cytoplasm of MEFs, despite their being lethally irradiated (which is a standard treatment for feeder cells). Our GoldEnhance data suggest that AuNP1.5 may not be taken up by hESCs, which points to a possibility that AuNP1.5 imposes its adverse effects on hESC survival and neuronal differentiation without significant intracellular uptake. In the literature, several mechanisms of AuNP cytotoxicity have been proposed. One attributes AuNP cytotoxicity to Au55 binding to the major groove of B-DNA thereby interfering with DNA transcription in the cell nucleus.[18] The second mechanism is the continuous generation of intracellular reactive oxygen species (ROS), such as superoxide radicals, hydrogen peroxide, and hydroxyl radicals, by Au55 reaction with dioxygen.[23] ROS subsequently can cause oxidation of biomolecules. The third mechanism is Au55’s interactions with cell membrane proteins, for example, by blocking membrane ion channel[58] or reducing free thiols on the cell membrane.[59] In another study of AuNP surface functionalities on zebrafish embryonic gene expression, 2-mercaptoethanesulfonic acid (MES)-capped 1.5 nm AuNPs were found to affect ion channel transport mechanism signaled by misregulated gene expression.[60] Our data of hydrodynamic sizes of AuNP1.4, G0.5, and G1.5 (i.e., difficulty in measuring their sizes even in the presence of BSA) suggest that BSA could not form a corona around these ultrasmall nanoparticles due to steric constraints. Taken all together, our data suggest that AuNP1.5 interferes with membrane functions due to lack of BSA surface modification as well as their larger surface area to volume ratio. Further studies are necessary in order to clearly elucidate the mechanism underlying Au55 toxicity to hESCs and their differentiation. Finally, the leaching of ionic gold is also a possibility that cannot be ruled out. However, others have shown that gold nanoparticles of smaller sizes, 0.8 nm and 1.2 nm in diameter, are much less toxic than 1.4 nm gold.[55] This seems to argue against a simple ion leaching mechanism.
A third major finding from this study is that AuNP4 causes greater than 20% decrease in DNA methylation at over 31,000 CpG sites, which represents over 6% of the probes on the HM450K array. There were no CpG sites that had a greater than 20% increase in DNA methylation, which suggests that AuNP4 somehow leads to a global decrease in DNA methylation by some unknown mechanism. This is surprising because AuNP4 showed no toxicity in our hESC assays, so it suggests that AuNP4 might actually be beneficial to these cells by decreasing the levels of DNA methylation. Stem cells generally exhibit a much lower level of DNA methylation level than somatic cells. Consistent with the idea that AuNP4 causes a global decrease in DNA methylation, colorimetric assaysusing 5mC antibodies showed a trend for a decrease in DNA methylation. Similarly, colorimetric assay with 5hmC antibodies showed a trend for an increase in DNA hydroxymethylation. Since 5hmC is generated by Ten-eleven-tranlocase (TET) family proteins, that are involved in DNA demethylation,[61] we speculate that AuNP4 stimulates the activity of one or more TET proteins and this ultimately leads to a global decrease in DNA methylation.
4. Conclusions
This work described AuNPs of 1.5 nm, 4 nm, and 14 nm in core size on hESC viability, cell uptake, pluripotency, neuronal differentiation into EBs and NPCs, and epigenetic modifications to its DNA. AuNPs were used as model nanoparticles due to their versatile surface chemistry, ease of imaging, and tunability for targeted drug/imaging agent delivery. We found that hESC colonies exposed to 1.5 nm MSA-capped AuNPs exhibited loss of cohesiveness, rounding up, and detachment suggesting ongoing cell death. The hESCs exposed to 1.5 nm AuNPs did not form EBs but rather rapidly disintegrated into single cells within 48 hours of treatment. Cell death caused by 1.5 nm AuNPs also occurred in the treatment hESC-derived neural precursor/progenitor cells. None of the other nanoparticles exhibited toxic effects on the hSECs or their derivatives in experiments lasting as long as 19 days. In contrast to some reports, we found that Au55 capped by the strongly binding thiolate could still be toxic to cells. Our data suggest that hESC uptake of nanoparticles may be different from other cell types as we did not observe significant amount of AuNPs inside the hESCs while AuNPs were clearly present inside the MEF feeder cells. Our data also show that 4 nm AuNPs lead to a global decrease in DNA methylation and may benefit cells by decreasing the levels of DNA methylation. In conclusion, this work identifies a type of AuNPs highly toxic to hESCs and demonstrates the potential of hESCs in predicting neurotoxicity of nanoparticles. This work could ultimately impact consumers, patients, and manufacturing workers of products containing nanoparticles.
5. Experimental Section
Reagents
Gold(III) chloride trihydrate (HAuCl4·3H2O, 99% metal trace), chloro(triphenylphosphine)gold(I) (AuCl(PPh3), 97%), sodium citrate tribasic dehydrate (99%), mercaptosuccinic acid (MSA, 97%), borane tert-butylamine complex (97%), sodium borohydride (NaBH4) (98%), polyamidoamine (PAMAM, ethylenediamine core) dendrimer G0.5 (powder) and G1.5 (methanol solution), and bovine serum albumin (BSA, 98%) were all purchased from Sigma-Aldrich. Sodium hydroxide (98%) was purchased from Fisher Scientific.
AuNP Synthesis
The chemical structures of the nanoparticles used in this study are shown in Scheme 1. The number corresponds to the average AuNP diameter in nanometer measured by TEM. Thiolate-capped 1.5 nm ultrasmall nanoparticles (AuNP1.5) were synthesized by modifying a literature procedure.[32] The Au(I) precursor AuCl(PPh3) (0.375 mmol) was partially dissolved in 21 mL ethanol at room temperature under vigorous stirring (600–800 rpm). Tert-butylamine-borane (3.75 mmol) was used as a mild reducing agent, and 2 h after its addition, MSA (0.15 mmol) was added to control the particles size.[33] Mercapto-alkyl acids have been found to be effective in stabilizing ultrasmall AuNPs in aqueous solution. The final AuNP ethanol solution was washed and purified by ultra-centrifugation using MW10000 Amicon® centrifuge tubes in deionized water repeatedly.
MSA-capped AuNP4 were synthesized as following by modifying a literature procedure.[34] A 500 mL aqueous solution containing 0.25 mM HAuCl4·3H2O and 0.25 mM trisodium citrate was prepared in an Erlenmeyer flask under vigorous stirring at room temperature. Then, 15 mL of ice cold 0.1 M NaBH4 solution were added to the solution, resulting in a change of the color solution to pink, indicative of the AuNP formation. After adjusting the pH of the solution to 11 with 0.1 M NaOH solution, 50 mg MSA was added and the solution was stirred for 12 h. The final solution was concentrated to 2.5 g (gold)/l by ultracentrifugation using MW10000 Millipore Amicon® centrifuging units and washed by deionized water several times.
Citrate-capped AuNP14 were synthesized by using the Turkevich method.[35–38] In a typical synthesis, a 500 mL aqueous solution containing 0.25 mM HAuCl4·3H2O was heated to boil, then 175 mg of sodium citrate dissolved 30 mL water solution was directly poured into the reaction mixture. The color of the reaction turned grey, pink, and finally wine-red. After adjusting the pH, AuNP14 were coated with BSA as described above for AuNP4.
To apply the BSA coating, 10 mg of the nanoparticle dssolved in 10 mL deionized water was incubated overnight with 400 mg BSA.
AuNP Characterization
Varian Cary 50 UV-vis Absorption Spectroscopy was used to determine AuNP sizes, based on the Beer-Lambert law, with a scan range of 200 to 800 nm.
TEM images of AuNPs were taken on JEOL JEM-2010. The samples were prepared by placing a droplet of the AuNP solution on a Formvar-coated copper TEM grid. Excess liquid was removed by a piece of filter paper under the grid and dried in the air. The working voltage was 200 keV and the current was 109 mA. At least 50 particles were measured on the same sample to yield an average particle size.
DLS and zeta potential were measured using a Malvern Nano-ZS. A 1 mL solution was transferred to a 2.0 mL polystyrene cuvette. The Z-average hydrodynamic diameter (HD), polydispersity index (PDI), and zeta potential were measured at 25°C. 15 scans were performed in each measurement. The backscattering angle Θ was fixed at 172° with a laser wavelength λ = 633 nm. The size measurement range was set between 1 nm and 6 µm. HD is a function of the diffusion coefficient (D), temperature (T), and viscosity (η) according to the Stokes-Einstein equation: , where k is Boltzmann constant, T is 25°C, and D was obtained from autocorrelation function via the cumulant fitting. The electrophoretic mobility of the nanoparticles was measured using the laser Doppler velocimetry and phase analysis light scattering technique of the Malvern Zetasizer. The electrophoretic mobility was converted into zeta potential using the Smoluchowski equation using the Malvern software.[62]
Human Embryonic Stem Cell Cultures
The NIH-approved and registered WA09 hESC line (passages 26–32; WiCell Research Institute, Madison, WI, USA)[63] was maintained in a humidified incubator at 37°C with 5% CO2 as previously described.[64] Briefly, cells were grown on a feeder layer of irradiated mouse embryonic fibroblasts (GlobalStem, Rockville, MD, USA) in hESC culture medium made of 20% knockout serum replacement, 2 mM L-glutamine, 1% non-essential amino acids, 1% penicillin/streptomycin (all products from Life Technologies Corporation, Carlsbad, CA), 4 ng/mL human fibroblast growth factor basic (bFGF, Peprotech, Rocky Hill, NJ), and 0.1 µM 2-mercaptoethanol (Sigma-Aldrich, St Louis, MO) in DMEM/F12 (Life Technologies Corporation, Carlsbad, CA). The active ingredient within the serum replacement is lipid-rich albumin. hESCs were regularly passaged by mechanical dissection and their pluripotent state assessed by immunofluorescence detection for pluripotency markers, such as Oct4, Nanog, and Lin28.
Trypan Blue Exclusion Assay
At the end of exposure, control and AuNP-treated hESCs were harvested and stained with 0.4% Trypan blue (Life Technologies Corporation, Carlsbad, CA). Viable (unstained) and non-viable (dark-blue stained) cells were counted with a hemacytometer. The number of viable cells was determined by subtracting the number of non-viable cells from the total number of cells counted and expressed as a percentage relative to the total number of cells. Data were collected from three independent experiments.
MTT Assay
hESCs were cultured in phenol red-free culture medium and plated at a density of 100,000 cells per well of a 12 well-plate. At the end of AuNP exposure, the medium was supplemented with 0.5 mg/mL of 3-(4,5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT, Sigma-Aldrich, St Louis, MO) freshly prepared in phosphate-buffered saline (PBS), and cells were incubated in the dark at 37°C for 3 h. MTT-formazan crystals were then solubilized for 15 min, at room temperature in dimethyl sulfoxide (DMSO). Absorbance was measured at 590 nm using a spectrophotometer, and cell survival was expressed as a percentage of absorbance relative to that of control samples. Data were obtained from three separate experiments.
Uptakes Studies by TEM
The cellular uptake of AuNPs was assessed on MEFs and undifferentiated WA09 cells (passage 32) exposed to vehicle or 10 µg/mL AuNP14 for 48 h. At the end of exposure, cells were washed and fixed in 2.5% glutaraldehyde (Electron Microscopy Sciences, Hatfield, PA) in PBS. Samples were processed at the MSU Center for Advanced microscopy where they were post-fixed in 1% osmium tetroxide, rinsed in distilled water, dehydrated through a graded series of acetone, and embedded in epoxy resin. Ultrathin 70 nm sections were obtained and then analyzed and photographed using a JEOL 100CXII electron microscope.
Pluripotency in vitro Studies
The effects of exposure to AuNPs on the in vitro pluripotency capabilities of hESCs were determined using spontaneous differentiation of embryoid bodies (EBs).[65] Briefly, EBs were generated by mechanical dissociation of hESC colonies and cultured for 2 weeks in FGF2-free hESC medium in presence of vehicle, 0.6 µg/mL AuNP1.5, or 10 µg/mL of AuNP4 and AuNP14. At the end of exposure, total RNA was extracted from EBs using Qiazol (Qiagen, Valencia, CA) according to the manufacturer’s protocol, quantified by optical density, and subjected to reverse transcription using Superscript II® Reverse Transcriptase and random primers (Life Technologies Corporation, Carlsbad, CA) for 50 min at 40°C followed by 15 min at 75°C. Quantitative PCR analyses of the well established lineage marker genes - endodermal (AFP and NODAL), ectodermal (NCAM and NESTIN), and mesodermal (BRACHYURY, PITX2, and LEFTY) - were performed using an ABI PRISM 7000 Sequence Detection System® and its software (Applied Biosystems) with 10 ng cDNA, 400 nM of each primers and SYBR Green PCR Master Mix® (Life Technologies Corporation, Carlsbad, CA). Resulting data were normalized against the expression of the internal control GAPDH gene. Data analysis was performed using the 2−ΔΔCt method (Livak and Schmittgen 2001) and standardized by log transformation, mean centering, and autoscaling as previously described.[66] Data were collected from three separate experiments.
Differentiation of hESCs into NPCs
Generation of NPCs from hESCs was performed as previously described.[64, 67] Following mechanical dissociation of hESC colonies, EBs were cultured in suspension for 4 days in 1:1 hESC/N2 medium (Life Technologies Corporation, Carlsbad, CA), followed by 4 days in N2 medium supplemented with 3 µM retinoic acid (RA; Sigma-Aldrich, St Louis, MO), and 3 days in N2 medium added with 20 ng/mL FGF2 (Peprotech, Rocky Hill, NJ). At this time, EBs were transferred to fibronectin-coated plates (20µg/mL fibronectin; Life Technologies Corporation, Carlsbad, CA) and cultured in N2/FGF2 medium for an additional 8 days to obtain neural rosettes. The latter were subsequently dissociated and plated in N2/FGF2 medium on 10 µg/mL polyornithine/5 µg/mL laminin-coated dishes. Cells were fixed in 4% paraformaldehyde and immunostained for the NPC marker Nestin (mouse anti-Nestin; 1:500; EMD Millipore Corporation, Billerica, MA)mouse anti-Nestin (1:500, EMD Millipore Corporation, Billerica, MA).
Exposure of hESC-Derived NPCs to AuNPs
hESCs-derived NPCs were cultured on fibronectin-coated 6-well dishes in N2/FGF2 medium and chronically exposed to vehicle or the different sized AuNPs for 72 h, at which time cells were fixed and immunostained for Nestin and the DNA marker DAPI. The total number of Nestin-positive cells and DAPI-stained nuclei were counted in five fields per well in three separate experiments, by an observer blind to the experimental conditions. The number of cells harboring pyknotic nuclei, was also calculated. Total numbers were expressed as a percentage of the total number of cells, as evaluated by DAPI or Hoescht staining.
Neuronal Differentiation of hESC-Derived NPCs
Neuronal differentiation of NPCs derived from control and AuNP-exposed hESCs was achieved as previously described.[50, 64] NPCs were cultured on polyornithine/laminin-coated 12-well dishes at a 5,105 cells/cm2 density in N2 medium supplemented with 10 ng/mL brain-derived neurotrophic factor (BDNF), 10 ng/mL glial cell-derived neurotrophic factor (GDNF), 10 ng/mL insulin-like growth factor (IGF) (all factors from Peprotech, Rocky Hill, NJ), 200 ng/mL ascorbic acid (Sigma-Aldrich, St Louis, MO) and 100 ng/mL cAMP (Sigma-Aldrich, St Louis, MO). Cells were fixed and stained for the neuronal marker microtubule-associated protein 2 (MAP2) (mouse antibody; 1:500, EMD Millipore Corporation, Billerica, MA).
Morphological Analysis of MAP2-positive Cells
Determination of neuritic complexity of MAP2-positive cells derived from control and AuNP-exposed hESCs was performed using Sholl analysis, as previously described.[64, 68, 69] We randomly selected neurons for which neurites could be clearly identified and distinguished from neighboring cells. For each experimental condition, analysis of 18–27 neurons was performed blindly to well treatment. For each neuron examined, the average sum of crossings, the average maximal neuritic distance, and the average total number of branchings were recorded.
Antibodies and Immunofluorescence Procedures
Immunofluorescence staining was performed as previously described.[70] Following fixation in 4% paraformaldehyde, cells were rinsed in PBS, blocked in 3% donkey serum (Sigma-Aldrich, St Louis, MO) in 0.3% Triton-X-100 (NDST3%), and incubated overnight at 4°C with the primary antibodies diluted in NDST1%. After several rinses in NDST1%, cells were incubated for 2 h in the dark with secondary anti-mouse antibodies conjugated to Alexa 488 (1:1000, Life Technologies Corporation, Carlsbad, CA). Following rinses, cells were stained with the nuclear marker DAPI (Sigma-Aldrich, St Louis, MO). Specificity of the immunostainings was assessed by replacing the primary antibodies with normal serum, and by omitting one immunoreagent of the immunostaining protocol.
Statistical Analysis
Statistical significance between experimental groups was determined by Students t-test or one-way analysis of variance (ANOVA) followed by a post-hoc Holm-Sidak test when F value was significant, or Kruskal-Wallis one way ANOVA on ranks followed by Dunn’s test using the SigmaPlot 12 software (SYSTAT® Software Inc., San Jose, CA). Statistical significance was defined at p ≤ 0.05. Data are expressed as the mean ± SEM.
Immunoprecipitation Based Colometric Assays
Global DNA methylation status of exposed and control hESCs were determined using MethylFlash™ Methylated DNA Quantification Kit (catalogue no: P-1034) and MethylFlash™ Hydroxymethylated DNA Quantification Kit (catalogue no: P-1036) from Epigentek. Briefly, 200 ng of DNA was bound to the bottom of pre-treated wells and 5hmC DNA was detected using sequential treatment with capture antibody and detection antibody. The final colorimetric signal was normalized to the standard input DNA to estimate the overall methylation percentage.
Statistical Analysis of HM450K Data
The human methylation 450K array (HM450K) is capable of measuring CpG methylation at over 480,000 CpG sites.[71, 72] The percent methylation at the CpG sites is represented as beta values ranging from 0 to 1.[73] The Beta or β for the ith interrogated CpG nucleotide is:
Where yi,methy and yi,unmethy are the intensities measured by the ith methylated and unmethylated probes, respectively. Illumina recommends adding a constant offset α (by default, α = 100) to the denominator to regularize β value when both methylated and unmethylated probe intensities are low. For preprocessing and normalization of the HM450K data the raw signal intensity files (.idat) for HM450K array were obtained. Quality control, background correction and pre-processing of the data were carried out using the pipeline proposed by Teschendorff et al., 2013.[74] The preprocessed beta values were further corrected for Type1/Type2 probe bias. The Type 1 probes are composed of an un-methylated bead and a methylated bead.[75] If the bead for methylated DNA matches the target site there is a single base extension which results in detection which signals into the red channel. Similarly if an un-methylated bead binds to the DNA it signals into the green channel. The Type 2 probes incorporate both the un-methylated and methylated bead type on a single probe and the ratio of incorporation of two differently-colored fluorescent nucleotides determines the methylation signal. The Type 1 probes have been shown to have more stable signal with less variability compared to the Type 2 probes.[76] Therefore to reconcile the differences between the Type 1 and Type 2 probes we used a 3-state beta mixture model (BMIQ) to normalize the data.[74] The preprocessing and the normalization pipeline was implemented using ChAMP package in R. The data was further corrected for batch effects using ComBat function in R and all known human SNP sites (n = 92,667) were removed prior to differential methylation analysis. Principle component analysis (PCA) was performed on the normalized beta matrix for visualization of differences in sample methylation profile. Differential methylation analysis was performed using a fixed effect model implemented using the R function “CpGassoc”.[77]
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the financial support of the National Institutes of Health grants R01HD031550, R01ES012933, R21ES021893, and the WSU-NIEHS Center P30ES020957. The authors are grateful to Dr. Jose Cibelli and all the members of the Cellular Reprogramming Laboratory (CRL) at Michigan State University for the use of CRL facilities, stem cell expertise, and insightful discussions. We would also like to thank Dr. Steven Suhr for the gifts of qRT-PCR primers and Dr. Alicia Woothrow from the MSU Center for Advanced Microscopy for skillful assistance with the TEM studies.
Contributor Information
Dr Marie-Claude Senut, Institute of Environmental Health and Sciences, Wayne State University, 275 East Hancock Road, Detroit, Michigan 48201, USA.
Dr Yanhua Zhang, Department of Chemical Engineering and Materials Science, Wayne State University, 5050 Anthony Wayne Drive, Detroit, Michigan 48202, USA.
Fangchao Liu, Department of Chemical Engineering and Materials Science, Wayne State University, 5050 Anthony Wayne Drive, Detroit, Michigan 48202, USA.
Arko Sen, Institute of Environmental Health and Sciences, Wayne State University, 275 East Hancock Road, Detroit, Michigan 48201, USA.
Dr Douglas M. Ruden, Institute of Environmental Health and Sciences, Wayne State University, 275 East Hancock Road, Detroit, Michigan 48201, USA.
Dr Guangzhao Mao, Department of Chemical Engineering and Materials Science, Wayne State University, 5050 Anthony Wayne Drive, Detroit, Michigan 48202, USA.
REFERENCES
- 1.Shvedova AA, Kagan VE, Fadeel B. Annu Rev Pharmacol. 2010;50:63–88. doi: 10.1146/annurev.pharmtox.010909.105819. [DOI] [PubMed] [Google Scholar]
- 2.Xia T, Li N, Nel AE. Annu Rev Publ Health. 2009;30:137–150. doi: 10.1146/annurev.publhealth.031308.100155. [DOI] [PubMed] [Google Scholar]
- 3.Maynard AD, Aitken RJ, Butz T, Colvin V, Donaldson K, Oberdorster G, Philbert MA, Ryan J, Seaton A, Stone V, Tinkle SS, Tran L, Walker NJ, Warheit DB. Nature. 2006;444(7117):267–269. doi: 10.1038/444267a. [DOI] [PubMed] [Google Scholar]
- 4.De Jong WH, Hagens WI, Krystek P, Burger MC, Sips AJ, Geertsma RE. Biomaterials. 2008;29(12):1912–1919. doi: 10.1016/j.biomaterials.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 5.Khlebtsov N, Dykman L. Chemical Society reviews. 2011;40(3):1647–1671. doi: 10.1039/c0cs00018c. [DOI] [PubMed] [Google Scholar]
- 6.Arvizo RR, Bhattacharyya S, Kudgus RA, Giri K, Bhattacharya R, Mukherjee P. Chemical Society reviews. 2012;41(7):2943–2970. doi: 10.1039/c2cs15355f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb JA, Bardhan R. Nanoscale. 2014;6(5):2502–2530. doi: 10.1039/c3nr05112a. [DOI] [PubMed] [Google Scholar]
- 8.Kogan MJ, Bastus NG, Amigo R, Grillo-Bosch D, Araya E, Turiel A, Labarta A, Giralt E, Puntes VF. Nano letters. 2006;6(1):110–115. doi: 10.1021/nl0516862. [DOI] [PubMed] [Google Scholar]
- 9.Olmedo I, Araya E, Sanz F, Medina E, Arbiol J, Toledo P, Alvarez-Lueje A, Giralt E, Kogan MJ. Bioconjugate chemistry. 2008;19(6):1154–1163. doi: 10.1021/bc800016y. [DOI] [PubMed] [Google Scholar]
- 10.Brambilla D, Le Droumaguet B, Nicolas J, Hashemi SH, Wu LP, Moghimi SM, Couvreur P, Andrieux K. Nanomed-Nanotechnol. 2011;7(5):521–540. doi: 10.1016/j.nano.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Gao N, Sun HJ, Dong K, Ren JS, Qu XG. Chem-Eur J. 2015;21(2):829–835. doi: 10.1002/chem.201404562. [DOI] [PubMed] [Google Scholar]
- 12.Connor EE, Mwamuka J, Gole A, Murphy CJ, Wyatt MD. Small (Weinheim an der Bergstrasse, Germany) 2005;1(3):325–327. doi: 10.1002/smll.200400093. [DOI] [PubMed] [Google Scholar]
- 13.Shukla R, Bansal V, Chaudhary M, Basu A, Bhonde RR, Sastry M. Langmuir : the ACS journal of surfaces and colloids. 2005;21(23):10644–10654. doi: 10.1021/la0513712. [DOI] [PubMed] [Google Scholar]
- 14.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Science (New York, N.Y.) 2006;312(5776):1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 15.Trickler WJ, Lantz SM, Murdock RC, Schrand AM, Robinson BL, Newport GD, Schlager JJ, Oldenburg SJ, Paule MG, Slikker W, Hussain SM, Ali SF. Nanotoxicology. 2011;5(4):479–492. doi: 10.3109/17435390.2010.540356. [DOI] [PubMed] [Google Scholar]
- 16.Goodman CM, McCusker CD, Yilmaz T, Rotello VM. Bioconjugate chemistry. 2004;15(4):897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- 17.Pernodet N, Fang XH, Sun Y, Bakhtina A, Ramakrishnan A, Sokolov J, Ulman A, Rafailovich M. Small (Weinheim an der Bergstrasse, Germany) 2006;2(6):766–773. doi: 10.1002/smll.200500492. [DOI] [PubMed] [Google Scholar]
- 18.Tsoli M, Kuhn H, Brandau W, Esche H, Schmid G. Small (Weinheim an der Bergstrasse, Germany) 2005;1(8–9):841–844. doi: 10.1002/smll.200500104. [DOI] [PubMed] [Google Scholar]
- 19.Nelson BC, Petersen EJ, Marquis BJ, Atha DH, Elliott JT, Cleveland D, Watson SS, Tseng IH, Dillon A, Theodore M, Jackman J. Nanotoxicology. 2013;7(1):21–29. doi: 10.3109/17435390.2011.626537. [DOI] [PubMed] [Google Scholar]
- 20.Cho WS, Cho MJ, Jeong J, Choi M, Cho HY, Han BS, Kim SH, Kim HO, Lim YT, Chung BH, Jeong J. Toxicol Appl Pharm. 2009;236(1):16–24. doi: 10.1016/j.taap.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 21.Truong L, Saili KS, Miller JM, Hutchison JE, Tanguay RL. Comp Biochem Phys C. 2012;155(2):269–274. doi: 10.1016/j.cbpc.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim KT, Zaikova T, Hutchison JE, Tanguay RL. Toxicol Sci. 2013;133(2):275–288. doi: 10.1093/toxsci/kft081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan Y, Leifert A, Ruau D, Neuss S, Bornemann J, Schmid G, Brandau W, Simon U, Jahnen-Dechent W. Small (Weinheim an der Bergstrasse, Germany) 2009;5(18):2067–2076. doi: 10.1002/smll.200900466. [DOI] [PubMed] [Google Scholar]
- 24.Soderstjerna E, Johansson F, Klefbohm B, Englund Johansson U. PloS one. 2013;8(3):e58211. doi: 10.1371/journal.pone.0058211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soenen SJ, Manshian B, Montenegro JM, Amin F, Meermann B, Thiron T, Cornelissen M, Vanhaecke F, Doak S, Parak WJ, De Smedt S, Braeckmans K. Acs Nano. 2012;6(7):5767–5783. doi: 10.1021/nn301714n. [DOI] [PubMed] [Google Scholar]
- 26.Chen YS, Hung YC, Lin LW, Liau I, Hong MY, Huang GS. Nanotechnology. Vol. 21. England: Size-dependent impairment of cognition in mice caused by the injection of gold nanoparticles; p. 485102. [DOI] [PubMed] [Google Scholar]
- 27.Siddiqi NJ, Abdelhalim MAK, El-Ansary AK, Alhomida AS, Ong WY. J Neuroinflamm. 2012;9 doi: 10.1186/1742-2094-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung S, Bang M, Kim BS, Lee S, Kotov NA, Kim B, Jeon D. PloS one. 2014;9(3) doi: 10.1371/journal.pone.0091360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science (New York, N.Y.) 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 30.Campagnolo L, Fenoglio I, Massimiani M, Magrini A, Pietroiusti A. Methods Mol Biol. 2013;1058:49–60. doi: 10.1007/7651_2013_11. [DOI] [PubMed] [Google Scholar]
- 31.Di Guglielmo C, Lopez DR, De Lapuente J, Mallafre JML, Suarez MB. Reprod Toxicol. 2010;30(2):271–276. doi: 10.1016/j.reprotox.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Liu S, Yao T, Sun Z, Jiang Z, Huang Y, Cheng H, Huang Y, Jiang Y, Xie Z, Pan G, Yan W, Wei S. Dalton transactions (Cambridge, England : 2003) 2012;41(38):11725–11730. doi: 10.1039/c2dt31270k. [DOI] [PubMed] [Google Scholar]
- 33.Woehrle GH, Brown LO, Hutchison JE. Journal of the American Chemical Society. 2005;127(7):2172–2183. doi: 10.1021/ja0457718. [DOI] [PubMed] [Google Scholar]
- 34.Jana NR, Gearheart L, Murphy CJ. The Journal of Physical Chemistry B. 2001;105(19):4065–4067. [Google Scholar]
- 35.Turkevich J, Stevenson PC, Hillier J. Discuss Faraday Soc. 1951;(11) 55-&. [Google Scholar]
- 36.Kimling J, Maier M, Okenve B, Kotaidis V, Ballot H, Plech A. J Phys Chem B. 2006;110(32):15700–15707. doi: 10.1021/jp061667w. [DOI] [PubMed] [Google Scholar]
- 37.Frens G. Kolloid Z Z Polym. 1972;250(7):736. [Google Scholar]
- 38.Frens G. Nature-Phys Sci. 1973;241(105):20–22. [Google Scholar]
- 39.Casals E, Pfaller T, Duschl A, Oostingh GJ, Puntes V. Acs Nano. 2010;4(7):3623–3632. doi: 10.1021/nn901372t. [DOI] [PubMed] [Google Scholar]
- 40.Dominguez-Medina S, Blankenburg J, Olson J, Landes CF, Link S. Acs Sustain Chem Eng. 2013;1(7):833–842. doi: 10.1021/sc400042h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen F, Englert U, Gutrath B, Simon U. Eur J Inorg Chem. 2008;2008(1):106–111. [Google Scholar]
- 42.Taylor U, Rehbock C, Streich C, Rath D, Barcikowski S. Nanomedicine-Uk. 2014;9(13):1971–1989. doi: 10.2217/nnm.14.139. [DOI] [PubMed] [Google Scholar]
- 43.Chithrani BD, Ghazani AA, Chan WCW. Nano Lett. 2006;6(4):662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 44.Bartova E, Galiova G, Krejci J, Harnicarova A, Strasak L, Kozubek S. Dev Dynam. 2008;237(12):3690–3702. doi: 10.1002/dvdy.21773. [DOI] [PubMed] [Google Scholar]
- 45.Kennedy AL, McBryan T, Enders GH, Johnson FB, Zhang RG, Adams PD. Cell Div. 2010:5. doi: 10.1186/1747-1028-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johkura K, Cui L, Asanuma K, Okouchi Y, Ogiwara N, Sasaki K. Journal of anatomy. 2004;205(4):247–255. doi: 10.1111/j.0021-8782.2004.00336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park SH, Park SH, Kook MC, Kim EY, Park S, Lim JH. Ultrastruct Pathol. 2004;28(4):229–238. doi: 10.1080/01913120490515595. [DOI] [PubMed] [Google Scholar]
- 48.Jacobsen JV, Andersen CY, Hyttel P, Møllgård K. From Pluripotency to Early Differentiation of Human Embryonic Stem Cell Cultures Evaluated by Electron Microscopy and Immunohistochemistry, Embryonic Stem Cells - Basic Biology to Bioengineering. 2011 [Google Scholar]
- 49.Konorov SO, Schulze HG, Atkins CG, Piret JM, Aparicio SA, Turner RFB, Blades MW. Anal Chem. 2011;83(16):6254–6258. doi: 10.1021/ac201581e. [DOI] [PubMed] [Google Scholar]
- 50.Hu BY, Zhang SC. Cellular Programming and Reprogramming: Methods and Protocols. 2010;636:123–137. [Google Scholar]
- 51.Osman AM, van Dartel DAM, Zwart E, Blokland M, Pennings JLA, Piersma AH. Reprod Toxicol. 2010;30(2):322–332. doi: 10.1016/j.reprotox.2010.05.084. [DOI] [PubMed] [Google Scholar]
- 52.Soenen SJ, Demeester J, De Smedt SC, Braeckmans K. Nano Today. 2013;8(2):121–125. [Google Scholar]
- 53.Naylor AM, Goddard WA, Kiefer GE, Tomalia DA. Journal of the American Chemical Society. 1989;111(6):2339–2341. [Google Scholar]
- 54.Paulo PMR, Lopes JNC, Costa SMB. J Phys Chem B. 2007;111(36):10651–10664. doi: 10.1021/jp072211x. [DOI] [PubMed] [Google Scholar]
- 55.Pan Y, Neuss S, Leifert A, Fischler M, Wen F, Simon U, Schmid G, Brandau W, Jahnen-Dechent W. Small (Weinheim an der Bergstrasse, Germany) 2007;3(11):1941–1949. doi: 10.1002/smll.200700378. [DOI] [PubMed] [Google Scholar]
- 56.Pan Y, Leifert A, Graf M, Schiefer F, Thoroe-Boveleth S, Broda J, Halloran MC, Hollert H, Laaf D, Simon U, Jahnen-Dechent W. Small (Weinheim an der Bergstrasse, Germany) 2013;9(6):863–869. doi: 10.1002/smll.201201173. [DOI] [PubMed] [Google Scholar]
- 57.Huang KY, Ma HL, Liu J, Huo SD, Kumar A, Wei T, Zhang X, Jin SB, Gan YL, Wang PC, He ST, Zhang XN, Liang XJ. Acs Nano. 2012;6(5):4483–4493. doi: 10.1021/nn301282m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leifert A, Pan Y, Kinkeldey A, Schiefer F, Setzler J, Scheel O, Lichtenbeld H, Schmid G, Wenzel W, Jahnen-Dechent W, Simon U. P Natl Acad Sci USA. 2013;110(20):8004–8009. doi: 10.1073/pnas.1220143110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor U, Barchanski A, Petersen S, Kues WA, Baulain U, Gamrad L, Sajti L, Barcikowski S, Rath D. Nanotoxicology. 2014;8:118–127. doi: 10.3109/17435390.2013.859321. [DOI] [PubMed] [Google Scholar]
- 60.Truong L, Tilton SC, Zaikova T, Richman E, Waters KM, Hutchison JE, Tanguay RL. Nanotoxicology. 2013;7(2):192–201. doi: 10.3109/17435390.2011.648225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delgado AV, Gonzalez-Caballero E, Hunter RJ, Koopal LK, Lyklema J. Pure Appl Chem. 2005;77(10):1753–1805. [Google Scholar]
- 63.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Science (New York, N.Y.) 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 64.Senut MC, Sen A, Cingolani P, Shaik A, Land SJ, Ruden DM. Toxicol Sci. 2014;139(1):142–161. doi: 10.1093/toxsci/kfu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ross PJ, Suhr ST, Rodriguez RM, Chang EA, Wang K, Siripattarapravat K, Ko T, Cibelli JB. Stem cells and development. 2010;19(8):1221–1229. doi: 10.1089/scd.2009.0459. [DOI] [PubMed] [Google Scholar]
- 66.Willems E, Leyns L, Vandesompele J. Anal Biochem. 2008;379(1):127–129. doi: 10.1016/j.ab.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 67.Zhang SC, Wernig M, Duncan ID, Brustle O, Thomson JA. Nature Biotechnology. 2001;19(12):1129–1133. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
- 68.Gensel JC, Schonberg DL, Alexander JK, McTigue DM, Popovich PG. J Neurosci Methods. Vol. 190. Netherlands: Elsevier B.V; 2010. Semi-automated Sholl analysis for quantifying changes in growth and differentiation of neurons and glia; pp. 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sholl DA. Journal of anatomy. 1953;87(4):387. [PMC free article] [PubMed] [Google Scholar]
- 70.Senut MC, Suhr ST, Kaspar B, Gage FH. Journal of Neuroscience. 2000;20(1):219–229. doi: 10.1523/JNEUROSCI.20-01-00219.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 72.Jin P, Warren ST. Trends Biochem Sci. 2003;28(3):152–158. doi: 10.1016/S0968-0004(03)00033-1. [DOI] [PubMed] [Google Scholar]
- 73.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. Bioinformatics. 2013;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 76.Touleimat N, Tost J. Epigenomics. 2012;4(3):325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 77.Barfield RT, Kilaru V, Smith AK, Conneely KN. Bioinformatics. 2012;28(9):1280–1281. doi: 10.1093/bioinformatics/bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.