Abstract
Purpose
5-Lipoxygenase (5-LOX) oxygenates arachidonic acid to form 5-hydroperoxyeicosatetraenoic acid, which is further converted into biologically detrimental leukotrienes, such as leukotriene B4 (LTB4). The RPE and retina express the PNPLA2 gene for pigment epithelium–derived factor receptor (PEDF-R), a lipase involved in cell survival. The purpose here was to investigate the role of PEDF-R on the 5-LOX pathway in oxidative stress of RPE.
Methods
Lipoxygenase activity assays were performed with soybean and potato lipoxygenase. Binding was evaluated by peptide-affinity chromatography and pull-down assays with PEDF-R–derived synthetic peptides or recombinant protein. Oxidative stress was induced in human ARPE-19 and primary pig RPE cells with indicated concentrations of H2O2/TNF-α. Reverse transcription–PCR of ALOX5 and PNPLA2 genes was performed. Cell viability and death rates were determined using respective biomarkers. Leukotriene B4 levels were measured by ELISA.
Results
Among five peptides spanning between positions Leu159 and Met325 of human PEDF-R polypeptide, only two overlapping peptides, E5b and P1, bound and inhibited lipoxygenase activity. Human recombinant 5-LOX bound specifically to peptide P1 and to His6/Xpress-tagged PEDF-R via ionic interactions. The two inhibitor peptides E5b and P1 promoted cell viability and decreased cell death of RPE cells undergoing oxidative stress. Oxidative stress decreased the levels of PNPLA2 transcripts with no effect on ALOX5 expression. Exogenous additions of P1 peptide or overexpression of the PNPLA2 gene decreased both LTB4 levels and death of RPE cells undergoing oxidative stress.
Conclusions
A novel peptide region of PEDF-R inhibits 5-LOX, which intersects with RPE cell death pathways induced by oxidative stress.
Keywords: 5-Lipoxygenase, RPE, oxidative stress, PNPLA2, LTB4
Oxidative stress has been implicated in the pathophysiology of numerous diseases. In the eye, upon oxidative stress–mediated injury of the RPE or retina, degeneration occurs, which leads to blindness (e.g., age-related macular degeneration [AMD] and photo-oxidative damage).1–4 Numerous reports provide evidence that protection of retina cells from oxidative damage slows the progressive degeneration of photoreceptors under photo-oxidative stress.2,4,5
Lipoxygenases (LOX) form a family of lipid-peroxidizing enzymes that catalyze the addition of oxygen to polyunsaturated fatty acids (PUFAs). Arachidonate 5-lipoxygenase (5-LOX), encoded by the ALOX5 gene, plays a central role in leukotriene biosynthesis. The overactivation of the 5-LOX pathway results in the formation of excess of leukotrienes and lipoxins, which are potent cytotoxic mediators6,7 involved in diverse pathophysiological processes, for example, cancer, psoriasis, and artherosclerosis.8,9 Studies have shown age-dependent increase in 5-LOX expression and oxidative stress.1,6,10,11 In rat retinas, light and trauma activate 5-LOX to elicit synthesis of one subtype of leukotriene, leukotriene B4 (LTB4), suggesting the involvement of LTB4 in the pathogenesis of retinal diseases due to light damage.12 Conversely, inhibition of 5-LOX by small molecules can protect RPE cells against oxidative stress (U.S. patent application no. 13/098,200, filed on 4/29/2011).
Recently, a group of genes encoding proteins with a common domain termed patatin-like phospholipase (PNPLA domain) was discovered. The nine members of the PNPLA family display lipase, phospholipase, and transacylase enzymatic activities, and have major roles in adipocyte differentiation, lipid metabolism, and signaling.13–15 We have identified a novel gene member of this family, PNPLA2, in the retina and RPE.16,17 Its protein product, pigment epithelium–derived factor receptor (PEDF-R) (a.k.a. PNPLA2, desnutrin, adipocyte triglyceride lipase, and calcium-independent phospholipase ζ) distributes to the RPE, photoreceptors, and the ganglion cell layer.16,17 This protein acts in retina survival via its phospholipase activity, likely acting on phospholipids of cell surfaces.16,18 It is also present intracellularly at higher levels associated with lipid droplets (e.g., in adipose cells), where it can also liberate free fatty acids but from triglycerides.19 The activities of this enzyme are modulated by several proteins; for example, they are stimulated by PEDF and CGI-5818,20 and inhibited by G(0)/G(1) switch gene 2 (G0S2) and perilipin 5.20,21 The binding region for CGI-58 and G0S2 is located within the amino terminal region spanning Met1–Leu254 residues.20 We have mapped the PEDF-binding region to Leu159-Met325 of the PEDF-R amino acid sequence and identified two synthetic peptides derived from this region, P1 and E5b, that retain the PEDF-binding affinity of PEDF-R.18 Of more interest for this study is the involvement of PNPLA2 in prevention of oxidative stress in the heart.22 While overexpression of this gene abolishes oxidative and inflammatory stress in cardiomyocytes, there is high cardiac oxidative stress in mice that lack PNPLA2 expression in the heart, likely due to cardiac lipotoxicity.22 However, the role of PEDF-R in RPE or retina undergoing oxidative stress remains unknown.
The purpose of this study was to investigate the relationship between PEDF-R and LOX under oxidative stress. Given that preliminary experiments revealed particular fragments of PEDF-R that inhibit LOX-V (a plant orthologue of mammalian lipoxygenase), we set out to characterize prospective LOX-binding region(s) and inhibitors in PEDF-R using human recombinant polypeptides and synthetic peptides. We also used RPE cells to test the protective activity of peptides on oxidative stress–induced death. We report the identification of a region in PEDF-R that contains a critical site for interaction with 5-LOX and for inhibiting oxidative stress.
Materials and Methods
Proteins and Peptides
Recombinant PEDF-R proteins were expressed by cell-free in vitro protein synthesis using expression plasmids pEXP1-PEDF-R as described previously.16,18 Soybean LOX-V was purchased from Sigma (St. Louis, MO, USA). Recombinant human 5-LOX and potato 5-LOX were from Cayman Chemical (Ann Arbor, MI, USA). Recombinant tumor necrosis factor alpha (TNF-α) was from Cell Sciences (Newbury, MA, USA). Peptides were designed from the human PEDF-R sequence and chemically synthesized (bioSYNTHESIS, Inc., Lewisville, TX, USA) as previously described,18 and the following sequence for scrambled: NH2-KRLQFEPRNYPSLLSTALPNILFRRLGGKFQDMRELCVYL-COOH.
Lipoxygenase Activity
The standard reaction mixture (1 mL) contained 25 μM linoleic acid and 8 μg/mL lipoxygenase in 50 mM Tris buffer, pH 9, containing 3 mM deoxycholate (DOC) and was at 25°C. The reaction was started by adding lipoxygenase to the assay mixture. Spectrophotometric measurements for product formation were performed every minute for 10 minutes using a Beckman DU 640 spectrophotometer (Beckman Coulter, Indianapolis, IN, USA).
Peptide-Affinity Chromatography
Peptide-affinity beads (Aves Labs, Tigard, OR, USA) were mixed with LOX-V or 5-LOX in 0.1 M sodium phosphate and 0.1% nonyl phenoxypolyethoxylethanol-40 (NP-40; binding buffer) and incubated with gentle rotation at 4°C for 1.5 hours, unless otherwise indicated. Bound proteins were separated by low-speed centrifugation from unbound materials, extracted with SDS-PAGE sample buffer, and resolved by SDS-PAGE.
His-Tag Pull-Down Assays
Binding of 5-LOX to His6-tagged PEDF-R polypeptides was assayed by pull-down with Ni-nitrilotriacetic acid (NTA) resin as indicated in the legends. Western blotting was against anti-5-LOX antibodies (Pierce, Waltham, MA, USA) diluted 1:10,000 in 1% BSA and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:100,000 in 1% BSA), and detection was with West Dura (Thermo Scientific, Waltham, MA, USA) followed by exposure of blot to X-ray film.
Cell Cultures
Human ARPE-19 cells (passages 27–40; American Type Culture Collection, Manassas, VA, USA) were cultured in DMEM/F-12 medium (cat. no. 11330-032; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cell line short tandem repeat (STR) analysis to validate ARPE-19 cell line was performed at bioSYNTHESIS, and no contamination was detected. Cultures were incubated at 37°C with 5% CO2. Primary RPE were isolated from freshly enucleated pig eyes following the protocol described previously.23 Confluence was achieved in 10 days, after which cells were trypsinized, counted, and plated.
Oxidative Stress
Cells were plated at a density of 2 × 104 cells/well in commercial tissue culture–treated plastic 96-well white plates with clear bottom or 2 × 105 cells/well in 24-well plates. The cells were cultured to 100% confluency (72 hours). These cultures showed that a well-established apical microvilli marker for RPE cells (ezrin) localized preferentially to the apical membrane as in polarized RPE (Supplementary Fig. S1). A combination of H2O2 and TNF-α was used to induce oxidative stress as described before.24,25 ARPE-19 cell medium was removed and replaced with medium containing 0.5% serum to starve the cells for 8 hours to sensitize cells to apoptotic cell death. Then, they were treated with H2O2 to achieve the desired final concentrations and TNF-α (10 ng/mL) plus indicated concentrations of PEDF-R–derived peptides for 16 hours. For Hoechst staining, cells were fixed with methanol for 15 minutes at room temperature followed by addition of 5 μM Hoechst stain in PBS for 15 minutes at room temperature. The percentage of pyknotic cell nuclei as seen by condensed morphology in the field was counted from the digital images.25 Pig RPE cells were starved in media without serum for 24 hours followed by treatment with indicated concentrations of H2O2 and TNF-α (10 ng/mL) plus indicated concentrations of PEDF-R–derived peptides for 2 hours.
Detection of Reactive Oxygen Species (ROS)
ARPE-19 cells were cultured in 96-well black plates with clear bottom. CellRox Green (Life Technologies, Carlsbad, CA, USA) was added 30 minutes before end point, and the fluorescence intensity of CellRox Green in each well was measured using SpectraMax 5 (Molecular Devices, Downington, PA, USA) following manufacturer's instructions.
Cell Viability and Death Assays
Live cells were measured by determining the relative levels of intracellular adenosine triphosphate (ATP) as a biomarker for live cells using a CellTiter-Glo (Promega, Madson, WI, USA) kit, as described previously.18
Dead cells were quantitated with CytoTox-Glo (Promega) kit following manufacturer's instructions. Lysis reagent was added to obtain the luminescent signal associated with the total number of cells in each assay well.
RNA Extraction, cDNA Synthesis, and RT-PCR
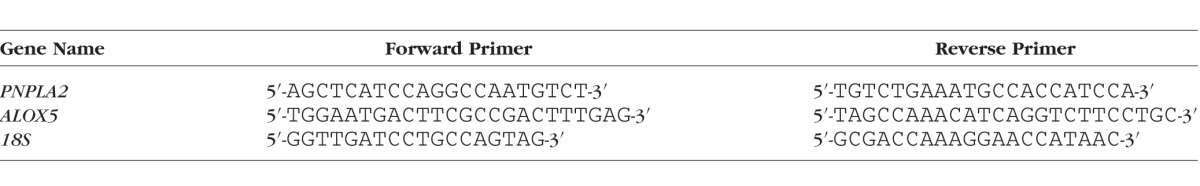
Total RNA was purified using RNeasy mini kits (Qiagen, Valencia, CA, USA), and 1 μg mRNA was reverse transcribed using SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA). PNPLA2 or ALOX5 mRNA levels were normalized to 18S levels by quantitative RT-PCR using SYBR Green mix (Qiagen) in the Bio-Rad Chromo4 real-time system (Hercules, CA, USA). The sequences of forward and reverse primers used are in the Table.
Table.
Primers Used for RT-PCR
Leukotriene B4 ELISA
Leukotriene B4 concentrations in the media were measured using a competitive LTB4 enzyme immunoassay kit (Cayman Chemical). Medium was collected from each well and spun briefly at low speed, and 50 μL was assayed using the kit following the manufacturer's instructions. Standard curves of LTB4 were used to determine the concentration per well and normalized against the total protein concentration per well.
Statistical Analyses
The data were analyzed by 2-tailed unpaired Student's t-test where indicated.
Results
PEDF-R Fragments Inhibit Lipoxygenase V Activity
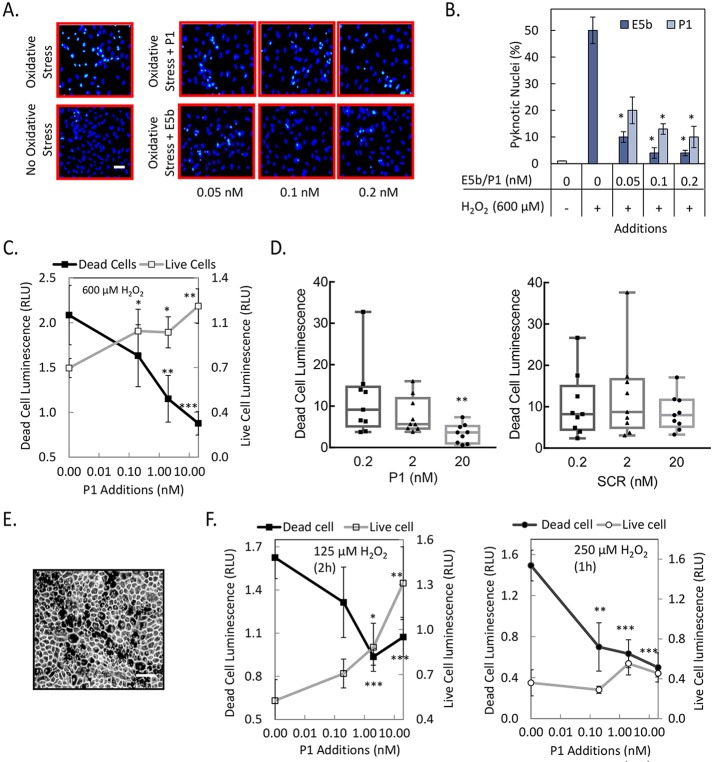
The human PEDF-R polypeptide has 504 amino acid residues, a PNPLA domain located toward its amino end (10–179), and a lipase active site formed by catalytic dyad of amino acids serine in position 47 (Ser47) and aspartic acid in position 166 (Asp166) (Fig. 1A).18,26,27 It also contains a region L4 spanning Leu159-Met325 involved in activating the lipase upon binding to PEDF.16 Previously, we assayed the phospholipase activity of PEDF-R in a reaction coupled to a LOX-V reaction.16,18 One of the control experiments in which the coupled reaction was performed hinted that P1, a L4-derived peptide containing residues Thr210-Leu249, can inhibit soybean LOX-V. Therefore we tested additional peptides spanning Ile193-Met325 for LOX inhibition and found that only the overlapping peptides P1 and E5b (Ile193-Leu232) decreased the activity of potato 5-LOX relative to no addition (Figs. 1B–D). A scrambled peptide (SCR), generated by randomly rearranging amino acids in P1, did not have any effect. The LOX-V activity in the presence of P1 at 10 or 100 nM P1 was 59% and 13%, respectively, of the one without inhibitor (Fig. 1C). Similarly, the LOX-V specific activity with 100 nM E5b was14% of the one without inhibitor (Fig. 1D). The observations show clearly that the PEDF-R fragments P1 and E5b inhibited LOX-V.
Figure 1.
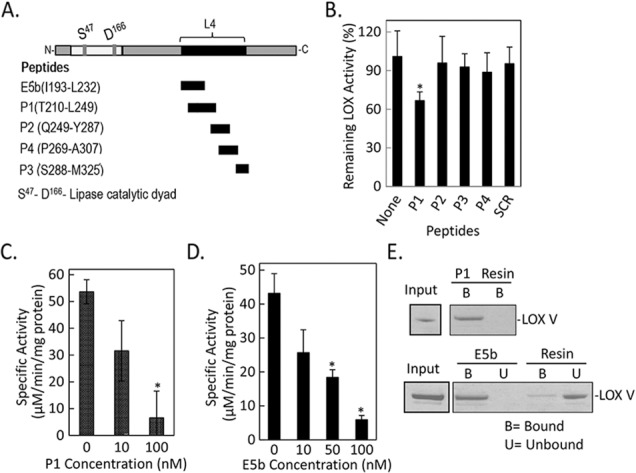
Effects of PEDF-R–derived peptides on LOX assays in vitro. (A) A linear representation of the amino acid sequence of PEDF-R. Black box indicates the location of L4. Residues that comprise the lipase catalytic site dyad (Ser47 and Asp166) are shown (gray). The names and the amino acid sequence of each synthetic peptide from L4 are given to the left. N- and C-, amino-, and carboxy-termini, respectively. (B) In vitro LOX activity was measured using linoleic acid (25 μM) substrate and 8 μg/mL 5-LOX (potato) enzyme in reaction buffer (50 mM Tris-HCl, pH 9, with 3 mM DOC) and 10 nM of the indicated peptide. The product linoleoyl hydroperoxide was measured spectrophotometrically. No peptide (None) and a scrambled peptide (SCR) were negative controls. The y-axis shows percentage of remaining LOX activity relative to no peptide addition (none). Shown is the average of four independent measurements (n = 4). Error bars: ±SD. *P ≤ 0.05. (C, D) In vitro LOX-V (soybean) activity measured as in (B) in the presence of P1 (C) and E5b (D). Shown is the average of four independent experiments (n = 4). Error bars: ±SD. *P ≤ 0.05 compared to no peptide. (E) Peptide-affinity chromatography of LOX-V (1 pmol) with resins containing 0.4 nmol P1 peptide (top) or E5b peptide (bottom) at 1:100 enzyme:peptide molar ratio in binding buffer (10 mM Tris-HCl, pH 7.5, and 3 mM DOC, 50 μL final volume). Flow-through containing the unbound material was collected. After extensive washes with binding buffer, bound protein was eluted from resins with SDS-PAGE sample buffer. Photos of SDS-PAGE gel stained with Coomassie Blue of LOX input, bound (B) protein for P1, and bound (B) and unbound (U) protein for E5b are shown. Resin, agarose without peptide (control).
PEDF-R Fragments Bind Plant Lipoxygenase V
We hypothesized that the inhibition requires an interaction between enzyme and peptide. Thus, we evaluated the direct binding of LOX-V to E5b- and P1-affinity agarose resins. Figure 1E shows that LOX-V was clearly detected in the bound fractions of both P1 and E5b resins, whereas no LOX-V detection was observed in the unbound ones. In controls using agarose resin without peptide, LOX-V was observed in the unbound fraction (lane U, Resin), with minimal detection in the bound material (lane B, Resin).
Taken together, these results show that PEDF-R fragments from the Ile193–Leu249 region can specifically bind and inhibit the activity of plant LOX.
Binding of Human Recombinant 5-LOX to Peptide P1 and Full-Length PEDF-R
Binding assays were performed to study the interaction of mammalian 5-LOX with PEDF-R. Human 5-LOX was mixed with P1-affinity agarose at 1:75 molar ratio under varying conditions to determine optimal binding. Figure 2A shows that 5-LOX bound to peptide P1 in the presence or absence of DOC, at 4°C or 25°C, and as early as 15 minutes. Figure 2B shows that with increasing ionic strength the binding decreased, to be completely abolished at 250 mM NaCl. Figure 2C shows pull-down assays of 5-LOX with human recombinant His6-tag-PEDF-R. Increasing volumes of in vitro synthesized human recombinant His6-tag-PEDF-R in the binding reaction resulted in a proportional increase of bound 5-LOX (Fig. 2C). These results show that mammalian 5-LOX interacted with PEDF-R when presented as an immobilized peptide P1 or full-length polypeptide in solution.
Figure 2.
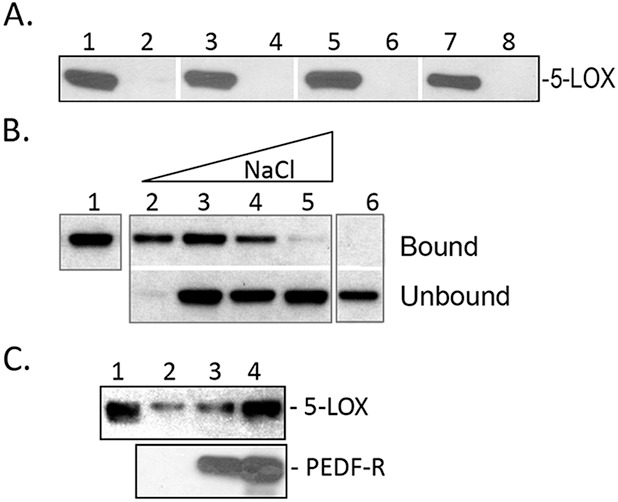
Binding of human recombinant 5-LOX to P1 peptide and PEDF-R. (A) Peptide-affinity chromatography of 5-LOX (300 ng) to P1 (0.4 nmol). Reactions were performed in 10 mM Tris-HCl, pH 7.5, under the following conditions: Lanes 1, 2, at 4°C, incubated for 60 minutes; lanes 3, 4, at 25°C, incubated for 15 minutes; lanes 5, 6, at 4°C, incubated for 60 minutes in buffer plus 3 mM DOC; lanes 7, 8, at 25°C, incubated for 15 minutes in buffer plus 3 mM DOC. After extensive washes with respective binding buffer, bound protein was eluted with SDS-PAGE sample buffer. Western blot versus anti-5-LOX is shown. Samples were loaded in gels as follows: Lanes 1, 3, 5, 7, bound to P1-agarose; lanes 2, 4, 6, 8, bound material to agarose without peptide. (B) Binding of 5-LOX (300 ng) to P1-affinity agarose column (0.4 nmol) in binding buffer (0.1 M sodium phosphate and 0.1% NP-40) with increasing NaCl concentrations. After extensive washes with binding buffer, bound protein was eluted with SDS-PAGE sample buffer. Lane 1, load; lane 2, 25 mM NaCl; lane 3, 50 mM NaCl; lane 4, 150 mM NaCl; lane 5, 250 mM NaCl; lane 6, 50 mM NaCl with control resin. Western blot versus anti-5-LOX is shown. (C) His-tag pull-down assays. Soluble fractions of cell-free expression reactions containing His6/Xpress-tagged PEDF-R at ∼350 and 700 ng (lanes 3, 4, respectively) were mixed with 5-LOX (250 ng) in binding buffer (0.1 M sodium phosphate, 50 mM NaCl, and 0.1% NP-40, 50 μL final volume) and incubated for 2 hours at 4°C with gentle rotation. Lane 2 was 5-LOX incubated with soluble fractions of cell-free expression reactions in the absence of PEDF-R. Then Ni-NTA resin beads (25 μL) were added, and the suspension was incubated for 1 hour at 4°C with gentle rotation. Bound 5-LOX (pull-down) was extracted with SDS sample buffer and analyzed by Western blot versus anti-5-LOX (top) and anti-PEDF-R (lower). Lane 1 was 5-LOX (250 ng) input.
ROS Generation in RPE Cells Under Oxidative Stress
We performed concentration–response curve studies with H2O2 on confluent RPE cells. Retinal pigment epithelium cells were serum starved to sensitize them to oxidative stress–triggered cell death induced by H2O2/TNF-α. Additions of increasing concentrations of H2O2 (between 100 and 1000 μM) to ARPE-19 cells resulted in a concentration-dependent decrease in cell numbers (Fig. 3A). Preliminary assays with a concentration range of H2O2 showed that primary pig RPE cells were more sensitive to oxidative stress than ARPE-19 cells. Therefore, they were treated with lower concentrations of H2O2. Additions of increasing concentration of H2O2 (between 50 and 250 μM) to pig RPE cells resulted in a concentration-dependent decrease in cell numbers (Fig. 3B). The concentration of H2O2 that induced half-maximum cell death was estimated to be 650 and 125 μM for ARPE-19 cells and pig RPE cells, respectively. Therefore, in subsequent experiments we used 650 to 750 μM H2O2 for ARPE-19 and 125 to 250 μM H2O2 for pig RPE to induce cell death. Treatment of ARPE cells with increasing H2O2 in the absence or presence of 10 ng/mL TNF-α increased the intracellular accumulation of ROS (Figs. 3C, 3D). Reactive oxygen species production was comparable under both conditions. Peptide P1 attenuated the ROS accumulation in a concentration-dependent manner, and this decrease of ROS generation was observed with or without TNF-α.
Figure 3.
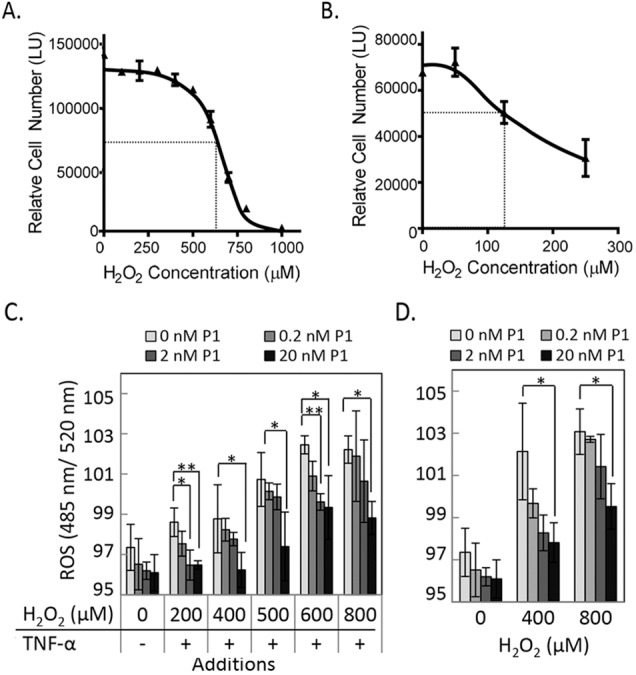
Cell viability and reactive oxygen species (ROS) levels with increasing oxidative stress. (A) ARPE-19 cells were treated with increasing concentrations of H2O2 in the presence of 10 ng/mL TNF-α for 16 hours. Relative cell numbers were determined at the end point using intracellular ATP as a live cell biomarker. A plot of relative cell numbers as a function of concentration was obtained using GraphPad Prism (La Jolla, CA, USA). Each point is the average of values from duplicate wells ± SD. The calculated half-maximal cell death occurred at 650 μM. (B) Oxidative stress treatment with increasing concentrations of H2O2 in the presence of 10 ng/mL TNF-α in pig RPE culture. Relative cell numbers were determined as in (A). The calculated half-maximal cell death occurred at 125 μM. (C) ARPE-19 cells were treated with increasing concentrations of H2O2 and 10 ng/mL TNF-α in the presence of indicated concentration of peptide P1 followed by ROS measurement. CellRox Green reagent was added 30 minutes before the end point. The fluorescence intensity (ex. 485 nm and em. 520 nm) for each condition was measured and plotted as a function of concentration. Each point is the average of triplicate wells ± SD. (D) ROS measurement of ARPE-19 cells treated as in (C) but in the absence of TNF-α. Each point is the average of triplicate wells ± SD. *P ≤ 0.05; **P ≤ 0.005. Each experiment was performed twice.
Protective Effect of Peptide P1 and E5b on RPE Cells
We next investigated whether peptides P1 and E5b can protect RPE cells from oxidative stress–induced cell death. Treatments with 600 μM H2O2 resulted in an increase, while additions of peptides P1 and E5b resulted in a decrease of pyknotic nuclei level (see Fig. 4A). Determination of the numbers of pyknotic nuclei as percentage of total number of cells showed that they decreased by 60%, 74%, and 80% for peptide P1 and 80%, 92%, and 92% for E5b, at concentrations of 0.05, 0.1, and 0.2 nM, respectively, and relative to no peptide additions under oxidative stress (Fig. 4B). Quantification of relative cell numbers using two different biomarkers for live and dead cells, intracellular ATP content, and dead cell protease activity, respectively, was also performed. Peptide P1 increased the ARPE-19 cell viability and decreased the number of dead cells under oxidative stress in a concentration-dependent fashion (Fig. 4C), while the scrambled peptide P1 (SCR) did not decrease the number of dead cells (Fig. 4D).
Figure 4.
Oxidative stress in ARPE-19 and pig primary RPE cells. (A) ARPE-19 cells were treated with 600 μM H2O2 and indicated concentrations of peptides at the same time. No oxidative stress and oxidative stress without peptides served as negative and positive controls, respectively. After 16 hours, cells were fixed with methanol and stained with Hoechst stain to visualize pyknotic nuclei. Representative images from each condition are shown. Scale bar: 100 μm. (B) Photos of five random fields per well were taken; the numbers of pyknotic and total number of cells were quantified using ImageJ software (http://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA), and the number of pyknotic cells was plotted as the percentage of total number of cells. (C) Plot of relative cell numbers determined using intracellular biomarkers–protease activity for dead cells (shown on left axis) and ATP for live cells (shown on right axis) as a function of P1 concentration is shown for ARPE-19 cells treated with indicated concentration of H2O2. Relative cell numbers are averages of duplicate wells ± SD (error bars). (D) Quantitation of relative dead cells as in (C) with increasing concentration of P1 and scrambled P1 (SCR) for ARPE-19 cells treated with 600 μM H2O2. Relative cell numbers of individual wells are shown in box-and-whiskers plots. The horizontal line inside the box is the median. (E) Bright-field photograph of primary pig RPE cells in culture. Scale bar: 100 μm. (F) Quantitation of live and dead cells as in (C) in primary pig RPE culture with indicated concentration of H2O2 and at indicated time. *P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0005. Each experiment was performed more than two times.
Retinal pigment epithelium cells from pig eye were cultured and used to test the protective effect of peptide P1. The primary cultures had pigmented cells, hexagonal in shape, which are distinctive characteristics of native RPE (see Fig. 4E). Peptide P1 increased pig RPE cell viability and decreased death at 125 μM H2O2 (Fig. 4F), as observed with ARPE-19. Peptide P1 decreased cell death at 250 μM H2O2, but did not increase the cell numbers (Fig. 4F). It is possible that oxidative stress with 250 μM H2O2 is too toxic for peptide P1 to rescue. Taken together, these results show that peptides E5b and P1 protected ARPE-19 and pig primary RPE cells from oxidative stress–induced cell death in a specific and concentration-dependent fashion.
PNPLA2 and ALOX5 Expression in RPE Under Oxidative Stress
Both ARPE-19 and pig RPE cells expressed ALOX5 and PNPLA2 genes (Fig. 5A). We evaluated the levels of these two genes in ARPE-19 cells exposed to varying H2O2 concentrations in combination with TNF-α (10 ng/mL). Additions of H2O2 at 200, 400, 500, 600, and 800 μM decreased the levels of PNPLA2 (Fig. 5B), while those of ALOX5 remained unchanged (Fig. 5C). The protein levels of PEDF-R and 5-LOX paralleled those for expression of their respective genes (Figs. 5D, 5E). The results show that oxidative stress conditions regulated the expression of PNPLA2 in RPE cells.
Figure 5.
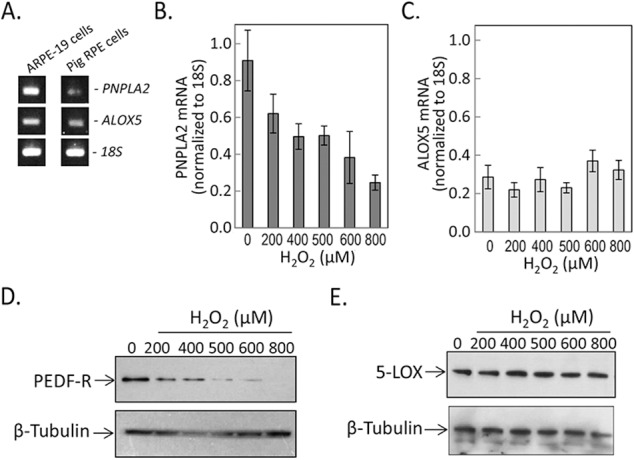
Levels of PNPLA2 and ALOX5 under oxidative stress. (A) Reverse transcription–PCR on mRNA from ARPE-19 and pig RPE with specific primers to PNPLA2 and ALOX5. Primers for 18S were used as internal control. PCR products were resolved by agarose gel electrophoresis and visualized with ethidium bromide under ultraviolet light. (B, C) Semiquantitative real-time reverse transcription–PCR to measure mRNA levels of PNPLA2 and ALOX5 in human ARPE-19 cells treated with increasing concentrations of H2O2 plus 10 ng/mL TNF-α for 8 hours. Values relative to 18S mRNA levels are shown on the y-axis. Each condition was tested in duplicate wells per experiment ± SD (error bars). (D, E) Total cell lysates from cells treated with indicated concentrations of H2O2 plus 10 ng/mL TNF-α for 8 hours. Samples were resolved by SDS-PAGE followed by immunoblotting with anti-PEDF-R (D) and anti-5-LOX (E). Both blots were stripped and immunoblotted with antibody to β-tubulin (loading control). Samples from duplicate wells were tested in each experiment. Each experiment was performed at least twice.
LTB4 in RPE Under Oxidative Stress
Leukotriene B4 is a product of the 5-LOX enzymatic activity. To determine 5-LOX activity in cell cultures, secreted LTB4 was measured in RPE culturing media. Treatments with 600 μM H2O2/TNF-α increased the levels of LTB4 in ARPE-19 by 3-fold, while additions of P1 peptide (20 nM) decreased LTB4 to basal levels (Fig. 6A). However, peptide P1 additions to ARPE-19 cells exposed to 750 μM H2O2/TNF-α only partially diminished LTB4 (Fig. 6A). Peptide P1 also decreased LTB4 levels in primary pig RPE cells treated at 125 μM or 250 μM H2O2/TNF-α in a concentration-dependent fashion (Fig. 6B). These results indicate that peptide P1 had an inhibitory effect on 5-LOX activity in RPE cells.
Figure 6.
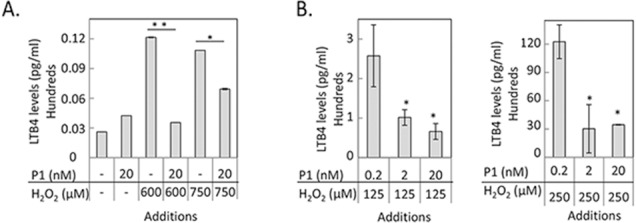
Effect of P1 on 5-LOX product leukotriene B4 (LTB4) in RPE cells. (A) Quantitation of LTB4 levels in ARPE-19 cells treated with indicated concentrations of H2O2 and P1. (B) Quantitation of LTB4 levels in primary pig RPE cells treated with indicated concentration of H2O2 and P1. The levels of LTB4 were normalized to the total number of cells per well. Each condition was tested in duplicate wells per experiment ± SD (error bars). LTB4 level in each well was measured in triplicates. Each experiment was performed twice. *P ≤ 0.05.
Effect of Silencing and Overexpression of PEDF-R Under Oxidative Stress
The above results suggest that peptide P1 can modulate the 5-LOX activity in RPE cells. To further explore this modulation, the PNPLA2 gene was silenced and overexpressed in ARPE-19. The cells were exposed to oxidative stress, followed by the determination of the levels of LTB4 in the culturing media. In the absence of oxidative stress, the levels of LTB4 in ARPE-19 cells showed no significant change. However, oxidative stress increased the LTB4 levels by >10-fold in cells that were transfected with siSCR, siPNPLA2, or those that were not transfected (Fig. 7A). Overexpression of PNPLA2 prevented such an increased LTB4, which was similar to observations in cells transfected to siPNPLA2 and treated with peptide P1 (20 nM). We also determined the death rates in the above cells and found that the pattern of values for cell death paralleled those for LTB4 (compare Figs. 7A, 7B). Overexpression of PNPLA2 decreased death of cells exposed to 600 μM H2O2/TNF-α, while silencing PNPLA2 did not unless P1 was supplemented. Confirmation of PNPLA2 silencing with siRNA and overexpression of PNPLA2 at the protein levels is shown in Figures 7C and 7D. From the three different siRNAs tested for PNPLA2 silencing, siPNPLA2-1 was used in the above experiments. The results indicate that PEDF-R decreased LTB4 levels and protected ARPE-19 cells from cell death.
Figure 7.
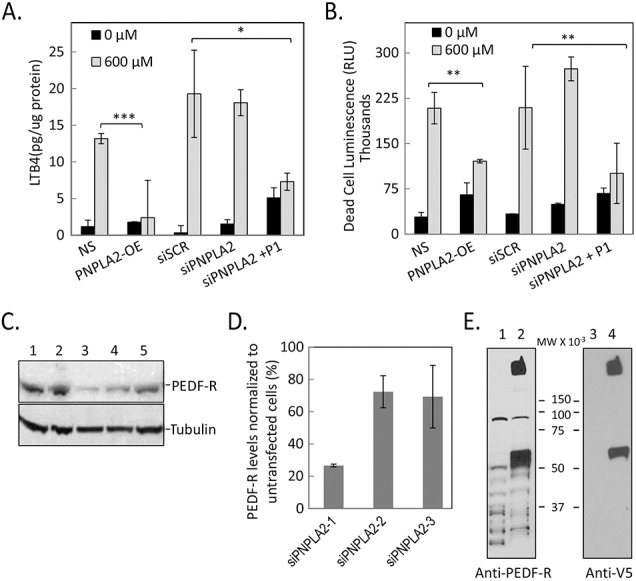
LTB4 levels and cell death in genetically manipulated ARPE-19 cells. (A) Quantitation of LTB4 levels in ARPE-19 cells untransfected and transfected with siScrambled (siSCR), siPNPLA2, PNPLA2 overexpression (PNPLA2-OE), or siPNPLA2 plus P1 and treated with indicated concentration of H2O2. The level of LTB4 is normalized to total protein per well. Each condition was tested in triplicate wells per experiment ± SD (error bars). (B) Plot of relative cell death determined at the end point using intracellular protease activity as biomarker corresponding to conditions shown in (A). Relative cell death values are averages of triplicate wells ± SD (error bars). *P < 0.05, **P < 0.005, and ***P < 0.0005. (C) Total protein from cells harvested 72 hours post transfection with three different siRNAs. Lane 1, no silencing; lane 2, siScrambled; lane 3, siPNPLA2-1; lane 4, siPNPLA2-2; lane 5, siPNPLA2-3. Samples were resolved by SDS-PAGE followed by immunoblotting with anti-PEDF-R. The blot was stripped and immunoreacted with antibody to β-tubulin as loading control. (D) Quantitation of PEDF-R levels in siPNPLA2 transfected cells shown as percentage remaining compared to untransfected cells. Data are average from three independent experiments. (E) Total protein from cells harvested 72 hours post transfection with PNPLA2-OE. Lane 1, nontransfected; lane 2, PNPLA2-OE. Samples were resolved by SDS-PAGE followed by immunoblotting with anti-PEDF-R. The blot was stripped and immunoreacted with anti-V5 to probe V5-tag-PEDF-R-OE. The experiments were repeated three times.
Discussion
We report the identification of a region of PEDF-R that inhibits 5-LOX activity. The conclusions point to a mechanism that requires binding of the inhibitor to the enzyme to inactivate it, which in RPE cells leads to a decrease of the oxidative stress–associated LTB4 and cell death. The implications apply equally to human ARPE-19 and primary RPE cells. Most importantly, there is evidence for the essential role of PEDF-R in protecting RPE cells from oxidative stress–induced cell death via inhibiting 5-LOX.
Given the limitations of obtaining enzymatically active 5-LOX from mammalian sources, we used plant orthologues that have similar catalytic mechanism to the mammalian enzyme and are used as an alternative source of the enzyme in screening inhibitors.28 Our findings show that 5-LOX specifically binds PEDF-R (full-length or fragments) in solution or when immobilized and that the interaction is ionic in nature. More interestingly, this is the first report showing that PEDF-R peptides E5b (Ile193-Leu232) and P1 (Thr210-Leu249) are inhibitors of LOX. The structure of their overlap suggests that a region spanning between Thr210and Leu232 in PEDF-R mediates the inhibition of 5-LOX. In physiological RPE cells, plasma membrane PEDF-R has this region facing extracellularly on the exposed loop L4 and available to bind extracellular ligands, such as PEDF.16,18 In RPE damaged by oxidative stress, the expression of PEDF-R might decrease, as implied from our findings of downregulation of PNPLA2 with H2O2 treatments, and thus the orientation of this region becomes irrelevant. However, in conditions in which PEDF-R is found intracellularly associated with lipid droplets, such as in adipose cells,29 the orientation of the LOX-inhibitory region and its ability to interact with LOX are still not known. Given that LOX is located in the cytosol, it is presumed that the PEDF-R/LOX interactions occur intracellularly. The fact that exogenous P1 or E5b peptide protects cells points to a yet unknown mechanism of peptide internalization preceding inhibition of LOX. Endocytosis or direct perturbation of the membrane by the peptides could facilitate cellular internalization of the peptides to interact with cytosolic 5-LOX.
The addition of H2O2 to cultured RPE cells is a classic model to study oxidative stress and effects of antiapoptotic effectors.30,31 Our preliminary experiments included treatment of cells with a wide range of H2O2 plus TNF-α (10 ng/mL). With ARPE-19 cells treated with H2O2 concentrations between 100 and 1000 μM, we noticed that peptide P1 had protective effects against 600 μM H2O2 but lacked effect with higher concentrations of H2O2 (data not shown). The ineffectiveness can be due to differences in cell death mechanism induced by oxidative stress, which P1 might not be able to counteract. In this regard, Kim et al.32 have reported that ARPE-19 cells undergo apoptosis at concentrations between 400 and 600 μM H2O2, whereas higher concentrations result in necrosis. Furthermore, Mukherjee et al.24 have reported an apoptotic mechanism via caspase activation with a combination of TNF-α (10 ng/mL) and 400 to 800 μM H2O2 in an ARPE-19 cell model of oxidative stress.24
Most striking are the implications of our results derived from the comparison between LTB4 levels and death rates of cells exposed to oxidative stress. Overexpression of PNPLA2 or exogenous addition of peptide P1 completely reverses the increase in LTB4 levels in cells undergoing oxidative stress, which they protect. Our observations imply a direct inhibition of 5-LOX enzymatic activity by PEDF-R. Moreover, in RPE under oxidative stress conditions, repression of PNPLA2 expression leads to lowering the potential to inhibit 5-LOX by PEDF-R and permitting an increase of LTB4 and cell death. Conversely, induction of PNPLA2 gene expression or additions of P1/E5b peptides can benefit RPE by counteracting the detrimental effect. The downregulation of PNPLA2 expression and increased LTB4 levels detected under oxidative stress support this idea. Our observation of decreased LTB4 being protective is complemented by studies done in mouse models of Alzheimer's disease that were genetically deficient in ALOX5. The deficiency of ALOX5 resulted in depletion of LTB4 levels in their brains,33 further confirming the direct role of 5-LOX for LTB4 production. Interestingly, ALOX5 deficiency was beneficial in that it slowed the progression of the disease.
Since protecting RPE cells from oxidative damage is an important consideration for treating AMD, our results demonstrating PEDF-R and peptides P1/E5b as novel inhibitors of 5-LOX suggest possible therapeutic approaches for diseases in which oxidative stress plays a role.
Supplementary Material
Acknowledgments
The authors thank Joseph Urban, U.S. Department of Agriculture, for generously providing pig eyes; Archana Balakrishnan for technical assistance in the preparation of pig primary RPE culture; and Robert Fariss for assistance and consultation on bioimaging under confocal microscopy. The authors thank Wai Wong, Ronald Bush, Federica Polato, and Luis Bonet-Ponce for critical proofreading of the manuscript.
Supported by the Intramural Research Program of the National Institutes of Health, National Eye Institute.
Disclosure: P. Subramanian, None; E.F. Mendez, None; S.P. Becerra, None
References
- 1. Beatty S,, Koh H,, Phil M,, Henson D,, Boulton M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv Ophthalmol. 2000; 45: 115–134. [DOI] [PubMed] [Google Scholar]
- 2. Cai J,, Nelson KC,, Wu M,, Sternberg P, Jr,, Jones DP. Oxidative damage and protection of the RPE. Prog Retin Eye Res. 2000; 19: 205–221. [DOI] [PubMed] [Google Scholar]
- 3. Nowak JZ. Age-related macular degeneration (AMD): pathogenesis and therapy. Pharmacol Rep. 2006; 58: 353–363. [PubMed] [Google Scholar]
- 4. Rex TS,, Tsui I,, Hahn P,, et al. Adenovirus-mediated delivery of catalase to retinal pigment epithelial cells protects neighboring photoreceptors from photo-oxidative stress. Hum Gene Ther. 2004; 15: 960–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong X,, Li Z,, Wang W,, et al. Protective effect of canolol from oxidative stress-induced cell damage in ARPE-19 cells via an ERK mediated antioxidative pathway. Mol Vis. 2011; 17: 2040–2048. [PMC free article] [PubMed] [Google Scholar]
- 6. Uz T,, Pesold C,, Longone P,, Manev H. Aging-associated up-regulation of neuronal 5-lipoxygenase expression: putative role in neuronal vulnerability. FASEB J. 1998; 12: 439–449. [DOI] [PubMed] [Google Scholar]
- 7. Werz O,, Steinhilber D. Therapeutic options for 5-lipoxygenase inhibitors. Pharmacol Ther. 2006; 112: 701–718. [DOI] [PubMed] [Google Scholar]
- 8. Radmark O. 5-lipoxygenase-derived leukotrienes: mediators also of atherosclerotic inflammation. Arterioscler Throm Vasc Biol. 2003; 23: 1140–1142. [DOI] [PubMed] [Google Scholar]
- 9. Steele VE,, Holmes CA,, Hawk ET,, et al. Lipoxygenase inhibitors as potential cancer chemopreventives. Cancer Epidemiol Biomarkers Prev. 1999; 8: 467–483. [PubMed] [Google Scholar]
- 10. Chinnici CM,, Yao Y,, Pratico D. The 5-lipoxygenase enzymatic pathway in the mouse brain: young versus old. Neurobiol Aging. 2007; 28: 1457–1462. [DOI] [PubMed] [Google Scholar]
- 11. Khandhadia S,, Lotery A. Oxidation and age-related macular degeneration: insights from molecular biology. Exp Rev Mol Med. 2010; 12: e34. [DOI] [PubMed] [Google Scholar]
- 12. Reinboth JJ,, Gautschi K,, Clausen M,, Reme CE. Lipid mediators in the rat retina: light exposure and trauma elicit leukotriene B4 release in vitro. Curr Eye Res. 1995; 14: 1001–1008. [DOI] [PubMed] [Google Scholar]
- 13. Jenkins CM,, Mancuso DJ,, Yan W,, Sims HF,, Gibson B,, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004; 279: 48968–48975. [DOI] [PubMed] [Google Scholar]
- 14. Kienesberger PC,, Oberer M,, Lass A,, Zechner R. Mammalian patatin domain containing proteins: a family with diverse lipolytic activities involved in multiple biological functions. J Lipid Res. 2009; 50 (suppl): S63–S68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilson PA,, Gardner SD,, Lambie NM,, Commans SA,, Crowther DJ. Characterization of the human patatin-like phospholipase family. J Lipid Res. 2006; 47: 1940–1949. [DOI] [PubMed] [Google Scholar]
- 16. Notari L,, Baladron V,, Aroca-Aguilar JD,, et al. Identification of a lipase-linked cell membrane receptor for pigment epithelium-derived factor. J Biol Chem. 2006; 281: 38022–38037. [DOI] [PubMed] [Google Scholar]
- 17. Subramanian P,, Notario PM,, Becerra SP. Pigment epithelium-derived factor receptor (PEDF-R): a plasma membrane-linked phospholipase with PEDF binding affinity. Adv Exp Med Biol. 2010; 664: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Subramanian P,, Locatelli-Hoops S,, Kenealey J,, DesJardin J,, Notari L,, Becerra SP. Pigment epithelium-derived factor (PEDF) prevents retinal cell death via PEDF Receptor (PEDF-R): identification of a functional ligand binding site. J Biol Chem. 2013; 288: 23928–23942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zimmermann R,, Strauss JG,, Haemmerle G,, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 2004; 306: 1383–1386. [DOI] [PubMed] [Google Scholar]
- 20. Cornaciu I,, Boeszoermenyi A,, Lindermuth H,, et al. The minimal domain of adipose triglyceride lipase (ATGL) ranges until leucine 254 and can be activated and inhibited by CGI-58 and G0S2, respectively. PLoS One. 2011; 6: e26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang H,, Bell M,, Sreenivasan U,, et al. Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J Biol Chem. 2011; 286: 15707–15715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schrammel A,, Mussbacher M,, Winkler S,, et al. Cardiac oxidative stress in a mouse model of neutral lipid storage disease. Biochim Biophys Acta. 2013; 1831: 1600–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Toops KA,, Tan LX,, Lakkaraju A. A detailed three-step protocol for live imaging of intracellular traffic in polarized primary porcine RPE monolayers. Exp Eye Res. 2014; 124: 74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mukherjee PK,, Marcheselli VL,, Serhan CN,, Bazan NG. Neuroprotectin D1: a docosahexaenoic acid-derived docosatriene protects human retinal pigment epithelial cells from oxidative stress. Proc Natl Acad Sci U S A. 2004; 101: 8491–8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian P,, Crawford SE,, Becerra SP. Assays for the antiangiogenic and neurotrophic serpin pigment epithelium-derived factor. Methods Enzymol. 2011; 499: 183–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duncan RE,, Wang Y,, Ahmadian M,, Lu J,, Sarkadi-Nagy E,, Sul HS. Characterization of desnutrin functional domains: critical residues for triacylglycerol hydrolysis in cultured cells. J Lipid Res. 2010; 51: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lake AC,, Sun Y,, Li JL,, et al. Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J Lipid Res. 2005; 46: 2477–2487. [DOI] [PubMed] [Google Scholar]
- 28. Aparoy P,, Reddy RN,, Guruprasad L,, Reddy MR,, Reddanna P. Homology modeling of 5-lipoxygenase and hints for better inhibitor design. J Comput Aided Mol Des. 2008; 22: 611–619. [DOI] [PubMed] [Google Scholar]
- 29. Smirnova E,, Goldberg EB,, Makarova KS,, Lin L,, Brown WJ,, Jackson CL. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 2006; 7: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geiger RC,, Waters CM,, Kamp DW,, Glucksberg MR. KGF prevents oxygen-mediated damage in ARPE-19 cells. Invest Ophthalmol Vis Sci. 2005; 46: 3435–3442. [DOI] [PubMed] [Google Scholar]
- 31. Kaczara P,, Sarna T,, Burke JM. Dynamics of H2O2 availability to ARPE-19 cultures in models of oxidative stress. Free Radic Biol Med. 2010; 48: 1064–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim MH,, Chung J,, Yang JW,, Chung SM,, Kwag NH,, Yoo JS. Hydrogen peroxide-induced cell death in a human retinal pigment epithelial cell line, ARPE-19. Korean J Ophthalmol. 2003; 17: 19–28. [DOI] [PubMed] [Google Scholar]
- 33. Joshi YB,, Giannopoulos PF,, Chu J,, et al. Absence of ALOX5 gene prevents stress-induced memory deficits, synaptic dysfunction and tauopathy in a mouse model of Alzheimer's disease. Hum Mol Genet. 2014; 23: 6894–6902. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.